Abstract
Xanthophyllomyces dendrorhous is a basidiomycete yeast known as a natural producer of astaxanthin, a carotenoid of commercial interest because of its antioxidant properties. Recent studies indicated that X. dendrorhous has a functional SREBP pathway involved in the regulation of isoprenoid compound biosynthesis, which includes ergosterol and carotenoids. SREBP is a major regulator of sterol metabolism and homeostasis in mammals; characterization in fungi also provides information about its role in the hypoxia adaptation response and virulence. SREBP protease processing is required to activate SREBP pathway functions in fungi. Here, we identified and described the STP1 gene, which encodes a metallopeptidase of the M50 family involved in the proteolytic activation of the transcription factor Sre1 of the SREBP pathway, in X. dendrorhous. We assessed STP1 function in Δstp1 strains derived from the wild-type and a mutant of ergosterol biosynthesis that overproduces carotenoids and sterols. Bioinformatic analysis of the deduced protein predicted the presence of characteristic features identified in homologs from mammals and fungi. The Δstp1 mutation decreased yeast growth in the presence of azole drugs and reduced transcript levels of Sre1-dependent genes. This mutation also negatively affected the carotenoid- and sterol-overproducing phenotype. Western blot analysis demonstrated that Sre1 was activated in the yeast ergosterol biosynthesis mutant and that the Δstp1 mutation introduced in this strain prevented Sre1 proteolytic activation. Overall, our results demonstrate that STP1 encodes a metallopeptidase involved in proteolytic activation of Sre1 in X. dendrorhous, contributing to our understanding of fungal SREBP pathways.
Keywords: isoprenoids, nuclear receptors/sterol regulatory element-binding protein, antioxidants, molecular biology, site-2 protease, astaxanthin, sterols, mevalonate pathway, Stp1, Sre1
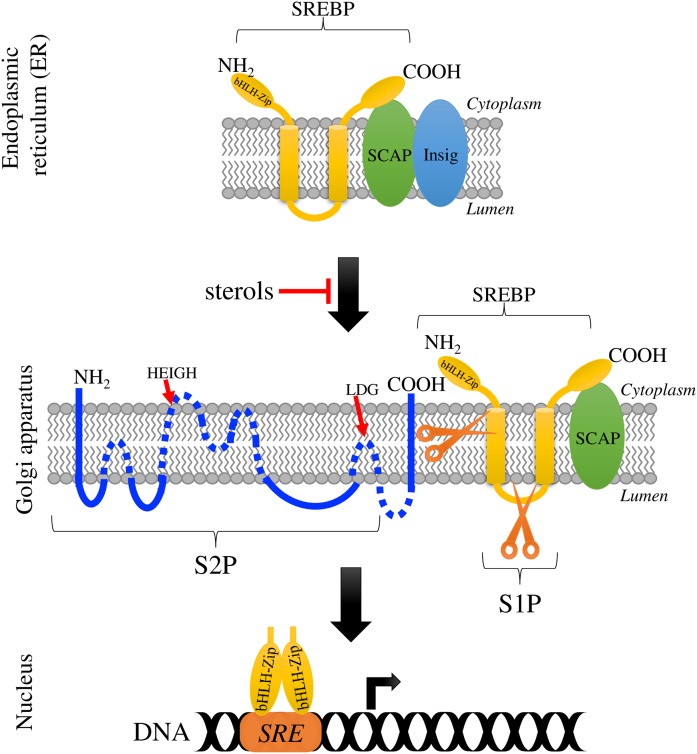
One of the major mechanisms regulating sterol metabolism and homeostasis in mammalian cells is the SREBP pathway (Fig. 1), which uses a family of ER membrane-bound transcription factors called SREBPs (1). This pathway has been well described in mammals, and two genes encode the three isoforms of the transcription factor SREBP designated SREBP-1a, SREBP-1c, and SREBP-2; the first two are encoded by a single gene with alternative transcription start sites (2). All three SREBPs are oriented in the ER membrane in a hairpin fashion with two transmembrane (TM) helices and with both domains, the N-terminal and the C-terminal domains, facing the cytoplasm (3). The N-terminal domain is a transcription factor that regulates genes involved in the uptake and synthesis of cholesterol, fatty acids, triglycerides, and phospholipids (4). SREBPs belong to the basic-helix-loop-helix (bHLH) leucine zipper family with a unique tyrosine residue in the bHLH motif that distinguishes these proteins from other bHLH transcription factors (1, 5). The C-terminal domain of SREBP is a regulatory domain that interacts with the SREBP cleavage activating protein (SCAP), which is an integral ER membrane protein (3). In the presence of sufficient cellular sterol, SCAP binds sterols and interacts with the protein Insig, which is an ER retention protein that holds the SREBP-SCAP complex within the ER membrane (6, 7). In sterol-depleted cells, the interaction between SCAP and Insig is disrupted, and the SREBP-SCAP complex is packaged into COPII vesicles and travels to the Golgi apparatus. At the Golgi apparatus, SREBP is a substrate of site-1 and site-2 proteases (S1P and S2P, respectively) that release the N-terminal domain of SREBP through a two-step proteolytic process (8). The activation of SREBP has been studied most thoroughly for SREBP-2 and begins when the subtilisin-related serine protease S1P cleaves SREBP at site 1 at the hydrophilic loop projected into the Golgi apparatus lumen (9). Then, the metallopeptidase S2P cleaves SREBP at site 2 within the first TM segment, releasing the N-terminal domain of SREBP (10, 11), which translocates to the nucleus to regulate gene transcription (4).
Fig. 1.
The SREBP pathway. Scheme of the mammalian SREBP pathway [adapted from (67)]. With sufficient sterol, the SREBP-SCAP complex is retained at the ER membrane due to SCAP interacting with Insig; when sterol levels decrease, the SREBP-SCAP complex is transported to the Golgi apparatus. In the Golgi apparatus, SREBP undergoes two sequential proteolytic cleavages by S1P and S2P. First, S1P (subtilisin-related serine protease) cleaves SREBP at the hydrophilic loop projected into the lumen of the Golgi apparatus, and then S2P (metallopeptidase) cuts SREBP within the first TM segment. This releases the N-terminal domain of SREBP (the activated transcription factor), which travels to the nucleus to regulate gene transcription of target genes, including genes required for the synthesis and uptake of sterols and other lipids. bHLH-ZIP represents the N-terminal domain of SREBP, containing the bHLH leucine zipper motif, and SRE denotes the sterol regulatory element. The illustrated S2P membrane topology was based on the results of Zelenski et al. (42), and conserved residues in the motifs HEIGH and LDG involved in the coordination of the zinc atom are shown.
More recently, the SREBP pathway has been studied in some fungi, organisms that produce ergosterol as the main sterol (12). Most studies of the SREBP pathway in fungi have been performed in the fission yeast Schizosaccharomyces pombe, where homologous genes of human proteins SREBP-1a, SCAP, and Insig named SRE1, SCP1, and INS1, respectively, have been identified (13). Sre1 is activated and cleaved in response to sterol depletion or under hypoxic conditions by a Scp1-dependent mechanism but is Ins1-independent (13). The fission yeast lacks identifiable homolog genes of S1P and S2P, and Sre1 is cleaved by a different mechanism that depends on the Golgi membrane-anchored Dsc E3 ligase complex, the AAA-ATPase Cdc48, and the Golgi-resident rhomboid protease Rbd2 (14–16). As in S. pombe, the SREBP homolog in Aspergillus fumigatus, named SrbA, is also involved in the regulation of ergosterol biosynthesis and is critical for growth under hypoxic conditions. Interestingly, srbA-null mutants fail to cause disease in invasive pulmonary aspergillosis murine models, demonstrating the role of SrbA in fungal pathogenesis (17). This ascomycete does not have SCAP or S1P and S2P homolog genes. The activation of SrbA would be mediated by the ER-resident aspartyl protease SppA and by homolog components of the Dsc E3 ligase complex and rhomboid protease Rbd2 described in S. pombe that are named DscA-D and RbdB in A. fumigatus. However, it is still not clear how and where exactly (ER or Golgi apparatus) SrbA is processed (18). Regarding basidiomycete fungi, the genes SRE1 and SCP1, encoding Sre1 and Scp1, were described in the yeast Cryptococcus neoformans, where Sre1 activation is also required for host adaptation and virulence in addition to sterol biosynthesis regulation and hypoxic response (19, 20). In C. neoformans, a homolog of the mammalian S2P protease gene was identified (named STP1), and its role in Sre1 activation was demonstrated (20, 21).
In the basidiomycetous yeast Xanthophyllomyces dendrorhous [formerly Phaffia rhodozyma (22)], an SREBP/Sre1 homolog encoded by the SRE1 gene has been recently identified and proven to be involved in the regulation of carotenoid and sterol synthesis (Fig. 2) (23). X. dendrorhous is one of the few organisms that synthesizes the carotenoid astaxanthin, which is of biotechnological interest due to its antioxidant properties and use as a dye in salmonid and crustacean aquaculture, and whose color is perceived as a key quality attribute by consumers (24, 25). Ergosterol biosynthesis mutants of X. dendrorhous overproduce carotenoids (26), and the carotenoid-overproducing phenotype reverts to producing wild-type levels of carotenoids by a mutation in the SRE1 gene (23). This result indicates that in addition to sterol biosynthesis, Sre1 favors carotenoid production in X. dendrorhous, supporting the SREBP pathway in this yeast as a target for obtaining carotenoid-overproducing strains. In this study, the X. dendrorhous STP1 gene was identified and characterized. Its predicted gene product has characteristic features of metallopeptidases of the M50 family (the family that includes the mammalian protein S2P). In several aspects, the Δstp1 mutation affects the yeast phenotype similarly to the sre1− mutation. The gene STP1 (as SRE1) is not essential for yeast viability under laboratory culture conditions, but Δstp1 (and sre1−) mutants are unable to grow in the presence of sterol synthesis inhibitors such as azole compounds. Additionally, the Δstp1 mutation reduced sterol and carotenoid production to wild-type levels in an ergosterol biosynthesis mutant that overproduces carotenoids, and it was confirmed that the Δstp1 mutation prevented Sre1 proteolytic activation in X. dendrorhous.
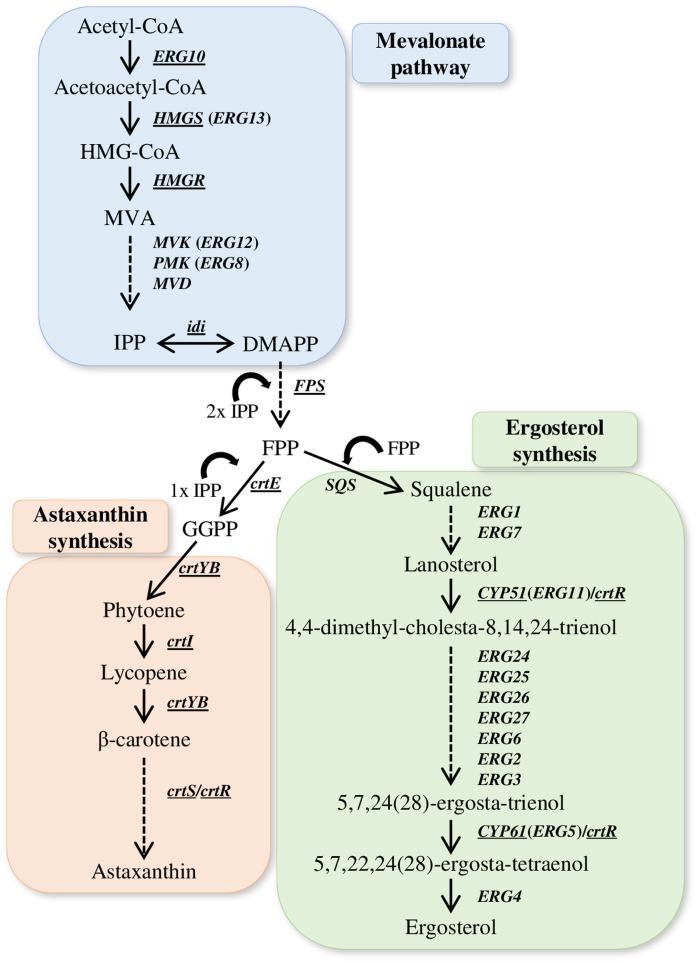
Fig. 2.
