Abstract
We previously described the expression of CD36 and LPL by breast cancer (BC) cells and tissues and the growth-promoting effect of VLDL observed only in the presence of LPL. We now report a model in which LPL is bound to a heparan sulfate proteoglycan motif on the BC cell surface and acts in concert with the VLDL receptor to internalize VLDLs via receptor-mediated endocytosis. We also demonstrate that gene-expression programs for lipid synthesis versus uptake respond robustly to triglyceride-rich lipoprotein availability. The literature emphasizes de novo FA synthesis and exogenous free FA uptake using CD36 as paramount mechanisms for lipid acquisition by cancer cells. We find that the uptake of intact lipoproteins is also an important mechanism for lipid acquisition and that the relative reliance on lipid synthesis versus uptake varies among BC cell lines and in response to VLDL availability. This metabolic plasticity has important implications for the development of therapies aimed at the lipid dependence of many types of cancer, in that the inhibition of FA synthesis may elicit compensatory upregulation of lipid uptake. Moreover, the mechanism that we have elucidated provides a direct connection between dietary fat and tumor biology..
Keywords: lipase, fatty acids, synthesis, metabolism
The dependence of several tumor cell types, including breast cancer (BC), on a supply of FAs to maintain proliferation is well established (1). The literature emphasizes de novo lipid synthesis, and more recently, FFA uptake via CD36, as the mechanisms used to satisfy this dependency. De novo lipogenesis produces the saturated long-chain FA palmitate (C16:0), most of which is used for plasma membrane phospholipid synthesis. The low rates of FA synthesis observed in most nonmalignant tissues has positioned fatty acid synthase (FASN) as a therapeutic target, prompting efforts to develop FASN inhibitors (2).
In this context, we and others have observed that the provision of exogenous FFAs to cancer cells permits evasion of the cytotoxic effects of FASN inhibition, revealing the ability of cancer cells to take up, as well as synthesize, lipids (3, 4). We further demonstrated that BC cells and clinical BC tissues robustly express LPL, the principal enzyme for the hydrolysis of triglyceride (TG) carried in lipoproteins, as well as the cell-surface channel for FFA uptake, CD36. Others have begun to highlight the presence of LPL in and on the surface of cancer cells, the most prominent example being in chronic lymphocytic leukemia, where evidence supports LPL as a strong prognostic biomarker (5, 6). The precise roles of LPL in cancer cells, however, are unresolved.
LPL is best known as the enzyme responsible for the extracellular hydrolysis of TG carried in lipoproteins. LPL is produced by myocytes and adipocytes, secreted into the interstitial space, and transported to the capillary lumen (7). For years, dogma held that secreted LPL was tethered to capillary endothelial cells by its heparin-binding domains and heparan sulfate proteoglycans (HSPGs) on the capillary surface (8). This belief was supported by the fact that LPL can be demarginated into plasma by heparin (9) and by in vitro studies showing that LPL binds to HSPGs and that this interaction can be disrupted by the desulfation of HSPGs or digestion with heparinase or heparitinase (10, 11).
An alternate model has come to light in recent years in which LPL is secreted into interstitial spaces, captured by glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 (GPIHBP1) on the antiluminal surface of capillary endothelial cells, and shuttled to the luminal surface (12). Here, GPIHBP1 facilitates LPL binding to the luminal surface of the capillary wall, creating a “platform for lipolysis.” On this platform LPL mediates TG hydrolysis, releasing glycerol and FFAs that can be taken up through the cell-surface channel CD36 on adipocytes and myocytes.
Apart from this lipolytic function, LPL may act as a noncatalytic bridge, promoting the uptake of lipoproteins via receptor-mediated endocytosis (13). In this role, LPL interacts with lipoproteins and a variety of different cell-surface proteins, including HSPGs and members of the LDL receptor family, including the VLDL receptor (VLDLR) (14). The ability of LPL to serve as a bridge has been supported by both in vitro and in vivo experiments, including the work of Merkel et al. (15), who showed that catalytically inactive LPL expressed in muscle could still bind to HSPGs and induce VLDL uptake. This function of LPL has not been previously reported in cancer cells.
We previously described the expression of CD36 and LPL by BC cells and tissues and the growth-promoting effect of VLDL supplementation observed in BC cell lines only in the presence of LPL. We now describe the deployment of LPL in BC cells. Our data support a model in which LPL is bound to a heparin-like HSPG motif on the cell surface and acts in concert with the VLDLR to rapidly internalize intact lipoproteins via receptor-mediated endocytosis. We further observe substantial alterations in patterns of gene expression related to pathways for lipid acquisition (synthesis vs. uptake) in response to the availability of lipoproteins in tissue culture (TC) media and cellular LPL expression status. These findings highlight the importance of lipoprotein uptake as a method of lipid acquisition for cancer cells and demonstrate BC cell metabolic plasticity in response to nutrient availability.
EXPERIMENTAL PROCEDURES
Cell lines and tissue culture
MCF-7, MDA-MB-231, BT-474, DU4475, SKBR3, and T47-D BC cells and HeLa cervical cancer cells were from the American Type Culture Collection and cultured in phenol red-containing HyClone RPMI-1640 media with 10% (v/v) heat-inactivated FBS (GE Healthcare Life Sciences) and 1% penicillin-streptomycin. MCF10A mammary epithelial cells were cultured in DMEM/F12 growth media (Invitrogen) supplemented with 5% horse serum (Invitrogen), 20 ng/ml epidermal growth factor (Peprotech), 0.5 mg/ml hydrocortisone, 100 ng/ml cholera toxin, 10 µg/ml insulin (Sigma-Aldrich), and 1% penicillin-streptomycin. Cells were maintained at 37°C in a humidified atmosphere containing 5% CO2. Characteristics of the cell lines are in supplemental Table S1. Lipoprotein-depleted serum (LPDS) and matched-control FBS were from Kalen Biomedical.
Manipulated cell lines
MDA-MB-231 LPL shRNA cells.
MDA-MB-231 shRNA cells were generated by lentiviral transduction using HuSH shRNA plasmid panels with pGFP-C-Lenti vectors and the Lenti-vpack packaging kit (TR30037) according to the manufacturer’s guidelines (OriGene Technologies). Plasmids included four LPL shRNAs and a negative control (TL311692). Cells transduced with shLPL or scrambled shRNA were selected in puromycin.
MDA-MB-231 siRNA cells.
siRNAs targeting LPL or VLDLR (Sigma-Aldrich) were transfected into MDA-MB-231 BC cells using Lipofectamine RNAiMAX (Thermo Fisher Scientific) according to the manufacturer’s guidelines. siRNAs used were LPL siRNA 1 (SASI_Hs01_00208454), LPL siRNA 2 (SASI_Hs01_00208455); VLDLR siRNA 1 (SASI_Hs02_00335553), VLDLR siRNA 2 (SASI_Hs01_00219062); and MISSION siRNA Universal Negative Control #1, SIC001 (Sigma-Aldrich). DiI-VLDL uptake, RNA expression by RT-PCR, and VLDLR protein expression by Western blot were assessed 96 h after transfection.
Immunoblotting.
Cells were lysed in RIPA buffer [20 mM Tris, 150 mM NaCl, 1% NP-40, 10% glycerol, 1 mM EDTA, 1 mM EGTA, 5 mM NaPPi, 50 mM NaF, 10 mM Na-β-glycerophosphate (pH 7.4), Halt protease inhibitor cocktail (Pierce), and 1 mM Na3VO4 (New England Biolabs)]. Lysates were sonicated for 15 s and centrifuged at 17,000 g for 10 min at 4°C, and protein was quantified by BCA assay (Pierce). Protein was denatured in LDS sample buffer (GenScript) with 1.25% β-mercaptoethanol and heated to 95°C for 1 min. Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane. Blots were probed with antibodies against VLDLR (6A6; Santa Cruz Biotechnology) and vinculin (Cell Signaling Technology).
MCF-7 pCMV Neo and LPL cells.
Human LPL was expressed from a pCMV-6AC vector from OriGene (Cat. No. SC322258). An empty vector (pCMV Neo) served as the control. Plasmids were transfected using Lipofectamine 2000 DNA transfection reagent (Thermo Fisher Scientific). Pooled and clonal cell lines were selected by limiting dilution in neomycin (G418) and propagated in G418-containing media. Total RNA was subjected to on-column DNase digestion with the RNase-free DNase set (Qiagen, Cat. No. 79254), cDNA synthesis, and RT-PCR, as previously described.
Materials
The following reagents were prepared as stock solutions in DMSO: GSK264220A, lipase inhibitor (50 mM; Cayman Chemical); Dynasore, inhibitor of dynamin 1 and 2 (80 mM; Sigma-Aldrich); and Hoechst 33342 nuclear stain (20 mM; Thermo Fisher Scientific). Other reagents were prepared and stored at 4°C until use: heparin sodium salt from porcine intestinal mucosa (50 mM stock solution in H2O; Sigma-Aldrich, Cat. No. H3149), human DiI-VLDLs (1 mg protein/ml; Alfa Aesar Chemicals), and LPL from bovine milk (Sigma-Aldrich, Cat. No. L2254). The protocol for preparing LPL from suspension (3.8 M ammonium sulfate, 0.02 M Tris HCl, pH 8.0) was adapted from Nishitsuji et al. (16). Heparinase I from Flavobacterium (Sigma-Aldrich) was dissolved at 1 mg/ml in 20 mM Tris-HCl (pH 7.5), 50 mM NaCl, 4 mM CaCl2, and 0.01% BSA and reconstituted in 20 mM Tris-HCl (pH 7.5), 4 mM CaCl2, and 0.01% BSA.
