Abstract
Background
Immunotherapy, including checkpoint inhibition, has remarkably improved prognosis in advanced melanoma. Despite this success, acquired resistance is still a major challenge. The T cell costimulatory receptor TNFRSF9 (also known as 4-1BB and CD137) is a promising new target for immunotherapy and two agonistic antibodies are currently tested in clinical trials. However, little is known about epigenetic regulation of the encoding gene. In this study we investigate a possible correlation of TNFRSF9 DNA methylation with gene expression, clinicopathological parameters, molecular and immune correlates, and response to anti-PD-1 immunotherapy to assess the validity of TNFRSF9 methylation to serve as a biomarker.
Methods
We performed a correlation analyses of methylation at twelve CpG sites within TNFRSF9 with regard to transcriptional activity, immune cell infiltration, mutation status, and survival in a cohort of N = 470 melanoma patients obtained from The Cancer Genome Atlas. Furthermore, we used quantitative methylation-specific PCR to confirm correlations in a cohort of N = 115 melanoma patients’ samples (UHB validation cohort). Finally, we tested the ability of TNFRSF9 methylation and expression to predict progression-free survival (PFS) and response to anti-PD-1 immunotherapy in a cohort comprised of N = 121 patients (mRNA transcription), (mRNA ICB cohort) and a case-control study including N = 48 patients (DNA methylation, UHB ICB cohort).
Findings
We found a significant inverse correlation between TNFRSF9 DNA methylation and mRNA expression levels at six of twelve analyzed CpG sites (P ≤ 0.005), predominately located in the promoter flank region. Consistent with its role as costimulatory receptor in immune cells, TNFRSF9 mRNA expression and hypomethylation positively correlated with immune cell infiltrates and an interferon-γ signature. Furthermore, elevated TNFRSF9 mRNA expression and TNFRSF9 hypomethylation correlated with superior overall survival. In patients receiving anti-PD-1 immunotherapy (mRNA ICB cohort), we found that TNFRSF9 hypermethylation and reduced mRNA expression correlated with poor PFS and response.
Interpretation
Our study suggests that TNFRSF9 mRNA expression is regulated via DNA methylation. The observed correlations between TNFRSF9 DNA methylation or mRNA expression with known features of response to immune checkpoint blockage suggest TNFRSF9 methylation could serve as a biomarker in the context of immunotherapies. Concordantly, we identified a correlation between TNFRSF9 DNA methylation and mRNA expression with disease progression in patients under immunotherapy. Our study provides rationale for further investigating TNFRSF9 DNA methylation as a predictive biomarker for response to immunotherapy.
Funding
AF was partly funded by the Mildred Scheel Foundation. SF received funding from the University Hospital Bonn BONFOR program (O-105.0069). DN was funded in part by DFG Cluster of Excellence ImmunoSensation (EXC 1023). The funders had no role in study design, data collection and analysis, interpretation, decision to publish, or preparation of the manuscript; or any aspect pertinent to the study.
Keywords: TNFRSF9, DNA methylation, Melanoma, Prognostic biomarker, Predictive biomarker, Immune cell infiltration, Immune checkpoint, Immunotherapy, Response prediction, 4-1BB
Abbreviations: 95% CI, 95% confidence interval; ARID2, AT-rich interactive domain-containing protein 2; BRAF, V-raf murine sarcoma viral oncogene homolog B1; IDH1, Isocitrate dehydrogenase 1; CTLA4, Cytotoxic T-lymphocyte associated protein 4; CR, Complete response; FFPET, Formalin-fixed and paraffin-embedded tissue; HIF1-α, Hypoxia-inducible factor 1-alpha; IFN-γ, Interferon gamma; IL-2, Interleukin 2; IRB, Institutional Review Board; LAG3, lymphocyte activating 3; LHR, Likelihood Ratio; MYC, MYC proto-oncogene; MAPK, Mitogen-activated protein kinase; MR, Mixed response; NA, Not assessable; NAT, Normal adjacent tissue; n.c., Normalized counts; NF-kB, Nuclear factor 'kappa-light-chain-enhancer' of activated B-cells; NK cells, Natural killer cells; OS, Overall survival; PBMCs, Peripheral blood mononucleated cells; PD, Progressive disease; PD-1, Programmed cell death 1; PD-L1, Programmed cell death 1 ligand 1; PFS, Progression-free survival; PR, Partial response; qMSP, quantitative methylation-specific PCR; SKCM, Skin cutaneous melanoma; TCGA, The Cancer Genome Atlas; TILs, Tumor infiltrating lymphocytes; TALs, Tumor-associated lymphocytes; TNFRS9, Tumor necrosis factor receptor super family 9; TPM, Transcripts per million; Tregs, Regulatory T cells
Research in context.
Evidence before this study
The immune checkpoint TNFRSF9 (Tumor necrosis factor receptor superfamily member 9) is an attractive new target for cancer immunotherapy and currently, two agonistic antibodies are tested in clinical trials including melanoma. So far, little is known about the regulation of immune checkpoint genes, in particularly on an epigenetic level, which is fundamental for the development of more accurate mechanism-based biomarkers. Based on the results and data of a current landscape paper provided by The Cancer Genome Atlas Network and a current multicenter study on predictive biomarkers for response to anti-PD 1 therapy, we investigated the prognostic and predictive significance of TNFRSF9 methylation in melanoma patients with and without PD-1 directed immunotherapy.
Added value of this study
Our present study suggests a high biological significance of TNFRSF9 gene methylation and strongly indicates that TNFRSF9 methylation plays a role in the transcriptional regulation of TNFRSF9. Our results demonstrate significant correlations between TNFRSF9 hypomethylation and patients’ survival, pointing to a prognostic significance of TNFRSF9 methylation. Finally, our independent validation analysis in melanoma patients treated with anti-PD-1 immune checkpoints provides first evidence of TNFRSF9 methylation as a potential predictive biomarker for response to immunotherapy.
Implications of all the available evidence
Our data provide rationale for further investigating TNFRSF9 DNA methylation as a predictive biomarker in melanoma to assist the identification of patients that might benefit from agonistic TNFRSF9 therapy as well as anti-PD-1 immune checkpoint blockade or a combination therapy of the two.
Alt-text: Unlabelled box
1. Introduction
The tumor necrosis factor receptor superfamily member 9 (TNFRSF9), also known as 4-1BB and CD137, is an immune costimulatory receptor [1]. TNFRSF9 is expressed on activated immune cells including natural killer (NK) cells, effector T cells and antigen presenting cells, among them dendritic cells, macrophages, and B cells [2], [3], [4], [5]. TNFRSF9 expression is tightly controlled and has been demonstrated to be upregulated from 12 h to up to 5 days, depending on the specific T cell stimulus [6], [7], [8] with a peak expression after 24 h [9].
In mouse models, in vivo effects of TNFRSF9 signaling activation were demonstrated to include CD8+ T cell activation and tumor eradication [1,10]. Induction of the TNFRSF9 signaling pathway, via receptor binding, recruits TNFR-associated factor 1 and 2, leading to activation of the transcription factor NF-kB and the mitogen-activated protein kinase (MAPK) cascade [3,11,12]. In CD8+ T cells, TNFRSF9 signaling promotes activation, proliferation and production of cytokines, interleukin 2 (IL-2) and interferon gamma (IFN-γ) [13], [14], [15]. Furthermore, TNFRSF9 signaling contributes to upregulation of members of the anti-apoptotic Bcl-2 family, thus protecting against activation-induced cell death [16], [17], [18], [19]. In regulatory T cells (Tregs), agonistic TNFRSF9 antibody treatment can lead to inhibition of immune suppressive functions, augmenting the antitumor response [20]. Yet, the influence of TNFRSF9 on Treg cells is controversial and TNFRSF9 has also been shown to maintain the suppressive capacity of Tregs [21,22]. As well as its expression on activated immune cells, TNFRSF9 is also expressed by inflamed or hypoxic endothelial cells [23] and has been detected on tumor endothelial cells [22]. Hypoxia-mediated TNFRSF9 signaling was shown to promote migration of tumor-infiltrating lymphocytes (TILs) into malignant tissue [24]. Overall, the mechanisms summarized above make TNFRSF9 an attractive target for immunotherapy and agonistic monoclonal antibodies are currently being tested in multiple clinical trials.
Preclinical evidence for the potential therapeutic relevance of TNFRSF9 in melanomas was shown in a B16.SIY model by Weigelin et al., which demonstrated that agonistic TNFRSF9 antibodies restored the function of CD8+ TILs to secrete IL-2. Furthermore, combined treatment with anti-LAG3 (lymphocyte activating 3) antibodies increased the amount of CD8+ effector TILs among rejuvenated exhausted TILs [25]. In another preclinical study, utomilumab, a human IgG2 agonistic antibody to TNFRSF9, demonstrated its ability to inhibit tumor growth in a human peripheral blood lymphocyte (PBL)-SCID xenograft tumor model [26]. Currently, there are several ongoing clinical trials investigating two TNFRSF9 agonists, urelumab (Bristol-Myers Squibb, NY, USA; ClinicalTrials.gov Identifiers: NCT02253992, NCT02845323, NCT02534506, NCT02451982, NCT02658981, NCT03431948, NCT02652455) and utomilumab (Pfizer, NY, USA; ClinicalTrials.gov Identifiers: NCT03258008, NCT03704298, NCT03440567, NCT03414658, NCT03318900, NCT03364348, NCT03636503, NCT03390296, NCT02554812, NCT03217747, NCT03290937, NCT02951156) in different tumor types including melanoma.
The rapidly evolving landscape of therapeutic options in advanced melanoma requires the development of companion diagnostics for patient stratification, as not all patients respond equally to a particular medication. Prognostic biomarkers might help to identify patients with localized but aggressive disease who may potentially benefit from an adjuvant treatment. Predictive biomarkers may guide the choice of the most promising therapy. Mechanism-driven biomarkers for immune checkpoint modulators include CD8+ TIL density, mutational load, and immune checkpoint gene expression [27]. A current multicenter study investigated clinical, genomic and transcriptomic data of melanoma patients to discover predictive features for individual response to anti-PD1 therapy [28]. However, to facilitate the development of more accurate mechanism-based biomarkers, precise knowledge on the regulation of immune checkpoint genes, specifically on an epigenetic level, is critical.
DNA methylation is an important epigenetic regulation mechanism, playing a fundamental role in T cell differentiation and T cell exhaustion [29], [30], [31]. A multitude of studies report on aberrant methylation of inhibitory immune checkpoint genes, i.e. PD-1, PD-L1, PD-1 ligand 2 (PD-L2), and cytotoxic T-lymphocyte associated protein 4 (CTLA4), in various malignancies [30,[32], [33], [34], [35], [36], [37], [38], [39], [40]]. Moreover, early data suggest CTLA4 methylation is a predictive biomarker for anti-PD-1 and anti-CTLA-4 antibodies in patients with metastatic melanoma [41].
TNFRFS9 expression has been suggested as a biomarker for TILs in ovarian cancer and melanoma [42]. A predictive significance for response to combined anti-PD-1/CTLA-4 therapy has been demonstrated experimentally for the expression of TNFRSF9 on blood and tumor CD8+ T cells and CD4+ T [43]. To our knowledge there is so far no study investigating TNFRFS9 methylation as a predictive biomarker in melanoma patients treated with immune checkpoint blockade. In this work we analyzed correlates of TNFRSF9 methylation with mRNA expression, clinicopathological parameters, and patient outcome as well as molecular and immune correlates. We investigated the prognostic and predictive significance of TNFRSF9 methylation in melanoma patients with and without anti-PD-1 directed immunotherapy. Our results provide rationale to further investigate TNFRSF9 methylation as a prognostic biomarker in melanoma and as a predictive biomarker for melanoma patients who may benefit from treatment with TNFRSF9 agonists.
