Abstract
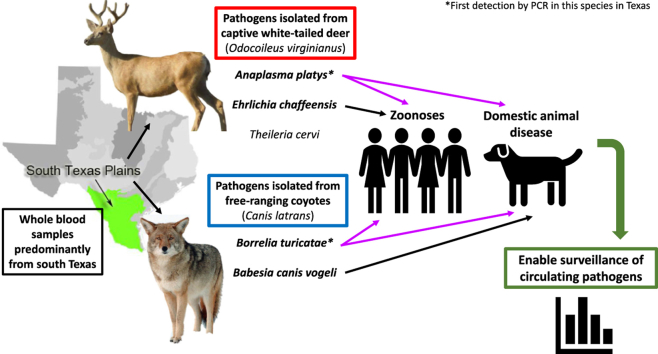
Determining which wildlife hosts are involved in the enzootic cycles of tick-borne diseases (TBD) enables enhanced surveillance and risk assessment of potential transmission to humans and domestic species. Currently, there is limited data to indicate which tick-borne pathogens (TBP) can infect coyotes. Additionally, limited surveillance data for white-tailed deer (WTD) in south Texas is available. The purpose of this study was to detect current infections of common TBP in coyotes and WTD in south Texas, which represents a transboundary region and common site for animal migrations across the U.S.-Mexico border. A patent pending real-time PCR assay, the TickPath layerplex test, was used to screen whole-blood samples for species from Borrelia, Rickettsia, Ehrlichia, Anaplasma, and Babesia genera. Conventional PCR and subsequent sequencing of positive samples confirmed the pathogen species. Of 122 coyote samples, 11/122 (9.0%) were positive for Babesia vogeli and 1/122 (0.8%) was positive for Borrelia turicatae. Of 245 WTD samples, 1/245 (0.4%) was positive for Anaplasma platys, 4/245 (1.6%) were positive for Ehrlichia chaffeensis, and 18/245 (7.3%) were positive for Theileria cervi. All positive samples from both species, except for one coyote, were collected from counties located in south Texas along the U.S.Mexico border. One coyote positive for B. vogeli originated from a county in northern Texas. The results from this study depicts the first known molecular detection of B. turicatae in a coyote, and demonstrates that coyotes and WTDs can potentially serve as sentinels for several zoonotic TBD as well as TBD that affect domestic animals.
Keywords: Anaplasma spp., Babesia spp., Borrelia turicatae, Theileria cervi, Wildlife, Zoonotic infections
Graphical abstract
Highlights
-
•
Coyote samples positive for Babesia vogeli and Borrelia turicatae.
-
•
WTD samples positive for Anaplasma platys, Ehrlichia chaffeensis, Theileria cervi.
-
•
First PCR detection of B. turicatae in a coyote.
-
•
First PCR detection of A. platys in WTD within Texas.
1. Introduction
Since year 2000, the incidence of tick-borne diseases (TBD) in the United States have consistently increased (Paddock et al., 2016). For example, cases of human granulocytic anaplasmosis caused by Anaplasma phagocytophilum have more than doubled from 2000 to 2007 (Dahlgren et al., 2011). Current data from the United States’ Center for Disease Control and Prevention (CDC) show that reported cases of Lyme disease (LD) have increased two-fold and cases of human monocytic ehrlichiosis caused by Ehrlichia chaffeensis have increased by at least five-fold from 2000 to 2016 (Rosenberg et al., 2018). While more sensitive and specific diagnostic tools and more accurate reports may contribute to the rise in recorded cases, an escalation in environmental disturbance may be influencing the rise in TBD. Specifically, increased anthropogenic environmental changes can favor increased abundance and density of wildlife populations, which encourages tick population expansion and increases in TBD (Paddock and Yabsley, 2007). Understanding how wildlife species may amplify TBD through propagating tick vectors and maintaining TBP in nature is pivotal to disease surveillance. To investigate the roles of wildlife in the enzootic cycles of zoonotic tick-borne pathogens (TBP) in Texas, this study evaluated molecular prevalence of TBP in coyotes (Canis latrans) and white-tailed deer (Odocoileus virginianus) (WTD). The study of TBP in these wildlife species has received little attention in areas of the southern U.S. and the state of Texas. Pathogens of interest include those responsible for Tick Borne Relapsing Fever (TBRF [Borrelia hermsii, B. parkeri, B. turicatae]), canine babesiosis (Babesia canis, B. gibsoni, B. conradae), and human/canine ehrlichiosis (E. canis, E. chaffeensis and E. ewingii) (Modarelli et al., 2019b).
Despite a paucity of data for WTD in Texas, the species has been widely studied in other areas of the U.S. and represents a clear example of wildlife dynamics directly impacting tick populations. WTD populations in the U.S. have boomed within the last few decades, with significant ecological effects (Hill, 2018; Nuttle et al., 2014). This population increase can be explained in part by the increase in available habitat for WTD due to human landscape modification (Paddock and Yabsley, 2007). Paddock and collaborators (Paddock et al., 2016) have also shown that increased WTD populations are closely linked with a rise in tick populations (e.g. Ixodes scapularis and Amblyomma americanum) that serve as competent vectors for many TBDs within the same habitat. In addition, WTD are the reservoir hosts for E. chaffeensis and are implicated as the reservoir hosts for E. ewingii (Lockhart et al., 1997; Yabsley et al., 2002). Both pathogens are transmitted by the lone star tick, Amblyomma americanum, which is found in southern and eastern Texas and feed on WTD throughout their life cycle (Kollars et al., 2000; Centers for Disease Control and Prevention, https://www.cdc.gov/ticks/geographic_distribution.html, Accessed August 26, 2019).
