Abstract
Background
Melanoma risk prediction models could be useful for matching preventive interventions to patients’ risk.
Objectives
To develop and validate a model for incident first-primary cutaneous melanoma using clinically-assessed risk factors.
Methods
We used unconditional logistic regression with backward selection on the Australian Melanoma Family Study (461 cases and 329 controls) in which age, sex and city of recruitment were kept in each step, and externally validated it using the Leeds Melanoma Case-Control Study (960 cases and 513 controls). Candidate predictors included clinically-assessed whole-body naevi and solar lentigines, and self-assessed pigmentation phenotype, sun exposure, family history and history of keratinocyte cancer. We evaluated the predictive strength and discrimination of the model risk factors using odds per age- and sex-adjusted standard deviation (OPERA) and the area under curve (AUC), and calibration using the Hosmer-Lemeshow (H-L) test.
Results
The final model included the number of naevi ≥2mm in diameter on the whole body, solar lentigines on the upper back (a 6-level scale), hair colour at age 18 years and personal history of keratinocyte cancer. Naevi was the strongest risk factor; the OPERA was 3.51(95%CI 2.71–4.54) in the Australian study and 2.56(95%CI 2.23–2.95) in the Leeds study. The AUC was 0.79(95%CI 0.76–0.83) in the Australian study and 0.73(95%CI 0.70–0.75) in the Leeds study. The H-L test p-value was 0.30 in the Australian study and <0.001 in the Leeds study.
Conclusions
This model had good discrimination, and could be used by clinicians to stratify patients by melanoma risk for the targeting of preventive interventions.
Keywords: melanoma, risk prediction, model, prevention
Introduction
Melanoma incidence has been increasing among fair-skinned populations, with the highest incidence rates in Australia, New Zealand, North America and Europe.(1) Risk factors include sun exposure, sunbed use, common and dysplastic naevi, Fitzpatrick skin type I and II, freckle density, skin colour, eye colour, hair colour, family history and a number of susceptibility genes, with sun exposure recognized as the major environmental risk factor.(2–5) Australian primary care prevention guidelines recommend a stratified approach to melanoma prevention, which includes: (1) sun protection for people at average melanoma risk, (2) sun protection and clinical-skin examinations for people at increased melanoma risk; and (3) sun protection, clinical-skin examinations and self-skin examinations for people at high melanoma risk.(6)
Risk prediction models provide a single personalised assessment of an individual’s risk based on a combination of melanoma risk factors rather than relying on multiple individual risk factors, and may assist clinicians in matching preventive interventions to risk levels.(7) Many melanoma prediction models use self-assessed risk factors for reasons of feasibility, time and cost.(8) However, individuals tend to underestimate their naevus counts,(9) and clinically-assessed dermatological risk factors may improve model performance.(10) We aimed to develop a model for incident first-primary cutaneous melanoma using both clinically- and self-assessed risk factors from the Australian Melanoma Family Study,(11) and externally validate the model using the Leeds Melanoma Case-Control Study.(12)
Materials and methods
Study participants
The Australian Melanoma Family Study is a population-based, case-control-family study.(11) Data were collected using self-administered and telephone-administered questionnaires, and skin examinations by dermatology trainees at 30 minute intervals on 461 incident first-primary cutaneous melanoma cases and 329 controls from Brisbane, Sydney and Melbourne, Australia. Cases, diagnosed between July 2000 and December 2002 at ages 18–39 years, were identified from state cancer registries. Controls were identified from the electoral roll (registration to vote is compulsory in Australia) or nominated by cases, and were frequency-matched to cases by city, age and sex.
The Leeds Melanoma Case-Control Study is a population-based case-control study.(12) Data were collected using self-administered and telephone administered questionnaires, and skin examinations by research nurses on 960 incident first-primary cutaneous melanoma cases and 513 controls from Yorkshire, United Kingdom. Cases, diagnosed between September 2000 and December 2005 at ages 18–76 years, were identified from clinicians, pathology registers and cancer registries. Controls were identified from the cases’ general practice (usually the practice nearest to their home residence), and were frequency-matched to cases by age and sex.
