Abstract
The study of hereditary cancer, which accounts for ~10% of cancer cases worldwide is an important subfield of oncology. Our understanding of hereditary cancers has greatly advanced with recent advances in sequencing technology, but as with any genetic trait, gene frequencies of cancer-associated mutations vary across populations, and most studies that have located hereditary cancer genes have been conducted on European or Asian populations. There is an urgent need to trace hereditary cancer genes across the Arab world. Hereditary disease is particularly prevalent among members of consanguineous populations, and consanguineous marriages are particularly common in the Arab world. There are also cultural and educational idiosyncrasies that differentiate Arab populations from other more thoroughly studied groups with respect to cancer awareness and treatment. Therefore, a review of the literature on hereditary cancers in this understudied population was undertaken. We report that BRCA mutations are not as prevalent among Arab breast cancer patients as they are among other ethnic groups, and therefore, other genes may play a more important role. A wide variety of germline inherited mutations that are associated with cancer are discussed, with particular attention to breast, ovarian, colorectal, prostate, and brain cancers. Finally, we describe the state of the profession of familial cancer genetic counselling in the Arab world, and the clinics and societies dedicated to its advances. We describe the complexities of genetic counselling that are specific to the Arab world. Understanding hereditary cancer is heavily dependent on understanding population-specific variations in cancer-associated gene frequencies.
Subject terms: Genetics, Cancer
Introduction
Cancer remains a leading cause of morbidity and mortality across the globe,1,2 with increases in mortality of approximately 25.0% since the 1990s and projections of ≥23 million cases annually by 2030.3,4 Hereditary causes account for ~10% of cancer cases, and an estimated 20% of cancer patients have a positive family history of cancer.5–7
Hereditary cancer syndrome is defined as an elevated risk of cancer that runs in the family. The risk originates from heritable mutations in specific genes.8,9 The type of cancer is dependent on the mutated gene. Hereditary breast and ovarian cancers originate from BRCA1 and/or BRCA2 gene mutations that significantly increase the likelihood of developing breast, ovarian, prostate and other types of cancer.1,10 Patients with Li-Fraumeni syndrome, characterised by TP53 mutations, have a heightened risk of cancer before age 30, and are almost guaranteed to suffer from cancer by the age of 60.8,11–13 Carriers of cancer syndrome associated genes also have a higher risk of multiple malignancies and rare cancers, and are more likely to develop cancer at a younger age.
Advances in sequencing technologies, particularly, high throughput sequencing have permitted the discovery of novel genes responsible for cancer heritability, facilitating efficient genetic screening.14–16 The major genetic changes in cancer include single nucleotide variants (SNVs); duplications, insertions, or deletions; exon and gene copy number changes; and structural variants (SVs).17 The molecular profiling of heritable cancer genes ranges from simple assessments of known hotspot mutations in single genes, to more complex tests that simultaneously detect all gene alterations using allele-specific PCR, Sanger sequencing, multiplex ligation-dependent probe amplification (MLPA), pyrosequencing or mass spectrometry (MS).17,18 Gene copy numbers and SVs can also be assessed through fluorescence in situ hybridisation (FISH). Next generation sequencing (NGS) technologies have revolutionised molecular profiling permitting whole exome sequencing (WES) that examines all protein-coding regions and whole genome sequencing (WGS) that profiles protein-coding and non-coding regions. Example NGS technologies include Illumina MiSeq and HiSeq and the Life Technologies Ion Torrent personal genome machine.17–19 These technologies can permit the identification of a family history of cancer and help identify those at-risk and likely to benefit from enhanced surveillance and early detection. Patients diagnosed with cancer syndromes do not necessarily develop cancer, but awareness of their status may enable early detection to prevent mortality.20,21 Accordingly, increased public awareness that cancer can be heritable, and that the heritable risk can be evaluated has increased as has the demand for genetic counselling and screening.14,22–24
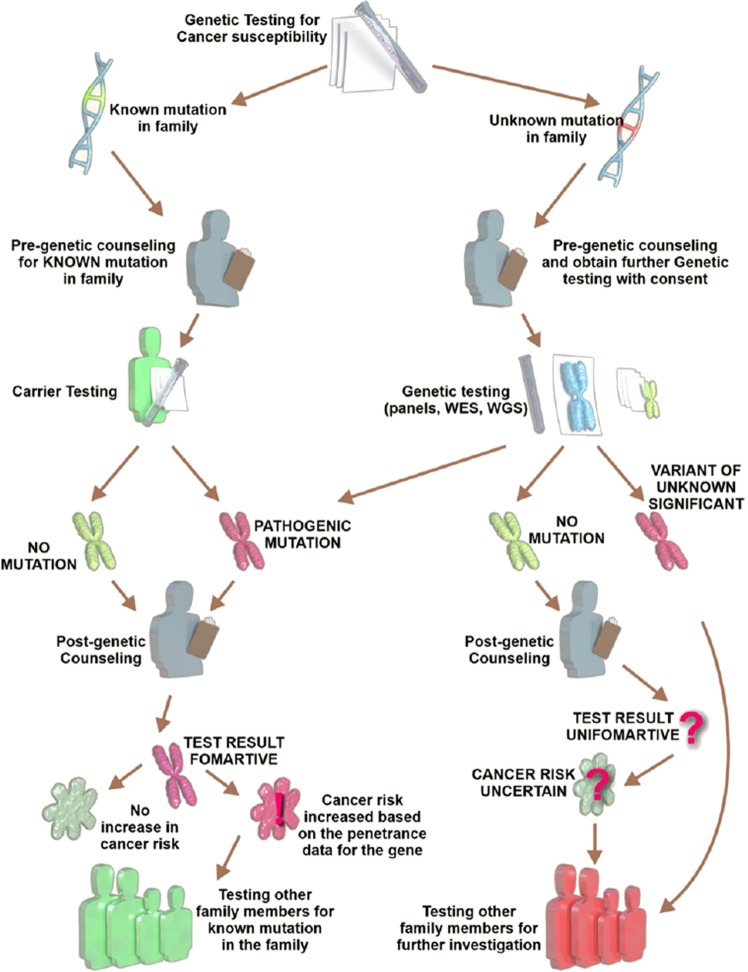
The incidence and prevalence of hereditary cancer amongst different ethnic populations is often distinct. Cancer is a major problem in the Arab world4,25–31 which is delimited by Lebanon and Syria to the north, Morocco to the west, south to Yemen, and Iraq in the east, accounting for >300 million people. The incidence of cancer in Arab countries has increased over the last 10 years, primarily due to lifestyle changes and obesity, as traditional foods are replaced with Western-diets.25 For example, SA, Qatar, Kuwait, UAE, Bahrain, and Saudi Arabia are amongst the top ten countries for obesity prevalence, which for breast cancer alone has increased the incidence rates by ~2% in adult males and ~7% in adult females.32 Barriers to cancer screening in addition to a lack of cancer education remain problems in the region.31 From the perspective of hereditary cancer, genetic disorders occur at a high frequency in several Arab communities due to high rates of inbreeding, with 25–60% of all marriages being consanguineous, with common first cousin marriages. Problems are compounded by the lack of public health measures directed towards the prevention of congenital and genetic disorders, due to cultural, legal, and religious restrictions.33 Overall, further research on familial cancer is needed in the Arab world, particularly large genetic screening programmes and improved genetic counselling.34 The Saudi Human Genome Project (SGHP) is an initiative that aims to describe the genetic distribution of cancer within Saudi Arabia, and this has been joined by programmes from Qatar, Egypt and the United Arab Emirates (Fig. 1). However, knowledge of targetable familial cancer genes in the Arab region remains sparse.
Fig. 1.
Genetic counselling process for cancer susceptibility.
In this review, we discuss the prevalence and roles of familial and sporadic genetic mutations and their ethnic-specific differences, with a focus on the Arab world. Knowledge of this genetic landscape will be important for determining the prevalence of regional familial genetic predisposition to cancer.
Epidemiology of familial cancer syndrome in the Arab world
Genetic cancer epidemiology represents the study of hereditary factors that are responsible for cancer initiation, metastasis, and prognosis.9,35 Knowledge of the regional genetic epidemiology of cancer can facilitate the development of suitable therapeutic treatments.9 Unfortunately, such studies in the Arab world have been lacking due to limited recognition that cancer health outcomes are influenced by genetic as well as social, economic and environmental, behaviours. However, progress has been made as using the American College of Medical Genetics and Genomics (ACMG) guidelines, Jastanahia et al. performed a multi-centre cross-sectional study on 1858 children with cancer in Saudi Arabia and found that 704 (40.4%) out of 1742 patients fulfilled criteria for hereditary cancer syndrome.36 Of these patients, consanguinity was reported in 629 (38%) of cases, with 50 (2.9%) first-degree, 535 (30.7%) second-degree, and 272 (15.6%) third-degree relatives afflicted. The data obtained in this study suggested that as many as 4/10 children with cancer in Saudi Arabia are afflicted with the hereditary form of the disease, due largely to consanguinity. This highlights the need for further genetic epidemiology testing across the Arab world.
Breast cancer is the most common malignancy amongst women in Arab countries, with 50% of cases presenting before the age of 50 years.37,38 From 2009 to 2012, El Saghir et al. assessed 250 Lebanese women with breast cancer, considered to be at high risk of carrying mutations in the BRCA1 or BRCA2 tumour suppressor genes, due to presentation of the disease at a young age and/or a positive family history of breast or ovarian cancer.10 The results showed that 14 of 250 patients (5.6%) carried deleterious BRCA mutations (7 BRCA1, 7 BRCA2) and 31 (12.4%) had variants of uncertain significance. These results were somewhat surprising and suggest that the prevalence of BRCA mutations is lower than predicted in Arab women.10 The notion that BRCA mutations alone cause the high incidence of breast cancer in young Arab women was not supported. Given that the rates of developing breast cancer vary amongst other racial and ethnic groups, and carriers vary by region, there is an urgent need for ethnic-specific genetic programmes.