Antibodies
Our mouse monoclonal anti-human LPL antibody (3) was conjugated to Alexa-Fluor 647 (AF-647) using the AF-647 antibody labeling kit (Thermo Fisher Scientific, Cat No. A20186). A monoclonal antibody directed against the HSNS4F5 HSPG motif, (GlcNS6S-IdoA2S)3, was supplied by Nicole Smits. The binding site from the single-chain NS4F5 antibody (NS4F5scFv) (17) was converted to a functional full-length human IgG2 antibody (NS4F5IgG). Briefly, the VH region of NS4F5scFv was linked to the human γ-2 constant region under the control of the human T lymphotropic virus elongation factor-1 promoter, whereas the VL region of NS4F5scFv was linked to the C-gene of the human lambda constant region under the control of a cytomegalovirus promoter. A nonfunctional isotype control (NS4F5null) was generated by modifying the VH sequence of NS4F5IgG (CARSGRKGRMR to CARSGSGGSGS). Heavy- and light-chain constructs of NS4F5 (NS4F5IgG, NS4F5null) were cotransfected into HEK-293 cells. Antibodies were purified by protein A-sepharose affinity chromatography and analyzed using SDS-PAGE and size-exclusion HPLC analysis using a TOSOH TSK gel SuperSW3000 column.
Quantitative RT-PCR
Total RNA was isolated using RNeasy minicolumns from extracts prepared with QiaShredder (Qiagen RNeasy Kit). Concentration and purity of RNA were assessed by a NanoDrop DM-1000 spectrophotometer (NanoDrop Technologies). cDNA was produced using iScript Reverse Transcription Supermix for RT-qPCR (Bio-Rad) with 1 μg RNA template. Quantitative RT-PCR was performed using TaqMan Gene Expression Master Mix and TaqMan Gene Expression Assays (Thermo Fisher Scientific). Primers are described in supplemental Table S2. qRT-PCR was performed on a Bio-Rad CFX 96 Real Time System C1000 Thermal Cycler with the following program: (1) 50°C × 2 min, (2) 90°C × 10 min, (3) 95°C × 15 s, (4) 60°C × 1 min + plate read, (5) Go to (3) 39 more cycles, (6) Melt curve 65°C to 95°C increment 0.5°C for 5 s + plate read. Relative gene expression is displayed in accordance with the 2−ΔΔCT method, normalized to cyclophilin. Where applicable, RT-PCR products were separated on 2% agarose gels prepared using 0.5× TBE with SYBR Safe DNA gel stain (Invitrogen) and visualized using a Bio-Rad Molecular Imager ChemiDoc XRS+ with Image Lab Software.
Flow cytometry
Cells were detached with CellStripper reagent (Thermo Fisher Scientific). Viability was assessed using the Molecular Probes LIVE/DEAD stain kit with excitation at 405 nm (20 min, 1:3000 dilution). Cells were blocked with human IgG (1.25 mg/ml, 15 min; Sigma Aldrich). Surface staining of unfixed/unpermeabilized cells used the following antibodies: mouse monoclonal LPL-AF-647 (1 µg/ml, 30 min) and Dylight 650-NS4F5IgG or Dylight 650-NS4F5null (3.2 µg/ml, 30 min). All incubations were carried out at 4°C with PBS washes between each step. Analysis of intracellular antigens was achieved using the Foxp3/Transcription Factor Staining Buffer Set (Thermo Fisher Scientific). Following staining, cells were fixed with 0.5% formalin (20 min). Fluorescence was measured using a FACScan system on a MacsQuant-10 Flow Cytometer (Miltenyi Biotec), and data were analyzed using FlowJo 10 software (FlowJo, LLC). For LPL staining, the unstained control condition was verified as having equivalent fluorescent reads as a mouse IgG2b K Isotype Control APC antibody (Thermo Fisher Scientific).
VLDL uptake experiments
DiI-VLDLs.
VLDLs labeled with DiI (1,1′-dioctadecyl-3,3,3′-tetramethyl-indocarbocyanine perchlorate) were from Alfa Aesar (Cat. No. J65568). VLDLs with protein- and lipid-specific labels were prepared using a technique adapted from Goulbourne et al. (18). Briefly, DiI-VLDLs were dialyzed into PBS containing 0.05 M sodium borate (pH 8.5) and incubated with DyLight 650 NHS-Ester (Thermo Fisher Scientific) at room temperature for 1.5 h. Nonreacted DyLight dye was removed by extensive dialysis against PBS.
DiI-VLDL plate-based uptake assay.
MDA-MB-231 cells were seeded subconfluently in 12-well plates. Growth media were replaced with RPMI-1640 containing 10% LPDS (Kalen) overnight, and cells were assayed for uptake of DiI-VLDL using methods adapted from Teupser (19). Briefly, cells were pretreated for 30 min in serum-free media prior to a 1 h incubation with 10 µg/ml DiI-VLDL. Surface-bound DiI-VLDL was removed with acid-wash buffer (0.5 M acetic acid with 150 mM NaCl, pH 2.5). Cells were washed with DPBS with calcium and magnesium, lysed in 1% SDS and 0.1 M NaOH, transferred to a black 96-well half-area plate (Greiner Bio-One), and assessed using a SpectraMax i3x microplate reader (Ex/Em: 520/580 nm; Molecular Devices). Fluorescence was corrected for protein.
DiI-VLDL flow cytometry-based uptake assay.
Cells were plated in complete media into 12-well plates, allowed to adhere overnight, and serum-starved for 1–8 h. Confluency was less than 70% at the time of treatment. All pretreatments (30 min) and VLDL uptake incubations (45 min) were carried out at 37°C in a humidified atmosphere containing 5% CO2, unless otherwise indicated. Cells were washed and then harvested by trypsinization. Samples were run on a ZE-5 flow cytometer (Bio-Rad). Data were analyzed using FlowJo 10 software (FlowJo, LLC).
Confocal microscopy
Lipoprotein uptake.
Cells were seeded subconfluently into an 8-chambered Lab-Tek coverglass (Nunc), allowed to adhere overnight, serum-starved for 1–8 h, and then assessed for DiI-VLDL uptake. Treatments with uptake-inhibiting reagents (30 min) were followed by a 45 min incubation with DiI-VLDL (5 µg/ml). Cells were rinsed with DPBS with calcium and magnesium and fixed with 3% paraformaldehyde in DPBS. Nuclei were stained with DAPI (Thermo Scientific) or Hoechst 33342 (Sigma-Aldrich). Images were acquired by confocal fluorescence microscopy with a Zeiss LSM 800 microscope equipped with a 63× oil-immersion objective with identical exposure settings for all experimental conditions. Images were processed uniformly across comparisons with Zen Lite 2.3 (Zeiss) and ImageJ 1.49 (Fiji).
Lipid droplet staining.
Cells were grown in chambered Lab-Tek coverglass wells (Nunc), washed with DPBS, stained with Hoechst 33342 (1 µg/ml), fixed in 3% paraformaldehyde in DPBS with calcium and magnesium, and incubated with LipidTOX Red Neutral Lipid Stain (1:800, 30 min at 37°C; Invitrogen).
Lipase assay
Lipase activity was measured using a fluorescent assay (20) using zwitterionic detergent [3-(N,N-dimethylmyristylammonio)propanesulfonate] and EnzChek lipase substrate, green fluorescent, 505/515 (250 µM stock; Sigma-Aldrich). Fluorescence was measured on a SpectraMax i3x plate reader and SoftMax Pro software from Molecular Devices (Ex/Em: 482/515 nm; 495 nm filter cutoff). Assays were carried out after a 10 min incubation at 37°C in black half-area 96-well plates (Greiner Bio-One) in 0.15 M NaCl, 20 mM Tris-HCl (pH 8.0), 0.0125% Zwittergent, and 1.5% FA-free BSA in a total volume of 100 µl. The average of no lipase controls was subtracted from raw values.
Cell viability assays
Cells were seeded into 96-well plates, allowed to adhere overnight, treated with drugs, and cultured for 72 h at 37°C before viability assay using the ATP-based CellTiter-Glo (CTG) 2.0 Assay (Promega). Luminescence was read with the LMAX II luminometer (Molecular Devices). Background luminescence was calculated from CTG alone wells. Luminescence values (RLUs) were normalized to vehicle and compiled as a percentage of DMSO control. IC50 determination was performed using a nonlinear regression curve-fitting algorithm: log(inhibitor) versus response-variable (four-parameter) slope with Graph Pad Prism 6.0.
Cell death and apoptosis assays
Apoptosis was assessed using the Dead Cell Apoptosis Kit with Annexin V Alexa Fluor 488 and propidium iodide (Thermo Fisher Scientific). Flow cytometry data obtained with the eight-color MACSQuant-10 (Miltenyi Biotec) were analyzed using FlowJo software. Percentage apoptosis was the percentage Annexin V+ cells in the population.