Table 1 details the Australian and Leeds population-based case-control studies,(11, 12) which shared measurement protocols. In both studies, participants answered questions on skin colour of the upper inside arm, eye colour, natural hair colour at age 18, sunburn response, naevi on the body based on a 4- level pictogram and freckling density on the face based on a 6-level pictogram, without reference to time of the year.(11, 12) Clinical assessment of naevi was conducted by dermatology trainees in Australia and research nurses in Leeds, with separate counts for melanocytic naevi between 2 to 5mm and ≥5mm, and dysplastic naevi. (11, 12) There was no agreement between naevus count and first-degree family history in the Australian study (Pearson correlation coefficient= −0.017, p=0.72) but good agreement in the Leeds study (Pearson correlation coefficient= 0.21, p=0.03). The prevalence for CDKN2A variants in melanoma cases was 2.3% in the Australian study and 2.0% in the Leeds study.(13)
Table 1.
Summary of the Australian Melanoma Family Study (development dataset) and Leeds Melanoma Case-Control Study (external validation dataset)
| Study | Study design | Geographical location | Cases/ Controls | Case ascertainment | Control ascertainment | Diagnosis age | Diagnosis years | Data collection |
|---|---|---|---|---|---|---|---|---|
| Australian Melanoma Family Study | Population-based case-control-family study | Brisbane, Sydney and Melbourne, Australia | 461/329 | State cancer registries | Electoral rolla and spouses or friends of cases | 18–39 years | 2000–2002 | Skin examinations by dermatology trainees, and self-administered and telephone-administered questionnaires |
| Leeds Melanoma Case-Control Study | Population-based case-control study | Yorkshire, United Kingdom | 960/513 | Clinicians, pathology registers and cancer registries | General practiceb | 18–76 years | 2000–2005 | Skin examinations by research nurses, and self-administered and telephone-administered questionnaires |
Population-based controls were identified from the electoral roll and frequency-matched to cases by geographical location, age and sex; and spouse/friend controls were nominated by the cases.
Controls were identified from the cases’ general practice patient lists and were frequency-matched to cases by age and sex
Model Development
The model was developed with the Australian study data.(11) All relevant candidate predictors were included: demographic factors- age, sex, city of recruitment; clinically-assessed factors- total number of naevi ≥2mm diameter, number of raised naevi, number of dysplastic naevi, and solar lentigines on the upper back (based on a 6-level pictogram); and self-assessed risk factors- freckle density (based on 6-level pictogram), country of birth, ethnicity, skin colour, eye colour, natural hair colour at age 18 years, skin response to sunlight, height, weight, blistering sunburn frequency (childhood, lifetime), sunbed use, sunscreen use, personal history of keratinocyte cancer and first-degree family history of melanoma. We have limited family history to first-degree relatives, which is reported with better accuracy.(14) Previous studies have shown fair to moderate agreement between clinically- and self-assessed skin colour, and good agreement between clinically-and self-assessed eye colour and hair colour.(9) We have previously shown that a ‘pigmentation phenotype score’ derived from self-assessed risk factors comprising childhood freckling, skin colour, eye colour, hair colour, ability to tan and propensity to sunburn, gave the same improvement to discrimination in a melanoma risk prediction model as did a pigmentation phenotype score that incorporated clinically-assessed skin reflectance, eye colour and hair colour (incremental improvement to the AUC was 0.053 for the self-assessed score and 0.047 for the clinically-assessed score).(10) For this reason we selected self-assessed pigmentation phenotype risk factors because it is more efficient and less costly for patients to complete risk factor information in the waiting room than for the clinician to complete during the consultation. However, it was important to include clinically-assessed naevi and solar lentigines in the risk prediction model as these are usually more difficult for patients to assess and our previous study showed that clinical assessment of naevi and solar lentigines gave much higher improvement in the AUC than self-assessed naevi (incremental improvement to the AUC was 0.048 for self-assessed naevi, 0.111 for clinically-assessed naevi and 0.063 for clinically-assessed solar lentigines).(10)
We used unconditional logistic regression with backward selection in which the study design variables age, sex and city of recruitment were kept in each step and other variables with p-values >0.05 were removed. Effect modification was tested by adding terms for the interaction between pairs of variables in the final model. Multiple imputation by chained equations with 10 imputed datasets was used to impute missing values.(15)
Relative risks and odds per age- and sex-adjusted standard deviation (OPERA)(16) were calculated as a way of comparing the predictive strengths, in terms of differentiating cases from controls, for variables included in the final model. As described elsewhere,(17) remaining lifetime (to 85 years of age) absolute risk was estimated using the Gail method(18) by combining relative risks with Australian melanoma incidence and competing mortality rates (Online Table 1).
