Abstract
We performed a transcriptome-wide meta-analysis and gene co-expression network analysis to identify genes and gene networks dysregulated in the peripheral blood of bipolar disorder (BD) cases relative to unaffected comparison subjects, and determined the specificity of the transcriptomic signatures of BD and schizophrenia (SZ). Nineteen genes and 4 gene modules were significantly differentially expressed in BD cases. Thirteen gene modules were shown to be differentially expressed in a combined case-group of BD and SZ subjects called “major psychosis”, including genes biologically linked to apoptosis, reactive oxygen, chromatin remodeling, and immune signaling. No modules were differentially expressed between BD and SZ cases. Machine-learning classifiers trained to separate diagnostic classes based solely on gene expression profiles could distinguish BD cases from unaffected comparison subjects with an area under the curve (AUC) of 0.724, as well as BD cases from SZ cases with AUC = 0.677 in withheld test samples. We introduced a novel and straightforward method called “polytranscript risk scoring” that could distinguish BD cases from unaffected subjects (AUC = 0.672) and SZ cases (AUC = 0.607) significantly better than expected by chance. Taken together, our results highlighted gene expression alterations common to BD and SZ that involve biological processes of inflammation, oxidative stress, apoptosis, and chromatin regulation, and highlight disorder-specific changes in gene expression that discriminate the major psychoses.
Keywords: Bipolar disorder, biomarker, peripheral blood, schizophrenia, transcriptome
Introduction
Bipolar disorder (BD) is a highly heritable psychiatric disorder (h2 ~ 68%, (Polderman et al., 2015)) with a lifetime prevalence near 1% (Ferrari et al., 2016). Although there are symptomological, neuropsychological, and brain-structural differences between BD and schizophrenia (SZ) (Murray et al., 2004); there is strong genetic evidence supporting the hypothesis that BD and SZ have shared molecular abnormalities (Bulik-Sullivan et al., 2015; Purcell et al., 2009). Dysregulation in immune and neurodevelopment genes have been associated with BD and SZ from postmortem brain studies (Chen et al., 2013; Darby et al., 2016; Hess et al., 2016; Iwamoto et al., 2005; Pacifico and Davis, 2017). Investigating gene expression profiles in the blood is a critical next step to uncovering biomarkers and molecular processes that may be common and discordant between BD and SZ. We sought to identify gene expression alterations associated with BD by meta-analyzing a collection of publicly available datasets from peripheral blood studies that assayed transcriptome-wide profiles. We then conducted a cross-disorder transcriptomic analysis of BD and SZ to find shared and discordant gene expression changes.
Some prior studies compared gene expression profiles between BD and SZ cases, but those have used small samples or examined only a few candidate genes (Cattane et al., 2015; Drexhage et al., 2010; Shao and Vawter, 2008; Tesli et al., 2016; Tsuang et al., 2005; Wirgenes et al., 2014). Although the brain is the most relevant tissue for investigating gene-expression abnormalities associated with psychopathology, there is evidence that gene-expression levels in the blood are significantly correlated with those in the brain (Kim et al., 2014; McKenzie et al., 2014; Tylee et al., 2013). We recently showed that SZ-associated networks of co-expressed genes detected in the prefrontal cortex were enriched with neurodevelopmental and immune signaling function, and showed greater than chance over-representation of SZ-associated networks detected in blood (Hess et al., 2016).
In this study, we obtained all publicly available transcriptome-wide gene expression data from peripheral blood for BD cases (n = 95) and unaffected comparison subjects (n = 111), and integrated transcriptomic data from 258 SZ cases and an additional 241 unaffected comparison subjects from our previous study of SZ (Hess et al., 2016). This study characterizes transcriptomic changes in peripheral blood that are shared and dissimilar between BD and SZ, which may shed light on biomarkers and pathogenic mechanisms of major psychosis.
Methods and Materials
Literature search for transcriptome studies of BD
An electronic literature search was conducted up to April 30, 2018 using the keywords “bipolar disorder”, “manic”, “mania”, “microarray”, “gene expression”, “transcriptom*”, and “blood” across the databases PubMed, Scopus, and Google Scholar, coupled with a query of published gene expression data sets in Gene Expression Omnibus (GEO) and ArrayExpress. Studies that met the following inclusion criteria were kept: (1) recruited affected cases with a clinical diagnosis of BD based on DSM-IV or DSM-IV-TR criteria, (2) recruited unrelated, unaffected comparison subjects, and (3) had available raw transcriptome-wide gene expression data (not quantitative PCR) generated from whole blood or peripheral blood leukocytes. One potentially useful study was excluded due to relatedness between cases and controls (Matigian et al., 2007). A total of seven studies were retained for analysis yielding 95 BD cases and 111 unaffected comparison subjects. Table 1 provides an overview of the demographics, sample sizes, and numbers of probes collapsed to genes for the seven studies included in our analysis (Beech et al., 2010; Bousman et al., 2010; Clelland et al., 2013; Padmos et al., 2008; Savitz et al., 2013; Tsuang et al., 2005; Witt et al., 2014).
Table 1.
Demographics of bipolar disorder cases and unaffected comparison subjects included in our transcriptome-wide meta-analysis.
| Study ID | Tissue | Predominant Ancestry (%) | BD Cases (n) | Controls (n) | % Male | Age (s.e.) | Genes analyzed (n) | Array |
|---|---|---|---|---|---|---|---|---|
| Witt et al. (2014) | Leukocytes | European (100%) | 11 | 10 | 100 | 48.19 (2.47) | 15,611 | Affymetrix Human Exon 1.0 ST Illumina |
| Beech et al. (2010) | Whole blood | European (65.7%) | 20 | 15 | 31.4 | 34.31 (1.78) | 14,564 | Human-6 v2 BeadChip Affymetrix |
| Bousman et al. (2010) | Leukocytes | European (70.5%) | 9 | 8 | 70.6 | 43.59 (1.75) | 18,629 | Human Exon 1.0 ST Affymetrix |
| Clelland et al. (2013) | Leukocytes | European (66.7%) | 26 | 25 | 100 | 36.65 (1.81) | 22,409 | Human Genome U133 Plus 2.0 |
| Padmos et al. (2008) | Monocytes | -- | 5 | 6 | 45.5 | 23.64 (2.90) | 9,702 | Affymetrix U95v2 Illumina |
| Savitz et al. (2013) | Monocytes | -- | 8 | 24 | 46.9 | 35.03 (1.96) | 20,504 | HT-12 v4 expression BeadChip Affymetrix |
| Tsuang et al. (2005) | Leukocytes | Asian (100%) | 16 | 23 | 41 | 42.38 (2.16) | 22,409 | Human Genome U133 Plus 2.0 |
| Total studies: 7 | 95 | 111 |
Data not reported designated as (--). Abbreviations: bipolar disorder (BD), standard error (s.e.).
Data import and quality control
A full description of our pre-processing, quality control, and normalization steps are provided in the Supplementary Methods. Briefly, raw array data from seven studies were imported into R (version 3.4.1) for pre-processing using a custom pipeline available on Github (https://github.com/hessJ/gxp_bipolar). Log2 and quantile normalized array data was obtained per study for between 9,702 and 22,409 genes (mean ~17,689 genes).
Transcriptome-wide meta-analysis
We fit a multivariate linear regression model that specified expression of each gene as a dependent variable (DV) and diagnosis as an independent variable (IV). Age, sex (except for the all-male sample from Witt et al., [2014]), and array batch (in the Tsuang et al., [2005] sample which featured both Affymetrix U133A and U133 Plus 2.0 arrays) and significant latent surrogate variables were included as covariates. We applied this model to every gene per study to compute the adjusted mean difference in gene expression between BD cases relative to unaffected comparison subjects. Genes that were present in at least four out of the seven studies were retained for meta-analysis (ngenes = 17,201). A conservative random effect model inverse-variance weighted meta-analysis was used to pool differential expression estimates per gene across studies. The R package metafor (Viechtbauer, 2010) was used to perform the meta-analysis with the DerSimonian-Laird estimator.
Differential expression of weighted gene co-expression networks and module preservation
We derived a weighted gene co-expression network using the R package WGCNA (Langfelder and Horvath, 2008) to investigate gene regulatory networks associated with BD and major psychosis as described in the Supplementary Methods.
Linear regressions were fit to estimate the effect of diagnostic group on module eigengenes while simultaneously adjusting for confounding effects of age, sex, study site, and significant surrogate variables. The following diagnostic groups were compared: (1) BD cases compared to unaffected subjects, (2) BD cases compared to SZ cases, and (3) BD and SZ (together called ‘major psychosis’) compared to the combined set of unaffected subjects. P-values were adjusted using the Benjamini-Hochberg false discovery rate (FDR) procedure within each network (BD and controls: 24 modules; BD, SZ, and controls: 33 modules × 2 tests = 66 total tests). A threshold of FDRp < 0.05 was used declare statistical significance for differential gene module expression.
Pathway and gene set analysis
A permutation-based gene set test for differential expression was performed using the piano package from R/Bioconductor (Väremo et al., 2013), which is described in the Supplementary Methods.
Discriminating diagnostic groups with machine learning classifiers
We evaluated classification performance of multiple machine learning (ML) algorithms in predicting diagnostic status of subjects based on expression levels for top differentially expressed autosomal genes. Details regarding the model training and testing procedures are described in the Supplementary Methods. The classification analysis was conducted using the R/CRAN package exprso (Quinn et al., 2016).
Polytranscript scoring
We developed an approach analogous to polygenic risk scoring for application to transcriptomic data (Purcell et al., 2009), which computes a score of the average expression level across multiple genes and weighted by their disease-associated expression changes. This score can serve as a predictor of disease that accounts for expression variation across many genes. An overview of the method is provided in Supplementary Figure 4 with details available in the Supplementary Methods. A logistic regression model was used in the training set with diagnostic status set as the outcome variable and polytranscript score as the predictor variable. After training, the model was then deployed in the test sample and classification accuracy was obtained (metric: ROC AUC). A 10-fold bootstrapping procedure was performed, each iteration producing a new training and test set in order to calculate mean and standard deviation of ROC AUC.
