Abstract
Asthma is the most common chronic disease and is characterized by airway remodeling and chronic inflammation. Increasingly, studies have found that the activation and M1 phenotypic transformation of macrophages play important roles in asthma progress, including airway remodeling. However, the reversal of M1 macrophages to the M2 phenotype has been shown to attenuate airway remodeling. Exosomes are nano-sized extracellular vesicles derived from endosomes; they play direct roles in governing physiological and pathological conditions by the intracellular transfer of bioactive cargo, such as proteins, enzymes, nucleic acids (microRNA [miRNA], mRNA, DNA), and metabolites. However, transfer mechanisms are unclear. To uncover potential therapeutic mechanisms, we constructed an ovalbumin-induced asthma mouse model and lipopolysaccharide-induced RAW264.7 macrophages cells. High-throughput sequencing showed that mmu_circ_0001359 was downregulated in asthmatic mice when compared with normal mice. Adipose-derived stem cell (ADSC)-exosome treatment suppressed inflammatory cytokine expression by the conversion of M1 macrophages to the M2 phenotype, under lipopolysaccharide-induced conditions. Exosomes from mmu_circ_0001359 overexpression in ADSCs increased therapeutic effects, in terms of cytokine expression, when compared with wild-type exosomes. Luciferase reporter assays confirmed that exosomes from mmu_circ_0001359-modified ADSCs attenuated airway remodeling by enhancing FoxO1 signaling-mediated M2-like macrophage activation, via sponging miR-183-5p. In conclusion, mmu_circ_0001359-enriched exosomes attenuated airway remodeling by promoting M2-like macrophages.
Keywords: asthma, mmu_circ_0001359, ADSCs, exosomes, FoxO1, miR-183-5p
Introduction
In recent years, the prevalence of allergic diseases has significantly increased.1 Asthma is a heterogeneous disease characterized by respiratory symptoms, including shortness of breath, wheezing, and chest tightness, resulting from variable airflow. Chronic airway inflammation is common in asthma, and its clinical characterization is central to the development of effective and efficient, personalized asthma care programs. As a result, airway inflammation is proposed as a core “treatable trait” in chronic respiratory disease; however, prolonged inflammation and associated organ damage can be lethal to asthma patients.2,3
Macrophages are the most abundant immune cells in the lung (approximately 70% of all immune cells), and they play important roles in environmental allergen-induced airway inflammation in asthma.4,5 There are two macrophage function extremes: the classically activated (M1) and the alternatively activated (M2) phenotypes. M1 macrophages are characterized by proinflammatory phenotypes and functions (M1 biomarkers include inducible nitric oxide synthase [iNOS], tumor necrosis factor alpha [TNF-α], and interferon-γ [IFN-γ]), whereas M2 macrophages (M2 biomarkers include Ym1, interleukin-10 [IL-10], IL-4, CD206, Fizz1, and Arg1) display anti-inflammatory functions.6,7 Increased M1 macrophage polarization and activation have been observed in asthma and might play important roles in allergic asthma,8,9 but they promoted the polarization of M2 macrophages through upregulating the expression of IL-4, contributing to its regulation by attenuated airway remodeling after asthma.10
Studies indicate that exosomes, which are small vesicles secreted by most cell types and body fluids, may have roles in both immune stimulation and tolerance because they are involved in several processes, such as immune signaling, inflammation, and angiogenesis.11 Adipose-derived stem cells (ADSCs), also known as adipose-derived stromal cells, possess a multipotency, differentiating them into different cell types, such as adipocytes, chondrocytes, and osteoblasts. As well as their potential application in tissue repair and regeneration, ADSCs have a great capacity for immune regulation, as evidenced by their ability to impact on inflammatory or autoimmune diseases, including obesity, diabetes, and acute myocardial ischemic injury.12,13 Previously, a study observed that exosomes from microRNA (miRNA)-126-modified ADSCs promoted a functional recovery after stroke in rats by improving neurogenesis and suppressing microglia activation.14 Exosomes secreted from ADSCs have been shown to attenuate diabetic nephropathy by promoting autophagy flux and inhibiting apoptosis in podocytes.15 However, the underlying mechanisms governing the cross talk between ADSCs and macrophages remain to be unveiled.
In this study, we used ADSC-derived exosomes to study the therapeutic effects of exosomes on asthma in an ovalbumin (OVA)-induced mouse model. Our data and results showed that mmu_circ_0001359-enriched exosomes had better therapeutic effects by reprogramming the microenvironment through enhancing FoxO1 signaling-mediated M2-like macrophage activation, with sponging miR-183-5p.
Results
The Downregulation of mmu_circ_0001359 Plays a Role in OVA-Induced Asthma in Mice
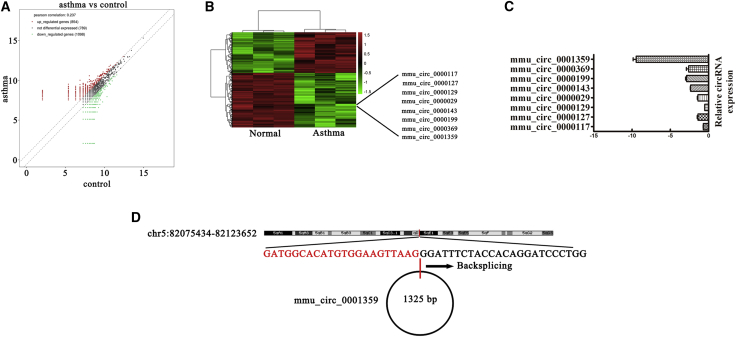
Increasing evidence has found that circular RNAs (circRNAs) are a class of non-coding RNAs that serve as novel biomarkers for the diagnosis and treatment of diseases. To investigate circRNA expression profiles and possible mechanisms in asthma, we constructed an OVA-induced asthma mouse model. High-throughput sequencing of mouse lung tissue found that 854 circRNAs were upregulated and 1,098 circRNAs were downregulated (Figures 1A and 1B). Quantitative real-time PCR was used to assess downregulation, and it was observed that mmu_circ_0001359 was significantly decreased in OVA-induced asthma mice when compared with normal mice. This observation suggested that mmu_circ_0001359 played an important role in OVA-induced asthma (Figure 1C). Nonetheless, the function of mmu_circ_0001359 in asthma remains unclear. Our data revealed that mmu_circ_0001359 was derived and cyclized by part of the exon from the Lphn3 gene; mmu_circ_0001359 was located at chr5:82075434-82123652 (Figure 1D).
