Abstract
Glioblastoma is the most common and malignant form of primary central nervous tumor in adults. Long noncoding RNAs (lncRNAs) have been reported to play a pivotal role in modulating gene expression and regulating human tumor’s malignant behaviors. In this study, we confirmed that lncRNA brain-derived neurotrophic factor antisense (BDNF-AS) was downregulated in glioblastoma tissues and cells, interacted and stabilized by polyadenylate-binding protein cytoplasmic 1 (PABPC1). Overexpression of BDNF-AS inhibited the proliferation, migration, and invasion, as well as induced the apoptosis of glioblastoma cells. In the in vivo study, PABPC1 overexpression combined with BDNF-AS overexpression produced the smallest tumor and the longest survival. Moreover, BDNF-AS could elicit retina and anterior neural fold homeobox 2 (RAX2) mRNA decay through STAU1-mediated decay (SMD), and thereby regulated the malignant behaviors glioblastoma cells. Knockdown of RAX2 produced tumor-suppressive function in glioblastoma cells and increased the expression of discs large homolog 5 (DLG5), leading to the activation of the Hippo pathway. In general, this study elucidated that the PABPC1-BDNF-AS-RAX2-DLG5 mechanism may contribute to the anticancer potential of glioma cells and may provide potential therapeutic targets for human glioma.
Subject terms: Targeted therapies, Long non-coding RNAs
Introduction
Glioma is the most common tumor type of the central nervous system with account for more than 50% of all primary brain tumors, and has been classified into I–IV grades of malignancy according to the World Health Organization (WHO)1. Glioblastoma is the most common and malignant type. The average survival of patients with glioblastoma is ~12–15 months due to the high invasion and proliferation2. Despite several efforts of surgical resection in combination with adjuvant chemoradiotherapy, patient prognosis remains poor. Currently, the field of glioblastoma research is focused on developing molecular targeted therapy that may provide new approaches for the treatment.
RNA-binding proteins have been identified as modulators of posttranscriptional mechanisms, including RNA splicing, transport, translation, and localization3. Numerous RBPs have been reported to be implicated in multiple tumor types, such as PCBP2 and Musashi 1 have been observed to contribute to glioma cells initiation and growth4,5. Polyadenylate-binding protein cytoplasmic 1 (PABPC1) is a cytoplasmic-nuclear shuttling protein expressed in most eukaryotes, which is important for protein translation initiation and decay. In addition, PABPC1 regulates mRNAs by binding to the poly(A) tails of them6. PABPC1 was corrected with tumor progression and patient's prognosis in esophageal cancer7. Besides, abnormal changes and functions of PABPC1 were found in cervical cancer (CC), colorectal cancer, and gastric cancer8. The expression and functional roles of PABPC1 in glioma remain unknown.
Long noncoding RNAs (LncRNAs) are defined as protein noncoding RNAs that, >200 nucleotides in length, have been shown to play important roles in various cancer development and progression. Multiple lncRNAs are identified as biomarkers for tumor diagnosis, treatment, or prognosis in recent years9,10. Brain-derived neurotrophic factor antisense (BDNF-AS) is a natural noncoding antisense of BDNF, locates at chromosome region 11p14.1, plays important roles in neuronal system together with BDNF11. Besides, BDNF-AS was lowly expressed in retinoblastoma (RB), non-small cell lung cancer (NSCLC) and CC, and suppressed cells proliferation and migration in these cancers12. The relationship between BDNF-AS and glioblastoma has not been found yet. The binding site between PABPC1 and BDNF-AS was predicted using the database of RBP specificities.
Stau1-mediated mRNA decay (SMD) is an upframeshift factor 1 (UPF1)-dependent mRNAs degradation mechanism, which is mediated by direct binding of STAU1 to STAU1-binding site (SBS) in mammalian cells13. STAU1 is a double-stranded RNA-binding protein, which can recognize SBSs in the 3′-UTR of target mRNAs and recruit UPF1 thereby mediated SMD14. SMD is a characteristic posttranscriptional regulation manner which has been reported to regulate a wide spectrum of physiological processes by degrading the expression of mRNAs involved. Cho et al. found that SMG1 can stimulate efficient adipogenesis by enhancing SMD15. In gastric cancer, lncRNA-TINCR caused KLF2 mRNA degradation via SMD thereby regulating gastric cancer cell proliferation and apoptosis16.
The retina and anterior neural fold homeobox 2 (RAX2) gene has 2428 bp in length and a cytogenetic location of 19p13.3, encodes 230 amino acids. As a member of transcription factor RAX, RAX2 is essential for eye development of several vertebrate species by modulating the proliferation and differentiation of retinal cells17. According to the prediction via software, an SBS were found between BDNF-AS and RAX2 mRNA 3'-UTR. Moreover, the JASPAR CORE database showed that RAX2 could bind to the promoter regions of discs large homolog 5 (DLG5). DLG5 belongs to the membrane associated guanylate kinase (MAGUK) family, participates in epithelial cell polarity maintenance and cancer development18. DLG5 expression was downregulated in many human cancers, such as bladder cancer, prostate cancer, breast cancer, and hepatocellular carcinoma, not only that, knockdown of DLG5 significantly increased cell migration and invasion in these cancers19,20.
In our study, we provided the endogenous expression and functions of PABPC1, BDNF-AS, RAX2, and DLG5 in human glioma tumor specimens and cells. Further, we explored their molecular mechanisms and effects on the biological behaviors of glioblastoma cells. The goal of this study was to demonstrate a novel molecular mechanism and experimental basis for the regulation of biological behaviors of glioblastoma cells, and provided promising therapeutic targets in human glioma.
Materials and methods
Cell culture
Normal human astrocyte (HA) cells were purchased from the ScienCell Research Laboratories (Carlsbad, CA, USA) and cultured in Roswell Park Memorial Institute (RPMI)–1640 supplemented with 10% fetal bovine serum (FBS, Gibco, Carlsbad, CA, USA). Human U87, U251 glioblastoma cell lines, and human embryonic kidney (HEK) 293T cells were purchased from Shanghai Institutes for Biological Sciences Cell Resource Center and cultured in high glucose Dulbecco’s modified Eagle’s medium supplemented with 10% FBS. All cells were maintained in a humidified incubator (5% CO2, at 37 °C).
Human tissue samples
Glioma tissues and normal brain tissues (NBTs) were obtained from the patients who underwent surgery at the Department of Neurosurgery of Shengjing Hospital of China Medical University. Each patient was provided informed consent and the use of the samples for this study was approved by the Ethics Committee of Shengjing Hospital of China Medical University. All tumors were quickly frozen into liquid nitrogen at the time of resection and classified according to the WHO classification by neuropathologists.
Cell transfections
The short-hairpin RNA directly against human BDNF-AS (NR_002832.2), PABPC1 (NM_002568.3), RAX2 (NM_001319074.1), or DLG5 (NM_004747.3) gene and their nontargeting sequences were ligated into pGPU6/GFP/Neo vectors (GenePharma, Shanghai, China), respectively. Human PABPC1 (NM_002568.3) mRNA sequence was ligated into the pcDNA3.1 vector. Human BDNF-AS gene and the negative control were ligated into pGCMV/MCS/IRES/EGFP/Neo vector (GenePharma, Shanghai, China). Full-length human DLG5 gene and respective nontargeting sequence were ligated into pIRES2-EGFP (GenScript, Piscataway, NJ, USA). The transfection of U87 and U251 cells was carried out using Lipofectamine 3000 and Opti-MEM (Life Technologies, Carlsbad, CA, USA). Geneticin (G418) and Puromycin (Sigma-Aldrich, St Louis, MO, USA) were used to select the stable-transfected cells.
Western blotting
Western blot procedures were performed as previously described21. Proteins were detected by specific antibodies included PABPC1 (1:1000, Abcam, Cambridge, MA, USA), RAX2 (1:800, Proteintech Group, Rosement, USA), DLG5 (1:800, Proteintech Group, Rosement, USA), STAU1 (1:1000, Proteintech Group, Rosement, USA), UPF1 (1:1000, Proteintech Group, Rosement, USA), GAPDH (1:5000, Cell Signaling Technology, Danvers, MA, USA), or Hippo Signaling Antibody Sampler Kit (Cell Signaling Technology, Danvers, MA, USA).
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total RNA was extracted using the Trizol reagent (Life Technologies Corporation, Carlsbad, CA, USA), and quantified using a NanoDropTM 3000 Spectrophotometer (Thermo Fisher Scientific, USA). QRT-PCR was conducted using One-Step SYBR PrimerScript RT-PCR Kit (TakataBio, Inc., Japan) according to the manufacturer’s protocol. See Additional file 1 for details and primers used.
LncRNAs microarray
LncRNAs analysis and microarray hybridization were performed by Kangchen Bio-tech (Shanghai, China).
Nascent RNA capture assay
To capture the newly synthesized RNA transcripts of BDNF-AS and RAX2 mRNA, the nascent RNA capture assay was carried out using the Click-iT Nascent RNA Capture Kit (Thermo Fisher Science, Waltham, MA, USA) according to the manufacturer’s protocol. Briefly, cells were seeded at 40–50% confluency and incubated with appropriate volume of 5-ethynyluridine (EU) solution at 37 °C at 5% CO2 for 24 h. Cells were harvest and then centrifuged and total RNA was isolated as described before. Dynabeads® MyOne™ Streptavidin T1 magnetic beads were used in biotinylated total RNA to pull out the biotin-labeled EU-RNA. After removing the supernatant and washing the beads using wash buffer, the nascent RNA captured on the beads was analyzed using qRT-PCR as described.
