Abstract
Aims
Glioblastoma (GBM) is the most common and lethal malignant brain tumor in adults. Glioma stem cells (GSCs) are implicated in this poor prognosis and in radio(chemo‐)resistance. We have previously demonstrated that among potentially highly specific GSC markers oligodendrocyte lineage transcription factor 2 (OLIG2) appears to be the most specific and cyclin D2 (CCND2) the only one related to cell cycle regulation. The purpose of this work was to investigate the clinical significance and the evolution of OLIG2 and CCND2 protein expression in GBM.
Methods and results
Immunohistochemical expression analysis of Olig2 and Ccnd2 was carried out on a cohort of human paired GBM samples comparing initial resections with local recurrent tumors after radiation therapy (RT) alone or radio‐chemotherapy with temozolomide (RT‐TMZ). Uni‐ and multivariate logistic regression analysis revealed that significant risk factors predicting early mortality (<12 months) are: subtotal surgery for recurrence, time to recurrence <6 months, Ccnd2 nuclear expression at initial surgery ≥30%, and Olig2 nuclear expression <30% at second surgery after RT alone and RT‐TMZ.
Conclusions
We demonstrated that patients for whom nuclear expression of Olig2 becomes low (<30%) after adjuvant treatments have a significantly shorter time to recurrence and survival reflecting most probably a proneural to mesenchymal transition of the GSCs population. We also highlighted the fact that at initial surgery, high nuclear expression (≥30%) of CCND2, a G1/S regulator specific of GSCs, has a prognostic value and is associated with early mortality (<12 months).
Keywords: cyclin D2, glioblastoma, immunohistochemistry, neoplastic stem cells, OLIG2, radiotherapy
We investigated by immunohistochemistry the clinical significance and the evolution of Oligodendrocyte lineage transcription factor 2 (OLIG2) and cyclin D2 (CCND2) protein expression, two markers highly specific of glioma stem cells, on a cohort of human paired glioblastoma samples comparing initial resections with recurrent tumors after radiation therapy alone or radio‐chemotherapy with temozolomide according to the Stupp regimen. Our study highlighted for the first time the prognostic value of high nuclear expression of CCND2 at initial surgery which is associated with early mortality and patients for whom nuclear expression of OLIG2 becomes low after adjuvant treatments have a significantly shorter time to recurrence and survival reflecting most probably a proneural to mesenchymal transition of the stem cells population.
1. INTRODUCTION
Glioblastoma (GBM) (World Health Organization [WHO] grade IV glioma) is the most common and lethal malignant brain tumor in adults.1 To better define GBM entities, the 2016 WHO classification now integrates to histological features the isocitrate dehydrogenase (IDH) mutation status, one of the most important genetic alterations found in GBM, as the few IDH‐mutant present a more favorable prognosis.1 However over the past decades despite some improvements in surgical and radio‐chemotherapeutics treatments and the multiplicity of clinical trials testing new therapies without major success so far,2 GBM remains incurable with a median survival of only 12‐18 months3 and up to 31 months for IDH‐mutant.4 Several factors could contribute to this poor prognosis; the most important appears to be related to the presence of a population of radio/chemoresistant cells with stem‐like properties.5, 6, 7, 8 The glioma stem cells (GSCs) subpopulation is capable of self‐renewal, persistent proliferation, dedifferentiation, multipotency and has the ability to be highly tumorigenic allowing for tumor regrowth after standard treatments.9 Therefore, developing additional therapeutic strategies targeting and eliminating the GSCs component is crucial to one day render GBM curable. The issue when studying GSCs comes from the fact that they represent a heterogeneous population difficult to identify by routine methods. Today, the gold standard for the determination of GSCs remains the capability of these cells to reshape the complexity of the initial patient tumor after serial orthotopic transplantation assays into mice brains.8 Many studies have explored known normal stem cell markers such as CD133 or CD44 to recognize and enrich GSC cultures with flawed results as other cells that do not express such markers also display tumorigenic capacities.5, 10, 11 A universal marker appearing illusory to obtain, a combination of biomarkers seems the best way to explore GSCs for potential routine identification. By performing a comparative analysis of differentially expressed genes between differentiated and GSC enriched cultures from similar DNA chip microarray platforms, we have previously determined a panel of eight genes potentially highly specific of GSCs. Among them, oligodendrocyte lineage transcription factor 2 (OLIG2) appears as the most specific and cyclin D2 (CCND2) as the only one related to cell cycle regulation.12 The purpose of this work was to investigate by immunohistochemistry (IHC) the clinical significance and the evolution of OLIG2 and CCND2 protein expression on a cohort of human paired GBM samples comparing initial resections with recurrent tumors after radiation therapy (RT) alone or radio‐chemotherapy with temozolomide (RT‐TMZ) according to the Stupp regimen.3
2. MATERIALS AND METHODS
2.1. Retrospective clinical series
We analyzed the protein product of OLIG2 and CCND2, two gene candidates for GSC biomarkers, in a retrospective paired samples cohort of 72 GBM. Uncommon histopathological subtypes of GBM were not included. The patients underwent an initial subtotal or macroscopically complete tumor resection at the Erasme University Hospital (Brussels, Belgium) between April 1990 and March 2014. The patients then received adjuvant therapy according to the standard guidelines in use at the time of the initial surgery: either RT alone (n = 37) or RT‐TMZ according to the Stupp regimen (n = 35). The Stupp regimen corresponds to fractionated conformal RT (60 Gy in 30 fractions given 5 days per week for 6 weeks) with continuous daily oral TMZ (75 mg/m2 of body‐surface area per day, 7 d/wk from the first to the last day of RT) followed by six cycles of adjuvant TMZ (150‐200 mg/m2 for five every 28 days).3 At recurrence in the same area of the primary tumor, the patients underwent a second subtotal or macroscopically complete tumor resection at the Erasme University Hospital between August 1991 and September 2014. All tissue samples (paraffin blocks) analyzed in this study came from the archives of the Department of Pathology of the Erasme University Hospital. This study was approved by the Erasme University Hospital Ethics Committee (P2014/290). The clinical data recorded for each patient are summarized in Table 1.
Table 1.