Figure 1.
Downregulation of mmu_circ_0001359 Plays a Role in OVA-Induced Asthma
(A) Scatterplots were used to evaluate the differential expression of circRNAs in asthma lung and normal lung tissues. (B) Heatmap of all differentially expressed circRNAs between normal and asthma lung tissues. (C) Relative expression of 10 circRNAs from asthma lung tissue and normal lung tissues as assessed by quantitative real-time PCR. (D) The genomic locus of mmu_circ_0001359. The arrow indicates back-splicing.
Characterization of ADSC Exosomes
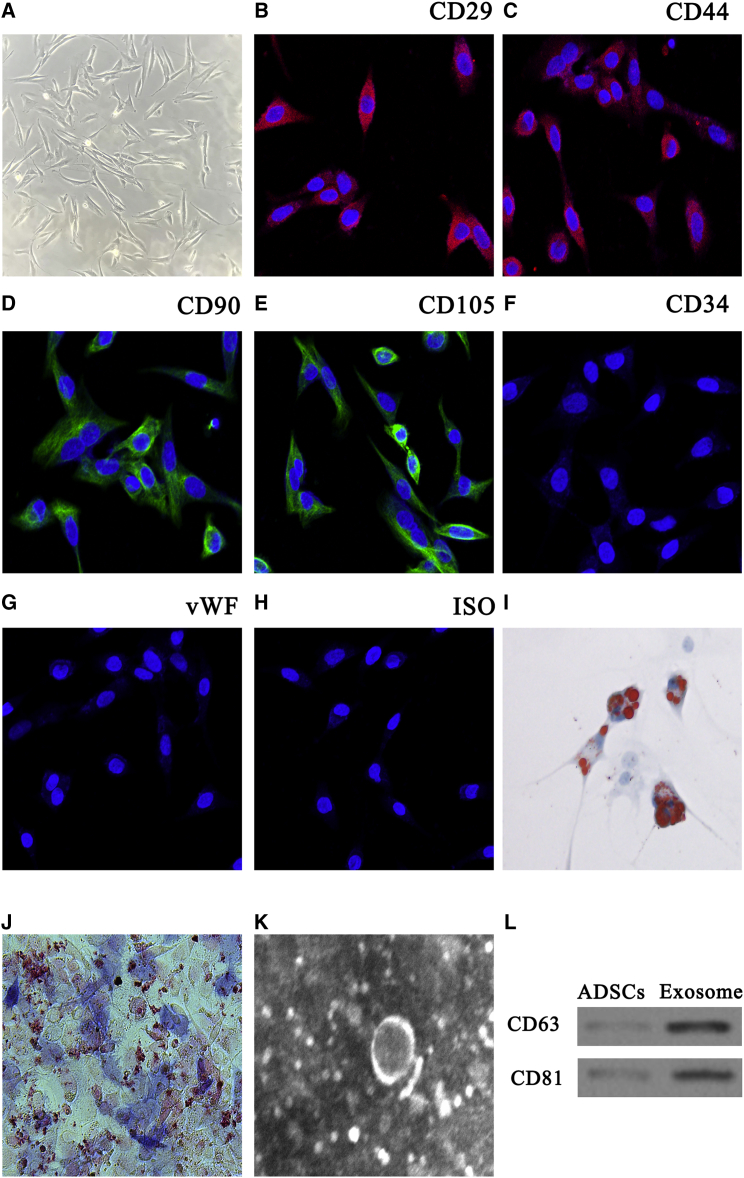
To assess whether mmu_circ_0001359 increased the therapeutic effects of ADSC exosomes, we isolated ADSCs from the adipose tissue of C57BL/6 mice. Our results showed that isolated ADSCs had a typical cobblestone-like morphology (Figure 2A). Immunofluorescence staining was positive for the expression of cell surface mesenchymal markers, CD29, CD90, CD44, and CD105, but negative for the endothelial markers, CD34 and von Willebrand Factor (vWF) (Figures 2B–2H). The results from oil red O and alkaline phosphatase (ALP) staining confirmed adipocyte and osteoblast differentiation (Figures 2I and 2J).
Figure 2.
Characterization of Exosomes Released by Adipose-Derived Mesenchymal Stem Cells
(A) ADSCs show a typical cobblestone-like morphology. Scale bar: 30 μm. (B–H) Immunofluorescence staining of cell surface markers. Antibodies were labeled with either fluorescein isothiocyanate (green) or phycoerythrin (red). (B–E) CD29 (B), CD90 (D), CD44 (C), and CD105 (E) are positive. (F and G) CD34 (F) and vWF (G) expression are negative. (H) FITC- and PE-labeled mouse IgG isotype controls are shown (original magnification ×200). Scale bar: 30 μm. (I and J) Differentiation potential of ADSCs using oil red O (I) and alkaline phosphatase staining (J). (K) Transmission electron micrographs showing ADSC exosome morphology. (L) Western blots of CD63 and CD81 expression in ADSCs and exosomes.
ADSC exosomes had cup- or sphere-shaped morphologies under transmission electron micrographs (Figure 2K), similar to previously described exosomes.16 The expression of the exosome markers CD63 and CD81 in ADSC exosomes was confirmed using western blotting (Figure 2L). The data showed that both ADSC exosomes and cellular components expressed CD63 and CD81, and that the nanoparticles were indeed exosomes.