Statistics
Statistical significance was evaluated using an unpaired two-tailed Student’s t-test with Welch’s correction or one-way ANOVA with correction for multiple comparisons (Dunnett’s multiple-comparisons test) where applicable. P < 0.05 was deemed significant. Errors are presented as mean ± SD or SEM, as indicated.
RESULTS
LPL is in and on the surface of cancer cells and clinical breast tumor tissue
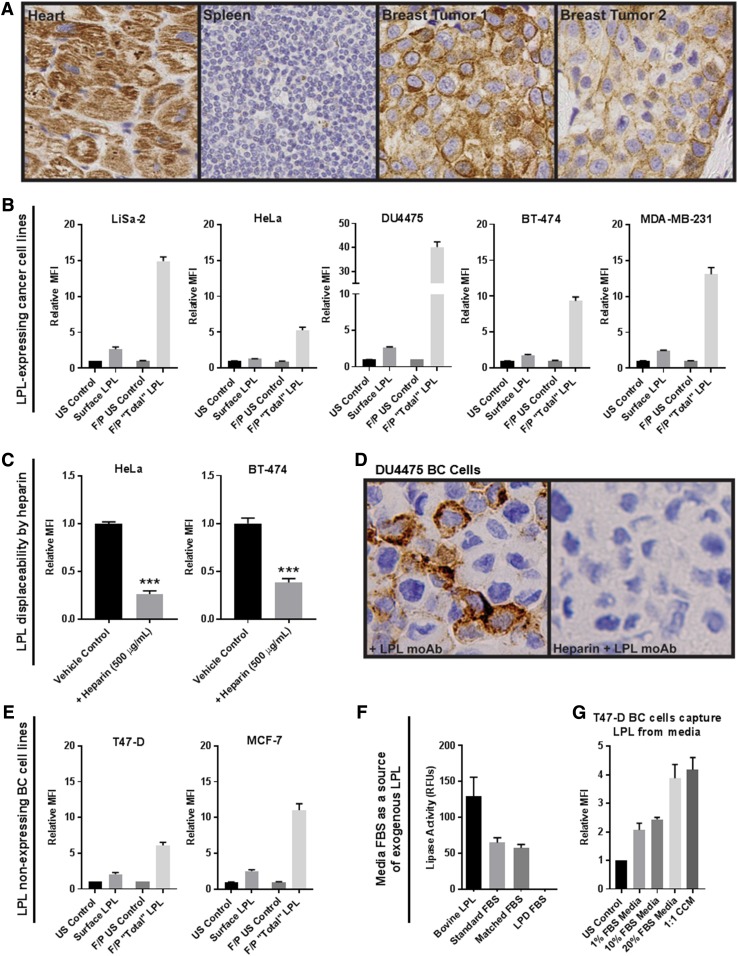
Breast tumor tissues stained with antibody directed against LPL displayed both cytoplasmic and cell-surface LPL staining (Fig. 1A). Using qRT-PCR, we established that LPL is variably expressed across a panel of human cancer cell lines (supplemental Fig. S1). We previously described LiSa-2 liposarcoma cells, HeLa cervical cancer cells, and DU4475 BC cells as high LPL-expressing and -secreting cell lines. Here, using flow cytometry with our AF-647-LPL antibody, we show that LPL is found to a variable degree on the surface of LPL-expressing cancer cells and at higher levels inside the cell, with the total LPL pool represented by the fixed/permeabilized condition (Fig. 1B). Cell-surface-bound LPL was displaceable by 500 µg/ml heparin, as quantified by flow cytometry (Fig. 1C) and immunocytochemistry using nonadherent DU4475 TNBC cells with a protocol designed to preserve cell-surface HSPGs (Fig. 1D).
Fig. 1.
LPL is in and on the surface of breast tumor cells and cancer cell lines and is displaceable by heparin. A: Control (heart and spleen) and breast tumor slides were stained for LPL. Breast tumor cells display cell surface and cytoplasmic LPL staining. B: The presence of LPL protein in and on the surface of cancer cells was assessed via flow cytometry. For all cell lines, the total LPL present in fixed/permeabilized (F/P) cells exceeded that on the cell surface. Note different scales; mean ± SEM of >3 experiments. C: Representative data from heparin displacement studies using LPL-expressing HeLa cervical cancer and BT-474 BC cells. Heparin displaces LPL from the cell surface, as detected by flow cytometry (***P < 0.001; two-tailed unpaired t-test with Welch’s correction). D: Immunocytochemical analysis of LPL on the surface of DU4475 nonadherent TNBC cells. Cells were incubated with LPL antibody ± heparin (40 µg/ml), embedded in agarose, formalin-fixed, embedded in paraffin, and sectioned. Cell-surface LPL antibody was detected with peroxidase-conjugated anti-IgG (brown pigment) with hematoxylin counterstain (blue). E: T47-D and MCF-7 BC cells had no detectable LPL expression by qRT-PCR. However, LPL was found on the cell surface and, to a greater extent, inside these cells by flow cytometry. F: Standard, LPD, and matched-control FBS were assessed for lipase activity. Activity was detected in bovine LPL and in standard and matched-control FBS but not LPD FBS. G: T47-D BC cells were incubated in different media overnight to determine whether BC cells can capture LPL from media. A FBS dose-dependent increase of cell-surface LPL was observed. LPL was also captured by cells incubated with culture media conditioned by LPL-secreting LiSa-2 liposarcoma cells.
We predicted that LPL would be present in cells that express the gene but were surprised to find it in and on the surface of T47-D and MCF-7 LPL low to nonexpressing BC cell lines (Fig. 1E). We hypothesized that without LPL mRNA expression, the protein must be acquired from an exogenous source, such as the FBS of the TC media. The lipase assay confirmed that lipase activity was present in standard and matched-control FBS but not in lipoprotein-depleted FBS (Fig. 1F). We assessed the ability of T47-D cells to capture LPL from TC media by incubation with different amounts of FBS overnight (Fig. 1G). A FBS concentration-dependent increase was observed in cell-surface LPL in T47-D cells. LPL was also detected on cells incubated with media conditioned for 3 days by LPL-secreting LiSa-2 liposarcoma cells.
HSPG motif HSNS4F5 is on the surface of BC cells and is a binding site for LPL
LPL has been reported to bind to HSPGs, the (GlcNS6S-IdoA2S)3 or HSNS4F5 motif, in particular (11, 17). Using qRT-PCR, we first determined that the cancer cell lines used in our studies did not express detectable levels of GPIHBP1, the one exception being DU4475 TNBC cells (not shown). Using flow cytometry and a Dylight 650-labeled antibody for HSNS4F5 (NS4F5IgG) we established that this binding site for LPL is present on the surface of cancer cells (supplemental Fig. S2A). This was confirmed visually using fluorescence confocal microscopy, shown by a representative image of HSNS4F5 staining on MDA-MB-231 BC cells (supplemental Fig. S2B). The abundance of HSNS4F5 on the cell surface was sensitive to confluence and nutrient supply (not shown). Knockdown of heparan sulfate 6-O-sulfotransferase 1 (HS6ST1), an enzyme responsible for 6-O-sulfation of heparan sulfate, reduced HSNS4F5 on the surface of MDA-MB-231 BC cells, supporting the specificity of the NS4F5IgG (supplemental Fig. S2C).
BC cells take up intact VLDL particles in a temperature-, concentration- and time-dependent manner by receptor-mediated endocytosis
Initial studies revealed that VLDL bind to and are rapidly internalized by cancer cells. This raised the question of whether the observed uptake represented the internalization of intact lipoproteins or that of FFA released by LPL-mediated hydrolysis of TG. To investigate this, the lipid and protein components of VLDLs were labeled with DiI and DyLight 650, respectively, and visualized using confocal microscopy (supplemental Fig. S3). As shown in the merged channel, the fluorescently labeled lipid and protein components of internalized VLDL particles coincide, indicating that intact lipoproteins are internalized.
