Abstract
Bladder cancer is the most common malignant tumor of the urinary system, and it has high incidence, high degree of malignancy, and easy recurrence after surgery. The etiology and pathogenesis of bladder cancer are not fully understood, but more and more studies have shown that its development may be regulated by some core molecules. To identify key molecules in bladder cancer, we constructed a three-layer network by merging lncRNA-miRNA regulatory network, miRNA-mRNA regulatory network, and lncRNA-mRNA coexpression network, and further analyzed the topology attributes of the network including the degree, betweenness centrality and closeness centrality of nodes. We found that miRNA-93 and miRNA-195 are controllers for a three-layer network and regulators of numerous target genes associated with bladder cancer. Functional enrichment analysis of their target mRNAs revealed that miRNA-93 and miRNA-195 may be closely related to bladder cancer by disturbing the homeostasis of the cell cycle or HTLV-I infection. In addition, since E2F1 and E2F2 are enriched in various KEGG signaling pathways, we conclude that they are important target genes of miRNA-93, and participate in the apoptotic process by forming a complex with a certain protein or transcription factor activity, sequence-specific DNA binding in bladder cancer. Similarly, AKT3 is an important target gene of miRNA-195, its expression is associated with PI3K-Akt-mTOR signaling pathway and AMPK-mTOR signaling pathway. Therefore, we speculate that AKT3 may participate in proliferation and apoptosis of bladder cancer cells through these pathways, and ultimately affect the biological behavior of tumor cells. Furthermore, through survival analysis, we found that miRNA-195 and miRNA-93 are associated with poor prognosis of bladder cancer. And the Kaplan-Meier curve showed that 24 mRNAs and nine lncRNAs are closely related to overall survival of bladder cancer.
Keywords: bladder cancer, three-layer network, topology, miRNA, TCGA database
Introduction
Bladder cancer (BC) is a common malignant tumor with the tenth incidence rate and the ninth mortality rate in the world (Bray et al., 2018). As the most common malignant tumor of the urinary system, its etiology and pathogenesis are still not very clear. It has some biological characteristics such as high incidence, high degree of malignancy and easy recurrence after surgery, and it shows a clear upward trend, which is a serious threat to human health and social development (Knollman et al., 2015; Tian et al., 2015). The etiology of BC is complex, including both intrinsic genetic factors and external environmental factors. At present, it has been clarified that the occurrence and development of BC are related to smoking, environmental pollution, and other factors (Kiriluk et al., 2012; Van Roekel et al., 2013; Smith et al., 2016; Wilcox et al., 2016). But its molecular mechanism is still not completely clear. Therefore, the research and clarification of its cause of disease and the specific molecular mechanism will effectively promote the diagnosis and treatment of BC.
In recent years, a large number of studies on single molecules of BC have found that dysregulated mRNA contributes to the occurrence of BC tumors by participating in abnormal activation of many pathways such as Akt signal pathway and NF-kB pathway (Jiang et al., 2016; Tsui et al., 2018). Abnormal expression of lncRNA UCA1 and H19 is closely related to the initiation, development, and metastasis of BC (Zhang et al., 2013). Studies on the relationship between urinary tumors and miR-145, miR-205 indicate that a large number of miRNAs have been involved in tumor progression (Catto et al., 2011; Simonato et al., 2013; Braicu et al., 2015; Chen et al., 2017). Some people also analyzed the differentially expressed miRNAs and their target genes in BC from the perspective of the network, and it is pointed out that miRNAs participate in the occurrence and development of BC by regulating their target genes, and are related to BC recurrence (Guancial et al., 2014; Jin et al., 2015; Li et al., 2017; Zhao et al., 2017). LncRNAs and mRNAs can form complex coexpression networks in various diseases including BC, and promote the development of BC by regulating protein-coding genes (Cao et al., 2018b; Li et al., 2018; Chen et al., 2019a; Chen et al., 2019b). In addition, the ceRNA hypothesis suggests that lncRNA can not only directly participate in the expression of target regulatory genes, but also may adsorb target miRNAs, affect the binding of miRNAs to mRNAs, and further intervene in mRNA expression (Wang et al., 2016; Lv et al., 2017; Wang et al., 2017; Miao et al., 2019; Teng et al., 2019).
Since the mRNA-lncRNA coexpression network and the miRNA-related regulatory network are in the common system, it is one-sided to study the key molecules of BC by the single-layer network method. In contrast, converged network analysis can reflect the importance of molecules in the network more accurately and more directly. Therefore, it is considered as a feasible method to further analyze the topology of the network according to the degree, betweenness centrality and closeness centrality of the nodes in the network to find and predict the core molecules potentially related to the development of BC (Greenbury et al., 2010). In this paper, we constructed a three-layer network by merging lncRNA-miRNA regulatory network, miRNA-mRNA regulatory network and lncRNA-mRNA coexpression network, and further analyzed the topology attributes of nodes in the network.
Materials and Methods
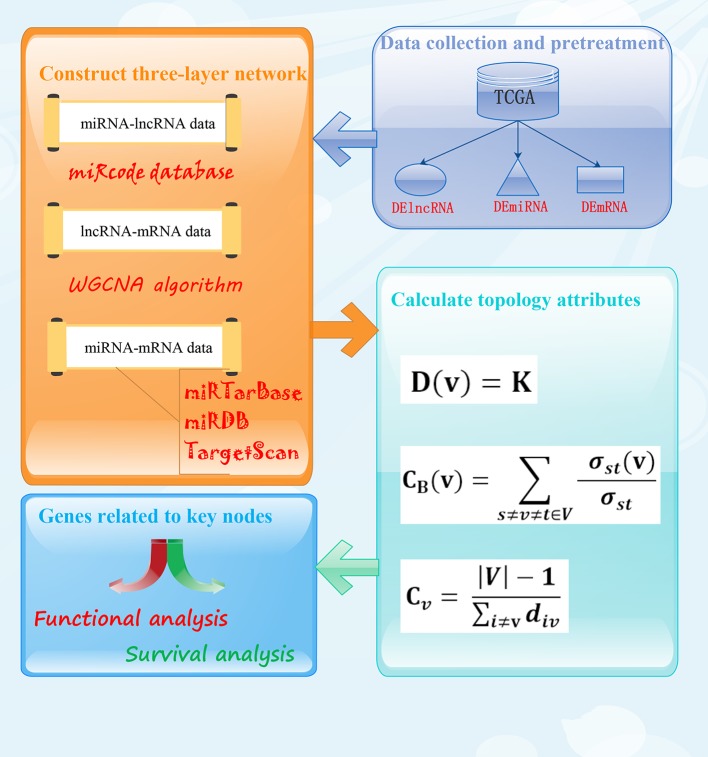
The main steps of identification of the key factors related to BC by lncRNA-miRNA-mRNA three-layer network include: data collection and pretreatment, construct three-layer network, calculate topology properties, identify the key nodes, functional analysis, and survival analysis of the key nodes. The flowchart of data collection and method implementation was shown in Figure 1 . Next, we will describe a more detailed process of identification of the key factors related to BC by lncRNA-miRNA-mRNA three-layer network.
Figure 1.
The flowchart of data collection and method implementation.
Data Resources and Pretreatment
RNA-Seq data, miRNA-Seq data, and clinical information of transitional cell papillomas and carcinomas in human BC were downloaded from the Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/) at the time of 2018-12-03. The RNA-Seq data includes lncRNA and mRNA, including 19 normal samples and 411 BC samples. The miRNA-Seq data contains 19 normal samples and 415 BC samples. We selected their common sample number, 19 normal samples and 405 BC samples, including 19645 mRNAs, 14176 lncRNAs, and 1881 miRNAs.
Identification of DERNAs
We used “EdgeR” package in R (version 3.5.1) (https://www.r-project.org/) software to identify differentially expressed mRNAs (DEmRNAs), differentially expressed lncRNAs (DElncRNAs) and differentially expressed miRNAs (DEmiRNAs) in BC with the thresholds of Fold change (FC) > 1.5 and adjusted P-value < 0.05.