More recently, Younes et al. performed a systematic review to estimate the genetic epidemiology of ovarian cancer in 22 Arab countries. These ranged from a low incidence of 0.9/100,000/year in Saudi Arabia to a high incidence of 8.0/100,000/year in Sudan.9 The total number of ovarian cancer patients identified were 802, of which 53 harboured mutations in BRCA1/2 genes. BRCA1 mutations were more frequent than BRCA2 mutations and eight of the identified mutations were unique to Arab populations. This highlighted the importance of BRCA1/2 mutations for the high prevalence of ovarian cancer observed across the Arab world.9 This was further supported by Siraj et al. who characterised the spectrum of BRCA1/2 mutations regarding prevalence and founder effects in Arab regions to advance genetic counselling.39 Despite clear progress in our understanding of ethnic-specific hereditary cancer, problems still remain that are specific to the Arab region. A major barrier to genetic epidemiology is the avoidance of cancer screening and lack of knowledge of the importance of cancer, its risk factors, and the benefits that could be gained through screening. Al Abdouli et al. examined the public’s understanding of colorectal cancer in the United Arab Emirates (UAE) in which when surveyed, 64% believed that colorectal cancer is uncommon, and 67% had no knowledge of colorectal cancer screening tests.40 This highlights the need for changes in cancer awareness, attitudes and practices across the UAE. Racial stigmas also present a barrier to genetic screening programmes in the Arab world. The diagnosis of breast cancer in the middle-east often occurs at a later stage and in a higher proportion of women aged 30–40. It has been reported that 90.7% of women are aware of breast cancer, but only 7.6% have basic knowledge of breast cancer screening activities. The major reason for these low numbers is that Arab society is fundamentally conservative, and Arab women rely on mothers and sisters for disease support and protection. It was thus suggested that breast cancer detection efforts in Arab regions should include men so that they can encourage their female family members to participate in breast cancer screening activities. It is likely that national genetic screening campaigns will have similar problems in the region. Improved education and the communication of the benefits of early cancer detection should be pursued to improve familial genetic screening programmes across the Arab world.
Cancer from the genetic perspective
Using advanced genetic methods, researchers have been able to determine the potency of gene expression and defective proteins, and the detection of novel cancer biomarkers in afflicted families. In addition, various studies have explored the epigenetic mechanisms and their relationship to the development, and progression of cancer. Whilst many aspects of epigenetic regulation remain unknown, identification of pivotal genes allows a comprehensive map for further efforts to reduce heritable cancer in future generations. This is particularly important for genetic counselling and the inheritance of genes that predispose individuals to cancer.41,42 Herein, from the brain to the colon, we review our genetic knowledge of specific cancers, and ethnic genetic predispositions.
Paediatric cancer
Approximately 10% of all new cancer diagnoses are due to inherited genetic traits.43 Individuals with specific germline mutations possess a higher likelihood of developing life-threatening cancers, typically at a young age. The genetic basis of numerous childhood, and adolescent cancers, and cancers in young adults have been reported and identified. The detection of mutations is critical for the lifelong management of these patients and can dictate genetic counselling, surveillance, and early therapeutic interventions.44–50 Although the prevalence of childhood cancer is rare, 100,000 children younger than 15 years of age die from cancer each year, and whilst a further understanding of the scale of the problem is required amongst Arab countries, the proportions are thought to be higher in developing countries.
Germline/inherited mutations can be either dominant or recessive. Similarly, the disease profiles of inherited cancer syndrome drastically vary, leading to both early-onset and late disease-onset, and marked variations in cancer presentation. The majority of cancer predisposition genes (up to 90%) are through the inhibition of tumour suppressors. Approximately 10% of cancer predisposition genes (including ALK, KIT, and MET) are gain-of-function mutations. Notable examples of known cancer syndromes and the genes involved include X-linked recessive Simpson Golabi Behmel syndrome (caused by GPC3 mutations), X-linked lymphoproliferative disease (caused by SH2D1A abnormalities), autosomal recessive ataxia telangiectasia (ATM mutations), Bloom syndrome (BLM mutations), Fanconi anemiam (FANC family), MUTYH-associated polyposis (MUTYH), Nijmegen breakage syndrome (NBN), Rothmund-Thomson syndrome (RECQL4) and Werners syndrome (WRN) (reviewed in the ref. 43). Notable autosomal dominant syndromes include Hereditary breast-ovarian cancer syndrome (BRCA1/2 mutations), hereditary diffuse gastric cancer (CDH1 mutations), Howel-Evans syndrome (RHBDF2), Li-Fraumeni syndrome (TP53 mutations), Lynch syndrome and constitutional mismatch repair deficiency syndrome (MLH1, MSH2, MSH6, PMS2 mutations), neurofibromatosis type 1/2 (NF1/2 mutations), prostate cancer (HPC1, BRCA1/2), retinoblastoma (RB1), and tuberous sclerosis (TSC1/2). Examples of these cancers and their associated mutations will be discussed herein.
Brain tumours
Neurofibromatoses leads to an increased likelihood of cancer development, particularly for peripheral nerve sheath tumours and gliomas. Type I the most common type is neurofibromatosis, is characterised by benign neurofibromas around the peripheral nerves. Neurofibromatosis type I is caused by autosomal dominant mutations in the NF1 gene. NF1 regulates cell division through its ability to regulate RAS and PI3K activity. Approximately 50% of neurofibromatosis type I cases have a recognised family history of NF1 mutations. Although rare, cases of hereditary neurofibromatosis type 1 have been reported in Arab children.
Glioblastomas are malignant brain tumours that develop from astrocytes. Most GBMs are not inherited and occur sporadically. However, glioblastomas can occur in those with neurofibromatosis type 1, Turcot syndrome (APC, MLH1, PMS2 mutations) and Li Fraumeni syndrome (TP53). In each of these conditions, mutations are inherited in an autosomal dominant manner. According to the WHO classification of glioblastomas based on histopathological origin, it is categorised as a primary brain tumour of neuroepithelial, glial origin.51 When glioblastoma is ranked according to its clinical and prognostic significance, it is within the highest grade IV and is highly aggressive.52 Glioblastoma can be a result of progression from less malignant glial tumours (secondary type) or can occur de novo (primary).53 Paediatric glioblastoma is another subtype of this disease. Primary tumours are the most common.
Glioblastomas have an incidence rate of 3.19 per 100,000 persons in the US, with rates 2.0 times higher in Caucasians than African–Americans.52 The incidence of glioblastoma is lower in Asians and Native-American.54 The current treatment strategy for glioblastoma patients combines maximal surgical resection, followed by radiotherapy with concomitant and adjuvant temozolomide (TMZ).55–57 Less than 5% of newly diagnosed glioblastoma patients however survive longer than 5 years after diagnosis. It is clear that improved diagnostic and therapeutic strategies are required.
A family clustering of glioblastoma is recognised but relevant hereditary factors still remain elusive. Primary, secondary, and paediatric glioblastomas do however have known distinct sporadic genetic and epigenetic alterations. The most common mutations influence PI3K/AKT/mTOR, Ras/RAF/MAPK, and p53/Rb signalling pathways.58–61 These include mutations in TP53, PTEN, and CDK4.62–68 TP53 signalling is altered in 87% of glioblastomas, mostly affecting p53, murine double minute-2 (MDM2), MDM4, and cyclin-dependent kinase (CDK) N2A genes. Around 78% of glioblastomas have disruptions in RB signalling and frequently show alterations in RB1, CDK4, CDK6, CCND2, and the cyclin-dependent kinase inhibitor 2 (CDKN2) family.63,67,69 RTK/RAS/PI3K activation is observed in approximately 88% of tumours, typically affecting known hereditary (NF1) and non-hereditary (PIK3R1, and PIK3CA) genes.70–72
A problem of glioblastoma is its presentation as a heterogeneity of altered genetic pathways, evidenced by The Cancer Genome Atlas Research Network’s study classification based on gene expression profiles.73 Glioblastoma can be genetically typed66; Classical glioblastoma, with its highest survival rates, harbours no TP53 mutations, but high rates of EGFR mutations. Mesenchymal glioblastoma has frequent mutations of NF1, TP53, and PTEN, and aggressive chemotherapy can increase survival. Proneural glioblastoma has the highest mutation rates typically in TP53, PDGFRA, and isocitrate dehydrogenase (IDH), usually afflicting young adults. The neural type is more common in older patients and is correlated with frequent mutations in IDH1. The identification of these genes has encouraged drug development efforts focused on these pathways with limited success.74–77 The heterogeneous nature of the disease suggests that multiple approaches may be more effective.
Understanding the genetics and epigenetics of glioblastoma can distinguish various subgroups, often histologically indistinguishable. This could lead to the development of genetic glioblastoma classification with clinical impact, subgroup-specific treatment regimens, and improved design of future clinical trials.78 Recently, large population-based studies in the US revealed a 13% decreased risk in Hispanics compared with white glioblastoma patients.79 Recent studies using data from 21,184 glioblastoma patients in adult Hispanics Americans also reported an increased survival for Hispanics compared with white non-Hispanics.52 From a hereditary perspective, is it unclear if the differences in survival among races are due to differences in genetic factors, environmental exposures, or differences in treatment? Wiencke and colleagues found that secondary glioblastomas have more TP53 mutations that occur more frequently in Black and Asian patients.80 Conversely, Whites tend to harbour EGFR amplifications.66 As both secondary glioblastomas and TP53 mutations have an improved prognosis and longer survival times compared to primary glioblastoma and EGFR amplification, this may explain why Blacks and Asians with glioblastoma live longer than Whites.