Overview of the MVA pathway and the synthesis of astaxanthin and ergosterol in X. dendrorhous. The MVA pathway (in blue), astaxanthin (in red), and ergosterol (in green) biosynthesis [adapted from (26)]. Arrows represent the catalytic step with the corresponding enzyme-encoding gene in X. dendrorhous (underlined) or in S. cerevisiae. MVA-P, mevalonate-5-phosphate; MVA-PP, mevalonate-5-pyrophosphate; IPP, isopentenyl-pyrophosphate; DMAPP, dimethylallyl-pyrophosphate; GPP, geranyl-pyrophosphate; FPP, farnesyl-pyrophosphate; GGPP, geranylgeranyl-pyrophosphate. The included X. dendrorhous genes, encoded enzymes and GenBank accession numbers (in square brackets) are ERG10, acetyl-CoA C-acetyltransferase [KX267759]; HMGS, HMG-CoA synthase [MK368600]; HMGR, [MK368599]; idi, IPP isomerase [DQ235686]; FPS, FPP synthase [KJ140284]; crtE, GGPP synthase [DQ012943]; crtYB, bifunctional lycopene cyclase/phytoene synthase [DQ016503]; crtI, phytoene desaturase [Y15007]; crtS, astaxanthin synthase [EU713462]; crtR, cytochrome p450 reductase [EU884133]; CYP51, lanosterol 14α-demethylase [KP317478]; and CYP61, C-22 sterol desaturase [JX183240].
MATERIALS AND METHODS
Microorganisms and culture conditions
The strains and plasmids used and created in this work are listed in Table 1.
TABLE 1.
Strains and plasmids used in this work
| Strain/Plasmid | Description | Reference or Source |
| Strain | ||
| E. coli | ||
| DH5α | AmpS. Used for molecular cloning and plasmid maintenance. | (27) |
| S. cerevisiae | ||
| S288c | Haploid reference strain used for plasmid construction by DNA assembler. | ATCC 204508 |
| X. dendrorhous | ||
| CBS 6938 | Wild-type strain (ZeoS, HygS, and NatS). | ATCC 96594 |
| CBS.cyp61− | Mutant (ZeoR, HygS, and NatS) derived from CBS 6938. The single CYP61 locus was interrupted by the zeocin resistance cassette. | This work |
| CBS.Δstp1 | Mutant (ZeoS, HygR, and NatS) derived from CBS 6938. The single STP1 locus was replaced by the hygromycin B resistance cassette. | This work |
| CBS.Δstp1/STP1 | Mutant (ZeoR, HygS, and NatS) derived from CBS.Δstp1, with the native STP1 allele reintegrated at the STP1 native locus and followed by the zeocin resistance cassette. | This work |
| CBS.cyp61−.Δstp1 | Mutant (ZeoR, HygR, and NatS) derived from CBS.cyp61-. The single STP1 locus was replaced by the hygromycin B resistance cassette. | This work |
| CBS.FLAG.SRE1 | Mutant (ZeoR, HygS, and NatS) derived from CBS 6938. The native SRE1 gene was replaced by a gene variant that expresses the Sre1 protein fused to the 3xFLAG epitope at its N-terminus, followed by the zeocin resistance cassette. | This work |
| CBS.FLAG.SRE1.Δstp1 | Mutant (ZeoR, HygR, and NatS) derived from CBS.FLAG.SRE1. The single STP1 locus was replaced by the hygromycin B resistance cassette. | This work |
| CBS.Δstp1.FLAG.SRE1N | Mutant (ZeoR, HygR, and NatS) derived from CBS.Δstp1. The native SRE1 gene was replaced by a gene variant that expresses the N-terminal domain of Sre1 (Sre1N) fused to the 3xFLAG epitope at its N-terminus, followed by the zeocin resistance cassette. | This work |
| CBS.FLAG.SRE1N | Mutant (ZeoR, HygS, and NatS) derived from CBS 6938. The native SRE1 gene was replaced by a gene version that expresses Sre1N fused to the 3xFLAG epitope at its N-terminal, followed by the zeocin resistance cassette. | (23) |
| CBS.cyp61−.FLAG.SRE1 | Mutant (ZeoR, HygR, and NatS) derived from CBS.cyp61−. The native SRE1 gene was replaced by a gene variant that expresses the Sre1 protein fused to the 3xFLAG epitope at its N-terminus, followed by the hygromycin B resistance cassette. | This work |
| CBS.cyp61−.FLAG.SRE1.Δstp1 | Mutant (ZeoR, HygR, and NatR) derived from CBS.cyp61−.FLAG.SRE1. The single STP1 locus was replaced by the nourseothricin resistance cassette. | This work |
| CBS.sre1− | Mutant (ZeoR, HygS, and NatS) derived from CBS 6938. Gene SRE1 was partially deleted (approximately 90% of the coding region was replaced by the zeocin resistance cassette). | (23) |
| CBS.ΔS1P-Cn | Mutant (ZeoS, HygR, and NatS) derived from CBS 6938. The single CDZ97435 locus (putative homolog of the potential S1P homolog identified in C. neoformans) was replaced by the hygromycin B resistance cassette. | This work |
| CBS.ΔS1P-Hs | Mutant (ZeoS, HygR, and NatS) derived from CBS 6938. The single CED82778 locus (putative homolog of H. sapiens S1P encoding gene) was replaced by the hygromycin B resistance cassette. | This work |
| CBS.cyp61−.ΔS1P-Cn | Mutant (ZeoS, HygR, and NatS) derived from CBS.cyp61−. The single CDZ97435 locus was replaced by the hygromycin B resistance cassette. | This work |
| CBS.cyp61−.ΔS1P-Hs | Mutant (ZeoS, HygR, and NatS) derived from CBS.cyp61−. The single CED82778 locus was replaced by the hygromycin B resistance cassette. | This work |
| Plasmid | ||
| pBluescript SK- (pBS) | Cloning vector (ColE1 ori, AmpR, blue-white colony selection). | Agilent Technologies Inc. |
| pMN-hph | pBS containing the hygromycin B resistance cassette (1.8 kb) used for X. dendrorhous transformant selection at the EcoRV site. | (30) |
| pIR-zeo | pBS containing the zeocin resistance cassette (1.2 kb) used for X. dendrorhous transformant selection at the EcoRV site. | (26) |
| pBS-nat | pBS containing the nourseothricin resistance cassette (1.4 kb) used for X. dendrorhous transformant selection at the EcoRV site. The constructed cassette contains the Streptomyces noursei nat1 gene regulated by the X. dendrorhous EF-1α gene promoter and GPD gene transcription terminator. | This work |
| pBS-ΔgSTP1hph | pBS containing the 715 bp upstream and 620 bp downstream of the STP1 gene and the hygromycin B resistance cassette between them at the EcoRV site. Used to delete the X. dendrorhous STP1 gene by homologous recombination. | This work |
| pBS-ΔgSTP1nat | pBS containing the 715 bp upstream and 620 bp downstream of the STP1 gene and the nourseothricin resistance cassette between them at the EcoRV site. Used to delete the X. dendrorhous STP1 gene by homologous recombination. | This work |
| pBS-gSTP1up-down | pBS containing at the SmaI site, the X. dendrorhous STP1 gene and the zeocin resistance cassette at the AgeI site of the downstream region of the STP1 gene. Used to integrate the X. dendrorhous STP1 gene by homologous recombination in Δstp1 mutants. | This work |
| pXd-gSRE1N-zeo | Plasmid constructed by DNA assembler used to replace the X. dendrorhous SRE1 gene with the gene variant that expresses the Sre1N (Sre1 N-terminal domain) 3xFLAG-tagged at its N-terminal end, followed by the zeocin resistance cassette for transformant selection. | (23) |
| pXd-gSRE1-zeo | Plasmid constructed by DNA assembler used to replace the X. dendrorhous SRE1 gene with the gene variant that expresses Sre1 (full-length Sre1 protein) 3xFLAG-tagged at its N-terminal end. It contains 2,816 bp (from the start to the stop codon) of the genomic SRE1 fused to the 3xFLAG-encoding sequence at the 5′ end. This sequence is flanked by the upstream and downstream regions of the SRE1 gene to direct its integration at the SRE1 locus including a zeocin resistance cassette for transformant selection. | This work |
| pXd-gSRE1-hph | Plasmid constructed by DNA assembler used to replace the X. dendrorhous SRE1 gene with the gene variant that expresses Sre1 (full-length Sre1 protein) 3xFLAG-tagged at its N-terminal end, followed by the hygromycin B resistance cassette for transformant selection. It was constructed in the same way as pXd-gSRE1-zeo, but instead of zeocin, it contains the hygromycin B resistance cassette for transformant selection. | This work |
| pYES2 | S. cerevisiae expression vector containing 2 μ origin. Used to amplify the 2 μ DNA by PCR, which was then used for plasmid construction by DNA assembler. | Thermo Fisher Scientific Inc. |
| pFA6 | Yeast plasmid with kanamycin/geneticin (G418) resistance marker. Used to amplify the G418 marker by PCR, which was then used for plasmid construction by DNA assembler. | (65) |
| pFlagTEM1 | Expression plasmid containing the FLAG-tagged lactamase-encoding sequence. Used to amplify the 3xFLAG epitope (3xDYKDDDDK)-encoding sequence, which was then used for plasmid construction by DNA assembler. | (66) |
| pBS.ΔS1P-Cn hph | pBS containing the 720 bp upstream and 659 bp downstream of the CDZ97435 locus and the hygromycin B resistance cassette between them at the EcoRV site. Used to delete the X. dendrorhous CDZ97435 locus by homologous recombination. | This work |
| pBS.ΔS1P-Hs hph | pBS containing the 725 bp upstream and 640 bp downstream of the CED82778 locus and the hygromycin B resistance cassette between them at the EcoRV site. Used to delete the X. dendrorhous CED82778 locus by homologous recombination. | This work |
| AmpS, sensitive to ampicillin; ZeoS/ZeoR, sensitive/resistant to zeocin; HygS/HygR, sensitive/resistant to hygromycin B; NatS/NatR, sensitive/resistant to nourseothricin; ColE1 ori, replication origin of E. coli ColE1 plasmid, AmpR, ampicillin resistance. | ||
X. dendrorhous strains were grown at 22°C with constant agitation in YM medium (0.3% yeast extract, 0.3% malt extract, and 0.5% peptone) supplemented with 1% glucose. Yeast transformant selection was performed on 1.5% agar YM plates with 35 μg/ml hygromycin B, 70 μg/ml zeocin, and/or 45 μg/ml nourseothricin. Yeast growth was evaluated on YM-agar plates (1.5%) supplemented with itraconazole (5.0 nM), clotrimazole (0.1 μg/ml), ketoconazole (0.1 μg/ml), or CoCl2 (400 μM). For this evaluation, 5 μl of serial dilutions of cultures of each strain were seeded on a YM-agar plate with the corresponding compound at a concentration that did not affect the wild-type CBS 6938 growth and incubated at 22°C for 5 days. For growth curves and phenotypic analysis, yeast strains were cultured in YM medium at 22°C with constant agitation in triplicate. The optical density of the cultures was registered at 600 nm using a V-630 JASCO spectrophotometer (JASCO Inc., Easton, MD). After 120 h of culture (stationary phase of growth), samples were collected to extract carotenoids, sterols, and RNA, and to determine the dry weight of the yeast.