Comparing significant results with previous mega-analyses
A list of 382 differentially expressed genes associated with BD was obtained from a previous transcriptome-wide meta-analysis of postmortem brain tissues (Seifuddin et al., 2013), which we cross-referenced to the lists of differentially expressed genes and gene modules associated with BD and SZ identified in the present study. We also compared results from our current meta-analysis of BD cases with our previous study of peripheral blood and postmortem brain gene expression changes for SZ cases (Hess et al., 2016). Hypergeometric tests of enrichment were performed when overlap was observed between gene lists. We tested whether differential gene expression values observed in BD were similar to changes in SZ cases using Pearson’s correlation coefficient.
Results
Leukocyte abundance and surrogate variables
BD cases and unaffected comparison subjects did not show a statistically significantly difference in estimated abundance of leukocytes (Supplementary Table 1). One significant surrogate variable was recovered from the data set of BD cases and unaffected comparison subjects. A second surrogate variable analysis (SVA) was performed after adding transcriptomic profiles for SZ cases and unaffected comparison subjects from our previous study (Hess et al., 2016), yielding 4 SVs which were correlated with estimated abundance of 16 out of 17 circulating leukocytes (Supplementary Figure 3).
Genes differentially expressed in BD
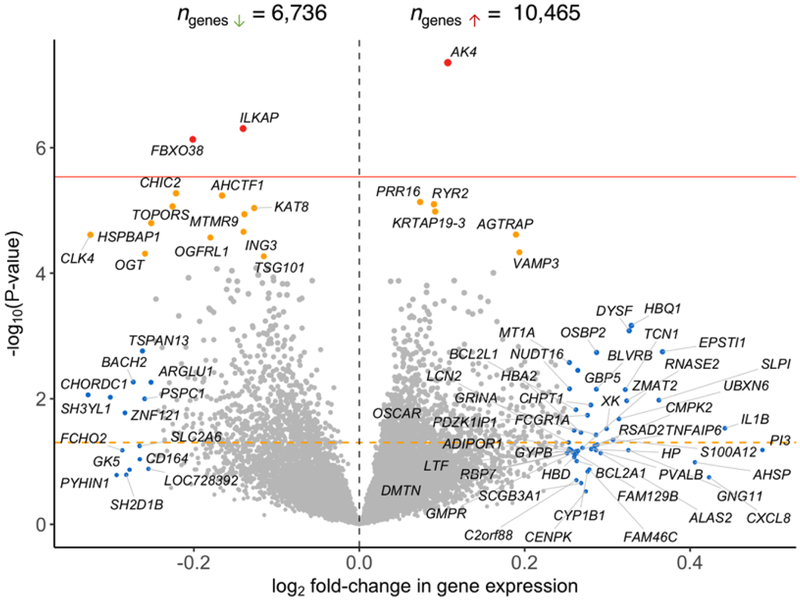
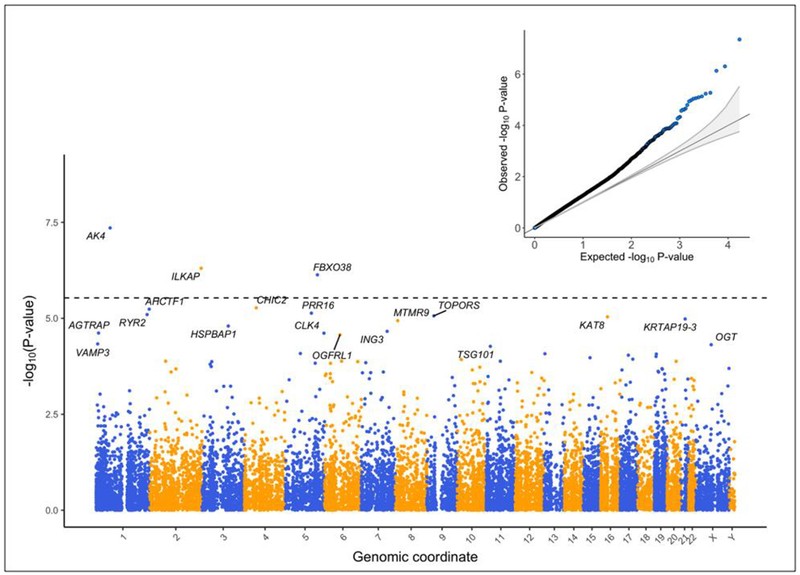
Our meta-analysis uncovered 19 genes significantly differentially expressed (six over-regulated and 13 under-regulated) in BD cases relative to unaffected comparison subjects (FDRp < 0.05, Figures 1 and 2). Summary statistics for the 19 dysregulated genes are provided in Table 2. These genes did not show significant evidence of heterogeneity across studies (Cochran’s Q-test, p > 0.05). Forest plots for the 19 significant genes are provided in Supplementary File 1 with study-wise and pooled estimates. A total of 60 gene sets were significantly differentially expressed in BD cases relative to unaffected comparison subjects (FDRp < 0.05) (Supplementary Table 2), from which 30 gene sets were over-expressed and 30 gene sets were under-expressed. Some of the biological gene sets over-expressed in BD included (all P-values = 0.0001, FDRp < 0.05): immune and cytokine signaling, cell adhesion, pro-apoptotic signaling, and positive regulation of reactive oxygen species. Conversely, some of the major biological processes down-regulated in BD cases included (all P-values = 0.0001, FDRp < 0.05): regulation of transcription, nuclear export of RNA and splicing, DNA repair, and histone modification. Leave-one-study-out sensitivity analyses for the 19 differentially expressed genes associated with BD indicated that pooled effects were relatively stable. The association for 18 genes with BD remained significant (p<0.05) in all leave-one-study-out analyses. The significance of the association between TSG101 and BD was lost when we removed Clelland et al., (2013) from the meta-analysis (Supplementary Figure 5).
Figure 1.
Volcano plot showing the log2 fold-change in gene expression between bipolar disorder (BD) cases and unaffected comparison subjects (horizontal axis) relative to p-value for the fold-change estimate (vertical axis). An orange horizontal dotted line denotes p = 0.05. A red dotted horizontal line denotes a Bonferroni significance threshold (adjusted p = 0.05/17201 = 2.91e–06). Genes shaded red showed an association with BD at a Bonferroni adjusted p < 0.05. Genes shaded in orange showed an association with BD at a Benjamini-Hochberg false discovery rate (FDR) p < 0.05. Genes shaded blue showed an absolute log2 fold-change > 0.25 in BD cases.
Figure 2.
The association significance (vertical axis) and physical location of each gene (horizontal axis) is depicted in this Manhattan plot. A horizontal dotted line denotes a Bonferroni adjusted significance threshold. The genes labeled in this plot reached a statistical significance in association with BD at a Benjamini-Hochberg false discovery rate adjusted p-value < 0.05. A quantile-quantile (QQ) plot in the inset shows the expected distribution of p-values under the null hypothesis of no association versus the p-values observed from our meta-analysis.
Table 2.
Summary statistics for 19 genes that were significant over- or under-expressed in bipolar disorder (BD) cases compared to unaffected subjects from our transcriptome-wide meta-analysis of 17,201 genes.
| Heterogeneity statistics | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| HGCN symbol | Effect direction per study | # of studies | # of cases | # of controls | Total n | Log 2 FC | Std. Error | p-value | FD Rp | Cochran’s Q | p-value |
| AK4 | -++++ | 5 | 64 | 86 | 150 | 0.107 | 0.020 | 4.42E-08 | 0.0008 | 2.004 | 0.735 |
| ILKAP | 6 | 90 | 105 | 195 | −0.140 | 0.028 | 4.96E-07 | 0.0042 | 4.571 | 0.470 | |
| FBXO38 | -+--- | 5 | 79 | 95 | 174 | −0.201 | 0.041 | 7.38E-07 | 0.0042 | 2.652 | 0.618 |
| CHIC2 | 6 | 90 | 105 | 195 | −0.222 | 0.049 | 5.33E-06 | 0.018 | 3.203 | 0.669 | |
| AHCTF1 | ---+--- | 7 | 95 | 111 | 206 | −0.166 | 0.037 | 5.79E-06 | 0.018 | 3.366 | 0.762 |
| PRR16 | +++++ | 5 | 70 | 90 | 160 | 0.074 | 0.016 | 7.34E-06 | 0.018 | 4.768 | 0.312 |
| RYR2 | ++++++ | 6 | 87 | 87 | 174 | 0.090 | 0.020 | 7.98E-06 | 0.018 | 4.472 | 0.484 |
| TOPORS | ------ | 6 | 87 | 87 | 174 | −0.226 | 0.051 | 8.63E-06 | 0.018 | 2.234 | 0.816 |
| KAT8 | ---- | 4 | 55 | 78 | 133 | −0.127 | 0.029 | 9.16E-06 | 0.018 | 0.195 | 0.978 |
| KRTAP19–3 | +-+++ | 5 | 74 | 82 | 156 | 0.092 | 0.021 | 1.04E-05 | 0.018 | 4.222 | 0.377 |
| MTMR9 | -+----- | 7 | 95 | 111 | 206 | −0.139 | 0.032 | 1.15E-05 | 0.018 | 4.838 | 0.565 |
| HSPBAP1 | ------ | 6 | 90 | 105 | 195 | −0.252 | 0.058 | 1.59E-05 | 0.023 | 7.633 | 0.178 |
| ING3 | -+----- | 7 | 95 | 111 | 206 | −0.140 | 0.033 | 2.19E-05 | 0.028 | 2.787 | 0.835 |
| AGTRAP | +++++ | 5 | 74 | 82 | 156 | 0.190 | 0.045 | 2.42E-05 | 0.028 | 4.000 | 0.406 |
| CLK4 | -+--- | 5 | 82 | 81 | 163 | −0.325 | 0.077 | 2.44E-05 | 0.028 | 2.355 | 0.671 |
| OGFRL1 | ------ | 6 | 90 | 105 | 195 | −0.180 | 0.043 | 2.70E-05 | 0.029 | 3.158 | 0.676 |
| VAMP3 | +++-+-+ | 7 | 95 | 111 | 206 | 0.194 | 0.048 | 4.65E-05 | 0.047 | 7.001 | 0.321 |
| OGT | ------- | 7 | 95 | 111 | 206 | −0.259 | 0.064 | 4.88E-05 | 0.047 | 11.005 | 0.088 |
| TSG101 | ---+-- | 6 | 87 | 87 | 174 | −0.115 | 0.029 | 5.39E-05 | 0.049 | 4.794 | 0.441 |
Abbreviations: Fold change (FC), false discovery rate adjusted p-value (FDRp)
Among genes reaching a nominally significant association (p < 0.05) with BD, 70.6% were found to be over-expressed, which was statistically greater than expected by chance (binomial p = 9.44×10−59). The quantile-quantile plot in Figure 2 shows that meta-analysis associations deviated from the null with a global inflation factor of λGC = 1.54. Three out of the 19 significant genes associated with BD were also found to be significantly differentially expressed in the peripheral blood of SZ cases (FDRp < 0.05; AK4, KAT8, and OGFRL1), but this was not more than expected by chance (one-tailed Fisher’s exact test p = 0.26) (Hess et al., 2016). AK4 was over-expressed in SZ cases while KAT8 was under-expressed in SZ cases (both results concordant with BD cases). While OGFRL1 was under-expressed in SZ cases (discordant with BD cases). We compared effect sizes for the BD-unaffected contrast with those observed for the SZ-unaffected contrast and found a small but statistically significant negative correlation 16,436 genes(Pearson’s r = −0.069, p = 3.69×10−19, 16,436 common genes). Filtering the list of genes down to 480 genes related to immune and cytokine signaling led to a significant positive correlation in differential expression effect sizes between SZ and BD (Pearson’s r = 0.105, p = 0.021). There was no overlap between genes presently associated with BD in blood and those reported in previous study of BD postmortem brain tissue (Seifuddin et al., 2013).