Glioblastoma in the Arab world is less well understood. To understand hereditary cancer in the region, Backes et al. performed exome sequencing in an Arab family in which both parents were healthy, whilst both children had glioblastoma.81 The study reported 85 homozygous non-synonymous single nucleotide polymorphisms (SNPs) in both siblings that were heterozygous in the parents, and thus represented potential hereditary glioblastoma genes. In addition to known glioblastoma genes including ERBB2, PMS2, or CHI3L1, over 50 genes new genes were identified. Of these, they identified an accumulation of effects that potentially increased the likelihood of glioblastoma in the siblings, including a clustering of multiple variants in single genes (PTPRB, CROCC), aggregation of genes that influence specific pathways (focal adhesion or ECM receptor interactions) and genomic proximity (chr22.q12.2, chr1.p36.33).81 The reported variants underlined the relevance of genetic predisposition and cancer development in this family.
Breast cancer is both complex and heterogeneous, encompassing many entities with variable clinical behaviours and biological features. High-throughput molecular methods have increased the ability to characterise the genetic landscape of breast cancer, some of which are now included in clinical practice. These include prognostic gene signatures for oestrogen receptor (ER)-positive and HER2-negative breast cancer patients, the assessment of HER2/neu status by FISH or IHC, OncoType Dx tests and MammaPrint assessments of 70 genes associated with tumour recurrence. Hereditary breast cancer is caused by germline mutations that occur in BRCA1, BRCA2, TP53, CHEK2, PTEN, ATM, and PPM1D.82 The discovery of these genes has permitted their classification into two groups, namely high-penetrance and low-penetrance that interact with other genes and/or environmental factors to cause disease. The two most common breast cancer genes are BRCA1 and BRCA2, both of which are required for homologous DNA repair.1,10,83,84 A loss of heterozygosity and hereditary mutations in BRCA1 or BRCA2 increase chromosomal instability, thus increasing the cancer risk. Mutations in these genes also stimulate malignant transformation. Pathologically, familial breast cancers due to BRCA1 mutations differ to those caused by BRCA2 mutations and non-familial breast cancer. Understanding these pathological differences along with the genetic history of the patient are required to offer individualised treatment regimens.85
Germline mutations in BRCA1 and BRCA2 are responsible for 90% of hereditary breast cancer cases in the Western world and their mutation therefore represents the most important marker for the early detection of breast cancer.86–88 The landscape of breast cancer in the Arab world is somewhat different. Although it is known that breast cancer is on the rise (Fig. 1), its genetic epidemiology was less well understood. From a disease perspective, Arab patients with breast cancer have an advanced stage disease and a younger age of onset compared to Western countries. The Arab genome project pioneered by Saudi Arabia has been tasked with the discovery of new hereditary biomarkers for breast cancer in the region.89 This need for this was highlighted by Rahman and Zayed, who suggested that unlike Western populations, BRCA1/2 mutations are not significantly involved in hereditary breast cancer in Bahrain, Kuwait, Oman, Qatar, Saudi Arabia, and the United Arab Emirates (UAE).90 In the Arab region, Al-Eitan et al. provided evidence that genetic variations in MMP9, TOX3, and DAPK1 genes contribute to the development of breast cancer in the Jordanian population,91 whilst Alshawati et al. identified single-nucleotide polymorphisms in TP53 and MDM-2 that increase the risk of breast cancer in ethnic Arab populations.12 Karakas et al. also reported the prevalence of PIK3CA mutations and the SNP rs17849079 in Arab breast cancer patients.2
Breast Cancer
For many years, the incomplete cataloguing of germline alterations in hereditary breast cancer cases led to a lack of consensus on those patients who should be tested. To overcome this issue, Siraj et al. designed the hereditary oncogenesis predisposition evaluation (HOPE) including genes with known association to breast cancer and other tumours in 1300 Arab cancer patients.92 Pathogenic or likely pathogenic alleles in genes other than BRCA2, ATM and PALB2 accounted for ~16.8% of mutation-positive breast cancers in which a family history was lacking in 63.7% of mutation-positive cases.92 This highlighted how germline mutations to breast cancer predisposition extend beyond the classic hereditary cancer genes. In similar studies, Crawford and colleagues assessed 300 high-risk women previously shown as negative for BRCA1/BRCA2 mutations, in which 26 women were found to harbour 28 pathogenic mutations in 19 sequenced genes. These included ATM, CDH1, CHEK2, and RAD51D in cases of bilateral breast cancer and CHK2, MHS6 and NBN in those with ovarian cancer.93 Lee and colleagues similarly reported the occurrence of MSH2, PMS2, and CHEK2 in four hereditary breast and ovarian cancer syndrome patients with a family history of cancer.94 This highlights the importance of multi-gene panel testing as a follow-on test for those with incomplete testing. Armed with the knowledge that hereditary cancer is not strictly familial, this highlights the need for more widespread screening programmes in Arab regions.
Regarding ovarian cancer (OvCa) over 20% of ovarian tumours possess hereditary susceptibility. OvCa is common amongst Arabs with one of the highest global incidences. The reported numbers are likely to be higher due to underdiagnoses and underreporting. On the genetic level, up to 85% of hereditary OvCa cases are thought be caused by germline mutations in the BRCA genes. However, OvCa is not limited to BRCA susceptibility and other suppressor genes and oncogenes have been reported. RAD51 truncating mutations confer a reported 6-fold increased risk of OvCa but cause only a small increase in BCa susceptibility.95 PALB2 mutations (a BRCA2-interacting protein) have been also been reported in families negative for BRCA mutations with OvCa. Several groups have assessed the role of CHEK2 mutations in OvCa, particularly the missense variant I157T.96 This mutation was identified as involved in cystadenomas, borderline tumours, but not high-grade OvCa.97 Other variants including del1100C and A252G have also been reported but their association with OvCa is controversial. The Mre11 complex consists of Mre11, NBS1, and RAD50 and is a critical component of the DNA repair machinery. Three OvCa associated mutations have been reported including Mre11 913C>T (Arg305Trp), NBS1 448C>T (Leu150Phe), and RAD50 687delT (stop codon at 234).98 BARD1 germline mutations (c.1690C>T, p.Gln564X; c.1315- 2A>G; c.1977A>G) have also been reported as pathogenic for familial OvCa and BCa.99,100
Colorectal cancer
Whilst most patients with Stage II/III colorectal cancer can be cured through combined surgery, radiotherapy and chemotherapy, treatment is costly and recurrence is frequent.40 There has been a remarkable improvement in our molecular understanding of colorectal cancer over the last three decades that has revolutionised numerous aspects of care. Lynch syndrome (LS) is the most common cause of hereditary colorectal cancer and its early detection provides an opportunity for preventive cancer approaches.101 Genetic mutations can make some tumours less responsive to treatment and the stratification of patients into genetic subgroups for targeted therapies represents an efficacious strategy to improve the clinical effects of treatment. It is now understood that genetic subsets of colorectal cancer carry different risk factors, disease prognoses, and responses to treatment.
In the case of inherited colorectal cancers, many are attributed to hereditary nonpolyposis (HNPCC), familial adenomatous polyposis (FAP) and other related but variable syndromes.102,103 Up to 30% of the patients fall into this category, with their first-line or second-line relatives having colorectal cancer. Novel and/or de novo germ-line mutations of the adenomatous polyposis coli (APC) occur in up to 25% of FAP patients when untreated, the incidence of colorectal cancer is high.11,104–106 The most frequent germ-line APC mutations occur on codons 1061 and 1309. Hereditary nonpolyposis is the product of mutations in the MMR genes, including MLH1, MSH2, MSH6, PMS1, and PMS2.107–115 Mutations in MLH1 and MSH2 are most common. Juvenile polyposis syndrome is caused by mutations in the bone morphogenetic protein receptor, type 1A (BMPR1A) or SMAD family member 4 (SMAD4), both of which are tumour suppressors.116–123 Cowden syndrome, results from mutations in PTEN. Homozygous mutations in the base excision repair (BER) pathway gene mutY DNA glycosylase (MUTYH) leads to MUTYH-associated polyposis syndrome, and heterozygous MUTYH mutations are observed in familial colorectal cancer
The molecular characterisation of colorectal cancer may help identify familial predispositions, permit the movement from conventional chemotherapy drugs to biomarker-driven treatments in advanced cases. Again, the identification of specific prognostic subgroups that can again be explored among the Arab population where known colorectal cancer drivers including KRAS, NRAS, BRAF, and PIK3CA mutations differ across populations. Al Shamsi et al. determined the mutational frequencies of these drivers in the Arab population11 in 198 cases (99 Arab patients and 99 Western patients). The frequency of KRAS, NRAS, BRAF, TP53, APC, and PIK3CA mutations were similar between Arab and Western populations, but SMAD4 mutations were of lower frequency whilst FBXW7 mutations were more frequent. Studies comparing Arab colorectal cancer occurrence to the Jewish population concluded that colorectal cancer is more advanced, aggressive and symptomatic in Arab populations.124,125 Since Arab patients are younger at the time of diagnosis, familial specific genetic variations are predicted to be involved. Armed with this information, screening amongst genetically high-risk groups in the Arab world in addition to policies designed to encourage healthier living are now required to reduce colorectal cancer incidence in the region.