Model performance and validation
The model was internally validated with the Australian study and externally validated with the Leeds study data.(11, 12) We assessed discrimination, the model’s ability to distinguish between individuals with and without melanoma, using the area under curve (AUC).(19) In age and sex-matched case-control studies, the distributions of risk factors among controls may be more similar to cases than the general population;(20) we therefore reweighted the age and sex distribution of the control participants to the general population (Online Method 1). Bootstrap procedures were used with 1000 repetitions to estimate the 95% confidence intervals.
We assessed overall calibration, the agreement between the model’s predicted and observed risk, using the Hosmer-Lemeshow (H-L) test, where the predicted and observed risks of melanoma were compared across deciles of predicted risk using a chi-square test, and a high test p-value indicates good overall calibration.(19) The predicted risk was applied to the logistic regression model as a fixed term, while the intercept could vary to account for the higher proportion with melanoma in the case-control studies.
The same variables were used in both studies except for solar lentigines, which was not measured in the Leeds study. Instead, we used freckle density as a proxy for solar lentigines in the Leeds analysis, as there was moderate agreement between these variables in the Australian study (Spearman correlation coefficient=0.28, 95% CI 0.21–0.35, p<0.001). Leeds study participants with missing values for any of the predictor variables were excluded. Online Table 2 shows the distributions of the predictor variables in the analyses.
Ethical approvals were obtained from the University of Sydney and the relevant UK Multi Centre Ethics Committee (MREC) and Patient Information Advisory Group (PIAG). All participants gave written informed consent. Statistical analyses were conducted using Stata version 12 for model development and SAS version 9.3 for model validation. Two-sided statistical significance was inferred at p<0.05, except for interaction terms where p<0.01 was used to allow for multiple testing.(21) We report methods and results in accordance with the Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis statement.(22)
Results
The final model included the total number of naevi ≥2mm, solar lentigines on the upper back, hair colour at age 18 years and personal history of keratinocyte cancer. There were no significant interactions between pairs of variables in the final model. In, addition, there was no significant correlation between nevus count and CDKN2A pathogenic variant carrier status, which might have been expected and influential in the model.(23) Only 2% of cases were mutation carriers (13), however, and model performance did not change when these carriers were excluded from the analysis. Relative risk and OPERA estimates are shown for both studies in Table 2, the number of body naevi ≥2mm in diameter was by far the strongest predictor in terms of differentiating cases from controls on a population basis.
Table 2.
Relative risk estimatesa and odds per adjusted standard deviation (OPERA)b for the multivariable melanoma risk prediction model on the Australian Melanoma Family Study (development dataset) and Leeds Melanoma Case-Control Study (external validation dataset)
| Australia Melanoma Family Study | Leeds Melanoma Case-Control Studyd | |||
|---|---|---|---|---|
| Risk factor in the modelc | Relative riska (95% CI) | Relative riska (95% CI) | ||
| Total number of whole-body naevi ≥2mm diametere,f | ||||
| < 28 | 1.00 | 1.00 | ||
| 28–61 | 3.10 (1.47 – 6.53) | 2.88 (2.15 – 3.86) | ||
| 62–143 | 6.74 (3.32 – 13.67) | 6.28 (4.18 – 9.43) | ||
| More than 144 | 20.03 (9.76 – 41.09) | 28.72 (8.79 – 93.81) | ||
| Solar Lentigines on upper backd,e | ||||
| <20% | 1.00 | 1.00 | ||
| 20% | 1.41 (0.68 – 2.92) | 2.71 (1.70– 4.31) | ||
| 40% | 2.08 (0.97 – 4.46) | 2.43 (1.52 – 3.87) | ||
| 60% | 2.81 (1.25 – 6.29) | 2.20 (1.34 – 3.61) | ||
| 80% | 3.08 (1.40 – 6.76) | 2.34 (1.36 – 4.03) | ||
| 100% | 3.49 (1.29 – 9.41) | 0.77 (0.35 −1.67) | ||
| Hair colour at age 18 | ||||
| Black/ dark brown | 1.00 | 1.00 | ||
| Light brown | 1.22 (0.80 – 1.85) | 1.62 (1.19 – 2.20) | ||
| Blonde | 2.29 (1.30 – 4.03) | 2.46 (1.64 – 3.68) | ||
| Red | 3.29 (1.39 – 7.78) | 4.17 (2.45 – 7.09) | ||
| Personal history of keratinocyte cancer | ||||
| No | 1.00 | |||
| Yes | 9.76 (2.09 – 45.54) | 1.31 (0.56 – 3.04) | ||
| Analysed as OPERAsb | Odds ratio (95% CI) | P value | Odds ratio (95% CI) | P value |
| Total number of body naevi ≥2mm diameter e,f | 3.51 (2.71–4.54) | <0.001 | 2.56 (2.23–2.95) | <0.0001 |
| Solar Lentigines on upper back d,e | 1.50 (1.19–1.89) | 0.001 | 0.95 (0.83–1.08) | 0.41 |
| Hair colour at age 18 years | 1.37 (1.15–1.63) | 0.001 | 1.44 (1.27–1.64) | <0.0001 |
| Personal history of keratinocyte cancer | 1.12 (0.93–1.34) | 0.236 | 0.98 (0.87–1.11) | 0.81 |
Odds ratios were used to estimate relative risks, and were adjusted for all other variables in the model as well as age, sex and city of recruitment.