The Effects of Exosomes on Proinflammatory Cytokine Generation and Phenotypic Transformation in RAW264.7 Cells
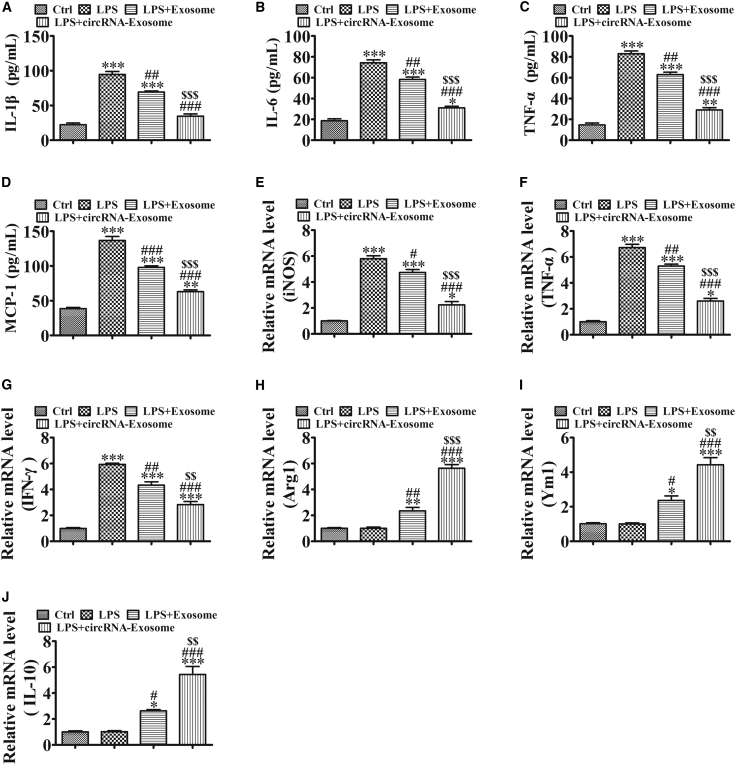
We also used enzyme-linked immunosorbent assay (ELISA) to assess the expression of the proinflammatory cytokines IL-1β, IL-6, TNF-α, and monocyte chemo-attractant protein-1 (MCP-1) in RAW264.7 cells. The expression of these markers was increased in lipopolysaccharide (LPS)-induced asthma mice when compared with normal mice. ADSC exosome treatment decreased LPS-induced inflammatory cytokines, and mmu_circ_0001359-enriched exosomes had greater therapeutic effects than exosomes from wild-type (WT) ADSCs in decreased inflammatory cytokines expression (Figures 3A–3D).
Figure 3.
The Effects of Exosomes on Proinflammatory Cytokines and the Phenotypic Transformation in RAW264.7 Cells
(A–D) The expression of inflammatory factors IL-1β (A), IL-6 (B), TNF-α (C), and MCP-1 (D) in RAW264.7 cell media was detected by ELISA after treatment with LPS (50 ng/mL), exosomes (100 μg/mL), or circRNA-exosomes (100 μg/mL). Results are expressed as the mean ± SD (n = 4). **p < 0.01, ***p < 0.001 versus the control group; ##p < 0.01, ###p < 0.001 versus the LPS group; $$$p < 0.001 versus the LPS+Exosome group. (E–J) qRT-PCR of M1 macrophage marker expression (E: iNOS; F: TNF-α; and G: IFN-γ) and M2 macrophage marker expression (H: Arg1; I: Ym1; and J: IL-10). Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 versus the control group; #p < 0.05, ##p < 0.01, ###p < 0.001 versus the LPS group; $p < 0.05, $$p < 0.01, $$$p < 0.001 versus the LPS+Exosome group.
Quantitative real-time PCR revealed that LPS induced the expression of M1 macrophage markers iNOS, TNF-α, and IFN-γ, but had no effects on the expression of the M2 macrophage markers Arg1, Ym1, and IL-10. ADSC exosome treatment, especially mmu_circ_0001359-enriched exosome treatment, significantly decreased the expression of M1 macrophage markers iNOS, TNF-α, and IFN-γ, but they appeared to promote the expression of the M2 macrophage markers Arg1, Ym1, and IL-10 (Figures 3E–3J). These observations suggested that mmu_circ_0001359 enhanced the transitional effects of M1 macrophages into M2 phenotypes, under LPS-inducing conditions.
FoxO1 and miR-183-5p Are the Downstream Targets of mmu_circ_0001359
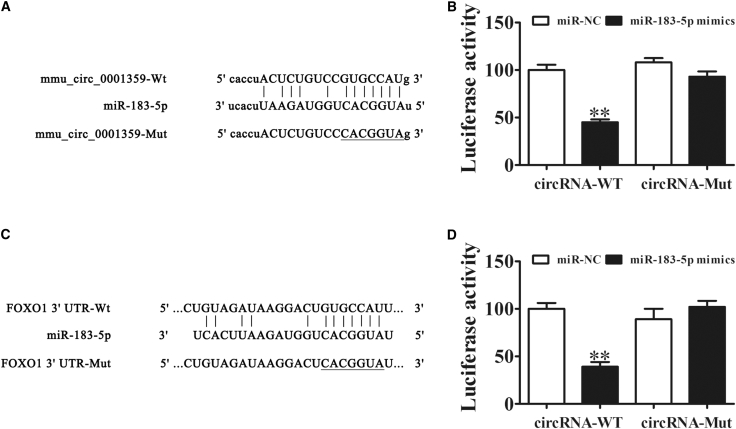
To uncover regulatory mechanisms, we used bioinformatics to predict downstream signals. This approach determined that both FoxO1 and miR-183-5p were the downstream targets of mmu_circ_0001359. To further establish miR-183-5p as a target of mmu_circ_0001359, we used a luciferase reporter assay system, which identified miR-183-5p as an mmu_circ_0001359 downstream binding target (Figure 4A). Notably, mmu_circ_0001359 inhibited luciferase activity in WT cell lines, but not in mutated cells (Figure 4B). To indicate whether FoxO1 was also a potential miR-183-5p target, bioinformatics analyses confirmed direct interactions between miR-183-5p and the 3′ UTR of FoxO1 and consequential suppression of FoxO1 mRNA (Figure 4C). In the luciferase reporter assay, miR-183-5p inhibited the activity of luciferase in WT cell lines, but not in mutated cells (Figure 4D). Our findings suggested that mmu_circ_0001359 overexpression reversed macrophages to an M2 phenotype via targeting of the miR-183-5p/FoxO1 axis.
Figure 4.