RNA stability assay
To measure the stability of BDNF-AS, cells were treated with 5 μg/ml actinomycin D (Sigma, MO, USA) which could block transcription. Cells were collected at 0–5 h after addition of actinomycin D, and the total cellular RNA was isolated. QRT-PCR was performed to measure the half-life of BDNF-AS and GAPDH mRNA was applied as an internal control.
RNA-protein immunoprecipitation (RIP) assay
RIP were carried out using the EZ-Magna RIP Kit (Millipore, Billerica, MA, USA) according to the manufacturer’s instruction. Cells were lysed by a complete RNA lysis buffer with protease inhibitor and RNase, and then incubated with RIP buffer containing magnetice beads conjugated with antibodies of STAU1 with negative control (IgG), respectively. Wash buffer was added to all RIP reactions and vortex gently, and repeat wash steps until magnetic beads have been washed a total of five times. The fold enrichments of immunoprecipitated RNAs with different antibodies were analyzed using qRT-PCRs when the immunoprecipitated RNA was isolated and purified.
Cell proliferation assay
Cell proliferation was examined with the Cell Counting Kit-8 (CCK-8) assay using a Cell Counting Kit (Beyotime, Jiangsu, China) as previously described21.
Apoptosis assay
Apoptosis rate was routinely determined by flow cytometry using Annexin V-PE/PI staining (Southern Biotech, Birmingham, AL). Briefly, cells were harvested and stained with Annexin V-FITC and PI according to the manufacturer’s instruction. Data analysis was performed using BD accuri C6 software.
Migration and invasion assay
The migration and invasion assays were carried out using transwell chambers (Corning, NY, USA, 3422, 8-μm pores) according to previously described21.
RNA pull-down assay
Biotin-labeled, full-length BDNF-AS RNA, or BDNF-AS with mutational SBS (BDNF-AS-SBS-Mut) fragment was prepared with the Biotin RNA Labeling Mix (GenePharma, Shanghai, China) and transfected into U87 and U251 cells. Biotinylated RNAs were treated with RNase-free DNase I and purified. RNA-protein complexes were isolated by streptavidin agarose beads (Invitrogen, Shanghai, China) and washed three times. The retrieved proteins were detected using a standard western blotting technique as described above.
Reporter vectors constructs and luciferase assay
BDNF-AS full-length and RAX2 3'-UTR sequences were amplified by PCR and cloned in the pmirGlo Dual-luciferase Vector (Promega, Madison, USA) to construct luciferase reporter vector (BDNF-AS-Wt and RAX2-Wt) (GenePharma, Shanghai, China). The sequence of putative binding site of BDNF-AS (ALU: 1982–2062bp) and RAX2 (ALU: 2116–2419 bp) was replaced as indicated (BDNF-AS-Mut, RAX2-Mut). The responsive RAX2-binding sites in the DLG5 promotor were predicted by bioinformatics tool JASPAR and were determined by dual-luciferase reporter system. Promoter fragments were subcloned into pGL3-Basic-Luciferase vector (Promega, WI, USA). Human full-length RAX2 was constructed in pEX3 vector (GenePharma, Shanghai, China). The assay was performed 48 h after transfection the indicated constructs into 2.4 × 104 HEK-293T cells per well seeded into 96-well plates. Cells were analyzed by the luciferase assay using the dual-luciferase reporter assay system. The relative luciferase activity was expressed as the ratio of firefly luciferase activity to renilla luciferase activity.
Nude mouse xenograft model
For the in vivo study, cells stably transfected with PABPC1(+), BDNF-AS(+), PABPC1(+) + BDNF-AS(+) were selected. Four weeks old female BALB/C nude mice were purchased from Cancer Institute of the Chinese Academy of Medical Science and divided into five groups by chance (n = 6, each group) for subcutaneous and intracranial orthotropic inoculation, respectively. A total of 3 × 105 cells were subcutaneously inoculated in the left flank of the mice. Tumor nodules were estimated every 5 days after injection. The following formula was used to calculated tumor volume: volume (mm3) = length × width2/2. For intracranial orthotropic inoculation, 3 × 105 cells were injected into the right striatum of nude mice. Then, the number of survived nude mice was recorded every 5 days. Survival analysis was carried out in the form of Kaplan–Meier survival curve. All animal experiments were complied with the Guide for the Care Committee of the Shengjing Hospital.
Chromatin immunoprecipitation (ChIP) assay
Chromatin was fixed and immunoprecipitated using the Simple ChIP Enzymatic Chromatin IP Kit (Cell signaling Technology, MA, USA) as recommended by the manufacturer. See Additional file 1 for details and primers used.
Statistical analysis
In this work, all data were described as mean ± SD from at least three times independent experiments. All statistical analysis was conducted using SPSS 18.0 statistical software with student’s t test (between two groups) or one-way ANOVA analysis (three or more groups) of variance. Differences were considered as statically significant when P < 0.05.
Results
PABPC1 acted as a tumor suppressor in glioblastoma cell lines
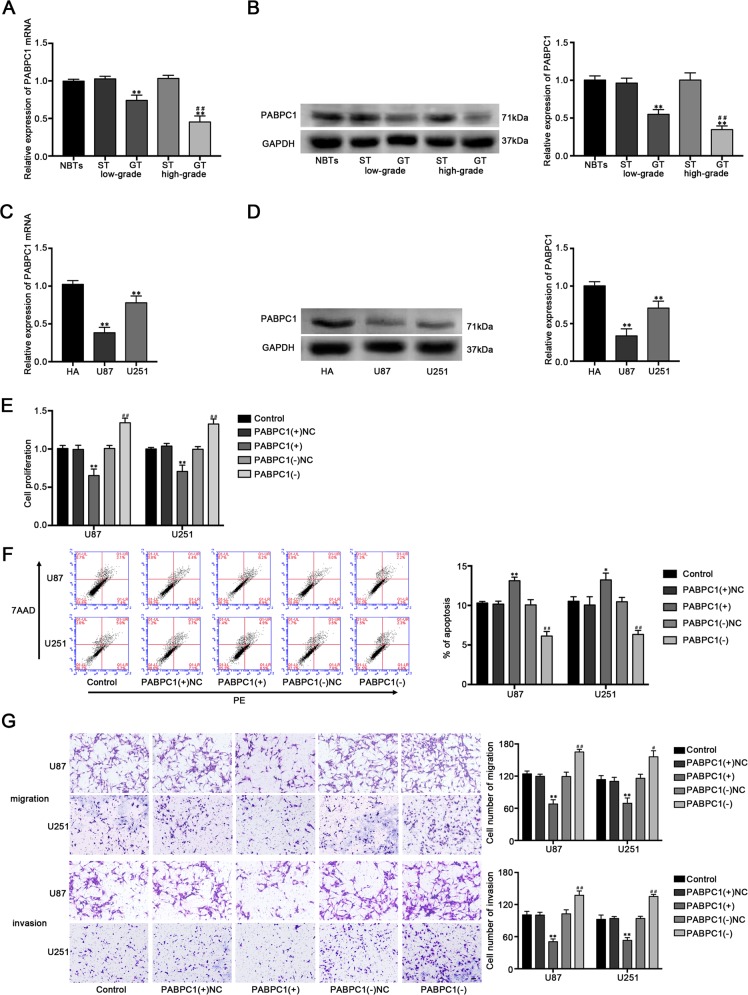
By using the Oncomine database (https://www.oncomine.org/resource/main.html), the lower expression of PABPC1 in glioblastoma tissues compared with neural stem cells were found (Fig. S1A). We further examined the expression levels of PABPC1 in human glioma tissues (GT) and cell lines by qRT-PCR and western blot. As shown in Fig. 1a–d, PABPC1 expressed lower in GT and cells than in surrounding nonneoplastic tissues (ST) and NBTs, and the expression level was negatively correlated with the histopathological grades of gliomas. Furthermore, PABPC1 expression was significantly lower in U87 and U251 cells than in HA cells. Stable PABPC1 overexpressed and silenced constructs were used to further evaluate the biological role (Fig. S1B). As shown in Fig. 1e, the proliferation ability of glioblastoma cells was decreased in the PABPC1(+) group, while increased in the PBAPC1(−) group compared with their nonspecific control (NC) group, respectively. Overexpression of PABPC1 significantly increased the apoptosis ratio of glioblastoma cells (Fig. 1f) and inhibited the migration and invasion capability in glioblastoma cells (Fig. 1g). These data suggested that PABPC1 functioned as a tumor suppressor in glioblastoma cells.
Fig. 1. The expression and effects of PABPC1 in glioblastoma cells.
a The PABPC1 mRNA expression levels in normal brain tissues (NBTs), low and high grades of human glioma tissues (GT), and homologous surrounding nonneoplastic tissues (ST). b The PABPC1 protein expression levels in NBTs, low and high grades of GT and homologous ST (n = 4, each group). **P < 0.01 vs. ST group; ##P < 0.01 vs. low-grade GT group. c The mRNA expression level of PABPC1 in human astrocytes (HA) and glioblastoma cell lines (U87 and U251). d The protein expression level of PABPC1 in human astrocytes (HA) and glioblastoma cell lines (U87 and U251). (n = 3, each group). **P < 0.01 vs. HA group. e The CCK-8 assay was used to measure the effect of PABPC1 on the proliferation of U87 and U251 cells. f The apoptotic percentages of U87 and U251 cells were detected after PABPC1 overexpression or knockdown. g The transwell assays were used to measure the effect of PABPC1 on cell migration and invasion of U87 and U251 cells. Scale bars represent 40 μm. (n = 5, each group). *P < 0.05 or **P < 0.01 vs. PABPC1(+) NC group; #P < 0.05 or ##P < 0.01 vs. PABPC1(−)NC group.