The OPERA scores are calculated for variables included in the prediction model to enable comparison of the predictive strengths, in terms of ability to differentiate cases from controls, of the risk factors across different diseases and populations using the formula OPERA= exp [In (RR)/A]=RRs; where RR is relative risk, and A=1/s adjusted standard deviations.(16) The standard deviations were adjusted for age (in 5 year intervals) and sex using the control data. Given the difference between the upper and lower quartiles of a normal distribution is approximately 2.54 standard deviations, the estimated OPERA risk gradients are equivalent to interquartile risk ratios of 3.512.54 ~ 24 (95% CI 13–47) for the total number of body naevi ≥ 2mm diameter, 1.502.54 ~ 2.8 (95% CI 1.6–5.0) for solar lentigines on the upper back, 1.372.54 ~ 2.2 (95%CI 1.4 −3.5) for hair colour at age 18 years, and 1.122.54 ~ 1.3 (95% CI 0.8 – 2.1) for personal keratinocyte cancer.
The model intercept was 0.53 (95% CI 0.05 – 5.79) and attributable fraction, as calculated from the distribution of relative risk among the cases,(30) was 0.96 (95% CI 0.95–0.97).
In the Leeds Melanoma Case-Control Study, freckle density categorized by the cut points 20%, 40%, 60%, 80%, 100% was used as a proxy for solar lentigines (Spearman correlation coefficient=0.28, 95% CI 0.21– 0.35, p<0.0001).
Assessed from clinical examinations in dermatology clinics.
Based on quartile cut-points in the Australian Melanoma Family Study controls.
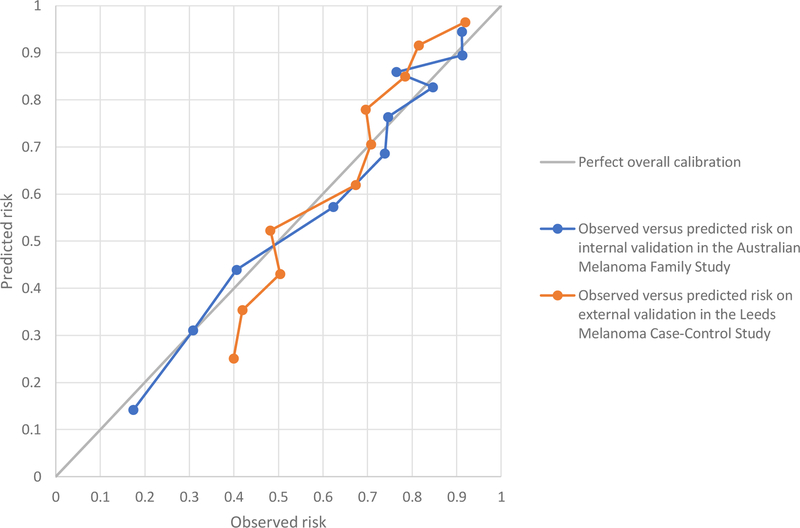
The AUC was 0.79 (95% CI 0.76–0.83) on internal validation in the Australian study (Online Figure 1) and was 0.73 (95% CI 0.70–0.75) on external validation in the Leeds study (Online Figure 2). The AUC did not increase when the age and sex distribution was reweighted to the general population (Online Table 3). The H-L test p-value was 0.30 on internal validation and <0.001 on external validation. Figure 1 shows that the poor calibration for the Leeds study was due to the model under-estimating risk at lower risk levels and over-estimating risk at higher levels. In sensitivity analyses, calibration in the Leeds study did not improve when we re-calculated lifetime absolute risks using Leeds incidence and mortality rates, or when using naevus quartile cut-points based on the Leeds rather than Australian dataset (results not shown).