Gene modules associated with BD
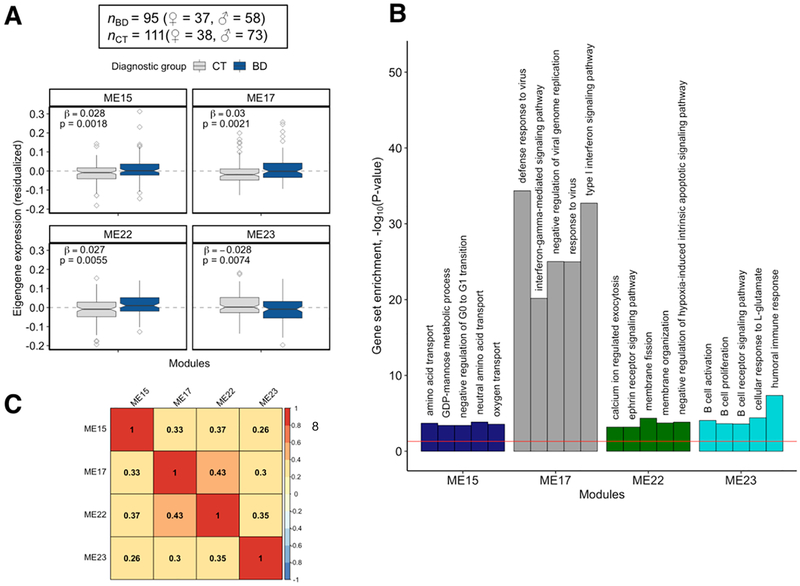
The first WGCNA network generated for this analysis clustered 14,443 genes into 24 modules. Four modules were significantly differentially expressed in BD cases at a FDRp < 0.05 (Figure 3A): ME15 (ngenes = 137, p = 0.0018), ME17 (ngenes = 109, p = 0.0021), ME22 (ngenes = 65, p = 0.006), and ME23 (ngenes = 61, p = 0.007).
Figure 3.
(A) Four WGCNA modules (ME15, ME17, ME22, and ME23) identified from transcriptome-wide peripheral blood gene expression data were found to be differentially expressed in bipolar disorder (BD) cases relative to unaffected comparison subjects. (B) The top five most significantly over-represented Gene Ontology (GO) gene sets found in the four BD-associated WGCNA modules. (C) A heat map showing the degree of similarity between pairs of modules based on the similarity of the sets of GO gene sets that were over-represented in the four BD-associated modules. Similarity index is on a 0 – 1 scale, with a value closer to 1.0 indicating high similarity.
Pathway analysis uncovered 23 gene sets significantly enriched in ME15, 66 enriched in ME17, 40 enriched in ME22, and 7 enriched in ME23 (FDRp < 0.05, Supplementary Table 3). The top five most significantly over-represented gene sets were found in ME17 and were related to immunity: defense response to virus (p = 4.32×10−35), type I interferon signaling pathway (p = 1.89×10−33), negative regulation of viral genome replication (p = 9.5×10−26), response to virus (p = 1.02×10−25), and interferon-gamma-mediated signaling pathway (p = 6.7×10−21) (Figure 3B). ME22 and ME23 had the highest degree of semantic similarity (i.e., graph-based measure of compute topological overlap between GO terms) based on over-represented GO gene sets found in these modules (semantic similarity index = 0.43, Figure 3C). Eight genes from ME17 were found to be differentially expressed in the postmortem brain of BD cases (hypergeometric p = 0.003) as reported by Seifuddin et al. 2013(Seifuddin et al., 2013) (Supplementary Table 4). A permutation-based preservation analysis found that 21 modules (87.5%) were strongly preserved (z-summary > 10) and two modules were moderately preserved (z-summary statistics = 6.25 and 8.15) between BD cases and unaffected comparison subjects (Supplementary Figure 6).
We downloaded the summary statistics from a recent pre-print study by Krebs et al. that performed RNA-sequencing of peripheral blood samples from 240 BD cases and 240 unaffected comparison subjects to compare with our gene-level and network association analyses of BD. Krebs et al. identified six genes significantly dysregulated in the blood of BD cases. Five of these six genes were assayed by studies included in our meta-analysis (BBS9, C6orf163, COG4, DOCK3, and PVT1). We found a strong concordance of effect sizes between these our meta-analysis and Krebs et al. for the five significant genes (Pearson’s r = 0.92, p-value = 0.028). However, the five genes were not statistically significant in our meta-analysis (p-values = 0.10 – 0.87), and these genes were not included in the four WGCNA modules associated with BD.
Gene modules associated with major psychosis
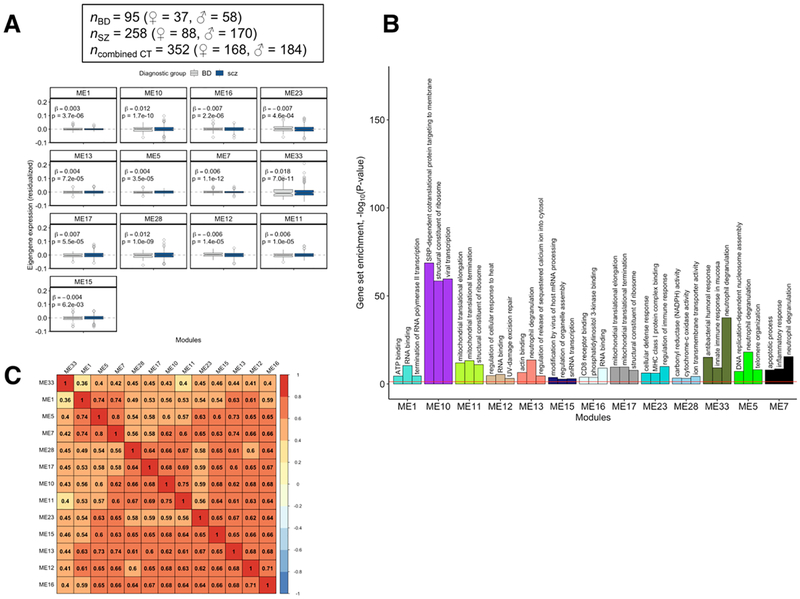
The WGNCA network generated from BD and SZ cases and unaffected comparison subjects comprised 33 gene modules comprising 11,033 genes along with 4,413 unclustered genes that were excluded from analysis. Thirteen gene modules had significant altered expression in cases with major psychosis relative to unaffected comparison subjects (FDRp < 0.05, Figure 4A), nine of which were over-expressed: ME1 (ngenes = 1,845), ME5 (ngenes = 596), ME7 (ngenes = 439), ME10 (ngenes = 331), ME11 (ngenes = 273), ME13 (ngenes = 260), ME17 (ngenes = 184), ME28 (ngenes = 107), and M33 (ngenes = 37). The four remaining modules were under-expressed in major psychosis: ME12 (ngenes = 262), ME15 (ngenes = 201), ME16 (ngenes = 185), and ME23 (ngenes = 139). No module showed a significant difference in expression between BD and SZ cases (Supplementary Table 5).
Figure 4.
(A) Thirteen WGCNA modules identified from transcriptome-wide peripheral blood gene expression data were found to be differentially expressed in cases with major psychosis (bipolar disorder + schizophrenia) relative to unaffected comparison subjects. (B) The top three most significantly over-represented Gene Ontology (GO) gene sets found in the 13 major psychosis-associated WGCNA modules. (C) A heat map showing the degree of similarity between pairs of modules based on the similarity of the sets of GO gene sets that were over-represented in the 13 major psychosis-associated modules. Similarity index is on a 0 – 1 scale, with a value closer to 1.0 indicating high similarity. Rows and columns in the heat map were ordered by performing hierarchical clustering on the pair-wise similarity values.