Limited data on the role of coyotes in TBD enzootic cycles in Texas hints at the species’ exposure to TBP, but has yet to implicate coyotes as TBP hosts. For example, it is unknown which species are the reservoir hosts of TBRF pathogens in Texas but Neotoma genus rats and wild canids such as coyotes are suspected (Bissett et al., 2018; Donaldson et al., 2016). Dworkin et al. (2008) and Lopez et al. (2016) suggest coyotes as a potential host species for B. turicatae based on seroprevalence studies in coyotes and infections in domestic dogs (Armstrong et al., 2018). Multiple cases of B. turicatae infection and exposure have been documented in domestic dogs in Texas, including dogs from south Texas (Esteve-Gasent et al., 2017; Modarelli et al., 2019b; Piccione et al., 2016; Whitney et al., 2007). The potential role of coyotes in TBRF pathogen cycles is supported by results from Armstrong et al. (2018) where 10.1% of sampled coyotes were seropositive for B. turicatae. Despite detectable seropositivity, active TBRF pathogen infections have not been molecularly detected in coyotes. Additional data surrounding coyotes and TBP are limited to seroprevalence studies. Coyotes sampled in Oklahoma and Texas have documented exposure to zoonotic TBP such as Rickettsia rickettsii, E. canis, E. ewingii, and E. chaffeensis; the latter pathogen has also been isolated from free-ranging coyotes by PCR (Kocan et al., 2000; Paras et al., 2012; Starkey et al., 2013). Other potentially domestic and wildlife significant TBP include those responsible for canine babesiosis. The high incidence of babesiosis in dogfighting rings suggests that transmission can occur through blood-blood exposure as a result of bites and not limited to tick vectors (Cannon et al., 2016; Yeagley et al., 2009). The established direct route of transmission may be of significance due to the potentially violent interaction of dogs and coyotes at the domestic-wildlife interface that could lead to cross-species infections.
Thus, in the context of current evidence, our overall hypothesis for this study was that WTD and coyotes are essential species in the maintenance of zoonotic TBD that have been previously reported in humans and domestic dogs in Texas. The objective for this study is to evaluate the molecular prevalence of TBP within WTD and coyote samples collected in southern Texas. As many TBDs that affect wildlife species are zoonotic, determining the prevalence of TBD in wildlife can enable more accurate predictions regarding transmission to human and domestic animal populations.
2. Methods
2.1. Sample collection
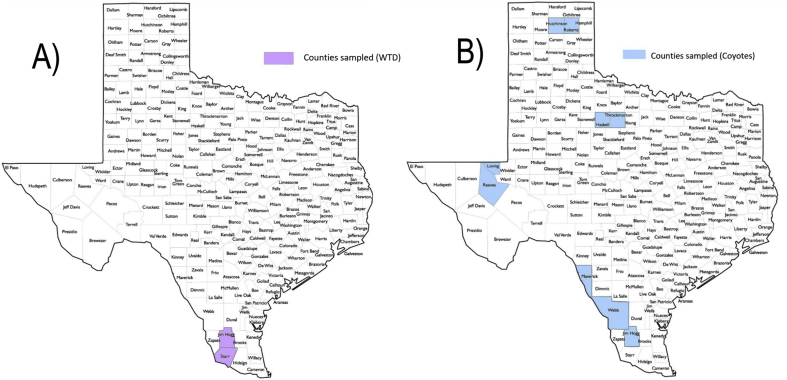
A total of 122 coyote and 245 WTD EDTA-whole blood samples were collected for testing. Coyote blood samples were collected from February–April 2016 and December 2016 from nine different counties located mainly in south Texas. Of the coyote samples, 100 were collected from Webb, Maverick, and Jim Hogg counties in south Texas, while 18 came from Haskell, Throckmorton, Hutchinson, and Roberts counties in north Texas, and 4 from Reeves and Loving county in west Texas (Fig. 1B). Deer blood samples were collected during November 2016 from the East Foundation's San Antonio Viejo Ranch in Jim Hogg and Starr counties (Fig. 1A). The ranch spans an area of 60,179 ha (148,705 acres). Blood samples from both species were collected by other researchers under their respective Animal Use Protocols and blood samples were provided for testing. No tick collection or analysis was performed. WTD samples were collected under Texas A&M Kingsville AUP # 2017-09-22-A2. All experiments were conducted under the Texas A&M University institutional biosafety permit IBC#2016–051.
Fig. 1.
Texas counties where samples were collected. (A) Purple denotes Jim Hogg and Starr counties where WTD samples were collected from the East Foundation's San Antonio Viejo Ranch while coyote samples originated from counties highlighted in blue (B). (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)
2.2. DNA extractions
Coyote EDTA-whole blood samples, in aliquots of 200 μL, were DNA purified using the High Pure PCR Template Preparation Kit (Roche, Indianapolis, IN) following manufacturer's instructions with the following modifications. Proteinase K was added to the sample and then vortexed before binding buffer was added. Furthermore, the final drying step was extended to 30 s to decrease potential ethanol contamination. The concentration and 260:280 ratios of extracted DNA were then determined using a microplate reader (Take 3, Synergy H1, BioTek, Shoreline, WA). WTD blood samples were DNA purified using the MagMAX™ Nucleic Acid Isolation Kit AMB1836 (ThermoFisher, Waltham, MA) following manufacturer's instructions from a previous publication (Schroeder et al., 2013). All extracted DNA was stored at −20 °C until use.
2.3. PCR analysis
The TickPath layerplex qPCR test (Patent Application Serial No. 16/130,177) was used to screen all samples (Modarelli et al., 2019a). Specifically, samples were screened for Borrelia turicatae, B. parkeri, B. hermsii (genomic groups I and II), B. burgdorferi sensu lato, Rickettsia rickettsii, Ehrlichia canis, E. chaffeensis, E. ewingii, Anaplasma phagocytophilum, and pan-specific Babesia spp (B. canis, B. vogeli, B. gibsoni, B. bovis, B. microti, B. caballi). The sensitivity and specificity values (and 95% confidence intervals) are 100% (86.8–100%) and 99.8% (99.4–99.9%) for the borrelial layer, 100% (90.5–100%) and 99.1% (98.4–99.5%) for the rickettsial layer, and 100% (47.8–100%) and 100% (99.7–100%) for the babesial layers. The qPCR was performed using the BioRad CFX 96 system (Bio-Rad). Cycling conditions (thermal profile) consisted of activation and denaturation at 95 °C for 3 min (1 cycle), and 40 cycles of amplification at 95 °C for 10 s and 60 °C for 45 s. Samples were considered positive if the quantification cycle (Cq) ≤ 38 and were confirmed through conventional PCR and Sanger sequencing.