2. Methods
2.1. Patients
2.1.1. TCGA cohort
The data we used for our analyses are partly based on datasets of The Cancer Genome Atlas Research Network (TCGA, http://cancergenome.nih.gov/). On the whole we included a total of N = 470 samples from the TCGA skin cutaneous melanoma (SKCM) cohort. We analyzed one sample per patient. Only data from primary solid and metastatic tumor tissue samples were extracted to our collective, whereas solid normal tissues and additional metastatic tumor tissues were excluded from our analyses. In patients providing primary solid as well as metastatic tumor tissue only primary solid tumor tissue was included. We obtained supplementary clinicopathological and molecular data (Supplemental Table 1) from a trial previously published by the TCGA Research Network [45]. Datasets containing information about sample purity and ploidy estimates calculated using the ABSOLUTE algorithm [73] were also adopted from the TCGA Research Network. The leukocyte fraction within the tumor samples was quantified by Saltz et al. and Thorsson et al. who used DNA methylation array data to identify pure leukocyte cells [46,74]. We additionally used the results provided by Thorsson et al. [46] who calculated RNASeq signatures as estimates for distinct immune cell infiltrates using the CIBERSORT algorithm [75]. Further data on infiltrating lymphocytes were again adopted from the TCGA Research Network [45]. We included data on lymphocyte distribution (0–3; 0 = no lymphocytes within the tissue, 1 = lymphocytes present involving <25% of the tissue cross sectional area, 2 = lymphocytes present in 25 to 50% of the tissue, 3 = lymphocytes present in >50% of tissue), lymphocyte density (0–3; 0 = absent, 1 = mild, 2 = moderate, 3 = severe), and a lymphocyte score (0–6, score defined as the sum of the lymphocyte distribution and density scores).
The TCGA Research Network obtained informed consent from all patients in accordance with the Helsinki Declaration of 1975.
2.1.2. Validation cohort
In the validation analysis tumor tissue samples of N = 115 melanoma patients of the University Bonn were included (UHB validation cohort). The cohort was composed of tissue obtained from primary melanomas, subcutaneous and cutaneous metastases, and lymph node metastases. The tumor tissue included was obtained from patients naïve to systemic antitumor treatment, including targeted therapies or immune checkpoint blockade. Our study was approved by the Institutional Review Board (IRB) of the University Hospital Bonn.
2.1.3. Anti-PD-1 treated patients
Methylation and mRNA expression was investigated in two groups of patients who received PD-1 directed immune checkpoint blockage. TNFRSF9 methylation levels were available from a case-control study group comprised of N = 48 anti-PD-1-treated patients who did not respond (N = 19) or responded (N = 29) to therapy (UHB ICB cohort). The study was approved by the local IRB. TNFRSF9 mRNA levels from samples of N = 121 anti-PD-1-treated patients were obtained from a recently published work by Liu and co-workers [28] (mRNA ICB cohort).
2.2. Primary cell lines and isolated immune cells
Primary melanocyte (N = 8) and melanoma cell lines (N = 44) were included from the Gene Expression Omnibus (GEO) database [76,77] (GEO accessions: GSE44662 and GSE122909) [78,79].
Peripheral blood mononuclear cells (PBMCs, N = 54), monocytes (N = 52), B cells (N = 60), CD8+ T cells (N = 28), and CD4+ T cells (N = 54) isolated from peripheral blood donated by healthy individuals was obtained from four published datasets (GSE82218, GSE71245, GSE87650, GSE59250) [80], [81], [82], [83].
2.3. mRNA expression analysis
The mRNA expression analyses are based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov/) using the Illumina HiSeq 2000 RNA Sequencing Version 2 analysis (Illumina, Inc., San Diego, CA, USA). Expression data of level 3 were obtained from the TCGA webpage and were available from N = 468 patient samples. Normalized counts (n.c.) per genes were calculated using the SeqWare framework via the RSEM (RNA-Seq by Expectation Maximization) algorithm [84]. In addition, we included whole-transcriptome sequencing data reported as expression in transcripts per million (TPM) provided by Liu et al. [28].
2.4. DNA preparation and bisulfite conversion
DNA from formalin-fixed and paraffin-embedded tissue (FFPET) specimens (UHB validation cohort and immunotherapy case-control study (UHB ICB cohort)) was conducted after macrodissection of tumor tissues from sections mounted on glass slides. Tissue lysis and bisulfite conversion was performed using the innuCONVERT Bisulfite All-In-One Kit (Analytik Jena, Jena, Germany) according the manufacturer's instructions.
2.5. Methylation analysis
Data on gene methylation (Infinium HumanMethylation450 BeadChip, Illumina, Inc., San Diego, CA, USA) from the TCGA Research Network were available from N = 470 patient samples and were downloaded from the UCSC Xena browser (www.xena.ucsc.edu). Methylation levels (β-values) were calculated: Beta-value = (Intensity_Methylated)/(Intensity_Methylated + Intensity_Unmethylated + α) [85]. The constant offset α was set to 0. HumanMethylation450 BeadChip data (β-values) from isolated immune cells were downloaded from GEO database (GSE44662, GSE122909, GSE82218, GSE71245, GSE87650, GSE59250). Methylation data from the case-control study including N = 48 anti-PD-1 treated patients’ samples was generated using the Infinium MethylationEPIC BeadChip. β-values (values between 0 and 1) were multiplied with the factor 100% in order to approximate percent methylation (0% to 100%).
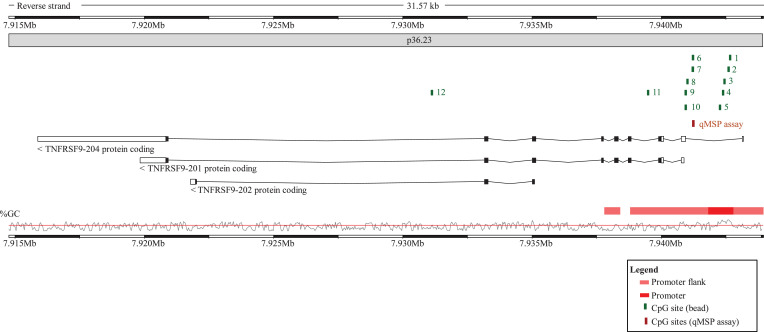
TNFRSF9 promoter methylation analysis of N = 115 melanomas from the University Hospital Bonn (UHB validation cohort) was performed using quantitative methylation-specific real-time PCR (qMSP) technology developed by Lehmann and Kreipe [86] in order to determine the promoter methylation levels at CpG site targeted by bead 6 (forward primer: actccataatcactataatacaataa, reverse primer: gtagtgtatttttgatgtttggta, probemethylated: 6-FAM-ccattacttaaacacaaccgata-BHQ-1, probeunmethylated: HEX-accattacttaaacacaaccaatat-BHQ-1). All oligonucleotides were purchased from biomers.net (Ulm, Germany). The qMSP assays amplifies the target sequence on chromosome 1: 7,941,202–7,941,277 (Genome Reference Consortium Human Build 38, GRCh38.p13, http://www.ensembl.org [87], Fig. 1). PCR reactions were performed in 20 µl volumes (buffer composition as previously described [88] containing 20 ng bisulfite converted DNA (quantified via UV–vis spectrophotometry) and 0.4 µM each primer and 0.2 µM each probe. qMSP was carried out using a 7900HT Fast Real-Time PCR system (Applied Biosystems, Waltham, MA, USA) with the following temperature profile: 10 min at 95 °C and 40 cycles with 15 s at 95 °C, 2 s at 62 °C, and 60 s at 52 °C. We calculated percentage methylation levels using cycle treshold (CT) values obtained from probes specifically binding to bisulfite-converted methylated (CTmethylated) and unmethylated (CTunmethylated) DNA, respectively (Methylation [%] = 100%/(1 + 2CTmethylated–CTunmethylated).
Fig. 1.
Genomic organization of the TNFRSF9 gene. Shown are regulatory elements, CG-density and target sites of HumanMethylation450 BeadChip beads and the quantitative methylation-specific PCR (qMSP assay). The modified illustration was exported from www.ensemble.org (release 98) [87] and is based on Genome Reference Consortium Human Build 38 patch release 13 (GRCh38.p13). Beads are numbered as follows: cg16839093 (1), cg27305704 (2), cg18859763 (3), cg07836592 (4), cg23959705 (5), cg06956444 (6), cg14614416 (7), cg18025409 (8), cg14153654 (9), cg08840010 (10), cg17123655 (11), cg16117781 (12).
2.6. Statistics
Statistical analyses were performed using SPSS, version 23.0 (SPSS Inc., Chicago, IL, USA). Analyses regarding potential correlations of characteristics were calculated using Spearman's rank correlation (Spearman's ρ). Significance levels for the Spearman's rank correlation coefficients were computed using a large sample normal theory approximation that utilizes a t-distribution. Mean value comparisons were performed using Wilcoxon–Mann–Whitney U (two groups) and Kruskal–Wallis (>2 groups) test. One-way ANOVA and post-hoc Bonferroni test were applied to perform multiple comparisons between groups. In order to reduce the influence of age-related deaths overall survival was censored after 5 years (1825 days). Kaplan–Meier method, likelihood ratios and Cox proportional hazards regression were used for the performance of survival analyses. For Kaplan–Meier analysis methylation levels and mRNA expression levels were dichotomized based on an optimized cut-off (lowest P-value). Cox proportional hazards analyses were performed with continuous methylation and log2-transformed mRNA expression data (mRNA expression levels of 0 n.c. were set to 0.1 and levels of 0 TPM were set to 0.01 prior to log2-transformation). P-values refer to log-rank and Wald tests, respectively. Two-sided P-values lower than 0.05 were considered statistically significant.
2.7. Role of the funding source
The funders had no role in study design, data collection and analysis, interpretation, decision to publish, or preparation of the manuscript; or any aspect pertinent to the study. DD confirms that he had full access to all the data in the study and had final responsibility for the decision to submit for publication.
3. Results
3.1. Methylation of TNFRSF9 inversely correlates with mRNA expression
The Infinium HumanMethylation450 BeadChip contains twelve beads targeting CpG sites within the TNFRSF9 gene locus (Fig. 1). Eleven CpG sites (targeted by beads one to eleven) were located in the promoter region (beads one to five are located in the promoter, beads six to eleven in the downstream promoter flank) and one in the gene body (bead twelve).
DNA methylation in promoters is frequently associated with transcriptional gene silencing [44]. We analyzed the correlation between methylation of the twelve CpG sites and mRNA expression in N = 468 malignant melanoma from the TCGA Research Network [45]. We found significant inverse correlations between TNFRSF9 DNA methylation and mRNA expression levels at six of twelve analyzed CpG sites indicating that TNFRSF9 mRNA expression may be regulated by gene methylation (Table 1). The inverse correlation was most pronounced in the promoter flank CpGs probed by bead six and seven, as well as in the gene body CpG targeted by bead twelve. Methylation of the CpGs within the promoter (targeted by beads one to five) showed no significant inverse correlation with mRNA expression.
Table 1.