Overexpression of BDNF-AS inhibited malignant behaviors of glioblastoma cells
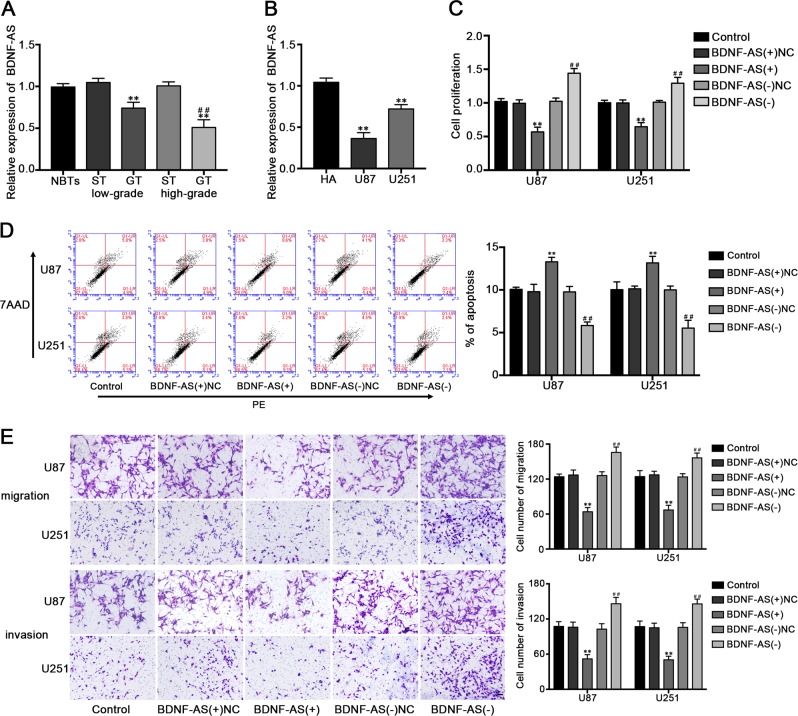
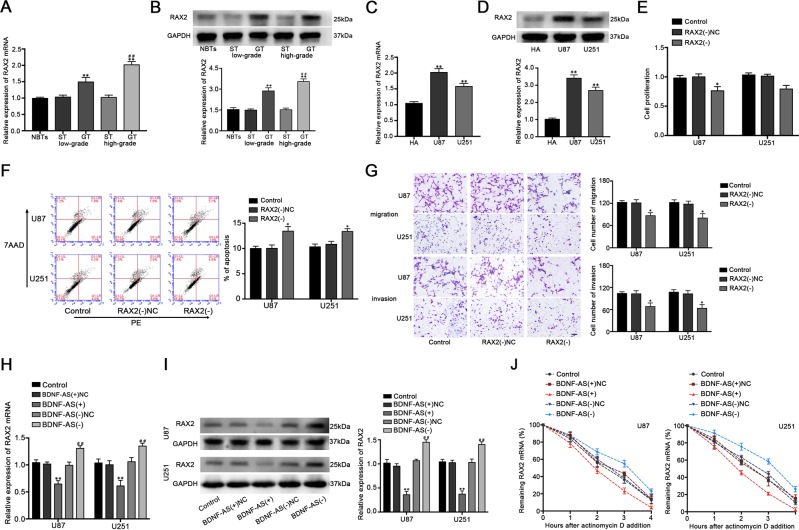
QRT-PCR was performed to evaluate BDNF-AS expression levels in GT and cells, and the results indicated that BDNF-AS was downregulated in GT and cell lines compared with NBTs and HA cells, respectively. Moreover, the expression level of BDNF-AS in GT was negatively correlated with histopathological grade in human GT (Fig. 2a, b). To determine the effects of BDNF-AS on glioblastoma cells, the stable overexpression and knockdown of BDNF-AS of U87 and U251 cell lines were established, the transfection efficiency were shown in Fig. S1C. The CCK-8 assay manifested that the overexpression of BDNF-AS inhibited the proliferation of U87 and U251 cells (Fig. 2c). Flow cytometry analysis results showed that the apoptosis of U87 and U251 cells was increased in BDNF-AS(+) group compared with the BDNF-AS(+)NC group (Fig. 2d). Moreover, as showed in Fig. 2e, BDNF-AS overexpression significantly inhibited the migration and invasion capabilities in glioblastoma cells. In the meantime, knockdown of BDNF-AS exerted opposite effects in same assays. We proposed that BDNF-AS exerted tumor-suppressive function in glioblastoma cells.
Fig. 2. The expression and effects of BDNF-AS in glioblastoma cells.
a The relative expression levels of BDNF-AS in NBTs, low and high grades of human glioma tissues. Data are presented as the mean ± SD (n = 4, each group). **P < 0.01 vs. ST group; ##P < 0.01 vs. low-grade GT group. b The relative expression levels of BDNF-AS in HA and glioblastoma cell lines. Data are presented as the mean ± SD (n = 3, each group). **P < 0.01 vs. HA group. c Effect of BDNF-AS on the cell proliferation, cell apoptosis (d), cell migration and invasion (e) of glioblastoma cells. Scale bars represent 40 μm. (n = 5, each group). **P < 0.01 vs. BDNF-AS(+)NC group; ##P < 0.01 vs. BDNF-AS(−)NC group.
PABPC1 increased the stability of BDNF-AS and interacted with BDNF-AS in glioblastoma cells
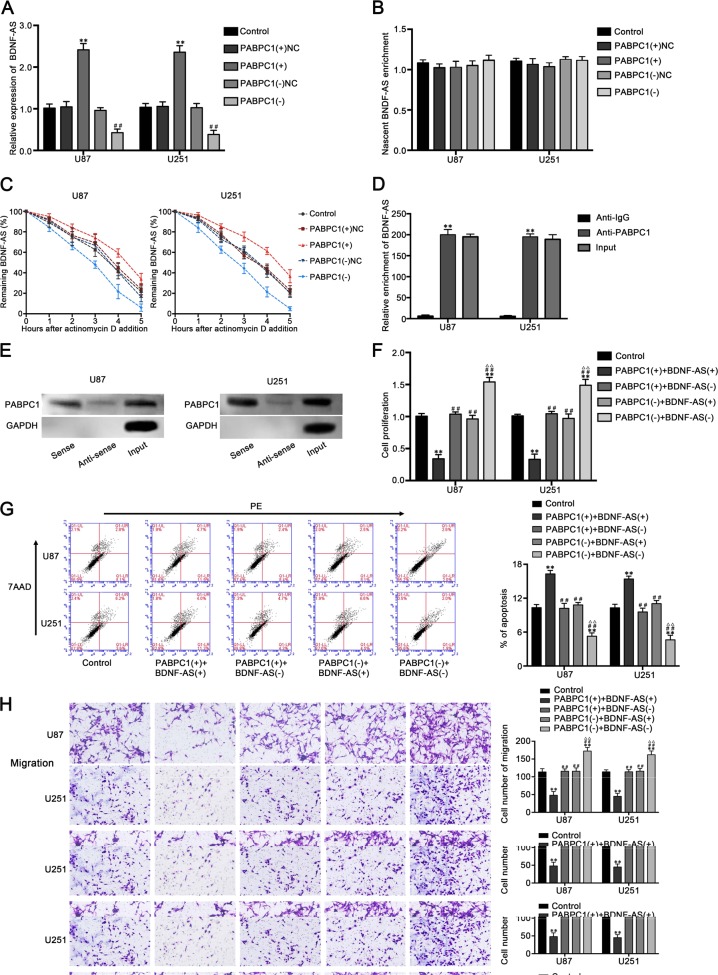
Using the RNA-binding protein database and lncRNAs microarray analysis, we found the putative binding between PABPC1 and many lncRNAs, and we found that BDNF-AS was significantly upregulated by the overexpression of PABPC1 (Fig. S1D). Therefore, we proposed that BDNF-AS might be involved in the regulation of PABPC1 on glioma cells. Our results showed that the expression of BDNF-AS was increased in PABPC1(+) group, whereas decreased in PABPC1(−) group (Fig. 3a). Since PABPC1 was confirmed to increase the stability of mRNAs or LncRNAs22, we hypothesized that PABPC1 increases the expression of BDNF-AS by increasing the stability of it. As shown in Fig. 3b, c, nascent RNA capture assays showed no significant effect of PABPC1 on BDNF-AS RNA synthesis, whereas PABPC1 overexpressing significantly increased the half-life of BDNF-AS. These results indicated that PABPC1 regulated BDNF-AS expression at a posttranscriptional level. To confirm whether PABPC1 interacts with BDNF-AS in U87 and U251 cells, RNA immunoprecipitation (RIP) and RNA pull-down assays were carried out. As show in Fig. 3d, RIP was performed with an antibody against PABPC1 using extracts from glioblastoma cells and BDNF-AS enrichment was observed to increase in anti-PABPC1 group than in anti-IgG group significantly. Then, western blot analysis using proteins form RNA pull-down assays confirmed that PABPC1 is associated with BDNF-AS compared with antisense group (Fig. 3e). Moreover, the stable BDNF-AS overexpressed and silenced constructs were transfected into PABPC1(+) group and PABPC1(−) group cells respectively to investigate the role of BDNF-AS on the function of PABPC1. As shown in Fig. 3f–h, silencing of BDNF-AS counteract the effect of the overexpression of PABPC1 on cell proliferation, migration, invasion, and apoptosis, implied that BDNF-AS is associated with the PABPC1-induced effects on glioblastoma cells.