The conventional PCR protocols were used for confirmation of any positive or suspected positive samples according to prior published protocols (Table 1) (Anderson et al., 1992; Bunikis et al., 2004; Davitkov et al., 2015; Dawson et al., 1994, 1996; Wen et al., 1997). To detect Theileria cervi, TcerviF (5′-TTCCCTTTGAGGGGT-3′) and TcerviR (5′-GAAGCCTATTCCCGTACCC-3′) primers targeting the 18S rRNA gene were used. PCR's for T. cervi were performed in 25 μL reactions containing 12.5 μL Accustart II Supermix Buffer (Quantabio, Beverly, MA), 7.5 μL of PCR-grade water, and 3 μL of template DNA and 1 μL of each primer (2.5 μM concentration). The PCR cycling parameters were: initial DNA denaturation of 3 min at 94 °C followed by 45 cycles of 30 s at 94 °C, 30 s at 55 °C, 1 min at 72 °C, and finished with a final extension step at 72 °C for 2 min. PCR's were performed using the Mastercycler™ pro (Eppendorf, Inc.). Positive DNA amplicons were then purified using the Wizard® SV Gel and PCR Clean-Up System (Promega, Madison, WI). Purified DNA amplicons were sequenced in both directions to produce consensus sequences (Eurofins Scientific, Louisville, KY). Sequences were then analyzed using MacVector (MacVector, Inc. Apex, NC) and identified by comparison with published sequences on the National Center for Biotechnology Information (NCBI) database using the Basic Local Alignment Search Tool (BLAST®). All identified sequences were uploaded to GenBank ®.
Table 1.
Primers utilized for confirmatory PCR testing.
| Pathogen | Gene Target | Primers | Primer Sequence | Reference |
|---|---|---|---|---|
| Ehrlichia/Anaplasma spp. | 16SrRNA | ECC/ECB | 5′-AGAACGAACGCTGGCGGCAAGCC-3′ 5′-CGTATTACCGCGGCTGCTGGC-3′ |
Dawson et al. (1994) |
| Ehrlichia canis | 16SrRNA | ECA/HE3 | 5′-CAATTATTTATAGCCTCTGGCTATAGGAA-3′ 5′-TATAGGTACCGTCATTATCTTCCCTAT-3′ |
Dawson et al. (1996) |
| Ehrlichia chaffeensis | 16SrRNA | HE1/HE3 | 5′-CAATTGCTTATAACCTTTTGGTTATAAAT-3′ 5′-TATAGGTACCGTCATTATCTTCCCTAT-3′ |
Anderson et al. (1992) |
| Anaplasma spp. | rrs | 16SANAF/16SANAR | 5′-CAGAGTTTGATCCTGGCTCAGAACG-3′ 5′-GAGTTTGCCGGGACTTCTTCTGTA-3′ |
Silaghi et al. (2017) |
| Borrelia spp. | rrs-rrlA (23S/5S rRNA genes) | rrs-rrlA-F/rrs-rrlA-R | 5′-GGTATTTAAGGTATGTTTAGTGAG-3′ 5′-GGATCATAGCTCAGGTGGTTAG-3′ |
Bunikis et al. (2004) |
| rrs-rrlA-Fn/rrs-rrlA-Rn | 5′-GGTGAAGTCGTAACAAGGTAG-3′ 5′-GTCTGATAAACCTGAGGTCGG A-3′ |
|||
| Babesia spp. | 18SrRNA | PIRO-A/PIRO-B | 5′-AATACCCAATCCTGACACAGGG-3′ 5′-TTAAATACGAAT GCCCCCAAC-3′ |
Davitkov et al. (2015) |
| Piroplasm genus | 18SrRNA | A/B | 5′-ACCTGGTTGATCCTGCCAG-3′ 5′-GATCCTTCTGCAGGTTCACCTAC-3′ |
Sogin (1990) |
| Theileria cervi | 18SrRNA | TcerviF/TcerviR | 5′-TTCCCTTTGAGGGGT-3′ 5′-GAAGCCTATTCCCGTACCC-3′ |
This study |
2.4. Space use analysis
Ten coyotes from the East Foundation's San Antonio Viejo Ranch that were sampled in this study were also GPS collared and their movements studied. Each of these coyotes was captured on December 10, 2016 via helicopter and net gun (Gese et al., 1987). They were each fit with a Vertex Plus GPS collar (Vectronic Aerospace GmbH, Berlin) programmed to record their location hourly for one year. The annual range for each coyote was estimated as the 75% isopleth of a fixed kerned density estimate of their utilization distribution (Worton, 1989). This demarks the area within which each coyote spent 75% of the monitoring period.
3. Results
3.1. Molecular findings
Out of 122 coyote samples, 11/122 (9.0%) were positive for Babesia vogeli and 1/122 (0.8%) was positive for Borrelia turicatae. No co-infections were detected. The identified B. vogeli 18S rRNA sequences were 100% identical to sequences published in GenBank® from domestic dog samples (KY290979.1, MF459002.1). The B. turicatae isolate was found to be 100% identical to strain BTE5EL, which was originally isolated from a human in Texas (CP015629.1) (Bissett et al., 2018). GenBank® accession numbers of the newly generated sequences are indicated in Table 2.
Table 2.
GenBank® accession numbers of sequences generated in this study.
| Host | Pathogen | Gene Target | GenBank® Accession | County (positive) |
|---|---|---|---|---|
| Coyote | Babesia vogeli | 18SrRNA | MK611613-MK611623 | Jim Hogg (1/11), Maverick (5/11), Webb (4/11), Throckmorton (1/11) |
| Borrelia turicatae | rrs-rrl (23S/5S rRNA genes) | MK615613 | Maverick | |
| WTD | Theileria cervi | 18SrRNA | MK611629-MK611644 | Jim Hogg/Starr |
| Ehrlichia chaffeensis | 16SrRNA | MK611625-MK611628 | ||
| Anaplasma platys | 16SrRNA | MK611624 |
From 245 WTD samples, 1/245 (0.41%) was positive for Anaplasma platys, 4/245 (1.6%) were positive for Ehrlichia chaffeensis, and 18/245 (7.3%) were positive for Theileria cervi. One deer indicated a co-infection with E. chaffeensis and T. cervi. The A. platys sequence was found to be 99% identical to isolate YY36 (MF289478.). All 4 E. chaffeensis positive samples were sequenced and found to be 100% identical to the Arkansas strain (NR_074500.2). Of the 18 T. cervi samples 8 positive samples were selected for sequencing and were found to be 100% identical to the 28S rRNA gene of Wisconsin elk 1 clone isolate (AY735135.1). GenBank® accession numbers of the newly generated sequences are indicated in Table 2.