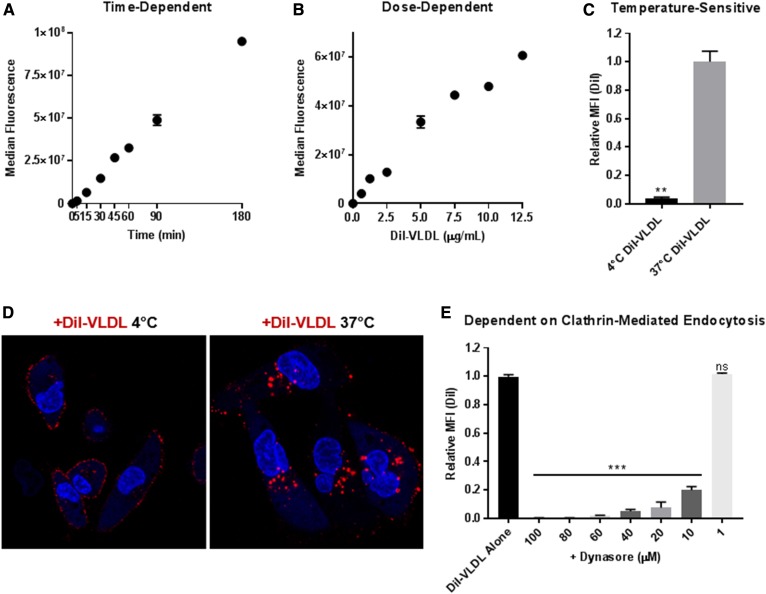
We characterized DiI-VLDL uptake using flow cytometry, confocal microscopy, and plate-based fluorescent assays. Using MDA-MB-231 BC cells, we show that DiI-VLDL uptake is time- and dose-dependent and temperature-sensitive (Fig. 2). MDA-MB-231 cells incubated at 4°C with DiI-labeled VLDL particles (5 µg/ml) bind VLDLs at the cell surface, while incubations at 37°C result in both binding and internalization (Fig. 2C, D). Binding of DiI-VLDL to the surface of MDA-MB-231 cells at 4°C was abrogated by excess unlabeled VLDL, indicating specificity consistent with a ligand-receptor interaction (supplemental Fig. S4). Rapid internalization of intact VLDL particles through a temperature-dependent process led us to investigate whether uptake was by receptor-mediated endocytosis. Treatment of MDA-MB-231 cells with Dynasore, an inhibitor of clathrin-coated pit-mediated endocytosis, resulted in a concentration-dependent decrease in DiI-VLDL uptake (Fig. 2E).
Fig. 2.
VLDL particles bind to and are internalized by cancer cells. DiI-VLDL uptake is temperature-, dose-, and time-dependent and is inhibited by treatment with Dynasore. Median fluorescence of MDA-MB-231 BC cells incubated with (A) DiI-VLDL (5 µg/ml) for durations of time ranging from 5 min to 3 h and (B) DiI-VLDLs for 45 min at increasing dosage. C, D: MDA-MB-231 BC cells were incubated with DiI-VLDL (5 µg/ml, 45 min) in 37°C or 4°C. C: Cells at 37°C displayed significantly more uptake than those incubated at 4°C, as quantified by flow cytometry (**P < 0.01; two-tailed unpaired t-test with Welch’s correction). Duplicate or triplicate for all data points; mean ± SD. D: Visualization via confocal microscopy shows that DiI-VLDLs remain bound at the cell surface at 4°C, whereas they are internalized at 37°C. E: Relative uptake of DiI-VLDL particles following treatment with Dynasore (30 min at 37°C) measured by flow cytometry. Concentration-dependent reduction in DiI-VLDL uptake with Dynasore treatment (***P < 0.001; one-way ANOVA with correction for multiple comparisons).
DiI-VLDL binding and uptake are abrogated by treatment with heparin, heparinase, or an antibody targeting HSNS4F5
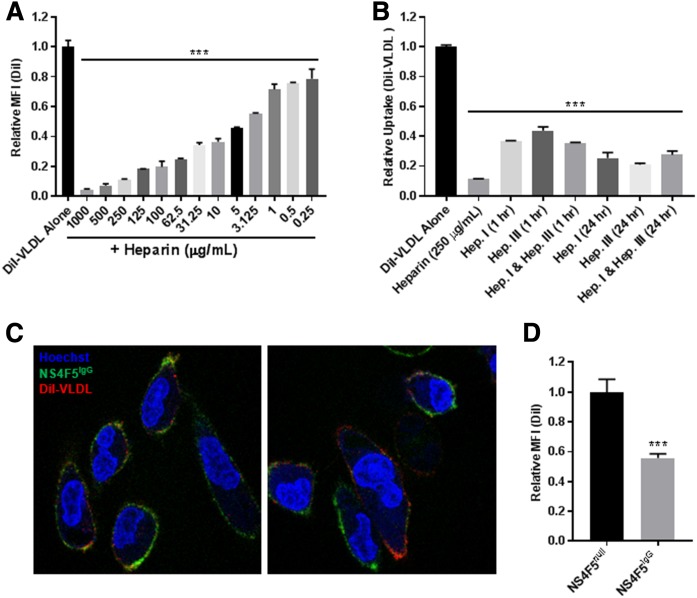
We hypothesized that VLDL uptake was mediated by cell-surface HSPG-bound LPL acting as a noncatalytic bridge to facilitate the uptake of intact lipoprotein particles through receptor-mediated endocytosis. We assessed the dependence on HSPGs (directly) and LPL (indirectly) by first testing the effect of heparin on DiI-VLDL uptake. Heparin caused a concentration-dependent decrease in DiI-VLDL uptake by MDA-MB-231 cells (Fig. 3A). Heparinase I and/or III likewise reduced DiI-VLDL uptake (Fig. 3B). Confocal microscopy using DiI-VLDLs and DyLight 650-NS4F5IgG shows that both VLDLs and HSNS4F5 localize to the cell surface (in cells incubated at 4°C) (Fig. 3C). Treating cells with unlabeled NS4F5 antibody reduced DiI-VLDL uptake, as measured by flow cytometry (Fig. 3D). Together, these data support a model in which HSPGs, particularly the HSNS4F5 motif, are integral for VLDL uptake by MDA-MB-231 cells.
Fig. 3.
Lipoprotein uptake is reliant on HSPGs. DiI-VLDL binding and internalization is abrogated by treatment with heparin, heparinase, or an antibody directed against the HSNS4F5 motif in MDA-MB-231 BC cells. A: Heparin treatment caused a dose-dependent reduction in DiI-VLDL uptake, as assessed by flow cytometry (***P < 0.001; one-way ANOVA with correction for multiple comparisons). B: Treatments with heparin or heparinase reduced DiI-VLDL uptake compared with the control, as assessed via a plate-based DiI-VLDL uptake assay (mean ± SD; ***P < 0.001; one-way ANOVA with correction for multiple comparisons). C: Representative confocal microscopy images of MDA-MB-231 colabeled with Hoechst 33342 nuclear stain (blue), NS4F5 (green), and DiI-VLDL (red). Both DiI-VLDLs and the HSNS4F5 antibody localize to the cell surface of cells incubated for 30 min at 4°C. D: DiI-VLDL uptake was reduced in MDA-MB-231 cells treated with unlabeled NS4F5IgG antibody (3.2 µg/ml) compared with those treated with the control NS4F5null antibody (***P < 0.001; two-tailed unpaired t-test with Welch’s correction).
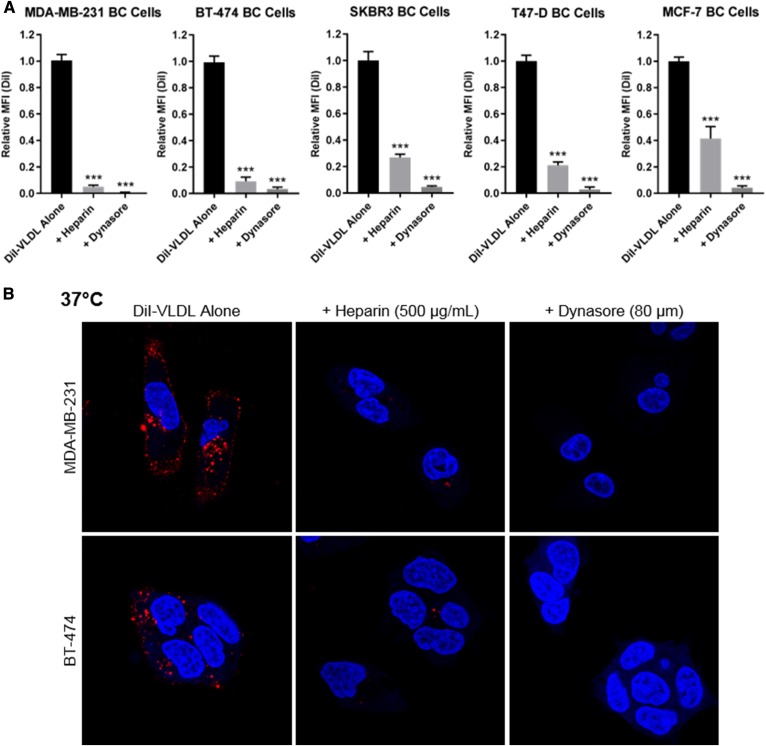
The effects of heparin and Dynasore were tested across an array of BC cell lines (Fig. 4). All cells rapidly took up DiI-VLDL. Binding and internalization were reduced by 500 µg/ml heparin or 80 µM Dynasore, as quantified via flow cytometry and visualized using confocal microscopy (Fig. 4).
Fig. 4.
DiI-VLDL uptake is abrogated by treatment with heparin or Dynasore across BC cell lines. A: Treatment with heparin (500 µg/ml) or Dynasore (80 µM) reduced DiI-VLDL uptake, as quantified by flow cytometry. Replicates from ≥3 experiments are shown (***P < 0.001; one-way ANOVA with correction for multiple comparisons). B: Effects of heparin (500 µg/ml) and Dynasore (80 µM) on Di-VLDL binding and uptake in adherent BC cells visualized by confocal microscopy. Heparin or Dynasore prevented both binding and internalization of DiI-VLDL particles at 37°C. Representative images shown: DiI-VLDL (red), Hoechst nuclear stain (blue).