Fig. 3. PABPC1 binds to BDNF-AS and stabilizes BDNF-AS.
a Expression of BDNF-AS after PABPC1 overexpression or knockdown (n = 3, each group). **P < 0.01 vs. PABPC1(+)NC group. ##P < 0.01 vs. PABPC1(−)NC group. b Identification of the newly synthesized RNA enrichment of BDNF-AS via RNA capture assay and qRT-PCR. c Half-life of BDNF-AS measured by qRT-PCR and normalized to the level of GAPDH. d RIP was performed in U87 and U251 cells and followed by qRT-PCR to detect BDNF-AS associated with PABPC1. **P < 0.01 vs. anti-IgG group. e Western blot analysis following RNA pull-down assays performed using U87 and U251 cellular exacts. f Effect of PABPC1 and BDNF-AS on cell proliferation, g apoptosis, h migration and invasion of glioblastoma cells. Scale bars represent 40 μm. (n = 5, each group). **P < 0.01 vs. control group; ##P < 0.01 vs. PABPC1(+) + BDNF-AS(+) group. ▵▵P < 0.01 vs. PABPC1(+) + BDNF-AS(−) group.
The combination of PABPC1 overexpression and BDNF-AS overexpression suppressed tumor growth and exhibited high survival time in nude mice
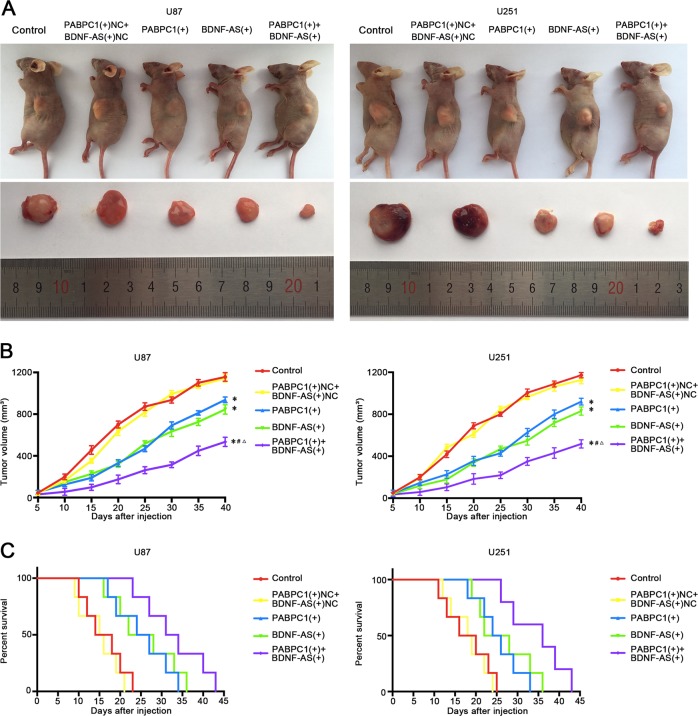
To further explore the antitumor function of PABPC1 and BDNF-AS in vivo, the stable expression cells were used in nude mice. As shown in Fig. 4a, b, PABPC1 overexpression, BDNF-AS overexpression, and combination of them produced lower tumors than the control group. In addition, PABPC1 overexpression combined with BDNF-AS overexpression led to the smallest tumor among all groups. Similarly, survival analysis indicated that mice in PABPC1(+) + BDNF(+) group had the longest survival time (Fig. 4c).
Fig. 4. Tumor xenograft experiments.
a The nude mice carrying tumors from respective groups were shown and the sample tumors from respective group were shown. b Tumor growth curves were shown. Tumor volume was calculated every 5 days after injection, and the tumor was taken after 40 days. c Survival curves from respective nude mice injected into the right striatum were shown (n = 6, each group).
Knockdown of RAX2 inhibited the cell proliferation, migration and invasion, and promoted apoptosis of glioblastoma cells
QRT-PCR and western blot were used to analyze the expression level of RAX2 in human GT and cells. The results showed that RAX2 was upregulated in GT and cell lines and the expression level in tissues was positively correlated with histopathological grade of GT (Fig. 5a–d). This increase of RAX2 expression prompted that RAX2 may have possible biological effects on glioblastoma cells, therefore the biological behaviors of U87 and U251 cells were detected after stably constructing RAX2 silenced cells (Fig. S1E). As shown in Fig. 5e–g, RAX2 knockdown inhibited cell proliferation, migration and invasion, and promoted the cell apoptosis of U87 and U251 cells. These results indicated that knockdown of RAX2 exerts tumor-suppressive effects in human glioblastoma cells.
Fig. 5. The expression and effects of RAX2 in glioblastoma cells.
a The mRNA and b protein expression level of RAX2 in NBTs, low and high grades of GT and homologous ST (n = 4, each group). **P < 0.01 vs. ST group; ##P < 0.01 vs. low-grade GT group. c The mRNA and d protein expression level of RAX2 in HA, U87, and U251 (n = 3, each group). **P < 0.01 vs. HA group. e Effect of RAX2 knockdown on the cell proliferation of U87 and U251 cells. f Effect of RAX2 knockdown on the cell apoptosis of U87 and U251 cells. g Effect of RAX2 knockdown on the cell migration and invasion of U87 and U251 cells. Scale bars represent 40 μm. (n = 5, each group). *P < 0.05 vs. RAX2(−)NC group. h Expression levels of RAX2 mRNA and I protein after BDNF-AS overexpression or knockdown (n = 3, each group). **P < 0.01 vs. BDNF-AS(+)NC group. ##P < 0.01 vs. BDNF-AS(−)NC group. j The half-life of RAX2 mRNA were estimated after BDNF-AS overexpression or knockdown (n = 3, each group).
BDNF-AS elicited RAX2 mRNA decay through SMD
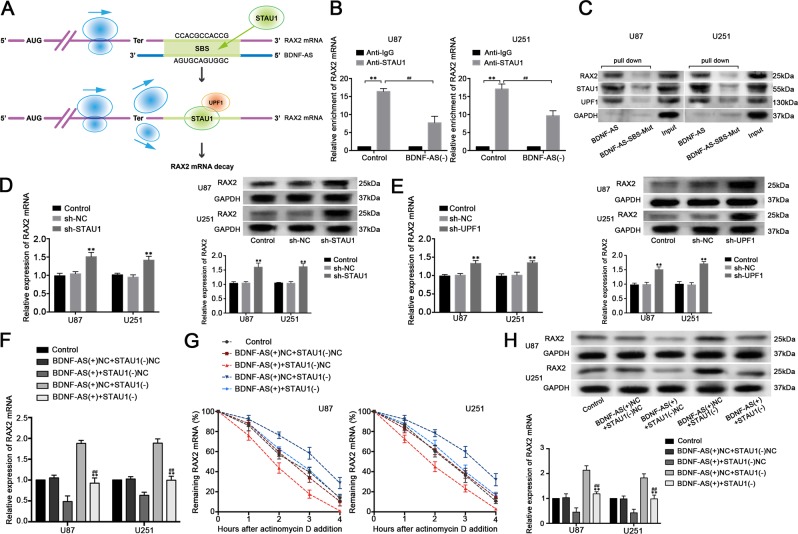
Several studies indicated that lncRNAs are functionally in regulating many fundamental cellular process including affecting mRNA expression23. We revealed that the overexpression of BDNF-AS decreased RAX2 expression and mRNA half-life, while the knockdown of BDNF-AS produced opposite effects (Fig. 5h–j). Moreover, we captured the nascent RAX2 mRNA and found no significant changes among each group (Fig. S2A). Recent studies reported that STAU1 could bind to the intermolecular base pairing in the 3'-UTR of the target mRNA and cause a decay at posttranscriptional level24. Interestingly, two Alu elements on BDNF-AS and 3'-UTR of RAX2 mRNA were found using bioinformatic software Repeatmasker (http://www.repeatmasker.org/cgi-bin/WEBRepeatMasker) respectively, and a putative imperfect base pairing between them was predicted by IntaRNA (http://rna.informatik.uni-freiburg.de/IntaRNA/Input.jsp, Fig. 2b). This made us speculated that whether BDNF-AS caused RAX2 mRNA degradation through SMD. RIP assays with IgG and STAU1 antibodies were used to investigate the association between BDNF-AS and RAX2 mRNA. As shown in Fig. S2C, BDNF-AS and RAX2 mRNA were enriched in anti-STAU1 groups compared with the anti-IgG control groups, proving exist of the binding between STAU1 with BDNF-AS and RAX2 mRNA, respectively. Moreover, knockdown of BDNF-AS decreased the association between STAU1 and RAX2 mRNA, suggesting that BDNF-AS is wanted for the combination between STAU1 with RAX2 mRNA (Fig. 6b). Cells were further transfected with biotinylated BDNF-AS and BDNF-AS with mutational SBS variants and extracted for pull-down assays. As expected, RAX2, STAU1, and UPF1 were enriched in BDNF-AS groups, while few were observed in BDNF-AS-SBS-Mut groups (Fig. 6c). This indicated that the relation among BDNF-AS, RAX2, and STAU1 is based on the predicted SBS sequence in BDNF-AS, and the SMD factor UPF1 is participated. Furthermore, as shown in Fig. 6d, e, loss of STAU1 and UPF1 increased RAX2 expression in both mRNA and protein levels. Consistently, knockdown of STAU1 restore the inhibiting effects of BDNF-AS on RAX2 mRNA expression and half-life time, including RAX2 protein expression (Fig. 6f–h). These results suggested that BDNF-AS affects RAX2 mRNA expression through SMD.