Supplementary Table 6 shows the number of genes dysregulated in the blood of BD cases, postmortem brain of BD cases, peripheral blood of SZ cases, or postmortem brain of SZ cases that overlapped the 13 gene modules associated with major psychosis. ME7 contained a significant amount of overlap with genes differentially expressed in the blood of BD and SZ, as well as a significant number of overlapping genes with those differentially expressed in the postmortem brain of BD cases. Seven modules showed significant overlap with the blood-based differential expression gene list for SZ. A total of 449 unique gene sets were found to be significantly over-represented in thirteen modules associated with major psychosis (Supplementary Table 7). The top three gene sets enriched in each module are presented in Figure 4B, which highlighted mitochondrial components, apoptosis, cellular respiration, transcription, translation, and inflammation and immune signaling. Gene sets related to histone modification were also found to be significantly enriched in modules that were significantly over-expressed in major psychosis (GO:0042393, p = 0.001; GO:0033522, p = 0.00761, GO:0008334, p = 0.00186; shown in Supplementary Table 7). The thirteen modules associated with major psychosis showed relatively high pair-wise similarity with respect to overlapping GO gene sets (Figure 4C). Genes involved in histone modification, apoptosis, and immunity were found to be similarly dysregulated in BD and SZ (Supplementary Figure 7).
Performance of machine learning methods
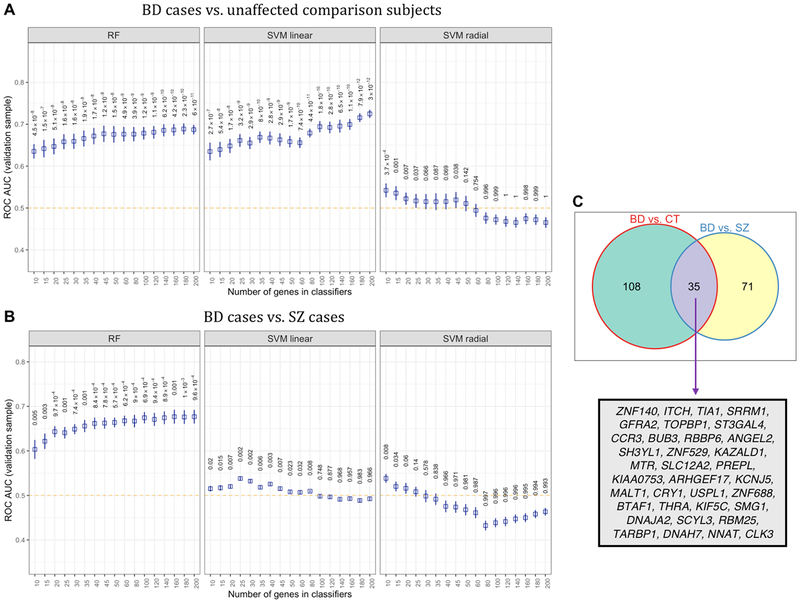
RF, linear SVM, and radial SVM yielded classification accuracies that were significantly above chance when distinguishing BD cases from unaffected comparison subjects in the test sets (Figure 5A). RF and linear SVM showed a typical pattern of classification accuracy increasing with the inclusion of genes into the models. In contrast, the classification accuracy of radial SVMs declined as more genes were included in the trained model. The highest average hold-out set classification performance for linear SVM was AUC = 0.724, (95% CI = 0.715 – 0.734, p = 2.98×10−12) using the top 200 differentially expressed genes associated with BD. RF outperformed linear SVM and radial SVM in terms of hold-out AUC in the classification task distinguishing BD and SZ cases (Figure 5B). RF reached an average hold-out set AUC of 0.677 (95% CI = 0.661 – 0.692). From the top 200 most commonly selected genes used to train ML models for Figure 5A and Figure 5B, 35 genes were found to be used in both classification tasks (Figure 5C). Our final validation analysis showed that all three classification models could significantly discriminate BD and SZ cases with better-than-chance accuracy (Supplementary Figure 8) in a pure hold-out sample wherein both case groups were profiled on the same microarray array in the same study (Tsuang et al., 2005). The best classification observed for the final validation analysis was for RF trained on the top 180 differentially expressed genes (AUC = 0.60, 95% CI = 0.586 – 0.622, p = 7.1×10−7), which was qualitatively on-par with the classification performance obtained when training models with balanced sub-sampling of the full BD-SZ data set. As our sample size was relatively small, there was a risk of overfitting the data in our classification analysis. We sought to determine if trained models could predict randomized sample labels within our 10-fold cross-validation test sets. We would not expect models trained to identify true disorder-related gene expression patterns to perform better than a random classifier (AUC ~50%) on a test data set with randomized phenotype labels. Our results showed that average AUCs dropped to a range of 0.49 to 0.52 (mean AUC = 0.51) for classification models applied to test sets with shuffled phenotype labels, suggesting that discrimination of groups was not biased from overfitting.
Figure 5.
Classification performance of three machine learning (ML) algorithms that were trained on peripheral blood gene expression profiles to distinguish bipolar disorder (BD) from unaffected comparison subjects, and BD cases from schizophrenia (SZ) cases. (A) Average ROC AUCs found in with-held test samples for random forests (RF), linear-kernel support vector machines (SVM), and radial-kernel SVM based on the classification of BD cases and unaffected comparison subjects. P-values for ROC AUCs were computed using a one-tailed t-test with a population mean of 0.5 (i.e., chance accuracy, designated by orange dotted line) (B) The performance of three ML models in classifying BD cases and SZ cases in test samples that were completely with-held from sample of BD and SZ cases used to train the ML classifiers. (C) A Venn diagram shows the number of genes used for classification in two or more bootstraps.
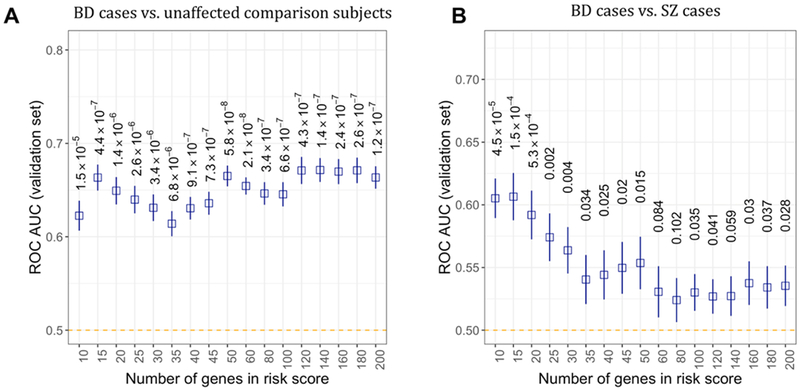
Polytranscript risk scores were able to distinguish hold-out set BD cases from unaffected comparison subjects, and BD cases from SZ cases significantly better than expected by chance (Figure 6A and Figure 6B). Numerous gene bins used for polytranscript risk scores outperformed radial SVMs in terms of AUC for the classification tasks depicted in Figure 5A and Figure 5B, as well as linear SVMs in Figure 5B. An average AUC of 0.672 (95% CI = 0.647 – 0.70, p = 1.4×10−7) and AUC of 0.607 (95% CI = 0.57 – 0.643, p = 4.5×10−5) was reached by polytranscript risk scores for the classification of BD cases from unaffected comparisons and BD from SZ cases, respectively.
Figure 6.
The average of classification accuracies (metric: receive operating characteristic area under the curve, ROC AUC) of polytranscript risk scores observed in test sets. (A) Logistic regression models trained using polytranscript scores for 10 – 200 genes could distinguish bipolar disorder (BD) case from unaffected comparison subjects, with better-than-chance performance (designated by orange dotted line).
Discussion
We performed a transcriptome-wide meta-analysis after rigorous pre-processing and quality control of gene expression data obtained from seven blood-derived microarray data sets, representing the largest collection of publicly available peripheral blood transcriptomic data (to the best of our knowledge and at the time of this writing) of BD cases and unaffected comparison subjects. Our study uncovered 19 genes, 60 biologically annotated gene sets, and four gene co-expression modules that were differentially expressed in BD cases relative to unaffected subjects. We further found that BD cases had abnormally increased expression levels of genes in involved in oxidative stress, immune signaling, and apoptosis. but significant down-regulation of genes involved in histone modification, DNA repair, and RNA processing and splicing.
Three genes were found to be differentially expressed in the blood of BD cases from our meta-analysis and in SZ cases from our previous study (Hess et al., 2016). Genes involved in immune signaling, histone modification, and apoptosis showed concordant dysregulation in the blood of BD and SZ cases. From WGCNA, thirteen gene modules were found to be differentially expressed in major psychosis. Although no significant differences in gene expression were found between BD and SZ cases based on our network analysis, ML classifiers and our novel method of polytranscript risk scoring were able to significantly discriminate BD and SZ.
Twenty-nine of 382 genes found by Seifuddin et al. to be dysregulated in the brain of BD cases displayed at least nominally significant association with BD from our meta-analysis (p < 0.05) (Seifuddin et al., 2013). Gene sets related to oxidative stress and mitochondrial components were associated with BD from both our study and Seifuddin et al. (Seifuddin et al., 2013). This highlights that some differential expression changes associated with BD from analysis of postmortem brain tissue can be identified in the blood of living BD patients. Eight genes were found in gene co-expression modules that were differentially expressed in the blood of BD cases and showed gene-level dysregulation in the postmortem brain of BD cases from the study by Seifuddin et al.: GLRX, IFI27, IFI44L, LIPA, MT1E, MT2A, MX1, and TNFSF10. Six of these genes belong to Gene Ontology pathways related to immune system signaling (IFI27, IFI44L, LIPA, MT2A, MX1, and TNFSF10). GLRX encodes a cytosolic enzyme glutaredoxin-1 that participates in redox-regulation and protects against buildup of excessive oxidative stress (Morgan et al., 2010). Abnormally increased expression of GLRX has been associated with pro-inflammatory signaling and was shown to promote neuronal cell death (Gorelenkova Miller et al., 2016). MT1E encodes a metallothionine that has high affinity for heavy metals and contributes to the migration of human glioma cells (Ryu et al., 2012), and was found to be differentially expressed in postmortem brain tissue of individuals with mood disorder that completed suicide compared to non-suicide cases (Sequeira et al., 2012).