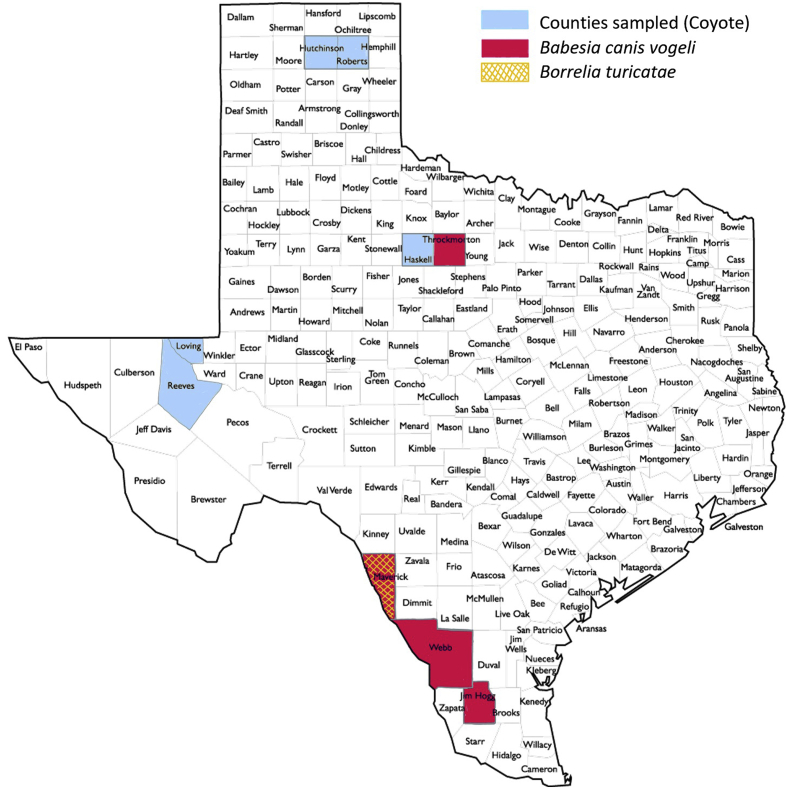
Positive samples for both species, except for one coyote, were collected from counties located in south Texas along the U.S.-Mexico border. One coyote positive for B. vogeli originated from northern Texas. The 1 coyote positive for B. turicatae originated from Maverick county, where 5/11 B. vogeli positive coyote were also located. The remaining 6/11 B. vogeli positive coyote originated from Webb (4/6), Jim Hogg (1/6), and Throckmorton (1/6) counties (Fig. 2).
Fig. 2.
Geographic representation of study area and molecular prevalence of tick-borne pathogens in coyotes of Texas.
3.2. Telemetry data
Although ten coyotes from the East Foundation's San Antonio Viejo Ranch were GPS collared, annual home ranges could not be calculated for two animals because they did not survive the year. In comparing annual ranges in Table 3, ranges varied from 210 to 5141 hectares (ha), with an average range of 1650 ha.
Table 3.
Calculated annual home ranges for eight coyotes sampled on the San Antonio Viejo Ranch, based on 75% fixed KDE.
| Coyote ID Number | KDE (hectares) |
|---|---|
| 20440-M01 | 2157.73 |
| 20441-F02 | 3745.51 |
| 20439-M03 | 210.48 |
| 20442-M04 | 310.37 |
| 20443-F06 | 1042.74 |
| 20444-F08 | 5141.86 |
| 20424-M09 | 288.09 |
| 20445-M10 | 287.75 |
4. Discussion
4.1. Significance
In this study multiple pathogens that affect humans and domestic animals were detected from coyotes and WTD, two important wildlife species in Texas. Detection of E. chaffeensis from WTD aligned with data that the species serve as reservoirs for the pathogen. However, detecting A. platys from a WTD in Texas was unexpected. Two prior studies have reported detection of A. platys or a closely related Anaplasma sp. from WTD, though neither population included Texas WTD (Munderloh et al., 2003; Rankins et al., 2017). In regard to coyotes, previous studies have mostly focused on seroprevalence, and existing molecular prevalence studies have not sampled coyotes from the south Texas region. Specifically, this is the first molecular detection of a TBRF pathogen from a coyote in a transboundary region of the U.S., and complements past serosurveillance studies conducted on coyotes in Texas (Armstrong et al., 2018). Thus, the current study revealed novel information about what pathogens coyotes and WTD may harbor in a region where exchange of pathogens and vectors across the U.S.-Mexico border is possible.
4.2. WTD and south Texas TBD enzootic cycles
To expand upon the findings in WTD, it is important to note that while the species has been studied extensively in other TBD enzootic cycles and different regions of the U.S., they have not been studied as closely in Texas. Texas, while not traditionally known as a TBD hotspot as compared to the Northeastern U.S., represents a state where WTD populations are prominent (https://tpwd.texas.gov/landwater/land/habitats/trans_pecos/big_game/wtd/, Accessed August 26, 2019). The extensive Texas WTD population may influence an increased TBD risk to humans and companion animals. Detection of two zoonotic TBDs, E. chaffeensis and A. platys, which have recently contributed to human and companion animal infections (Arraga-Alvarado et al., 2014; Breitschwerdt et al., 2014; Maggi et al., 2013; Modarelli et al., 2019b), suggests that WTD are involved in TBD enzootic cycles in south Texas.