Fig. 6. BDNF-AS affects RAX2 mRNA expression and stability through SMD.
a Diagrams of the mechanism of SMD. A predicted imperfect base pairing (green) between Alu elements of RAX2 mRNA (purple) and BDNF-AS forms SBS. STAU1 (green ellipse) can recognize SBS and recruit UPF1 (orange ellipse) to trigger SMD and cause degradation of RAX2 mRNA. b Effects of BDNF-AS knockdown on the relative enrichment of RAX2 mRNA in IgG or STAU1 precipitates (n = 3, each group). **P < 0.01 vs. Anti-IgG control group. ##P < 0.01 vs. Anti-STAU1 control group. c U87 and U251 cells were transfected with biotinylated BDNF-AS (BDNF-AS) and BDNF-AS with mutational SBS (BDNF-AS-SBS-Mut), and western blot analysis following RNA pull-down assays performed using cellular exacts. d Effect of STAU1 and e UPF1 on the mRNA and protein expression of RAX2 in U87 and U251 cells (n = 3, each group). **P < 0.01 vs. sh-NC group. f Effects of the combination of overexpressed BDNF-AS and the decreased STAU1 on mRNA expression level, g stability and h protein expression level (n = 5, each group). **P < 0.01 vs. BDNF-AS(+)+STAU1(−)NC group. ##P < 0.01 vs. BDNF-AS(+)NC+STAU1(−) group.
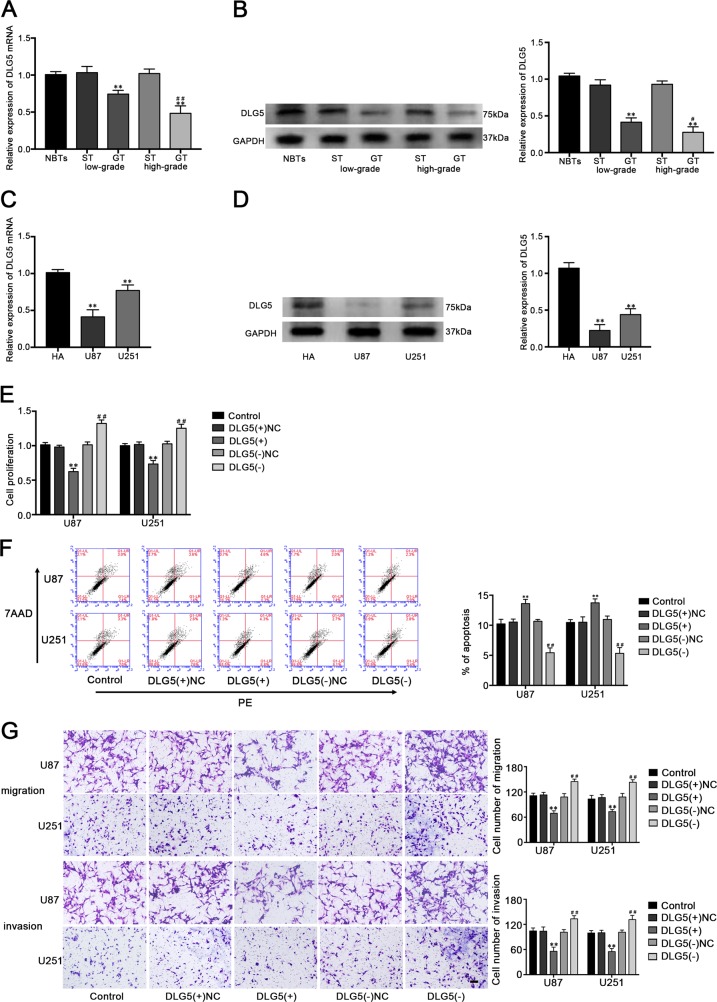
DLG5 was downregulated in glioma tissues and cell lines, and functioned as tumor suppressor in glioblastoma cells
QRT-PCR and western blot assays were used to measure the expression level of DLG5 and it was significantly decreased in GT and cell lines, and more downregulated in high-grade GT compared with low-grade GT (Fig. 7a–d). To further explore the impact of DLG5 on glioblastoma cell lines, a series of assays were carried out after DLG5 overexpression or knockdown. CCK-8 assay indicated that overexpression of DLG5 inhibited the proliferation of U87 and U251 cells, whereas DLG5 knockdown exerted the opposite effect (Fig. 7e). Cell apoptosis in the DLG5(+) group was promoted, and inhibited in the DLG5(−) group compared with their respective NC group (Fig. 7f). Transwell assays showed that the migration and invasion U87 and U251 cell numbers were apparently decreased in DLG5(+) group and increased in DLG5(−) group than in respective NC group (Fig. 7g). We proposed that DLG5 may exert tumor-suppressive function in glioblastoma cells.
Fig. 7. Expression of DLG5 in glioma tissues and cell lines and the effects of DLG5 on glioblastoma cells.
a The mRNA and b protein expression level of DLG5 in normal and glioma tissues (n = 4, each group). **P < 0.01 vs. ST group; ##P < 0.01 vs. low-grade GT group. c The mRNA and d protein expression level of DLG5 in HA, U87 and U251 (n = 3, each group). **P < 0.01 vs. HA group. e Effect of DLG5 on the cell proliferation, f cell apoptosis, g cell migration and invasion of glioblastoma cells (n = 5, each group). **P < 0.01 vs. DLG5(+)NC group; ##P < 0.01 vs. DLG5(−)NC group.
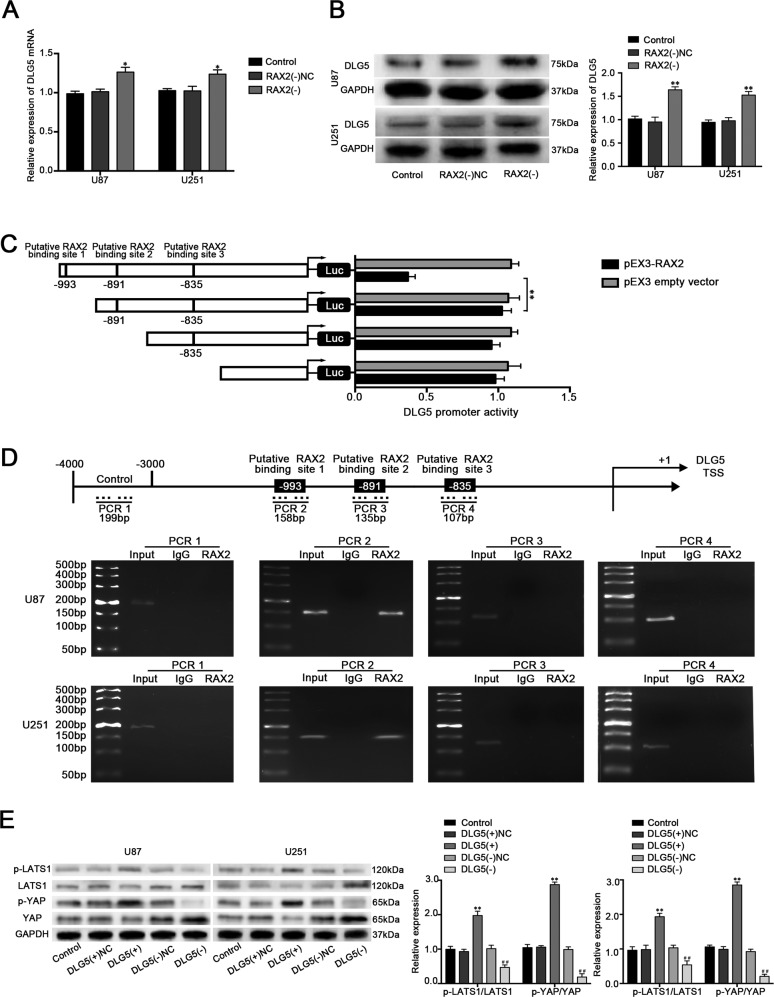
RAX2 suppressed DLG5 expression by directly binding to its promoter
Our previous results showed that RAX exerted an oncogenic factor in glioblastoma cells. Additionally, RAX2 as a transcription factor may transcriptionally regulated the downstream gene in controlling behaviors of glioblastoma cells. DLG5 promoter was predicted that it has binding sites of RAX2 by bioinformatic database JASPAR (Fig. S2D). As shown in Fig. 8a–b, DLG5 expression was increased in the RAX2(−) group compared with RAX2(−)NC group. By analyzing the DNA sequence of DLG5 promoter in the region from 1000 bp upstream to 100 bp downstream of transcription start site, three putative binding sites of RAX2 were found. As shown in Fig. 8c, deletion of the putative binding site 1 (−993 bp site region) significantly upregulated the promoter activities of DLG5. ChIP assay was performed subsequently and the results showed that there was an interaction of RAX2 with putative binding site 1 of DLG5 and there was no association of RAX2 with the control region (Fig. 8d). DLG5 was identified to have connection with the Hippo pathway25. In addition, loss of DLG5 promoted cancer malignancy by inactivating the Hippo pathway in breast cancer26. To elucidate whether DLG5 could suppress glioblastoma progression by affecting Hippo pathway, the activity of Hippo pathway was detected. As shown in Fig. 8e, p-LATS1 and p-YAP expression was increased in DLG5(+) group while reduced in DLG5(−) group. These results indicated that DLG5 could up-regulate the activity of the Hippo pathway in glioblastoma cells.