Sample description according to adjuvant treatment received (RT alone or RT‐TMZ)
| Whole cohort | RT alone | RT‐TMZ | P‐value | |
|---|---|---|---|---|
| CCND2 nuclear expression (%) | P < .001 | P = .013 | P = .001 | |
| Before adjuvant treatment | 31.00 ± 19.00 | 32.00 ± 19.00 | 31.00 ± 20.00 | .893a |
| After adjuvant treatment | 20.00 ± 18.00 | 20.00 ± 19.00 | 19.00 ± 18.00 | .731a |
| OLIG2 nuclear expression (%) | P = .003 | P = .027 | P = .044 | |
| Before adjuvant treatment | 43.00 ± 23.00 | 44.00 ± 22.00 | 41.00 ± 24.00 | .707a |
| After adjuvant treatment | 31.00 ± 22.00 | 32.00 ± 23.00 | 30.00 ± 22.00 | .764a |
| Age at diagnosis | 54.23 ± 11.63 | 54.69 ± 11.22 | 55.04 ± 11.69 | .897a |
| Gender (male) | 62.50% | 59.50% | 65.70% | .584b |
| Number of lesions (multiple) | 12.50% | 16.22% | 8.87% | .327b |
| Preoperative corticosteroids (yes) | 80.56% | 81.08% | 80.00% | .908b |
| MGMT methylation status | NA | |||
| Unknown | 72.22% | 100% (n = 37) | 42.85% (n = 15) | |
| Methylated | 11.11% | 0% | 22.86% (n = 8) | |
| Not methylated | 16.67% | 0% | 34.29% (n = 12) | |
| GBM | NA | |||
| NOS | 72.22% | 100% (n = 37) | 42.85% (n = 15) | |
| IDH‐mutant | 4.17% | 0% | 8.57% (n = 3) | |
| IDH‐wildtype | 23.61% | 0% | 48.57% (n = 17) | |
| Initial type of surgery (subtotal) | 40.28% | 37.84% | 42.86% | .664b |
| Time to first recurrence (mo) | 6.90 (4.54‐10.90) | 6.00 (4.20‐8.67) | 9.03 (4.83‐16.37) | .018c |
| Type of surgery at recurrence (subtotal) | 43.06% | 56.76% | 57.14% | .974b |
| Time between the two surgeries (mo) | 8.28 (5.52‐12.00) | 6.93 (5.20‐10.30) | 9.60 (7.10‐16.37) | .038c |
| Overall survival (mo) | 15.69 (11.63‐23.14) | 14.04 (11.28‐20.42) | 15.83 (13.10‐28.3) | .111c |
| (n = 72) | (n = 37) | (n = 35) |
Abbreviations: CCND2, cyclin D2; OLIG2, oligodendrocyte lineage transcription factor 2; GBM, glioblastoma; IDH, isocitrate dehydrogenase; NA, not applicable; NOS, not otherwise specified; RT, radiotherapy; RT‐TMZ, radio‐chemotherapy with temozolomide.
t Test.
Chi‐squared.
Wilcoxon test.
2.2. IHC and semi‐quantitative analysis
Standard IHC was applied to 5‐μm thick sections to display OLIG2 and CCND2 expression using respectively a specific antibody provided by Chemicon‐Millipore (ab9610, dilution 1:500) and ProteintechGroup Inc (Rabbit polyclonal, 10934‐1‐AP, dilution 1:150). Immunohistochemistry was performed on the BONDMAX. Briefly, as previously described,13 the immunohistochemical expression was visualized by means of streptavidin‐biotin‐peroxidase complex kit reagents (BioGenex) with diaminobenzidine/H2O2 as chromogenic substrate. Finally, the sections were counterstained with hematoxylin. IHC method was chosen instead of RNA sequencing analysis to allow precise in situ localization of the protein expression within the analyzed tissue.14 Semi‐quantitative analysis was performed by two independent observers (CB and ALT). The staining was assessed by means of two features: staining intensity (absent, low, moderate or strong) and labeling index (0: no staining; low nuclear expression <30% and high nuclear expression ≥30%). The labeling index was determined by random selection of five fields of representative tumor blocks at 40× power magnification. For the few cases where there was a discrepancy between the two scores obtained, a third observer (PD) assessed the final index.
2.3. Statistics
Statistical analyses were performed using Stata 14. The normal distribution of the data was verified using histograms, boxplots, and quantile‐quantile plots, and the equality of variances was checked using the Levene's test.
Since our study included a combination of GBM patients treated with surgical resection plus RT alone or RT‐TMZ, we chose 12 months as cut‐off for early mortality because it is the shortest median survival obtained in the major randomized phase III trial by Stupp et al.3 Thus, we divided our sample (n = 72) into a group with late mortality (≥12 months, from the date of the initial surgery) (n = 51) and a group with early mortality (<12 months, from the date of the initial surgery) (n = 21).
Categorical data were described with percentages and numbers, and continuous data were described with means and SD or median and interquartile range. Normally distributed variables were analysed with a t test. A Wilcoxon test or chi‐squared test was used on asymmetric distributed or dichotomous variables.
Univariate and multivariate binary logistic regression models were used to study the effects of risk factors on the occurrence of early mortality. Risk factor variables included number of lesions (categorical: unique, multiple), type of initial surgery (categorical: total, subtotal), type of adjuvant treatment (categorical: radiotherapy alone, radio‐chemotherapy), type of surgery at recurrence (categorical: total, subtotal), age (categorical: <50 years, ≥50 years), time to recurrence (categorical: <6 months, ≥6 months), nuclear expression of CCND2 before at initial surgery (categorical: <30%, ≥30%), nuclear expression of CCND2 after adjuvant treatment (categorical: <30%, ≥30%), nuclear expression of OLIG2 at initial surgery (categorical: <30%, ≥30%), nuclear expression of OLIG2 after adjuvant treatment (categorical: <30%, ≥30%), and as binary variables for gender and preoperative corticosteroids. Cut‐off values of 30% for the protein expression of CCND2 and OLIG2 were chosen because these expression levels were associated with the best sensitivity and specificity for the prediction of early mortality in our GBM cohort.
The automatic selection of risk factors in the model was performed by a stepwise backward method with an entry threshold of 0.05 and an exit threshold of 0.1. The adequacy of the model was verified by the Hosmer‐Lemeshow test, and the specificity of model was verified by the Link test. The other conditions of application of the multivariate logistic regression (number of subjects by risk factors, outliers, and collinearity between risk factors) were also verified. A P‐value of less than .05 was considered significant.