Correlations of TNFRSF9 methylation with TNFRSF9 mRNA expression, lymphocyte score and overall survival.TNFRSF9 methylation was determined at 12 different CpG sites targeted by HumanMethylation450 BeadChip beads (Fig. 1). TNFRSF9 expression was analyzed as log2-transformed variable. Significant data are shown in boldface.
| Correlation with mRNA expressiona |
Correlation with lymphocyte scorea |
Correlation with total mutationsa |
Overall survival |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Analyte | Bead no. | Spearman's ρ | P-value | Spearman's ρ | P-value | Spearman's ρ | P-value | Hazard Ratio [95% CI] | P-value |
| mRNA | NA | NA | NA | 0.51 | <0.001 | 0.04 | 0.43 | 0.92 [0.87–0.97] | 0.002 |
| cg16839093 | 1 | 0.06 | 0.23 | 0.08 | 0.13 | 0.12 | 0.039 | 0.20 [0.03–1.33] | 0.096 |
| cg27305704 | 2 | 0.01 | 0.89 | −0.07 | 0.21 | 0.15 | 0.007 | 0.69 [0.31–1.53] | 0.36 |
| cg18859763 | 3 | −0.06 | 0.22 | −0.02 | 0.67 | 0.12 | 0.034 | 0.51 [0.15–1.76] | 0.29 |
| cg07836592 | 4 | −0.01 | 0.91 | −0.03 | 0.58 | 0.13 | 0.020 | 0.47 [0.15–1.53] | 0.21 |
| cg23959705 | 5 | 0.10 | 0.025 | 0.14 | 0.012 | 0.14 | 0.013 | 0.59 [0.17–2.10] | 0.42 |
| cg06956444 | 6 | −0.47 | <0.001 | −0.31 | <0.001 | −0.01 | 0.91 | 3.42 [1.28–9.14] | 0.014 |
| cg14614416 | 7 | −0.32 | <0.001 | −0.22 | <0.001 | −0.04 | 0.51 | 6.28 [2.06–19.2] | 0.001 |
| cg18025409 | 8 | −0.19 | <0.001 | −0.04 | 0.44 | −0.01 | 0.84 | 1.80 [0.84–3.85] | 0.13 |
| cg14153654 | 9 | −0.02 | 0.62 | 0.07 | 0.20 | −0.03 | 0.66 | 3.03 [1.01–9.07] | 0.048 |
| cg08840010 | 10 | −0.27 | <0.001 | −0.23 | <0.001 | −0.03 | 0.57 | 2.85 [1.23–6.63] | 0.015 |
| cg17123655 | 11 | −0.32 | <0.001 | −0.18 | 0.001 | 0.05 | 0.41 | 3.89 [1.25–12.1] | 0.019 |
| cg16117781 | 12 | −0.43 | <0.001 | −0.23 | <0.001 | −0.04 | 0.52 | 3.57 [1.65–7.76] | 0.001 |
NA: Not Applicable.
Correlations were performed including N = 468 (TNFRSF9 methylation and mRNA expression), N = 317 (total mutations and TNFRSF9 mRNA expression), N = 318 (total mutations and TNFRSF9 methylation), N = 328 (lymphocyte score and TNFRSF9 mRNA expression), N = 329 (lymphocyte score and TNFRSF9 methylation) samples.
We found significantly different TNFRSF9 mRNA expression levels among four types of tumor tissue sites (Supplemental Table 1). Regional lymph node metastases showed a higher level of mRNA expression compared to tissue obtained from primary tumors, cutaneous or distant metastases. Accordingly, significant differences in TNFRSF9 methylation levels were found at eight of twelve CpG sites (targeted by beads one, five to eight, and ten to twelve). We further analyzed correlations of TNFRSF9 methylation with TNFRSF9 mRNA expression with regard to tumor tissue site (Supplemental Tables 1 and 2). Inverse correlations between TNFRSF9 methylation and TNFRSF9 mRNA expression in lymph nodes were similar to the general analysis including all tumor sites. However, we observed inverse correlations between TNFRSF9 methylation and mRNA expression in primary tumor tissue, distant and cutaneous/subcutaneous metastases to be more pronounced than in samples from lymph node metastases.
3.2. Methylation of TNFRSF9 correlates with immune cell infiltrates
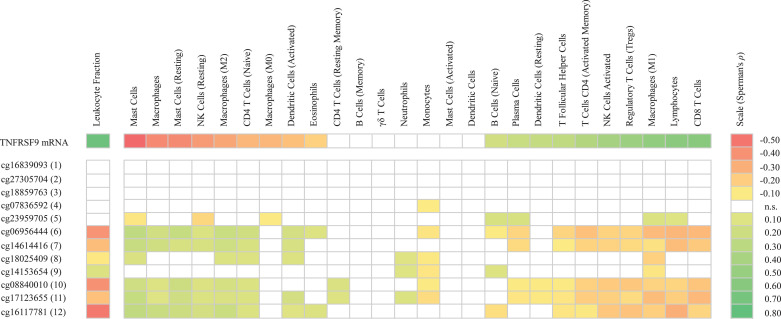
As TNFRSF9 is an immune stimulatory gene we predicted that its expression could augment activity of tumor infiltrating immune cells. Therefore, we investigated correlations between TNFRSF9 mRNA levels and methylation with lymphocyte score, leukocyte fraction, and RNASeq signatures of tumor infiltrating immune cell subsets. As expected, we found a significant correlation between TNFRSF9 mRNA expression with lymphocyte score and leukocyte fraction within the analyzed tumor infiltrating immune cells (Table 1, Supplemental Table 1, and Fig. 2). Accordingly, we observed significant inverse correlations between TNFRSF9 methylation and lymphocyte score at CpG sites within the gene body and promoter flank regions probed by beads six, seven and ten to twelve (Table 1). Furthermore, there were significant inverse correlations between CpG methylation in several regions of the promoter flank and the gene body (bead target sites six to eight, ten to twelve) with the leukocyte fraction (Fig. 2). In line with the observed positive correlation between TNFRSF9 mRNA expression and leukocyte fraction, tumor cell content (percentage tumor cell nuclei in sample) showed a significant negative correlation with TNFRSF9 mRNA expression (Supplemental Table 1).
Fig. 2.
Correlation of TNFRSF9 methylation and mRNA expression with immune cell infiltrates. Shown are Spearman's ρ correlation coefficients of significance (P<0.05) between TNFRSF9 methylation and mRNA expression with leukocyte fraction (mRNA: N = 468; methylation: N = 470) and distinct immune cell infiltrate signatures (mRNA: N = 468; methylation: N = 469). Immune cell infiltrates include RNA signatures of lymphocytes (including naive B cells, memory B cells, naive CD4+ T cells, activated and resting CD4+ memory T cells, T follicular helper cells, regulatory T cells, CD8+ T cells, γδ T cells, activated and resting NK cells, and plasma cells), macrophages (including monocytes and M0/M1/M2 macrophages), dendritic cells (including resting and activated dendritic cells), mast cells (including activated and resting mast cells), CD4+ T cells (including naive, activated memory, and resting memory CD4+ T cells), eosinophils, and neutrophils. P-values and Spearman's ρ correlation coefficients can be found in Supplemental Table 1; n.s.: not significant.
To follow up on our results based on the TCGA data we set up a validation cohort composed of N = 115 melanoma samples (UHB validation cohort). We designed a methylation-specific qPCR (qMSP) assay targeting CpG site six, located within the promoter flank region (Fig. 1), which had shown highly significant correlations between methylation and lymphocyte score in the TCGA cohort. In a histopathological examination we quantified leukocyte and tumor cell content as well as lymphocyte score. Mean lymphocyte score was 1.2 [95% CI, 0.9–1.4], mean percentage leukocytes in the tumor made up 5.8% [95% CI, 4.6−7.0%] and mean percentage of tumor cells were 93.7% [95% CI, 92.4−95.0%]. We observed significant inverse correlations between methylation of the CpG site six targeted by the qMSP assay and both lymphocyte score (ρ =−0.239, P = 0.010) and the leukocyte content (ρ =−0.247, P = 0.008). Accordingly, we observed a positive correlation between methylation of CpG site six with tumor cell content (ρ = 0.279, P = 0.003) in our validation cohort (UHB validation cohort).
We further performed correlative analyses stratified by tumor tissue types contained in the TCGA cohort in order to exclude the influence of sample type specific features, e.g. higher numbers of lymphocytes in lymph node metastases potentially leading to higher immune scores (Supplemental Table 3). Inverse correlations between TNFRSF9 methylation and lymphocyte score in the promoter flank regions were confirmed in four CpG sites for lymph node metastases, three CpG sites in distant metastases and two CpG sites in subcutaneous/cutaneous metastases. Methylation at CpG sites targeted by beads 6 and 10 correlated inversely with lymphocyte score for all metastatic sites. However, primary tumors showed a positive correlation between TNFRSF9 methylation and mRNA expression in the promotor region. Accordingly, a significant correlation between TNFRSF9 mRNA expression and lymphocyte score, was confirmed in the subgroup analysis for tissue obtained from lymph nodes metastases, distant metastases, and cutaneous/subcutaneous but not for primary tumors.
3.3. TNFRSF9 methylation differs significantly between subsets of leukocytes
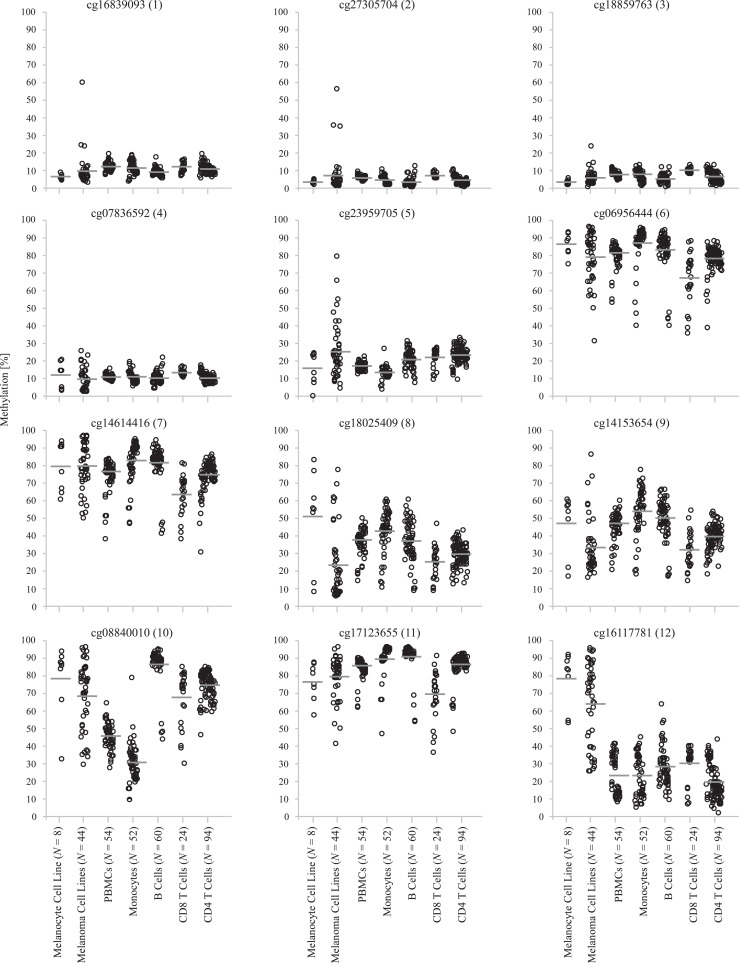
The strongest inverse correlation between methylation and tumor leukocyte fraction was found at CpG site targeted by bead twelve (Spearman's ρ = −0.484, P < 0.001). Hence, we expected significant methylation differences between melanoma cells compared to immune cells and potentially among immune cell subtypes. In order to test our hypothesis, we analyzed primary melanoma cell lines, primary melanocyte cell lines, and isolated leukocytes from peripheral blood donated by healthy individuals. Data on the isolated leukocytes were obtained from published datasets, data for primary melanocyte and melanoma cell lines were included from the Gene Expression Omnibus (GEO) database (Fig. 3). We were able to confirm methylation differences between all analyzed leukocytes (PBMCs, monocytes, B cells, CD4+ and CD8+ T cells) and melanoma cell lines as well as melanocytes (Fig. 3). As expected, melanoma cell lines showed high levels of TNFRFS9 methylation at the gene body CpG site targeted by bead 12.