Fig. 8. RAX2 increased the expression of DLG5 by binding to the promoter of DLG5.
a The DLG5 mRNA expression after RAX2 knockdown was detected via qRT-PCR. b Western blot assay was used to detect the protein expression of DLG5 after RAX2 knockdown (n = 3, each group). **P < 0.01 vs. RAX2(−)NC group. c RAX2 affect on DLG5 promoter activity. d RAX2 bound to the promoter of DLG5 in U87 and U251 cells. Schematic representation of the human DLG5 promoter region 4000 bp upstream of the transcription start site (TSS), which was designated as +1. Putative RAX2-binding site was indicated. PCR was conducted with the resulting precipitated DNA. e Western blot assay of the p-LATS1/LATS and p-YAP/YAP expression regulated by DLG5. **P < 0.01 vs. DLG5(+)NC group; ##P < 0.01 vs. DLG5(−)NC group.
Discussion
In this study, we identified PABPC1 and BDNF-AS the significantly downregulated expression in GT and cells. Overexpression of PABPC1 and BDNF-AS inhibited the cell proliferation, migration and invasion, and promoted apoptosis of glioblastoma cells. PABPC1 interacted with BDNF-AS and increased the expression of BDNF-AS by stabilizing it. BDNF-AS bound to the 3'UTR of RAX2 mRNA and reduced RAX2 mRNA through SMD. RAX2 expression was elevated in glioma and knockdown RAX2 inhibited the malignant biological behaviors of glioblastoma cells. Moreover, RAX2 could increase DLG5 expression through the level of transcription. Overexpression of DLG5 inhibited glioblastoma tumor malignancy by activating the Hippo signaling pathway. We demonstrated for the first time that the PABPC1/BDNF-AS/RAX2/DLG5 pathway plays the vital role in regulating the malignant biological behaviors of glioblastoma cells.
PABPC1 is a classical complex cytoplasmic polyadenylate-binding protein involved in mRNA biogenesis, mRNA turnover and mRNA deadenylation. In addition, PABPC1 plays important roles in promoting protein translation initiation and ribosome recruitment27. In eukaryotic cytoplasm, PABPC1 stabilizes RNA by covering the 3' poly (A) tail of mRNA through RNA recognition motifs and interacting with eukaryotic initiation factor 4G28. PABPC1 is located at chromosome region 8q22.3, and the loss of heterozygosity at this region is associated with occurrence of oral cancer and breast cancer29,30. The abnormally expression of PABPC1 was found in many human malignancies, such as esophageal squamous cell carcinoma (ESCC), CC, colorectal cancer, and gastric cancer8,31,32. In this study, we found that PABPC1 was downregulated in human GT and cell lines, especially in high grade. Overexpression of PABPC1 inhibited cell proliferation, migration, and invasion, while promoted apoptosis in glioblastoma cells; knockdown PABPC1 produced the opposite effect. These results suggested that PABPC1 acted as a tumor suppressor in glioblastoma cells. Similarly, the decreased expression of PABPC1 was found in ESCC, which is correlated with poorer prognosis of patients and local invasiveness in tumor tissues7.
Increasing evidence has shown lncRNAs can act as either tumor oncogenes or suppressors by regulating tumor progression including glioma33,34. BDNF-AS is a naturally conserved lncRNA that inhibits the expression of BDNF by decreasing BDNF mRNA levels and functions opposite to BDNF35,36. BDNF has been reported to be upregulated and act as a predominant oncogenic factor in a variety human cancers37,38. Interestingly, BDNF-AS was demonstrated to be reversely associated with BDNF and downregulated expressed in RB, NSCLC, and CC12,39,40. Xiong et al. found that mature BDNF induces glioma cells in vitro41. However, little is known about the expression or functions of BDNF-AS in human glioma. We evaluated that BDNF-AS was significantly downregulated in GT and cell lines, and found that BDNF-AS overexpression resulted in the inhibition of malignant biological behaviors of glioblastoma cells; knockdown BDNF-AS had the opposite effect. Similarly, overexpression of BDNF-AS induced cell cycle arrest at G0/G1 phase and inhibited cell proliferation and migration in RB39. Moreover, BDNF-AS inhibited cell viability by forced overexpression in NSCLC and CC12,40.
Since PABPC1 and BDNF-AS were both downregulated and act as tumor suppressors in glioblastoma cells, we wondered whether PABPC1 would exert direct action on BDNF-AS. Based on predictions of bioinformatics database, we firstly demonstrated that PABPC1 could bind to BDNF-AS and increased the expression of BDNF-AS by stabilizing it at a posttranscriptional level. Moreover, we revealed that tumor-suppressive effects of PABPC1 were mediated by BDNF-AS in glioblastoma cell lines by a series of assays. PABPC1 is known to be involved in regulating stabilities of mRNA and also lncRNAs42. For example, PABPC1 stabilized and increased the half-life of lncRNA-PAN by interacting with lncRNA-PAN through forming a complex with the Kaposi’s sarcoma-associated herpesvirus ORF57 protein beforehand43. In gallbladder cancer (GBC), PABPC1 was demonstrated to regulate tumorigenesis in GBC cells by interacting and stabilizing lncRNA-PAGBC22.
Furthermore, the in vivo studies also demonstrated that PABPC1 overexpression, BDNF-AS overexpression, and the combination of the above observably suppressed tumor growth and prolonged the survival days in nude mice. Among them, the mice carrying the combined treatment produced the significantly smallest tumors and had the highest survival, suggesting that the combination of PABPC1 overexpression and BDNF-AS overexpression could be potentially applied in the treatment of glioma.
Rax is a novel homeobox gene which controls development of the eye and forebrain44. Rax encoded transcription factor RAX is essential for vertebrate eye development, which also has been shown to act with cone-rod homeobox to control gene expression in retinal photoreceptors during vertebrate eye development45. RAX2 is a member of RAX family, whose gene maps to chromosome 19p13.3 and is composed of three exons. The RAX2 gene (previously known as Q50-type retinal homeobox) has previously been related to cone-rod dystrophy46. Yang et al. found that the frameshift heterozygous mutation in RAX2 was related to mixed cone and rod dysfunction47. But the role of RAX2 in cancer is unclear. In the present study, we first characterized RAX2 as an oncogenic factor in glioblastoma cells. The expression level of RAX2 was upregulated in GT and increased as the pathological grade increased. Moreover, RAX2 knockdown inhibited the proliferation, migration and invasion, and promoted apoptosis in glioblastoma cells.
There are approximately more than one million copies of Alu elements in the human genome, which are mostly situated in lncRNAs and the 3'-UTR of mRNAs48. Alu elements have been reported to be associated with molecular mechanisms, such as silencing gene expression, gene conversion, and alternative splicing49. Recently, Alu elements were found to have functions in the transactivation of SMD. SMD is an mRNA decay process of mammalian cells which is mediated by STAU1 binding to a SBS within the 3'-UTR of target mRNAs50. STAU1 was first identified in drosophila which described to be ubiquitously expressed and involved in mRNA stability and translation in mammalian cells51. The SBSs can be formed by intramolecular base pairing within the target mRNA 3'-UTR or imperfect base pairing between an Alu element in the 3'-UTR of target mRNAs and a complementary Alu element in lncRNAs called ½-sbsRNAs52. The SBS-bound STAU1 recruits UPF1 and this event triggers rapid mRNAs degradation53. As a posttranscriptional regulatory pathway, SMD has modest effects on mRNA abundance and the diversity of cell-types. For instance, SMD has effects on the expression of target mRNAs, such as PAX3, ARF1, KLF2, and SERPINE1, which can regulate cellular homeostasis like myogenesis, adipogenesis and cutaneous wound healing in mammalian24,54–56. We found that an Alu element sequence in the 3'-UTR of RAX2 and another Alu sequence in BDNF-AS were found and predicted to have binding site with each other. With a variety of assays, we confirmed that BDNF-AS could bind to STAU1 and reduce the expression and stability of RAX2 mRNA via SMD, thereby further inhibited the malignant behaviors of glioblastoma cells. In a similar way, lncRNA-TINCR can degrade KLF2 mRNA expression through SMD, and promote cell cycle progression, and tumorigenicity subsequently in gastric cancer cells16.
DLG5 belongs to the MAGUK family, participates in epithelial cell polarity maintenance and cancer development57. Our results proved that DLG5 had a low expression level in glioma tissues and cells. Overexpression of DLG5 inhibited glioma cells malignant biological behaviors of glioblastoma cells. Similar results were reported in many human cancers, such as bladder cancer, prostate cancer, breast cancer, and hepatocellular carcinoma19,20,58. We also found that knockdown of RAX2 could increase DLG5 expression. Moreover, DLG5 promoter was predicted that it has RAX2-binding sites. Subsequently, we confirmed that RAX2 could bind to the promoter region of DLG5 and downregulate the promoter activities. The results indicated that DLG5 is involved in RAX2 regulation in glioblastoma cells.