We found that immune signaling genes were abnormally expressed in major psychosis, which is in line with previous studies. There are clinical data suggesting that BD cases exhibit a higher incidence of autoimmunity than the general population (Perugi et al., 2015), and that autoimmune disease may even contribute risk for BD as well as SZ (Barbosa et al., 2014; Eaton et al., 2010). In a recently published study, we identified significant positive genetic correlations between a cluster of gastrointestinal inflammatory diseases and BD and SZ (Tylee et al., 2018), indicating that there is shared genetic risk between major psychosis and autoimmune disorders. Pathway enrichment analysis of GWAS results for BD, SZ, and major depression revealed significant enrichment of immune signaling genes in risk loci (The Network and Pathway Analysis Subgroup of the Psychiatric Genomics Consortium, 2015). The strongest GWAS signal for SZ identified by the Psychiatric Genomics Consortium was located in the major histocompatibility (MHC) region (Ripke et al., 2014), which was later found to be driven, in part, by a structural variant in the complement component C4 gene that alters the copy number and, in turn, abnormally elevates expression levels of the gene in brain leading to synaptic pruning deficits in mice (Sekar et al., 2016). The first cross-disorder GWAS by the PGC implicated the MHC region with BD and four other disorders (SZ, autism, depression, and attention-deficit/hyperactivity disorder) (Smoller et al., 2013). Although BD and SZ etiology remains elusive, there are several genetic studies supporting the hypothesis that genes involved in immune regulation may play a role in pathophysiology.
Upstream regulators of pro-inflammatory cytokines may also play an important role in pathophysiology. Our study found that toll-like receptor gene sets were differentially expressed in major psychosis cases. Toll-like receptors can induce inflammation by mediating the secretion of pro-inflammatory cytokines (e.g., IL1β, IL-6, IL-8, and TNF-α), which have been also shown to stimulate production of reactive oxygen species (García Bueno et al., 2016; McKernan et al., 2011; Troutman et al., 2012; Wieck et al., 2016). The biological link between inflammation and oxidative stress may explain some of the associations we uncovered for BD and major psychosis. Genes involved in oxidative stress were found to be significantly increased in BD cases relative to unaffected comparison subjects, as well as in cases with major psychosis. We also found significant enrichment of toll-like receptor genes in ME7 and ME13 which were significantly over-expressed in major psychosis cases relative to unaffected comparison subjects. Over-expression of the oxidative stress pathway in major psychosis might be a consequence of increased expression of toll-like receptors stimulating pro-inflammatory cytokines, which may result in a reciprocal feedback loop (Biswas, 2016), whereby accumulation of reactive oxygen species may promote inflammation, impair cellular repair mechanisms, and exacerbate DNA damage and apoptosis. Aberrant inflammation and oxidative stress has been linked with white matter hyper-intensities in BD patients (Beyer et al., 2009), and may induce neuronal and glial loss (Gigante et al., 2011).
In our study, pro-apoptotic genes showed abnormally increased expression in BD and major psychosis. Abnormal expression of pro-apoptotic machinery in the blood of cases with major psychosis has been previously reported (Fries et al., 2014; Lowthert et al., 2012; Pietruczuk et al., 2018; Scaini et al., 2017). Evidence of apoptotic dysregulation has also been reported in postmortem brain studies of BD cases (Benes et al., 2006; Kim et al., 2010). An increased production of pro-apoptotic genes has been associated with SZ (Batalla et al., 2015; Catts and Weickert, 2012; Hess et al., 2016). A biological link between the pro-inflammatory cytokine TNF-α and apoptosis has been reported (Wang et al., 2008; Zhao et al., 2001), which could be a pathogenic mechanism.
At least 47% of the BD cases and 84% of the SZ cases in our sample had at some point received or were being actively treated with mood stabilizers and/or antipsychotics; however, these rates could be higher but are obscured due to missing data. We can make some inferences whether the differential expression patterns we observed in major psychosis were congruent with known medication effects based on findings reported in the literature for lithium and antipsychotics. Lithium may provide neuroprotective effects through inhibition of pro-apoptotic signals (Li et al., 2010)and reduction of oxidative stress and inflammation (Beurel et al., 2010; Guloksuz et al., 2012; Nassar and Azab, 2014). Antipsychotics were reported to diminish levels of circulating pro-inflammatory cytokines in SZ patients (Tourjman et al., 2013). Higher levels of apoptotic markers were seen in fibroblast cells from treatment-naïve patients with SZ compared to unaffected comparison subjects (Gassó et al., 2014). Antipsychotics were also associated with a reduction in oxidative stress markers among treatment-naïve first-episode patients (Kriisa et al., 2016; Noto et al., 2015). Though definitive conclusions cannot be drawn, the abnormal expression changes in immune, oxidative stress, and apoptotic genes found in BD and major psychosis cases in our study do not appear to be consistent with reported medication effects.
Histone tail modification is a dynamic regulatory process that can produce short-term changes to gene expression (Reik, 2007). Histone modifying genes were found to be significantly under-expressed in BD and major psychosis relative to unaffected comparison subjects. Genes involved in histone acetylation – a process associated with a transcriptionally active state of chromatin (Eberharter and Becker, 2002) – were significant under-expressed in BD and SZ, more so than genes related to histone deacetylation and methylation (Supplementary Figure 6). Histone acetylation and transcriptional activation may be perturbed in BD and SZ, and this might hold promise as a therapeutic target. Lithium and valproate were found to increase histone acetylation in cellular and animal models through inhibition of histone deacetylases (HDACs) (Leng et al., 2008; Wu et al., 2013). Histone modification may be linked with the variety of neuroprotective effects associated with lithium and valproate such as anti-inflammation, reduction of oxidative stress, and enhanced synaptic plasticity (Chiu et al., 2013).
The values of ROC AUC achieved by our classification models were too modest to be considered clinically significant (72% for BD vs. UCs, and ~60% for BD vs. SCZ). However, classification performance of our models was significantly better than a random classifier, supporting the idea that blood-based gene expression classifiers could be further optimized and eventually have diagnostic utility. Our results of our machine learning analysis should be interpreted in light of limitations. We sought to build a machine learning classification model only on blood-based gene expression levels, yet there are advantages to integrating multiple modalities (i.e., behavioral, brain imaging, ‘omics data, environmental data) to capture underlying biology of neuropsychiatric disorders that would otherwise be missed by a single modality. In this vein, it is possible that one single biomarker test will never be sufficient for BD and SZ because of obstacles presented by clinical and etiology heterogeneity. An increasingly common theme in psychiatry is improving precision medicine by parsing heterogenous disorders into homogenous sub-groups, which could conceivably benefit classification performances for BD and SCZ (Lombardo et al., 2019). The concept of serial biomarker testing (i.e., one test to screen for high-risk individuals followed by additional tests to discriminate related disorders) has been proposed in context of autism spectrum disorders and related neurodevelopmental disorders, and could be an effective strategy for the pursuit of biomarkers for BD and SZ (Glatt, 2013). Our sample size was relatively small, therefore had the risk of overfitting the data when training classification models. We evaluated classification models for bias from overfitting by shuffling test set phenotype labels. Our results showed that classification performance was no better than random chance (AUC ~ 50%) suggesting that performance of our machine learning models were not driven by overfitting. Another limitation of our analysis was that we did not perform laboratory-based validation of our differential expression results. However, we used summary statistics to evaluate the concordance between the top differentially expressed genes associated with BD in the study by Krebs et al. compared to our meta-analysis, and found strong concordance of effect sizes (Pearson’s r = 0.92, p-value = 0.028), suggesting that our findings are not driven by random chance (Krebs et al., 2018).
We performed an electronic literature search for studies that trained machine learning classification models on gene expression profiles to discriminate BD cases from unaffected controls (the present study excluded) and reported classification performance as area-under-the-curve to allow results to be comparable to our study. We identified four studies that met our inclusion criteria (Kato et al., 2011; Munkholm et al., 2019, 2015; Struyf et al., 2008). The classification performance for these studies ranged from AUCs of 0.704 to 0.920. The study with the highest AUC utilized postmortem brain gene expression profiles in addition to phenotypic data to achieve a high classification performance, which does not have the same utility as a classifier based on ex vivo samples of mRNA. Two of the four studies utilized gene expression data from ex vivo peripheral blood samples (Munkholm et al., 2019, 2015), but those studies evaluated potential biomarkers that were selected based on a candidate gene approach which may have skewed classification performance relative to unbiased feature selection. Candidate biomarker genes were also evaluated by (Kato et al., 2011) within lymphoblastoid cells. In sum, we have identified limitations at various levels of existing gene expression biomarker literature for BD. The first proof-of-concept study was published about 14 years ago that demonstrated the utility of ‘liquid biopsies’ for BD (Tsuang et al., 2005). There has not been another transcriptomic study published in that 14 year period to have evaluated the classification performance of blood-based gene expression biomarkers to discriminate BD cases from unaffected comparison subjects. Our study provides a critical benchmark in the pursuit of blood-based biomarkers for BD.
There were important technical factors that could impact the interpretation of our results. First, the number of samples varied per gene due to missing data caused by platform differences. We used an approach to adjust for differences between platforms as described in our previous transcriptome-wide studies of autism and SZ 9,79 that can handle missing values. Lifetime and current medication use was not reported in all subjects included in our study, therefore we used surrogate variable analysis (SVA) to indirectly control for potentially confounding effects of medication on gene expression. ComBat is a popular approach to cope with batch effects, however, applying that approach to our data set would have unnecessarily excluded more than two-thirds of genes with usable data due to a minor amount of missingness. Second, the only studies that met our inclusion criteria at the time of writing were microarray-based studies. RNA-sequencing has the potential to uncover small RNAs and other non-coding genes, exons, alternative splicing isoforms, and novel transcripts which were poorly represented or absent in our analysis. Lastly, causality could not be determined due to the cross-sectional nature of this study.