Prostate cancer
The incidence and mortality rates of prostate cancer show significant discrepancies among countries and ethnicities. Among people of European descent, prostate cancer is the most common cancer,126 but its prevalence is lower in men in the Middle East and North Africa.127 Prostate cancer is extremely heterogeneous compared to other tumours, and accordingly, various familial and sporadic mutations have been identified that increase its risk.128,129 Those with multiple single-gene polymorphisms and a family history of prostate cancer are at the highest risk.126,128–132 These include BRCA1 and BRCA2, MMR mutations including MLH1, MSH2 and MSH6, PMS2, HOXB13, checkpoint kinase 2 CHEK2, NBN, BRCA1-interacting protein C-terminal helicase 1 (BRIP1), and ATM.129–131 The potential of these findings is underscored by the recent approval of olaparib for BRCA1/BRCA2 or ATM-mutated, metastatic, castrate-resistant prostate cancer. This highlights the impact of genetic testing in therapeutic strategies.133–136 It should be noted that, genetic profiling has not replaced PSA monitoring, prostate gland MRIs, or biopsies for screening. The presence of mutations associated with prostate cancer encourages patients to undergo earlier and more frequent screening with the hope of earlier diagnosis and treatment.
The molecular changes involved in prostate cancer have not been broadly explored across the Arab world.127 Prostate cancer is more frequent amongst men of African descent, which is attributed to ethnic-specific differences in genotype frequencies for both SRD5A2 and CYP3A4.137 The CAG repeat length of the androgen receptor also differs according to population and is associated with prostate cancer.138–141 Progress in this area is required in the Arab world.
Familial cancer genetic counselling clinic in Saudi Arabia
The worldwide demand for cancer genetic counselling is growing.41 Studies investigating cancer susceptibility within families have suggested genetic links to an array of malignancies at the population level.14,142–144 Genetic screening offers increased monitoring and surveillance of those with a risk of cancer, in addition to prophylactic, risk-reducing interventions.144 Fifteen relevant guidelines were developed to provide recommendations on genetic counselling (Supplementary Material) and were in general agreement of the importance of genetic counselling prior to BRCA testing, including breast cancer risk-reduction procedures including mastectomy and oophorectomy.37 Genetic counselling is a “process of helping people understand and adapt to the medical, psychological and familial implications of genetic contributions to disease”.22,145–148 Genetic counselling concerning cancer risks has benefited individuals and their relatives through improved adherence to risk management, increased knowledge of genetics, improved patient satisfaction, and cost.149 Conversely, negative outcomes, such as test result misinterpretation, incorrect medical management, and psychological distress can arise when genetic testing is performed in the absence of adequate genetic counselling.150 For example, many societies struggle to deal with cultural stigmas associated with harbouring a genetic disease, ultimately disadvantaging the afflicted families through social events including marriage refusal.151 This can lead to a refusal to participate in some regions.
Two of the first clinics in Saudi Arabia (both in Riyadh) that offered genetic counselling for familial cancer are King Fahad Medical City (KFMC) at the comprehensive cancer centre, and the Saudi Diagnostic Laboratory (SDL), Cancer Genetic Counselling clinic at King Faisal Specialist Hospital & Research Centre. Patients referred to this process must complete a minimum of two in-person visits (Fig. 1). At the initial visit, risk assessments are performed, and if applicable, informed consent for genetic testing is obtained. At the initial visit, a family history is obtained and the life risk for specific cancers is calculated. If a patient’s life risk is ≥20%, they are considered “high risk” and eligible for higher-level cancer screening processes, including more frequent mammograms. Furthermore, risk-reduction procedures (mastectomy, and oophorectomy) are also offered. For those who consent to genetic testing, a post-test visit is arranged for the interpretation of test results. During this visit, implications for other family members will are discussed, in addition to the patient’s future management of their familial cancer risk. Medications, surgeries, and lifestyle changes are discussed.
A challenge in familial cancer genetic counselling in Saudi Arabia is the pervasive belief that cancer is not genetic, as many attributes the aetiology of cancer to non-biological causes.42 A further challenge is navigating client fears regarding genetic counselling sessions.23,152 In Saudi Arabia, “’counselling” is not a term in general use and patients have no prior understanding of the process. When creating appointments, patients are asked an extensive panel of questions regarding their family history of cancer, which can cause distress. A further stress is that families receiving genetic counselling are provided negative information on a family member’s health.50 The decision to select the correct genetic test and how the data are interpreted and communicated the patient remain ongoing issues in these areas.22,23,41,42
An additional challenge is determining the goals of testing and the information required. Furthermore, genetic counsellors must successfully communicate risk perception to their patients.143,153 Genetic counsellors provide significant levels of long-term psychosocial support to patients and their families, termed “support counselling”.154 This allows genetic counsellors to help the patients cope with the test data.
The state of the profession in the Middle East
Due to increased demand, the genetic counselling profession, led by Saudi Arabia, is expanding in the Middle East, and requires increased professionalism to accommodate future patients. Saudi Arabia established a master’s degree training programme in 2005,155 and the establishment of a professional society that focuses specifically on genetic counselling is planned. Active societies for medical genetics in the Middle East include the Saudi Society of Medical Genetics155 the National society of Human genetics in Egypt156 and the African Society of human genetics.157 Medical societies outside the middle-East have also been developed158,159
In the Middle East, the societies need to evolve to provide a distinct and coordinated forum for genetic counselling networking. The SSMG has proposed serves as an umbrella organisation and sponsors the formation of a Saudi-led genetic counsellor professional society. This society serves to integrate Middle-Eastern genetic counselling and could represent a nexus from which training and education can occur. To-date, those in the field of medical genetics know relatively little about genetic counselling. The development of professional societies can serve an educational role for both patients and colleagues.
Currently, Saudi Arabia is implementing a national strategy termed Vision 2030.160 This broad vision entails commercial and governmental goals, including improved public health and healthcare delivery, and the expansion of Saudi Arabia’s educational and research infrastructure.161 Organising and professionalizing the genetic counselling field in the Middle East is not only necessary to serve patients, but provides an opportunity for the Middle East to excel in this area globally.
Conclusions
Familial cancer syndromes due to inherited mutations that increase the risk of tumour development account for ~5–10% of all malignancies and are generally characterised by early-onset cancers.6 In the majority of familial tumours, the risk is associated with monogenic hereditary disease. The ability to identify germline variants in familial cancer has been challenging due to incomplete cataloguing of cancer-mutations and disagreements on those who should be tested. What is clear is that before we can understand familial cancers, we must first identify relevant cancerous mutations that show prevalence in individual ethnicities. This is being increasingly recognised in the Arab world and our genetic understanding of cancer in the region is growing. This must now be combined with familial testing to define novel hereditary cancer drivers that can provide genetic counselling to families in the face of high consanguinity.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Supplementary information
Acknowledgements
The author thanks King Abdulaziz City for Science and Technology and the Saudi Human Genome Project for technical support. This study was funded by King Fahad Medical City (IRF 17-17) (M.A.).
Author contributions
F.H., A.E. and A.Q. edited the manuscript. M.A. designed the study and wrote the manuscript. All co-authors read and approved the final version of the article and accept accountability for it.
Data availability
No datasets were generated or analyzed during the current study.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information is available for this paper at 10.1038/s41525-019-0110-y.