The Hippo pathway is a evolutionarily conserved regulator of diverse biological processes including cell proliferation, tissue growth, and cancer development59. In mammalian, the core Hippo pathway consists of a kinase cascade and associated proteins including macrophage stimulating 1/2 kinase, Salvador homolog 1, and large tumor suppressor kinase 1/2 (LATS1/2)60. Yes-associated protein (YAP) is the major downstream effector of the Hippo pathway, which is identified as an oncogene. Activated YAP can translocate into the nucleus and bind to the TEAD transcription factor family to regulate cell proliferation, migration, epithelial–mesenchymal transition, and organ growth. Activation of the Hippo pathway results in the cytoplasmic retention and degradation of YAP mediated by LATS1/2 direct phosphorylation on YAP Ser12761. It had been demonstrated that inhibition of Hippo/YAP signaling promotes cell proliferation motility and invasion in glioma62, and the downstream effectors of Hippo pathway could be biomarkers of the prognosis of glioma patients63. Moreover, Liu et al. suggested that loss of DLG5 expression inhibited the Hippo pathway thus promoted breast cancer malignancy26. In this study, we first showed that overexpression of DLG5 activated the Hippo pathway by increasing the phosphorylation of YAP and suppressed the malignant behaviors of glioblastoma cells consequently.
In summary, we first found that PABPC1, BDNF-AS, and DLG5 were downregulated in human glioma tumors and cells, while RAX2 was upregulated conversely. Overexpression of PABPC1, BDNF-AS, and DLG5 along with depletion of RAX2 inhibited malignant biological behaviors of glioblastoma cells. PABPC1 could bind to and stabilize BDNF-AS, which promoted RAX2 mRNA degradation through SMD. Knockdown of RAX2 increased DLG5 expression thereby inhibited the malignant biological behaviors of glioblastoma cells by activating the Hippo pathway. Our findings first revealed that PABPC1/BDNF-AS/RAX2/DLG5 module plays an important role in glioblastoma cells, provided insight into potential therapeutic targets to treat glioma.
Supplementary information
Acknowledgements
We thank the Department of Neurobiology of China Medical University for providing equipment and assistance to support this research, You Li for illustrating the diagrams and all individuals who participates in this study. This work is supported by grants from the Natural Science Foundation of China (81872503 and 81872073), Liaoning Science and Technology Plan Project (No.2017225020, 2015225007), Project of Key Laboratory of Neuro-oncology in Liaoning Province (112–2400017005), special developmental project guided by central government of Liaoning Province (No. 2017011553–301).
Data availability
All data generated in this study are included within the article and its supplementary materials.
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All experiments of this study were approved by the Ethics Committee of Shengjing Hospital of China Medical University.
Informed consent
Each patient was provided informed consent.
Footnotes
Edited by N. Barlev
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information accompanies this paper at (10.1038/s41419-020-2267-9).
References
- 1.Agnihotri S, Aldape KD, Zadeh G. Isocitrate dehydrogenase status and molecular subclasses of glioma and glioblastoma. Neurosurg. Focus. 2014;37:E13. doi: 10.3171/2014.9.FOCUS14505. [DOI] [PubMed] [Google Scholar]
- 2.Alifieris C, Trafalis DT. Glioblastoma multiforme: pathogenesis and treatment. Pharmacol. Therap. 2015;152:63–82. doi: 10.1016/j.pharmthera.2015.05.005. [DOI] [PubMed] [Google Scholar]
- 3.Koster T, Staiger D. RNA-binding protein immunoprecipitation from whole-cell extracts. Methods Mol. Biol. 2014;1062:679–695. doi: 10.1007/978-1-62703-580-4_35. [DOI] [PubMed] [Google Scholar]
- 4.Chen X, et al. The histone deacetylase SIRT6 suppresses the expression of the RNA-binding protein PCBP2 in glioma. Biochem. Biophys. Res. Commun. 2014;446:364–369. doi: 10.1016/j.bbrc.2014.02.116. [DOI] [PubMed] [Google Scholar]
- 5.Uren PJ, et al. RNA-binding protein musashi1 is a central regulator of adhesion pathways in glioblastoma. Mol. Cell. Biol. 2015;35:2965–2978. doi: 10.1128/MCB.00410-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stupfler B, Birck C, Seraphin B, Mauxion F. BTG2 bridges PABPC1 RNA-binding domains and CAF1 deadenylase to control cell proliferation. Nat. Commun. 2016;7:10811. doi: 10.1038/ncomms10811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Takashima N, et al. Expression and prognostic roles of PABPC1 in esophageal cancer: correlation with tumor progression and postoperative survival. Oncol. Rep. 2006;15:667–671. [PubMed] [Google Scholar]
- 8.Zhu J, Ding H, Wang X, Lu Q. PABPC1 exerts carcinogenesis in gastric carcinoma by targeting miR-34c. Int. J. Clin. Exp. Pathol. 2015;8:3794–3802. [PMC free article] [PubMed] [Google Scholar]
- 9.Cheng WS, et al. Both genes and lncRNAs can be used as biomarkers of prostate cancer by using high throughput sequencing data. Eur. Rev. Med. Pharmacol. Sci. 2014;18:3504–3510. [PubMed] [Google Scholar]
- 10.Yarmishyn AA, et al. HOXD-AS1 is a novel lncRNA encoded in HOXD cluster and a marker of neuroblastoma progression revealed via integrative analysis of noncoding transcriptome. BMC Genom. 2014;15(Suppl. 9):S7. doi: 10.1186/1471-2164-15-S9-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li, Y., Xu, F., Xiao, H. & Han, F. Long noncoding RNA BDNF-AS inversely regulated BDNF and modulated high-glucose induced apoptosis in human retinal pigment epithelial cells. J. Cell Biochem. 119, 817–823 10.1002/jcb.26245 (2018). [DOI] [PubMed]
- 12.Zhang, H., Liu, C., Yan, T., Wang, J. & Liang, W. Long noncoding RNA BDNF-AS is downregulated in cervical cancer and has anti-cancer functions by negatively associating with BDNF. Arch. Biochem. Biophys.10.1016/j.abb.2018.03.023 (2018). [DOI] [PubMed]
- 13.Park E, Maquat LE. Staufen-mediated mRNA decay. Wiley Interdiscip. Rev. RNA. 2013;4:423–435. doi: 10.1002/wrna.1168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kretz M, et al. Control of somatic tissue differentiation by the long non-coding RNA TINCR. Nature. 2013;493:231–235. doi: 10.1038/nature11661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cho H, Han S, Park OH, Kim YK. SMG1 regulates adipogenesis via targeting of staufen1-mediated mRNA decay. Biochimica. Biophys. Acta. 2013;1829:1276–1287. doi: 10.1016/j.bbagrm.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 16.Xu TP, et al. SP1-induced upregulation of the long noncoding RNA TINCR regulates cell proliferation and apoptosis by affecting KLF2 mRNA stability in gastric cancer. Oncogene. 2015;34:5648–5661. doi: 10.1038/onc.2015.18. [DOI] [PubMed] [Google Scholar]
- 17.Bailey TJ, et al. Regulation of vertebrate eye development by Rx genes. Int. J. Dev. Biol. 2004;48:761–770. doi: 10.1387/ijdb.041878tb. [DOI] [PubMed] [Google Scholar]
- 18.Liu J, Li J, Ren Y, Liu P. DLG5 in cell polarity maintenance and cancer development. Int. J. Biol. Sci. 2014;10:543–549. doi: 10.7150/ijbs.8888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou Z, et al. Methylation-mediated silencing of Dlg5 facilitates bladder cancer metastasis. Exp. Cell Res. 2015;331:399–407. doi: 10.1016/j.yexcr.2014.11.015. [DOI] [PubMed] [Google Scholar]
- 20.Ke Y, et al. Discs large homolog 5 decreases formation and function of invadopodia in human hepatocellular carcinoma via Girdin and Tks5. Int. J. Cancer. 2017;141:364–376. doi: 10.1002/ijc.30730. [DOI] [PubMed] [Google Scholar]
- 21.Su R, et al. Knockdown of SOX2OT inhibits the malignant biological behaviors of glioblastoma stem cells via up-regulating the expression of miR-194-5p and miR-122. Mol. Cancer. 2017;16:171. doi: 10.1186/s12943-017-0737-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu, X. S. et al. LncRNA-PAGBC acts as a microRNA sponge and promotes gallbladder tumorigenesis. EMBO Rep. 18, 1837–1853 10.15252/embr.201744147 (2017). [DOI] [PMC free article] [PubMed]