Construction of the Three-Layer Network
The target lncRNAs of miRNAs were obtained from miRcode database (http://www.mircode.org/). And miRDB (http://www.mirdb.org/), miRTarBase (http://mirtarbase.mbc.nctu.edu.tw/), and TargetScan database (http://www.targetscan.org/) were used to predict the target mRNAs of miRNAs. Then, the intersection of the target mRNAs predicted by above three databases and DEmRNAs was considered as final target mRNAs of DEmiRNAs.
Using R software, the DEmRNAs and DElncRNAs coexpression data were integrated into a matrix. Then, the lncRNA-mRNA interactions were predicted according to the method of weighted gene coexpression network by using the WGCNA package in R and the correlation coefficient is set to 0.8.
Finally, we merged all miRNA-mRNA pairs and miRNA-lncRNA pairs and mRNA-lncRNA pairs to construct a converged network. We defined the miRNA, mRNA, and lncRNA as the nodes of the network. If they interacted each other, we added an edge between them, then the converged three-layer network was constructed. Cytoscape software (https://cytoscape.org/) was used to visualize the network.
Calculation of Topology Attributes
Further calculate the topology attribute of the network according to the degree, betweenness centrality, and closeness centrality of the nodes in the three-layer network. For a given graph, G = (V, E), V and E represent a set of nodes and edges, respectively, and degree (D) is usually used to reflect the basic topology attributes of the network, indicating the number of nodes connected to neighboring nodes, assuming K is the number of edges connected to node v, and described as follows:
The betweenness centrality is one of the most effective methods for evaluating the importance of a node in the network. It is defined as the proportion of the shortest path through a node in the network to the total number of shortest paths, reflecting the influence of the node in the network. It plays an important role in the process of information transfer. And it can be seen from the definition that if a node is the only way for other nodes in the network to communicate, the node has an important position in the network. The higher the node betweenness centrality value, the greater its influence, the expression is as follows:
where ‘σst (v)’ represents the number of the shortest path between nodes ‘s’ and ‘t’ passing through node ‘v,’ and ‘σst’ represents the number of shortest paths between node pairs ‘s’ and ‘t.’
Closeness centrality reflects the proximity of a node in the network to other nodes. Closeness centrality of a node ‘v’ is the reciprocal of the average shortest path distance from node ‘v’ to all other nodes. That is, the closer a node is to other nodes, the greater its closeness centrality. Described as follows:
where ‘div’ is the shortest-path distance between ‘i’ and ‘v,’ and ‘|V|’ is the number of nodes in the network.
Extract a Subnetwork From Three-Layer Network
Find important nodes whose topological attributes including degree, betweenness centrality, and closeness centrality ranked in the top ten in the network, namely hub nodes. Then extract the hub nodes and their directly connected RNAs in a three-layer network, and construct a subnetwork related to hub nodes.
Enrichment Analysis
To achieve enrichment analysis of candidate genes in BC, we used DAVID 6.8(https://david.ncifcrf.gov/) for gene ontology(GO) analysis, including biological process (BP) analysis, cellular component (CC) analysis, and molecular function (MF) analysis, with p < 0.05 as a screening condition to find significantly enriched GO terms. Then, in order to more fully understand the situation of pathway enrichment, KOBAS 3.0 (http://kobas.cbi.pku.edu.cn/) was used as the metabolic pathway KEGG enrichment analysis, at P < 0.01, the number of genes enriched in the pathway was greater than or equal to 2 as a screening condition. By analyzing the significantly enriched GO biological process classification and KEGG pathways, we can further predict the biological function of DEmRNAs from gene function and pathway function, respectively.
Survival Analysis
According to the clinical information downloaded from TCGA database and the expression level of differentially expressed RNAs, the median was used as the cut-off point, and the RNAs expression was divided into two groups: high expression and low expression. Then, we performed Kaplan-Meier survival analysis on the differentially expressed RNAs, the RNAs in the three-layer converged network, and the RNAs in the subnetwork. The “survival” Package in R software was used to plot the survival curves and view RNAs that is significantly associated with the patient’s survival prognosis. P-value < 0.05 was considered statistically significant.
Results
Differentially Expressed RNA
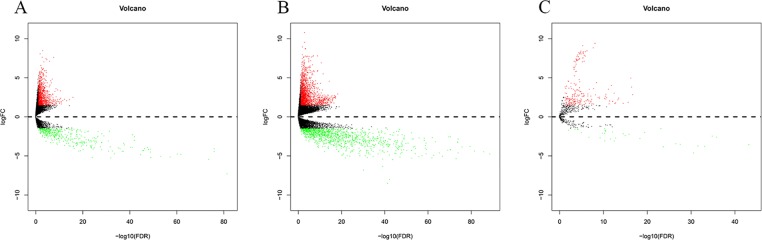
By comparing normal and cancer samples of BC, 1,457(1,036 upregulated, 421 downregulated, Figure 2A ) DElncRNAs, 3,047(1,823 upregulated, 1224 downregulated, Figure 2B ) DEmRNAs, and 247(214 upregulated, 33 downregulated, Figure 2C ) DEmiRNAs were obtained.
Figure 2.
Differentially expressed RNA in bladder cancer (|logFC| > 1.5 and P-value < 0.05). The Panels (A–C) are the volcano plot of 1457 DElncRNAs, 3047 DEmRNAs (B) and 247 DEmiRNAs. The red denotes upregulations and green denotes downregulations. The x-axis stands for the value of –log10(FDR) and the y-axis stands for the value of logFC.
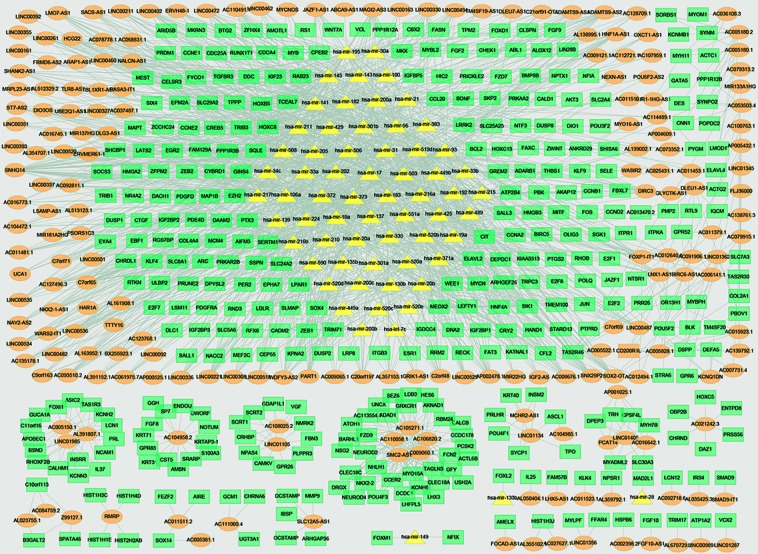
Construction of a Three-Layer Network
The lncRNA-mRNA coexpression network included 75 lncRNAs, 169 mRNAs, and 380 edges. The lncRNA-miRNA regulatory network constructed in the present study exhibited 129 lncRNAs and 35 miRNAs and 882 edges. And the regulatory network between miRNAs and target genes was established, which included 201 mRNAs, 56 miRNAs, and 487 edges.
Then, we constructed a three-layer network by merging lncRNA-miRNA regulatory network, miRNA-mRNA regulatory network, and lncRNA-mRNA coexpression network, which included 367 mRNAs, 65 miRNAs, 197 lncRNAs, and 1749 edges. The three-layer network was visualized with Cytoscape software and was shown in Figure 3 .
Figure 3.
Three-layer network in bladder cancer. Orange ellipse denotes DElncRNAs, green rectangle denotes DEmRNAs, and yellow triangle denotes DEmiRNAs.
Analysis of Topology Attributes
Further analyze the topology of the network and find out the important nodes in the network according to the degree, closeness centrality, and betweenness centrality of the nodes. There are two miRNAs (miRNA-93 and miRNA-195) appeared in each dimension, indicating that they have higher degree, betweenness centrality, and closeness centrality and played crucial roles in the three-layer network. The topology attributes of the top 10 genes were shown in Table 1 .