FoxO1 and miR-183-5p Are Downstream Targets of mmu_circ_0001359
(A) Complementary sequences in miR-183-5p and mmu_circ_0001359 were obtained using publicly available algorithms. A mutated version of mmu_circ_0001359 is also shown. (B) The mmu_circ_0001359 was fused to the luciferase coding region (pMIR-mmu_circ_0001359) and co-transfected into 293T cells with miR-183-5p mimics, to confirm that mmu_circ_0001359 interacts with miR-183-5p. Both pMIR-mmu_circ_0001359 and miR-183-5p mimic constructs were co-transfected into 293T cells with a control vector, and relative luciferase activity was determined 48 h after co-transfection. Data are mean ± SD. **p < 0.01. (C) Complementary sequences in miR-183-5p and the 3′ UTR of FoxO1 mRNA were obtained using publicly available algorithms. The mutated version of the FoxO1 3′ UTR is also shown. (D) The 3′ UTR of FoxO1 was fused to the luciferase coding region (pMIR-FoxO1 3′ UTR) and co-transfected into 293T cells with miR-183-5p mimics, to confirm that FoxO1 was the target of miR-183-5p. The pMIR-FoxO1 3′ UTR and miR-183-5p mimic constructs were co-transfected into 293T cells with a control vector, and relative luciferase activity was determined 48 h after co-transfection. Data are mean ± SD. **p < 0.01.
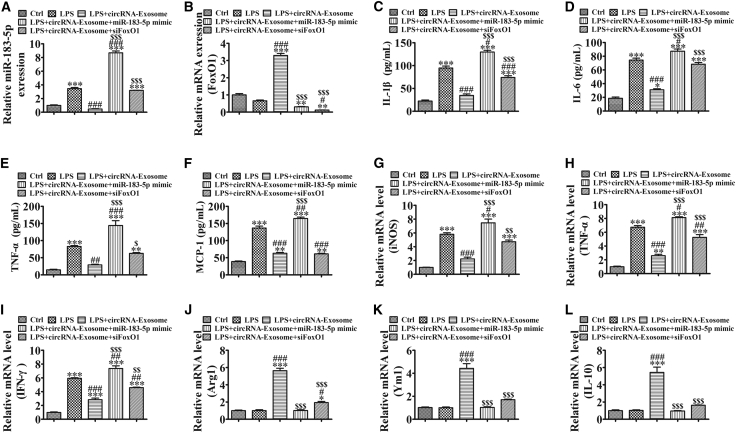
Overexpression of miR-183-5p or Downregulation of FoxO1 in RAW264.7 Cells Reverses Exosome-mmu_circ_0001359-Induced Proinflammatory Cytokines and M2-like Macrophage Activation under LPS-Inducing Conditions
To illuminate whether mmu_circ_0001359 overexpression reversed macrophage to the M2 phenotype via targeting of the miR-183-5p/FoxO1 axis, we constructed an miR-183-5p overexpression (Figure 5A) or FoxO1 silence (Figure 5B) RAW264.7 cells. An ELISA showed that mmu_circ_0001359-enriched exosomes significantly decreased inflammatory cytokine expression, but overexpression of miR-183-5p or silencing of FoxO1 significantly reversed the protective effects of mmu_circ_0001359-enriched exosomes (Figures 5C–5F). Quantitative real-time PCR showed that mmu_circ_0001359-enriched exosome treatment significantly decreased the expression of the M1 macrophage markers iNOS, TNF-α, and IFN-γ, but promoted the expression of the M2 macrophage markers Arg1, Ym1, and IL-10. These observations were reversed by the overexpression of miR-183-5p or FoxO1 silencing (Figures 5G–5L). These data suggested that mmu_circ_0001359 overexpression reversed macrophage to the M2 phenotype via targeting of the miR-183-5p/FoxO1 axis under LPS-inducing conditions.
Figure 5.
Overexpression of miR-183-5p or Downregulation of FoxO1 in RAW264.7 Cells Reverses Exosome-mmu_circ_0001359-Induced Proinflammatory Cytokines and M2-like Macrophage Activation under LPS-Inducing Condition
(A and B) Quantitative real-time PCR shows the expression of miR-183-5p (A) and FoxO1 (B). Data are presented as the mean ± SEM. **p < 0.01, ***p < 0.001 versus the control group; #p < 0.05, ###p < 0.001 versus the LPS group; $$$p < 0.001 versus the LPS+circRNA-Exosome group. (C–F) The expression of inflammatory factors IL-1β (C), IL-6 (D), TNF-α (E), and MCP-1 (F) in RAW264.7 cell media was detected by ELISA. Results are expressed as mean ± SD (n = 4). **p < 0.01, ***p < 0.001 versus the control group; ##p < 0.01, ###p < 0.001 versus the LPS group; $$$p < 0.001 versus the LPS+circRNA-Exosome group. (G–L) qRT-PCR expression of M1 macrophage markers iNOS (G), TNF-α (H), and IFN-γ (I), and M2 macrophage markers Arg1 (J), Ym1 (K), and IL-10 (L). Data are presented as the mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001 versus the control group; #p < 0.05, ##p < 0.01, ###p < 0.001 versus the LPS group; $p < 0.05, $$p < 0.01, $$$p < 0.001 versus the LPS+circRNA-Exosome group.