3. RESULTS
3.1. Semi‐quantitative analysis of OLIG2 and CCND2 protein expression
Olig2 expression was predominantly nuclear and cytoplasmic in fewer cases. Almost all cases were positive, only one case presented negative paired tumor samples and two cases became negative at second surgery. The staining intensity was moderate‐to‐strong in all positive tumors. Ccnd2 expression was nuclear and cytoplasmic. All cases were positive for nuclear staining at initial surgery; only one case became negative at second surgery. The nuclear staining intensity was moderate to strong in all positive tumors. (Figure 1).
Figure 1.
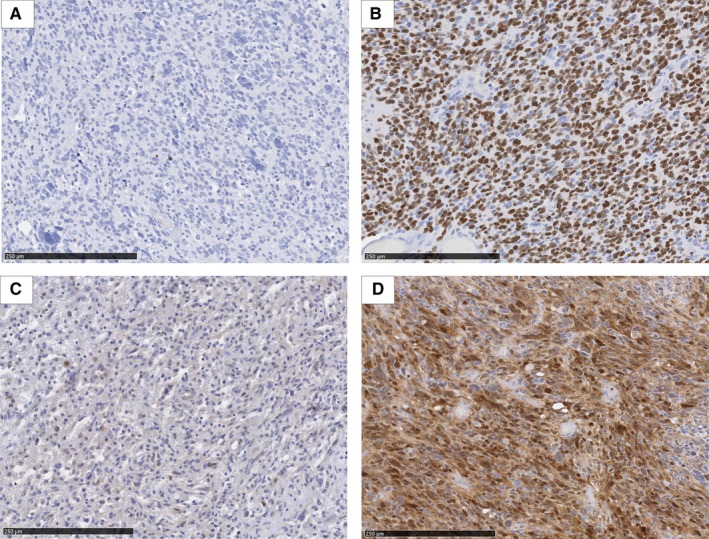
Cyclin D2 (CCND2) and oligodendrocyte lineage transcription factor 2 (OLIG2) expression profile in glioblastoma. CCND2 and OLIG2 present a nuclear and a cytoplasmic expression in glioblastoma. A, Low nuclear expression of OLIG2 (<30% of tumor cells nuclei stained). B, High nuclear expression of OLIG2 (≥30% of tumor cells nuclei stained). C, Low nuclear expression of CCND2 (<30% of tumor cells nuclei stained). D, High nuclear expression of CCND2 (≥30% of tumor cells nuclei stained)
3.2. Sample description according to adjuvant treatment received
Compared to initial samples, Ccnd2 and Olig2 expressions were significantly decreased in GBM recurrences. The decrease in expression observed did not differ significantly according to the treatment used. The RT alone group relapsed faster and had to be re‐operated more quickly than the RT‐TMZ group. There were no significant differences for other parameters (Table 1).
3.3. Sample description according to mortality
Subtotal second surgery for recurrence, time to recurrence <6 months, Ccnd2 nuclear expression at initial surgery ≥30%, and Olig2 nuclear expression <30% at second surgery after RT alone and RT‐TMZ were more frequent in patients with early mortality. Patients with early mortality presented shorter time to recurrence and lower Olig2 expression after adjuvant treatment than the patients with late mortality. There were no significant differences for other parameters (Table 2).
Table 2.
Sample description according to mortality (n = 72)
| Variables | Whole cohort | Categories | % | Late mortality ≥12 mo (n = 51) | Early mortality <12 mo (n = 21) |
P‐value Chi‐squared |
|---|---|---|---|---|---|---|
| Gender | Male (n = 45) | 62.50 | 62.75% (n = 32) | 61.90% (n = 13) | .947 | |
| Female (n = 27) | 37.50 | 37.25% (n = 19) | 38.10% (n = 8) | |||
| Number of lesions | Unique (n = 63) | 87.50 | 87.27% (n = 44) | 90.48% (n = 19) | .624 | |
| Multiple (n = 9) | 12.50 | 13.73% (n = 7) | 9.52% (n = 2) | |||
| Preoperative corticosteroids | No (n = 14) | 19.44 | 21.57% (n = 11) | 14.29% (n = 3) | .478 | |
| Yes (n = 58) | 80.56 | 78.43% (n = 40) | 85.71% (n = 18) | |||
| Type of initial surgery | Total (n = 43) | 59.72 | 66.67% (n = 34) | 42.86% (n = 9) | .061 | |
| Subtotal (n = 29) | 40.28 | 33.33% (n = 17) | 57.14% (n = 12) | |||
| Adjuvant treatment | RT alone (n = 37) | 51.39 | 47.06% (n = 24) | 61.90% (n = 13) | .252 | |
| RT‐TMZ (n = 35) | 48.61 | 52.94% (n = 27) | 38.10% (n = 8) | |||
| Type of surgery at recurrence | Total (n = 31) | 43.06 | 50.98% (n = 26) | 23.81% (n = 5) | .034 | |
| Subtotal (n = 41) | 56.94 | 49.02% (n = 25) | 76.19% (n = 16) |
| Mean ± SD | t Test | |||||
|---|---|---|---|---|---|---|
| Age at diagnosis (y) | 54.23 ± 11.63 | 54.23 ± 11.63 | 56.39 ± 10.85 | .469 | ||
| <50 (n = 20) | 27.78 | 25.49% (n = 13) | 33.33% (n = 7) | .499a | ||
| ≥50 (n = 52) | 72.22 | 74.51% (n = 38) | 66.67% (n = 14) |
| Median (P25‐P75) | Wilcoxon test | |||||
|---|---|---|---|---|---|---|
| Overall survival (mo) | 15.69 (11.63‐23.14) | 19.40 (15.19‐25.90) | 9.94 (8.43‐11.13) | <.001 | ||
| Time to recurrence (mo) | 6.90 (4.54‐10.90) | 9.03 (6.00‐13.43) | 4.33 (2.97‐5.00) | <.001 | ||
| >6 (n = 42) | 58.33 | 19.05% (n = 4) | <.001a | |||
| ≤6 (n = 30) | 41.67 | 74.51% (n = 38) | 80.95% (n = 17) | |||
| 25.49% (n = 13) | ||||||
| CCND2 nuclear expression (%) at T0 | 30 (10‐50) | 25 (10‐45) | 40 (30‐50) | .086 | ||