Escherichia coli strains were grown with constant agitation at 37°C in lysogeny broth medium. Lysogeny broth-agar plates were supplemented with 100 μg/ml ampicillin for plasmid selection and 32 μg/ml X-gal (5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside) for recombinant clone selection (27). Recombinant clones bearing plasmids constructed in this work were selected by direct colony PCR.
Saccharomyces cerevisiae strains were cultured at 30°C in YPD medium (1% yeast extract, 2% peptone, 2% glucose). When necessary, geneticin (G418) was supplemented at 200 μg/ml.
Nucleic acid purification and analysis
DNA from X. dendrorhous was obtained by mechanical rupture of cell pellets suspended in 600 μl of TE buffer (25 mM Tris-HCl, 10 mM EDTA, pH 8.0) and 600 μl of phenol:chloroform:isoamyl alcohol mixture (25:24:1, v/v/v) with 100 μl of 0.5 mm glass beads. Then, the mixture was agitated using a Mini-beadbeater-16 (BioSpec Products Inc., Bartlesville, OK) for 3 min, followed by centrifugation for 5 min at 18,440 g to recover the aqueous phase. Next, 1 vol of chloroform:isoamyl alcohol (24:1, v/v) was added to remove phenol traces, followed by centrifugation for 5 min at 18,440 g. The aqueous phase was recovered, and DNA was precipitated with 1 ml of cold absolute ethanol and incubation at −20°C for 1 h. The mixture was centrifuged for 10 min at 18,440 g, the supernatant was eliminated, and the DNA pellet was allowed to dry at 37°C before being suspended in 100 μl of sterile water.
Purification of plasmid DNA from E. coli was performed using the GeneJET Plasmid Miniprep Kit (Thermo Fisher Scientific Inc., Waltham, MA) according to the supplier’s instructions.
RNA from X. dendrorhous was extracted from cell pellets obtained from 500 to 1,000 μl of a yeast culture that were suspended in 200 μl of lysis buffer (sodium acetate 2 mM pH 5.5, 0.5% SDS, 1 mM EDTA, in water 0.1% DEPC) with 100 μl of 0.5 mm glass beads. Cells were broken using a Mini-beadbeater-16 for 3 min, followed by the addition of 800 μl of TRI Reagent™ solution (Thermo Fisher Scientific Inc.) and another 3 min of mechanical rupture. Then, 200 μl of chloroform were added, followed by incubation at room temperature for 10 min and centrifugation at 18,440 g for 10 min at 4°C. The aqueous phase was recovered and deposited in two Eppendorf tubes, and 250 μl of sterile water and 550 μl of cold isopropanol were added to each; then, the tubes were incubated at room temperature for 10 min and centrifuged for 10 min at 18,440 g at 4°C. The supernatant was removed, and the pellet was washed with 1 ml of 70% ethanol solution (v/v) and centrifuged at 18,440 g for 6 min at 4°C. The pellet was allowed to dry and suspended in 20–30 μl of sterile water.
DNA amplification and cDNA synthesis for RT-qPCR analysis
All primers used in this work are included in supplemental Table S1.
Plasmid construction and DNA fragment integration in the X. dendrorhous genome were evaluated by PCR. In general, each reaction included 1× PCR buffer (500 mM KCl, 200 mM Tris-HCl pH 8.4), 2 mM MgCl2, 0.2 μM each dNTP, 1 μM each primer, 1 U of Taq or Pfu DNA polymerase and approximately 10 ng of template DNA. Reactions were carried out in a 2720 thermal cycler (Applied Biosystems, Foster City, CA) using the following program: initial denaturation at 94°C for 3 min; 35 cycles of denaturation at 94°C for 30 s, annealing at 55°C for 30 s, and elongation at 72°C for a period whose length was adjusted on the basis of the size of the amplified fragment; followed by 10 min at 72°C and a final stage held at 4°C.
cDNA was synthesized according to the enzyme provider’s protocol using 5 μg of total RNA as a template, M-MLV reverse transcriptase (Thermo Fisher Scientific Inc.), Oligo(dT), and dNTPs in a final reaction volume of 20 μl. For quantitative (q)PCR, the SensiMix™ SYBR® Hi-ROX kit (Bioline Reagents Ltd., London, UK) was used according to specifications. The samples were prepared in triplicate and loaded into an Mx3000P qPCR system (Agilent Technologies Inc., Santa Clara, CA). To evaluate the relative transcript levels of a particular gene, the Ct (threshold cycle) values were normalized to the Ct values of the X. dendrorhous actin-encoding gene [GenBank: X89898.1], and the 2−ΔΔCt method (28) was used to compare transcript levels of strains. The obtained decimal values were converted to negative values according to Schmittgen and Livak (29).
Plasmid construction and yeast transformation
All plasmids used and constructed in this work are detailed in Table 1.
To delete the STP1 gene in X. dendrorhous, plasmids pBS-ΔgSTP1hph and pBS-ΔgSTP1nat were constructed. To obtain pBS-ΔgSTP1hph, the upstream (715 bp) and downstream (620 bp) regions of the STP1 locus were PCR-amplified from genomic DNA of X. dendrorhous strain CBS 6938 (supplemental Fig. S1). These regions were then joined by overlap extension PCR introducing an HpaI site between them, and the resulting fragment was inserted at the EcoRV site of pBluescript SK-. Then, at the HpaI site, a hygromycin B resistance cassette was inserted that had been previously PCR-amplified from pMN-hph (30). In the case of pBS-ΔgSTP1nat, the nourseothricin resistance cassette was amplified from the plasmid pBS-nat. This plasmid was constructed by inserting a cassette of 1.4 kb that carried the Streptomyces noursei nourseothricin resistance nat1 gene under the regulation of the X. dendrorhous EF-1α gene promoter and the GPD gene terminator at the EcoRV site of pBluescript SK-. Both plasmids, pBS-ΔgSTP1hph and pBS-ΔgSTP1nat, were digested with ApaI and XbaI to transform and replace the STP1 gene with the corresponding antibiotic resistance module in strains CBS.6938, CBS.cyp61−, which derives from CBS 6938 and was constructed with the plasmid pBS-cyp61/Zeo reported in (26), and the 3xFLAG-tagged strains, CBS.FLAG.SRE1 and CBS.cyp61−.FLAG.SRE1, whose construction is described below. Thus, through a double homologous recombination event (supplemental Fig. S1), the strains CBS.Δstp1, CBS.cyp61−.Δstp1, CBS.FLAG.SRE1.Δstp1, and CBS.cyp61−.FLAG.SRE1.Δstp1 were obtained, respectively (Table 1).
To replace the SRE1 gene by a version of it that expresses the Sre1 protein fused to a 3xFLAG epitope at its N-terminal end, plasmids pXd.gSRE1-zeo and pXd.gSRE1-hph were constructed in vivo in S. cerevisiae by DNA assembler methodology (31) (supplemental Fig. S2). First, nine DNA fragments were PCR-amplified using primer pairs designed to allow their assembly in vivo through homologous recombination in S. cerevisiae. The amplified fragments were: i) the S. cerevisiae 2 μ origin (amplified from pYES2 with primers B.Fw + B.Rv); ii) 829 bp of the upstream SRE1 gene region (amplified from CBS 6938 genomic DNA with primers C.Fw + C_2.Rv); iii) a 3xFLAG epitope-encoding sequence (amplified from pFlagTEM1 with primers D_2.Fw + D_2.Rv); iv) the SRE1 gene (amplified from CBS 6938 genomic DNA with primers SRE1_ATG.Fw + SRE1-stop/term.Rv); v) 338 bp of the SRE1 gene terminator region (amplified from CBS 6938 genomic DNA with primers E-2all.Fw + E.Rv); vi) a zeocin or hygromycin B resistance cassette for X. dendrorhous transformant selection (amplified from pIR-zeo or pMN-hph with primers F.Fw + F.Rv); vii) 500 bp of the SRE1 gene downstream region (amplified from CBS 6938 genomic DNA with primers G.Fw + G.Rv); viii) a geneticin resistance cassette (amplified from pFA6 with primers H.Fw + H.Rv); and ix) the ColE1 origin and ampicillin resistance for E. coli replication and selection (amplified from pBluescript SK- with primers A.Fw + A.Rv). S. cerevisiae was transformed with approximately 100 ng of each PCR product, and the assembled plasmids were recovered from yeast transformants by extracting total DNA and subsequent E. coli transformation. Plasmids were recovered from E. coli and evaluated by PCR, restriction analyses, and DNA sequencing. Through HpaI digestion of the resulting plasmid, the X. dendrorhous transformant DNA was released (approximately 6 kb), which was used to replace the native SRE1 gene with a version of it fused to the coding sequence of the 3xFLAG epitope through homologous recombination in X. dendrorhous. Thus, the wild-type strain CBS 6938 and strain CBS.cyp61−, were transformed with pXd.gSRE1-zeo and pXd.gSRE1-hph to obtain mutants CBS.FLAG.SRE1 and CBS.cyp61−.FLAG.SRE1, respectively (Table 1).
To reintegrate the STP1 gene in strain CBS.Δstp1, plasmid pBS-gSTP1up-down was constructed. This plasmid was obtained by inserting a 3,918 bp DNA fragment containing the STP1 gene, which was PCR-amplified from genomic DNA of X. dendrorhous strain CBS 6938, at the SmaI site of pBluescript SK-. Then, a zeocin resistance cassette (PCR-amplified from pIR-zeo) was introduced at the AgeI site of the downstream region of the STP1 gene, resulting in plasmid pBS-gSTP1up-down, which, once digested with BamHI, released a DNA fragment that allowed reintegration of the STP1 gene at its native locus by a double homologous recombination event. In this way, strain CBS.Δstp1/STP1 was obtained.
X. dendrorhous transformation was performed by electroporation (32, 33). Electrocompetent cells were prepared from cultures at the exponential phase of growth, and cells were electroporated using a Gene Pulser Xcell™ (Bio-Rad Laboratories Inc., Hercules, CA) under the following conditions: 125 mF, 600 Ω, and 0.45 kV. Transformations were performed using 10–15 μg of linear donor DNA prepared by digestion of the constructed plasmids. Transformant selection was performed on YM plates supplemented with hygromycin B, zeocin and/or nourseothricin. To confirm the designed gene replacements through homologous recombination and the presence of the corresponding resistance cassette used for transformant selection, all the X. dendrorhous strains obtained in this work were verified by PCR analysis using comprehensive sets of primers (supplemental Fig. S1).
S. cerevisiae was transformed by electroporation. Cells from exponential yeast cultures were harvested by centrifugation, washed twice with sterile cold water and one time with cold sterile 1 M sorbitol, and finally suspended in 1 M sorbitol. Cells were electroporated under the conditions 25 μF, 200 Ω, and 1.5 kV, and then, liquid YPD medium was added. The cells were incubated at 30°C for 2 h, and transformant selection was performed on YPD medium supplemented with geneticin at 200 μg/ml.