A role for LPL and VLDLR in the uptake of VLDLs through receptor-mediated endocytosis
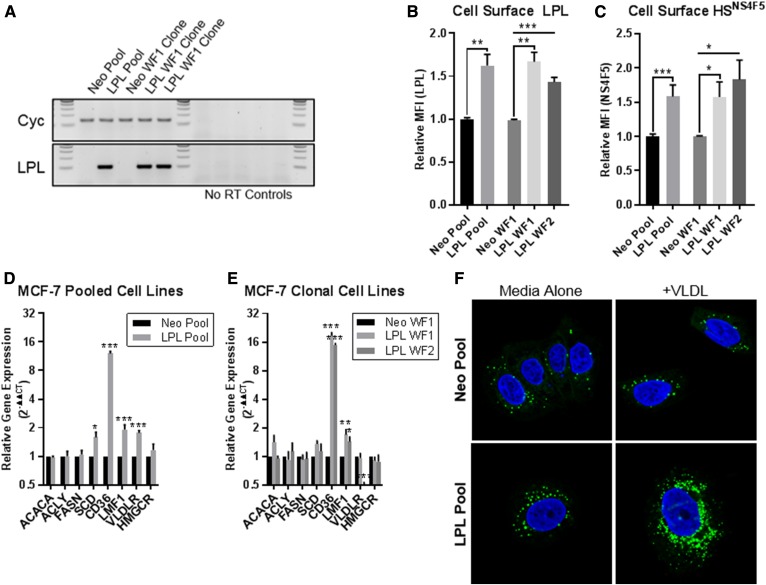
We used several approaches to directly implicate LPL in the uptake of lipoprotein particles by BC cells. MCF-7 (LPL-nonexpressing) BC cells were engineered to overexpress LPL. MCF-7 pooled and clonal LPL-overexpressing cell lines displayed upregulated LPL mRNA expression (Fig. 5A) and cell-surface LPL protein (Fig. 5B) compared with pCMV Neo controls. Cell-surface HSNS4F5 was significantly elevated in LPL-overexpressing MCF-7 cells, suggesting a direct relationship between the expression of LPL and display of this LPL-binding motif (Fig. 5C). Several other genes associated with lipid uptake, including CD36, LMF1, and VLDLR were also upregulated, with no differences observed in the expression of the FA synthesis genes ACACA, ACLY, and FASN or cholesterol synthesis gene HMGCR (Fig. 5D, E). The lipid droplet (LD) content of LPL-overexpressing and Neo control MCF-7 cells was investigated both at baseline and with VLDL supplementation using LipidTox Neutral Red Lipid Stain. LPL-overexpressing cells displayed higher basal LD staining than pCMV Neo control cells, as shown by confocal microscopy using the MCF-7 pooled cell-line pair (Fig. 5F). VLDL supplementation (72 h, 100 µg/ml) increased the size and abundance of LDs present in pCMV Neo and pCMV LPL cells, with the change in LPL-overexpressing cells exceeding that of the Neo control (Fig. 5F). This trend was consistent across media, including matched FBS and LPD FBS RPMI-1640, and was mirrored in the clonal cell lines (not shown). Together, these data indicate that overexpressing LPL alters the metabolism of MCF-7 BC cells by augmenting their capacity to take up lipids and increasing intracellular LD content.
Fig. 5.
MCF-7 pCMV LPL cell lines overexpress LPL (RNA and protein), display increased cell-surface HSNS4F5, and upregulate mRNA expression of genes involved in FA uptake. A: RT-PCR products visualized on agarose gels. Increased LPL product was observed in MCF-7 pCMV LPL pooled and clonal cell lines compared with the pCMV Neo controls. B: Increased cell-surface staining of LPL in MCF-7 LPL-overexpressing cells quantified by flow cytometry. C: MCF-7 LPL-overexpressing lines display increased cell-surface HSNS4F5. D, E: qRT-PCR of FA metabolism genes; the relative gene expression was calculated using the 2−ΔΔCT method. Data are from ≥3 experiments; mean ± SEM. Statistical significance was determined using two-tailed unpaired t-tests. *P < 0.05, **P < 0.01, and ***P < 0.001. F: MCF-7 pCMV Neo and pCMV LPL cell pools were cultured for 72 h standard media ± VLDL (100 µg/ml). MCF-7 LPL-overexpressing cells exhibited higher LD content both at baseline and with VLDL supplementation. LipidTox (LD stain; green), Hoechst 3342 nuclear stain (blue).
We also investigated the effects of LPL knockdown by generating MDA-MB-231 LPL shRNA cell lines. As shown by qRT-PCR, LPL shRNA A cells exhibit a complete knockdown of LPL mRNA, whereas LPL shRNA D cells were partially knocked down compared with the control (supplemental Fig. S5A, B). Knockdown of LPL in the LPL shRNA A cells resulted in the transcriptional upregulation of FA synthesis genes, as well as an increase in the expression of CD36, VLDLR, LMF1, and HMGCR (supplemental Fig. S5B). Partial knockdown of LPL mRNA (in LPL shRNA D cells) was associated with upregulation of the VLDLR mRNA alone. MDA-MB-231 control and LPL shRNA cells were compared for DiI-VLDL uptake and LD composition basally and with VLDL supplementation (100 µg/ml). To our surprise, LPL shRNA D cells (with partial knockdown of LPL mRNA and upregulation of VLDLR mRNA) displayed a significant increase in DiI-VLDL uptake (supplemental Fig. S5C), as well as a higher LD content both basally and with VLDL supplementation, compared with LPL shRNA A cells or the Scr control (not shown).
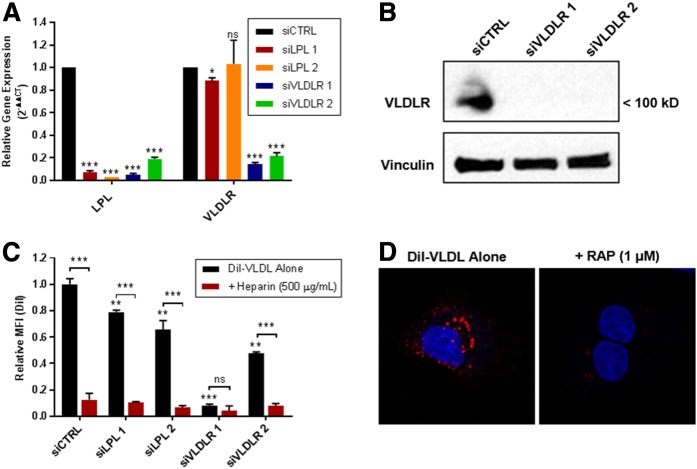
We used acute knockdown of LPL using siRNA to avoid the compensatory changes in FA metabolism-related gene expression that occurred with stable LPL knockdown. We compared the effects of two siRNA constructs directed against both LPL and VLDLR in MDA-MB-231 cells to that of a scrambled control. As shown by qRT-PCR, both LPL siRNAs efficiently reduced the expression of LPL (Fig. 6A). VLDLR siRNA constructs 1 and 2 significantly knocked down VLDLR gene expression compared with the control and unexpectedly caused a reduction in LPL gene expression (a finding that was repeated in multiple experiments). All MDA-MB-231 siRNA-treated cells had significantly reduced DiI-VLDL uptake compared with the control (Fig. 6B). These effects were less dramatic in LPL siRNA 1 and 2 cells, where LPL alone was knocked down (and presumably present in the original TC medium), and most significant in VLDLR siRNA 1 cells, which had the most complete knockdown of both LPL and the VLDLR. We used the receptor-associated protein (RAP), an inhibitor of VLDLR and other members of the LDL receptor family, to independently assess the role of VLDLR in VLDL endocytosis by MDA-MB-231 cells. Exposure to RAP prior to the addition of DiI-VLDL abolished VLDL uptake (Fig. 6D). Taken together, these data indicate that both LPL and VLDLR are required for efficient lipoprotein uptake in MDA-MB-231 BC cells.
Fig. 6.
LPL and VLDLR are involved in VLDL uptake in MDA-MB-231 BC cells. A: qRT-PCR verification of LPL and VLDLR knockdown in MDA-MB-231 BC cells treated with LPL or VLDLR siRNA. LPL and VLDLR gene expression of siRNA cells is presented relative to that of the Scr siRNA control. LPL siRNAs knocked down LPL mRNA, while VLDLR siRNAs significantly reduced both VLDLR and LPL mRNAs. B: Western blot shows VLDLR protein knockdown by siRNAs. C: siRNA knockdown of LPL or VLDLR reduces DiI-VLDL uptake in MDA-MB-231 BC cells. The greatest decreases were observed in VLDLR siRNA 1 cells, which had the highest efficiency knockdown of both VLDLR and LPL mRNAs. DiI-VLDL uptake was reduced by heparin by all treatments except VLDLR siRNA, where the reduction from VLDL alone was not significant. P ≥ 0.05 (ns), *P < 0.05, **P < 0.01, and ***P < 0.001. D: RAP (1 µM) inhibits DiI-VLDL binding and uptake at 37°C by MDA-MB-231 cells, as visualized by confocal microscopy. DiI-VLDL (red), DAPI (blue).