Fig. 3.
TNFRSF9 methylation in distinct cell types.TNFRSF9 methylation at twelve CpG sites within TNFRSF9 in primary melanoma and melanocyte cell lines as well as in isolated PBMCs, monocytes, B cells, CD8+ T cells, and CD4+ T cells from healthy donors. All P < 0.001 (Kruskal–Wallis test).
We found a positive correlation between TNFRFS9 methylation and leukocyte fraction at the promoter flank CpG site targeted by bead nine (Spearman's ρ = 0.108, P = 0.019) (Fig. 2), whereas there were no correlations with the lymphocyte score (Table 1). We therefore hypothesized there would be high methylation of total PBMCs compared to melanoma cells but no difference between melanoma cells and lymphocytes. As expected, the analysis showed no distinct differences between methylation of melanoma cells and CD4+/CD8+ T cells from blood of healthy donors. We did not find correlations between TNFRFS9 methylation and leukocyte fraction in the promoter region targeted by beads one to four. Accordingly, there were no significant differences between methylation in melanoma cells and immune cells under investigation (Fig. 3). Our analysis revealed differences in TNFRSF9 methylation within the single immune cell subtypes analyzed in the blood of healthy donors (Fig. 3).
3.4. Methylation of TNFRSF9 correlates with the infiltration signature of immune cell subsets
To gain further information concerning the composition of the immune cell infiltrates of the tumor samples contained in the TCGA cohort we used RNA signatures of tumor infiltrating leukocyte subgroups which we obtained from a study published by Thorsson et al. [46] (Fig. 2). The detailed analysis of the RNA signatures of infiltrating leukocyte subgroups revealed an infiltration signature of CD8+ T cells, activated NK cells, activated CD4+ T memory cells, proinflammatory M1 macrophages, Tregs and resting dendritic cells positively correlated with TNFRSF9 mRNA expression. In contrast we found an inverse correlation of infiltration signatures of resting NK cells, naïve CD4+ T cells and anti-inflammatory M2 macrophages with TNFRSF9 mRNA expression (Fig. 2). Similarly, correlations between TNFRSF9 methylation, in the promoter flank and gene body regions, and RNASeq signatures of tumor infiltrating immune cells depended on differentiation, activation status and inflammatory potential of the different leukocyte subsets. Infiltration signatures in the subgroups of activated NK cells, activated CD4+ memory T cells and proinflammatory M1 macrophages demonstrated inverse correlations with TNFRSF9 methylation, whereas significant correlations between TNFRSF9 methylation and infiltration signatures were found in the corresponding subgroups of resting, naïve or anti-inflammatory leukocytes. Furthermore, we found the infiltration signature of Tregs to be inversely correlated to TNFRSF9 methylation (Fig. 2).
3.5. Methylation of TNFRSF9 inversely correlates with an interferon-γ signature
TNFRSF9 signaling has been shown to promote the secretion IFN-γ and the activation of CD8+ T cells [3]. CD8+ T cells within the tumor microenvironment are associated with the activation of IFN-γ pathways [27,47]. We therefore investigated correlations between TNFRSF9 methylation and mRNA expression levels with an IFN-γ signature defined by the expression of IFN-γ and IFN-γ–regulated genes (STAT1, STAT2, JAK2, and IRF9; Table 2). TNFRSF9 mRNA expression levels positively correlated with an IFN-γ signature. Concordantly, we observed an inverse correlation of TNFRSF9 methylation levels (predominantly in the regions of the promoter flank and in the gene body) with an IFN-γ signature (Table 2).
Table 2.
Correlations of TNFRSF9 methylation and mRNA expression with IFN-γ-signature. Correlations of TNFRSF9 methylation and mRNA expression with IFN-γ and IFN-γ-regulated genes (STAT1, STAT2, JAK2, and IRF9). DNA methylation was determined at twelve different loci targeted by HumanMethylation450 BeadChip beads (Fig. 1). Significant data are shown in boldface. Data were obtained from N = 468 (methylation) and N = 467 (mRNA expression) tumor samples, respectively.
| IFN-γ |
STAT1 |
STAT2 |
JAK2 |
IRF9 |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Analyte | Bead no. | Spearman's ρ | P-value | Spearman's ρ | P-value | Spearman's ρ | P-value | Spearman's ρ | P-value | Spearman's ρ | P-value |
| mRNA | NA | 0.85 | <0.001 | 0.74 | <0.001 | 0.31 | <0.001 | 0.57 | <0.001 | 0.47 | <0.001 |
| cg16839093 | 1 | 0.04 | 0.42 | 0.06 | 0.18 | 0.04 | 0.40 | 0.13 | 0.006 | 0.03 | 0.57 |
| cg27305704 | 2 | 0.01 | 0.88 | 0.05 | 0.30 | 0.06 | 0.22 | 0.10 | 0.035 | 0.01 | 0.81 |
| cg18859763 | 3 | −0.08 | 0.084 | −0.10 | 0.035 | 0.00 | 0.98 | 0.02 | 0.61 | −0.06 | 0.24 |
| cg07836592 | 4 | −0.02 | 0.64 | −0.02 | 0.74 | 0.01 | 0.80 | 0.10 | 0.026 | −0.02 | 0.62 |
| cg23959705 | 5 | 0.07 | 0.15 | 0.09 | 0.064 | 0.05 | 0.27 | 0.13 | 0.005 | 0.08 | 0.082 |
| cg06956444 | 6 | −0.44 | <0.001 | −0.40 | <0.001 | −0.07 | 0.12 | −0.23 | <0.001 | −0.20 | <0.001 |
| cg14614416 | 7 | −0.33 | <0.001 | −0.24 | <0.001 | −0.11 | 0.020 | −0.14 | 0.002 | −0.20 | <0.001 |
| cg18025409 | 8 | −0.17 | <0.001 | −0.21 | <0.001 | −0.02 | 0.61 | −0.10 | 0.035 | −0.09 | 0.051 |
| cg14153654 | 9 | −0.07 | 0.13 | −0.13 | 0.006 | −0.02 | 0.70 | 0.00 | 0.99 | −0.05 | 0.24 |
| cg08840010 | 10 | −0.32 | <0.001 | −0.31 | <0.001 | −0.14 | 0.002 | −0.05 | 0.24 | −0.27 | <0.001 |
| cg17123655 | 11 | −0.35 | <0.001 | −0.36 | <0.001 | −0.07 | 0.12 | −0.18 | <0.001 | −0.19 | <0.001 |
| cg16117781 | 12 | −0.39 | <0.001 | −0.33 | <0.001 | −0.14 | 0.002 | −0.24 | <0.001 | −0.19 | <0.001 |
NA: Not Applicable.
3.6. Methylation of TNFRSF9 and mRNA expression is associated with age
A detailed analysis of associations between TNFRSF9 methylation and TNFRSF9 mRNA expression with clinicopathological parameters was performed in order to identify prognostically and biologically relevant correlates (Supplemental Table 1). We found a significant positive correlation between age and TNFRSF9 methylation in four out of twelve analyzed loci (targeted by beads one, six, eight, and eleven). There were no relevant gender-specific differences in TNFRSF9 methylation and mRNA expression.
3.7. Methylation of TNFRSF9 and mRNA expression is associated with genomic aberrations
We further investigated correlations between TNFRSF9 methylation and mRNA expression with genomic alterations. Methylation of CpG sites in the promoter (targeted by beads one to five) significantly correlated with the number of total mutations. In particular, we found a significant correlation between methylation of CpG sites in the promoter with mutations in ARID2 (AT-rich interactive domain-containing protein 2, targeted by beads one to five) and IDH1 (Isocitrate dehydrogenase 1, targeted by beads two, three and five). Furthermore, we found a significant correlation between TNFRSF9 methylation and BRAF mutation status in five CpG sites under investigation (bead target sites five, eight and ten to twelve). Regarding TNFRSF9 mRNA expression, we found significant correlations with a UV signature, indicated by C > T transitions at dipyrimidines, but no significant correlation with mutational load or BRAF, ARID2 or IDH1 mutation status.
3.8. Association of TNFRSF9 methylation and mRNA expression with patients’ survival
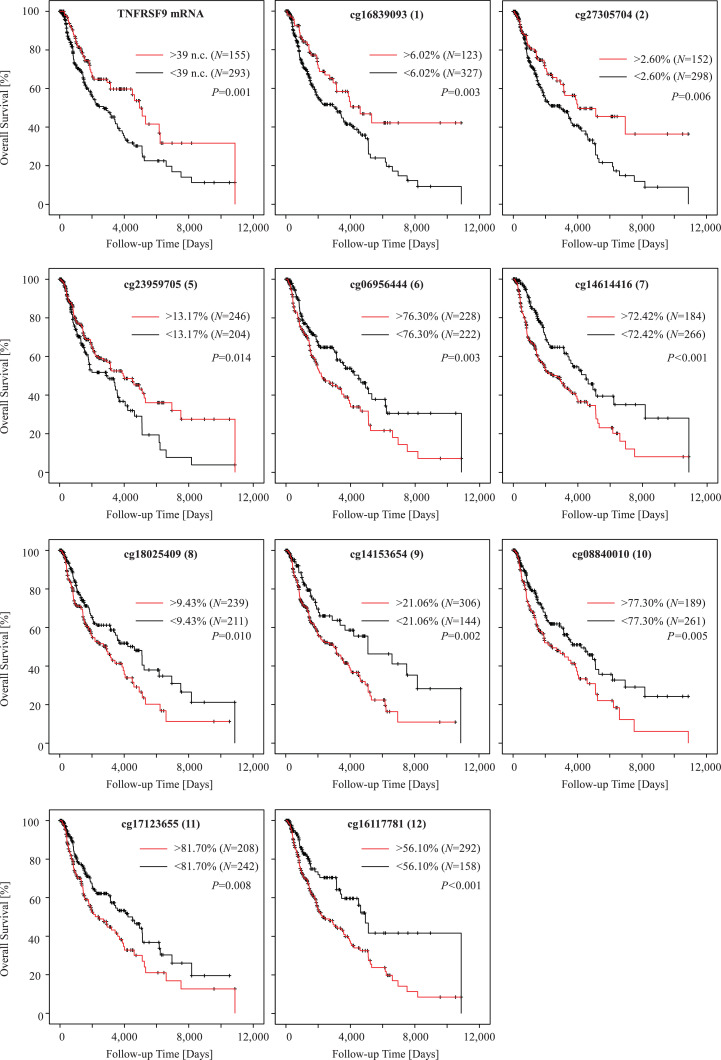
Finally, we studied the association of TNFRSF9 methylation and mRNA expression with patients’ survival. Methylation and mRNA expression levels were tested as continuous log2-transformed variates without prior dichotomization in order to avoid biases due to the introduction of cutoffs for patient sample classification. In univariate Cox proportional hazards analysis, elevated TNFRSF9 mRNA expression showed a significant correlation with better patient survival (Hazard ratio [HR] = 0.92, [95% CI: 0.87–0.97], P = 0.002, Wald test; Table 1). Concordantly, elevated methylation levels at six out of twelve CpG sites located in the promoter flank region and the gene body region (bead target sites six, seven, and nine to twelve) were significantly correlated with poor outcome (Table 1). We further dichotomized mRNA levels and methylation levels based on optimized cut-offs for patient classification. Kaplan–Meier survival analyses confirmed the better prognosis of patients with high mRNA-expressing (above cutoff) tumors and tumors showing hypomethylation (below cutoff) of the CpG sites located in the promoter flank region and the gene body region (Fig. 4). Of note, and in contrast to the CpG sites in the promoter flank and gene body region, hypermethylation of CpG sites located in the promoter region targeted by beads one, two, and five was associated with better survival. Finally, we investigated the influence of melanoma sample type on the associations of TNFRSF9 methylation and mRNA expression with overall survival (Supplemental Table 4). We observed significant correlations of TNFRSF9 methylation and survival not only in lymph node metastases but also in distant metastases samples (Supplemental Table 4). Hence, survival differences are unlikely to be biased by the different analyzed sample types.