The detection of E. chaffeensis within this study reinforces prior findings that WTD act as the main reservoir host species for the pathogen and confirms presence of this pathogen within Texas (Paddock and Childs, 2003). Further, this study establishes the first detection of A. platys from a WTD in the state of Texas. Prior to this study, WTD infection with A. platys has only been shown in one other study conducted in Alabama (Rankins et al., 2017). An additional study conducted in Georgia, detected an Anaplasma spp. pathogen most closely related to A. platys from wild WTD (Munderloh et al., 2003). This uncharacterized Anaplasma spp. pathogen has also been detected from several states that border Texas, such as Oklahoma, Louisiana, and Arkansas. Thus, geographically it was noteworthy to confirm A. platys in a WTD from south Texas. Currently, domestic dogs are implicated as the natural host of A. platys; several human infections have been documented, though it should be noted that the host range of the pathogen is largely unknown (Arraga-Alvarado et al., 2014; Breitschwerdt et al., 2014; Maggi et al., 2013). Modarelli et al., 2019a, Modarelli et al., 2019b demonstrated the first molecular detection of A. platys in Texas after detecting the pathogen in 2/1171 domestic dogs sampled. This study noted that all A. platys infections were incidentally found alongside coinfections with Ehrlichia canis, and that the TickPath layerplex assay utilized in the study does not test specifically for A. platys. Additional screening of WTD samples with an A. platys specific assay is recommended for future studies to alleviate any coinfection discrepancies and to more accurately determine the pathogen prevalence.
These recent findings may signify that the pathogen has a wider host range than previously reported and detection of the pathogen in south Texas may represent an expanded pathogen range. The potential of WTD as a competent host for A. platys has many implications, however further studies need to be performed before the potential risks can be assessed. Overall, detecting A. platys in a wildlife species from a region along the U.S.-Mexico border may have future implications for zoonotic pathogen surveillance in this international zone (Esteve-Gassent et al., 2014, 2016).
The hemoparasite Theileria cervi, was detected from 18/245 (7.35%) WTD. While the TickPath assay was not initially validated to screen for T. cervi, the assay's ability to consistently detect the hemoparasite was demonstrated through this study due to genetic similarity to other Babesia spp. These findings also corroborate prior studies that established T. cervi infections in Texas WTD populations, and primers specific for the parasite were used for confirmation testing (Waldrup et al., 1992). T. cervi is currently understood to be a non-pathogenic hemoparasite that is transmitted by A. americanum ticks (Kuttler et al., 1967). While T. cervi is non-pathogenic, its presence in the WTD samples demonstrates that A. americanum are present in the south Texas region and are actively feeding on deer. This information is noteworthy as A. americanum can transmit additional pathogens previously discussed, both zoonotic and those affecting domestic animals.
4.3. Novel findings in coyotes from south Texas
This study represents the first molecular detection of a TBRF pathogen in a coyote. The one coyote positive for Borrelia turicatae originated from Maverick county. Maverick county is adjacent to Webb county, which appears to be a consistent area of exposure, as B. turicatae seropositive domestic dogs and coyotes have also been sampled from within the county (Esteve-Gasent et al., 2017; Armstrong et al., 2018). In the case of seropositive coyotes, Armstrong et al. (2018) found that the majority of seropositive coyotes came from Webb county and the counties directly adjacent (Zapata and Dimmit counties). The repeated positive findings in coyotes implicates this geographical region of southern Texas as a potentially higher risk area for exposure to B. turicatae.
In addition to detecting B. turicatae, it was also important to detect Babesia vogeli, in the sampled coyotes. However, only B. gibsoni was detected in domestic dogs sampled in Texas (Modarelli et al., 2019a, Modarelli et al., 2019b). It is probable that the difference in Babesia species infecting wild versus domestic canids is due to the limited number of coyotes sampled for this study, and that most samples were collected in south Texas. These findings warrant further testing of coyotes and sample collection from different regions of Texas. A more comprehensive view of TBP in coyotes is required before the difference between coyote and domestic dog Babesia infections can be determined. Therefore, a hypothesized reason for the difference will be discussed. It is possible that the mode of transmission can play a role in why coyotes may be infected with B. vogeli instead of B. gibsoni and vice versa for dogs. B. vogeli is vectored by Rhipicephalus sanguineus while the tick vectors of B. gibsoni are not endemic to the U.S. (Baneth, 2018). It should be noted that in 2017, Haemaphysalis longicornis tick, one of the established vectors for this pathogen, was identified in New Jersey; nevertheless, the implications for B. gibsoni ecology are undetermined (Burtis et al. (2018); USDA-APHIS, 2019). However, B. gibsoni can be transmitted by direct contact with blood such as trans-placentally, blood transfusions, dog bites, and sharing surgical equipment (Cannon et al., 2016; Yeagley et al., 2009). B. vogeli, on the other hand, is not commonly isolated from dogs with babesiosis with those histories. Coyotes are more likely to be in close contact with ticks rather than other forms of horizontal transmission that domestic dogs are exposed to, although in-fighting between coyotes, such as over carrion, has been documented (Bekoff and Wells, 1986). Thus, it is possible that B. vogeli is more restricted to vector-borne transmission and B. gibsoni is maintained within the dog population through non-vectored transmission. The transmission of either Babesia spp. between wild coyotes and domestic dogs is possible as R. sanguineus ticks readily feed on domestic dogs in rural and urban settings (Dantas-Torres et al., 2012). Direct routes of transmission between coyotes and domestic dogs is also probable, as coyotes are a constant presence in urban landscapes. It is possible that coyotes highly habituated to urban areas would facilitate vector transfer between hosts through indirect routes such as shared common spaces for urination/defecation as domestic dogs (Gehrt et al., 2009). Although infrequently, fighting with domestic dogs may also occur, leading to direct transmission through bite wounds (Greenspan et al., 2018). Direct contact between these two species occurs more frequently in rural communities and ranches than urban landscapes and it is important to note that direct contact between the species may be instigated by either coyotes or domestic dogs. Therefore, coyotes may play a key role in the direct and indirect transmission and propagation of canine TBP. Future studies should aim to clarify routes of transmission for each pathogen species and to further elucidate the role of coyotes in TBP enzootic cycles in Texas.