Table 1.
The top 10 genes in degree, betweenness centrality, and closeness centrality. Genes that appear in each column are shown in bold texts.
| Gene | Degree | Gene | Betweenness centrality | Gene | Closeness centrality |
|---|---|---|---|---|---|
| hsa-mir-93 | 77 | hsa-mir-93 | 18,473.228 | SNHG14 | 0.26729 |
| hsa-mir-195 | 67 | hsa-mir-195 | 17,484.967 | hsa-mir-93 | 0.25661 |
| hsa-mir-17 | 56 | FOXP1-IT1 | 10,830.214 | MAGI2-AS3 | 0.24637 |
| hsa-mir-106a | 54 | hsa-mir-211 | 7,355.569 | FOXP1-IT1 | 0.24375 |
| hsa-mir-211 | 52 | hsa-mir-21 | 7,301.496 | hsa-mir-195 | 0.24228 |
| hsa-mir-519d | 51 | SNHG14 | 7,076.982 | LINC00472 | 0.24083 |
| hsa-mir-373 | 48 | hsa-mir-182 | 6,246.760 | SNX29P2 | 0.23868 |
| hsa-mir-182 | 45 | hsa-mir-200a | 6,215.666 | ADAMTS9-AS2 | 0.23727 |
| hsa-mir-372 | 45 | hsa-mir-19a | 6,010.876 | hsa-mir-106a | 0.23518 |
| hsa-mir-145 | 42 | hsa-let-7c | 5,751.381 | LINC00520 | 0.23313 |
Functional Analysis
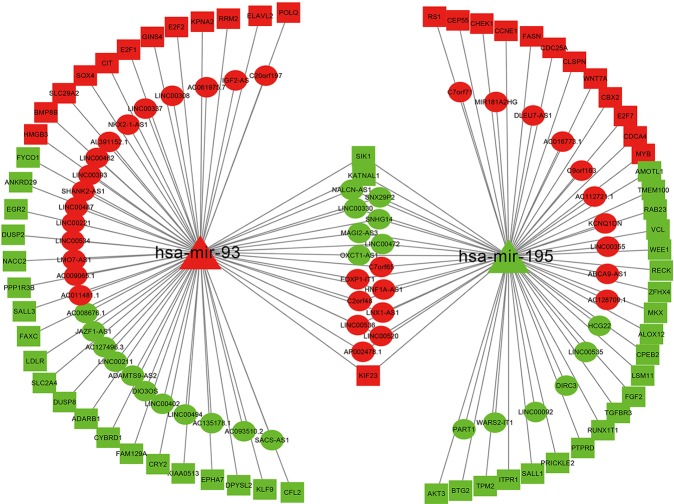
We extracted two key miRNAs and their linked mRNAs and lncRNAs from a three-layer network. Then rebuilt a new subnetwork, which included 71 mRNAs, 2 miRNAs, 73 lncRNAs, and 144 edges. The subnetwork was visualized with Cytoscape software and was shown in Figure 4 .
Figure 4.
Network related to miRNA-195 and miRNA-93 in bladder cancer. Ellipse represents for DElncRNAs, triangle represents for DEmiRNAs, and rectangle represents for DEmRNAs. Red denotes upregulations; green denotes downregulations.
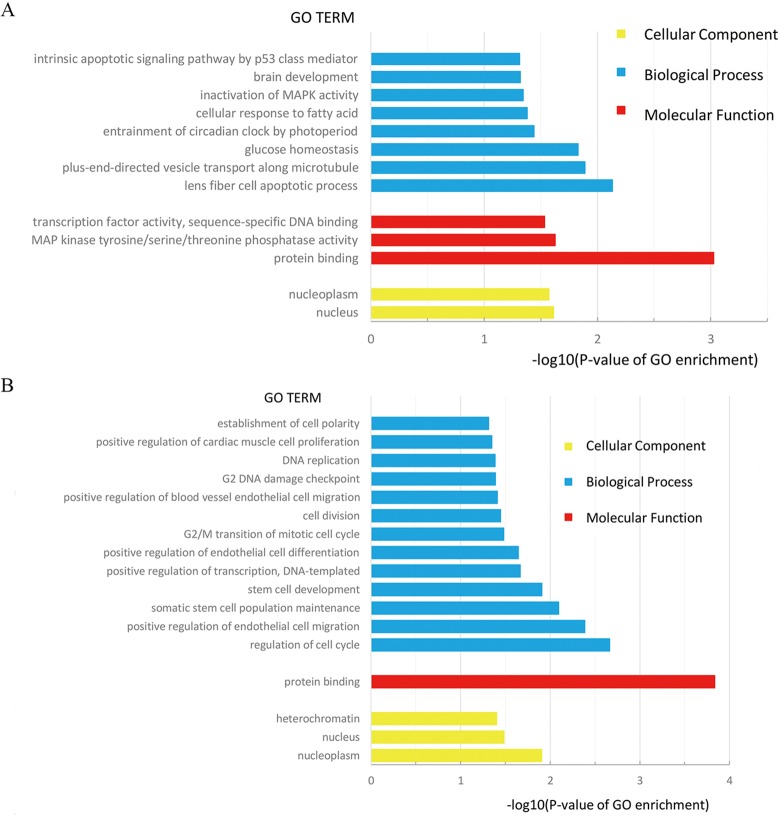
Through GO analysis of 35 target genes of miRNA-93 ( Figure 5A ), we found that the dysregulated mRNAs associated with several -BP-, including lens fiber cell apoptotic process and plus-end-directed vesicle transport along microtubule. And they are also related to nucleus and nucleoplasm in -CC-. Furthermore, based on -MF- analysis, we conclude that miRNA-93 may be involved in the biological processes of BC by regulating the expression of its target genes. The functions of its target gene are mainly glucose homeostasis, protein binding and MAP kinase tyrosine/serine/threonine phosphatase activity and transcription factor activity, sequence-specific DNA binding. Among them, the discovery of MAP kinase serine phosphatase activity in molecular function further confirms the previously reported results that miRNA-93 promotes BC cell proliferation and invasion by targeting PEDF, a member of the serine protease inhibitor superfamily (Jiang et al., 2019). The details of the GO term associated with 35 DEmRNAs are shown in Table 2 .
Figure 5.
(A) GO Term of 35 target genes of miRNA-93; (B) GO Term of 36 target genes of miRNA-195.
Table 2.
Gene ontology (GO) analysis of 35 target genes of miRNA-93 (list by PValue).