Figure 1. Calibration plot showing the observed versus the model’s predicted risk of incident melanomas across on internal and external validation.
This graph compares the observed and the model’s predicted risk of incident melanomas on internal validation in the Australian Melanoma Family Study (blue line) and on external validation in the Leeds Melanoma Case-Control Study (orange line), with perfect overall calibration (black line).
Discussion
This melanoma risk prediction model incorporating clinically-assessed naevi and solar lentigines had good discrimination that was maintained on external validation; and good overall calibration internally but less so externally. Consistent with previous melanoma risk prediction models, we observed higher melanoma risks and greater predictive strength from naevus counts equivalent to 10–20 fold interquartile risk ratios.(8) Solar lentigines mostly arise from prolonged sun exposure, and are a recognised melanoma risk factor,(24) but have not been included in previous models.(8)
This model’s discrimination compares well with previous melanoma risk prediction models, which reported AUCs from 0.62 to 0.93, although most of these were not externally validated.(8, 25, 26) There is higher discriminative performance for models using clinically-assessed than self-assessed dermatologic risk factors such as naevus counts, probably because they are more accurately measured by clinicians.(9) Our previous model using only self-assessed risk factors(17) reported lower AUCs, ranging from 0.63 to 0.70 on external validation, although this can be improved by incorporating data on common genomic variants.(27) The low p-value for the H-L test indicated poor overall calibration on external validation, with poorer agreement between the observed versus the model’s predicted lifetime absolute risk estimates by decile in the Leeds study. Poor calibration indicates that measures of lifetime risk generated using the model may be less accurate for non-Australian populations even though the model has good discrimination. However, the H-L test is sensitive to sample size, which may lead to a low p-value in situations of slight deviation from perfect fit, especially with a large number of participants in the external validation dataset.(19, 28) Unfortunately, in the absence of cohort studies with clinically-assessed melanoma risk factors, we were unable to estimate absolute measures of calibration or evaluate net benefit.
Strengths of our study include the multi-centred, population-based design, external validation, comprehensive assessment of risk factors and inclusion of clinically-assessed dermatologic risk factors. The Australian and Leeds study shared measurement protocols to ensure that risk factors were measured consistently across the studies and that clinical assessments were conducted with the same protocol. We used robust statistical approaches, including multiple imputation to impute missing data and external validation of the model in an independent population.
Despite the similar protocols, solar lentigines was not measured in the Leeds study, and as a consequence freckle density was used as a proxy for solar lentigines for the external validation. This is a limitation as they have different aetiologies; freckle density is an inherited characteristic associated with MC1R that has a similar distribution across both populations whereas solar lentigines is associated with prolonged sun exposure and thus less common in the UK population.(24, 29) This proxy was selected due to its moderate correlation with solar lentigines in the Australian study; however, as it was a stronger predictor than freckle density, the estimated external validation discriminative performance is probably an underestimate. Clinical assessment of naevi is more accurate than self-report(9) and our model has better discriminative performance compared with models that rely on only self-assessed risk factors(8) but is associated with more consultation time and costs. We have also not included genetic risk factors in the model. Other potential limitations of case-control studies include possible selection bias and recall bias.(11)
In summary, this melanoma risk prediction model incorporating clinically-assessed risk factors has good discrimination, with ability to distinguish between individuals with and without melanoma across two populations with different ambient sun exposure. The model may be useful for offering tailored preventive interventions such as sun protection advice and skin screening based on personal risk level, in primary care and other clinical settings where dermatologic risk factors can be assessed. Prospective evaluation of the clinical risk prediction model will be important to estimate absolute calibration, net benefit, cost-effectiveness, and impact on risk behaviours or melanoma outcomes.
Supplementary Material
What is already known about this topic?
Melanoma risk prediction models may be useful in prevention by tailoring interventions to personalised risk levels. For reasons of feasibility, time and cost, many melanoma prediction models use self-assessed risk factors. However, individuals tend to underestimate their nevus numbers.
What does this study add?