In conclusion, our study points to specific genes and gene sets with abnormal expression levels in the blood of cases with BD and major psychosis. The classification performance of our ML models were on par with past findings from much larger combined-sample transcriptomic analyses of autism, PTSD, and SZ (Breen et al., 2018; Hess et al., 2016; Tylee et al., 2017). Our classifiers should be further tested and potentially refined with longitudinal data to ensure that these models can yield reliable, clinically useful information. In all, our study found evidence that gene expression changes are shared between BD and SZ. Our results further support the notion that dysregulation in immune signaling, apoptosis, oxidative stress, and chromatin remodeling might be of central importance to BD and SZ. Further investigation of these pathways and their role in pathophysiology is warranted.
Pre-processed data and code availability
Pre-processed expression data and full summary statistics are available upon request. The custom R scripts used to analyze the data are available at: (https://github.com/hessJ/gxp_bipolar). Software is available for our PTRS method at: https://github.com/hessJ/ptrs.
Supplementary Material
Acknowledgements
SJG received funding from National Institute on Aging (5R01AG054002–02), the National Institute of Mental Health (5R01MH101519–04), and the Brain & Behavioral Research Foundation.
Role of Funding
The study sponsors did not influence the study design, collection of data, analysis, interpretation of findings, drafting of the manuscript, or decision to submit the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- Barbosa IG, Machado-Vieira R, Soares JC, Teixeira AL, 2014. The immunology of bipolar disorder. Neuroimmunomodulation. 10.1159/000356539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batalla A, Bargalló N, Gassó P, Molina O, Pareto D, Mas S, Roca JM, Bernardo M, Lafuente A, Parellada E, 2015. Apoptotic markers in cultured fibroblasts correlate with brain metabolites and regional brain volume in antipsychotic-naive first-episode schizophrenia and healthy controls. Transl. Psychiatry 5, e626–e626. 10.1038/tp.2015.122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beech RD, Lowthert L, Leffert JJ, Mason PN, Taylor MM, Umlauf S, Lin A, Lee JY, Maloney K, Muralidharan A, Lorberg B, Zhao H, Newton SS, Mane S, Epperson CN, Sinha R, Blumberg H, Bhagwagar Z, 2010. Increased peripheral blood expression of electron transport chain genes in bipolar depression. Bipolar Disord 12, 813–24. 10.1111/j.1399-5618.2010.00882.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benes FM, Matzilevich D, Burke RE, Walsh J, 2006. The expression of proapoptosis genes is increased in bipolar disorder, but not in schizophrenia. Mol. Psychiatry 10.1038/sj.mp.4001758 [DOI] [PubMed] [Google Scholar]
- Beurel E, Michalek SM, Jope RS, 2010. Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3). Trends Immunol. 10.1016/j.it.2009.09.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beyer JL, Young R, Kuchibhatla M, Krishnan KRR, 2009. Hyperintense MRI lesions in bipolar disorder: A meta-analysis and review. Int. Rev. Psychiatry 10.1080/09540260902962198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas SK, 2016. Does the Interdependence between Oxidative Stress and Inflammation Explain the Antioxidant Paradox? Oxid. Med. Cell. Longev 10.1155/2016/5698931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bousman CA, Chana G, Glatt SJ, Chandler SD, Lucero GR, Tatro E, May T, Lohr JB, Kremen WS, Tsuang MT, Everall IP, 2010. Preliminary evidence of ubiquitin proteasome system dysregulation in schizophrenia and bipolar disorder: convergent pathway analysis findings from two independent samples. Am. J. Med. Genet. B. Neuropsychiatr. Genet 153B, 494–502. 10.1002/ajmg.b.31006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breen MS, Tylee DS, Maihofer AX, Neylan TC, Mehta D, Binder EB, Chandler SD, Hess JL, Kremen WS, Risbrough VB, Woelk CH, Baker DG, Nievergelt CM, Tsuang MT, Buxbaum JD, Glatt SJ, 2018. PTSD Blood Transcriptome Mega-Analysis: Shared Inflammatory Pathways across Biological Sex and Modes of Trauma. Neuropsychopharmacology 43, 469–481. 10.1038/npp.2017.220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulik-Sullivan B, Finucane HK, Anttila V, Gusev A, Day FR, Consortium R, Genomics Consortium P, of the Wellcome Trust Consortium, G.C. for A., Perry JRB, Patterson N, Robinson E, Daly MJ, Price AL, Neale BM, 2015. An Atlas of Genetic Correlations across Human Diseases and Traits. bioRxiv 1–44. 10.1101/014498 [DOI] [Google Scholar]
- Cattane N, Minelli A, Milanesi E, Maj C, Bignotti S, Bortolomasi M, Bocchio Chiavetto L, Gennarelli M, 2015. Altered gene expression in schizophrenia: findings from transcriptional signatures in fibroblasts and blood. PLoS One 10, e0116686 10.1371/journal.pone.0116686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catts VS, Weickert C, 2012. Gene expression analysis implicates a death receptor pathway in schizophrenia pathology. PLoS One 10.1371/journal.pone.0035511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Cheng L, Grennan K, Pibiri F, Zhang C, Badner JA, Gershon ES, Liu C, 2013. Two gene co-expression modules differentiate psychotics and controls. Mol. Psychiatry 18, 1308–14. 10.1038/mp.2012.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiu C-T, Wang Z, Hunsberger JG, Chuang D-M, 2013. Therapeutic Potential of Mood Stabilizers Lithium and Valproic Acid: Beyond Bipolar Disorder. Pharmacol. Rev 10.1124/pr.111.005512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clelland CL, Read LL, Panek LJ, Nadrich RH, Bancroft C, Clelland JD, 2013. Utilization of never-medicated bipolar disorder patients towards development and validation of a peripheral biomarker profile. PLoS One 8, e69082 10.1371/journal.pone.0069082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darby MM, Yolken RH, Sabunciyan S, 2016. Consistently altered expression of gene sets in postmortem brains of individuals with major psychiatric disorders. Transl. Psychiatry 6, e890 10.1038/tp.2016.173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drexhage RC, van der Heul-Nieuwenhuijsen L, Padmos RC, van Beveren N, Cohen D, Versnel M. a, Nolen W. a, Drexhage H. a, 2010. Inflammatory gene expression in monocytes of patients with schizophrenia: overlap and difference with bipolar disorder. A study in naturalistically treated patients. Int. J. Neuropsychopharmacol 13, 1369–1381. 10.1017/S1461145710000799 [DOI] [PubMed] [Google Scholar]
- Eaton WW, Pedersen MG, Nielsen PR, Mortensen PB, 2010. Autoimmune diseases, bipolar disorder, and non-affective psychosis. Bipolar Disord 10.1111/j.1399-5618.2010.00853.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eberharter A, Becker PB, 2002. Histone acetylation: A switch between repressive and permissive chromatin. Second in review on chromatin dynamics. EMBO Rep 10.1093/embo-reports/kvf053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari AJ, Stockings E, Khoo J-P, Erskine HE, Degenhardt L, Vos T, Whiteford HA, 2016. The prevalence and burden of bipolar disorder: findings from the Global Burden of Disease Study 2013. Bipolar Disord. 18, 440–450. 10.1111/bdi.12423 [DOI] [PubMed] [Google Scholar]
- Fries GR, Vasconcelos-Moreno MP, Gubert C, Santos B.T.M.Q. do, da Rosa ALST, Eisele B, Sartori J, Pfaffenseller B, Kapczinski F, Kauer-Sant’Anna M, 2014. Early apoptosis in peripheral blood mononuclear cells from patients with bipolar disorder. J. Affect. Disord 152–154, 474–477. 10.1016/j.jad.2013.07.027 [DOI] [PubMed] [Google Scholar]
- García Bueno B, Caso JR, Madrigal JLM, Leza JC, 2016. Innate immune receptor Toll-like receptor 4 signalling in neuropsychiatric diseases. Neurosci. Biobehav. Rev 10.1016/j.neubiorev.2016.02.013 [DOI] [PubMed] [Google Scholar]
- Gassó P, Mas S, Molina O, Lafuente A, Bernardo M, Parellada E, 2014. Increased susceptibility to apoptosis in cultured fibroblasts from antipsychotic-naïve first-episode schizophrenia patients. J. Psychiatr. Res 10.1016/j.jpsychires.2013.09.017 [DOI] [PubMed] [Google Scholar]
- Gigante AD, Young LT, Yatham LN, Andreazza AC, Nery FG, Grinberg LT, Heinsen H, Lafer B, 2011. Morphometric post-mortem studies in bipolar disorder: Possible association with oxidative stress and apoptosis. Int. J. Neuropsychopharmacol 10.1017/S146114571000146X [DOI] [PubMed] [Google Scholar]
- Guloksuz S, Altinbas K, Aktas Cetin E, Kenis G, Bilgic Gazioglu S, Deniz G, Timucin Oral E, Van Os J, 2012. Evidence for an association between tumor necrosis factor-alpha levels and lithium response. J. Affect. Disord 10.1016/j.jad.2012.04.044 [DOI] [PubMed] [Google Scholar]