Carotenoid and sterol extraction and RP-HPLC analysis
Metabolites were extracted from cell pellets of 120-h-old yeast cultures. Analyses were performed in triplicate, and metabolites were normalized relative to the dry weight of the yeast. Carotenoids were extracted using the acetone extraction method (34) and were spectrophotometrically quantified at 474 nm using an absorption coefficient of A1% 1cm = 2,100. The method of sterol extraction was adapted from those of previous works (35, 36). Briefly, a cell pellet from a 5 ml sample of a 120 h culture (stationary phase of growth) was mixed with 4 g of KOH and 16 ml 60% ethanol solution (v/v). For saponification, the mixture was incubated at 80°C in a water bath for 2 h. Then, sterols were extracted with 10 ml of petroleum ether. The concentration of sterols was spectrophotometrically determined at 282 nm using the molar extinction coefficient Em = 11,900 M−1 cm−1 according to (37). Carotenoids and sterols were separated by RP-HPLC using a RP-18 LiChroCART® 125-4 (Merck KGaA, Darmstadt, Germany) column with mobile phases of acetonitrile:methanol:isopropyl (75:20:5, v/v) for carotenoids and methanol:water (97:3, v/v) for sterols with a 1 ml/min flux under isocratic conditions. Elution spectra were recorded using a diode array detector. Carotenoids were identified by their spectra and retention time according to standards (38), and ergosterol was identified by its spectra and retention time according to standard ergosterol (Sigma-Aldrich, Saint Louis, MO).
Protein extraction and Western blot assays
Protein extraction from X. dendrorhous strains was carried out from cellular pellets from 120-h-old yeast cultures using an adapted protocol. In brief, the cell pellet from 3,000 μl of culture was suspended in 250 μl of lysis buffer [NaHCO3 100 mM, Triton X-100 0.5%, PMSF 1 mM, protease inhibitor cocktail (Promega, Madison, WI) 1×, TCEP 2 mM] with 100 μl of 0.5 mm glass beads. Cells were broken by mechanical rupture using a Mini-beadbeater-16 for 30 s followed by 2 min on ice (seven cycles in total). Total protein extract was subjected to Western blot analysis using the monoclonal antibody ANTI-FLAG® M2 (catalog number F1804; Sigma-Aldrich) at a dilution of 1:1,000 and the Anti-Mouse IgG (whole molecule)-Peroxidase antibody (catalog number A9044; Sigma-Aldrich) at a dilution of 1:5,000. As a loading control, the monoclonal anti-ubiquitin antibody was used at a dilution of 1:1,000 (catalog number SAB2702288; Sigma-Aldrich). Protein transfer was performed under semidry conditions at 15 V for 30 min using a Trans-Blot® SD transfer cell (Bio-Rad Laboratories Inc.).
Bioinformatic characterization of the X. dendrorhous STP1 gene
The X. dendrorhous STP1 genomic and cDNA variants were identified using the genomic and transcriptomic database of strain UCD 67-385 (39) with STP1 homolog genes from other organisms, such as C. neoformans (19, 20) and Homo sapiens, (11) as queries. For domain and TM segment prediction, tools available online and indicated in the legend of Fig. 3 were used. General sequence analysis was performed using Geneious software (https://www.geneious.com).
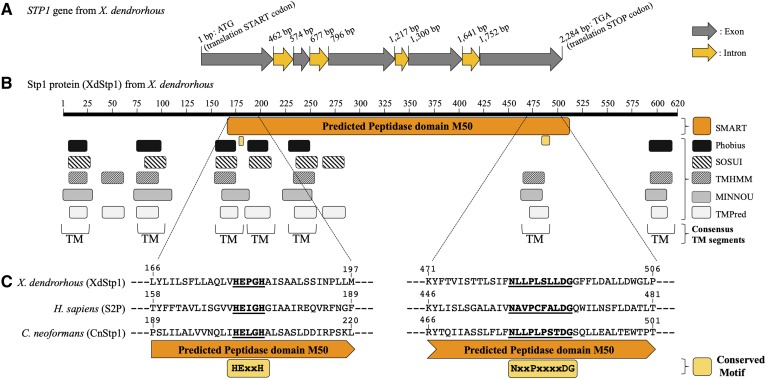
Fig. 3.
Bioinformatic analysis of the X. dendrorhous STP1 gene. A: Sequence analysis of the X. dendrorhous STP1 gene. The gene consists of five exons and encodes a predicted protein of 621 amino acids. B: Scheme of the predicted X. dendrorhous Stp1 protein in which the M50 peptidase domain (in orange) and the H-E-(X)2-H and N-(X)2-P-(X)4-D-G motifs (in yellow) are shown. The Stp1 protein TM segments were predicted with bioinformatic tools available online, and the consensus TM segments predicted by at least three tools are indicated. Protein domains were predicted with SMART (68) and TM segments with Phobius (69), SOSUI (70), TMHMM v. 2.0 (71), MINNOU (72), and TMpred (73). C: A sequence alignment of the M50 peptidase domain containing the conserved motifs H-E-(X)2-H and N-(X)2-P-(X)4-D-G from H. sapiens S2P protease [GenBank: AAC51937.1] and Stp1 from C. neoformans (CnStp1) [GenBank: XP_571333.1] and X. dendrorhous (XdStp1) [GenBank: MN380032] is shown.
RESULTS
Identification and bioinformatic characterization of the X. dendrorhous STP1 gene
To identify the STP1 gene from X. dendrorhous, BLAST searches using homologous STP1 nucleotide and amino acid sequences were performed on X. dendrorhous genomic and transcriptomic databases. The potential X. dendrorhous STP1 gene [GenBank: MN380032] consists of 2,284 bp (from translation start to stop codon) with five exons, giving an ORF of 1,863 bp in length (Fig. 3). STP1 encodes a 620 amino acid Stp1 protein with a predicted size of 66.7 kDa. The deduced protein sequence from X. dendrorhous was compared with sequences of characterized proteases of the SREBP pathway, such as the Stp1 protease [GenBank: XP_571333.1] from C. neoformans (19, 20), which shares 25.5% identity with 95% sequence coverage (E value 5e-30), and S2P protease [GenBank: AAC51937.1] from H. sapiens (11), which shares 21.7% identity with 78% sequence coverage (E value 3e-20). The potential X. dendrorhous Stp1 protease topology and features were studied using tools available online.
The H. sapiens S2P protease is an unusually hydrophobic integral membrane protease and has the peptidase domain M50 [InterPro identifier: IPR008915]. Proteins with the peptidase domain M50 [MEROPS identifier: M50] are metallopeptidases characterized by the presence of the hallmark H-E-(X)2-H motif (40), where the histidine residues (H171 and H175 in S2P from H. sapiens) coordinate the active-site zinc atom and the glutamic acid residue (E172 in S2P from H. sapiens) has a catalytic function (41). Additionally, metallopeptidases with the peptidase domain M50 contain an additional coordinating residue in the N-(X)2-P-(X)4-D-G motif (40), where the aspartic acid residue (D467 in S2P from H. sapiens) appears to be a third zinc-coordinating residue (42). In most S2P metallopeptidase orthologs, the H-E-(X)2-H motif lies inside a TM segment, while the N-(X)2-P-(X)4-D-G motif is located at the end or even outside of another TM segment; between the two TM segments containing these motifs, there is usually another TM segment forming a compact three TM segment structural core (43). Seven possible TM segments were predicted by at least three TM prediction tools in the potential Stp1 protease from X. dendrorhous (Fig. 3). Both conserved motifs in the M50 peptidase domain were also identified in Stp1, corresponding to residues H179-E180-(X)2-H183 and N484-(X)2-P487-(X)4-D492-G493, and these motifs were located at the third and sixth predicted TM segments, respectively (Fig. 3). These results support the hypothesis that the identified X. dendrorhous STP1 gene encodes a functional Stp1 metallopeptidase.
Functional studies of the X. dendrorhous STP1 gene
The SRE1 gene that encodes the transcription factor Sre1 of the X. dendrorhous SREBP pathway was previously identified and described (23). It was demonstrated that sre1− mutants were unable to grow in the presence of azole drugs [compounds that inhibit ergosterol biosynthesis, specifically by inhibiting cytochrome P450 monooxygenases (44)] or chloride cobalt [CoCl2, a hypoxia-mimicking agent (45)]. In addition, the sre1− mutation reduced sterol and carotenoid production in the X. dendrorhous ergosterol biosynthesis mutant strain CBS.cyp61− (derived from wild-type strain CBS 6938), which overproduces sterols and carotenoids (26). Then, if the identified STP1 gene encodes a protease involved in Sre1 activation in X. dendrorhous, its mutation should affect the yeast phenotype in a similar way as the sre1− mutation does. To evaluate this possibility, Δstp1 mutants CBS.Δstp1 and CBS.cyp61−.Δstp1 were obtained (Fig. 4), which were derived from strains CBS 6938 and CBS.cyp61−, respectively. In addition, the STP1 gene was reintegrated in strain CBS.Δstp1, resulting in strain CBS.Δstp1/STP1.
Fig. 4.
Growth phenotype of X. dendrorhous strains. Serial dilutions of X. dendrorhous strain cultures were seeded on YM-agar plates (1.5%) with different supplements and incubated for 5 days at 22°C. A: Control (YM medium) and YM medium supplemented with itraconazole (5.0 nM) (B), ketoconazole (0.1 μg/ml) (C), clotrimazole (0.1 μg/ml) (D), or CoCl2 (400 μM) (E).
The biosynthesis of ergosterol in fungi is a highly oxygen-demanding process in which cytochrome P450 monooxygenases are involved (46). Accordingly, in S. pombe (13) and C. neoformans (19, 45), it was observed that Sre1 proteolytic activation was induced by azole drugs or by CoCl2 supplementation, and sre1− mutants were unable to grow in the presence of these compounds. Therefore, to evaluate the potential role of STP1 in the X. dendrorhous SREBP pathway, the growth of strains constructed in this work in the presence of azole drugs or CoCl2 was evaluated. Unlike both parental strains, the two X. dendrorhous Δstp1 mutants (CBS.Δstp1 and CBS.cyp61−.Δstp1) were unable to grow in YM medium supplemented with either type of compound (Fig. 4). However, reintegration of the STP1 gene in strain CBS.Δstp1 (strain CBS.Δstp1/STP1) restored the wild-type growth phenotype under these conditions. Together, these results support that X. dendrorhous has a functional SREBP pathway that is activated in the presence of azole drugs and CoCl2 that involves the STP1 gene product.