| Type | Term | Count | PValue | Genes |
|---|---|---|---|---|
| Molecular Function | protein binding | 26 | 0.000942 | E2F1, KIF23, E2F2, ADARB1, NACC2, HMGB3, EGR2, LDLR, GINS4, SOX4, ELAVL2, DPYSL2, EPHA7, DUSP2, CRY2, SLC2A4, CFL2, RRM2, CYBRD1, FAM129A, POLQ, KATNAL1, SIK1, CIT, FYCO1, KPNA2 |
| Biological Process | lens fiber cell apoptotic process | 2 | 0.007364701 | E2F1, E2F2 |
| Biological Process | plus-end-directed vesicle transport along microtubule | 2 | 0.012853753 | KIF23, FYCO1 |
| Biological Process | glucose homeostasis | 3 | 0.014869797 | CRY2, SLC2A4, SOX4 |
| Molecular Function | MAP kinase tyrosine/serine/threonine phosphatase activity | 2 | 0.023620021 | DUSP2, DUSP8 |
| Cellular Component | nucleus | 17 | 0.024386573 | E2F1, KIF23, NACC2, ADARB1, HMGB3, EGR2, KLF9, GINS4, SOX4, SALL3, DUSP2, CRY2, RRM2, SIK1, KATNAL1, DUSP8, KPNA2 |
| Cellular Component | nucleoplasm | 11 | 0.026806886 | KIF23, E2F1, E2F2, ADARB1, KLF9, RRM2, GINS4, SOX4, ELAVL2, POLQ, KPNA2 |
| Molecular Function | transcription factor activity, sequence-specific DNA binding | 6 | 0.029303045 | SALL3, E2F1, E2F2, EGR2, KLF9, SOX4 |
| Biological Process | entrainment of circadian clock by photoperiod | 2 | 0.036302096 | CRY2, SIK1 |
| Biological Process | cellular response to fatty acid | 2 | 0.041636205 | E2F1, LDLR |
| Biological Process | inactivation of MAPK activity | 2 | 0.045176392 | DUSP2, DUSP8 |
| Biological Process | brain development | 3 | 0.047788991 | EPHA7, EGR2, DPYSL2 |
| Biological Process | intrinsic apoptotic signaling pathway by p53 class mediator | 2 | 0.048703923 | E2F1, E2F2 |
We also performed GO analysis on 36 target genes of miRNA-195 ( Figure 5B ), and functional enrichment results showed that these differentially expressed genes mainly (30/36) focused on protein binding in molecular function. At the same time, we found that the -CC- associated with mRNA is nucleoplasm (p = 0.012459971), and the -BP- most strongly associated with mRNA is the regulation of cell cycle (p = 0.002164798). In addition, the most relevant and only relevant -MF- associated with mRNA is the protein binding (P = 0.000145982). Based on these data, it can be speculated that the target genes of miRNA-195 can form complexes with different proteins to participate in regulating the cell cycle and positive regulation of endothelial cell migration, somatic stem cell population maintenance, stem cell development, accordingly, have certain effects on the occurrence and development of BC. The details of the GO term associated with 36 DEmRNAs are displayed in Table 3 .
Table 3.
Gene ontology (GO) analysis of 36 target genes of miRNA-195 (list by PValue).
| Type | Term | Count | PValue | Genes |
|---|---|---|---|---|
| Molecular Function | protein binding | 30 | 0.000145982 | KIF23, CLSPN, E2F7, CBX2, CHEK1, CEP55, AMOTL1, VCL, CCNE1, RAB23, FASN, TMEM100, MYB, SIK1, FGF2, CDCA4, AKT3, RECK, PTPRD, RUNX1T1, CDC25A, ITPR1, WEE1, BTG2, SALL1, TGFBR3, LSM11, KATNAL1, WNT7A, ALOX12 |
| Biological Process | regulation of cell cycle | 4 | 0.002164798 | CCNE1, FGF2, CDC25A, WEE1 |
| Biological Process | Positive regulation of endothelial cell migration | 3 | 0.004123996 | WNT7A, FGF2, ALOX12 |
| Biological Process | somatic stem cell population maintenance | 3 | 0.008085217 | SALL1, WNT7A, FGF2 |
| Biological Process | stem cell development | 2 | 0.012442813 | WNT7A, FGF2 |
| Cellular Component | nucleoplasm | 12 | 0.012459971 | KIF23, CLSPN, CCNE1, E2F7, SALL1, RUNX1T1, LSM11, CHEK1, CBX2, AKT3, CDC25A, WEE1 |
| Biological Process | positive regulation of transcription, DNA-templated | 5 | 0.021591723 | CCNE1, SALL1, MYB, WNT7A, FGF2 |
| Biological Process | positive regulation of endothelial cell differentiation | 2 | 0.022696818 | TMEM100, ALOX12 |
| Cellular Component | nucleus | 17 | 0.032803617 | KIF23, CPEB2, E2F7, CHEK1, CBX2, WEE1, CDC25A, CCNE1, ZFHX4, SALL1, LSM11, MKX, SIK1, MYB, KATNAL1, FGF2, CDCA4 |
| Biological Process | G2/M transition of mitotic cell cycle | 3 | 0.032988025 | CHEK1, CDC25A, WEE1 |
| Biological Process | cell division | 4 | 0.035939287 | CCNE1, CDC25A, CDCA4, WEE1 |
| Biological Process | positive regulation of blood vessel endothelial cell migration | 2 | 0.038888456 | AMOTL1, FGF2 |
| Cellular Component | heterochromatin | 2 | 0.039587501 | SALL1, CBX2 |
| Biological Process | G2 DNA damage checkpoint | 2 | 0.040893995 | CLSPN, CHEK1 |
| Biological Process | DNA replication | 3 | 0.041293616 | CLSPN, CHEK1, CDC25A |
| Biological Process | positive regulation of cardiac muscle cell proliferation | 2 | 0.044892884 | TGFBR3, FGF2 |
| Biological Process | establishment of cell polarity | 2 | 0.048875575 | WNT7A, WEE1 |
KEGG enrichment analysis of metabolic pathways for miRNA-93 and miRNA-195 targeted mRNAs showed that a total of 28 genes are enriched in 39 pathways, of which 10 target genes of miRNA-93 are mainly involved in the regulation of 16 KEGG signaling pathways including cancer pathways and Hepatitis B and Axon guidance and other pathways, the 19 target genes of miRNA195 are mainly involved in the regulation of 31 KEGG signaling pathways including many cancer pathways and Cell cycle, HTLV-I infection ( Figure 6 ). Demonstrating that miRNAs can play a role in a variety of BC signaling pathways. In addition, we found that the signaling pathways “MicroRNAs in cancer,” “HTLV-I infection,” and “cell cycle” are significantly enriched KEGG pathways, indicating that these three signaling pathways are closely related to BC. The details of the KEGG pathway enrichment are displayed in Table 4 .
Figure 6.
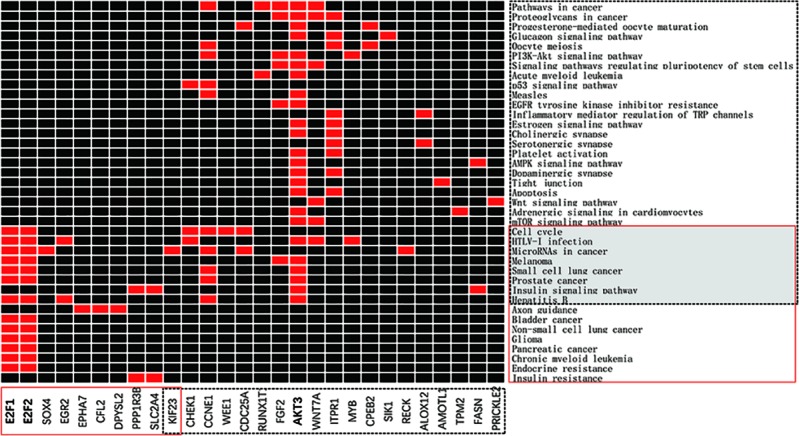
miRNA-targeted mRNA enrichment pathway. The x-axis represents the genes in the network, in which the red solid line is the target genes of miRNA-93, and the black dotted line is the target genes of miRNA-195. The y-axis represents the KEGG pathway, the red solid box is the enrichment pathway of the miRNA-93 target genes, and the black dotted box is the enrichment pathway of the miRNA-195 target genes.
Table 4.