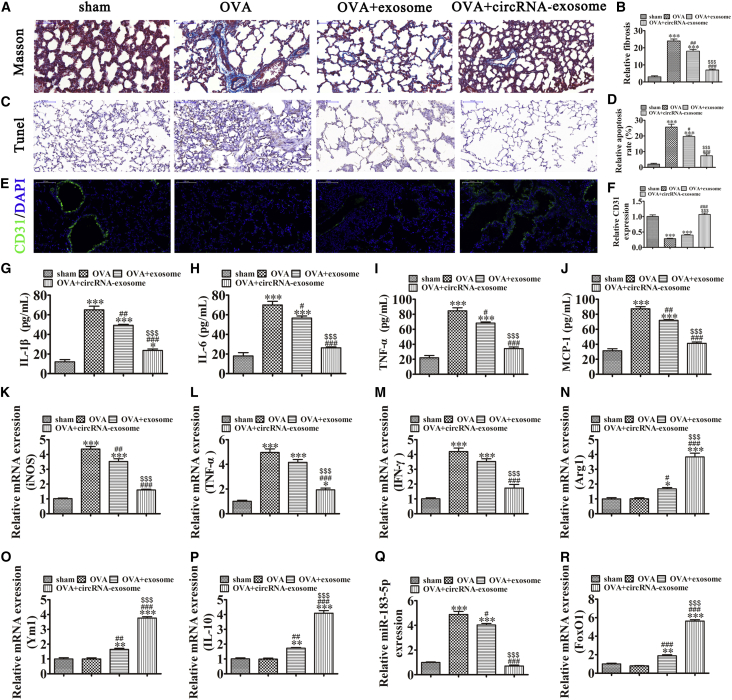
Exosomes from mmu_circ_0001359-Modified ADSCs Attenuate Airway Remodeling by Decreasing M1-like Macrophage Activation-Induced Inflammatory Cytokines and Pulmonary Fibrosis
The OVA-induced asthma in vivo mouse model showed that ADSC exosome treatment significantly decreased OVA-induced pulmonary fibrosis and apoptosis, but appeared to promote angiogenesis. Exosomes from mmu_circ_0001359-modified ADSCs increased therapeutic effectiveness (Figures 6A–6F). Our ELISA experiments showed that exosomes, notably mmu_circ_0001359-enriched exosomes, significantly decreased the expression of OVA-induced inflammatory cytokines, IL-1β, IL-6, TNF-α, and MCP-1 (Figures 6G–6J). ADSC exosomes, notably mmu_circ_0001359-enriched exosomes, significantly decreased the expression of M1 macrophage markers iNOS, TNF-α, and IFN-γ, but promoted the expression of M2 macrophage markers Arg1, Ym1, and IL-10 (Figures 6K–6P). Quantitative real-time PCR showed that the expression of miR-183-5p was increased in OVA-induced asthma mice, but downregulated FoxO1 expression. mmu_circ_0001359-enriched exosome treatment significantly decreased miR-183-5p expression, but promoted FoxO1 expression. These observations suggested that exosomes from mmu_circ_0001359-modified ADSCs attenuated airway remodeling by enhancing FoxO1 signaling-mediated M2-like macrophage activation via sponging miR-183-5p.
Figure 6.
Exosomes from mmu_circ_0001359-modified ADSCs attenuate airway remodeling by decreasing M1-like macrophage activation induced inflammatory cytokines and pulmonary fibrosis
(A) Collagen deposition and tissue fibrosis were assessed by Masson trichrome staining. (B) The relative fibrosis in different group were analysis. Data are presented as the mean ± SEM. ∗∗∗p < 0.001 versus control group. ##p < 0.01, ###p < 0.001 versus OVA group. $$$p < 0.001 versus the exosome treatment group. (C) Immunohistochemical staining of apoptosis. (D) The relative apoptosis in different group were analysis. Data are presented as the mean ± SEM. ∗∗∗p < 0.001 versus control group. #p < 0.05, ###p < 0.001 versus OVA group. $$$p < 0.001 versus exosome treatment group. (E) Immunofluorescence shows angiogenesis using CD31 staining. (F) The relative CD31 expression in different group were analysis. Data are presented as the mean ± SEM. ∗∗∗p < 0.001 versus Sham group. ###p < 0.001 versus OVA group. $$$p < 0.001 versus the exosome treatment group. (G–J) ELISA showing the expression of inflammatory markers IL-1β (G), IL-6 (H), TNF-α (I), and MCP-1 (J). Data are presented as the mean ± SEM. ∗p < 0.05, ∗∗∗p < 0.001 versus Sham group. #p < 0.05, ##p < 0.01, ###p < 0.001 versus OVA group. $$$p < 0.001 versus the exosome treatment group. (K–P) qRT-PCR expression of M1 macrophage markers [iNOS (K), TNF-α (L), and IFN-γ (M)] and M2 macrophage markers [Arg1 (N), Ym1 (O), and IL-10 (P)]. Data are presented as the mean ± SEM. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001 versus Sham group. #p < 0.05, ##p < 0.01, ###p < 0.001 versus OVA group. $$$p < 0.001 versus the exosome treatment group. (Q and R) RT-qPCR expression of miR-183-5p (Q) and FoxO1 (R). Data are presented as the mean ± SEM. ∗∗p < 0.01, ∗∗∗p < 0.001 versus Sham group. #p < 0.05, ###p < 0.001 versus OVA group. $$$p < 0.001 versus the exosome treatment group.
Discussion
Asthma is a chronic airway inflammatory disease associated with airway inflammation. Macrophages are key leukocytes participating in many inflammatory and infectious diseases, including asthma.17 In this study, we found that OVA-induced asthma or LPS induction promoted macrophage activation and M1 polarization. While attempting to identify non-coding RNAs that caused the polarity switch through high-throughput sequencing, we found that the expression of mmu_circ_0001359 was decreased in lung tissues of asthma. circRNAs are a class of non-coding RNAs that may serve as novel biomarkers for the diagnosis and treatment of diseases.18 This study is the first to observe that overexpression of mmu_circ_0001359 increases the therapeutic effects of exosome secretion from ADSCs.
Previous studies have demonstrated that mesenchymal stem cells (MSCs) are therapeutic candidates for halting the progression of different diseases, including diabetic nephropathy, cerebral ischemia, and acute myocardial infarction.14,15,19 In this study, we found that ADSC exosome treatment significantly decreased inflammatory cytokines, especially in mmu_circ_0001359-rich exosome. Furthermore, our study revealed that ADSC exosome delivery of mmu_circ_0001359 decreased OVA- or LPS-induced M1 macrophages, but promoted M2 macrophages phenotype.
Our study further found that the expression of mmu_circ_0001359 promoted FoxO1 expression by sponging miR-183-5p, which was confirmed by luciferase reporter assay. The forkhead box proteins, O (FoxO) family of transcription factors, play key roles in a number of cellular processes, including cell growth, metabolism, survival, and inflammation.20, 21, 22 In mammals, the FoxO subclass consists of four members, including FoxO1, FoxO3, FoxO4, and FoxO6.23 Although several studies have focused on the role of FoxO3 in hematopoietic and immune cells, a mechanistic role for FoxO1 in allergic asthmatic inflammation in activated macrophages has not been reported. This study showed that FoxO1 linked the alternatively activated macrophages into M2 phenotype and played an important role in ADSCs exosome-induced and mmu_circ_0001359/miR-183-5p signal axis-mediated remodeling restraint after asthma. By inhibiting the expression of macrophage-mediated inflammatory cytokines, OVA-induced apoptosis and fibrosis in mouse lung tissues was decreased, but angiogenesis was promoted.