To the naked eye, no color difference was found between CBS.Δstp1 and the wild-type when cultured in YM plates. However, strain CBS.cyp61−.Δstp1 was paler than its parental strain CBS.cyp61−, which has similar pigmentation as the wild-type strain. These results suggest that under this condition (growth in YM medium), Sre1 is only activated in strain CBS.cyp61, favoring carotenoid overproduction, and the Δstp1 mutation in this strain probably prevents Sre1 proteolytic activation; therefore, carotenoid production returns to wild-type levels. Thus, to evaluate whether carotenoid production is indeed affected in Δstp1 mutants, X. dendrorhous wild-type, CBS.Δstp1, CBS.Δstp1/STP1, CBS.cyp61−, and CBS.cyp61−.Δstp1 strains were cultured in YM medium until the stationary phase, and carotenoids were extracted after 120 h of culture. The five strains showed similar growth curves (supplemental Fig. S3), indicating that the introduced mutations did not greatly affect yeast growth under the studied conditions. No significant differences were observed in total carotenoid content between the wild-type and strains CBS.Δstp1 and CBS.Δstp1/STP1, and carotenoid overproduction was confirmed in strain CBS.cyp61−. In effect, introducing the Δstp1 mutation in strain CBS.cyp61− (strain CBS.cyp61−.Δstp1) reduced carotenoid production to similar levels as in the wild-type (Table 2). Carotenoid composition was also affected in strain CBS.cyp61−.Δstp1 compared with CBS.cyp61−: the astaxanthin fraction decreased from 74.5% to 66.3%, while the fraction of intermediary carotenoids between β-carotene and astaxanthin increased from 8.9% to 24.4% in strain CBS.cyp61−.Δstp1. Similar results were observed when sterol production was analyzed: The Δstp1 mutation decreased total sterol production in strain CBS.cyp61−, as the double mutant CBS.cyp61−.Δstp1 reached similar sterol levels as the wild-type (Table 2). As strain CBS.cyp61−, strain CBS.cyp61−.Δstp1 does not produce ergosterol due to the cyp61− mutation, as the CYP61 gene encodes a cytochrome P450 involved in the second to last step of ergosterol biosynthesis. The sterol profile in strain CBS.cyp61−.Δstp1 was analyzed by RP-HPLC, which was evaluated by co-injecting a sterol sample of this strain with standard ergosterol or sterols extracted from strain CBS.cyp61−. These phenotypic analysis assays confirmed the importance of the identified STP1 gene in isoprenoid biosynthesis in X. dendrorhous.
TABLE 2.
Production of carotenoids and sterols in X. dendrorhous strains
| Metabolite | Strain | ||||
| CBS 6938 (wild-type) | CBS.Δstp1 | CBS.cyp61− | CBS.cyp61−.Δstp1 | CBS.Δstp1/STP1 | |
| Total carotenoids (μg/g; 100%) | 287 ± 15a | 260 ± 9a | 442 ± 34b | 264 ± 16a | 292 ± 10a |
| Astaxanthin (%) | 70.4 | 70.4 | 74.5 | 66.3 | 70.1 |
| β-carotene (%) | 1.4 | 1.2 | 1.0 | 1.9 | 0.5 |
| Intermediary carotenoids from β-carotene to astaxanthin (%) | 15.9 | 18.1 | 8.9 | 24.4 | 14.7 |
| Other carotenoids (%) | 12.2 | 10.3 | 15.5 | 7.4 | 14.7 |
| Total sterols (mg/g; 100%) | 3.7 ± 0.1a | 3.5 ± 0.1a | 5.0 ± 0.2b | 3.6 ± 0.1a | 3.8 ± 0.4a |
| Ergosterol, peak at 16.5 min (%) | 96.1 | 97.4 | ND | ND | 97.5 |
| Peak 1 at 7.2 min (%) | 3.9 | 2.6 | 3.6 | 4.4 | 2.5 |
| Peak 2 at 13.5 min (%) | ND | ND | 27.3 | 14.8 | ND |
| Peak 3 at 19.5 min (%) | ND | ND | 69.1 | 80.8 | ND |
Total carotenoids and sterols were extracted after 120 h of culture and normalized to the yeast dry weight in grams. The table shows the mean ± standard deviation of three independent cultures of each strain. Intermediate carotenoids from β-carotene to astaxanthin include phoenicoxanthin, OH-echinenone, and echinenone; other carotenoids include torulene, OH-k-torulene, and other unidentified carotenoids. Peaks 1, 2, and 3 correspond to metabolites having a sterol spectrum, which were observed in chromatograms after approximately 7.2, 13.5, and 19.5 min of retention time, respectively. Data were evaluated with one-way ANOVA and the Tukey post hoc test to compare metabolite production between strains. Superscript letters indicate statistical comparisons: having the same letter denotes no statistically significant differences, and having different letters indicates significant differences between strains with P < 0.05. ND, not detected.
Sre1 gene targets are downregulated in Δstp1 mutants
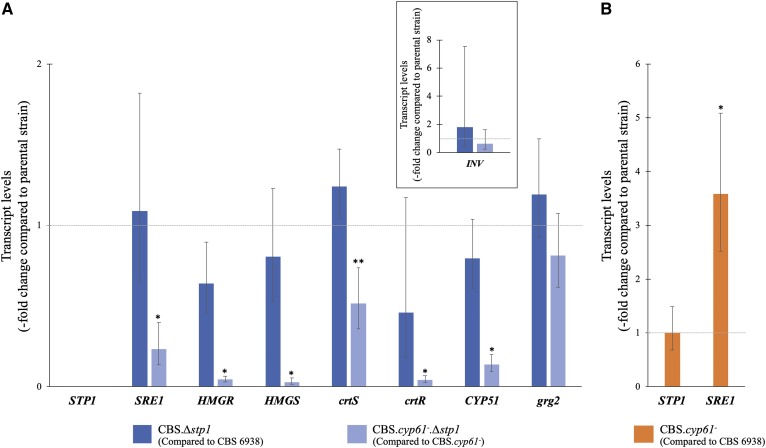
Carotenogenesis and the synthesis of sterols derive from the isoprenoid precursor isopentenyl pyrophosphate, which in nonphotosynthetic eukaryotes such as yeasts is synthesized through the mevalonate (MVA) pathway (Fig. 2). Many aspects of isoprenoid biosynthesis are well conserved (47), and genes of the MVA pathway are known targets of the SREBP pathway in mammals (48, 49). In vitro and in vivo evidence indicates that the HMG-Coa reductase (HMGR) and HMG-CoA synthase (HMGS) genes of the MVA pathway are Sre1 targets in X. dendrorhous (23); therefore, the relative transcript levels of these genes were evaluated in X. dendrorhous strains studied in this work by qPCR (Fig. 5). Genes INV (invertase encoding gene [GenBank: FJ539193.2]) and grg2 (glucose repressible gene 2 [GenBank: JN043364.1]) were selected as negative controls, as potential Sre1-binding sequences were not identified at their promoter regions, and there is no evidence supporting that these genes could be SREBP/Sre1 targets. Indeed, no significant differences in the transcript levels of these genes were observed in the comparisons made in this work. Additionally, the STP1 gene was included as a control, and as expected, no STP1 transcripts were detected in the Δstp1 mutant strains, confirming the correct construction of these strains. Regarding the genes HMGR and HMGS, a slight decrease in their relative transcript levels was observed in strain CBS.Δstp1 compared with the wild-type; however, the difference was not statistically significant. On the other hand, a significant relative transcript level difference was observed in strain CBS.cyp61−.Δstp1 compared with its parental strain, CBS.cyp61−, as HMGR and HMGS transcript levels decreased approximately 23- and 35-fold (Fig. 5A), respectively.
Fig. 5.
Relative expression of selected genes at the transcript level. Relative transcript levels in strain CBS.Δstp1 in relation to those in CBS 6938 (blue) and in strain CBS.cyp61−.Δstp1 in relation to those in CBS.cyp61− (light blue) (A) and in strain CBS.cyp61− compared with those in CBS 6938 (orange) (B). The transcript levels were evaluated by RT-qPCR after 120 h of culture in YM medium with constant agitation. The analyzed genes and GenBank numbers accession were HMGS [MK368600] and HMGR [MK368599] (MVA pathway), CYP51 [KP317478] (ergosterol biosynthesis), crtS [EU713462] (astaxanthin biosynthesis), crtR [EU884133] (ergosterol and astaxanthin biosynthesis), and SRE1 [MK368598] and STP1 [MN380032] (SREBP pathway). The genes grg2 [JN043364.1] and INV [FJ539193.2] (in an enclosed square), which are not likely regulated by Sre1N, were included as controls. Transcript levels were normalized to the housekeeping gene encoding β-actin [X89898.1] and expressed as fold-change compared with the levels in the corresponding parental strain using the 2−ΔΔCt method. Values are the average ± standard deviation of three independent cultures (*P < 0.01, **P < 0.05; Student’s t-test).
Considering that the astaxanthin fraction was reduced and that the fraction of intermediary carotenoids between β-carotene and astaxanthin was increased in strain CBS.cyp61−.Δstp1 compared with strain CBS.cyp61−, the relative transcript level of genes crtS and crtR, encoding the cytochrome P450 system (astaxanthin synthase and cytochrome P450 reductase, respectively) involved in these steps, was evaluated. The transcript levels of both crtS and crtR decreased in strain CBS.cyp61−.Δstp1 compared with CBS.cyp61−, showing an approximately 2- and 23-fold reduction, respectively (Fig. 5A), suggesting that the transcript level reduction of these two genes could be responsible for the carotenoid composition difference observed between these two strains. The reduced crtR transcript levels could also be responsible, at least in part, for the sterol composition changes observed in strain CBS.cyp61−.Δstp1 compared with CBS.cyp61− (Table 2), as crtR is also involved in ergosterol synthesis (50). Two cytochrome P450-encoding genes involved in ergosterol biosynthesis, CYP61 (26) and CYP51 (51), were identified and functionally described in X. dendrorhous; therefore, the CYP51 transcript level was also evaluated in the Δstp1 mutants. The transcript levels of this gene also decreased (approximately 7-fold) in strain CBS.cyp61−.Δstp1 compared with its parental strain. These results are in accordance with previous RNA-seq data analysis, where most genes of the MVA pathway (including HMGR and HMGS) and of sterol biosynthesis (including CYP51 and crtR) decreased in the sre1− mutant compared with the wild-type when cultured under standard conditions (23).
In S. pombe, Sre1 stimulates its own gene transcription (13). SRE1 relative transcript levels were thus evaluated and had been reduced approximately 4-fold in strain CBS.cyp61−.Δstp1 compared with CBS.cyp61−. Finally, SRE1 and STP1 relative transcript levels were studied in strain CBS.cyp61− in relation to the wild-type (Fig. 5B). Indeed, higher SRE1 transcript levels were observed in strain CBS.cyp61− (approximately 4-fold higher than the wild-type), but no differences were observed for the STP1 transcript, suggesting that STP1 expression is not directly regulated by Sre1 in X. dendrorhous, similar to the behavior reported for S2P in mammalian cells (10).
Together, these results support that the STP1 gene studied in this work is involved in Sre1 activation in X. dendrorhous. Sre1 is probably activated by Stp1 in strain CBS.cyp61−, as introducing the Δstp1 mutation in this strain reduces transcript levels of Sre1 target genes.
Gene STP1 is required for Sre1 cleavage
In C. neoformans, the Stp1 protease, a homolog of mammalian S2P, is involved in Sre1 activation (21). In this work, we also studied the role of the identified STP1 gene in Sre1 proteolytic activation by Western blot. For this, the SRE1 native gene was replaced by two different gene versions expressing the Sre1 protein fused to the 3xFLAG epitope at its N-terminal end (full-length Sre1 protein: FLAG.SRE1 or N-terminal domain of Sre1: FLAG.SRE1N) in different genetic backgrounds. In this way, strains CBS.FLAG.SRE1, CBS.FLAG.SRE1.Δstp1, CBS.Δstp1.FLAG.SRE1N, CBS.FLAG.SRE1N, CBS.cyp61−.FLAG.SRE1, and CBS.cyp61−.FLAG.SRE1.Δstp1 were obtained (Table 1). First, it was confirmed that the introduced SRE1 gene version did not majorly affect yeast growth (supplemental Fig. S3) under the studied conditions or carotenoid and sterol production compared with those of the equivalent untagged strains (supplemental Table S2). All strains containing either one of the two 3xFLAG-tagged Sre1 protein versions and lacking the Δstp1 mutation grew normally in the presence of azole drugs or CoCl2 (supplemental Fig. S4), indicating that the 3xFLAG-tagged Sre1 proteins functioned properly.