- Hess JL, Tylee DS, Barve R, de Jong S, Ophoff RA, Kumarasinghe N, Tooney P, Schall U, Gardiner E, Beveridge NJ, Scott RJ, Yasawardene S, Perera A, Mendis J, Carr V, Kelly B, Cairns M, Tsuang MT, Glatt SJ, Glatt SJ, 2016. Transcriptome-wide mega-analyses reveal joint dysregulation of immunologic genes and transcription regulators in brain and blood in schizophrenia. Schizophr. Res 176, 114–124. 10.1016/j.schres.2016.07.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamoto K, Bundo M, Kato T, 2005. Altered expression of mitochondria-related genes in postmortem brains of patients with bipolar disorder or schizophrenia, as revealed by large-scale DNA microarray analysis. Hum. Mol. Genet 14, 241–53. 10.1093/hmg/ddi022 [DOI] [PubMed] [Google Scholar]
- Kim HW, Rapoport SI, Rao JS, 2010. Altered expression of apoptotic factors and synaptic markers in postmortem brain from bipolar disorder patients. Neurobiol. Dis 10.1016/j.nbd.2009.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Xia K, Tao R, Giusti-Rodriguez P, Vladimirov V, van den Oord E, Sullivan PF, 2014. A meta-analysis of gene expression quantitative trait loci in brain. Transl. Psychiatry 4, e459 10.1038/tp.2014.96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriisa K, Haring L, Vasar E, Koido K, Janno S, Vasar V, Zilmer K, Zilmer M, 2016. Antipsychotic treatment reduces indices of oxidative stress in first episode psychosis patients. Oxid. Med. Cell. Longev 10.1155/2016/9616593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langfelder P, Horvath S, 2008. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9, 559 10.1186/1471-2105-9-559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leng Y, Liang M-H, Ren M, Marinova Z, Leeds P, Chuang D-M, 2008. Synergistic Neuroprotective Effects of Lithium and Valproic Acid or Other Histone Deacetylase Inhibitors in Neurons: Roles of Glycogen Synthase Kinase-3 Inhibition. J. Neurosci 10.1523/JNEUROSCI.5467-07.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Li H, Roughton K, Wang X, Kroemer G, Blomgren K, Zhu C, 2010. Lithium reduces apoptosis and autophagy after neonatal hypoxia-ischemia. Cell Death Dis 10.1038/cddis.2010.33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowthert L, Leffert J, Lin A, Umlauf S, Maloney K, Muralidharan A, Lorberg B, Mane S, Zhao H, Sinha R, Bhagwagar Z, Beech R, 2012. Increased ratio of anti-apoptotic to pro-apoptotic Bcl2 gene-family members in lithium-responders one month after treatment initiation. Biol. Mood Anxiety Disord. 2, 15 10.1186/2045-5380-2-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matigian N, Windus L, Smith H, Filippich C, Pantelis C, McGrath J, Mowry B, Hayward N, 2007. Expression profiling in monozygotic twins discordant for bipolar disorder reveals dysregulation of the WNT signalling pathway. Mol. Psychiatry 12, 815–25. 10.1038/sj.mp.4001998 [DOI] [PubMed] [Google Scholar]
- McKenzie M, Henders AK, Caracella A, Wray NR, Powell JE, 2014. Overlap of expression Quantitative Trait Loci (eQTL) in human brain and blood. BMC Med. Genomics 7, 31 10.1186/1755-8794-7-31 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKernan DP, Dennison U, Gaszner G, Cryan JF, Dinan TG, 2011. Enhanced peripheral toll-like receptor responses in psychosis: Further evidence of a pro-inflammatory phenotype. Transl. Psychiatry 1 10.1038/tp.2011.37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray RM, Sham P, Van Os J, Zanelli J, Cannon M, McDonald C, 2004. A developmental model for similarities and dissimilarities between schizophrenia and bipolar disorder. Schizophr. Res 71, 405–16. 10.1016/j.schres.2004.03.002 [DOI] [PubMed] [Google Scholar]
- Nassar A, Azab AN, 2014. Effects of lithium on inflammation. ACS Chem. Neurosci 10.1021/cn500038f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noto C, Ota VK, Gadelha A, Noto MN, Barbosa DS, Bonifácio KL, Nunes SO, Cordeiro Q, Belangero SI, Bressan RA, Maes M, Brietzke E, 2015. Oxidative stress in drug naïve first episode psychosis and antioxidant effects of risperidone. J. Psychiatr. Res 10.1016/j.jpsychires.2015.07.003 [DOI] [PubMed] [Google Scholar]
- Pacifico R, Davis RL, 2017. Transcriptome sequencing implicates dorsal striatum-specific gene network, immune response and energy metabolism pathways in bipolar disorder. Mol. Psychiatry 22, 441–449. 10.1038/mp.2016.94 [DOI] [PubMed] [Google Scholar]
- Padmos RC, Hillegers MHJ, Knijff EM, Vonk R, Bouvy A, Staal FJT, de Ridder D, Kupka RW, Nolen WA, Drexhage HA, 2008. A discriminating messenger RNA signature for bipolar disorder formed by an aberrant expression of inflammatory genes in monocytes. Arch. Gen. Psychiatry 65, 395–407. 10.1001/archpsyc.65.4.395 [DOI] [PubMed] [Google Scholar]
- Perugi G, Quaranta G, Belletti S, Casalini F, Mosti N, Toni C, Dell’Osso L, 2015. General medical conditions in 347 bipolar disorder patients: Clinical correlates of metabolic and autoimmune-allergic diseases. J. Affect. Disord 10.1016/j.jad.2014.08.052 [DOI] [PubMed] [Google Scholar]
- Pietruczuk K, Lisowska KA, Grabowski K, Landowski J, Witkowski JM, 2018. Proliferation and apoptosis of T lymphocytes in patients with bipolar disorder. Sci. Rep 8, 3327 10.1038/s41598-018-21769-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polderman TJC, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, Posthuma D, 2015. Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nat. Genet 47, 702–709. 10.1038/ng.3285 [DOI] [PubMed] [Google Scholar]
- Purcell SM, Wray NR, Stone JL, Visscher PM, O’Donovan MC, Sullivan PF, Sklar P, 2009. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752. https://doi.org/nature08185 [pii] 10.1038/nature08185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn T, Tylee D, Glatt S, 2016. exprso: an R-package for the rapid implementation of machine learning algorithms. F1000Research 5, 2588 10.12688/f1000research.9893.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reik W, 2007. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature 10.1038/nature05918 [DOI] [PubMed] [Google Scholar]
- Ripke S, Neale BM, Corvin A, Walters JTR, Farh K-H, Holmans PA, Lee P, Bulik-Sullivan B, Collier DA, Huang H, Pers TH, Agartz I, Agerbo E, Albus M, Alexander M, Amin F, Bacanu SA, Begemann M, Belliveau RA Jr, Bene J, Bergen SE, Bevilacqua E, Bigdeli TB, Black DW, Bruggeman R, Buccola NG, Buckner RL, Byerley W, Cahn W, Cai G, Campion D, Cantor RM, Carr VJ, Carrera N, Catts SV, Chambert KD, Chan RCK, Chen RYL, Chen EYH, Cheng W, Cheung EFC, Ann Chong S, Robert Cloninger C, Cohen D, Cohen N, Cormican P, Craddock N, Crowley JJ, Curtis D, Davidson M, Davis KL, Degenhardt F, Del Favero J, Demontis D, Dikeos D, Dinan T, Djurovic S, Donohoe G, Drapeau E, Duan J, Dudbridge F, Durmishi N, Eichhammer P, Eriksson J, Escott-Price V, Essioux L, Fanous AH, Farrell MS, Frank J, Franke L, Freedman R, Freimer NB, Friedl M, Friedman JI, Fromer M, Genovese G, Georgieva L, Giegling I, Giusti-Rodríguez P, Godard S, Goldstein JI, Golimbet V, Gopal S, Gratten J, de Haan L, Hammer C, Hamshere ML, Hansen M, Hansen T, Haroutunian V, Hartmann AM, Henskens FA, Herms S, Hirschhorn JN, Hoffmann P, Hofman A, Hollegaard MV, Hougaard DM, Ikeda M, Joa I, Julià A, Kahn RS, Kalaydjieva L, Karachanak-Yankova S, Karjalainen J, Kavanagh D, Keller MC, Kennedy JL, Khrunin A, Kim Y, Klovins J, Knowles JA, Konte B, Kucinskas V, Ausrele Kucinskiene Z, Kuzelova-Ptackova H, Kähler AK, Laurent C, Lee Chee Keong J, Hong Lee S, Legge SE, Lerer B, Li M, Li T, Liang K-Y, Lieberman J, Limborska S, Loughland CM, Lubinski J, Lönnqvist J, Macek M Jr, Magnusson PKE, Maher BS, Maier W, Mallet J, Marsal S, Mattheisen M, Mattingsdal M, McCarley RW, McDonald C, McIntosh AM, Meier S, Meijer CJ, Melegh B, Melle I, Mesholam-Gately RI, Metspalu A, Michie PT, Milani L, Milanova V, Mokrab Y, Morris DW, Mors O, Murphy KC, Murray RM, Myin-Germeys I, Müller-Myhsok B, Nelis M, Nenadic I, Nertney DA, Nestadt G, Nicodemus KK, Nikitina-Zake L, Nisenbaum L, Nordin A, O’Callaghan E, O’Dushlaine C, O’Neill FA, Oh S-Y, Olincy A, Olsen L, Van Os J, Endophenotypes International Consortium P, Pantelis C, Papadimitriou GN, Papiol S, Parkhomenko E, Pato MT, Paunio T, Pejovic-Milovancevic M, Perkins DO, Pietiläinen O, Pimm J, Pocklington AJ, Powell J, Price A, Pulver AE, Purcell SM, Quested D, Rasmussen HB, Reichenberg A, Reimers MA, Richards AL, Roffman JL, Roussos P, Ruderfer DM, Salomaa V, Sanders AR, Schall U, Schubert CR, Schulze TG, Schwab SG, Scolnick EM, Scott RJ, Seidman LJ, Shi J, Sigurdsson E, Silagadze T, Silverman JM, Sim K, Slominsky P, Smoller JW, So H-C, Spencer CA, Stahl EA, Stefansson H, Steinberg S, Stogmann E, Straub RE, Strengman E, Strohmaier J, Scott Stroup T, Subramaniam M, Suvisaari J, Svrakic DM, Szatkiewicz JP, Söderman E, Thirumalai S, Toncheva D, Tosato S, Veijola J, Waddington J, Walsh D, Wang D, Wang Q, Webb BT, Weiser M, Wildenauer DB, Williams NM, Williams S, Witt SH, Wolen AR, Wong EHM, Wormley BK, Simon Xi H, Zai CC, Zheng X, Zimprich F, Wray NR, Stefansson K, Visscher PM, Trust Case-Control Consortium W, Adolfsson R, Andreassen OA, Blackwood DHR, Bramon E, Buxbaum JD, Børglum AD, Cichon S, Darvasi A, Domenici E, Ehrenreich H, Esko T, Gejman PV, Gill M, Gurling H, Hultman CM, Iwata N, Jablensky AV, Jönsson EG, Kendler KS, Kirov G, Knight J, Lencz T, Levinson DF, Li QS, Liu J, Malhotra AK, McCarroll SA, McQuillin A, Moran JL, Mortensen PB, Mowry BJ, Nöthen MM, Ophoff RA, Owen MJ, Palotie A, Pato CN, Petryshen TL, Posthuma D, Rietschel M, Riley BP, Rujescu D, Sham PC, Sklar P, St Clair D, Weinberger DR, Wendland JR, Werge T, Daly MJ, Sullivan PF, O’Donovan MC, 2014. Biological insights from 108 schizophrenia-associated genetic loci. Nature 511, 421–427. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savitz J, Frank MB, Victor T, Bebak M, Marino JH, Bellgowan PSF, McKinney BA, Bodurka J, Kent Teague T, Drevets WC, 2013. Inflammation and neurological disease-related genes are differentially expressed in depressed patients with mood disorders and correlate with morphometric and functional imaging abnormalities. Brain. Behav. Immun 31, 161–71. 10.1016/j.bbi.2012.10.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaini G, Fries GR, Valvassori SS, Zeni CP, Zunta-Soares G, Berk M, Soares JC, Quevedo J, 2017. Perturbations in the apoptotic pathway and mitochondrial network dynamics in peripheral blood mononuclear cells from bipolar disorder patients. Transl. Psychiatry 10.1038/tp.2017.83 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifuddin F, Pirooznia M, Judy JT, Goes FS, Potash JB, Zandi PP, 2013. Systematic review of genome-wide gene expression studies of bipolar disorder. BMC Psychiatry 13, 213 10.1186/1471-244X-13-213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sekar A, Bialas AR, de Rivera H, Davis A, Hammond TR, Kamitaki N, Tooley K, Presumey J, Baum M, Van Doren V, Genovese G, Rose SA, Handsaker RE, Daly MJ, Carroll MC, Stevens B, McCarroll SA, 2016. Schizophrenia risk from complex variation of complement component 4. Nature advance on. 10.1038/nature16549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao L, Vawter MP, 2008. Shared gene expression alterations in schizophrenia and bipolar disorder. Biol. Psychiatry 64, 89–97. 10.1016/j.biopsych.2007.11.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smoller JW, Kendler K, Craddock N, Writing JW, Smoller N, Craddock K, Kendler PH, Lee BM, Neale JI, Nurnberger S, Ripke S, Santangelo PF, Statistical S, Ripke S, Lee PH, Neale BM, Editorial SP, Anney R, Buitelaar J, Fanous A, Faraone SV, Hoogendijk W, Lesch K-P, Levinson DF, Perlis RH, Purcell S, Rietschel M, Riley B, Sonuga-Barke E, Schachar R, Schulze TG, Pgc AT, Group C-D, Kendler KS, Neale B, Neale M, Nurnberger JI, Perlis R, Santangelo S, Sullivan PF, Bender P, Cichon S, Daly MJ, Kelsoe J, Lehner T, Levinson D, O ‘donovan M, Gejman P, Sebat J, Sklar P, 2013. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis Cross-Disorder Group of the Psychiatric Genomics Consortium *. Lancet 3811016, 1371–1379. 10.1016/S0140-6736(12)62129-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesli M, Wirgenes KV, Hughes T, Bettella F, Athanasiu L, Hoseth ES, Nerhus M, Lagerberg TV, Steen NE, Agartz I, Melle I, Dieset I, Djurovic S, Andreassen OA, 2016. VRK2 gene expression in schizophrenia, bipolar disorder and healthy controls. Br. J. Psychiatry 209, 114–120. 10.1192/bjp.bp.115.161950 [DOI] [PubMed] [Google Scholar]
- Network The and Pathway Analysis Subgroup of the Psychiatric Genomics Consortium, 2015. Psychiatric genome-wide association study analyses implicate neuronal, immune and histone pathways. Nat. Neurosci 18, 199–209. 10.1038/nn.3922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tourjman V, Kouassi É, Koué M-È, Rocchetti M, Fortin-Fournier S, Fusar-Poli P, Potvin S, 2013. Antipsychotics’ effects on blood levels of cytokines in schizophrenia: A meta-analysis. Schizophr. Res 151, 43–47. 10.1016/j.schres.2013.10.011 [DOI] [PubMed] [Google Scholar]
- Troutman TD, Bazan JF, Pasare C, 2012. Toll-like receptors, signaling adapters and regulation of the pro-inflammatory response by PI3K. Cell Cycle 11, 3559–3567. 10.4161/cc.21572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuang MT, Nossova N, Yager T, Tsuang M-M, Guo S-C, Shyu KG, Glatt SJ, Liew CC, 2005. Assessing the validity of blood-based gene expression profiles for the classification of schizophrenia and bipolar disorder: a preliminary report. Am. J. Med. Genet. B. Neuropsychiatr. Genet 133B, 1–5. 10.1002/ajmg.b.30161 [DOI] [PubMed] [Google Scholar]
- Tylee DS, Hess JL, Quinn TP, Barve R, Huang H, Zhang-James Y, Chang J, Stamova BS, Sharp FR, Hertz-Picciotto I, Faraone SV, Kong SW, Glatt SJ, 2017. Blood transcriptomic comparison of individuals with and without autism spectrum disorder: A combined-samples mega-analysis. Am. J. Med. Genet. Part B Neuropsychiatr. Genet 174, 181–201. 10.1002/ajmg.b.32511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tylee DS, Kawaguchi DM, Glatt SJ, 2013. On the outside, looking in: a review and evaluation of the comparability of blood and brain “-omes”. Am. J. Med. Genet. B. Neuropsychiatr. Genet 162B, 595–603. 10.1002/ajmg.b.32150 [DOI] [PubMed] [Google Scholar]
- Tylee DS, Sun J, Hess JL, Tahir MA, Sharma E, Malik R, Worrall BB, Levine AJ, Martinson JJ, Nejentsev S, Speed D, Fischer A, Mick E, Walker BR, Crawford A, Grant SFA, Polychronakos C, Bradfield JP, Sleiman PMA, Hakonarson H, Ellinghaus E, Elder JT, Tsoi LC, Trembath RC, Barker JN, Franke A, Dehghan A, Faraone SV, Glatt SJ, 2018. Genetic correlations among psychiatric and immune-related phenotypes based on genome-wide association data. Am. J. Med. Genet. Part B Neuropsychiatr. Genet 10.1002/ajmg.b.32652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Väremo L, Nielsen J, Nookaew I, 2013. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 41, 4378–91. 10.1093/nar/gkt111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viechtbauer W, 2010. Conducting Meta-Analyses in R with the metafor Package. J. Stat. Softw 36, 1–48. 10.1103/PhysRevB.91.121108 [DOI] [Google Scholar]
- Wang L, Du F, Wang X, 2008. TNF-α Induces Two Distinct Caspase-8 Activation Pathways. Cell 10.1016/j.cell.2008.03.036 [DOI] [PubMed] [Google Scholar]
- Wieck A, Grassi-Oliveira R, do Prado CH, Viola TW, Petersen LE, Porto B, Teixeira AL, Bauer ME, 2016. Toll-like receptor expression and function in type I bipolar disorder. Brain. Behav. Immun 54, 110–121. 10.1016/j.bbi.2016.01.011 [DOI] [PubMed] [Google Scholar]
- Wirgenes KV, Tesli M, Inderhaug E, Athanasiu L, Agartz I, Melle I, Hughes T, Andreassen OA, Djurovic S, 2014. ANK3 gene expression in bipolar disorder and schizophrenia. Br. J. Psychiatry 205, 244–5. 10.1192/bjp.bp.114.145433 [DOI] [PubMed] [Google Scholar]
- Witt SH, Juraeva D, Sticht C, Strohmaier J, Meier S, Treutlein J, Dukal H, Frank J, Lang M, Deuschle M, Schulze TG, Degenhardt F, Mattheisen M, Brors B, Cichon S, Nöthen MM, Witt CC, Rietschel M, 2014. Investigation of manic and euthymic episodes identifies state-and trait-specific gene expression and STAB1 as a new candidate gene for bipolar disorder. Transl. Psychiatry 4, e426 10.1038/tp.2014.71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Zheng S. Di, Huang HL, Yan LC, Yin XF, Xu HN, Zhang KJ, Gui JH, Chu L, Liu XY, 2013. Lithium down-regulates histone deacetylase 1 (HDAC1) and induces degradation of mutant huntingtin. J. Biol. Chem 10.1074/jbc.M113.479865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Bausano B, Pike BR, Newcomb-Fernandez JK, Wang KKW, Shohami E, Ringger NC, DeFord SM, Anderson DK, Hayes RL, 2001. TNF-? stimulates caspase-3 activation and apoptotic cell death in primary septo-hippocampal cultures. J. Neurosci. Res 64, 121–131. 10.1002/jnr.1059 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Pre-processed expression data and full summary statistics are available upon request. The custom R scripts used to analyze the data are available at: (https://github.com/hessJ/gxp_bipolar). Software is available for our PTRS method at: https://github.com/hessJ/ptrs.