Taken together, our findings showed that FoxO1 had a crucial role in upregulating the alternative activation of alveolar macrophages. Exosomes from mmu_circ_0001359-modified ADSCs appeared to attenuate airway remodeling by enhancing FoxO1 signaling-mediated M2-like macrophage activation via sponging miR-183-5p.
Materials and Methods
Ethics Statement
Male C57BL/6 mice (6–8 weeks) were purchased from the Shanghai Slac Laboratory Animal (Shanghai, China). All mice were housed under controlled room temperatures (22°C) and photoperiods (12-h light/12-h darkness cycles) in a specific pathogen-free conditioned animal care facility. The study was approved by the Ethics Committee of the Changhai Hospital, Second Military Medical University.
Expression Profile Analysis of circRNAs
Lung samples from asthmatic mice and normal mice were used for circRNA microarray analysis. Tissue specimens were obtained during surgery and immediately frozen at −80°C for further use. The circRNAs chip (ArrayStar Human circRNAs chip; ArrayStar, Rockville, MD, USA), containing 5,639 probes specific for mice circRNAs splicing sites, was used. Following hybridization and washing of samples, one pair of epileptic samples and control samples was analyzed on the circRNAs chips. Exogenous RNAs, developed by the External RNA Controls Consortium (ERCC), were used as controls. The circRNAs were enriched by digesting linear RNA with RNase R (Epicenter, Madison, WI, USA). Labeled RNAs were scanned using an Agilent Scanner G2505C (Agilent Technologies, Santa Clara, CA, USA). The circRNA microarray process was performed by KangChen Biotech (Shanghai, China).
Isolation, Culture, and Identification of ADSCs
In brief, adipose tissue was harvested from C57BL/6 mice. Tissues were washed in phosphate-buffered saline (PBS) and minced before digestion in 0.2% collagenase I (Sigma-Aldrich, St. Louis, MO, USA) for 1 h at 37°C, with intermittent shaking. The digested tissue was washed in Dulbecco’s modified Eagle’s medium (DMEM; Sigma-Aldrich) containing 15% fetal bovine serum (FBS; GIBCO BRL, Frederick, MD, USA) and centrifuged at 1,000 rpm for 10 min, to remove mature adipocytes. The resulting pellet was resuspended in DMEM, supplemented with 15% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin, and cultured at 37°C in 5% CO2. ADSCs reaching 80%–90% confluency were detached using 0.02% ethylenediaminetetraacetic acid (EDTA)/0.25% trypsin (Sigma-Aldrich) for 5 min at room temperature and replated. For phenotypic analyses, fluorescein isothiocyanate (FITC)- or phycoerythrin (PE)-conjugated CD29, CD90, CD44, CD105, and vWF antibodies were used. An IgG-matched isotype served as the internal control for each antibody. Normoxic ADSC cultures were grown in 95% air (20% O2) and 5% CO2.
Multilineage Differentiation of ADSCs
To evaluate multilineage differentiation of ADSCs, we cultured third-passage mouse ADSCs in adipogenic differentiation medium (Sigma-Aldrich) and stained them with oil red O after 14 days, or cultured them in osteogenic differentiation medium (Sigma-Aldrich) and stained with Alizarin red after 21 days.
Isolation and Identification of ADSC-Derived Exosomes
After reaching 80%–90% confluency, ADSCs were rinsed in PBS and cultured in FBS-free endothelial cell growth medium (EGM)-2MV, supplemented with 1× serum replacement solution (PeproTech, NJ, USA) for an additional 48 h. The conditioned culture medium was then removed and centrifuged at 300 × g for 10 min, and again at 2,000 × g for 10 min to remove dead cells and other cellular debris. After a further centrifugation step at 10,000 × g for 30 min, the supernatant was filtered through a 0.22-μm (Millipore, Billerica, MA, USA) filter, and 15 mL supernatant was transferred to an Amicon Ultra-15 Centrifugal Filter Unit (100 kDa; Millipore) and centrifuged at 4,000 × g to concentrate the volume to ∼1 mL. The ultrafiltration unit was then washed twice in PBS and filtered again at 4,000 × g to achieve a volume of 1 mL. Approximately one-fifth the volume of Exoquick exosome precipitation solution (System Biosciences, Palo, Alto, CA, USA) was added to the ultrafiltration liquid and mixed by inversion. After incubation for 12 h at 4°C, the mixture was centrifuged at 1,500 × g for 30 min and the supernatant aspirated. The exosome pellet was resuspended in 500 μL PBS. All procedures were performed at 4°C. Exosome protein concentrations were determined using a Pierce bicinchoninic acid (BCA) Protein Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). Exosomes were stored at −80°C until required. Exosomes were characterized by transmission electron microscopy (TEM) and western blotting to determine exosome size.
Cell Culture
RAW264.7 macrophages (BCRC No. 60001) derived from leukemic mouse monocytes were obtained from the Wuhan Biofavor Biotechnology Service (Wuhan, China). Cells were grown in DMEM, supplemented with 10% FBS, 100 U/mL penicillin, 100 μg/mL streptomycin, and 0.25 μg/mL Fungizone. Cells were cultured at 5% CO2, 95% air at 37°C. The growth medium was changed every 3 days until cells reached approximately 90% confluence, after which cells were passaged.
To study the effects of ADSC exosomes to macrophages phenotype transition, small interfering RNAs (siRNAs) against FoxO1 (siFoxO1) and miR-183-5p mimics were purchased from GeneCopoeia (Shanghai, China) and transfected into RAW264.7 cells using Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions, before treatment with 100 μg/mL exosomes. Cells were used for experimentation 48 h later.
Murine Model of OVA-Induced Asthma
To generate the asthma model, we treated male C57BL/6 mice (6–8 weeks old, weighing 18–24 g) with OVA. The mice were then sensitized on days 0, 7, and 14 by intraperitoneal injection of 20 μg OVA, emulsified in 1 mg aluminum hydroxide in a total volume of 0.2 mL. Seven days after the last sensitization, mice were exposed to a 1% OVA aerosol for up to 1 h every day for 7 days. The 1% OVA aerosol was generated using a compressed air atomizer driven by filling a Perspex cylinder chamber (diameter, 50 cm; height, 50 cm) with a nebulized solution. Saline was used in the control group instead of OVA. Once the study course was completed, mice were euthanized under anesthesia using sodium pentobarbital (50 mg/kg). The trachea was cannulated, and both lungs and airways were rinsed in 1 mL PBS for the collection of Bronchoalveolar Lavage Fluid (BALF). The right lung was collected, frozen in liquid nitrogen, and kept at −80°C for western blotting. The left lung was preserved and fixed in 4% paraformaldehyde, and used for immunohistochemical analyses. All animal studies were performed in accordance with the Guide for the Care and Use of Laboratory Animals. All study protocols were approved by the Ethics Committee of Changhai Hospital Affiliated to the Second Military Medical University.