LPL supplementation increases DiI-VLDL uptake in BC cell lines
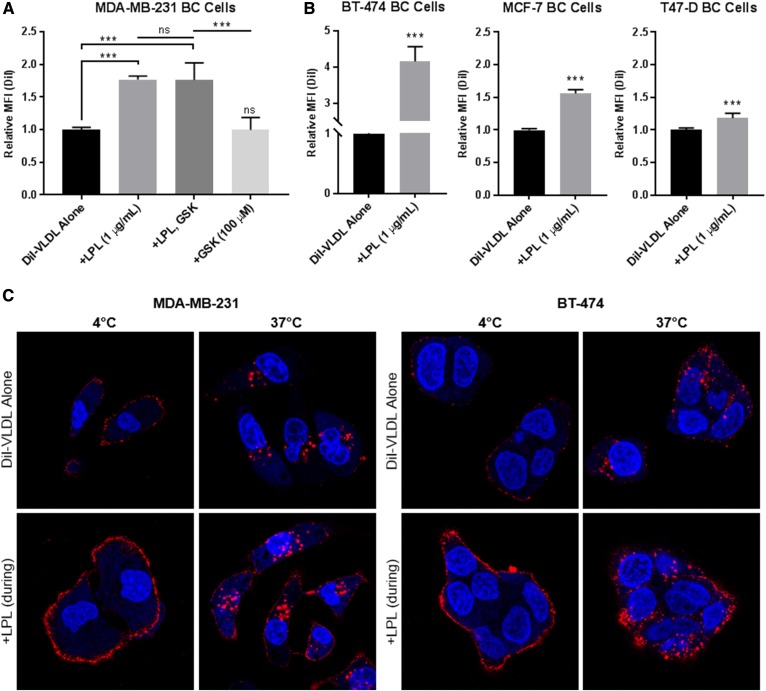
Our flow cytometry studies demonstrated that BC cells can take up exogenous LPL. We hypothesized that supplemental LPL would increase lipoprotein binding and internalization by BC cells and found that this was in fact the case. The addition of 1 µg/ml bovine LPL significantly increased DiI-VLDL uptake by MDA-MB-231 cells and was unaffected by treatment with GSK264220A, an inhibitor of the catalytic activity of LPL (7, 21) (Fig. 7A). Increased DiI-VLDL uptake upon LPL supplementation was observed across BC cell lines (Fig. 7B), albeit with variability potentially related to differences in basal LPL expression (BT-474 highest, MDA-MB-231; MCF-7 and T47-D as nonexpressing cells). Visualization of MDA-MB-231 and BT-474 cells using fluorescence confocal microscopy revealed that the addition of LPL increased both binding and internalization of VLDL particles (Fig. 7C).
Fig. 7.
LPL supplementation increases DiI-VLDL binding and uptake in BC cells. A: Quantification of DiI-VLDL uptake by flow cytometry. LPL (1 µg/ml) was supplied to MDA-MB-231 BC cells with DiI-VLDL (45 min, 5 µg/ml) at 37°C. LPL increased DiI-VLDL uptake (***P < 0.001; one-way ANOVA with correction for multiple comparisons). Treatment with the lipase inhibitor GSK264220A did not affect DiI-VLDL uptake. Biological replicates are displayed (mean ± SD). B: LPL supplementation significantly increased VLDL uptake in LPL-expressing and nonexpressing BC cell lines, as assessed by flow cytometry (*P < 0.05, **P < 0.01, and ***P < 0.001; two-tailed unpaired t-test with Welch’s correction). C: LPL supplementation increases DiI-VLDL binding and internalization across BC cell lines. Representative confocal microscopy images of MDA-MB-231 and BT-474 BC cells incubated with DiI-VLDLs at 4°C or 37°C with or without LPL supplementation (1 µg/ml). DiI-VLDL (red), Hoechst 33342 nuclear stain (blue).
Expression of FA metabolic genes in BC cells is affected by the availability of lipoproteins in media
MDA-MB-231 cells were cultured in media containing lipoprotein-depleted or matched-control FBS for 96 h. qRT-PCR was used to assess the expression of a panel of FA metabolism genes, including those involved with FA synthesis (ACACA, ACLY, ACSS2, FASN, and SCD, lipoprotein/FA uptake (CD36, LPL, LMF1, VLDLR), cholesterol synthesis and uptake (HMGCR; LDLR), and lipid storage (DGAT1, PLIN2).
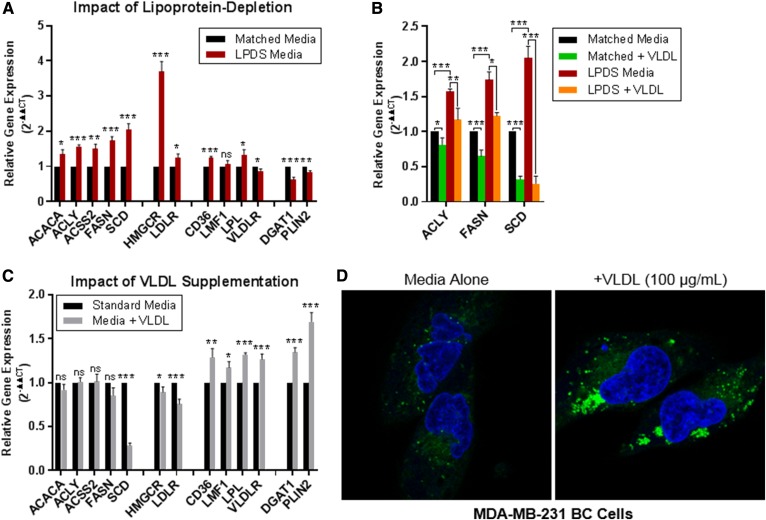
MDA-MB-231 cells cultured in LPDS media displayed significantly increased expression of genes involved in lipid synthesis (ACACA, ACLY, ACSS2, FASN, and SCD) and cholesterol metabolism (HMGCR and LDLR). Expression of the FA/lipoprotein uptake genes CD36 and LPL was likewise increased following prolonged culture in LPDS media, while the expression of the lipid-storage genes DGAT1 and PLIN2 was reduced (Fig. 8A). While some cell-line-specific changes were observed, these trends were largely consistent across BC cell lines (supplemental Fig. S6). We selected three FA synthesis genes (ACLY, FASN, SCD) to address the question of whether VLDL addback to LPDS media could restore gene expression to the levels observed in the matched-media control and found that this was the case. VLDL addback reduced the expression of all three FA synthesis genes back toward the level of the control baseline. MDA-MB-231 cells cultured in matched media supplemented with VLDL (100 μg/ml, 96 h) likewise exhibited decreased expression of these genes (Fig. 8B).
Fig. 8.
Lipoproteins in media affect the expression of FA metabolic genes and abundance of LDs in MDA-MB-231 BC cells. A–C: qRT-PCR was used to assess the impact of media lipoproteins on the expression of FA metabolism-related genes. Gene expression is displayed as 2−ΔΔCT (relative to the control). Data are mean ± SEM of >3 experiments. Two-tailed unpaired t-tests: P ≥ 0.05 (ns), *P < 0.05, **P < 0.01, and ***P < 0.001. A: MDA-MB-231 BC cells cultured in LPDS media for 96 h displayed increased expression of FA synthesis and cholesterol metabolism genes and significantly decreased the expression of FA storage genes (DGAT1, PLIN2) compared with cells grown in matched-control media. The expression of FFA/lipoprotein uptake genes was variably affected by lipoprotein depletion. B: BC cells were grown for 96 h in matched or LPDS media ± VLDLs (100 µg/ml) to assess the impact of “addback” on the expression of FA synthesis genes. Data are normalized to the matched media control. Culturing cells in LPDS media resulted in upregulation of ACLY, FASN, and SCD (as shown in A). The addition of VLDLs (100 μg/ml) reduced the expression of these genes back to or below the matched-control media baseline. C: qRT-PCR of cells grown in standard media ± VLDLs (100 μg/ml, 72 h). VLDL supplementation increased the expression of FFA/lipoprotein uptake and FA storage genes; there were no significant changes or decreases in FA synthesis and cholesterol metabolism genes. D: Increased size and abundance of LDs observed in cells grown in media supplemented with VLDLs (100 μg/ml, 72 h) compared with the media alone control. Cells were stained with Hoechst 33342 nuclear stain (blue) and LipidTOX Red Neutral Lipid Stain (green) and visualized using fluorescence confocal microscopy.
We next explored the impact of VLDL supplementation (100 μg/ml, 96 h) on the gene-expression profile of MDA-MB-231 BC cells cultured in standard RPMI-1640 media. Here, the addition of VLDLs to the media caused increased expression of genes involved in lipid uptake (CD36, LMF1, LPL, VLDLR) and storage (DGAT1, PLIN2), significant reductions in the expression of SCD, HMGCR, and LDLR, and no significant changes in the expression of FA synthesis genes (ACACA, ACLY, ACSS2, FASN) (Fig. 8C). We hypothesized that the induction of lipid uptake and storage genes may be reflected in the abundance of LDs. Visualization of LDs via confocal microscopy revealed that MDA-MB-231 BC cells cultured in media supplemented with VLDLs (100 μg/ml, 72 h) displayed a stark increase in the size and abundance of LDs compared with that of cells grown in media alone (Fig. 8D). This was recapitulated in multiple cell lines (BT-474, MCF-7, T47-D; not shown). These data indicate that BC cells can respond to lipoprotein availability and shift their metabolism-related gene-expression programs (lipogenesis vs. uptake) to meet their FA requirements.