Fig. 4.
Kaplan–Meier analysis of overall survival in melanoma patients stratified according to TNFRSF9 methylation and mRNA expression. Patient samples were dichotomized based on optimized cutoffs. Analysis of CpG sites targeted by beads 3 and 4 was omitted, as generation of an optimized cutoff did not result in significant survival differences. Follow-up data was available from N = 448 (mRNA expression) and N = 450 (methylation) patients, respectively. P-values refer to log-rank tests.
3.9. Association of TNFRSF9 methylation and mRNA expression with anti-PD-1 response and progression-free survival
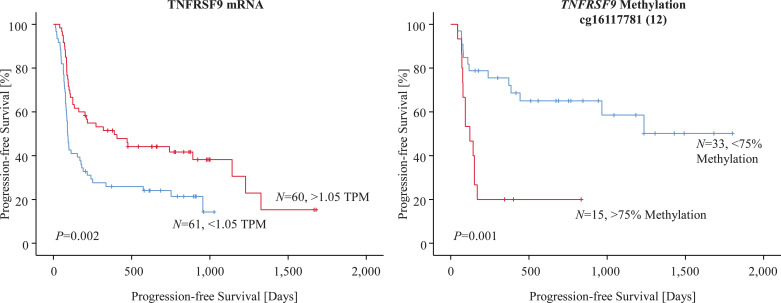
Finally, we analyzed TNFRSF9 methylation and mRNA expression in FFPE tumor tissues with regard to response and progression in melanoma patients who received anti-PD-1 directed immunotherapy. TNFRSF9 mRNA expression levels were available from N = 121 anti-PD-1 treated melanoma patients (mRNA ICB cohort) included in a study recently published by Liu et al. [28]. Our analysis demonstrated continuous log2-transformed mRNA levels to be significantly associated with progression-free survival (PFS) (HR = 0.92, [95% CI: 0.85–0.99], P = 0.022, Wald test). This result was confirmed in Kaplan–Meier survival analysis after result dichotomization based on an optimized cut-off (1.05 TPM, Fig. 5). Moreover, TNFRSF9 mRNA expression levels were significantly associated with response to anti-PD-1 immunotherapy (Fig. 6).
Fig. 5.
Kaplan–Meier analysis of progression-free survival in two cohorts of anti-PD-1 treated melanoma patients stratified according to TNFRSF9 methylation and mRNA expression. Methylation of CpG site targeted by bead 12 was investigated in a case-control study comprised of N = 19 patients with progressive disease and N = 29 patients with response (partial and complete), respectively, to PD-1 directed immunotherapy. TNFRSF9 mRNA expression was evaluated in N = 121 samples from a recently published cohort of patients who received anti-PD-1 directed immunotherapy [28]. Patient samples were dichotomized based on optimized cutoffs. P-values refer to log-rank tests.
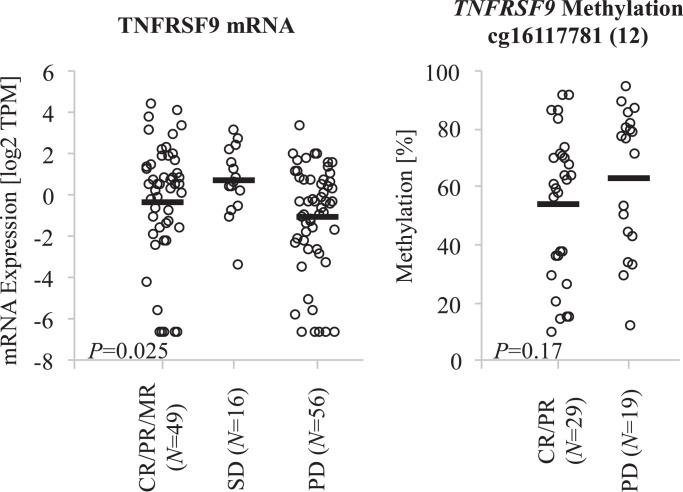
Fig. 6.
TNFRSF9 methylation and mRNA expression with regard to response to anti-PD-1 immunotherapy. TNFRSF9 mRNA expression levels in samples from a recently published anti-PD-1-treated cohort comprised of N = 49 responding patients (partial, mixed, and complete), N = 56 non-responding patients, and N = 16 patients with stable disease [28]. TNFRSF9 methylation at CpG targeted by bead 12 in melanoma samples from N = 19 responding (complete and partial responses) and N = 29 non-responding patients. P-values refer to Wilcoxon Mann–Whitney U and Kruskal–Wallis test, respectively. Mean levels are indicated by bars.
We additionally correlated continuous methylation levels with PFS in a case-control study comprised of N = 29 responding and N = 19 non-responding anti-PD-1 treated melanoma patients (UHB ICB cohort). Among the CpG sites under investigation we found methylation of CpG site targeted by bead 12 to be significantly correlated with PFS (HR = 8.34, [95% CI: 1.24–56.1], P = 0.029, Wald test). Again, we confirmed this result in Kaplan–Meier survival analysis after introduction of an optimized cut-off (75% methylation) for patient sample classification (Fig. 5). As expected, we also found higher mean methylation levels in samples from non-responding tumors (62.7% methylation) in comparison to samples from responding (53.4% methylation) tumors (Fig. 6), which, however, did not reach statistical significance (P = 0.17, Wilcoxon Mann–Whitney U).
4. Discussion
Epigenetic alterations, including changes in DNA methylation, have already been identified as a characteristic of T cell differentiation [29,31,41,48,49]. Accordingly, DNA methylation is being discussed as a quantitative surrogate biomarker for T cell exhaustion [30]. Expression of the immune checkpoint CTLA-4 for example has been shown to correlate inversely with promoter methylation in diverse malignancies, supporting the use of CTLA4 hypomethylation as a biomarker for T cell exhaustion [37,39,50] and for response to immunotherapy [41]. In our present study we investigated a possible association of TNFRSF9 DNA methylation with mRNA expression, clinicopathological, molecular and immune parameters, survival, and response to immune checkpoint blockade in melanoma patients. The rationale behind our study was to investigate the prognostic and predictive value of TNFRSF9 methylation in melanoma and to provide a rational for further testing TNFRSF9 methylation as a predictive biomarker in patients treated with TNFRSF9 agonists alone or in combination with PD-1 antagonists. We observed inverse correlations between TNFRSF9 mRNA expression levels and TNFRSF9 methylation. In addition, we found significant correlations between TNFRSF9 methylation and mRNA expression with lymphocyte score, leukocyte fraction, and signatures of tumor infiltrating leukocytes. Our results strongly support the hypothesis that TNFRSF9 DNA methylation regulates TNFRSF9 mRNA expression in tumor infiltrating immune cells. Finally, we showed that TNFRSF9 DNA methylation and expressions allows for the prediction of response to anti-PD-1 targeted immunotherapy.
TNFRSF9 is expressed on both T cells and antigen presenting cells and possesses the capacity to enhance effector functions of activated T lymphocytes, to augment cytokine production and promote expansion of TILs [21]. In addition, TNFRSF9 is expressed on tumor endothelial cells and stimulation of TNFRSF9 has been shown to mediate leukocyte extravasation resulting in augmented migration of TILs into malignant tissue [22]. In a mouse study, Palazón et al. identified hypoxia to be the microenvironmental factor inducing tumor endothelial TNFRSF9 expression via hypoxia-inducible factor 1-alpha (HIF1-α). The authors also demonstrated that HIF1-α induces TNFRSF9 upregulation in TILs, resulting in an augmented antitumor response [24]. TNFRSF9 expression on T cells is activation dependent. In healthy donors, this characteristic enables identification and isolation of small numbers of antigen-specific CD8+ T cells [9]. Further studies demonstrated that TILs expressing TNFRSF9 represented a tumor-experienced T cell lineage, leading to the conclusion that TNFRSF9 expression could be used to identify tumor antigen-experienced T cells, providing the rationale for adoptive T cell therapy [51,52]. Correspondingly, our data demonstrated a significant correlation between TNFRSF9 mRNA expression and an infiltration signature of activated CD4+ memory cells and a negative correlation with a signature of naïve CD4+ T cells. Conversely, TNFRSF9 methylation correlated positively with the signature of naïve CD4+ T cells but negatively with the signature of activated memory cells. We observed a similar pattern for NK cells and macrophages in different stages of activation and differentiation. That is, TNFRSF9 mRNA expression correlated with infiltration signatures of the activated and pro-inflammatory leukocyte subset, whereas inverse correlations were observed for resting NK cells and anti-inflammatory macrophages. The results are in line with the reported biological function of TNFRSF9 as a co-stimulatory receptor on activated immune cells. Furthermore, our data reflect the complex regulatory functions of TNFRSF9, as indicated by the inverse correlations of TNFRSF9 mRNA expression with an infiltration signature of dendritic cells and the significant correlation with a signature of Tregs. TNFRFS9 signaling has been reported to promote proliferation and survival of Tregs [53]. In the development of dendritic cells however, TNFRSF9 signaling was suggested to play a negative functional role [54].