- 23.Kugel JF, Goodrich JA. The regulation of mammalian mRNA transcription by lncRNAs: recent discoveries and current concepts. Epigenomics. 2013;5:95–102. doi: 10.2217/epi.12.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cho H, et al. Staufen1-mediated mRNA decay functions in adipogenesis. Mol. Cell. 2012;46:495–506. doi: 10.1016/j.molcel.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 25.Kwan J, et al. DLG5 connects cell polarity and Hippo signaling protein networks by linking PAR-1 with MST1/2. Genes Dev. 2016;30:2696–2709. doi: 10.1101/gad.284539.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu J, et al. Loss of DLG5 promotes breast cancer malignancy by inhibiting the Hippo signaling pathway. Sci. Rep. 2017;7:42125. doi: 10.1038/srep42125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang KL, Chadee AB, Chen CY, Zhang Y, Shyu AB. Phosphorylation at intrinsically disordered regions of PAM2 motif-containing proteins modulates their interactions with PABPC1 and influences mRNA fate. RNA. 2013;19:295–305. doi: 10.1261/rna.037317.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kini HK, Silverman IM, Ji X, Gregory BD, Liebhaber SA. Cytoplasmic poly(A) binding protein-1 binds to genomically encoded sequences within mammalian mRNAs. RNA. 2016;22:61–74. doi: 10.1261/rna.053447.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Garnis C, Coe BP, Zhang L, Rosin MP, Lam WL. Overexpression of LRP12, a gene contained within an 8q22 amplicon identified by high-resolution array CGH analysis of oral squamous cell carcinomas. Oncogene. 2004;23:2582–2586. doi: 10.1038/sj.onc.1207367. [DOI] [PubMed] [Google Scholar]
- 30.Miyazaki S, et al. The chromosome location of the human homolog of the mouse mammary tumor-associated gene INT6 and its status in human breast carcinomas. Genomics. 1997;46:155–158. doi: 10.1006/geno.1997.4996. [DOI] [PubMed] [Google Scholar]
- 31.Katzenellenbogen RA, Vliet-Gregg P, Xu M, Galloway DA. Cytoplasmic poly(A) binding proteins regulate telomerase activity and cell growth in human papillomavirus type 16 E6-expressing keratinocytes. J. Virol. 2010;84:12934–12944. doi: 10.1128/JVI.01377-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu D, et al. Cytoplasmic poly(A) binding protein 4 is highly expressed in human colorectal cancer and correlates with better prognosis. J. Genet. Genom. 2012;39:369–374. doi: 10.1016/j.jgg.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 33.Zhao X, et al. Gas5 exerts tumor-suppressive functions in human glioma cells by targeting miR-222. Mol. Ther. 2015;23:1899–1911. doi: 10.1038/mt.2015.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yao Y, et al. Knockdown of long non-coding RNA XIST exerts tumor-suppressive functions in human glioblastoma stem cells by up-regulating miR-152. Cancer Lett. 2015;359:75–86. doi: 10.1016/j.canlet.2014.12.051. [DOI] [PubMed] [Google Scholar]
- 35.Porritt MJ, Batchelor PE, Howells DW. Inhibiting BDNF expression by antisense oligonucleotide infusion causes loss of nigral dopaminergic neurons. Exp. Neurol. 2005;192:226–234. doi: 10.1016/j.expneurol.2004.11.030. [DOI] [PubMed] [Google Scholar]
- 36.Zheng X, et al. Long noncoding RNA BDNF-AS regulates ketamine-induced neurotoxicity in neural stem cell derived neurons. Biomed. Pharmacother. 2016;82:722–728. doi: 10.1016/j.biopha.2016.05.050. [DOI] [PubMed] [Google Scholar]
- 37.de Farias CB, et al. BDNF/TrkB signaling protects HT-29 human colon cancer cells from EGFR inhibition. Biochem. Biophys. Res. Commun. 2012;425:328–332. doi: 10.1016/j.bbrc.2012.07.091. [DOI] [PubMed] [Google Scholar]
- 38.Lai PC, Chiu TH, Huang YT. Overexpression of BDNF and TrkB in human bladder cancer specimens. Oncol. Rep. 2010;24:1265–1270. doi: 10.3892/or_00000981. [DOI] [PubMed] [Google Scholar]
- 39.Shang W, Yang Y, Zhang J, Wu Q. Long noncoding RNA BDNF-AS is a potential biomarker and regulates cancer development in human retinoblastoma. Biochem. Biophys. Res. Commun. 2018;497:1142–1148. doi: 10.1016/j.bbrc.2017.01.134. [DOI] [PubMed] [Google Scholar]
- 40.Shen M, et al. Long noncoding nature brain-derived neurotrophic factor antisense is associated with poor prognosis and functional regulation in non-small cell lung caner. Tumour Biol. 2017;39:1010428317695948. doi: 10.1177/1010428317695948. [DOI] [PubMed] [Google Scholar]
- 41.Xiong J, et al. Mature brain-derived neurotrophic factor and its receptor TrkB are upregulated in human glioma tissues. Oncol. Lett. 2015;10:223–227. doi: 10.3892/ol.2015.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fatscher T, Boehm V, Weiche B, Gehring NH. The interaction of cytoplasmic poly(A)-binding protein with eukaryotic initiation factor 4G suppresses nonsense-mediated mRNA decay. RNA. 2014;20:1579–1592. doi: 10.1261/rna.044933.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Massimelli MJ, Majerciak V, Kruhlak M, Zheng ZM. Interplay between polyadenylate-binding protein 1 and Kaposi’s sarcoma-associated herpesvirus ORF57 in accumulation of polyadenylated nuclear RNA, a viral long noncoding RNA. J. Virol. 2013;87:243–256. doi: 10.1128/JVI.01693-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rohde, K., Bering, T., Furukawa, T. & Rath, M. F. A modulatory role of the Rax homeobox gene in mature pineal gland function: investigating the photoneuroendocrine circadian system of a Rax conditional knockout mouse. J. Neurochem. 143, 100–111 10.1111/jnc.14120 (2017). [DOI] [PubMed]
- 45.Rohde K, Klein DC, Moller M, Rath MF. Rax: developmental and daily expression patterns in the rat pineal gland and retina. J. Neurochem. 2011;118:999–1007. doi: 10.1111/j.1471-4159.2011.07385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang QL, et al. QRX, a novel homeobox gene, modulates photoreceptor gene expression. Hum. Mol. Genet. 2004;13:1025–1040. doi: 10.1093/hmg/ddh117. [DOI] [PubMed] [Google Scholar]
- 47.Yang P, Chiang PW, Weleber RG, Pennesi ME. Autosomal dominant retinal dystrophy with electronegative waveform associated with a novel RAX2 mutation. JAMA Ophthalmol. 2015;133:653–661. doi: 10.1001/jamaophthalmol.2015.0357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim EZ, Wespiser AR, Caffrey DR. The domain structure and distribution of Alu elements in long noncoding RNAs and mRNAs. RNA. 2016;22:254–264. doi: 10.1261/rna.048280.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen LL, DeCerbo JN, Carmichael GG. Alu element-mediated gene silencing. EMBO J. 2008;27:1694–1705. doi: 10.1038/emboj.2008.94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim MY, et al. Staufen1-mediated mRNA decay induces Requiem mRNA decay through binding of Staufen1 to the Requiem 3'UTR. Nucleic Acids Res. 2014;42:6999–7011. doi: 10.1093/nar/gku388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dugre-Brisson S, et al. Interaction of Staufen1 with the 5' end of mRNA facilitates translation of these RNAs. Nucleic Acids Res. 2005;33:4797–4812. doi: 10.1093/nar/gki794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kretz M. TINCR, staufen1, and cellular differentiation. RNA Biol. 2013;10:1597–1601. doi: 10.4161/rna.26249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gong C, Maquat L. E. lncRNAs transactivate STAU1-mediated mRNA decay by duplexing with 3' UTRs via Alu elements. Nature. 2011;470:284–288. doi: 10.1038/nature09701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim YK, Furic L, Desgroseillers L, Maquat LE. Mammalian Staufen1 recruits Upf1 to specific mRNA 3'UTRs so as to elicit mRNA decay. Cell. 2005;120:195–208. doi: 10.1016/j.cell.2004.11.050. [DOI] [PubMed] [Google Scholar]
- 55.Kim YK, et al. Staufen1 regulates diverse classes of mammalian transcripts. EMBO J. 2007;26:2670–2681. doi: 10.1038/sj.emboj.7601712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yamaguchi Y, Oohinata R, Naiki T, Irie K. Stau1 negatively regulates myogenic differentiation in C2C12 cells. Genes Cells. 2008;13:583–592. doi: 10.1111/j.1365-2443.2008.01189.x. [DOI] [PubMed] [Google Scholar]
- 57.Nechiporuk T, Klezovitch O, Nguyen L, Vasioukhin V. Dlg5 maintains apical aPKC and regulates progenitor differentiation during lung morphogenesis. Dev. Biol. 2013;377:375–384. doi: 10.1016/j.ydbio.2013.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tomiyama L, Sezaki T, Matsuo M, Ueda K, Kioka N. Loss of Dlg5 expression promotes the migration and invasion of prostate cancer cells via Girdin phosphorylation. Oncogene. 2015;34:1141–1149. doi: 10.1038/onc.2014.31. [DOI] [PubMed] [Google Scholar]
- 59.Chen L, et al. USF1-induced upregulation of LINC01048 promotes cell proliferation and apoptosis in cutaneous squamous cell carcinoma by binding to TAF15 to transcriptionally activate YAP1. Cell Death Dis. 2019;10:296. doi: 10.1038/s41419-019-1516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yu FX, Zhao B, Guan KL. Hippo pathway in organ size control, tissue homeostasis, and cancer. Cell. 2015;163:811–828. doi: 10.1016/j.cell.2015.10.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.He C, et al. The Hippo/YAP pathway interacts with EGFR signaling and HPV oncoproteins to regulate cervical cancer progression. EMBO Mol. Med. 2015;7:1426–1449. doi: 10.15252/emmm.201404976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, H. et al. Expression and significance of Hippo/YAP signaling in glioma progression. Tumour Biol.10.1007/s13277-016-5318-1 (2016). [DOI] [PubMed]
- 63.Guichet PO, et al. Fatal correlation between YAP1 expression and glioma aggressiveness: clinical and molecular evidence. J. Pathol. 2018;246:205–216. doi: 10.1002/path.5133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are included within the article and its supplementary materials.