References
- 1.Alhuqail AJ, et al. High prevalence of deleterious BRCA1 and BRCA2 germline mutations in arab breast and ovarian cancer patients. Breast Cancer Res. Treat. 2018;168:695–702. doi: 10.1007/s10549-017-4635-4. [DOI] [PubMed] [Google Scholar]
- 2.Karakas B, et al. Prevalence of PIK3CA mutations and the SNP rs17849079 in Arab breast cancer patients. Cancer Biol. Ther. 2013;14:888–896. doi: 10.4161/cbt.25945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ghafoor M, Schuyten R, Bener A. Epidemiology of prostate cancer in United Arab Emirates. Med. J. Malays. 2003;58:712–716. [PubMed] [Google Scholar]
- 4.Hilal L, et al. Prostate cancer in the Arab world: a view from the inside. Clin. Genitourin. Cancer. 2015;13:505–511. doi: 10.1016/j.clgc.2015.05.010. [DOI] [PubMed] [Google Scholar]
- 5.Aghassi-Ippen M, Green MS, Shohat T. Familial risk factors for breast cancer among Arab women in Israel. Eur. J. Cancer Prev. 2002;11:327–331. doi: 10.1097/00008469-200208000-00003. [DOI] [PubMed] [Google Scholar]
- 6.Rahner N, Steinke V. Hereditary cancer syndromes. Dtsch Arztebl. Int. 2008;105:706–714. doi: 10.3238/arztebl.2008.0706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mayer DK, et al. American Society of Clinical Oncology clinical expert statement on cancer survivorship care planning. J. Oncol. Pract. 2014;10:345–351. doi: 10.1200/JOP.2014.001321. [DOI] [PubMed] [Google Scholar]
- 8.Al-Qasem AJ, et al. TP53 genetic alterations in Arab breast cancer patients: novel mutations, pattern and distribution. Oncol. Lett. 2011;2:363–369. doi: 10.3892/ol.2011.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Younes N, Zayed H. Genetic epidemiology of ovarian cancer in the 22 Arab countries: a systematic review. Gene. 2019;684:154–164. doi: 10.1016/j.gene.2018.10.044. [DOI] [PubMed] [Google Scholar]
- 10.El Saghir NS, et al. BRCA1 and BRCA2 mutations in ethnic Lebanese Arab women with high hereditary risk breast cancer. Oncologist. 2015;20:357–364. doi: 10.1634/theoncologist.2014-0364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Al-Shamsi HO, et al. Molecular spectrum of KRAS, NRAS, BRAF, PIK3CA, TP53, and APC somatic gene mutations in Arab patients with colorectal cancer: determination of frequency and distribution pattern. J. Gastrointest. Oncol. 2016;7:882–902. doi: 10.21037/jgo.2016.11.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Alshatwi AA, et al. A single-nucleotide polymorphism in the TP53 and MDM-2 gene modifies breast cancer risk in an ethnic Arab population. Fundam. Clin. Pharm. 2012;26:438–443. doi: 10.1111/j.1472-8206.2011.00939.x. [DOI] [PubMed] [Google Scholar]
- 13.Zick A, et al. Recurrent TP53 missense mutation in cancer patients of Arab descent. Fam. Cancer. 2017;16:295–301. doi: 10.1007/s10689-016-9951-z. [DOI] [PubMed] [Google Scholar]
- 14.Al-Aama JY, et al. Whole exome sequencing of a consanguineous family identifies the possible modifying effect of a globally rare AK5 allelic variant in celiac disease development among Saudi patients. PLoS ONE. 2017;12:e0176664. doi: 10.1371/journal.pone.0176664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bondagji NS, et al. Replication of GWAS loci revealed the moderate effect of TNRC6B locus on susceptibility of Saudi women to develop uterine leiomyomas. J. Obstet. Gynaecol. Res. 2017;43:330–338. doi: 10.1111/jog.13217. [DOI] [PubMed] [Google Scholar]
- 16.Fehringer G, et al. Cross-cancer genome-wide analysis of lung, ovary, breast, prostate, and colorectal cancer reveals novel pleiotropic associations. Cancer Res. 2016;76:5103–5114. doi: 10.1158/0008-5472.CAN-15-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sokolenko AP, Imyanitov EN. Molecular diagnostics in clinical oncology. Front Mol. Biosci. 2018;5:76. doi: 10.3389/fmolb.2018.00076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baraniskin A, et al. Clinical relevance of molecular diagnostics in gastrointestinal (GI) cancer: European Society of Digestive Oncology (ESDO) expert discussion and recommendations from the 17th European Society for Medical Oncology (ESMO)/World Congress on Gastrointestinal Cancer, Barcelona. Eur. J. Cancer. 2017;86:305–317. doi: 10.1016/j.ejca.2017.09.021. [DOI] [PubMed] [Google Scholar]
- 19.Van Heertum RL, Scarimbolo R, Ford R, Berdougo E, O'Neal M. Companion diagnostics and molecular imaging-enhanced approaches for oncology clinical trials. Drug Des. Dev. Ther. 2015;9:5215–5223. doi: 10.2147/DDDT.S87561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Al-Omran, H. Measurement of the knowledge, attitudes, and beliefs of Arab-American adults toward cancer screening and early detection: development of a survey instrument. Ethn. Dis. 15, S1–15 (2005). [PubMed]
- 21.Cohen M, Azaiza F. Early breast cancer detection practices, health beliefs, and cancer worries in Jewish and Arab women. Prev. Med. 2005;41:852–858. doi: 10.1016/j.ypmed.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 22.National Society of Genetic Counselors' Definition Task, F. et al. A new definition of Genetic Counseling: National Society of Genetic Counselors' Task Force report. J. Genet. Couns. 2006;15:77–83. doi: 10.1007/s10897-005-9014-3. [DOI] [PubMed] [Google Scholar]
- 23.Desrosiers, L. R., Quinn, E., Cramer, S. & Dobek, W. Integrating genetic counseling and testing in the pediatric oncology setting: parental attitudes and influencing factors. Pediatr Blood Cancer66, e27907 (2019). [DOI] [PubMed]
- 24.Korngiebel DM, et al. Practice implications of expanded genetic testing in oncology. Cancer Invest. 2019;37:39–45. doi: 10.1080/07357907.2018.1564926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arafa MA, Farhat K. Colorectal cancer in the Arab world–screening practices and future prospects. Asian Pac. J. Cancer Prev. 2015;16:7425–7430. doi: 10.7314/APJCP.2015.16.17.7425. [DOI] [PubMed] [Google Scholar]
- 26.Arafa MA, Rabah DM. With increasing trends of prostate cancer in the Saudi Arabia and Arab World: Should we start screening programs? World J. Clin. Oncol. 2017;8:447–449. doi: 10.5306/wjco.v8.i6.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.El-Attar, I. A. Cancer databases in the Arab world. Ethn. Dis.15, S1–3 (2005). [PubMed]
- 28.Hamadeh RR, Borgan SM, Sibai AM. Cancer research in the Arab World: a review of publications from seven countries between 2000–2013. Sultan Qaboos Univ. Med. J. 2017;17:e147–e154. doi: 10.18295/squmj.2016.17.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan G, Hashim MJ. Burden of virus-associated liver cancer in the Arab world, 1990-2010. Asian Pac. J. Cancer Prev. 2015;16:265–270. doi: 10.7314/APJCP.2015.16.1.265. [DOI] [PubMed] [Google Scholar]
- 30.Lakkis NA, Adib SM, Hamadeh G, El Jarrah R, Osman MH. Sociological transition and breast cancer in the Arab world: the experience of Lebanon. Asian Pac. J. Cancer Prev. 2017;18:1357–1364. doi: 10.22034/APJCP.2017.18.5.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salim EI, et al. Cancer epidemiology and control in the arab world—past, present and future. Asian Pac. J. Cancer Prev. 2009;10:3–16. [PubMed] [Google Scholar]
- 32.Taha Z, Eltom SE. The role of diet and lifestyle in women with breast cancer: an update review of related research in the Middle East. Biores. Open Access. 2018;7:73–80. doi: 10.1089/biores.2018.0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haddou Rahou B, et al. Quality of life in Arab women with breast cancer: a review of the literature. Health Qual. Life Outcomes. 2016;14:64. doi: 10.1186/s12955-016-0468-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cicero G, et al. Risk perception and psychological distress in genetic counselling for hereditary breast and/or ovarian cancer. J. Genet. Couns. 2017;26:999–1007. doi: 10.1007/s10897-017-0072-0. [DOI] [PubMed] [Google Scholar]
- 35.Radwan H, Hasan H, Ballout RA, Rizk R. The epidemiology of cancer in the United Arab Emirates: a systematic review. Medicine. 2018;97:e13618. doi: 10.1097/MD.0000000000013618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jastaniah W, et al. Prevalence of hereditary cancer susceptibility syndromes in children with cancer in a highly consanguineous population. Cancer Epidemiol. 2018;55:88–95. doi: 10.1016/j.canep.2018.05.006. [DOI] [PubMed] [Google Scholar]
- 37.Alaofi RK, Nassif MO, Al-Hajeili MR. Prophylactic mastectomy for the prevention of breast cancer: review of the literature. Avicenna J. Med. 2018;8:67–77. doi: 10.4103/ajm.AJM_21_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elobaid Y, Aw TC, Lim JNW, Hamid S, Grivna M. Breast cancer presentation delays among Arab and national women in the UAE: a qualitative study. SSM Popul. Health. 2016;2:155–163. doi: 10.1016/j.ssmph.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siraj AK, et al. Prevalence, spectrum, and founder effect of BRCA1 and BRCA2 mutations in epithelial ovarian cancer from the Middle East. Hum. Mutat. 2019;40:729–733. doi: 10.1002/humu.23736. [DOI] [PubMed] [Google Scholar]
- 40.Al Abdouli L, Dalmook H, Akram Abdo M, Carrick FR, Abdul Rahman M. Colorectal cancer risk awareness and screening uptake among adults in the United Arab Emirates. Asian Pac. J. Cancer Prev. 2018;19:2343–2349. doi: 10.22034/APJCP.2018.19.8.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hooker GW, et al. Cancer genetic counseling and testing in an Era of rapid change. J. Genet. Couns. 2017;26:1244–1253. doi: 10.1007/s10897-017-0099-2. [DOI] [PubMed] [Google Scholar]