In Western blot analysis, protein bands were only observed in lanes loaded with protein samples obtained from strains bearing either version of the 3xFLAG-tagged Sre1 protein, and no proteins were detected in protein samples from untagged control strains, indicating that there is no nonspecific antibody reaction with X. dendrorhous proteins. In general, two protein bands were observed: a larger band of approximately 130 kDa and a smaller band of approximately 95 kDa, which correspond to the full-length Sre1 protein (Sre1) and to the N-terminal domain of Sre1 (Sre1N, the active protein), both fused to the 3xFLAG epitope. In the wild-type genetic context, the ∼130 kDa protein band is mainly observed, but a slight ∼95 kDa band can also be observed (Fig. 6, lane 4). This indicates that under standard laboratory conditions, Sre1 is activated at some level in X. dendrorhous. In strain CBS.FLAG.SRE1.Δstp1, the smaller band is no longer visible, supporting that the STP1 gene product is involved in Sre1 cleavage. In control strains, CBS.Δstp1.FLAG.SRE1N and CBS.FLAG.SRE1N, only the ∼95 kDa protein band was observed, confirming the expected size of Sre1N fused to the 3xFLAG epitope. Although the same amount of total protein was loaded in each gel well, protein samples from strains expressing Sre1N showed a more intense band than the full-length Sre1 band observed in the wild-type genetic context or in strain CBS.FLAG.SRE1.Δstp1 (Fig. 6, compare lanes 6 and 7 versus lanes 4 and 5). Strain CBS.cyp61−.FLAG.SRE1 carried the full-length SRE1 gene, and both protein bands were observed (Fig. 6, lane 8); however, the Sre1N protein band was much more intense than the full-length Sre1 band and more intense than the full-length Sre1 band observed in the wild-type genetic background. Finally, the Sre1N protein band was no longer detectable by Western blot analysis in strain CBS.cyp61−.FLAG.SRE1.Δstp1 (Fig. 6, lane 9), confirming that the Δstp1 mutation avoids Sre1 activation. These data demonstrate that Sre1 is activated in strain CBS.cyp61− when cultured under standard conditions and that the STP1 gene is essential for Sre1 proteolytic activation in X. dendrorhous.
Fig. 6.
Sre1 cleavage dependency on STP1. A: Western blot assay of protein extracts obtained from X. dendrorhous strains carrying Sre1 or Sre1N labeled with 3xFLAG at its N-terminus after 48 h of culture in YM medium with constant agitation. Strains CBS 6938, CBS.sre1−, and CBS.Δstp1, without FLAG-tagged Sre1/Sre1N proteins, were included as controls (lanes 1, 2, and 3, respectively). Lanes 4–9 correspond to protein extracts from FLAG-Sre1- or FLAG-Sre1N-labeled strains: CBS.FLAG.SRE1 (lane 4), CBS.FLAG.SRE1.Δstp1 (lane 5), CBS.Δstp1.FLAG.SRE1N (lane 6), CBS.FLAG.SRE1N (lane 7), CBS.cyp61−.FLAG.SRE1 (lane 8), and CBS.cyp61−.FLAG.SRE1.Δstp1 (lane 9). Anti-ubiquitin was included as a control. FLAG-Sre1 (∼130 kDa) and FLAG-Sre1N (∼95 kDa) bands are indicated. M, PageRuler Plus prestained protein ladder (Thermo Fisher Scientific Inc.). B: Amino acid sequence alignment of the first TM segment of H. sapiens SREBP-2 [GenBank: Q12772.2] and the predicted first TM segment of Sre1 from C. neoformans [GenBank: XP_012052219.1] (CnSre1) and X. dendrorhous [GenBank: MK368598] (XdSre1). The first TM segment predicted in each protease is shown in blue. The arrow in SREBP-2 indicates the S2P cleavage site between a leucine and a cysteine residue (53), and asterisks in CnSre1 indicate the end of two truncated constructions of CnSre1 (residues 501 and 535), which suggests that Sre1 is cleaved by CnStp1 at the first TM segment (21). Potential helix-destabilizing residues are highlighted in yellow.
DISCUSSION
In this study, we identified and described a new gene involved in the SREBP pathway of the carotenogenic yeast X. dendrorhous. The identified STP1 gene is homologous to genes encoding the S2P protease in H. sapiens and Stp1 in C. neoformans, which are required for proteolytic activation of the transcription factor SREBP/Sre1. In the deduced protein sequence encoded by the X. dendrorhous STP1 gene, an M50 peptidase domain with the characteristic H-E-(X)2-H and N-(X)2-P-(X)4-D-G motifs was predicted, similar to that in the S2P protease from H. sapiens (40–42). The hydrophobic nature of Stp1 and the fact that the two motifs are characteristically embedded in highly hydrophobic segments of the protein as in the mammalian S2P (40) is consistent with the idea that the predicted cleavage site of Stp1 in Sre1 lies within a TM segment because S2P and its characterized orthologs utilize a unique proteolytic mechanism in which substrate cleavage occurs within the lipid bilayer (52). In the case of mammalian S2P, SREBP cleavage occurs between a leucine and cysteine residue in the first TM segment (53); however, it was demonstrated that these residues can be mutated without affecting SREBP cleavage by S2P (54). These residues are not present at the first predicted TM segment of the X. dendrorhous Sre1 protein (Fig. 6), similar to the case of the first predicted TM segment of Sre1 in C. neoformans; nevertheless, experimental data strongly support that Stp1 cuts Sre1 within its first TM segment in C. neoformans (21). Interestingly, asparagine and proline residues that are critical for Sre1 cleavage by Stp1 were found at the end of the first predicted TM segment of the X. dendrorhous Sre1 protein, as described for the mammalian S2P protease (54). Therefore, it is likely that Sre1 cleavage by Stp1 in X. dendrorhous occurs within the first TM segment of Sre1, but further studies are required to confirm this possibility. The subcellular localization prediction of X. dendrorhous Stp1 did not give clear results. The mammalian S2P protein resides in the Golgi apparatus (55), and SREBP cleavage depends on a first cut made at the luminal loop of SREBP by S1P, which is also in the Golgi apparatus (56). In A. fumigatus, it is not clear where exactly SrbA is processed, as ER and Golgi apparatus proteins are involved in the proteolytic activation of this transcription factor (18). Considering these results, Stp1 in X. dendrorhous could be located in any of these organelles.
A functional SREBP pathway has been described in several fungal model organisms, such as S. pombe (13), C. neoformans (19, 45), and A. fumigatus (17), and is essential for growth in the presence of azole drugs or CoCl2. Similar to X. dendrorhous sre1− mutants (23), Δstp1 mutants failed to grow in the presence of these compounds. However, reintegration of the gene STP1 in strain CBS.Δstp1 (Fig. 4) or the replacement of gene SRE1 by the version that only expresses Sre1N (23) rescued the phenotype resistant to azole drugs or CoCl2. In addition, the Δstp1 mutation in a wild-type genetic context did not majorly affect sterol or carotenoid production under the studied conditions. However, this mutation reduced the production of both to wild-type levels in strain CBS.cyp61−, which is an ergosterol biosynthesis mutant that overproduces sterols and carotenoids (26). The same results were obtained when the SRE1 gene was mutated (23). Together, these data indicate that the STP1 gene is also responsible for the carotenoid-overproducing phenotype observed in cyp61− X. dendrorhous mutants (26), probably because STP1 encodes a protease involved in the proteolytic activation of the Sre1 transcription factor. In support of this last statement, a sterol- and carotenoid-overproducing phenotype similar to that observed for cyp61− mutants was obtained by replacing the native SRE1 gene by a version that only encodes Sre1N (23), which corresponds to the active form of the transcription factor, independent of proteolytic cleavage. Finally, relative transcript levels of genes HMGR and HMGS (of the MVA pathway) and CYP51 and crtR (of ergosterol biosynthesis) significantly decreased in the Δstp1 mutant constructed in the cyp61− genetic context, which could contribute to the reduced carotenoid and sterol production and metabolite composition observed in strain CBS.cyp61−.Δstp1 in relation to strain CBS.cyp61−. These genes, among others, were reported as Sre1-dependent genes after RNA-seq analysis, and by ChIP-PCR assays, it was found that genes HMGR and HMGS were direct Sre1 targets (23). Microarray assays carried out in C. neoformans that compared the wild-type strain versus the Δstp1 mutant and the strain Δsre1+SRE1 versus the Δsre1 mutant showed that the expression of the genes HMGS, HMGR, and CYP51 was similarly affected between these strains (21). Therefore, our results support the assumption that Stp1 indirectly affects the expression of these genes as Stp1 activates Sre1 in X. dendrorhous. As gene crtR is also involved in carotenogenesis, particularly in the synthesis of astaxanthin from β-carotene (57), the observed changes in its transcript level could also contribute to the observed changes in carotenoid composition in strain CBS.cyp61−.Δstp1 with respect to that in CBS.cyp61−. In summary, the Δstp1 mutation studied in this work mimics the X. dendrorhous sre1− mutation (23) in several aspects. First, Δstp1 mutants are unable to grow in the presence of azole drugs or CoCl2. Second, the Δstp1 mutation reduced sterol and carotenoid production in the X. dendrorhous ergosterol biosynthesis mutant CBS.cyp61−, which overproduces sterols and carotenoids. Third, relative transcript levels of SREBP pathway gene targets were reduced in Δstp1 mutants. Together, these results support that the identified STP1 gene is involved in the X. dendrorhous SREBP pathway, which is required for Sre1 proteolytic activation.
Western blot analysis confirmed that Sre1 was activated in strain CBS.cyp61− when cultured under standard conditions. This result indicates that strain CBS.cyp61− provides the conditions that activate the SREBP pathway in X. dendrorhous, which are probably related to the altered sterol composition in relation to that of the wild-type. It was also demonstrated that Sre1 proteolytic activation depends on the STP1 gene because the Δstp1 mutant derived from CBS.cyp61− failed to accumulate Sre1N. In the wild-type strain, a weak band coinciding with the size of the Sre1N band was observed, suggesting that Sre1 is activated at some level in X. dendrorhous under the same conditions. Supporting this statement, RNA-seq data comparisons between X. dendrorhous wild-type and sre1− strains showed a low transcriptional effect in response to the absence of Sre1, including changes in the transcript levels of genes of the MVA and sterol biosynthesis pathways (23). In addition, similar results were reported in S. pombe (13) and C. neoformans (19), where low levels of the nuclear form of Sre1 were observed by Western blot assays in wild-type strains when cultured under aerobic conditions. Strains expressing only Sre1N showed a more intense band than the full-length Sre1 band observed in the other strains. This could result from the Sre1N regulation of its own gene expression increasing Sre1N protein levels, which is supported by the increased SRE1 transcript levels observed in strain CBS.cyp61−, which has an active SREBP pathway.