| <30 (n = 30) | 41.67 | 50.98% (n = 26) | 19.05% (n = 4) | .012a | ||
| ≥30 (n = 42) | 58.33 | 49.02% (n = 25) | 80.95% (n = 17) | |||
| CCND2 nuclear expression (%) at T1 | 12.5 (5‐30) | 15 (5‐35) | 10 (5‐25) | .528 | ||
| <30 (n = 53) | 73.71 | 70.59% (n = 36) | 80.95% (n = 17) | .364a | ||
| ≥30 (n = 19) | 26.39 | 29.41% (n = 15) | 19.05% (n = 4) | |||
| OLIG2 nuclear expression (%) at T0 | 50 (20‐60) | 50 (25‐70) | 30 (20‐50) | .119 | ||
| <30 (n = 20) | 27.78 | 25.49% (n = 13) | 33.33% (n = 7) | .499a | ||
| ≥30 (n = 52) | 72.22 | 74.51% (n = 38) | 66.67% (n = 14) | |||
| OLIG2 nuclear expression (%) at T1 | 30 (10‐50) | 30 (20‐50) | 20 (10‐25) | .022 | ||
| <30 (n = 35) | 48.61 | 37.25% (n = 19) | 76.19% (n = 16) | .003a | ||
| ≥30 (n = 37) | 51.39 | 62.75% (n = 32) | 23.81% (n = 5) |
Abbreviations: CCND2, cyclin D2; OLIG2, oligodendrocyte lineage transcription factor 2; RT, radiotherapy; RT‐TMZ, radio‐chemotherapy with temozolomide; SD, standard deviation; T0, at initial surgery; T1, at recurrence after adjuvant treatment.
Chi‐squared.
3.4. Univariate analysis and Multivariate analysis
Similar to the univariate analysis (Data S1), risk factors obtained by the method of automatic selection (stepwise backward) and significantly associated with an increased risk of early mortality were: subtotal surgery at recurrence, time to recurrence <6 months, Ccnd2 nuclear expression at initial surgery ≥30%, and Olig2 nuclear expression <30% at second surgery after RT alone and RT‐TMZ (Table 3).
Table 3.
Summary of logistic regression for screening variables predicting early mortality in glioblastoma patients (n = 72)
| Variables | Adjusted OR (CI 95%) | P‐value |
|---|---|---|
| Time to recurrence (mo) | .002 | |
| >6 (n = 42) | 1 | |
| ≤6 (n = 30) | 16.85 (2.78‐102.2) | |
| Type of surgery at recurrence | .006 | |
| Total (n = 31) | 1 | |
| Subtotal (n = 41) | 16.54 (2.20‐124.23) | |
| CCND2 nuclear expression (%) at T0 | .012 | |
| <30 (n = 30) | 1 | |
| ≥30 (n = 42) | 14.33 (1.80‐114.27) | |
| OLIG2 nuclear expression (%) at T1 | .023 | |
| <30 (n = 35) | 1 | |
| ≥30 (n = 37) | 0.14 (0.03‐0.76) |
Not included in the model because not significant: gender, age, number of lesions, preoperative corticosteroids, initial lesion surgery, adjuvant treatment, CCND2 nuclear expression after adjuvant treatment and OLIG2 expression at initial surgery.
Adequacy of model: Hosmer‐Lemeshow chi2 (P = .947).
Specificity of model: Linktest (linear component P = .001 and nonlinear component P = .899).
Abbreviations: CI, confidence interval; CCND2, cyclin D2; OLIG2, oligodendrocyte lineage transcription factor 2; OR, odd ratio; T0, at initial surgery; T1, at recurrence after adjuvant treatment.
3.5. Characteristics of the sample according to the evolution of Olig2 expression in individuals with initial Olig2 labeling index ≥30%
In individuals with initial nuclear Olig2 expression ≥30%, the decrease in Olig2 expression <30% at recurrence is associated with earlier relapse, shorter time between the two surgeries and poorer survival. There were no significant differences for other parameters (Table 4).
Table 4.
Characteristics of the sample according to the post‐treatment evolution of OLIG2 expression in glioblastoma patients with initial OLIG2 labeling index ≥30% (n = 52)
| Whole sample | OLIG2 < 30% (n = 23) | OLIG 2 ≥ 30% (n = 29) | P‐value | |
|---|---|---|---|---|
| Age at diagnosis | 56.44 ± 11.13 | 55.64 ± 13.06 | 57.08 ± 9.53 | .119a |
| Gender (male) | 57.69% | 52.17% | 62.07% | .473b |
| Number of lesions (multiple) | 9.62% | 8.70% | 10.34% | .841b |
| Preoperative corticosteroids (yes) | 75% | 73.91% | 75.86% | .872b |
| Time to recurrence (mo) | 6.74 (4.57‐10.65) | 5.37 (4.20‐7.27) | 8.67 (6.17‐13.43) | .007c |
| Time between surgery (mo) | 7.96 (5.62‐11.79) | 6.30 (4.72‐7.84) | 10.03 (7.37‐13.37) | .001c |
| Type of surgery at recurrence (subtotal) | 51.92% | 47.83% | 55.17% | .598b |
| Overall survival (mo) | 15.52 (11.84‐23.14) | 12.57 (9.90‐15.37) | 19.45 (14.83‐27.38) | <.001c |
Abbreviations: OLIG2, oligodendrocyte lineage transcription factor 2.
t Test.
Chi‐squared.
Wilcoxon test.