KEGG pathways enriched by 71 DEmRNAs involved in the Hub network.
| ID | Term description | Count | P-Value | Genes |
|---|---|---|---|---|
| mRNAs targeted by hsa-mir-93 | ||||
| hsa05206 | MicroRNAs in cancer | 4 | 0.000143218 | E2F1, KIF23, SOX4, E2F2 |
| hsa05161 | Hepatitis B | 3 | 0.000308336 | E2F1, E2F2, EGR2 |
| hsa04360 | Axon guidance | 3 | 0.000526831 | EPHA7, CFL2, DPYSL2 |
| hsa05219 | Bladder cancer | 2 | 0.000663363 | E2F1, E2F2 |
| hsa05223 | Nonsmall cell lung cancer | 2 | 0.001204323 | E2F1, E2F2 |
| hsa05166 | HTLV-I infection | 3 | 0.001580128 | E2F1, E2F2, EGR2 |
| hsa05214 | Glioma | 2 | 0.00160289 | E2F1, E2F2 |
| hsa05212 | Pancreatic cancer | 2 | 0.001650553 | E2F1, E2F2 |
| hsa05218 | Melanoma | 2 | 0.00189891 | E2F1, E2F2 |
| hsa05220 | Chronic myeloid leukemia | 2 | 0.002002919 | E2F1, E2F2 |
| hsa05222 | Small cell lung cancer | 2 | 0.002743232 | E2F1, E2F2 |
| hsa05215 | Prostate cancer | 2 | 0.002929728 | E2F1, E2F2 |
| hsa01522 | Endocrine resistance | 2 | 0.003455352 | E2F1, E2F2 |
| hsa04931 | Insulin resistance | 2 | 0.00431996 | PPP1R3B, SLC2A4 |
| hsa04110 | Cell cycle | 2 | 0.005526668 | E2F1, E2F2 |
| hsa04910 | Insulin signaling pathway | 2 | 0.006869918 | PPP1R3B, SLC2A4 |
| mRNAs targeted by hsa-mir-195 | ||||
| hsa04110 | Cell cycle | 4 | 0.00000555 | CHEK1, CCNE1, WEE1, CDC25A |
| hsa05200 | Pathways in cancer | 5 | 0.0000299 | CCNE1, RUNX1T1, FGF2, AKT3, WNT7A |
| hsa05205 | Proteoglycans in cancer | 4 | 0.0000382 | FGF2, AKT3, WNT7A, ITPR1 |
| hsa05166 | HTLV-I infection | 4 | 0.000093 | CHEK1, AKT3, WNT7A, MYB |
| hsa04914 | Progesterone-mediated oocyte maturation | 3 | 0.00010659 | CDC25A, AKT3, CPEB2 |
| hsa04922 | Glucagon signaling pathway | 3 | 0.000119603 | AKT3, ITPR1, SIK1 |
| hsa05206 | MicroRNAs in cancer | 4 | 0.000160144 | CCNE1, CDC25A, KIF23, RECK |
| hsa04114 | Oocyte meiosis | 3 | 0.000204986 | CCNE1, ITPR1, CPEB2 |
| hsa04151 | PI3K-Akt signaling pathway | 4 | 0.000265539 | CCNE1, FGF2, AKT3, MYB |
| hsa04550 | Signaling pathways regulating pluripotency of stem cells | 3 | 0.00030972 | FGF2, AKT3, WNT7A |
| hsa05221 | Acute myeloid leukemia | 2 | 0.001317859 | RUNX1T1, AKT3 |
| hsa04115 | p53 signaling pathway | 2 | 0.001901017 | CHEK1, CCNE1 |
| hsa05218 | Melanoma | 2 | 0.00200813 | FGF2, AKT3 |
| hsa01521 | EGFR tyrosine kinase inhibitor resistance | 2 | 0.002585615 | FGF2, AKT3 |
| hsa05222 | Small cell lung cancer | 2 | 0.002900294 | CCNE1, AKT3 |
| hsa05215 | Prostate cancer | 2 | 0.003097313 | CCNE1, AKT3 |
| hsa04750 | Inflammatory mediator regulation of TRP channels | 2 | 0.00372495 | ITPR1, ALOX12 |
| hsa04915 | Estrogen signaling pathway | 2 | 0.003798048 | AKT3, ITPR1 |
| hsa04725 | Cholinergic synapse | 2 | 0.004726968 | AKT3, ITPR1 |
| hsa04726 | Serotonergic synapse | 2 | 0.004808654 | ITPR1, ALOX12 |
| hsa04611 | Platelet activation | 2 | 0.005661192 | AKT3, ITPR1 |
| hsa04152 | AMPK signaling pathway | 2 | 0.005929498 | AKT3, FASN |
| hsa04728 | Dopaminergic synapse | 2 | 0.006389399 | AKT3, ITPR1 |
| hsa05162 | Measles | 2 | 0.0069621 | CCNE1, AKT3 |
| hsa04530 | Tight junction | 2 | 0.007256895 | AKT3, AMOTL1 |
| hsa04910 | Insulin signaling pathway | 2 | 0.007256895 | AKT3, FASN |
| hsa04210 | Apoptosis | 2 | 0.007356404 | AKT3, ITPR1 |
| hsa04310 | WNT signaling pathway | 2 | 0.007658643 | WNT7A, PRICKLE2 |
| hsa05161 | Hepatitis B | 2 | 0.00796643 | CCNE1, AKT3 |
| hsa04261 | Adrenergic signaling in cardiomyocytes | 2 | 0.008279735 | AKT3, TPM2 |
| hsa04150 | mTOR signaling pathway | 2 | 0.008814086 | AKT3, WNT7A |
Since E2F1 and E2F2 are enriched in various KEGG signaling pathways, they may be important target genes of miRNA-93. Similarly, AKT3 may be an important target gene of miRNA-195. From the functions annotated in the genes E2F1 and E2F2 in GO, we can speculate that the genes E2F1 and E2F2 are present on the nucleoplasm, and participate in the lens fiber cell apoptosis process or intrinsic apoptotic signaling pathway by p53 class mediator by forming a complex with a certain protein or transcription factor activity, sequence-specific DNA binding, which is consistent with the results reported previously (Castillo et al., 2015). In addition, PI3K-Akt and AMPK are reported to be positive and negative regulators of mTOR activity, respectively, and mTOR is a well-known sensor for extracellular nutrients and growth factors, which converges many key signals and is widely involved in cell cycle, cell proliferation, and cellular metabolic processes, and has important biological roles in regulating apoptosis, physiological, and pathological processes (Ciuffreda et al., 2010). The PI3K-Akt-mTOR signaling pathway has been shown to play a central role in several cellular functions, including survival, proliferation, adhesion, migration, differentiation, and metabolism (Gao et al., 2003). Activated AMPK inhibits mTOR and further inhibits protein synthesis, cell proliferation, cell cycle progression, and angiogenesis (Jalving et al., 2010).This study found that the expression of AKT3 is associated with these pathways, suggesting that AKT3 may participate in important processes such as cell proliferation and apoptosis through these pathways, ultimately affecting the biological behavior of tumor cells.
KEGG analysis of the downregulated target genes of miRNA-93 revealed that EPHA7, CFL2, and DPYSL2 are significantly enriched in axon guidance pathway (P = 0.000129528), and PPP1R3B and SLC2A4 are significantly attached to insulin resistance pathway (p = 0.001718367), we hypothesized that the activated axon guidance pathway and the insulin resistance pathway may be responsible for the downregulation of miRNA-93 in BC. KEGG analysis of the upregulated target genes of miRNA-195 revealed that CHEK1, CCNE1, and CDC25A are significantly enriched in the cell cycle pathway (p = 0.00000888), and CHEK1, MYB, and WNT7A are enriched in HTLV-I infection pathway (p = 0.000077), CCNE1, KIF23, and CDC25A enriches microRNAs in cancer (p = 0.000117168), CHEK1 and CCNE1 enriches p53 signaling pathway (p = 0.000241969). Therefore, we believe that “cell cycle, HTLV-I infection, MicroRNAs in cancer, p53 signaling pathway” may be related to downregulation of miRNA-195 in BC.
Survival Analysis
We performed Kaplan-Meier survival analysis to predict overall survival (OS) of DERNAs in BC. As a result, we found that 253 of the 1457 differentially expressed lncRNAs were related to survival, 38 of the 247 differentially expressed miRNAs were associated with survival, and 625 of the 3047 differentially expressed mRNAs were related to survival ( Supplementary Material Tables S1–S3 ). Similarly, Kaplan-Meier survival analysis was performed on miRNAs/lncRNAs and mRNAs in the three-layer converged network, and we found that 39 of the 197 lncRNAs were related to survival, 15 of the 65 miRNAs were related to survival, and 90 of the 367 mRNAs were related to survival ( Supplementary Material Tables S4–S6 ).