4.4. Conclusion
Detection of the TBP's discussed above suggests that the tick vectors are also present in the regions where the animals were sampled. While it does not identify the ticks themselves, this information could be useful towards constructing a preliminary guide to what ticks and pathogens may be circulating in south Texas. However, because wild animals can roam, it cannot be assumed that the collection location of blood samples is the same as the area where animals were infected. This is especially relevant in the case of coyotes. Annual ranges of our collared coyotes varied between 210 and 5141 hectares (ha) with an average range of 1650 ha (Table 3). Thus, the potential geographical range of the detected pathogens and their tick vectors may be larger than just the counties where positive samples originated. The risk of exposure within the counties where positive samples were collected warrants further research. Additionally, as most of the positive coyote samples were collected from counties along the U.S.-Mexico border, the free-roaming coyotes may have acquired or carried the pathogens across the border. This emphasizes that wildlife studies may impact zoonotic pathogen surveillance in border areas, as highlighted by novel findings in WTD from our study (Esteve-Gassent et al., 2014, 2016).
Furthermore, the novel molecular detection of pathogens such as B. turicatae and A. platys demonstrates that wildlife may harbor more pathogens than previously expected. Detection of multiple zoonotic TBPs highlights the connectedness of wildlife, domestic animal, and human health. In the case of coyotes found in Webb County, active surveillance for TBRF pathogens would better inform our understanding of multi-species infection risks. This study represents preliminary molecular data which future studies should build upon to increase surveillance of TBD not only in wildlife species, but also in humans and domestic species.
Declaration of competing interest
The authors do not have any conflicts of interest to declare.
Acknowledgements
Funding: Research reported in this publication was supported by the United States Department of Health and Human Services, Public Health Service of the National Institutes of Health under award number T35OD010991. Thank you to Dr. Randy DeYoung for the opportunity to collect blood samples from the deer that were captured under his own research project. We would also like to thank Dr. Walt Cook for transporting collected blood samples to TVMDL and Dr. Patricia Holman for her recommendations in the detection of Babesia and Theileria species. This is manuscript number 042 of the East Foundation.
References
- Anderson B.E., Greene C.E., Jones D.C., Dawson J.E. Ehrlichia ewingii sp. nov., the etiologic agent of canine granulocytic ehrlichiosis. Int. J. Syst. Bacteriol. 1992;42:299–302. doi: 10.1099/00207713-42-2-299. [DOI] [PubMed] [Google Scholar]
- Armstrong B.A., Kneubehl A., Krishnavajhala A., Wilder H.K., Boyle W., Wozniak E., Phillips C., Hollywood K., Murray K.O., Donaldson T.G., Teel P.D., Waldrup K., Lopez J.E. Seroprevalence for the tick-borne relapsing fever spirochete Borrelia turicatae among small and medium sized mammals of Texas. PLoS Neglected Trop. Dis. 2018;12 doi: 10.1371/journal.pntd.0006877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraga-Alvarado C.M., Qurollo B.A., Parra O.C., Berrueta M.A., Hegarty B.C., Breitschwerdt E.B. Case report: molecular evidence of Anaplasma platys infection in two women from Venezuela. Am. J. Trop. Med. Hyg. 2014;91:1161–1165. doi: 10.4269/ajtmh.14-0372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baneth G. Antiprotozoal treatment of canine babesiosis. Vet. Parasitol. 2018;254:58–63. doi: 10.1016/j.vetpar.2018.03.001. [DOI] [PubMed] [Google Scholar]
- Bekoff M., Wells M.C. Social ecology and behavior of coyotes. Adv. Stud. Behav. 1986;16:251–338. [Google Scholar]
- Bissett J.D., Ledet S., Krishnavajhala A., Armstrong B.A., Klioueva A., Sexton C., Replogle A., Schriefer M.E., Lopez J.E. Detection of tickborne relapsing fever spirochete, austin, Texas, USA. Emerg. Infect. Dis. 2018;24 doi: 10.3201/eid2411.172033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitschwerdt E.B., Hegarty B.C., Qurollo B.A., Saito T.B., Maggi R.G., Blanton L.S., Bouyer D.H. Intravascular persistence of Anaplasma platys, Ehrlichia chaffeensis, and Ehrlichia ewingii DNA in the blood of a dog and two family members. Parasites Vectors. 2014;7 doi: 10.1186/1756-3305-7-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bunikis J., Tsao J., Garpmo U., Berglund J., Fish D., Barbour A.G. Typing of Borrelia relapsing fever group strains. Emerg. Infect. Dis. 2004;10:1661–1664. doi: 10.3201/eid1009.040236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burtis J., Egizi A., Occi J., Mader E., Lejeune M., Stafford K., Harrington L. 2018. Intruder Alert: Longhorned Tick. [Google Scholar]
- Cannon S.H., Levy J.K., Kirk S.K., Crawford P.C., Leutenegger C.M., Shuster J.J., Liu J., Chandrashekar R. Infectious diseases in dogs rescued during dogfighting investigations. Vet. J. 2016;211:64–69. doi: 10.1016/j.tvjl.2016.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlgren F.S., Mandel E.J., Krebs J.W., Massung R.F., McQuiston J.H. Increasing incidence of Ehrlichia chaffeensis and Anaplasma phagocytophilum in the United States, 2000-2007. Am. J. Trop. Med. Hyg. 2011;85:124–131. doi: 10.4269/ajtmh.2011.10-0613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dantas-Torres F., Chomel B.B., Otranto D. Ticks and tick-borne diseases: a One Health perspective. Trends Parasitol. 2012;28:437–446. doi: 10.1016/j.pt.2012.07.003. [DOI] [PubMed] [Google Scholar]