For exosome treatment, 200 μg ADSC exosomes in 100 μL PBS or the same volume of PBS was used for tail intravenous injections (once a week). After 2 weeks of treatment, mice were euthanized for pathological examination.
ELISA
The expressions of IL-1β, IL-6, MCP-1, and TNF-α were assayed using ELISA kits (Abcam, Cambridge Science Park, UK) in accordance with the manufacturer’s instructions.
Western Blotting
Cells or exosomes were lysed, and lysates were centrifuged at 12,000 rpm at 4°C, after the addition of protease inhibitors. Protein concentrations were determined with a Pierce BCA assay kit (Thermo Fisher). Proteins were separated on a 10% SDS-PAGE and then transferred to polyvinylidene fluoride (PVDF) membranes. Primary antibodies were against CD63 (1:600) and CD81 (1:600). Horseradish peroxidase-conjugated secondary antibody (1:1,000; Abcam, Burlingame, CA, USA) was also used. An ECL chemiluminescent kit (Millipore, Burlington, MA, USA) was used to detect and size protein bands.
Quantitative Real-Time PCR
Total RNA was extracted using TRIzol (Invitrogen; Thermo Fisher Scientific) according to manufacturers’ instructions. RNA was then reverse transcribed using the One Step SYBR PrimeScript RT-PCR kit (Takara Biotechnology, Mountain View, CA, USA). The reaction was performed using an ABI PRISM 7500 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) at 42°C for 5 min, 95°C for 10 s; then 40 cycles of 95°C for 5 s, 55°C for 30 s, and 72°C for 30 s. Three independent experiments were conducted for each sample. Data were analyzed by comparing 2−ΔΔCt values. Glyceraldehyde phosphate dehydrogenase (GAPDH) and U6 were used as internal controls.
Immunohistochemistry and Immunofluorescence Assays
Samples of lung tissue were fixed in 10% formalin solution, embedded in paraffin, and sectioned at 5 μm. Tissue sections were stained with Masson trichrome for histological evaluation. Immunofluorescence staining of CD31 was performed to evaluate histopathological angiogenesis pathology. A terminal deoxynucleotidyl transferase dUTP nick end labeling (TUNEL) kit was used to identify apoptotic cells. Tissue sections were examined with an Axiophot light microscope (Zeiss, Oberkochen, Germany) or fluorescence microscope (Nikon, Tokyo, Japan) and photographed using a digital camera.
Luciferase Reporter Assay
The 3′ UTR target sequence for the miR-183-5p miRNA in the FoxO1 gene or mmu_circ_0001359 was predicted with the TargetScan online tool. WT and 3′ UTR mutant FoxO1 or mmu_circ_0001359 were performed and cloned into the pMIR firefly luciferase-expression vector. For luciferase assays, human embryonic kidney cells (HEK293T cells) at 70% confluence were co-transfected with 500 ng pMIR-FoxO1-WT/pMIR-FoxO1-Mut or pMIR-circRNA-WT/pMIR-circRNA-Mut and 50 nM miR-183-5p mimics using a Lipofectamine 2000 transfection kit (Thermo Scientific). Luciferase activity was monitored using a Dual-Luciferase Reporter System (Promega). Five independent assays were performed.
Statistical Analysis
Data are presented as the mean ± standard deviation (SD). GraphPad Prism software, version 5.0 (GraphPad, La Jolla, CA, USA) was used to compare the differences between groups. A p value ≤0.05 indicated a statistically significant difference.
Author Contributions
Y.S. and C.B. designed the studies and prepared the manuscript with comments from all authors. Y.S., J.X. and X.G. performed all the experiments and analyzed the data. Z.H., J.X., Y.N. and Y.D. carried out in all experiments and revised the manuscript. All authors read and approved the final manuscript.
Acknowledgments
This work was supported by Science and Technology Development Fund of Pudong New Area (grant PKJ2016-Y49); National Natural Science Foundation of China (grants 81570020, 81873405, 81300018, and 81800018); Zhejiang Province Public Welfare Technology Application Research Project Foundation (grant 2016C33216); Key Research Project of Educational Commission of Hunan Province (grant 16A152); and Changhai Hospital Foundation (grants 2019SLZ002 and 2019YXK018).
Contributor Information
Yan Shang, Email: shangyan751200@163.com.
Chong Bai, Email: bc7878@sohu.com.