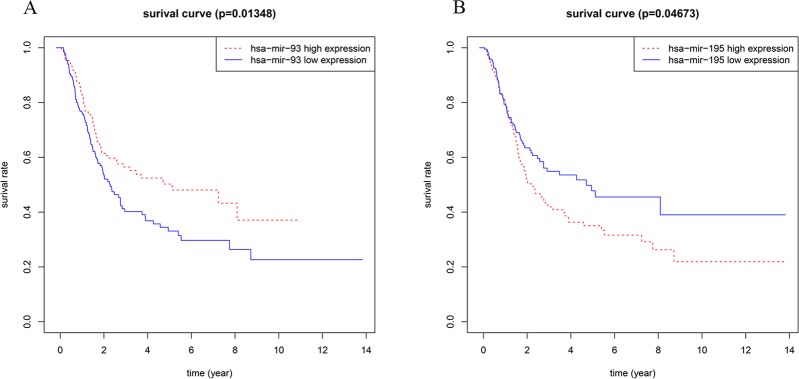
In addition, we also performed Kaplan-Meier survival analysis on miRNAs/lncRNAs and mRNAs in the subnetwork. Among them, miRNA-93 is highly correlated with overall survival of BC ( Figure 7A ). A large number of studies have shown that miRNA-93 can be widely involved in various biological behaviors of tumor cells and affect the development of tumors (Li et al., 2014). Therefore, in BC, the high expression of miRNA-93 may promote the proliferation of cancer cells and inhibit its apoptosis, which can lead to excessive proliferation and deterioration of cancer cells. miRNA-195 also has a correlation with the overall survival of BC ( Figure 7B ). Related studies (Fei et al., 2012) showed that miRNA-195 is a tumor suppressor gene, which inhibits cell proliferation and promotes apoptosis when normal expression, and downregulation leads to cancer, while miRNA -195 is downregulated in BC, so it may be closely related to the occurrence and development of BC. And we also found that nine lncRNAs (AC127496.3, ADAMTS9-AS2, C7orf65, LINC00536, MAGI2-AS3, SACS-AS1, AC016773.1, AC112721.1, and HCG22) were significantly associated with survival outcomes in patients with BC, among which MAGI2-AS3, C7orf65, and LINC00536 are lncRNAs linked to both miRNAs in the network. Twenty-four mRNAs (KATNAL1, ANKRD29, CFL2, DPYSL2, DUSP2, EGR2, FAM129A, KPNA2, LDLR, RRM2, AKT3, AMOTL1, BTG2, CPEB2, FASN, FGF2, LSM11, PTPRD, RAB23, RS1, RUNX1T1, TMEM100, VCL, and ZFHX4) are closely related to the survival prognosis of patients, 10 of which are target genes of miRNA-93, and 15 are target genes of miRNA-195. And KATNAL1 is a mRNA that is simultaneously regulated by two miRNAs in a network related to BC ( Supplementary Material Figure S1 ).
Figure 7.
(A) Survival curves of OS for miRNA-93; (B) Survival curves of OS for miRNA-195. The x-axis denotes the time of OS and y-axis denotes the survival rate.
Discussion
A large number of studies have shown that the development of BC is highly correlated with the abnormal expression of noncoding RNA and protein-coding genes. However, there are still few studies on the key molecules of BC and abnormal gene expression patterns and pathways in BC. In this study, we constructed a three-layer network by merging lncRNA-miRNA regulatory network, miRNA-mRNA regulatory network, and lncRNA-mRNA coexpression network, and calculated the topology attributes of each node in the network, including degree, closeness centrality, and betweenness centrality. Our study suggested that miRNA-93 and miRNA-195 are controllers of the three-layer network associated with BC and a regulator of numerous target genes, and their dysregulation may be closely related to the pathogenesis of BC. A recent study also showed that miRNA-93 is upregulated in BC and is associated with tumor stage and lymph node metastasis (Jiang et al., 2019). And studies have reported that miRNA-195 inhibits glucose uptake and proliferation of BC T24 cells by inhibiting GLUT3 expression (Fei et al., 2012), another study found that miRNA-195 inhibits BC cell proliferation by inhibiting Cdc42 / STAT3 signaling (Zhao et al., 2015).
To clarify the significance of miRNA-93 and miRNA-195 in BC as accurately as possible, we extracted mRNAs and lncRNAs directly linked to miRNA-195 and miRNA-93 from a three-layer network, and built a new subnetwork. Then, we assigned miRNA-targeted mRNA to terms in GO and known pathways in the KEGG database. Through GO analysis, we found that DEmRNAs in the network is mainly concentrated in the protein binding in molecular function, suggesting that the protein-coding genes play vital roles in the development of BC. What’s more, the result from KEGG pathway analysis showed that dysregulated mRNAs contribute to the development of BC through abnormal activation of many pathways. Therefore, we concluded that miRNA-93 and miRNA-195 may participate in various biological processes by regulating the expression of target genes with different functions in BC, which may have an important impact on the occurrence and development of BC. In addition, in recent years, a large number of studies have shown that miRNA-93 and miRNA-195 play an important role in the development of tumors. miRNA-93 overexpression was associated with tumor progression, metastasis and poor prognosis in patients with head and neck squamous cell carcinoma (Li et al., 2015).And inhibiting the expression of miRNA-93 can inhibit the proliferation and migration of gastric cancer and liver cancer cells (Xu et al., 2012; Zhang et al., 2016). In addition, studies have shown that miR-195 has tumor suppressive properties in gastric cancer and glioma (Deng et al., 2013; Hui et al., 2013). Some evidences indicate that miRNA-195 can inhibit the ability of liver cancer cells to promote endothelial cell migration and angiogenesis, and can directly inhibit the migration and invasion of liver cancer cells, and inhibit liver cancer. Some evidences indicate that miRNA-195 can inhibit the ability of hepatoma cells to promote endothelial cell migration and angiogenesis, and can directly inhibit the migration and invasion of hepatoma cells, thereby inhibiting liver cancer (Wang et al., 2013).In osteosarcoma, miRNA-195 inhibits tumor migration and invasion by acting on fatty acid synthase (Mao et al., 2012).Based on these reports and current findings, we believe that the study of the mechanisms of action of miRNA-93 and miRNA-195 has profound implications for the clinical treatment of tumors.
Through survival analysis, we identified several RNAs associated with the prognosis of BC, some of which have not been reported. It is worth noting that the miRNAs of forming the hub nodes are related to survival. Previous studies have shown that the abnormal expression of miRNA-93 and miRNA-195 can promote the proliferation of cancer cells and inhibit their apoptosis (Fei et al., 2012; Li et al., 2014). Therefore, they may be involved in and influence the occurrence and development of BC. In addition, some of the genes in the subnetwork are often present in different cancers. For example, studies have found that ADAMTS9-AS2 is significantly downregulated in glioma tissues compared with normal tissues, and negatively correlated with prognosis (Yao et al., 2014). Downregulation of ADAMTS9-AS2 is associated with poor prognosis in patients with gastric cancer (Cao et al., 2018a). Low expression of ADAMTS9-AS2 is associated with overall survival in patients with ovarian cancer (Wang et al., 2018a). And ADAMTS9-AS2 is significantly upregulated in TSCC tissues of patients with lymph node metastasis and is closely related to poor prognosis (Li et al., 2019b). Besides, there are reports that the downregulation of MAGI2-AS3 in BC patients is associated with poor prognosis (Wang et al., 2018b) and the high expression of LINC00536 is negatively correlated with the survival rate of patients with BC(Li et al., 2019a), this is consistent with our results. These reports help identify and improve the accuracy of prognostic predictions.
In this work, we considered the relations of differentially expressed genes to construct the networks. As we know, the human body is a complex system. Maybe, there are some genes that do not differentially express have effects on the oncogenesis and progression of BC. In addition, we analyzed how the key genes affect other genes. It is meaningful to study the relation between key genes with proteins or other chemical molecules from the perspective of pathway function. We are currently working to integrate various valid data sets and try to develop more reliable prediction methods to provide reasonable and valuable candidate factors for the early diagnosis and treatment of BC.