4. DISCUSSION
Glioma stem cells are critical cells implicated into GBM recurrence and radio(chemo)resistance through multiple and not fully elucidated mechanisms.15 The GSCs need urgently to be targeted and eliminated to one day offer a chance for cure for GBM patients. Unfortunately, the GSC population is highly heterogeneous even within a same tumor and shows phenotypic plasticity abilities between subpopulations.7, 16, 17, 18, 19, 20, 21 Two clinically relevant molecular subtypes signatures of GBM have consistently been highlighted between studies based on genetic and RNA expression profiles, namely the proneural (PN) and the mesenchymal (MES) which is associated with the worst prognosis.22, 23, 24, 25, 26 These two subtypes have also been identified for GSCs and seemed to be mutually exclusive.7, 16, 21, 27 Some data suggest that all GSC subtypes evolve from the PN phenotype, which is closely linked to OLIG2.20, 28 Olig2 is a bHLH transcriptional repressor protein that plays a critical role during the central nervous system development by maintaining glial progenitor cells in a competent proliferation state and allowing their specification.29, 30, 31, 32, 33, 34, 35 Olig2 is ubiquitously expressed in gliomas, irrespectively of grade, in various extents and is implicated in the gliomagenesis.36, 37, 38, 39, 40, 41 Previous studies have recognized Olig2 as a potential GSCs biomarker and it is one of the four transcription factors (with Sox2, Pouf3f2 and Sall2) that are sufficient to reprogram differentiated cells into GSCs.42, 43, 44 In the same way, we have recently highlighted the fact that Olig2 seems to be the best GSCs biomarker when comparing differentially expressed genes between differentiated and GSCs enriched cultures. Furthermore, Olig2 can easily be studied by IHC as we previously demonstrated in a small cohort of GBM.12 Olig2 is also a surrogate marker of the PN subtype of GSC, used instead of CD133 as this key PN marker can be studied by FACS but not by IHC.17, 21, 26 The PN phenotype seems to be predominant at the tumor edge based on gene expression profile.17, 21, 45, 46 However, in the present study, the nuclear labeling of Olig2 in the tumor core of initial surgery samples was found in majority of the cases (n = 71/72) with high nuclear expression (≥30%, n = 52/72). PN phenotype is sometimes associated with a better survival than MES subtype as illustrated by Pinel et al where high level of Olig2 expression tended to be associated with a better overall survival on TMA samples of 80 GBM.47 In our cohort, initial nuclear expression of Olig2 ≥30% is not associated with a significant improvement in survival. For patients with initial Olig2 expression ≥30%, a decrease in Olig2 expression (<30%) at second surgery is significantly associated with a shorter time to recurrence (5.37 months vs 8.67 months, P = .007) and poorer overall survival (12.57 months vs 19.45 months, P < .001). This could be explained by the fact that initial PN phenotype shifts into a more aggressive MES subtype induced by treatments, whether RT alone or RT‐TMZ.22 There is now more evidence that GBM and particularly GSCs exhibit phenotypic plasticity, particularly following (chemo‐)RT.16, 18, 19, 21, 48 Moreover, the maintenance of the oligodendrocyte precursor cells signature associated with the PN phenotype is linked to Olig2 expression.43, 49 Preclinical data showed that once OLIG2 was knocked down in PN GSCs lines, an upregulation of CD44 and others markers associated with MES phenotype was observed.50 The mechanisms behind this transition are not fully known but a beginning of response was described by Minata et al21 Following RT an upregulation of CD109, another MES marker, happens rapidly in PN GSCs (within 24 hours) with at the same time a reduction of CD133+ and Olig2+ GSCs. Radiation therapy induces the activation of nuclear factor κB through ATM activation by RT‐mediated DNA damage51, 52, 53, 54 resulting in the upregulation of CD109. The treatment‐naive or RT‐induced CD109+ GSCs are both highly tumorigenic and radioresistant.21 Other studies showed that PN to MES transition is also associated with global multidrug resistance.16, 17, 44, 55 All these data could therefore explain the poor prognosis associated with the decrease in Olig2 expression that we showed in GBM after adjuvant treatments. However, this shift appears to be not systematic after RT(‐TMZ) and further studies should investigate why and how PN‐to‐MES transition could be avoided.
We also studied another potential GCS marker, Ccnd2. The three D‐type cyclins (Cyclin D1, D2, and D3) are key checkpoint regulators of the mammalian cell cycle. Under the control of distinct intracellular pathway, they promote transition from G1 to S phase through activation of the cyclin‐dependent kinase Cdk4/6, phosphorylation of retinoblastoma suppressor protein (pRB), and suppression of pRB inhibitory function on E2F transcription factors ending in cell proliferation.56, 57 Cyclin D2 is necessary for normal gonadal cell proliferation and is crucial for neurogenesis.58, 59 Cyclin D2 is the predominant cyclin D in the human subventricular zone allowing expansion of the cortical intermediate progenitor cell population at embryonic stage and neurospheres of expanded neuronal precursors of adult hippocampi express only Ccnd2.59, 60 Cyclin D2 is rarely expressed in normal brain or low grade glioma and is significantly upregulated in GBM.61, 62, 63 Regarding GSCs, we have previously shown that CCND2 is part of the most differentially expressed genes between differentiated and GSC enriched cultures from similar DNA chip microarray platforms.12 In the same way, Koyama‐Nasu et al63 demonstrated by immunoblotting analysis and RT‐PCR that Ccnd2 is abundantly expressed only in undifferentiated GBM cell lines, which was not the case for Cyclin D1 and D3. Using siRNA, solely the knockdown of CCND2 resulted in a significant increase of G1 arrest of GSCs. Furthermore, Ccnd2 seems to have a critical role in the tumorigenicity as mice transplanted with GSCs in which Ccnd2 expression was repressed survived significantly longer than those maintaining an expression of Ccnd2.63 In the present study, we identified Ccnd2 nuclear expression at initial surgery ≥30% to be significantly associated with early mortality (<12 months), confirming the prognostic value of this GSCs marker in human. At recurrence, Ccnd2 expression is reduced and has no longer a prognostic impact. Therefore, targeting CCND2 before adjuvant RT‐TMZ could be a promising way to explore for GBM therapy.
Multivariate analysis and logistic regression also highlighted the fact that a short time to recurrence (≤6 months) and a subtotal surgery at recurrence are variables predicting early mortality (<12 months). It should be noted that subtotal initial surgery was not identified as a predictive variable as classically described but our cohort only included patients who presented a local recurrence and who could be re‐operated.64, 65, 66, 67 It may be therefore logically conceivable that we found only a predominant impact on survival for the type of surgery at recurrence (subtotal or not).