Our results demonstrate a correlation between TNFRSF9 expression and immune cell infiltrates which is consistent with the reported function of TNFRSF9 to enhance lymphocyte activation and infiltration in malignancies. In melanoma and ovarian cancer TNFRFS9 expression was suggested as a biomarker for TILs [42]. Beyond, our results indicate that methylation of TNFRSF9 might serve as a surrogate biomarker for tumor infiltrating immune cells. Tumor infiltrating lymphocytes are known to be associated with favorable prognosis in primary and advanced melanoma [48,55]. The recent landscape paper of the TCGA cohort further suggested that T cell signaling has prognostic significance, which was in particular true for CD8+ effector T cells [45]. Even though in vitro signaling of TNFRSF9 was shown to support both CD4+ and CD8+ T cell response, in vivo signaling was demonstrated to predominantly affect CD8+ T cell response [56]. Infiltration of CD8+ T cells is associated with an activation of the IFN-γ pathways [27,47]. We observed a significant correlation of TNFRSF9 mRNA expression with an IFN-γ signature. Correspondingly, TNFRSF9 methylation correlated inversely with mRNA expression of IFN-γ and IFN-γ-regulated genes. Previous studies demonstrated the ability of TNFRSF9 to induce high levels of IFN-γ, thereby improving tumor eradication [1]. A phase I trial combining the TNFRSF9 agonist, utomilumab, with anti PD-1 antibody, pembrolizumab, demonstrated antitumor activity in patients with advanced malignancies [57]. Although the study was not designed to detect relationships between clinical outcome and potential biomarkers, the authors observed a correlation between clinical benefit and elevated CD8+ memory cells and IFN-γ in the peripheral blood. IFN-γ signature correlates with immune cell infiltration, and both variables are known to correlate with patient survival in melanoma and many other types of cancers [58]. As such, a correlation between elevated levels of IFN-γ, immune cell infiltration and TNFRSF9 expression seems predictable, given the fact that TNFRSF9 is predominantly expressed on immune cells. The question of causality remains; whether TNFRSF9 expression is the source or result of immune cell infiltration and proinflammatory antitumor response. An experimental study following up on the role of TNFRSF9 in the interaction between TILs and tumors cells in melanoma and ovarian cancer demonstrated that TNFRSF9 was preferentially expressed on tumor-reactive subsets of TILs and that direct contact with tumor cells stimulated the expression of TNFRSF9 [42]. These results indeed support the idea that initially TNFRSF9 expression is the result of immune cell infiltration. Based on their results the authors suggested TNFRSF9 as a biomarker for tumor-reactive TILs, assuming that TNFRSF9 expression might predict improved prognosis in ovarian cancer and melanoma. Our results support this idea.
As discussed in our study, TNFRFS9 signaling has been shown to support CD4+ and CD8+ T cell responses. In CD8+ T cells TNFRFS9 signaling promotes cytokine production, including IFN-γ [1]. We therefore assume that TNFRFS9 expression might indicate increased cytokine levels. Ye et al. demonstrated that CD137 positive TILs and tumor-associated lymphocytes (TALs) secreted IFN-γ in response to autologous tumor cell stimulation, whereas CD137 negative TILs and TALs did not. The study showed that increased CD137 expression among CD8+ TILs in the presence of tumor cells was MHC-dependent, ruling out activation-independent effects mediated by cytokines alone [42]. It has been highly documented that IFN-γ correlates with immune cell infiltration and both variables serve as generally accepted prognostic biomarkers in malignancies. Our results, in conjunction with published knowledge on TNFRFS9, support the idea to further investigate the prognostic value of TNFRSF9 expression and its epigenetic regulation.
Beyond the prognostic significance of TNFRSF9 methylation, our study primarily aims at supplying a rational for further testing of TNFRSF9 methylation as a predictive biomarker in a cohort of patients treated with TNFRSF9 agonists. Despite its value as prognostic biomarkers, immune checkpoint mRNA expression and IFN-γ have only insufficiently proven their suitability as predictive biomarker for patients treated with immunotherapy [59]. Whereas TILs or IFN-γ are general biomarker, the identification of a specific biomarker of concomitant prognostic and predictive significance could extend the possibilities of patient-tailored therapy and be of great value in particular in the adjuvant setting. A current trial studied predictive biomarkers for response to immune checkpoint blockade in 37 lymph node metastases after ex vivo exposure to immune-checkpoint blockade in resected stage III melanoma [43]. The experimental study demonstrated TNFRSF9 expression on CD8+ peripheral blood T cells to be associated with progression-free survival in resected stage III melanoma patients who received adjuvant treatment with ipilimumab + nivolumab combination therapy, but not nivolumab alone. So far, agonistic TNFRSF9 antibodies have only been used in clinical trials, limiting the availability of data and samples from TNFRSF9 antibody treated patients. To investigate the feasibility of TNFRSF9 methylation as a predictive biomarker, we therefore investigated patients who received anti-PD-1 directed immune checkpoint blockage as a representative immunotherapy study group (mRNA ICB cohort). In this group, we demonstrated TNFRSF9 mRNA expression levels to be significantly associated with increased PFS and with response to anti-PD-1 immunotherapy. Furthermore, we found methylation to be significantly correlated with PFS and a marked trend towards higher mean methylation levels in non-responding tumors in comparison to tumors responding to anti-PD-1 therapy (UHB ICB cohort). These results provide first evidence of a predictive value of TNFRSF9 methylation in melanoma patients treated with immunotherapy. Given that mechanisms of anti-tumor response, including immune cell infiltration and IFN-γ expression, and immune checkpoint signaling are interlinked, we assume that the predictive significance of TNFRSF9 will also hold true for patients treated with agonistic TNFRSF9 antibodies. Analysis of TILs and TIL signature demands immunohistochemistry and RNAseq, with the latter being limited in FFPET. Beyond, TNFRSF9 expression has been shown to be stimulation dependent and temporally limited, whereas DNA methylation constitutes a rather robust marker. Here, methylation analysis could present an additional diagnostic tool. We therefore assume methylation of TNFRSF9 to be a sensible prognostic and predictive biomarker reflecting the complex molecular interplay of tumor microenvironment.
A recent study demonstrated that TNFRSF9 co-stimulation in CD8+ T cells leads to changes in DNA methylation and chromatin reprogramming in diverse immune-related genes [60]. In our study we investigated associations of methylation in TNFRSF9 with genomic alterations. We found a positive correlation between TNFRSF9 promoter methylation and the presence of somatic mutations in ARID2 and IDH1. Mutations in ARID2, encoding a component of the chromatin-remodeling complex, and the epigenetic regulator IDH1 are known to play a role in melanomagenesis [61]. The association of mutations in epigenetic enzymes and TNFRSF9 methylation could point to a possible significance of TNFRSF9 promoter methylation during malignant transformation. To our knowledge a tumor cell-intrinsic role of TNFRSF9 in melanoma has not been described yet and should be followed up in functional experiments.
We found significant higher levels of TNFRSF9 methylation in BRAF wildtype compared to mutated melanoma. A recent study identified a network of BRAF-regulated transcription factors, including HIF1-α, that controls glycolysis in melanoma cells, is critical for response to BRAF inhibition and is modulated by BRAF inhibition in melanoma [62]. A strong link between BRAF mutation and DNA hypermethylation, also referred to as CpG island methylator phenotype (CIMP), has been described in colorectal cancers [36,63]. A similar association in melanoma, however, could not be confirmed [64]. Comprehensive analyses of the data provided by TCGA identified a negative correlation between BRAF mutations and methylation changes in melanoma [45,46]. These results are in line with previous work demonstrating that mutations in BRAF [65] enhance the immune infiltrate, while those in IDH1 reduce immune cell infiltration [66]. The possibility of TNFRSF9 playing a role in the cross-talk of transcriptional regulators in melanoma needs to be followed up in future experimental studies.
We found significant correlations between age and TNFRSF9 methylation which is in line with published studies. Age-associated methylation changes of regulatory elements have been shown to predominantly affect genes involved in immune processes [67,68], e.g. immune checkpoint genes [69,70] or genes controlling T cell immune response [71], which might serve as a possible explanation for carcinogenesis in elderly patients. A recent study revealed age-related genome-wide changes in DNA methylation in human PBMCs (peripheral blood mononucleated cells), with a functional analysis showing a strong enrichment of genes involved in cancer in older subjects [72].
In conclusion, our results suggest that TNFRSF9 mRNA expression is regulated via DNA methylation. The observed correlations between TNFRSF9 DNA methylation, TNFRSF9 mRNA expression and known features of response to immune checkpoint blockage suggest that TNFRSF9 methylation could be a biomarker in the context of immunotherapies. Our study provides first evidence of TNFRSF9 as a potential predictive biomarker for response to anti-PD-1 checkpoint blockade. Based on our results, we recommend testing TNFRSF9 DNA methylation as a predictive biomarker in patients treated with TNFRSF9 agonists and PD-1 antagonists.
Acknowledgments
Acknowledgments
We thank the BioBank Bonn of the Bonn University Medical Faculty and the University Hospital Bonn for the support of this study.
Funding source
AF was partly funded by the Mildred Scheel Foundation. SF received funding from the University Hospital Bonn BONFOR program (O-105.0069). DN was funded in part by DFG Cluster of Excellence ImmunoSensation (EXC 1023).
Disclosure statement
DD owns patents and patent applications on methylation of immune checkpoint genes (including TNFRSF9) as predictive and prognostic biomarkers (DE102017125780, DE102016005947, DE102015009187, WO2019086642, US2019249258, CN109715829, EP3458602, JP2019516383, WO2017198617, EP3322819, WO2017008912, CN108138242). The patents are licensed to Qiagen GmbH (Hilden, Germany) and DD receives royalties from Qiagen. DD is a consultant for AJ Innuscreen GmbH (Berlin, Germany), a 100% daughter company of Analytik Jena AG (Jena, Germany), and receives royalties from product sales (innuCONVERT kits). AF has received speaker's honoraria or travel expense reimbursements from the following companies: Novartis, BMS, Almirall, and Eli Lilly Pharma. DN has received speakers’ honoraria or travel expense reimbursements from the following companies: BMS, Novartis, GSK, Celgene, and MSD. JS received speaker's honoraria from Novartis, BMS, MSD, and Roche. JL is a consultant/advisory board member of Bristol-Myers Squibb, Merck, Novartis, and Roche. No potential conflicts of interest were disclosed by the other authors.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2020.102647.