We present a melanoma risk prediction model, which includes clinically-assessed whole-body naevi and solar lentigines, and self-assessed risk factors including pigmentation phenotype, sun exposure, family history and history of keratinocyte cancer. This model performs well on discrimination, the model’s ability to distinguish between individuals with and without melanoma, and may assist clinicians to stratify patients by melanoma risk for targeted preventive interventions.
Acknowledgments
Funding sources
Kylie Vuong was supported by a University of Sydney Postgraduate Scholarship in Cancer Epidemiology (funded through a Cancer Institute NSW fellowship to AEC) and a Sydney Catalyst Top-Up Research Scholar Award. Anne E Cust was supported by fellowships from the Cancer Institute NSW (15/CDF/1-14) and the National Health and Medical Research Council (NHMRC) (1147843). The studies received funding from the NHMRC (project grants 107359, 211172 and Program Grant 402761 to GJM); project grants from the Cancer Councils New South Wales (77/00, 06/10), Victoria and Queensland (371); the US National Institutes of Health (via RO1 grant CA-83115 to the international Melanoma Genetics Consortium, GenoMEL, www.genomel.eu); and Cancer Research UK (Project Grant C8216/A6129 and Programme awards C588/A4994, C588/A10589 and C588/A19167).
Abbreviations used:
- OPERA
odds per age- and sex-adjusted standard deviation
- AUC
area under curve
- H-L
Hosmer-Lemeshow
- CI
confidence interval
Footnotes
Conflict of interest
None reported.
References
- 1.Erdmann F, Lortet-Tieulent J, Schuz J, Zeeb H, Greinert R, Breitbart EW, et al. International trends in the incidence of malignant melanoma 1953–2008--are recent generations at higher or lower risk? International Journal of Cancer 2013;132(2):385–400. [DOI] [PubMed] [Google Scholar]
- 2.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Picconi O, Boyle P, et al. Meta-analysis of risk factors for cutaneous melanoma: II. Sun exposure. Eur J Cancer 2005;41(1):45–60. [DOI] [PubMed] [Google Scholar]
- 3.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Abeni D, Boyle P, et al. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. European Journal of Cancer 2005;41(1):28–44. [DOI] [PubMed] [Google Scholar]
- 4.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Zanetti R, Masini C, et al. Meta-analysis of risk factors for cutaneous melanoma: III. Family history, actinic damage and phenotypic factors. European Journal of Cancer 2005;41(14):2040–59. [DOI] [PubMed] [Google Scholar]
- 5.Fang S, Han J, Zhang M, Wang LE, Wei Q, Amos CI, et al. Joint effect of multiple common SNPs predicts melanoma susceptibility. PLoS One 2013;8(12):e85642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.RACGP. Guidelines for preventive activities in general practice 9th ed. East Melbourne, Victoria: The Royal Australian College of General Practitioners; 2016. [Google Scholar]
- 7.Freedman A, Seminara D, Mitchell G, Hartge P, Colditz G, Ballard-Barbash R, et al. Cancer risk prediction models: a workshop on development, evaluation and application. Journal of the National Cancer Institute 2005:715–23. [DOI] [PubMed]
- 8.Vuong K, McGeechan K, Armstrong BK, Cust AE. Risk prediction models for incident primary cutaneous melanoma: a systematic review. JAMA Dermatol 2014;150(4):434–44. [DOI] [PubMed] [Google Scholar]
- 9.Cust AE, Pickles KM, Goumas C, Vu T, Schmid H, Nagore E, et al. Accuracy of self-reported nevus and pigmentation phenotype compared with clinical assessment in a population-based study of young Australian adults. Cancer Epidemiol Biomarkers Prev 2015;24(4):736–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cust AE, Goumas C, Vuong K, Davies JR, Barrett JH, Holland EA, et al. MC1R genotype as a predictor of early-onset melanoma, compared with self-reported and physician-measured traditional risk factors: an Australian case-control-family study. BMC Cancer 2013;13:406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cust AE, Schmid H, Maskiell JA, Jetann J, Ferguson M, Holland EA, et al. Population-based, case-control-family design to investigate genetic and environmental influences on melanoma risk: Australian Melanoma Family Study. Am J Epidemiol 2009;170(12):1541–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Newton-Bishop JA, Chang YM, Iles MM, Taylor JC, Bakker B, Chan M, et