4.1. Limitations
Our study presents limitations given that the molecular data (MGMT methylation and IDH mutation status) are missing in older cases as these analyses were not yet implemented routinely at that time. Additional molecular sequencing could not be performed due to the retrospective nature of our study. Therefore, these data could not be included in our analyses. Furthermore, our study included a relatively small cohort of 72 patients with paired samples from a single center. However due to the low frequency of GBM patients eligible for repeat surgery,68 our paired cohort is to our knowledge one of the larger described in the literature.69, 70, 71, 72, 73, 74 Finally, we studied only the marker expression on the tumor core. In future studies, it might be interesting to analyse the expression of OLIG2 and CCND2 also at the invasive tumor edge and to perform validation of our biomarkers with different techniques such as RNA sequencing.
5. CONCLUSIONS
To our knowledge, we are the first to demonstrate on a paired cohort of human GBM that:
A comparative IHC of OLIG2 realized at initial and recurrence surgery has prognostic value. The patients for whom nuclear expression of OLIG2 becomes low (<30%) after adjuvant treatments have a significantly shorter time to recurrence and survival reflecting most probably a PN to MES transition of the GSCs population.
At initial surgery, high nuclear expression (≥30%) of CCND2, a G1/S regulator specific of GSCs, has prognostic value and is associated with early mortality (<12 months).
In the future, prospective studies should be conducted with GBM patient to validate the risk factors for early mortality highlighted in our study and explore the relationships of OLIG2 and CCND2 stem cell markers with the molecular status of GBM.
CONFLICT OF INTEREST
None declared.
AUTHOR CONTRIBUTIONS
CB contributed to acquisition, analysis and interpretation of data, and drafted the manuscript; ALT contributed to acquisition, analysis, and interpretation of data; MH contributed to statistical analysis and interpretation of data; DVG: contributed to interpretation of data; PD contributed to acquisition, analysis, and interpretation of data, conceptualization, and supervision of the study. All the authors reviewed and edited the manuscript, gave critical input on interpretation of results, and gave the final approval for publication.
Supporting information
ACKNOWLEDGMENTS
The authors thank Professor Isabelle Salmon, head of department of Pathology at Erasmus University Hospital for her support and the technical team of DIAPATH, Center of Microscopy and Molecular Imaging (CMMI), Gosselies, Belgium.
Bouchart C, Trépant A‐L, Hein M, Van Gestel D, Demetter P. Prognostic impact of glioblastoma stem cell markers OLIG2 and CCND2. Cancer Med. 2020;9:1069–1078. 10.1002/cam4.2592
Funding information
This work was supported by a doctoral grant from the Fonds Erasme (ALT) and by the Fonds Yvonne Boël (Brussels, Belgium). The CMMI is supported by the European Regional Development Fund (FEDER) and Wallonia.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are available from the corresponding author upon reasonable request. The data are not publicly available due to privacy or ethical restrictions.
REFERENCES
- 1. Louis N, Perry A, Reifenberge RG, et al. The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol. 2016;131(6):803‐820. [DOI] [PubMed] [Google Scholar]
- 2. Ghosh D, Nandi S, Bhattacharjee S. Combination therapy to checkmate glioblastoma: clinical challenges and advances. Clin Transl Med. 2018;7(1):33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomized phase III study: 5‐year analysis of the EORTC‐NCIC trial. Lancet Oncol. 2009;10(5):459‐466. [DOI] [PubMed] [Google Scholar]
- 4. Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765‐773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Singh SK, Hawkins C, Clarke ID, et al. Identification of human brain tumour initiating cells. Nature. 2004;432(7015):396‐401. [DOI] [PubMed] [Google Scholar]
- 6. Galli R, Binda E, Orfanelli U, et al. Isolation and characterization of tumorigenic, stem‐like neural precursors from human glioblastoma. Can Res. 2004;64(19):7011‐7021. [DOI] [PubMed] [Google Scholar]
- 7. Bao S, Wu Q, McLendon RE, et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444(7120):756‐760. [DOI] [PubMed] [Google Scholar]
- 8. Chen J, Li Y, Yu TS, et al. A restricted cell population propagates glioblastoma growth after chemotherapy. Nature. 2012;488(7412):522‐526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lathia JD, Mack SC, Mulkearns‐Hubert EE, Valentim C, Rich J. Cancer stem cells in glioblastoma. Genes Dev. 2015;29(12):1203‐1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Pietras A, Katz AM, Ekström EJ, et al. Osteopontin‐CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014;14(3):357‐369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beier D, Hau P, Proescholdt M, et al. CD133(+) and CD133(−) glioblastoma‐derived cancer stem cells show differential growth characteristics and molecular profiles. Cancer Res. 2007;67(9):4010‐4015. [DOI] [PubMed] [Google Scholar]
- 12. Trépant AL, Bouchart C, Rorive S, et al. Identification of OLIG2 as the most specific glioblastoma stem cell marker starting from comparative analysis of data from similar DNA chip microarray platforms. Tumour Biol. 2015;36(3):1943‐1953. [DOI] [PubMed] [Google Scholar]
- 13. D'Haene N, Maris C, Sandras F, et al. The differential expression of Galectin‐1 and Galectin‐3 in normal lymphoid tissue and non‐Hodgkin's and Hodgkin's lymphomas. Int J Immunopathol Pharmacol. 2005;18(3):431‐443. [DOI] [PubMed] [Google Scholar]