- Davitkov D., Vucicevic M., Stevanovic J., Krstic V., Tomanovic S., Glavinic U., Stanimirovic Z. Clinical babesiosis and molecular identification of Babesia canis and Babesia gibsoni infections in dogs from Serbia. Acta Vet. Hung. 2015;63:199–208. doi: 10.1556/AVet.2015.017. [DOI] [PubMed] [Google Scholar]
- Dawson J.E., Biggie K., Warner C., Cookson K., Jenkins S., Levine J., Olson J. Polymerase chain reaction evidence of Ehrlichia chaffeensis, an etiologic agent of human ehrlichiosis, in dogs from southeast Virginia. Am. J. Vet. Res. 1996;57:1175–1179. [PubMed] [Google Scholar]
- Dawson J.E., Stallknecht D.E., Howerth E.W., Warner C., Biggie K., Davidson W.R., Lockhart J.M., Nettles V.F., Olson J., Childs J.E. Susceptibility of white-tailed deer (Odocoileus virginianus) to infection with Ehrlichia chaffeensis, the etiologic agent of human ehrlichiosis. J. Clin. Microbiol. 1994;32:2725–2728. doi: 10.1128/jcm.32.11.2725-2728.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson T.G., Perez de Leon A.A., Li A.Y., Castro-Arellano I., Wozniak E., Boyle W.K., Hargrove R., Wilder H.K., Kim H.J., Teel P.D., Lopez J.E. Assessment of the geographic distribution of ornithodoros turicata (argasidae): climate variation and host diversity. PLoS Neglected Trop. Dis. 2016;10 doi: 10.1371/journal.pntd.0004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin M.S., Schwan T.G., Anderson D.E., Jr., Borchardt S.M. Tick-borne relapsing fever. Infect. Dis. Clin. N. Am. 2008;22:449–468. doi: 10.1016/j.idc.2008.03.006. viii. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Gassent M.D., Perez de Leon A.A., Romero-Salas D., Feria-Arroyo T.P., Patino R., Castro-Arellano I., Gordillo-Perez G., Auclair A., Goolsby J., Rodriguez-Vivas R.I., Estrada-Franco J.G. Pathogenic landscape of transboundary zoonotic diseases in the Mexico-US border along the Rio Grande. Front Public Health. 2014;2:177. doi: 10.3389/fpubh.2014.00177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Gassent M.D., Castro-Arellano I., Feria-Arroyo T.P., Patino R., Li A.Y., Medina R.F., de Leon A.A., Rodriguez-Vivas R.I. Translating ecology, physiology, biochemistry, and population genetics research to meet the challenge of tick and tick-borne diseases in north America. Arch. Insect Biochem. Physiol. 2016;92:38–64. doi: 10.1002/arch.21327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esteve-Gasent M.D., Snell C.B., Adetunji S.A., Piccione J. Serological detection of tick-borne relapsing fever in texan domestic dogs. PLoS One. 2017;12 doi: 10.1371/journal.pone.0189786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehrt S.D., Anchor C., White L.A. Home range and landscape use of coyotes in a metropolitan landscape: conflict or coexistence? J. Mammal. 2009;90:1045–1057. [Google Scholar]
- Gese E.M., Rongstad O.J., Mytton W.R. Manual and net-gun capture of coyotes from helicopters. Wildl. Soc. Bull. 1987;15:444–445. [Google Scholar]
- Greenspan E., Nielsen C.K., Cassel K.W. 2018. Potential Distribution of Coyotes (Canis latrans), Virginia Opossums (Didelphis virginiana), Striped Skunks (Mephitis mephitis), and Raccoons (Procyon lotor) in the Chicago Metropolitan Area. Urban Ecosystems. [Google Scholar]
- Hill O.R. Texas A&M University-Kingsville, ProQuest Dissertations Publishing; 2018. White-tailed Deer Population and Supplemental Feeder Density Effects on Woody Vegetation in South Texas, Range and Wildlife Management. [Google Scholar]
- Kocan A., Crowder Levesque G., Whitworth L.C., Murphy G.L., Ewing S.A., Barker R.W. Naturally occurring Ehrlichia chaffeensis infection in coyotes from Oklahoma. Emerg. Infect. Dis. 2000;6:477–480. doi: 10.3201/eid0605.000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kollars T.M., Oliver J.H., Durden L.A., Kollars P.G. Host associations and seasonal activity of Amblyomma americanum (Acari: ixodidae) in Missouri. J. Parasitol. 2000;86:1156–1159. doi: 10.1645/0022-3395(2000)086[1156:HAASAO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Kuttler K.L., Robinson R.M., Bell R.R. Tick transmission of theileriasis in a white-tailed deer. J. Wildl. Dis. 1967;3:182–183. [Google Scholar]
- Lockhart J.M., Davidson W.R., Dawson J.E., Stallknecht D.E., Howerth E.W. Isolation of Ehrlichia chaffeensis from wild white-tailed deer (Odocoileus virginianus) confirms their role as natural reservoir hosts. J. Clin. Microbiol. 1997;35:1681–1686. doi: 10.1128/jcm.35.7.1681-1686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J.E., Krishnavahjala A., Garcia M.N., Bermudez S. Tick-borne relapsing fever spirochetes in the americas. Vet Sci. 2016;3 doi: 10.3390/vetsci3030016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggi R.G., Mascarelli P.E., Havenga L.N., Naidoo V., Breitschwerdt E.B. Co-infection with Anaplasma platys, Bartonella henselae and Candidatus Mycoplasma haematoparvum in a veterinarian. Parasites Vectors. 2013;6 doi: 10.1186/1756-3305-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarelli J.J., Ferro P.J., de Leon A.A.P., Esteve-Gasent M.D. TickPath Layerplex: adaptation of a real-time PCR methodology for the simultaneous detection and molecular surveillance of tick-borne pathogens. Sci. Rep. 2019;9:6950. doi: 10.1038/s41598-019-43424-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modarelli J.J., Tomecek J.M., Piccione J., Ferro P.J., Esteve-Gasent M.D. Molecular prevalence and ecoregion distribution of select tick-borne pathogens in Texas dogs. Transbound Emerg Dis. 2019;66:1291–1300. doi: 10.1111/tbed.13145. [DOI] [PubMed] [Google Scholar]