al. Melanocytic nevi, nevus genes, and melanoma risk in a large case-control study in the United Kingdom. Cancer Epidemiol Biomarkers Prev 19 United States: 2010 Aacr; 2010. p. 2043–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harland M, Cust AE, Badenas C, Chang YM, Holland EA, Aguilera P, et al. Prevalence and predictors of germline CDKN2A mutations for melanoma cases from Australia, Spain and the United Kingdom. Hered Cancer Clin Pract 2014;12(1):20. doi: 10.1186/897-4287-12-20 eCollection 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Aitken JF, Youl P, Green A, MacLennan R, Martin NG. Accuracy of case-reported family history of melanoma in Queensland, Australia. Melanoma Res 1996;6(4):313–7. [DOI] [PubMed] [Google Scholar]
- 15.Vergouwe Y, Royston P, Moons KG, Altman DG. Development and validation of a prediction model with missing predictor data: a practical approach. J Clin Epidemiol 63 United States: 2010 Elsevier Inc; 2010. p. 205–14. [DOI] [PubMed] [Google Scholar]
- 16.Hopper JL. Odds per adjusted standard deviation: comparing strengths of associations for risk factors measured on different scales and across diseases and populations. Am J Epidemiol 2015;182(10):863–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vuong K, Armstrong BK, Weiderpass E, Lund E, Adami HO, Veierod MB, et al. Development and External Validation of a Melanoma Risk Prediction Model Based on Self-assessed Risk Factors. JAMA Dermatol 2016;152(8):889–96. [DOI] [PubMed] [Google Scholar]
- 18.Gail MH, Brinton LA, Byar DP, Corle DK, Green SB, Schairer C, et al. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. Journal of the National Cancer Institute 1989;81(24):1879–86. [DOI] [PubMed] [Google Scholar]
- 19.Steyerberg EW. Clinical prediction models Gail MH, editor. New York: Springer; 2010. [Google Scholar]
- 20.Pepe MS, Fan J, Seymour CW, Li C, Huang Y, Feng Z. Biases introduced by choosing controls to match risk factors of cases in biomarker research. Clin Chem 58 United States2012 p. 1242–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bender R, Lange S. Adjusting for multiple testing—when and how? Journal of Clinical Epidemiology 2001;54(4):343–9. [DOI] [PubMed] [Google Scholar]
- 22.Collins GS, Reitsma JB, Altman DG, Moons KG. Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. Bmj 2015;350:g7594. [DOI] [PubMed] [Google Scholar]
- 23.Demenais F, Mohamdi H, Chaudru V, Goldstein AM, Newton Bishop JA, Bishop DT, et al. Association of MC1R variants and host phenotypes with melanoma risk in CDKN2A mutation carriers: a GenoMEL study. Journal of the National Cancer Institute 2010;102(20):1568–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kvaskoff M, Siskind V, Green AC. Risk factors for lentigo maligna melanoma compared with superficial spreading melanoma: a case-control study in Australia. Arch Dermatol 2012;148(2):164–70. [DOI] [PubMed] [Google Scholar]
- 25.Olsen CM, Pandeya N, Thompson BS, Dusingize JC, Webb PM, Green AC, et al. Risk Stratification for Melanoma: Models Derived and Validated in a Purpose-Designed Prospective Cohort. J Natl Cancer Inst 2018. [DOI] [PubMed]
- 26.Olsen CM, Neale RE, Green AC, Webb PM, the QS, the Epigene S, et al. Independent Validation of Six Melanoma Risk Prediction Models. J Invest Dermatol 2015;135(5):1377–84. [DOI] [PubMed] [Google Scholar]
- 27.Cust AE, Drummond M, Kanetsky PA, Goldstein AM, Barrett JH, MacGregor S, et al. Assessing the incremental contribution of common genomic variants to melanoma risk prediction in two population-based studies. J Invest Dermatol 2018;8(18):32046–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kramer AA, Zimmerman JE. Assessing the calibration of mortality benchmarks in critical care: The Hosmer-Lemeshow test revisited. Crit Care Med 2007;35(9):2052–6. [DOI] [PubMed] [Google Scholar]
- 29.Cust AE, Goumas C, Holland EA, Agha-Hamilton C, Aitken JF, Armstrong BK, et al. MC1R genotypes and risk of melanoma before age 40 years: a population-based case-control-family study. International Journal of Cancer 2012;131(3):E269–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bruzzi P, Green SB, Byar DP, Brinton LA, Schairer C. Estimating the population attributable risk for multiple risk factors using case-control data. American Journal of Epidemiology 1985;122(5):904–14. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.