- 14. O'Hurley G, Sjötedt E, Rahman A, et al. Garbage in, garbage out: a critical evaluation of strategies used for validation of immunohistochemical biomarkers. Mol Oncol. 2014;8(4):783‐798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Han X, Xue X, Zhou H, Zhang G. A molecular view of the radioresistance of gliomas. Oncotarget. 2017;8(59):100931‐100941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bhat K, Balasubramaniyan V, Vaillant B, et al. Mesenchymal differentiation mediated by NF‐κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24(3):331‐346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mao P, Joshi K, Li J, et al. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci USA. 2013;110(21):8644–8649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kim SH, Ezhilarasan R, Phillips E, et al. Serine/threonine kinase MLK4 determines mesenchymal identity in glioma stem cells in an NF‐κB‐dependent manner. Cancer Cell. 2016;29(2):201‐213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Halliday J, Helmy K, Pattwell S, et al. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural‐mesenchymal shift. Proc Natl Acad Sci USA. 2014;111(14):5248‐5253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Patel AP, Tirosh I, Trombetta JJ, et al. Single‐cell RNA‐seq highlights intratumoral heterogeneity in primary glioblastoma. Science. 2014;344(6190):1396‐1401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Minata M, Audia A, Shi J, et al. Phenotypic plasticity of invasive edge glioma stem‐like cells in response to ionizing radiation. Cell Rep. 2019;26(7):1893‐1905.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Verhaak RG, Hoadley KA, Purdom E, et al; Cancer Genome Atlas Research Network . Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma char‐acterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98‐110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Philips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high‐grade glioma predict prognosis, delineate a pattern of disease progression and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157‐173. [DOI] [PubMed] [Google Scholar]
- 24. Cooper LA, Gutman DA, Long Q, et al. The proneural molecular signature is enriched in oligodendrogliomas and predicts improved survival among diffuse gliomas. PLoS ONE. 2010;5(9):e12548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Colman H, Zhang L, Sulman EP, et al. A multigene predictor of outcome in glioblastoma. Neuro Oncol. 2010;12(1):49‐57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Popova SN, Bergqvist M, Dimberg A, et al. Subtyping of gliomas of various WHO grades by the application of immunohistochemistry. Histopathology. 2014;64(3):365‐379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Chandran UR, Luthra S, Santana‐Santos L, et al. Gene expression profiling distinguishes proneural glioma stem cells from mesenchymal glioma stem cells. Genom Data. 2015;5:333‐336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ozawa T, Riester M, Cheng YK, et al. Most human non‐GCIMP glioblastoma subtypes evolve from a common proneural‐like precursor glioma. Cancer Cell. 2014;26(2):288‐300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Furusho M, Ono K, Takebayashi H, et al. Involvement of the Olig2 transcription factor in cholinergic neuron development of the basal forebrain. Dev Biol. 2006;293(2):348‐357. [DOI] [PubMed] [Google Scholar]
- 30. Lu QR, Sun T, Zhu Z, et al. Common developmental requirement for Olig function indicates a motor neuron/oligodendrocyte connection. Cell. 2002;109(1):75‐86. [DOI] [PubMed] [Google Scholar]
- 31. Takebayashi H, Nabeshima Y, Yoshida S, Chisaka O, Ikenaka K, Nabeshima Y. The basic helix‐loop‐helix Olig2 is essential for the development of motoneuron and oligodendrocyte lineages. Curr Biol. 2002;12(13):1157‐1163. [DOI] [PubMed] [Google Scholar]
- 32. Zhou D, Anderson DJ. The bHLH transcription factors OLIG2 and OLIG1 couple neuronal and glial subtype specification. Cell. 2002;109(1):61‐73. [DOI] [PubMed] [Google Scholar]
- 33. Lee SK, Lee B, Ruiz EC, Pfaff SL. Olig2 and Ngn2 function in opposition to modulate gene expression in motor neuron progenitor cells. Genes Dev. 2005;19(2):282‐294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Arnett HA, Fancy SP, Alberta JA, et al. bHLH transcription factor Olig1 is required to repair demyelinated lesions in the CNS. Science. 2004;306(5704):2111‐2115. [DOI] [PubMed] [Google Scholar]
- 35. Xin M, Yue T, Ma Z, Wu FF, Gow A, Lu QR. Myelinogenesis and axonal recognition by oligodendrocytes in brain are uncoupled in Olig1‐null mice. J Neurosci. 2005;25(6):1354‐1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Azzarelli B, Miravalle L, Vidal R. Immunolocalization of the oligodendrocyte transcription factor 1 (Olig1) in brain tumors. J Neuropathol Exp Neurol. 2004;63(2):170‐179. [DOI] [PubMed] [Google Scholar]
- 37. Bouvier C, Bartoli C, Aguirre‐Cruz L, et al. Shared oligodendrocyte lineage gene expression in gliomas and oligodendrocyte progenitor cells. J Neurosurg. 2003;99(2):344‐350. [DOI] [PubMed] [Google Scholar]
- 38. Ligon KL, Alberta JA, Kho AT, et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63(5):499‐509. [DOI] [PubMed] [Google Scholar]
- 39. Lu QR, Park JK, Noll E, et al. Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. Proc Natl Acad Sci USA. 2001;98(19):10851‐10856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Marie Y, Sanson M, Mokhtari K, et al. OLIG2 as a specific marker of oligodendroglial tumour cells. Lancet. 2001;358(9278):298‐300. [DOI] [PubMed] [Google Scholar]
- 41. Ohnishi A, Sawa H, Tsuda M, et al. Expression of the oligodendroglial lineage‐associated markers Olig1 and Olig2 in different types of human gliomas. J Neuropathol Exp Neurol. 2003;62(10):1052‐1059. [DOI] [PubMed] [Google Scholar]
- 42. Ligon KL, Huillard E, Mehta S, et al. Olig2‐regulated lineage‐restricted pathway controls replication competence in neural stem cells and malignant glioma. Neuron. 2007;53(4):503‐517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kosty J, Lu F, Kupp R, Mehta S, Lu QR. Harnessing OLIG2 function in tumorigenicity and plasticity to target malignant gliomas. Cell Cycle. 2017;16(18):1654‐1660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Suvà ML, Rheinbay E, Gilepsie SM, et al. Reconstructing and reprogramming the tumor propagating potential of glioblastoma stem‐like cells. Cell. 2014;157(3):580‐594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Hubert CG, Rivera M, Spangler LC, et al. A three‐dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found in vivo. Cancer Res. 2016;76(8):2465‐2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Segerman A, Niklasson M, Haglund C, et al. Clonal variation in drug and radiation response among glioma‐initiating cells is linked to proneural‐mesenchymal transition. Cell Rep. 2016;17(11):2994‐3009. [DOI] [PubMed] [Google Scholar]