In mammalian cells, SREBP cleavage by S2P depends on a first cut made at the luminal loop of SREBP by S1P (56). However, our results indicate that Sre1 cleavage performed by Stp1 in X. dendrorhous does not depend on a previous cleavage on Sre1, because in the absence of the STP1 gene, only the full-length Sre1 protein band was observed. Similar results were observed in C. neoformans (21), suggesting that basidiomycetes have a different mechanism than mammalian cells to release the N-terminal domain of Sre1. Our results do not exclude the possibility of a second cut in the X. dendrorhous Sre1 protein after a first cut performed by Stp1. Considering this, we identified two potential S1P gene homologs in the X. dendrorhous yeast genome [GenBank: CED82778 and CDZ97435]. The deduced protein from CED82778 shared 30.16% identity with 44% coverage with S1P from H. sapiens [GenBank: AAI14962.1], and the protein deduced from CDZ97435 shared 53.29% identity with 89% coverage with the putative S1P homolog identified in C. neoformans (UniProtKB: CNH01120). However, the deletion of both genes did not affect growth in the presence of azole drugs or CoCl2 or the color phenotype in strain CBS.cyp61− (supplemental Fig. S5), indicating that these genes are not involved in the SREBP pathway in X. dendrorhous. Similar to our results, mutants of the potential S1P homolog identified in C. neoformans were resistant to CoCl2 (58). Further studies are required to confirm whether Stp1 is the only protease involved in Sre1 proteolytic activation in X. dendrorhous.
Metallopeptidases with the peptidase domain M50 are not only involved in the regulation of sterol levels in the cell. For example, in C. neoformans, it was indicated that Stp1 should have other substrates besides Sre1, as Stp1 is required for both Sre1-dependent and Sre1-independent gene transcription (21). The S2P from H. sapiens has at least two other substrates besides SREBP: the transcription factors, activating transcription factor 6 (ATF6) [GenBank: AAB64434.1] and cAMP-responsive element binding protein H (CREBH) [GenBank: AAI43610.1]. The first activates the unfolded protein response (59, 60), and the second regulates the systemic glucose and the lipid metabolism antagonistic to SREBP (61, 62). Then, to identify other potential Stp1 substrates in X. dendrorhous, a BLASTp search for ATF6 and CREBH homologs in the yeast genome was performed using the named GenBank identifiers as query. Although with low sequence coverage, potential Stp1 substrates besides Sre1 were identified in the X. dendrorhous genome. Hypothetical proteins having a basic leucine zipper domain were among them: CDZ97125.1, which is a potential ATF6 homolog (sharing 46% identity with 4% coverage, E value 0.026), and CEB83119.1, which is a potential CREBH homolog (sharing 28.3% identity with 11% coverage, E value 0.001). The range of action of Stp1 in X. dendrorhous remains to be elucidated.
Finally, this work provides new information regarding the SREBP pathway and contributes knowledge about the regulation of isoprenoid synthesis in X. dendrorhous. The results described in this work demonstrate that Stp1 is involved in the activation of Sre1 but do not rule out other proteases whose action could depend on the first Sre1 cleavage made by Stp1 being involved in this process. To date, two genes of the SREBP pathway in X. dendrorhous have been described: SRE1 (23), which encodes the transcription factor Sre1, and STP1 (this work), which encodes the protease Stp1 involved in Sre1 proteolytic activation. However, X. dendrorhous lacks identifiable homologs of other characteristic components of the SREBP pathway, such as SCAP. SCAP plays an important role in the SREBP pathway because it senses sterol levels through its sterol-sensing domain, which is an important step in regulating the activation of SREBP. SCAP homologs have been studied in S. pombe (13) and in C. neoformans (58), but SCAP has been lost in several fungal species (63) having a functional SREBP pathway, such as A. fumigatus (64). The ER-resident enzyme HMGR of the MVA pathway also has an sterol-sensing domain and fulfills an important role as a protein sensor for membrane sterols (3). However, in X. dendrorhous, it is unknown how HMGR is regulated in the ER because no Insig homologs have been identified. In this way, a future challenge is to identify a potential sterol-sensing protein that could fulfill a similar role as SCAP in the SREBP pathway in X. dendrorhous and to determine whether other proteins are involved in Sre1 processing in this yeast.
Supplementary Material
Footnotes
Abbreviations:
- ATF6
- activating transcription factor 6
- bHLH
- basic-helix-loop-helix
- CREBH
- cAMP-responsive element binding protein H
- Ct
- threshold cycle
- HMGR
- HMG-CoA reductase
- HMGS
- HMG-CoA synthase
- Insig
- insulin-induced gene
- MVA
- mevalonate
- SCAP
- SREBP cleavage activating protein
- S1P
- site-1 protease
- S2P
- site-2 protease
- TM
- transmembrane
This work was supported by FONDECYT Grant 1160202 and by the graduate scholarships CONICYT 21170613 to M.G. and CONICYT 21130708 to M.S.G. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Brown M. S., and Goldstein J. L.. 1997. The SREBP pathway: regulation of cholesterol metabolism by proteolysis of a membrane-bound transcription factor. Cell. 89: 331–340. [DOI] [PubMed] [Google Scholar]
- 2.Shimomura I., Shimano H., Horton J. D., Goldstein J. L., and Brown M. S.. 1997. Differential expression of exons 1a and 1c in mRNAs for sterol regulatory element binding protein-1 in human and mouse organs and cultured cells. J. Clin. Invest. 99: 838–845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goldstein J. L., DeBose-Boyd R. A., and Brown M. S.. 2006. Protein sensors for membrane sterols. Cell. 124: 35–46. [DOI] [PubMed] [Google Scholar]
- 4.Horton J. D., Goldstein J. L., and Brown M. S.. 2002. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 109: 1125–1131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Párraga A., Bellsolell L., Ferré-D’Amaré A. R., and Burley S. K.. 1998. Co-crystal structure of sterol regulatory element binding protein 1a at 2.3 A resolution. Structure. 6: 661–672. [DOI] [PubMed] [Google Scholar]
- 6.Yang T., Espenshade P. J., Wright M. E., Yabe D., Gong Y., Aebersold R., Goldstein J. L., and Brown M. S.. 2002. Crucial step in cholesterol homeostasis: sterols promote binding of SCAP to INSIG-1, a membrane protein that facilitates retention of SREBPs in ER. Cell. 110: 489–500. [DOI] [PubMed] [Google Scholar]
- 7.Yabe D., Brown M. S., and Goldstein J. L.. 2002. Insig-2, a second endoplasmic reticulum protein that binds SCAP and blocks export of sterol regulatory element-binding proteins. Proc. Natl. Acad. Sci. USA. 99: 12753–12758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown M. S., and Goldstein J. L.. 1999. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA. 96: 11041–11048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Duncan E. A., Brown M. S., Goldstein J. L., and Sakai J.. 1997. Cleavage site for sterol-regulated protease localized to a leu-ser bond in the lumenal loop of sterol regulatory element-binding protein-2. J. Biol. Chem. 272: 12778–12785. [DOI] [PubMed] [Google Scholar]
- 10.Sakai J., Duncan E. A., Rawson R. B., Hua X., Brown M. S., and Goldstein J. L.. 1996. Sterol-regulated release of SREBP-2 from cell membranes requires two sequential cleavages, one within a transmembrane segment. Cell. 85: 1037–1046. [DOI] [PubMed] [Google Scholar]
- 11.Rawson R. B., Zelenski N. G., Nijhawan D., Ye J., Sakai J., Hasan M. T., Chang T. Y., Brown M. S., and Goldstein J. L.. 1997. Complementation cloning of S2P, a gene encoding a putative metalloprotease required for intramembrane cleavage of SREBPs. Mol. Cell. 1: 47–57. [DOI] [PubMed] [Google Scholar]
- 12.Parks L. W., and Casey W. M.. 1995. Physiological implications of sterol biosynthesis in yeast. Annu. Rev. Microbiol. 49: 95–116. [DOI] [PubMed] [Google Scholar]
- 13.Hughes A. L., Todd B. L., and Espenshade P. J.. 2005. SREBP pathway responds to sterols and functions as an oxygen sensor in fission yeast. Cell. 120: 831–842. [DOI] [PubMed] [Google Scholar]
- 14.Stewart E. V., Nwosu C. C., Tong Z., Roguev A., Cummins T. D., Kim D. U., Hayles J., Park H. O., Hoe K. L., Powell D. W., et al. 2011. Yeast SREBP cleavage activation requires the Golgi Dsc E3 ligase complex. Mol. Cell. 42: 160–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stewart E. V., Lloyd S. J., Burg J. S., Nwosu C. C., Lintner R. E., Daza R., Russ C., Ponchner K., Nusbaum C., and Espenshade P. J.. 2012. Yeast sterol regulatory element-binding protein (SREBP) cleavage requires Cdc48 and Dsc5, a ubiquitin regulatory X domain-containing subunit of the Golgi Dsc E3 ligase. J. Biol. Chem. 287: 672–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hwang J., Ribbens D., Raychaudhuri S., Cairns L., Gu H., Frost A., Urban S., and Espenshade P. J.. 2016. A Golgi rhomboid protease Rbd2 recruits Cdc48 to cleave yeast SREBP. EMBO J. 35: 2332–2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Willger S. D., Puttikamonkul S., Kim K. H., Burritt J. B., Grahl N., Metzler L. J., Barbuch R., Bard M., Lawrence C. B., and Cramer R. A. J.. 2008. A sterol-regulatory element binding protein is required for cell polarity, hypoxia adaptation, azole drug resistance, and virulence in Aspergillus fumigatus. PLoS Pathog. 4: e1000200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dhingra S., and Cramer R. A.. 2017. Regulation of sterol biosynthesis in the human fungal pathogen Aspergillus fumigatus: opportunities for therapeutic development. Front. Microbiol. 8: 92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang Y. C., Bien C. M., Lee H., Espenshade P. J., and Kwon-Chung K. J.. 2007. Sre1p, a regulator of oxygen sensing and sterol homeostasis, is required for virulence in Cryptococcus neoformans. Mol. Microbiol. 64: 614–629. [DOI] [PubMed] [Google Scholar]
- 20.Chun C. D., Liu O. W., and Madhani H. D.. 2007. A link between virulence and homeostatic responses to hypoxia during infection by the human fungal pathogen Cryptococcus neoformans. PLoS Pathog. 3: e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bien C. M., Chang Y. C., Nes W. D., Kwon-Chung K. J., and Espenshade P. J.. 2009. Cryptococcus neoformans Site-2 protease is required for virulence and survival in the presence of azole drugs. Mol. Microbiol. 74: 672–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Golubev W. I. 1995. Perfect state of Rhodomyces dendrorhous (Phaffia rhodozyma). Yeast. 11: 101–110. [DOI] [PubMed] [Google Scholar]
- 23.Gutiérrez M. S., Campusano S., González A. M., Gómez M., Barahona S., Sepúlveda D., Espenshade P. J., Fernández-Lobato M., Baeza M., Cifuentes V., et al. 2019. Sterol regulatory element-binding protein (Sre1) promotes the synthesis of carotenoids and sterols in Xanthophyllomyces dendrorhous. Front. Microbiol. 10: 586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mortensen A., Skibsted L. H., and Truscott T. G.. 2001. The interaction of dietary carotenoids with radical species. Arch. Biochem. Biophys. 385: 13–19. [DOI] [PubMed] [Google Scholar]
- 25.Higuera-Ciapara I., Felix-Valenzuela L., and Goycoolea F. M.. 2006. Astaxanthin: a review of its chemistry and applications. Crit. Rev. Food Sci. Nutr. 46: 185–196. [DOI] [PubMed] [Google Scholar]