- 42.Ormond KE, et al. Genetic counseling globally: Where are we now? Am. J. Med. Genet. C. 2018;178:98–107. doi: 10.1002/ajmg.c.31607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Saletta F, Dalla Pozza L, Byrne JA. Genetic causes of cancer predisposition in children and adolescents. Transl. Pediatr. 2015;4:67–75. doi: 10.3978/j.issn.2224-4336.2015.04.08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burgess KR, Carmany EP, Trepanier AM. A comparison of telephone genetic counseling and in-person genetic counseling from the genetic counselor's perspective. J. Genet. Couns. 2016;25:112–126. doi: 10.1007/s10897-015-9848-2. [DOI] [PubMed] [Google Scholar]
- 45.Voils CI, Venne VL, Weidenbacher H, Sperber N, Datta S. Comparison of telephone and televideo modes for delivery of genetic counseling: a randomized trial. J. Genet. Couns. 2018;27:339–348. doi: 10.1007/s10897-017-0189-1. [DOI] [PubMed] [Google Scholar]
- 46.al Talabani J, Shubbar AI, Mustafa KE. Major congenital malformations in United Arab Emirates (UAE): need for genetic counselling. Ann. Hum. Genet. 1998;62:411–418. doi: 10.1046/j.1469-1809.1998.6250411.x. [DOI] [PubMed] [Google Scholar]
- 47.Albar MA. Counselling about genetic disease: an Islamic perspective. East Mediterr. Health J. 1999;5:1129–1133. [PubMed] [Google Scholar]
- 48.Saffi M, Howard N. Exploring the effectiveness of mandatory premarital screening and genetic counselling programmes for beta-Thalassaemia in the Middle East: a scoping review. Public Health Genom. 2015;18:193–203. doi: 10.1159/000430837. [DOI] [PubMed] [Google Scholar]
- 49.Saleem R, Gofin R, Ben-Neriah Z, Boneh A. Variables influencing parental perception of inherited metabolic diseases before and after genetic counselling. J. Inherit. Metab. Dis. 1998;21:769–780. doi: 10.1023/A:1005453301377. [DOI] [PubMed] [Google Scholar]
- 50.Abacan M, et al. The global state of the genetic counseling profession. Eur. J. Hum. Genet. 2019;27:183–197. doi: 10.1038/s41431-018-0252-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Stoyanov GS, Dzhenkov DL. On the concepts and history of glioblastoma multiforme—morphology, genetics and epigenetics. Folia Med. 2018;60:48–66. doi: 10.1515/folmed-2017-0069. [DOI] [PubMed] [Google Scholar]
- 52.Shabihkhani M, et al. Incidence, survival, pathology, and genetics of adult Latino Americans with glioblastoma. J. Neurooncol. 2017;132:351–358. doi: 10.1007/s11060-017-2377-0. [DOI] [PubMed] [Google Scholar]
- 53.Liu A, Hou C, Chen H, Zong X, Zong P. Genetics and epigenetics of glioblastoma: applications and overall incidence of IDH1 mutation. Front. Oncol. 2016;6:16. doi: 10.3389/fonc.2016.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Villanueva MT. Genetics: united colors of glioblastoma. Nat. Rev. Clin. Oncol. 2012;9:68. doi: 10.1038/nrclinonc.2011.202. [DOI] [PubMed] [Google Scholar]
- 55.Hutchinson L. Genetics: new paediatric glioblastoma treatment option. Nat. Rev. Clin. Oncol. 2017;14:4. doi: 10.1038/nrclinonc.2016.182. [DOI] [PubMed] [Google Scholar]
- 56.Desai V, et al. Combination of biochanin A and temozolomide impairs tumor growth by modulating cell metabolism in glioblastoma multiforme. Anticancer Res. 2019;39:57–66. doi: 10.21873/anticanres.13079. [DOI] [PubMed] [Google Scholar]
- 57.Robins HI, et al. Molecular evolution of a glioblastoma controlled with tumor treating fields and concomitant temozolomide. Front. Oncol. 2018;8:451. doi: 10.3389/fonc.2018.00451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brennan CW, et al. The somatic genomic landscape of glioblastoma. Cell. 2013;155:462–477. doi: 10.1016/j.cell.2013.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cancer Genome Atlas Research, N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Verhaak RG, et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17:98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vitucci M, et al. Cooperativity between MAPK and PI3K signaling activation is required for glioblastoma pathogenesis. Neuro. Oncology. 2013;15:1317–1329. doi: 10.1093/neuonc/not084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Qi Q, et al. Co-amplification of phosphoinositide 3-kinase enhancer A and cyclin-dependent kinase 4 triggers glioblastoma progression. Oncogene. 2017;36:4562–4572. doi: 10.1038/onc.2017.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Furgason JM, et al. Whole genome sequence analysis links chromothripsis to EGFR, MDM2, MDM4, and CDK4 amplification in glioblastoma. Oncoscience. 2015;2:618–628. doi: 10.18632/oncoscience.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Rickert CH, et al. Glioblastoma with adipocyte-like tumor cell differentiation–histological and molecular features of a rare differentiation pattern. Brain Pathol. 2009;19:431–438. doi: 10.1111/j.1750-3639.2008.00199.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Purow B, Schiff D. Advances in the genetics of glioblastoma: are we reaching critical mass? Nat. Rev. Neurol. 2009;5:419–426. doi: 10.1038/nrneurol.2009.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Benjamin R, Capparella J, Brown A. Classification of glioblastoma multiforme in adults by molecular genetics. Cancer J. 2003;9:82–90. doi: 10.1097/00130404-200303000-00003. [DOI] [PubMed] [Google Scholar]
- 67.Backlund LM, et al. Short postoperative survival for glioblastoma patients with a dysfunctional Rb1 pathway in combination with no wild-type PTEN. Clin. Cancer Res. 2003;9:4151–4158. [PubMed] [Google Scholar]
- 68.Tortosa A, et al. Molecular genetics of radiographically defined de novo glioblastoma multiforme. Neuropathol. Appl. Neurobiol. 2000;26:544–552. doi: 10.1046/j.0305-1846.2000.00290.x. [DOI] [PubMed] [Google Scholar]
- 69.Li M, et al. CDK4/6 inhibition is more active against the glioblastoma proneural subtype. Oncotarget. 2017;8:55319–55331. doi: 10.18632/oncotarget.19429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Venkatesan S, Lamfers ML, Dirven CM, Leenstra S. Genetic biomarkers of drug response for small-molecule therapeutics targeting the RTK/Ras/PI3K, p53 or Rb pathway in glioblastoma. CNS Oncol. 2016;5:77–90. doi: 10.2217/cns-2015-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Venkatesan S, et al. TP53 mutated glioblastoma stem-like cell cultures are sensitive to dual mTORC1/2 inhibition while resistance in TP53 wild type cultures can be overcome by combined inhibition of mTORC1/2 and Bcl-2. Oncotarget. 2016;7:58435–58444. doi: 10.18632/oncotarget.11205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Smardova J, et al. High frequency of temperature-sensitive mutants of p53 in glioblastoma. Pathol. Oncol. Res. 2013;19:421–428. doi: 10.1007/s12253-012-9596-7. [DOI] [PubMed] [Google Scholar]
- 73.Shergalis A, Bankhead A, 3rd, Luesakul U, Muangsin N, Neamati N. Current challenges and opportunities in treating glioblastoma. Pharm. Rev. 2018;70:412–445. doi: 10.1124/pr.117.014944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kreisl TN, et al. A pilot study of everolimus and gefitinib in the treatment of recurrent glioblastoma (GBM) J. Neuro. Oncol. 2009;92:99–105. doi: 10.1007/s11060-008-9741-z. [DOI] [PubMed] [Google Scholar]
- 75.Chang SM, et al. Phase II study of CCI-779 in patients with recurrent glioblastoma multiforme. Invest New Drugs. 2005;23:357–361. doi: 10.1007/s10637-005-1444-0. [DOI] [PubMed] [Google Scholar]
- 76.Reardon DA, et al. Phase 2 trial of erlotinib plus sirolimus in adults with recurrent glioblastoma. J. Neuro. Oncol. 2010;96:219–230. doi: 10.1007/s11060-009-9950-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Galanis E, et al. Phase II trial of temsirolimus (CCI-779) in recurrent glioblastoma multiforme: a North Central Cancer Treatment Group Study. J. Clin. Oncol. 2005;23:5294–5304. doi: 10.1200/JCO.2005.23.622. [DOI] [PubMed] [Google Scholar]
- 78.Amar D, Hait T, Izraeli S, Shamir R. Integrated analysis of numerous heterogeneous gene expression profiles for detecting robust disease-specific biomarkers and proposing drug targets. Nucleic Acids Res. 2015;43:7779–7789. doi: 10.1093/nar/gkv810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bohn A, Braley A, Rodriguez de la Vega P, Zevallos JC, Barengo NC. The association between race and survival in glioblastoma patients in the US: a retrospective cohort study. PLoS ONE. 2018;13:e0198581. doi: 10.1371/journal.pone.0198581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Wiencke JK, et al. Methylation of the PTEN promoter defines low-grade gliomas and secondary glioblastoma. Neuro. Oncology. 2007;9:271–279. doi: 10.1215/15228517-2007-003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Backes C, et al. New insights into the genetics of glioblastoma multiforme by familial exome sequencing. Oncotarget. 2015;20:5918–5931. doi: 10.18632/oncotarget.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ngene SO, Adedokun B, Adejumo P, Olopade O. Breast cancer genetics knowledge and testing intentions among Nigerian professional women. J. Genet. Couns. 2018;27:863–873. doi: 10.1007/s10897-017-0194-4. [DOI] [PubMed] [Google Scholar]
- 83.Sidaway P. Genetics: BRCA-mutant breast/ovarian cancer revealed. Nat. Rev. Clin. Oncol. 2017;14:524. doi: 10.1038/nrclinonc.2017.104. [DOI] [PubMed] [Google Scholar]