Conclusion
Taken together, we found that miRNA-93 and miRNA-195 are key factors for the development of BC by calculating the topology attributes of the nodes in the three-layer network. In addition, many mRNAs with different functions were found from KEGG and GO analysis, of which E2F1 and E2F2 are important target genes of miRNA-93, and AKT3 is an important target gene of miRNA-195, their dysregulation may be closely related to cell proliferation and apoptosis. Further the Kaplan-Meier curve analysis revealed that miRNA-93 and miRNA-195 are associated with poor prognosis of BC. In summary, our results have great significance in clinical practice, which can provide a reference for a better understanding of the occurrence and development of BC.
Data Availability Statement
The datasets analyzed for this study can be found in the Supplementary Material .
Author Contributions
XW and YD designed the research ideas. XW collected and organized the data. XW, YD, JW, and YW analyzed the data. XW wrote the manuscript.
Funding
This work was funded by The National Natural Science Foundation of China (grant numbers 21541006, 61772241).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2019.01398/full#supplementary-material
References
- Braicu C., Cojocneanu-Petric R., Chira S., Truta A., Floares A., Petrut B., et al. (2015). Clinical and pathological implications of miRNA in bladder cancer. Int. J Nanomed. 10, 791–800. 10.2147/IJN.S72904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray F., Ferlay J., Soerjomataram I., Siegel R. L., Torre L. A., Jemal A. (2018). Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 68, 394–424. 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- Cao B., Liu C., Yang G. (2018. a). Down-regulation of lncRNA ADAMTS9-AS2 contributes to gastric cancer development via activation of PI3K/Akt pathway. Biomed. Pharmacother. 107, 185–193. 10.1016/j.biopha.2018.06.146 [DOI] [PubMed] [Google Scholar]
- Cao Y. P., Zhou J., Li W. J., Shao Y., Zheng S. Y., Tian T., et al. (2018. b). Long non-coding RNA expression profiles for the characterization of different bladder cancer grade. Cell. Physiol. Biochem. 50, 1154–1163. 10.1159/000494542 [DOI] [PubMed] [Google Scholar]
- Castillo D. S., Campalans A., Belluscio L. M., Carcagno A. L., Radicella J. P., Canepa E. T., et al. (2015). E2F1 and E2F2 induction in response to DNA damage preserves genomic stability in neuronal cells. Cell Cycle 14, 1300–1314. 10.4161/15384101.2014.985031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catto J. W., Alcaraz A., Bjartell A. S., De Vere White R., Evans C. P., Fussel S., et al. (2011). MicroRNA in prostate, bladder, and kidney cancer: a systematic review. Eur. Urol. 59, 671–681. 10.1016/j.eururo.2011.01.044 [DOI] [PubMed] [Google Scholar]
- Chen X., Yan C. C., Zhang X., You Z. H. (2017). Long non-coding RNAs and complex diseases: from experimental results to computational models. Brief. Bioinform. 18, 558–576. 10.1093/bib/bbw060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Sun Y. Z., Guan N. N., Qu J., Huang Z. A., Zhu Z. X., et al. (2019. a). Computational models for lncRNA function prediction and functional similarity calculation. Brief. Funct. Genomics 18, 58–82. 10.1093/bfgp/ely031 [DOI] [PubMed] [Google Scholar]
- Chen X., Xie D., Zhao Q., You Z. H. (2019. b). MicroRNAs and complex diseases: from experimental results to computational models. Brief. Bioinform. 20, 515–539. 10.1093/bib/bbx130 [DOI] [PubMed] [Google Scholar]
- Ciuffreda L., Di Sanza C., Incani U. C., Milella M. (2010). The mTOR pathway: a new target in cancer therapy. Curr. Cancer Drug Targets 10, 484–495. 10.2174/156800910791517172 [DOI] [PubMed] [Google Scholar]
- Deng H., Guo Y., Song H., Xiao B., Sun W., Liu Z., et al. (2013). MicroRNA-195 and microRNA-378 mediate tumor growth suppression by epigenetical regulation in gastric cancer. Gene 518, 351–359. 10.1016/j.gene.2012.12.103 [DOI] [PubMed] [Google Scholar]
- Fei X., Qi M., Wu B., Song Y., Wang Y., Li T. (2012). MicroRNA-195-5p suppresses glucose uptake and proliferation of human bladder cancer T24 cells by regulating GLUT3 expression. FEBS Lett. 586, 392–397. 10.1016/j.febslet.2012.01.006 [DOI] [PubMed] [Google Scholar]
- Gao N., Zhang Z., Jiang B. H., Shi X. (2003). Role of PI3K/AKT/mTOR signaling in the cell cycle progression of human prostate cancer. Biochem. Biophys. Res. Commun. 310, 1124–1132. 10.1016/j.bbrc.2003.09.132 [DOI] [PubMed] [Google Scholar]
- Greenbury S. F., Johnston I. G., Smith M. A., Doye J. P., Louis A. A. (2010). The effect of scale-free topology on the robustness and evolvability of genetic regulatory networks. J. Theor. Biol. 267, 48–61. 10.1016/j.jtbi.2010.08.006 [DOI] [PubMed] [Google Scholar]
- Guancial E. A., Bellmunt J., Yeh S., Rosenberg J. E., Berman D. M. (2014). The evolving understanding of microRNA in bladder cancer. Urol. Oncol. 32, 41e31–e40. 10.1016/j.urolonc.2013.04.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui W., Yuntao L., Lun L., Wensheng L., Chaofeng L., Haiyong H., et al. (2013). MicroRNA-195 inhibits the proliferation of human glioma cells by directly targeting cyclin D1 and cyclin E1. PLoS One 8, e54932. 10.1371/journal.pone.0054932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalving M., Gietema J. A., Lefrandt J. D., De Jong S., Reyners A. K., Gans R. O., et al. (2010). Metformin: taking away the candy for cancer? Eur. J. Cancer 46, 2369–2380. 10.1016/j.ejca.2010.06.012 [DOI] [PubMed] [Google Scholar]
- Jiang Y., Han Y., Sun C., Han C., Han N., Zhi W., et al. (2016). Rab23 is overexpressed in human bladder cancer and promotes cancer cell proliferation and invasion. Tumour Biol. 37, 8131–8138. 10.1007/s13277-015-4590-9 [DOI] [PubMed] [Google Scholar]
- Jiang H., Bu Q., Zeng M., Xia D., Wu A. (2019). MicroRNA-93 promotes bladder cancer proliferation and invasion by targeting PEDF. Urol. Oncol. 37, 150–157. 10.1016/j.urolonc.2018.08.001 [DOI] [PubMed] [Google Scholar]
- Jin N., Jin X., Gu X., Na W., Zhang M., Zhao R. (2015). Screening biomarkers of bladder cancer using combined miRNA and mRNA microarray analysis. Mol. Med. Rep. 12, 3170–3176. 10.3892/mmr.2015.3739 [DOI] [PubMed] [Google Scholar]
- Kiriluk K. J., Prasad S. M., Patel A. R., Steinberg G. D., Smith N. D. (2012). Bladder cancer risk from occupational and environmental exposures. Urol. Oncol. 30, 199–211. 10.1016/j.urolonc.2011.10.010 [DOI] [PubMed] [Google Scholar]
- Knollman H., Godwin J. L., Jain R., Wong Y. N., Plimack E. R., Geynisman D. M. (2015). Muscle-invasive urothelial bladder cancer: an update on systemic therapy. Ther. Adv. Urol. 7, 312–330. 10.1177/1756287215607418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li F., Liang X., Chen Y., Li S., Liu J. (2014). Role of microRNA-93 in regulation of angiogenesis. Tumour Biol. 35, 10609–10613. 10.1007/s13277-014-2605-6 [DOI] [PubMed] [Google Scholar]