DISCUSSION
We previously showed that most breast tumors display both LPL and CD36 and that the provision of VLDL to BC cells (in the presence of LPL) stimulates cell growth and enables cells to evade the cytotoxic effects of FA synthesis inhibition (3). These results, in concert with the observation that exogenous FFA can rescue cancer cells from the cytotoxic effects of FA synthesis inhibitors (22, 23), prompted the idea that BC cells might deploy LPL as do nonmalignant LPL-secreting cells, such as adipocytes and cardiomyocytes. Our findings support an alternative model wherein LPL is bound to the cell surface, both in BC cell lines and in clinical BC tissue. Evidence supporting the identity of the LPL-binding site as a HSPG molecule includes displacement by heparin and significant reductions in DiI-VLDL binding and internalization following heparin or heparinase treatment. Decreased VLDL uptake by BC cells in the presence of an antibody directed against HSNS4F5 implicates this specific HSPG motif as the binding moiety. Notably, the HSNS4F5 sequence (GlcNS6S-IdoA2S)3 is repeated in heparin and was previously identified as an LPL-binding sequence (11) found on the surface of MCF-7 BC cells (24). Though MCF-7 and several other BC cell lines do not express detectable LPL mRNA, they do display HSNS4F5 on their cell surface. Our data indicate that this motif acts as a “bait” to acquire exogenous LPL and that this LPL can facilitate the uptake of lipoproteins. Acquisition of LPL in this manner could be important in anatomic areas in which primary or metastatic BC reside, such as the mammary fat pad or fatty bone marrow. We previously demonstrated an inverse correlation between the expression of the LPL and heparanase genes in BC cells, presaging a role for nonsecreted LPL adherent to the cell surface (3).
Our data do not exclude a hydrolytic role for LPL on the BC cell surface. Indeed, the cell lines that we have examined express CD36 and thus are equipped to take up FFA. On the other hand, our experimental findings clearly establish a noncatalytic role for the cell-surface LPL-HSNS4F5 complex in the binding and internalization of VLDLs. The rapid and temperature-sensitive uptake of intact lipoproteins and its prevention by Dynasore support receptor-mediated endocytosis as the mechanism observed in our studies.
Many attempts were undertaken to implicate LPL in the endocytosis of VLDLs. We learned through flow cytometry that cell lines that do not express LPL can acquire it from FBS in the media. Attempts to remove LPL from the system proved challenging and ultimately altered the metabolic phenotype of cells. While some variations were observed among BC cell lines, removal of lipoproteins generally induced the expression of FA and cholesterol synthesis-related genes, mirroring the metabolic plasticity observed by Daniels and coworkers in prostate cancer cells (23).
We generated LPL-overexpressing and shRNA knockdown cell lines in order to isolate a specific LPL phenotype. Enforced LPL expression in nonexpressing MCF-7 cells increased the expression of CD36 and LMF1 mRNAs, as well as the abundance of HSNS4F5 on the cell surface, an apparent FA uptake program to capitalize on the availability of endogenously produced LPL. These changes were associated with increased size and abundance of LDs. On the other hand, shRNA knockdown of LPL in MDA-MB-231 BC cells induced the expression of mRNAs coding lipogenic enzymes (ACACA, ACLY, FASN, SCD), as well as VLDLR and CD36. We interpret this as a shift toward FA synthesis and compensatory upregulation of other genes in the lipid-uptake program. Acute knockdown of LPL by siRNA caused a modest reduction of VLDL uptake, likely owing to the presence of residual LPL from FBS. Ultimately, the clearest evidence of LPL involvement came from experiments showing that LPL supplementation causes significant increases in DiI-VLDL binding and internalization. This trend was consistent across all cell lines tested, directly implicating LPL and further validating the ability of cancer cells to use exogenous LPL to facilitate lipoprotein uptake.
While it was feasible that HSPG-bound LPL performed this noncatalytic function alone, our data provide evidence that the VLDLR is also involved. Like LPL, the VLDLR is variably expressed by cancer cell lines; its expression is affected by both exogenous lipid supply and LPL expression. siRNA knockdown of VLDLR reduced LPL gene expression (a surprising finding that was repeated with different siRNAs over multiple experiments), and in the most efficacious dual-knockdown, nearly eliminated VLDL uptake. RAP also abrogated DiI-VLDL uptake, further implicating VLDLR. While the exact nature of the VLDLR-LPL interaction warrants future study, associations between LPL and the VLDLR are well established in other contexts.
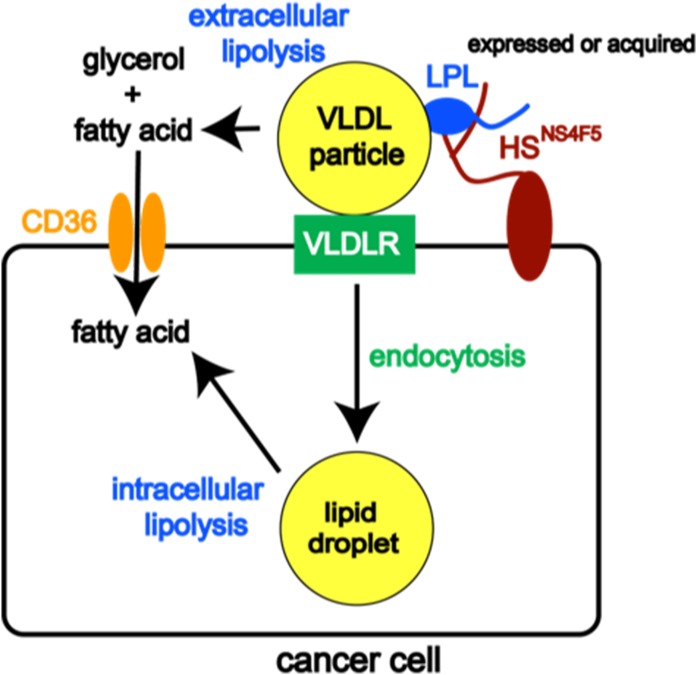
There are at least three nonmutually exclusive configurations in which LPL and VLDLR can interact: 1) LPL can bind directly to the VLDLR, increasing the binding affinity for VLDL; 2) HSPG-bound LPL can facilitate interactions between lipoprotein and the VLDLR; and 3) LPL-mediated hydrolysis of VLDLs can lead to the formation of remnant particles, which may remain bound to LPL and be endocytosed (25). The high-capacity model for lipoprotein uptake in BC cells outlined herein involves HSPG-bound cell-surface LPL binding to VLDLs and facilitating the uptake of intact lipoproteins via receptor-mediated endocytosis in concert with the VLDLR (Fig. 9).
Fig. 9.
Model of lipoprotein uptake in BC cells. Our previous and current work has shown that LPL binds to a specific HSPG motif (HSNS4F5) on the cancer cell surface, where it serves two distinct roles: 1) catalyzing the hydrolysis of TG carried by VLDL, releasing FFAs that can enter the cell through the FA uptake channel CD36, or 2) acting noncatalytically with VLDLR to facilitate the uptake of VLDL particles by receptor-mediated endocytosis. The latter mechanism has not been reported previously in cancer cells. LPL can be synthesized or acquired from exogenous sources. By functioning in this dual capacity, LPL plays a prominent role in mediating the uptake of FFAs and TGs required to sustain the growth and proliferation of cancer cells.
We recently pointed out that the literature related to lipid metabolism and cancer has long emphasized the role of FA synthesis and only more recently recognized the importance of CD36-mediated uptake of exogenous FFA (2). Here we have elucidated a novel mechanism through which BC cells take up exogenous lipid in the form of intact VLDL. Our data support the involvement of LPL, the VLDLR, and HSPGs in this mechanism and leave the potential for LPL to also act enzymatically at the cell surface. Our findings have implications for the development of cancer therapeutics aimed at lipid metabolism. We and others have shown that cancer cells display increased sensitivity to FA synthesis inhibitors when cultured in lipid-reduced media (23, 26, 27). It has likewise been demonstrated that cancer cells adjust their reliance on FA synthesis versus lipolysis/uptake in response to nutrient availability (23). Our data add to the mounting evidence that targeting FA synthesis may not achieve maximal efficacy in certain tumors because they may use both FA synthesis and uptake (of both FFA and VLDL), and reliance on these pathways may shift in response to nutrient availability or therapeutic inactivation of only one of them.
Supplementary Material
Acknowledgments
The authors thank Cary N. Mariash for his critical reading of this manuscript. The authors also acknowledge the following shared resource facilities at the Dartmouth-Hitchcock Norris Cotton Cancer Center: the Immune Monitoring and Flow Cytometry Resource and the Irradiation, Pre-clinical Imaging and Microscopy Resource.