Appendix. Supplementary materials
References
- 1.Vinay D.S., Kwon B.S. 4-1BB (CD137), an inducible costimulatory receptor, as a specific target for cancer therapy. BMB Rep. 2014;47(3):122–129. doi: 10.5483/BMBRep.2014.47.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alderson M.R., Smith C.A., Tough T.W., Davis-Smith T., Armitage R.J., Falk B. Molecular and biological characterization of human 4-1BB and its ligand. Eur J Immunol. 1994;24(9):2219–2227. doi: 10.1002/eji.1830240943. [DOI] [PubMed] [Google Scholar]
- 3.Chester C., Sanmamed M.F., Wang J., Melero I. Immunotherapy targeting 4-1BB: mechanistic rationale, clinical results, and future strategies. Blood. 2018;131(1):49–57. doi: 10.1182/blood-2017-06-741041. 04. [DOI] [PubMed] [Google Scholar]
- 4.Futagawa T., Akiba H., Kodama T., Takeda K., Hosoda Y., Yagita H. Expression and function of 4-1BB and 4-1BB ligand on murine dendritic cells. Int Immunol. 2002;14(3):275–286. doi: 10.1093/intimm/14.3.275. [DOI] [PubMed] [Google Scholar]
- 5.Pollok K.E., Kim Y.J., Hurtado J., Zhou Z., Kim K.K., Kwon B.S. 4-1BB T-cell antigen binds to mature B cells and macrophages, and costimulates anti-mu-primed splenic B cells. Eur J Immunol. 1994;24(2):367–374. doi: 10.1002/eji.1830240215. [DOI] [PubMed] [Google Scholar]
- 6.Cannons J.L., Lau P., Ghumman B., DeBenedette M.A., Yagita H., Okumura K. 4-1BB ligand induces cell division, sustains survival, and enhances effector function of CD4 and CD8 T cells with similar efficacy. J Immunol. 2001;167(3):1313–1324. doi: 10.4049/jimmunol.167.3.1313. [DOI] [PubMed] [Google Scholar]
- 7.Dawicki W., Watts T.H. Expression and function of 4-1BB during CD4 versus CD8 T cell responses in vivo. Eur J Immunol. 2004;34(3):743–751. doi: 10.1002/eji.200324278. [DOI] [PubMed] [Google Scholar]
- 8.Watts T.H. TNF/TNFR family members in costimulation of T cell responses. Annu Rev Immunol. 2005;23:23–68. doi: 10.1146/annurev.immunol.23.021704.115839. [DOI] [PubMed] [Google Scholar]
- 9.Wolfl M., Kuball J., Ho W.Y., Nguyen H., Manley T.J., Bleakley M. Activation-induced expression of CD137 permits detection, isolation, and expansion of the full repertoire of CD8+ T cells responding to antigen without requiring knowledge of epitope specificities. Blood. 2007;110(1):201–210. doi: 10.1182/blood-2006-11-056168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seo S.K., Choi J.H., Kim Y.H., Kang W.J., Park H.Y., Suh J.H. 4-1BB-mediated immunotherapy of rheumatoid arthritis. Nat Med. 2004;10(10):1088–1094. doi: 10.1038/nm1107. [DOI] [PubMed] [Google Scholar]
- 11.Sabbagh L., Pulle G., Liu Y., Tsitsikov E.N., Watts T.H. ERK-dependent BIM modulation downstream of the 4-1BB-TRAF1 signaling axis is a critical mediator of CD8 T cell survival in vivo. J Immunol. 2008;180(12):8093–8101. doi: 10.4049/jimmunol.180.12.8093. [DOI] [PubMed] [Google Scholar]
- 12.Vinay D.S., Kwon B.S. Role of 4-1BB in immune responses. Semin Immunol. 1998;10(6):481–489. doi: 10.1006/smim.1998.0157. [DOI] [PubMed] [Google Scholar]
- 13.Lynch D.H. The promise of 4-1BB (CD137)-mediated immunomodulation and the immunotherapy of cancer. Immunol Rev. 2008;222:277–286. doi: 10.1111/j.1600-065X.2008.00621.x. [DOI] [PubMed] [Google Scholar]
- 14.Segal N.H., He A.R., Doi T., Levy R., Bhatia S., Pishvaian M.J. Phase I study of single-agent utomilumab (PF-05082566), a 4-1BB/CD137 Agonist, in patients with advanced cancer. Clin Cancer Res. 2018;24(8):1816–1823. doi: 10.1158/1078-0432.CCR-17-1922. [DOI] [PubMed] [Google Scholar]
- 15.Vinay D.S., Kwon B.S. Therapeutic potential of anti-CD137 (4-1BB) monoclonal antibodies. Exp Opin Ther Targets. 2016;20(3):361–373. doi: 10.1517/14728222.2016.1091448. [DOI] [PubMed] [Google Scholar]
- 16.Hurtado J.C., Kim Y.J., Kwon B.S. Signals through 4-1BB are costimulatory to previously activated splenic T cells and inhibit activation-induced cell death. J Immunol. 1997;158(6):2600–2609. [PubMed] [Google Scholar]
- 17.Labiano S., Teijeira Á., Azpilikueta A., Aznar Á., Bolaños E., Melero I. Abstract 639: morphological changes in mitochondria induced by CD137 (4-1BB) co-stimulation on CD8 T cells. Cancer Res. 2017;77(13 Supplement):639. –639. [Google Scholar]
- 18.Maus M.V., Thomas A.K., Leonard D.G.B., Allman D., Addya K., Schlienger K. Ex vivo expansion of polyclonal and antigen-specific cytotoxic T lymphocytes by artificial APCs expressing ligands for the T-cell receptor, CD28 and 4-1BB. Nat Biotechnol. 2002;20(2):143–148. doi: 10.1038/nbt0202-143. [DOI] [PubMed] [Google Scholar]
- 19.Takahashi C., Mittler R.S., Vella A.T. Cutting edge: 4-1BB is a bona fide CD8 T cell survival signal. J Immunol. 1999;162(9):5037–5040. [PubMed] [Google Scholar]
- 20.Smith S.E., Hoelzinger D.B., Dominguez A.L., Van Snick J., Lustgarten J. Signals through 4-1BB inhibit T regulatory cells by blocking IL-9 production enhancing antitumor responses. Cancer Immunol Immunother. 2011;60(12):1775–1787. doi: 10.1007/s00262-011-1075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bartkowiak T., Curran M.A. 4-1BB Agonists: multi-Potent potentiators of tumor immunity. Front Oncol. 2015;5:117. doi: 10.3389/fonc.2015.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palazón A., Teijeira A., Martínez-Forero I., Hervás-Stubbs S., Roncal C., Peñuelas I. Agonist Anti-CD137 mAb act on tumor endothelial cells to enhance recruitment of activated T lymphocytes. Cancer Res. 2011;71(3):801–811. doi: 10.1158/0008-5472.CAN-10-1733. [DOI] [PubMed] [Google Scholar]
- 23.Teijeira Á., Palazón A., Garasa S., Marré D., Aubá C., Rogel A. CD137 on inflamed lymphatic endothelial cells enhances CCL21-guided migration of dendritic cells. FASEB J. 2012;26(8):3380–3392. doi: 10.1096/fj.11-201061. [DOI] [PubMed] [Google Scholar]
- 24.Palazón A., Martínez-Forero I., Teijeira A., Morales-Kastresana A., Alfaro C., Sanmamed M.F. The HIF-1α hypoxia response in tumor-infiltrating T lymphocytes induces functional CD137 (4-1BB) for immunotherapy. Cancer Discov. 2012;2(7):608–623. doi: 10.1158/2159-8290.CD-11-0314. [DOI] [PubMed] [Google Scholar]
- 25.Weigelin B., Bolaños E., Teijeira A., Martinez-Forero I., Labiano S., Azpilikueta A. Focusing and sustaining the antitumor CTL effector killer response by agonist anti-CD137 mAb. PNAS. 2015;112(24):7551–7556. doi: 10.1073/pnas.1506357112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fisher T.S., Kamperschroer C., Oliphant T., Love V.A., Lira P.D., Doyonnas R. Targeting of 4-1BB by monoclonal antibody PF-05082566 enhances T-cell function and promotes anti-tumor activity. Cancer Immunol Immunother. 2012;61(10):1721–1733. doi: 10.1007/s00262-012-1237-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Topalian S.L., Taube J.M., Anders R.A., Pardoll D.M. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16(5):275–287. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu D., Schilling B., Liu D., Sucker A., Livingstone E., Jerby-Amon L. Integrative molecular and clinical modeling of clinical outcomes to PD1 blockade in patients with metastatic melanoma. Nat Med. 2019;25(12):1916–1927. doi: 10.1038/s41591-019-0654-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Durek P., Nordström K., Gasparoni G., Salhab A., Kressler C., de Almeida M. Epigenomic profiling of human CD4+ T cells supports a linear differentiation model and highlights molecular regulators of memory development. Immunity. 2016 15;45(5):1148–1161. doi: 10.1016/j.immuni.2016.10.022. [DOI] [PubMed] [Google Scholar]
- 30.Ghoneim H.E., Fan Y., Moustaki A., Abdelsamed H.A., Dash P., Dogra P. De novo epigenetic programs inhibit PD-1 blockade-mediated t cell rejuvenation. Cell. 2017;170(1):142–157. doi: 10.1016/j.cell.2017.06.007. e19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scharer C.D., Barwick B.G., Youngblood B.A., Ahmed R., Boss J.M. Global DNA methylation remodeling accompanies CD8 T cell effector function. J Immunol. 2013;191(6):3419–3429. doi: 10.4049/jimmunol.1301395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chen Y.-.P., Zhang J., Wang Y.-.Q., Liu N., He Q.-.M., Yang X.-.J. The immune molecular landscape of the B7 and TNFR immunoregulatory ligand-receptor families in head and neck cancer: a comprehensive overview and the immunotherapeutic implications. Oncoimmunology. 2017;6(3) doi: 10.1080/2162402X.2017.1288329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gevensleben H., Holmes E.E., Goltz D., Dietrich J., Sailer V., Ellinger J. PD-L1 promoter methylation is a prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncotarget. 2016;7(48):79943–79955. doi: 10.18632/oncotarget.13161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goltz D., Gevensleben H., Dietrich J., Ellinger J., Landsberg J., Kristiansen G. Promoter methylation of the immune checkpoint receptor PD-1 (PDCD1) is an independent prognostic biomarker for biochemical recurrence-free survival in prostate cancer patients following radical prostatectomy. Oncoimmunology. 2016;5(10) doi: 10.1080/2162402X.2016.1221555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Goltz D., Gevensleben H., Dietrich J., Schroeck F., de Vos L., Droege F. PDCD1 (PD-1) promoter methylation predicts outcome in head and neck squamous cell carcinoma patients. Oncotarget. 2017;8(25):41011–41020. doi: 10.18632/oncotarget.17354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goltz D., Gevensleben H., Dietrich J., Dietrich D. PD-L1 (CD274) promoter methylation predicts survival in colorectal cancer patients. Oncoimmunology. 2017;6(1) doi: 10.1080/2162402X.2016.1257454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Goltz D., Gevensleben H., Grünen S., Dietrich J., Kristiansen G., Landsberg J. PD-L1 (CD274) promoter methylation predicts survival in patients with acute myeloid leukemia. Leukemia. 2017;31(3):738–743. doi: 10.1038/leu.2016.328. [DOI] [PubMed] [Google Scholar]
- 38.Micevic G., Thakral D., McGeary M., Bosenberg M. PD-L1 methylation regulates PD-L1 expression and is associated with melanoma survival. Pigment Cell Melanoma Res. 2019;32(3):435–440. doi: 10.1111/pcmr.12745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Röver L.K., Gevensleben H., Dietrich J., Bootz F., Landsberg J., Goltz D. PD-1 (PDCD1) promoter methylation is a prognostic factor in patients with diffuse lower-grade gliomas harboring isocitrate dehydrogenase (IDH) mutations. EBioMedicine. 2018;28:97–104. doi: 10.1016/j.ebiom.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lingohr P., Dohmen J., Semaan A., Branchi V., Dietrich J., Bootz F. Clinicopathological, immune and molecular correlates of PD-L2 methylation in gastric adenocarcinomas. Epigenomics. 2019;11(6):639–653. doi: 10.2217/epi-2018-0149. [DOI] [PubMed] [Google Scholar]
- 41.Goltz D., Gevensleben H., Vogt T.J., Dietrich J., Golletz C., Bootz F. CTLA4 methylation predicts response to anti-PD-1 and anti-CTLA-4 immunotherapy in melanoma patients. JCI Insight. 2018;3(13):e96793. doi: 10.1172/jci.insight.96793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye Q., Song D.-.G., Poussin M., Yamamoto T., Best A., Li C. CD137 accurately identifies and enriches for naturally-occurring tumor-reactive T cells in tumor. Clin Cancer Res. 2014;20(1):44–55. doi: 10.1158/1078-0432.CCR-13-0945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jacquelot N., Roberti M.P., Enot D.P., Rusakiewicz S., Ternès N., Jegou S. Predictors of responses to immune checkpoint blockade in advanced melanoma. Nat Commun. 2017;8(1):1–13. doi: 10.1038/s41467-017-00608-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jones P.A. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012;13(7):484–492. doi: 10.1038/nrg3230. [DOI] [PubMed] [Google Scholar]