- Munderloh U.G., Tate C.M., Lynch M.J., Howerth E.W., Kurtti T.J., Davidson W.R. Isolation of an Anaplasma sp. organism from white-tailed deer by tick cell culture. J. Clin. Microbiol. 2003;41:4328–4335. doi: 10.1128/JCM.41.9.4328-4335.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuttle T., Ristau T.E., Royo A.A., Gilliam F. Long-term biological legacies of herbivore density in a landscape-scale experiment: forest understoreys reflect past deer density treatments for at least 20 years. J. Ecol. 2014;102:221–228. [Google Scholar]
- Paddock C.D., Childs J.E. Ehrlichia chaffeensis: a prototypical emerging pathogen. Clin. Microbiol. Rev. 2003;16:37–64. doi: 10.1128/CMR.16.1.37-64.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paddock C.D., Lane R.S., Staples J.E., Labruna M.B. Workshop Summary; Washington (DC): 2016. Changing Paradigms for Tick-Borne Diseases in the Americas, Global Health Impacts of Vector-Borne Diseases; pp. 245–282. [Google Scholar]
- Paddock C.D., Yabsley M.J. Ecological havoc, the rise of white-tailed deer, and the emergence of Amblyomma americanum -associated Zoonoses in the United States. In: Childs James E., M J.S., Richt Jürgen A., editors. Curr Top Microbiol Immunol. 2007. pp. 289–324. [DOI] [PubMed] [Google Scholar]
- Paras K.L., Little S.E., Reichard M.V., Reiskind M.H. Detection of Dirofilaria immitis and Ehrlichia species in coyotes (Canis latrans), from rural Oklahoma and Texas. Vector Borne Zoonotic Dis. 2012;12:619–621. doi: 10.1089/vbz.2011.0815. [DOI] [PubMed] [Google Scholar]
- Piccione J., Levine G.J., Duff C.A., Kuhlman G.M., Scott K.D., Esteve-Gassent M.D. Tick-borne relapsing fever in dogs. J. Vet. Intern. Med. 2016;30:1222–1228. doi: 10.1111/jvim.14363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rankins S., Zohdy S., Ditchkoff S., Kaltenboeck B., Gao D., Zikeli S. Auburn University Journal of Undergraduate Scholarship; 2017. High Prevalence of Anaplasma Platys Infection in Alabama White-Tailed Deer. [Google Scholar]
- Rosenberg R., Lindsey N.P., Fischer M., Gregory C.J., Hinckley A.F., Mead P.S., Paz-Bailey G., Waterman S.H., Drexler N.A., Kersh G.J., Hooks H., Partridge S.K., Visser S.N., Beard C.B., Petersen L.R. 2018. Vital Signs: Trends in Reported Vectorborne Disease Cases — United States and Territories, 2004–2016, Morbidity and Mortality Weekly Report; pp. 496–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroeder M.E., Johnson D.J., Ostlund E.N., Meier J., Bounpheng M.A., Clavijo A. Development and performance evaluation of a streamlined method for nucleic acid purification, denaturation, and multiplex detection of Bluetongue virus and Epizootic hemorrhagic disease virus. J. Vet. Diagn. Investig. 2013;25:709–719. doi: 10.1177/1040638713503654. [DOI] [PubMed] [Google Scholar]
- Silaghi C., Santos A.S., Gomes J., Christova I., Matei I.A., Walder G., Domingos A., Bell-Sakyi L., Sprong H., von Loewenich F.D., Oteo J.A., de la Fuente J., Dumler J.S. Guidelines for the direct detection of Anaplasma spp. in diagnosis and epidemiological studies. Vector Borne Zoonotic Dis. 2017;17:12–22. doi: 10.1089/vbz.2016.1960. [DOI] [PubMed] [Google Scholar]
- Sogin M.L. Amplification of ribosomal RNA genes for molecular evolution studies. In: Innis M.A., Gelfand D.H., Suninsky J.J., White T.H., editors. PCR Protocols: a Guide to Methods and Applications. Academic Press; New York: 1990. pp. 307–314. [Google Scholar]
- Starkey L.A., West M.D., Barrett A.W., Saucier J.M., O'Connor T.P., Paras K.L., Reiskind M.H., Reichard M.V., Little S.E. Prevalence of antibodies to spotted fever group Rickettsia spp. and Ehrlichia spp. in coyotes (Canis latrans) in Oklahoma and Texas, USA. J. Wildl. Dis. 2013;49:670–673. doi: 10.7589/2012-08-215. [DOI] [PubMed] [Google Scholar]
- USDA-APHIS . In: Longhorned Tick: Information for Livestock and Pet Owners Factsheet. U.S.D.o.A. Animal and Plant Health Inspection Service, editor. 2019. [Google Scholar]
- Waldrup K.A., Moritz J., Baggett D., Magyar S., Wagner G.G. Monthly incidence of Theileria cervi and seroconversion to Babesia odocoilei in white-tailed deer (Odocoileus virginianus) in Texas. J. Wildl. Dis. 1992;28:457–459. doi: 10.7589/0090-3558-28.3.457. [DOI] [PubMed] [Google Scholar]
- Wen B., Rikihisa Y., Mott J.M., Greene R., Kim H.-y., Zhi N., Couto G.C., Unver A., Bartsch R. Comparison of nested PCR with immunofluorescent-antibody assay for detection of Ehrlichia canis infection in dogs treated with doxycycline. J. Clin. Microbiol. 1997;35:1852–1855. doi: 10.1128/jcm.35.7.1852-1855.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney M.S., Schwan T.G., Sultemeier K.B., McDonald P.S., Brillhart M.N. Spirochetemia caused by Borrelia turicatae infection in 3 dogs in Texas. Vet. Clin. Pathol. 2007;36:212. doi: 10.1111/j.1939-165x.2007.tb00213.x. [DOI] [PubMed] [Google Scholar]
- Worton B.J. Kernel methods for estimating the utilization distribution in home-range studies. Ecology. 1989;70:164–168. [Google Scholar]
- Yabsley M.J., Varela A.S., Tate C.M., Dugan V.G., Stallknecht D.E., Little S.E., Davidson W.R. Ehrlichia ewingii infection in white-tailed deer (Odocoileus virginianus) Emerg. Infect. Dis. 2002;8:668–671. doi: 10.3201/eid0807.020018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeagley T.J., Reichard M.V., Hempstead J.E., Allen K.E., Parsons L.M., White M.A., Little S.E., Meinkoth J.H. Detection of Babesia gibsoni and the canine small Babesia ‘Spanish isolate’ in blood samples obtained from dogs confiscated from dogfighting operations. J. Am. Vet. Med. Assoc. 2009;235:535–539. doi: 10.2460/javma.235.5.535. [DOI] [PubMed] [Google Scholar]