References
- 1.Burbank A.J., Sood A.K., Kesic M.J., Peden D.B., Hernandez M.L. Environmental determinants of allergy and asthma in early life. J. Allergy Clin. Immunol. 2017;140:1–12. doi: 10.1016/j.jaci.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gibson P.G., McDonald V.M., Marks G.B. Asthma in older adults. Lancet. 2010;376:803–813. doi: 10.1016/S0140-6736(10)61087-2. [DOI] [PubMed] [Google Scholar]
- 3.Agusti A., Bel E., Thomas M., Vogelmeier C., Brusselle G., Holgate S., Humbert M., Jones P., Gibson P.G., Vestbo J. Treatable traits: toward precision medicine of chronic airway diseases. Eur. Respir. J. 2016;47:410–419. doi: 10.1183/13993003.01359-2015. [DOI] [PubMed] [Google Scholar]
- 4.Cai Y., Sugimoto C., Arainga M., Alvarez X., Didier E.S., Kuroda M.J. In vivo characterization of alveolar and interstitial lung macrophages in rhesus macaques: implications for understanding lung disease in humans. J. Immunol. 2014;192:2821–2829. doi: 10.4049/jimmunol.1302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee Y.G., Jeong J.J., Nyenhuis S., Berdyshev E., Chung S., Ranjan R., Karpurapu M., Deng J., Qian F., Kelly E.A. Recruited alveolar macrophages, in response to airway epithelial-derived monocyte chemoattractant protein 1/CCl2, regulate airway inflammation and remodeling in allergic asthma. Am. J. Respir. Cell Mol. Biol. 2015;52:772–784. doi: 10.1165/rcmb.2014-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gordon S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 7.Li H., Ciric B., Yang J., Xu H., Fitzgerald D.C., Elbehi M., Fonseca-Kelly Z., Yu S., Zhang G.X., Rostami A. Intravenous tolerance modulates macrophage classical activation and antigen presentation in experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2009;208:54–60. doi: 10.1016/j.jneuroim.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melgert B.N., ten Hacken N.H., Rutgers B., Timens W., Postma D.S., Hylkema M.N. More alternative activation of macrophages in lungs of asthmatic patients. J. Allergy Clin. Immunol. 2011;127:831–833. doi: 10.1016/j.jaci.2010.10.045. [DOI] [PubMed] [Google Scholar]
- 9.Girodet P.O., Nguyen D., Mancini J.D., Hundal M., Zhou X., Israel E., Cernadas M. Alternative macrophage activation is increased in asthma. Am. J. Respir. Cell Mol. Biol. 2016;55:467–475. doi: 10.1165/rcmb.2015-0295OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y., Gao X., Miao Y., Wang Y., Wang H., Cheng Z., Wang X., Jing X., Jia L., Dai L. NLRP3 regulates macrophage M2 polarization through up-regulation of IL-4 in asthma. Biochem. J. 2018;475:1995–2008. doi: 10.1042/BCJ20180086. [DOI] [PubMed] [Google Scholar]
- 11.Xu H., Jia S., Xu H. Potential therapeutic applications of exosomes in different autoimmune diseases. Clin. Immunol. 2019;205:116–124. doi: 10.1016/j.clim.2019.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Zhao H., Shang Q., Pan Z., Bai Y., Li Z., Zhang H., Zhang Q., Guo C., Zhang L., Wang Q. Exosomes from adipose-derived stem cells attenuate adipose inflammation and obesity through polarizing M2 macrophages and beiging in white adipose tissue. Diabetes. 2018;67:235–247. doi: 10.2337/db17-0356. [DOI] [PubMed] [Google Scholar]
- 13.Luo Q., Guo D., Liu G., Chen G., Hang M., Jin M. Exosomes from MiR-126-overexpressing adscs are therapeutic in relieving acute myocardial ischaemic injury. Cell. Physiol. Biochem. 2017;44:2105–2116. doi: 10.1159/000485949. [DOI] [PubMed] [Google Scholar]
- 14.Geng W., Tang H., Luo S., Lv Y., Liang D., Kang X., Hong W. Exosomes from miRNA-126-modified ADSCs promotes functional recovery after stroke in rats by improving neurogenesis and suppressing microglia activation. Am. J. Transl. Res. 2019;11:780–792. [PMC free article] [PubMed] [Google Scholar]
- 15.Jin J., Shi Y., Gong J., Zhao L., Li Y., He Q., Huang H. Exosome secreted from adipose-derived stem cells attenuates diabetic nephropathy by promoting autophagy flux and inhibiting apoptosis in podocyte. Stem Cell Res. Ther. 2019;10:95. doi: 10.1186/s13287-019-1177-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hu Y., Rao S.S., Wang Z.X., Cao J., Tan Y.J., Luo J., Li H.M., Zhang W.S., Chen C.Y., Xie H. Exosomes from human umbilical cord blood accelerate cutaneous wound healing through miR-21-3p-mediated promotion of angiogenesis and fibroblast function. Theranostics. 2018;8:169–184. doi: 10.7150/thno.21234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shapouri-Moghaddam A., Mohammadian S., Vazini H., Taghadosi M., Esmaeili S.A., Mardani F., Seifi B., Mohammadi A., Afshari J.T., Sahebkar A. Macrophage plasticity, polarization, and function in health and disease. J. Cell. Physiol. 2018;233:6425–6440. doi: 10.1002/jcp.26429. [DOI] [PubMed] [Google Scholar]
- 18.Cardamone G., Paraboschi E.M., Rimoldi V., Duga S., Soldà G., Asselta R. The characterization of GSDMB splicing and backsplicing profiles identifies novel isoforms and a circular RNA that are dysregulated in multiple sclerosis. Int. J. Mol. Sci. 2017;18:e576. doi: 10.3390/ijms18030576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pan J., Alimujiang M., Chen Q., Shi H., Luo X. Exosomes derived from miR-146a-modified adipose-derived stem cells attenuate acute myocardial infarction-induced myocardial damage via downregulation of early growth response factor 1. J. Cell. Biochem. 2019;120:4433–4443. doi: 10.1002/jcb.27731. [DOI] [PubMed] [Google Scholar]
- 20.Dejean A.S., Hedrick S.M., Kerdiles Y.M. Highly specialized role of Forkhead box O transcription factors in the immune system. Antioxid. Redox Signal. 2011;14:663–674. doi: 10.1089/ars.2010.3414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Savai R., Al-Tamari H.M., Sedding D., Kojonazarov B., Muecke C., Teske R., Capecchi M.R., Weissmann N., Grimminger F., Seeger W. Pro-proliferative and inflammatory signaling converge on FoxO1 transcription factor in pulmonary hypertension. Nat. Med. 2014;20:1289–1300. doi: 10.1038/nm.3695. [DOI] [PubMed] [Google Scholar]
- 22.O’Sullivan I., Zhang W., Wasserman D.H., Liew C.W., Liu J., Paik J., DePinho R.A., Stolz D.B., Kahn C.R., Schwartz M.W., Unterman T.G. FoxO1 integrates direct and indirect effects of insulin on hepatic glucose production and glucose utilization. Nat. Commun. 2015;6:7079. doi: 10.1038/ncomms8079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chung S., Lee T.J., Reader B.F., Kim J.Y., Lee Y.G., Park G.Y., Karpurapu M., Ballinger M.N., Qian F., Rusu L. FoxO1 regulates allergic asthmatic inflammation through regulating polarization of the macrophage inflammatory phenotype. Oncotarget. 2016;7:17532–17546. doi: 10.18632/oncotarget.8162. [DOI] [PMC free article] [PubMed] [Google Scholar]