- 45.Cancer Genome Atlas Network Genomic classification of cutaneous melanoma. Cell. 2015;161(7):1681–1696. doi: 10.1016/j.cell.2015.05.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thorsson V., Gibbs D.L., Brown S.D., Wolf D., Bortone D.S., Ou Yang T.-.H. The immune landscape of cancer. Immunity. 2018;48(4):812–830. doi: 10.1016/j.immuni.2018.03.023. e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu X., Zhang H., Xing Q., Cui J., Li J., Li Y. PD-1(+) CD8(+) T cells are exhausted in tumours and functional in draining lymph nodes of colorectal cancer patients. Br J Cancer. 2014;111(7):1391–1399. doi: 10.1038/bjc.2014.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Azimi F., Scolyer R.A., Rumcheva P., Moncrieff M., Murali R., McCarthy S.W. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30(21):2678–2683. doi: 10.1200/JCO.2011.37.8539. [DOI] [PubMed] [Google Scholar]
- 49.Youngblood B., Hale J.S., Kissick H.T., Ahn E., Xu X., Wieland A. Effector CD8 T cells dedifferentiate into long-lived memory cells. Nature. 2017;552(7685):404–409. doi: 10.1038/nature25144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marwitz S., Scheufele S., Perner S., Reck M., Ammerpohl O., Goldmann T. Epigenetic modifications of the immune-checkpoint genes CTLA4 and PDCD1 in non-small cell lung cancer results in increased expression. Clin Epigenetics. 2017;9:51. doi: 10.1186/s13148-017-0354-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Choi B.K., Lee S.C., Lee M.J., Kim Y.H., Kim Y.-.W., Ryu K.-.W. 4-1BB-based Isolation and expansion of CD8+ T cells specific for self-tumor and non-self-tumor antigens for adoptive T-cell therapy. J Immunother. 2014;37(4):225. doi: 10.1097/CJI.0000000000000027. [DOI] [PubMed] [Google Scholar]
- 52.Melero I., Johnston J.V., Shufford W.W., Mittler R.S., Chen L. NK1.1 cells express 4-1BB (CDw137) costimulatory molecule and are required for tumor immunity elicited by Anti-4-1BB monoclonal antibodies. Cell Immunol. 1998;190(2):167–172. doi: 10.1006/cimm.1998.1396. [DOI] [PubMed] [Google Scholar]
- 53.So T., Lee S.-.W., Croft M. Immune regulation and control of regulatory T cells by OX40 and 4-1BB. Cytokine Growth Factor Rev. 2008;19(3–4):253–262. doi: 10.1016/j.cytogfr.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee S.-.W., Park Y., So T., Kwon B.S., Cheroutre H., Mittler R.S. Identification of regulatory functions for 4-1BB and 4-1BBL in myelopoiesis and the development of dendritic cells. Nat Immunol. 2008;9(8):917–926. doi: 10.1038/ni.1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Espinosa E., Márquez-Rodas I., Soria A., Berrocal A., Manzano J.L., Gonzalez-Cao M. Predictive factors of response to immunotherapy—a review from the Spanish Melanoma Group (GEM) Ann Transl Med. 2017;5(19) doi: 10.21037/atm.2017.08.10. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5653511/ [cited 2018 Oct 3]Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Vinay D.S., Cha K., Kwon B.S. Dual immunoregulatory pathways of 4-1BB signaling. J Mol Med. 2006;84(9):726–736. doi: 10.1007/s00109-006-0072-2. [DOI] [PubMed] [Google Scholar]
- 57.Tolcher A.W., Sznol M., Hu-Lieskovan S., Papadopoulos K.P., Patnaik A., Rasco D.W. Phase IB study of utomilumab (PF-05082566), a 4-1BB/CD137 agonist, in combination with pembrolizumab (MK-3475) in patients with advanced solid tumors. Clin Cancer Res. 2017;23(18):5349–5357. doi: 10.1158/1078-0432.CCR-17-1243. [DOI] [PubMed] [Google Scholar]
- 58.Ni L., Lu J. Interferon gamma in cancer immunotherapy. Cancer Med. 2018;7(9):4509–4516. doi: 10.1002/cam4.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ayers M., Lunceford J., Nebozhyn M., Murphy E., Loboda A., Kaufman D.R. IFN-γ–related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Aznar M.A., Labiano S., Diaz-Lagares A., Molina C., Garasa S., Azpilikueta A. CD137 (4-1BB) costimulation modifies DNA methylation in CD8+ T cell-relevant genes. Cancer Immunol Res. 2018;6(1):69–78. doi: 10.1158/2326-6066.CIR-17-0159. [DOI] [PubMed] [Google Scholar]
- 61.Hodis E., Watson I.R., Kryukov G.V., Arold S.T., Imielinski M., Theurillat J.-.P. A landscape of driver mutations in melanoma. Cell. 2012;150(2):251–263. doi: 10.1016/j.cell.2012.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parmenter T.J., Kleinschmidt M., Kinross K.M., Bond S.T., Li J., Kaadige M.R. Response of BRAF-mutant melanoma to BRAF inhibition is mediated by a network of transcriptional regulators of glycolysis. Cancer Discov. 2014;4(4):423–433. doi: 10.1158/2159-8290.CD-13-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Weisenberger D.J., Siegmund K.D., Campan M., Young J., Long T.I., Faasse M.A. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38(7):787–793. doi: 10.1038/ng1834. [DOI] [PubMed] [Google Scholar]
- 64.Jin S.-.G., Xiong W., Wu X., Yang L., Pfeifer G.P. The DNA methylation landscape of human melanoma. Genomics. 2015;106(6):322–330. doi: 10.1016/j.ygeno.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ilieva K.M., Correa I., Josephs D.H., Karagiannis P., Egbuniwe I.U., Cafferkey M.J. Effects of BRAF mutations and BRAF inhibition on immune responses to melanoma. Mol Cancer Ther. 2014;13(12):2769–2783. doi: 10.1158/1535-7163.MCT-14-0290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amankulor N.M., Kim Y., Arora S., Kargl J., Szulzewsky F., Hanke M. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev. 2017;31(8):774–786. doi: 10.1101/gad.294991.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kim J., Kim K., Kim H., Yoon G., Lee K. Characterization of age signatures of DNA methylation in normal and cancer tissues from multiple studies. BMC Genom. 2014;15:997. doi: 10.1186/1471-2164-15-997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peters M.J., Joehanes R., Pilling L.C., Schurmann C., Conneely K.N., Powell J. The transcriptional landscape of age in human peripheral blood. Nat Commun. 2015;6:8570. doi: 10.1038/ncomms9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Acevedo N., Reinius L.E., Vitezic M., Fortino V., Söderhäll C., Honkanen H. Age-associated DNA methylation changes in immune genes, histone modifiers and chromatin remodeling factors within 5 years after birth in human blood leukocytes. Clin Epigenetics. 2015;7:34. doi: 10.1186/s13148-015-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dozmorov M.G., Coit P., Maksimowicz-McKinnon K., Sawalha A.H. Age-associated DNA methylation changes in naive CD4+ T cells suggest an evolving autoimmune epigenotype in aging T cells. Epigenomics. 2017;9(4):429–445. doi: 10.2217/epi-2016-0143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tserel L., Kolde R., Limbach M., Tretyakov K., Kasela S., Kisand K. Age-related profiling of DNA methylation in CD8+ T cells reveals changes in immune response and transcriptional regulator genes. Sci Rep. 2015;5:13107. doi: 10.1038/srep13107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Steegenga W.T., Boekschoten M.V., Lute C., Hooiveld G.J., de Groot P.J., Morris T.J. Genome-wide age-related changes in DNA methylation and gene expression in human PBMCs. Age. 2014;36(3):9648. doi: 10.1007/s11357-014-9648-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Carter S.L., Cibulskis K., Helman E., McKenna A., Shen H., Zack T. Absolute quantification of somatic DNA alterations in human cancer. Nat Biotechnol. 2012;30(5):413–421. doi: 10.1038/nbt.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Saltz J., Gupta R., Hou L., Kurc T., Singh P., Nguyen V. Spatial organization and molecular correlation of tumor-infiltrating lymphocytes using deep learning on pathology images. Cell Rep. 2018;23(1):181–193. doi: 10.1016/j.celrep.2018.03.086. e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Newman A.M., Liu C.L., Green M.R., Gentles A.J., Feng W., Xu Y. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi: 10.1038/nmeth.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M. NCBI GEO: archive for functional genomics data sets–update. Nucleic Acids Res. 2013;41(Database issue):D991–D995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Marzese D.M., Scolyer R.A., Huynh J.L., Huang S.K., Hirose H., Chong K.K. Epigenome-wide DNA methylation landscape of melanoma progression to brain metastasis reveals aberrations on homeobox D cluster associated with prognosis. Hum Mol Genet. 2014;23(1):226–238. doi: 10.1093/hmg/ddt420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fujiwara S., Nagai H., Jimbo H., Jimbo N., Tanaka T., Inoie M. Gene expression and methylation analysis in melanomas and melanocytes from the same patient: loss of NPM2 expression is a potential immunohistochemical marker for melanoma. Front Oncol. 2018;8:675. doi: 10.3389/fonc.2018.00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Zhu H., Mi W., Luo H., Chen T., Liu S., Raman I. Whole-genome transcription and DNA methylation analysis of peripheral blood mononuclear cells identified aberrant gene regulation pathways in systemic lupus erythematosus. Arthritis Res Ther. 2016;18:162. doi: 10.1186/s13075-016-1050-x. 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mamrut S., Avidan N., Staun-Ram E., Ginzburg E., Truffault F., Berrih-Aknin S. Integrative analysis of methylome and transcriptome in human blood identifies extensive sex- and immune cell-specific differentially methylated regions. Epigenetics. 2015;10(10):943–957. doi: 10.1080/15592294.2015.1084462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ventham N.T., Kennedy N.A., Adams A.T., Kalla R., Heath S., O'Leary K.R. Integrative epigenome-wide analysis demonstrates that DNA methylation may mediate genetic risk in inflammatory bowel disease. Nat Commun. 2016;7:13507. doi: 10.1038/ncomms13507. 25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Absher D.M., Li X., Waite L.L., Gibson A., Roberts K., Edberg J. Genome-wide DNA methylation analysis of systemic lupus erythematosus reveals persistent hypomethylation of interferon genes and compositional changes to CD4+ T-cell populations. PLoS Genet. 2013;9(8) doi: 10.1371/journal.pgen.1003678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li B., Dewey C.N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Du P., Zhang X., Huang C.-.C., Jafari N., Kibbe W.A., Hou L. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010;11:587. doi: 10.1186/1471-2105-11-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lehmann U., Kreipe H. Real-time PCR-based assay for quantitative determination of methylation status. Methods Mol Biol. 2004;287:207–218. doi: 10.1385/1-59259-828-5:207. [DOI] [PubMed] [Google Scholar]
- 87.Zerbino D.R., Achuthan P., Akanni W., Amode M.R., Barrell D., Bhai J. Ensembl 2018. Nucleic Acids Res. 2018;46(D1):D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jung M., Uhl B., Kristiansen G., Dietrich D. Bisulfite conversion of DNA from tissues, cell lines, buffy coat, FFPE tissues, microdissected cells, swabs, sputum, aspirates, lavages, effusions, plasma, serum, and urine. Methods Mol Biol. 2017;1589:139–159. doi: 10.1007/7651_2015_260. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.