- 84.Kadouri L, et al. A novel BRCA-1 mutation in Arab kindred from east Jerusalem with breast and ovarian cancer. BMC Cancer. 2007;7:14. doi: 10.1186/1471-2407-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Bakhuizen, J. J., Velthuizen, M. E., Stehouwer, S., Bleiker, E. M. & Ausems, M. G. Genetic counselling of young women with breast cancer for Li-Fraumeni syndrome: a nationwide survey on the experiences and attitudes of genetics professionals. Fam. Cancer18, 231–239 (2018). [DOI] [PMC free article] [PubMed]
- 86.Ricks-Santi LJ, et al. BRCA1 polymorphisms and breast cancer epidemiology in the Western New York exposures and breast cancer (WEB) study. Genet. Epidemiol. 2013;37:504–511. doi: 10.1002/gepi.21730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Einbeigi Z, Enerback C, Wallgren A, Nordling M, Karlsson P. BRCA1 gene mutations may explain more than 80% of excess number of ovarian cancer cases after breast cancer—a population based study from the Western Sweden Health Care region. Acta Oncol. 2010;49:361–367. doi: 10.3109/02841860903521095. [DOI] [PubMed] [Google Scholar]
- 88.Cao AY, Hu Z, Yin WJ, Jin W, Shao ZM. Some common mutations of RAD50 and NBS1 in western populations do not contribute significantly to Chinese non-BRCA1/2 hereditary breast cancer. Breast Cancer Res. Treat. 2010;121:247–249. doi: 10.1007/s10549-009-0629-1. [DOI] [PubMed] [Google Scholar]
- 89.Abedalthagafi MS. Precision medicine of monogenic disorders: lessons learned from the Saudi human genome. Front. Biosci. 2019;24:870–889. doi: 10.2741/4757. [DOI] [PubMed] [Google Scholar]
- 90.Rahman S, Zayed H. Breast cancer in the GCC countries: a focus on BRCA1/2 and non-BRCA1/2 genes. Gene. 2018;668:73–76. doi: 10.1016/j.gene.2018.05.045. [DOI] [PubMed] [Google Scholar]
- 91.Al-Eitan LN, Jamous RI, Khasawneh RH. Candidate gene analysis of breast cancer in the Jordanian population of Arab descent: a case–control study. Cancer Invest. 2017;35:256–270. doi: 10.1080/07357907.2017.1289217. [DOI] [PubMed] [Google Scholar]
- 92.Siraj AK, et al. Expanding the spectrum of germline variants in cancer. Hum. Genet. 2017;136:1431–1444. doi: 10.1007/s00439-017-1845-0. [DOI] [PubMed] [Google Scholar]
- 93.Crawford B, et al. Multi-gene panel testing for hereditary cancer predisposition in unsolved high-risk breast and ovarian cancer patients. Breast Cancer Res. Treat. 2017;163:383–390. doi: 10.1007/s10549-017-4181-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yoo J, et al. Clinical validity of next-generation sequencing multi-gene panel testing for detecting pathogenic variants in patients With hereditary breast-ovarian cancer syndrome. Ann. Lab. Med. 2020;40:148–154. doi: 10.3343/alm.2020.40.2.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Loveday C, et al. Germline mutations in RAD51D confer susceptibility to ovarian cancer. Nat. Genet. 2011;43:879–882. doi: 10.1038/ng.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Szymanska-Pasternak J, et al. CHEK2 variants predispose to benign, borderline and low-grade invasive ovarian tumors. Gynecol. Oncol. 2006;102:429–431. doi: 10.1016/j.ygyno.2006.05.040. [DOI] [PubMed] [Google Scholar]
- 97.Baysal BE, et al. Analysis of CHEK2 gene for ovarian cancer susceptibility. Gynecol. Oncol. 2004;95:62–69. doi: 10.1016/j.ygyno.2004.07.015. [DOI] [PubMed] [Google Scholar]
- 98.Heikkinen K, Karppinen SM, Soini Y, Makinen M, Winqvist R. Mutation screening of Mre11 complex genes: indication of RAD50 involvement in breast and ovarian cancer susceptibility. J. Med. Genet. 2003;40:e131. doi: 10.1136/jmg.40.12.e131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Ratajska M, et al. Cancer predisposing BARD1 mutations affect exon skipping and are associated with overexpression of specific BARD1 isoforms. Oncol. Rep. 2015;34:2609–2617. doi: 10.3892/or.2015.4235. [DOI] [PubMed] [Google Scholar]
- 100.Ratajska M, et al. Cancer predisposing BARD1 mutations in breast-ovarian cancer families. Breast Cancer Res. Treat. 2012;131:89–97. doi: 10.1007/s10549-011-1403-8. [DOI] [PubMed] [Google Scholar]
- 101.Jiang Wu, Cai Mu-Yan, Li Shi-Yong, Bei Jin-Xin, Wang Fang, Hampel Heather, Ling Yi-Hong, Frayling Ian M., Sinicrope Frank A., Rodriguez-Bigas Miguel A., Dignam James J., Kerr David J., Rosell Rafael, Mao Mao, Li Ji-Bin, Guo Yun-Miao, Wu Xiao-Yan, Kong Ling-Heng, Tang Jing-Hua, Wu Xiao-Dan, Li Chao-Feng, Chen Jie-Rong, Ou Qing-Jian, Ye Ming-Zhi, Guo Feng-Ming, Han Peng, Wang Qi-Wei, Wan De-Sen, Li Li, Xu Rui-Hua, Pan Zhi-Zhong, Ding Pei-Rong. Universal screening for Lynch syndrome in a large consecutive cohort of Chinese colorectal cancer patients: High prevalence and unique molecular features. International Journal of Cancer. 2019;144(9):2161–2168. doi: 10.1002/ijc.32044. [DOI] [PubMed] [Google Scholar]
- 102.Yang Y, You YN. Comprehensive clinical genetics care for patients with inherited colorectal cancer associated with Lynch syndrome: Western and Asian perspectives. Chin. Clin. Oncol. 2018;7:9. doi: 10.21037/cco.2018.01.06. [DOI] [PubMed] [Google Scholar]
- 103.Gryfe R. Clinical implications of our advancing knowledge of colorectal cancer genetics: inherited syndromes, prognosis, prevention, screening and therapeutics. Surg. Clin. N. Am. 2006;86:787–817. doi: 10.1016/j.suc.2006.05.007. [DOI] [PubMed] [Google Scholar]
- 104.De Queiroz Rossanese LB, De Lima Marson FA, Ribeiro JD, Coy CS, Bertuzzo CS. APC germline mutations in families with familial adenomatous polyposis. Oncol. Rep. 2013;30:2081–2088. doi: 10.3892/or.2013.2681. [DOI] [PubMed] [Google Scholar]
- 105.Nielsen M, et al. Germline mutations in APC and MUTYH are responsible for the majority of families with attenuated familial adenomatous polyposis. Clin. Genet. 2007;71:427–433. doi: 10.1111/j.1399-0004.2007.00766.x. [DOI] [PubMed] [Google Scholar]
- 106.O'Sullivan Maureen J., Mulcahy Tanya M., Cambell J., O'Suilleabhain Christoir B., Kirwan William O., Doyle Cuimin T., McCarthy Tommie V. Detection of five novel germline mutations of the APC gene in Irish familial adenomatous polyposis families. Human Mutation. 1998;11(S1):S251–S253. doi: 10.1002/humu.1380110180. [DOI] [PubMed] [Google Scholar]
- 107.Tanyi M, et al. MLH1 and MSH2 mutation screening in HNPCC families of Hungary—two new MMR gene mutations. Eur. J. Surg. Oncol. 2014;40:1445–1452. doi: 10.1016/j.ejso.2014.07.032. [DOI] [PubMed] [Google Scholar]
- 108.Giedl J, et al. Low frequency of HNPCC-associated microsatellite instability and aberrant MMR protein expression in early-onset bladder cancer. Am. J. Clin. Pathol. 2014;142:634–639. doi: 10.1309/AJCPVTCJ4VU5HKVZ. [DOI] [PubMed] [Google Scholar]
- 109.Hirai Y, et al. Molecular epidemiological and mutational analysis of DNA mismatch repair (MMR) genes in endometrial cancer patients with HNPCC-associated familial predisposition to cancer. Cancer Sci. 2008;99:1715–1719. doi: 10.1111/j.1349-7006.2008.00886.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Shibata D. When does MMR loss occur during HNPCC progression? Cancer Biomark. 2006;2:29–35. doi: 10.3233/CBM-2006-21-204. [DOI] [PubMed] [Google Scholar]
- 111.Schiemann U, et al. Detection of occult high graded microsatellite instabilities in MMR gene mutation negative HNPCC tumors by addition of complementary marker analysis. Eur. J. Med. Res. 2005;10:23–28. [PubMed] [Google Scholar]
- 112.Ashton KA, Meldrum CJ, McPhillips ML, Kairupan CF, Scott RJ. Frequency of the common MYH mutations (G382D and Y165C) in MMR mutation positive and negative HNPCC patients. Hered. Cancer Clin. Pract. 2005;3:65–70. doi: 10.1186/1897-4287-3-2-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Apessos A, et al. hMSH2 is the most commonly mutated MMR gene in a cohort of Greek HNPCC patients. Br. J. Cancer. 2005;92:396–404. doi: 10.1038/sj.bjc.6602260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Di Fiore F, et al. Screening for genomic rearrangements of the MMR genes must be included in the routine diagnosis of HNPCC. J. Med. Genet. 2004;41:18–20. doi: 10.1136/jmg.2003.012062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Papadopoulos N, Lindblom A. Molecular basis of HNPCC: mutations of MMR genes. Hum. Mutat. 1997;10:89–99. doi: 10.1002/(SICI)1098-1004(1997)10:2<89::AID-HUMU1>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 116.Schwenter F, et al. Juvenile polyposis syndrome, SMAD4 mutations, and hereditary hemorrhagic telangiectasia. J. Pediatr. Gastroenterol. Nutr. 2012;54:120–122. doi: 10.1097/MPG.0b013e3182238514. [DOI] [PubMed] [Google Scholar]
- 117.Gallione C, et al. Overlapping spectra of SMAD4 mutations in juvenile polyposis (JP) and JP-HHT syndrome. Am. J. Med. Genet. A. 2010;152A:333–339. doi: 10.1002/ajmg.a.33206. [DOI] [PubMed] [Google Scholar]
- 118.Gallione CJ, et al. A combined syndrome of juvenile polyposis and hereditary haemorrhagic telangiectasia associated with mutations in MADH4 (SMAD4) Lancet. 2004;363:852–859. doi: 10.1016/S0140-6736(04)15732-2. [DOI] [PubMed] [Google Scholar]
- 119.Fogt F, Brown CA, Badizadegan K, Zimmerman RL, Odze R. Low prevalence of loss of heterozygosity and SMAD4 mutations in sporadic and familial juvenile polyposis syndrome-associated juvenile polyps. Am. J. Gastroenterol. 2004;99:2025–2031. doi: 10.1111/j.1572-0241.2004.30502.x. [DOI] [PubMed] [Google Scholar]