- 47. Pinel B, Duschesne M, Godet J, et al. Mesenchymal subtype of glioblastomas with high DNA‐PKcs expression is associated with better response to radiotherapy and temozolomide. J Neurooncol. 2017;132(2):287‐294. [DOI] [PubMed] [Google Scholar]
- 48. Brown DV, Filiz G, Daniel PM, et al. Expression of CD133 and CD44 in glioblastoma stem‐cells correlates with cell proliferation, phenotype stability and intra‐tumor heterogeneity. PLoS ONE. 2017;12(2):e0172791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lu F, Chen Y, Zhao C, et al. Olig‐2dependant reciprocal shift in PDGF and EGF receptor signaling regulates tumor phenotype and mitotic growth in malignant glioma. Cancer Cell. 2016;29(5):669‐683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Kupp R, Shtayer L, Tien AC, et al. Lineage‐restricted OLIG2‐RTK signaling governs the molecular subtype of glioma stem‐like cells. Cell Rep. 2016;16(11):2838‐2845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bhattacharjee S, Nandi S. Choices have consequences: the nexus between DNA repair pathways and genomic instability in cancer. Clin Transl Med. 2016;5(1):45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Bhattacharjee S, Nandi S. DNA damage response and cancer therapeutics through the lens of the Fanconi Anemia DNA repair pathway. Cell Commun Signal. 2017;15(1):41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bhattacharjee S, Nandi S. Synthetic lethality in DNA repair network: a novel avenue in targeted cancer therapy and combination therapeutics. IUBMB Life. 2017;69(12):929‐937. [DOI] [PubMed] [Google Scholar]
- 54. Bhattacharjee S, Nandi S. Rare genetic diseases with defects in DNA repair: opportunities and challenges in orphan drug development for targeted cancer therapy. Cancers (Basel). 2018;10(9):298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Piao Y, Liang J, Holmes L, Henry V, Sulman E, de Groot JF. Acquired resistance to anti‐VEGF therapy in glioblastoma is associated with a mesenchymal transition. Clin Cancer Res. 2013;19(16):4392‐4403. [DOI] [PubMed] [Google Scholar]
- 56. Sherr CJ. D‐type cyclins. Trends Biochem Sci. 1995;20(5):187‐190. [DOI] [PubMed] [Google Scholar]
- 57. Yang K, Hitomi M, Stacey DW. Variations in cyclin D1 levels through the cell cycle determine the proliferative fate of a cell. Cell Div. 2006;1:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Sicinski P, Donaher JL, Geng Y, et al. Cyclin D2 is an FSH‐responsive gene involved in gonadal cell proliferation and oncogenesis. Nature. 1996;384(6608):470‐474. [DOI] [PubMed] [Google Scholar]
- 59. Kowalczyk A, Filipkowski RK, Rylski M, et al. The critical role of cyclin D2 in adult neurogenesis. J Cell Biol. 2004;167(2):209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Glickstein SB, Monaghan JA, Koeller HB, Jones TK, Ross ME. Cyclin D2 is critical for intermediate progenitor cell proliferation in the embryonic cortex. J Neurosci. 2009;29(30):9614‐9624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Zhang X, Zhao M, Huang AY, Fei Z, Zhang W, Wang XL. The effect of cyclin D expression on cell proliferation in human gliomas. J Clin Neurosci. 2005;12(2):166‐168. [DOI] [PubMed] [Google Scholar]
- 62. Kheirollahi M, Mehr‐Azin M, Kamalian N, Mehdipour P. Expression of cyclin D2, P53, Rb and ATM cell cycle genes in brain tumors. Med Oncol. 2011;28(1):7‐14. [DOI] [PubMed] [Google Scholar]
- 63. Koyama‐Nasu R, Nasu‐Nishimura Y, Todo T, et al. The critical role of cyclin D2 in cell cycle progression and tumorigenicity of glioblastoma stem cells. Oncogene. 2013;32(33):3840‐3845. [DOI] [PubMed] [Google Scholar]
- 64. McGirt MJ, Chaichana KL, Gathinji M, et al. Independent association of extent of resection with survival in patients with malignant brain astrocytoma. J Neurosurg. 2008;110(1):156‐162. [DOI] [PubMed] [Google Scholar]
- 65. Chaichana KL, Jusue‐Torres I, Navarro‐Ramirez R, et al. Establishing percent resection and residual volume thresholds affecting survival and recurrence for patients with newly diagnosed intracranial glioblastoma. Neuro Oncol. 2014;16(1):113‐122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Brown TJ, Brennan MC, Li M, et al. Association of the extent of resection with survival in glioblastoma: a systematic review and meta‐analysis. JAMA Oncol. 2016;2(11):1460‐1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Trifiletti DM, Alonso C, Grover S, Fadul CE, Sheehan JP, Showalter TN. Prognosis implications of extent of resection in glioblastoma: analysis from a large database. World Neurosurg. 2017;103:330‐340. [DOI] [PubMed] [Google Scholar]
- 68. Weller M, Cloughesy T, Perry JR, Wick W. Standards of care for treatment of recurrent glioblastoma – are we there yet? Neuro Oncol. 2013;15(1):4‐27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hersh DS, Harder BG, Roos A, et al. The TNF receptor family member Fn14 is highly expressed in recurrent glioblastoma and in GBM patient‐derived xenografts with acquired temozolomide resistance. Neuro Oncol. 2018;20(10):1321‐1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Hudson AL, Parker NR, Khong P, et al. Glioblastoma recurrence correlates with increased APE1 and polarization toward an immuno‐suppressive microenvironment. Front Oncol. 2018;8:314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Cioca A, Olteanu EG, Gisca MD, Morosanu CO, Marin I, Florian IS. Expression of EGFR in paired new and recurrent glioblastomas. Asian Pac J Cancer Prev. 2016;17(9):4205‐4208. [PubMed] [Google Scholar]
- 72. Schäfer N, Gielen GH, Rauschenbach L, et al. Longitudinal heterogeneity in glioblastoma: moving targets in recurrent versus primary tumors. J Transl Med. 2019;17(1):96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Azoulay M, Santos F, Shenouda G, et al. Benefit of re‐operation and salvage therapies for recurrent glioblastoma multiforme: results from a single institution. J Neurooncol. 2017;132(3):419‐426. [DOI] [PubMed] [Google Scholar]
- 74. Kubelt C, Hattermann K, Sebens S, Mehdorn HM, Held‐Feindt J. Epithelial‐to‐mesenchymal transition in paired human primary and recurrent glioblastomas. Int J Oncol. 2015;46(6):2515‐2525. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon reasonable request. The data are not publicly available due to privacy or ethical restrictions.