- 26.Loto I., Gutierrez M. S., Barahona S., Sepulveda D., Martinez-Moya P., Baeza M., Cifuentes V., and Alcaíno J.. 2012. Enhancement of carotenoid production by disrupting the C22-sterol desaturase gene (CYP61) in Xanthophyllomyces dendrorhous. BMC Microbiol. 12: 235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sambrook J., and Russell D. W.. 2001. A Laboratory Manual. 3rd edition. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 28.Livak K. J., and Schmittgen T. D.. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 29.Schmittgen T. D., and Livak K. J.. 2008. Analyzing real-time PCR data by the comparative CT method. Nat. Protoc. 3: 1101–1108. [DOI] [PubMed] [Google Scholar]
- 30.Niklitschek M., Alcaíno J., Barahona S., Sepulveda D., Lozano C., Carmona M., Marcoleta A., Martinez C., Lodato P., Baeza M., et al. 2008. Genomic organization of the structural genes controlling the astaxanthin biosynthesis pathway of Xanthophyllomyces dendrorhous. Biol. Res. 41: 93–108. [DOI] [PubMed] [Google Scholar]
- 31.Kuijpers N. G., Solis-Escalante D., Bosman L., van den Broek M., Pronk J. T., Daran J. M., and Daran-Lapujade P.. 2013. A versatile, efficient strategy for assembly of multi-fragment expression vectors in Saccharomyces cerevisiae using 60 bp synthetic recombination sequences. Microb. Cell Fact. 12: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adrio J. L., and Veiga M.. 1995. Transformation of the astaxanthin-producing yeast Phaffia rhodozyma. Biotechnol. Tech. 9: 509–512. [DOI] [PubMed] [Google Scholar]
- 33.Kim I. G., Nam S. K., Sohn J. H., Rhee S. K., An G. H., Lee S. H., and Choi E. S.. 1998. Cloning of the ribosomal protein L41 gene of Phaffia rhodozyma and its use a drug resistance marker for transformation. Appl. Environ. Microbiol. 64: 1947–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.An G-H., Schuman D. B., and Johnson E. A.. 1989. Isolation of Phaffia rhodozyma mutants with increased astaxanthin content. Appl. Environ. Microbiol. 55: 116–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shang F., Wen S., Wang X., and Tan T.. 2006. Effect of nitrogen limitation on the ergosterol production by fed-batch culture of Saccharomyces cerevisiae. J. Biotechnol. 122: 285–292. [DOI] [PubMed] [Google Scholar]
- 36.Cheng B., Yuan Q. P., Sun X. X., and Li W. J.. 2010. Enhanced production of coenzyme Q10 by overexpressing HMG-CoA reductase and induction with arachidonic acid in Schizosaccharomyces pombe. Appl. Biochem. Biotechnol. 160: 523–531. [DOI] [PubMed] [Google Scholar]
- 37.Venkateswarlu K., Kelly D. E., Manning N. J., and Kelly S. L.. 1998. NADPH cytochrome P-450 oxidoreductase and susceptibility to ketoconazole. Antimicrob Agents Chemother. 42: 1756–1761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercadante A. Z., and Egeland E. S.. 2004. Carotenoids. Handbook. Birkhäuser Verlag, Basel - Boston - Berlin. [Google Scholar]
- 39.Baeza M., Alcaíno J., Barahona S., Sepúlveda D., and Cifuentes V.. 2015. Codon usage and codon context bias in Xanthophyllomyces dendrorhous. BMC Genomics. 16: 293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rudner D. Z., Fawcett P., and Losick R.. 1999. A family of membrane-embedded metalloproteases involved in regulated proteolysis of membrane-associated transcription factors. Proc. Natl. Acad. Sci. USA. 96: 14765–14770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rawlings N. D., and Barrett A. J.. 1995. Evolutionary families of metallopeptidases. Methods Enzymol. 248: 183–228. [DOI] [PubMed] [Google Scholar]
- 42.Zelenski N. G., Rawson R. B., Brown M. S., and Goldstein J. L.. 1999. Membrane topology of S2P, a protein required for intramembranous cleavage of sterol regulatory element-binding proteins. J. Biol. Chem. 274: 21973–21980. [DOI] [PubMed] [Google Scholar]
- 43.Chen G., and Zhang X.. 2010. New insights into S2P signaling cascades: regulation, variation, and conservation. Protein Sci. 19: 2015–2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lamb D. C., Baldwin B. C., Kwon-Chung K. J., and Kelly S. L.. 1997. Stereoselective interaction of the azole antifungal agent SCH39304 with the cytochrome P-450 monooxygenase system isolated from Cryptococcus neoformans. Antimicrob. Agents Chemother. 41: 1465–1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee H., Bien C. M., Hughes A. L., Espenshade P. J., Kwon-Chung K. J., and Chang Y. C.. 2007. Cobalt chloride, a hypoxia-mimicking agent, targets sterol synthesis in the pathogenic fungus Cryptococcus neoformans. Mol. Microbiol. 65: 1018–1033. [DOI] [PubMed] [Google Scholar]
- 46.Rosenfeld E., and Beauvoit B.. 2003. Role of the non-respiratory pathways in the utilization of molecular oxygen by Saccharomyces cerevisiae. Yeast. 20: 1115–1144. [DOI] [PubMed] [Google Scholar]
- 47.Miziorko H. M. 2011. Enzymes of the mevalonate pathway of isoprenoid biosynthesis. Arch. Biochem. Biophys. 505: 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Osborne T. F., Gil G., Goldstein J. L., and Brown M. S.. 1988. Operator constitutive mutation of 3-hydroxy-3-methylglutaryl coenzyme A reductase promoter abolishes protein binding to sterol regulatory element. J. Biol. Chem. 263: 3380–3387. [PubMed] [Google Scholar]
- 49.Smith J. R., Osborne T. F., Brown M. S., Goldstein J. L., and Gil G.. 1988. Multiple sterol regulatory elements in promoter for hamster 3-hydroxy-3-methylglutaryl-coenzyme A synthase. J. Biol. Chem. 263: 18480–18487. [PubMed] [Google Scholar]
- 50.Gutiérrez M. S., Rojas M. C., Sepúlveda D., Baeza M., Cifuentes V., and Alcaíno J.. 2015. Molecular characterization and functional analysis of cytochrome b5 reductase (CBR) encoding genes from the carotenogenic yeast Xanthophyllomyces dendrorhous. PLoS One. 10: e0140424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Leiva K., Werner N., Sepulveda D., Barahona S., Baeza M., Cifuentes V., and Alcaíno J.. 2015. Identification and functional characterization of the CYP51 gene from the yeast Xanthophyllomyces dendrorhous that is involved in ergosterol biosynthesis. BMC Microbiol. 15: 89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Urban S., and Shi Y.. 2008. Core principles of intramembrane proteolysis: comparison of rhomboid and site-2 family proteases. Curr. Opin. Struct. Biol. 18: 432–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Duncan E. A., Davé U. P., Sakai J., Goldstein J. L., and Brown M. S.. 1998. Second-site cleavage in sterol regulatory element-binding protein occurs at transmembrane junction as determined by cysteine panning. J. Biol. Chem. 273: 17801–17809. [DOI] [PubMed] [Google Scholar]
- 54.Ye J., Davé U. P., Grishin N. V., Goldstein J. L., and Brown M. S.. 2000. Asparagine-proline sequence within membrane-spanning segment of SREBP triggers intramembrane cleavage by site-2 protease. Proc. Natl. Acad. Sci. USA. 97: 5123–5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rawson R. B., and Li W. P.. 2007. The site-2 protease at ten. In Intramembrane-Cleaving Proteases (I-CLiPs). N. M. Hooper and U. Lendeckel, editors. Springer Science & Business Media, Dordrecht. 1–15. [Google Scholar]
- 56.DeBose-Boyd R. A., Brown M. S., Li W. P., Nohturfft A., Goldstein J. L., and Espenshade P. J.. 1999. Transport-dependent proteolysis of SREBP: relocation of site-1 protease from Golgi to ER obviates the need for SREBP transport to Golgi. Cell. 99: 703–712. [DOI] [PubMed] [Google Scholar]
- 57.Alcaíno J., Barahona S., Carmona M., Lozano C., Marcoleta A., Niklitschek M., Sepulveda D., Baeza M., and Cifuentes V.. 2008. Cloning of the cytochrome P450 reductase (crtR) gene and its involvement in the astaxanthin biosynthesis of Xanthophyllomyces dendrorhous. BMC Microbiol. 8: 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chang Y. C., Ingavale S. S., Bien C., Espenshade P., and Kwon-Chung K. J.. 2009. Conservation of the sterol regulatory element-binding protein pathway and its pathobiological importance in Cryptococcus neoformans. Eukaryot. Cell. 8: 1770–1779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haze K., Yoshida H., Yanagi H., Yura T., and Mori K.. 1999. Mammalian transcription factor ATF6 is synthesized as a transmembrane protein and activated by proteolysis in response to endoplasmic reticulum stress. Mol. Biol. Cell. 10: 3787–3799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ye J., Rawson R. B., Komuro R., Chen X., Davé U. P., Prywes R., Brown M. S., and Goldstein J. L.. 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell. 6: 1355–1364. [DOI] [PubMed] [Google Scholar]
- 61.Zhang K., Shen X., Wu J., Sakaki K., Saunders T., Rutkowski D. T., Back S. H., and Kaufman R. J.. 2006. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 124: 587–599. [DOI] [PubMed] [Google Scholar]
- 62.Nakagawa Y., and Shimano H.. 2018. CREBH regulates systemic glucose and lipid metabolism. Int. J. Mol. Sci. 19: E1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Butler G. 2013. Hypoxia and gene expression in eukaryotic microbes. Annu. Rev. Microbiol. 67: 291–312. [DOI] [PubMed] [Google Scholar]
- 64.Chung D., Barker B. M., Carey C. C., Merriman B., Werner E. R., Lechner B. E., Dhingra S., Cheng C., Xu W., Blosser S. J., et al. 2014. ChIP-seq and in vivo transcriptome analyses of the Aspergillus fumigatus SREBP SrbA reveals a new regulator of the fungal hypoxia response and virulence. PLoS Pathog. 10: e1004487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Wach A., Brachat A., Pöhlmann R., and Philippsen P.. 1994. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 10: 1793–1808. [DOI] [PubMed] [Google Scholar]
- 66.Raffatellu M., Sun Y. H., Wilson R. P., Tran Q. T., Chessa D., Andrews-Polymenis H. L., Lawhon S. D., Figueiredo J. F., Tsolis R. M., Adams L. G., et al. 2005. Host restriction of Salmonella enterica serotype Typhi is not caused by functional alteration of SipA, SopB, or SopD. Infect. Immun. 73: 7817–7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bien C. M., and Espenshade P. J.. 2010. Sterol regulatory element binding proteins in fungi: hypoxic transcription factors linked to pathogenesis. Eukaryot. Cell. 9: 352–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Letunic I., and Bork P.. 2018. 20 years of the SMART protein domain annotation resource. Nucleic Acids Res. 46: D493–D496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Käll L., Krogh A., and Sonnhammer E. L. L.. 2007. Advantages of combined transmembrane topology and signal peptide prediction–the Phobius web server. Nucleic Acids Res. 35: W429–W432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hirokawa T., Boon-Chieng S., and Mitaku S.. 1998. SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics. 14: 378–379. [DOI] [PubMed] [Google Scholar]
- 71.Krogh A., Larsson B., von Heijne G., and Sonnhammer E. L. L.. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305: 567–580. [DOI] [PubMed] [Google Scholar]
- 72.Cao B., Porollo A., Adamczak R., Jarrell M., and Meller J.. 2006. Enhanced recognition of protein transmembrane domains with prediction-based structural profiles. Bioinformatics. 22: 303–309. [DOI] [PubMed] [Google Scholar]
- 73.Hofmann K., and Stoffel W.. 1993. TMbase - a database of membrane spanning protein segments. Biol. Chem. Hoppe Seyler. 374: 166 Accessed December 16, 2019, at: https://embnet.vital-it.ch/software/TMPRED_form.html [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.