- 120.Zhou XP, et al. Germline mutations in BMPR1A/ALK3 cause a subset of cases of juvenile polyposis syndrome and of Cowden and Bannayan-Riley-Ruvalcaba syndromes. Am. J. Hum. Genet. 2001;69:704–711. doi: 10.1086/323703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Woodford-Richens KL, et al. CDX2 mutations do not account for juvenile polyposis or Peutz-Jeghers syndrome and occur infrequently in sporadic colorectal cancers. Br. J. Cancer. 2001;84:1314–1316. doi: 10.1054/bjoc.2001.1800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bevan S, et al. Screening SMAD1, SMAD2, SMAD3, and SMAD5 for germline mutations in juvenile polyposis syndrome. Gut. 1999;45:406–408. doi: 10.1136/gut.45.3.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Houlston R, et al. Mutations in DPC4 (SMAD4) cause juvenile polyposis syndrome, but only account for a minority of cases. Hum. Mol. Genet. 1998;7:1907–1912. doi: 10.1093/hmg/7.12.1907. [DOI] [PubMed] [Google Scholar]
- 124.Rainis T, Halloun L, Keren D, Shuv-ami I, Lavy A. Colorectal cancer among Arab-Israeli women–possible reasons for increased incidence and mortality. J. Gastrointest. Cancer. 2010;41:130–134. doi: 10.1007/s12029-009-9128-1. [DOI] [PubMed] [Google Scholar]
- 125.Glushko Y, Basher W, Barchana M, Zidan J. Differences in clinical and pathological characteristics of colorectal cancer in Arab as compared to Jewish patients in Northern Israel. Fam. Cancer. 2010;9:327–330. doi: 10.1007/s10689-010-9337-6. [DOI] [PubMed] [Google Scholar]
- 126.Cremers RG, et al. The clinical phenotype of hereditary versus sporadic prostate cancer: HPC definition revisited. Prostate. 2016;76:897–904. doi: 10.1002/pros.23179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Al-Abdin OZ, Al-Beeshi IZ. Prostate cancer in the Arab population. An overview. Saudi Med. J. 2018;39:453–458. doi: 10.15537/smj.2018.5.21986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Boehm K, Thomas C, Tsaur I. Genetic profiling in the diagnosis of hereditary prostate cancer: Where do we stand? Aktuelle Urol. 2018;49:525–529. doi: 10.1055/a-0755-7360. [DOI] [PubMed] [Google Scholar]
- 129.Zhen JT, et al. Genetic testing for hereditary prostate cancer: current status and limitations. Cancer. 2018;124:3105–3117. doi: 10.1002/cncr.31316. [DOI] [PubMed] [Google Scholar]
- 130.Cremers RG, et al. Known susceptibility SNPs for sporadic prostate cancer show a similar association with "hereditary" prostate cancer. Prostate. 2015;75:474–483. doi: 10.1002/pros.22933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.San Francisco IF, et al. Association of RNASEL and 8q24 variants with the presence and aggressiveness of hereditary and sporadic prostate cancer in a Hispanic population. J. Cell Mol. Med. 2014;18:125–133. doi: 10.1111/jcmm.12171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zuhlke KA, et al. Identification of a novel germline SPOP mutation in a family with hereditary prostate cancer. Prostate. 2014;74:983–990. doi: 10.1002/pros.22818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ma Y, et al. Response to olaparib in metastatic castration-resistant prostate cancer with germline BRCA2 mutation: a case report. BMC Med. Genet. 2018;19:185. doi: 10.1186/s12881-018-0703-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Feiersinger GE, et al. Olaparib is effective in combination with, and as maintenance therapy after, first-line endocrine therapy in prostate cancer cells. Mol. Oncol. 2018;12:561–576. doi: 10.1002/1878-0261.12185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Clarke N, et al. Olaparib combined with abiraterone in patients with metastatic castration-resistant prostate cancer: a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2018;19:975–986. doi: 10.1016/S1470-2045(18)30365-6. [DOI] [PubMed] [Google Scholar]
- 136.Antonarakis ES. Abiraterone plus olaparib in prostate cancer: a new form of synthetic lethality? Lancet Oncol. 2018;19:860–861. doi: 10.1016/S1470-2045(18)30409-1. [DOI] [PubMed] [Google Scholar]
- 137.Hatcher D, Daniels G, Osman I, Lee P. Molecular mechanisms involving prostate cancer racial disparity. Am. J. Transl. Res. 2009;1:235–248. [PMC free article] [PubMed] [Google Scholar]
- 138.Paz YMC, et al. Positive association of the androgen receptor CAG repeat length polymorphism with the risk of prostate cancer. Mol. Med. Rep. 2016;14:1791–1798. doi: 10.3892/mmr.2016.5414. [DOI] [PubMed] [Google Scholar]
- 139.Mao X, et al. Involvement of different mechanisms for the association of CAG repeat length polymorphism in androgen receptor gene with prostate cancer. Am. J. Cancer Res. 2014;4:886–896. [PMC free article] [PubMed] [Google Scholar]
- 140.Figg WD, et al. Androgen receptor CAG repeat length and TMPRSS2:ETS prostate cancer risk: results from the Prostate Cancer Prevention Trial. Urology. 2014;84:127–131. doi: 10.1016/j.urology.2014.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Powell IJ. Germline CAG repeat length of the androgen receptor and time to progression in patients with prostate cancer treated with androgen deprivation therapy. BJU Int. 2011;108:1092. doi: 10.1111/j.1464-410X.2011.10541.x. [DOI] [PubMed] [Google Scholar]
- 142.Statement of the American Society of Clinical Oncology: genetic testing for cancer susceptibility. J. Clin. Oncol.14, 1730–1736 (1996). Adopted on February 20, 1996. discussion 1737–1740. [DOI] [PubMed]
- 143.Eisler I, et al. Training genetic counsellors to deliver an innovative therapeutic intervention: their views and experience of facilitating multi-family discussion groups. J. Genet. Couns. 2017;26:199–214. doi: 10.1007/s10897-016-0008-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Socio-Psychological Research in Genomics, C. et al. Developing an intervention to facilitate family communication about inherited genetic conditions, and training genetic counsellors in its delivery. Eur. J. Hum. Genet. 2016;24:794–802. doi: 10.1038/ejhg.2015.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.MacDonald C. Commercialisation of genetic services: the role of genetic counsellors. Hum. Reprod. Genet. Ethics. 2002;8:1–3. doi: 10.1179/hrge.8.1.jl0184m51pt22576. [DOI] [PubMed] [Google Scholar]
- 146.Middleton A, et al. The role of genetic counsellors in genomic healthcare in the United Kingdom: a statement by the Association of Genetic Nurses and Counsellors. Eur. J. Hum. Genet. 2017;25:659–661. doi: 10.1038/ejhg.2017.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Middleton A, et al. Position statement on opportunistic genomic screening from the Association of Genetic Nurses and Counsellors (UK and Ireland) Eur. J. Hum. Genet. 2014;22:955–956. doi: 10.1038/ejhg.2013.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Newman WG, Murphy BF, Callard A, Payne K. A role for genetic counsellors and clinical geneticists in pharmacogenetics? Clin. Genet. 2012;82:201–202. doi: 10.1111/j.1399-0004.2012.01872.x. [DOI] [PubMed] [Google Scholar]
- 149.African genetics for human society. Nat. Genet.50, 1495. https://www.nature.com/articles/s41588-018-0277 (2018). [DOI] [PubMed]
- 150.Donnai D, et al. Tensions in implementing the new genetics. Genetic counsellors could be based in genetic centres but be formally linked to general practice. BMJ. 2000;321:241. [PubMed] [Google Scholar]
- 151.El Shanti H, Chouchane L, Badii R, Gallouzi IE, Gasparini P. Genetic testing and genomic analysis: a debate on ethical, social and legal issues in the Arab world with a focus on Qatar. J. Transl. Med. 2015;13:358. doi: 10.1186/s12967-015-0720-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Alsaeed ES, et al. Distribution of hemoglobinopathy disorders in Saudi Arabia based on data from the premarital screening and genetic counseling program, 2011–2015. J. Epidemiol. Glob. Health. 2018;7(Suppl 1):S41–S47. doi: 10.1016/j.jegh.2017.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Begleiter ML. Training for genetic counsellors. Nat. Rev. Genet. 2002;3:557–561. doi: 10.1038/nrg844. [DOI] [PubMed] [Google Scholar]
- 154.Harris J, Ward SA. UK collaborative 1-day pilot information and support forum facilitated by a national breast cancer charity and NHS cancer genetic counsellors, for women at high risk, BRCA 1/2 gene carriers and hereditary breast cancer. Eur. J. Cancer Care. 2011;20:818–824. doi: 10.1111/j.1365-2354.2011.01273.x. [DOI] [PubMed] [Google Scholar]
- 155.Saudi Society of Medical Genetics. https://www.ssmg.org.sa/en/ (2019).
- 156.National society of Human Genetics. https://www.afshg.org/about/member-societies/egypt/ (2019).
- 157.National Society of Human Genetics—Egypt (NSHG). https://www.afshg.org/about/member-societies/egypt/ (2019).
- 158.Genetic Society of Israel. http://genetics-il.org/ (2019).
- 159.Canadian Association of Genetic Counsellors. https://www.cagc-accg.ca/ (2019).
- 160.Al-Dossary RN. The Saudi Arabian 2030 vision and the nursing profession: the way forward. Int. Nurs. Rev. 2018;65:484–490. doi: 10.1111/inr.12458. [DOI] [PubMed] [Google Scholar]
- 161.Bah S. How feasible is the life expectancy target in the Saudi Arabian vision for 2030? East Mediterr. Health J. 2018;24:401–404. doi: 10.26719/2018.24.4.401. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
No datasets were generated or analyzed during the current study.