- Li G., Ren S., Su Z., Liu C., Deng T., Huang D., et al. (2015). Increased expression of miR-93 is associated with poor prognosis in head and neck squamous cell carcinoma. Tumour Biol. 36, 3949–3956. 10.1007/s13277-015-3038-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Liu J., Zou D., Cai X., Wang J., Wang J., et al. (2017). Exploration of bladder cancer molecular mechanisms based on miRNA-mRNA regulatory network. Oncol. Rep. 37, 1461–1468. 10.3892/or.2017.5433 [DOI] [PubMed] [Google Scholar]
- Li M., Liu Y., Zhang X., Liu J., Wang P. (2018). Transcriptomic analysis of high-throughput sequencing about circRNA, lncRNA and mRNA in bladder cancer. Gene 677, 189–197. 10.1016/j.gene.2018.07.041 [DOI] [PubMed] [Google Scholar]
- Li R., Zhang L., Qin Z., Wei Y., Deng Z., Zhu C., et al. (2019. a). High LINC00536 expression promotes tumor progression and poor prognosis in bladder cancer. Exp. Cell Res. 378, 32–40. 10.1016/j.yexcr.2019.03.009 [DOI] [PubMed] [Google Scholar]
- Li Y., Wan Q., Wang W., Mai L., Sha L., Mashrah M., et al. (2019. b). LncRNA ADAMTS9-AS2 promotes tongue squamous cell carcinoma proliferation, migration and EMT via the miR-600/EZH2 axis. Biomed. Pharmacother. 112, 108719. 10.1016/j.biopha.2019.108719 [DOI] [PubMed] [Google Scholar]
- Lv M., Zhong Z., Huang M., Tian Q., Jiang R., Chen J. (2017). lncRNA H19 regulates epithelial-mesenchymal transition and metastasis of bladder cancer by miR-29b-3p as competing endogenous RNA. Biochim. Biophys. Acta Mol. Cell Res. 1864, 1887–1899. 10.1016/j.bbamcr.2017.08.001 [DOI] [PubMed] [Google Scholar]
- Mao J. H., Zhou R. P., Peng A. F., Liu Z. L., Huang S. H., Long X. H., et al. (2012). microRNA-195 suppresses osteosarcoma cell invasion and migration in vitro by targeting FASN. Oncol. Lett. 4, 1125–1129. 10.3892/ol.2012.863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao L., Liu H. Y., Zhou C., He X. (2019). LINC00612 enhances the proliferation and invasion ability of bladder cancer cells as ceRNA by sponging miR-590 to elevate expression of PHF14. J. Exp. Clin. Cancers Res. 38, 143. 10.1186/s13046-019-1149-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonato F., Ventura L., Sartori N., Cappellesso R., Fassan M., Busund L. T., et al. (2013). Detection of microRNAs in archival cytology urine smears. PLoS One 8, e57490. 10.1371/journal.pone.0057490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith N. D., Prasad S. M., Patel A. R., Weiner A. B., Pariser J. J., Razmaria A., et al. (2016). Bladder cancer mortality in the United States: a geographic and temporal analysis of socioeconomic and environmental factors. J. Urol. 195, 290–296. 10.1016/j.juro.2015.07.091 [DOI] [PubMed] [Google Scholar]
- Teng J., Ai X., Jia Z., Wang K., Guan Y., Guo Y. (2019). Long non-coding RNA ARAP1-AS1 promotes the progression of bladder cancer by regulating miR-4735-3p/NOTCH2 axis. Cancer Biol. Ther. 20, 552–561. 10.1080/15384047.2018.1538613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian Y. F., Zhou H., Yu G., Wang J., Li H., Xia D., et al. (2015). Prognostic significance of lymphovascular invasion in bladder cancer after surgical resection: a meta-analysis. J. Huazhong Univ. Sci. Technol. Med. Sci. 35, 646–655. 10.1007/s11596-015-1484-4 [DOI] [PubMed] [Google Scholar]
- Tsui K. H., Chiang K. C., Lin Y. H., Chang K. S., Feng T. H., Juang H. H. (2018). BTG2 is a tumor suppressor gene upregulated by p53 and PTEN in human bladder carcinoma cells. Cancer Med. 7, 184–195. 10.1002/cam4.1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Roekel E. H., Cheng K. K., James N. D., Wallace D. M., Billingham L. J., Murray P. G., et al. (2013). Smoking is associated with lower age, higher grade, higher stage, and larger size of malignant bladder tumors at diagnosis. Int. J. Cancer 133, 446–454. 10.1002/ijc.28017 [DOI] [PubMed] [Google Scholar]
- Wang R., Zhao N., Li S., Fang J. H., Chen M. X., Yang J., et al. (2013). MicroRNA-195 suppresses angiogenesis and metastasis of hepatocellular carcinoma by inhibiting the expression of VEGF, VAV2, and CDC42. Hepatology 58, 642–653. 10.1002/hep.26373 [DOI] [PubMed] [Google Scholar]
- Wang H., Niu L., Jiang S., Zhai J., Wang P., Kong F., et al. (2016). Comprehensive analysis of aberrantly expressed profiles of lncRNAs and miRNAs with associated ceRNA network in muscle-invasive bladder cancer. Oncotarget 7, 86174–86185. 10.18632/oncotarget.13363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Chen L., Yang Y., Zhang M., Wong G. (2017). Identification of bladder cancer prognostic biomarkers using an ageing gene-related competitive endogenous RNA network. Oncotarget 8, 111742–111753. 10.18632/oncotarget.22905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A., Jin C., Li H., Qin Q., Li L. (2018. a). LncRNA ADAMTS9-AS2 regulates ovarian cancer progression by targeting miR-182-5p/FOXF2 signaling pathway. Int. J. Biol. Macromol. 120, 1705–1713. 10.1016/j.ijbiomac.2018.09.179 [DOI] [PubMed] [Google Scholar]
- Wang F., Zu Y., Zhu S., Yang Y., Huang W., Xie H., et al. (2018. b). Long noncoding RNA MAGI2-AS3 regulates CCDC19 expression by sponging miR-15b-5p and suppresses bladder cancer progression. Biochem. Biophys. Res. Commun. 507, 231–235. 10.1016/j.bbrc.2018.11.013 [DOI] [PubMed] [Google Scholar]
- Wilcox A. N., Silverman D. T., Friesen M. C., Locke S. J., Russ D. E., Hyun N., et al. (2016). Smoking status, usual adult occupation, and risk of recurrent urothelial bladder carcinoma: data from The Cancer Genome Atlas (TCGA) Project. Cancer Causes Control 27, 1429–1435. 10.1007/s10552-016-0821-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D., He X. X., Chang Y., Sun S. Z., Xu C. R., Lin J. S. (2012). Downregulation of MiR-93 expression reduces cell proliferation and clonogenicity of HepG2 cells. Hepatogastroenterology 59, 2367–2373. 10.5754/hge12458 [DOI] [PubMed] [Google Scholar]
- Yao J., Zhou B., Zhang J., Geng P., Liu K., Zhu Y., et al. (2014). A new tumor suppressor LncRNA ADAMTS9-AS2 is regulated by DNMT1 and inhibits migration of glioma cells. Tumour Biol. 35, 7935–7944. 10.1007/s13277-014-1949-2 [DOI] [PubMed] [Google Scholar]
- Zhang Q., Su M., Lu G., Wang J. (2013). The complexity of bladder cancer: long noncoding RNAs are on the stage. Mol. Cancer 12, 101. 10.1186/1476-4598-12-101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Li F., Wang W., Wang X., Li S., Liu J. (2016). The effect of antisense inhibitor of miRNA 106b approximately 25 on the proliferation, invasion, migration, and apoptosis of gastric cancer cell. Tumour Biol. 37, 10507–10515. 10.1007/s13277-016-4937-x [DOI] [PubMed] [Google Scholar]
- Zhao C., Qi L., Chen M., Liu L., Yan W., Tong S., et al. (2015). microRNA-195 inhibits cell proliferation in bladder cancer via inhibition of cell division control protein 42 homolog/signal transducer and activator of transcription-3 signaling. Exp. Ther. Med. 10, 1103–1108. 10.3892/etm.2015.2633 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Zhao F., Ge Y. Z., Zhou L. H., Xu L. W., Xu Z., Ping W. W., et al. (2017). Identification of hub miRNA biomarkers for bladder cancer by weighted gene coexpressions network analysis. Onco Targets Ther. 10, 5551–5559. 10.2147/OTT.S146479 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets analyzed for this study can be found in the Supplementary Material .