Footnotes
Abbreviations:
- BC
- breast cancer
- FASN
- FA synthase
- GPIHBP1
- glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1
- HSPG
- heparan sulfate proteoglycan
- LD
- lipid droplet
- LPDS
- lipoprotein-depleted serum
- RAP
- receptor-associated protein
- TC
- tissue culture
- TG
- triglyceride
- VLDLR
- VLDL receptor
This work was supported by National Institutes of Health Grants RO1CA58961 (W.B.K.), R01CA200994 (T.W.M.), and R01CA211869 (T.W.M.); KU Leuven Grants C16/15/073 and C32/17/052 (J.V.S.); the Interreg V-A EMR23 EURLIPIDS program (J.V.S.); Dartmouth-Hitchcock Norris Cotton Cancer Center Grant P30CA023108 (W.B.K./T.W.M.); National Cancer Institute Grant 5P30CA023108-37; and National Institute of General Medical Sciences COBRE Grant P30GM103415-15. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. The authors declare that they have no conflicts of interest with the contents of this article.
The online version of this article (available at https://www.jlr.org) contains a supplement.
REFERENCES
- 1.Menendez J. A., and Lupu R.. 2007. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat. Rev. Cancer. 7: 763–777. [DOI] [PubMed] [Google Scholar]
- 2.Kinlaw W. B., Baures P., Lupien L., Davis W., and Kuemmerle N.. 2016. Fatty acids and breast cancer: make them on site or have them delivered. J. Cell. Physiol. 231: 2128–2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuemmerle N. B., Rysman E., Lombardo P. S., Flanagan A. J., Lipe B. C., Wells W. A., Pettus J. R., Froehlich H. M., Memoli V. A., Morganelli P. M., et al. 2011. Lipoprotein lipase links dietary fat to solid tumor cell proliferation. Mol. Cancer Ther. 10: 427–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zaidi N, Lupien L, Kuemmerle N, Kinlaw W, Swinnen J, Smans K.. 2013. Lipogenesis and lipolysis: the pathways exploited by the cancer cells to acquire fatty acids. Prog. Lipid Res. 52: 585–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kristensen L., Kristensen T., Abildgaard N., Royo C., Frederiksen M., Mourits-Andersen T., Campo E., and Moller M. B.. 2016. LPL gene expression is associated with poor prognosis in CLL and closely related to NOTCH1 mutations. Eur. J. Haematol. 97: 175–182. [DOI] [PubMed] [Google Scholar]
- 6.Heintel D., Kienle D., Shehata M., Krober A., Kroemer E., Schwarzinger I., Mitteregger D., Le T., Gleiss A., Mannhalter C., et al. 2005. High expression of lipoprotein lipase in poor risk B-cell chronic lymphocytic leukemia. Leukemia. 19: 1216–1223. [DOI] [PubMed] [Google Scholar]
- 7.Davies B. S., Beigneux A., Barnes R., Tu Y., Gin P., Weinstein M., Nobumori C., Nyren R., Olivecrona G., Bensadoun A., et al. 2010. GPIHP1 is responsible for the entry of lipoprotein lipase into capillaries. Cell Metab. 12: 42–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goldberg I. J. 1996. Lipoprotein lipase and lipolysis: central roles in lipoprotein metabolism and atherogenesis. J. Lipid Res. 37: 693–707. [PubMed] [Google Scholar]
- 9.Korn E. D. 1955. Clearing factor, a heparin-activated lipoprotein lipase. I. Isolation and characterization of the enzyme from normal rat heart. J. Biol. Chem. 215: 1–14. [PubMed] [Google Scholar]
- 10.Hoogewerf A. J., Cisar L. A., Evans D. C., and Bensadoun A.. 1991. Effect of chlorate on the sulfation of lipoprotein lipase and heparan sulfate proteoglycans. Sulfation of heparan sulfate proteoglycans affects lipoprotein lipase degradation. J. Biol. Chem. 266: 16564–16571. [PubMed] [Google Scholar]
- 11.Parthasarathy N., Goldberg I., Sivaram P., Mulloy B., Flory D., and Wagner W.. 1994. Oligosaccharide sequences of endothelial cell surface heparan sulfate proteoglycan with affinity for lipoprotein lipase. J. Biol. Chem. 269: 22391–22396. [PubMed] [Google Scholar]
- 12.Beigneux A. P., Davies B., Gin P., Weinstein M., Farber E., Qiao X., Peale F., Bunting S., Walzem R., Wong J., et al. 2007. Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 5: 279–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blain J. F., Aumont N., Theroux L., Dea D., and Poirier J.. 2006. A polymorphism in lipoprotein lipase affects the severity of Alzheimer’s disease pathophysiology. Eur. J. Neurosci. 24: 1245–1251. [DOI] [PubMed] [Google Scholar]
- 14.Accioly M. T., Pacheco P., Maya-Monteiro C. M., Carrossini N., Robbs B. K., Oliveira S. S., Kaufmann C., Morgado-Diaz J. A., Bozza P. T., and Viola J. P.. 2008. Lipid bodies are reservoirs of cyclooxygenase-2 and sites of prostaglandin-E2 synthesis in colon cancer cells. Cancer Res. 68: 1732–1740. [DOI] [PubMed] [Google Scholar]
- 15.Merkel M., Kako Y., Radner H., Cho I. S., Ramasamy R., Brunzell J. D., Goldberg I. J., and Breslow J. L.. 1998. Catalytically inactive lipoprotein lipase expression in muscle of transgenic mice increases very low density lipoprotein uptake: direct evidence that lipoprotein lipase bridging occurs in vivo. Proc. Natl. Acad. Sci. USA. 95: 13841–13846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nishitsuji K., Hosono T., Uchimura K., and Michikawa M.. 2011. Lipoprotein lipase is a novel amyloid beta (Abeta)-binding protein that promotes glycosaminoglycan-dependent cellular uptake of Abeta in astrocytes. J. Biol. Chem. 286: 6393–6401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smits N. C., Kurup S., Rops A. L., Ten Dam G. B., Massuger L. F., Hafmans T., Turnbull J. E., Spillmann D., Li J. P., Kennel S. J., et al. 2010. The heparan sulfate motif (GlcNS6S-IdoA2S)3, common in heparin, has a strict topography and is involved in cell behavior and disease. J. Biol. Chem. 285: 41143–41151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goulbourne C. N., Gin P., Tatar A., Nobumori C., Hoenger A., Jiang H., Grovenor C., Adeyo O., Esko J., Goldberg I., et al. 2014. The GPIHBP1-LPL complex is responsible for the margination of triglyceride-rich lipoproteins in capillaries. Cell Metab. 19: 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Teupser D., Thiery J., Walli A., and Seidel D.. 1996. Determination of LDL- and scavenger-receptor activity in adherent and non-adherent cultured cells with a new single-step fluorometric assay. Biochim. Biophys. Acta. 1303: 193–198. [DOI] [PubMed] [Google Scholar]
- 20.Basu D., Manjur J., and Jin W.. 2011. Determination of lipoprotein lipase activity using a novel fluorescent lipase assay. J. Lipid Res. 52: 826–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller P. M., Rust T., Murphy D. J., Matico R., Trill J. J., Krawiec J. A., Jurewicz A., Jaye M., Harpel M., Thrall S., et al. 2008. A high-throughput screen for endothelial lipase using HDL as substrate. J. Biomol. Screen. 13: 468–475. [DOI] [PubMed] [Google Scholar]
- 22.Olsen A. M., Eisenberg B., Flanagan A., Kuemmerle N., Morganelli P., Lombardo P., and Kinlaw W.. 2010. Fatty acid synthesis is a therapeutic target in human liposarcoma. Int. J. Oncol. 36: 1309–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daniels V, Smans K, Royaux I, Chypre M, Swinnen J, Zaidi N. 2014. Cancer cells differentially activate and thrive on de novo lipid synthesis pathways in a low-lipid environment. PLoS One. 9: e106913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Parthasarathy N., Gotow L., Bottoms J., Kute T., Wagner W., and Mulloy B.. 1998. Oligosaccharide sequence of human breast cancer cell heparan sulfate with high affinity for laminin. J. Biol. Chem. 273: 21111–21114. [DOI] [PubMed] [Google Scholar]
- 25.Kamataki A., Takahashi S., Masamura K., Iwasaki T., Hattori H., Naiki H., Yamada K., Suzuki J., Miyamori I., Sakai J., et al. 2002. Remnant lipoprotein particles are taken up into myocardium through VLDL receptor—a possible mechanism for cardiac fatty acid metabolism. Biochem. Biophys. Res. Commun. 293: 1007–1013. [DOI] [PubMed] [Google Scholar]
- 26.Zaidi N., Royaux I., Swinnen J. V., and Smans K.. 2012. ATP citrate lyase knockdown induces growth arrest and apoptosis through different cell- and environment-dependent mechanisms. Mol. Cancer Ther. 11: 1925–1935. [DOI] [PubMed] [Google Scholar]
- 27.Lupien L. E., Dunkley E. M., Maloy M. J., Lehner I. B., Foisey M. G., Ouellette M. E., Lewis L. D., Pooler D. B., Kinlaw W. B., and Baures P. W.. 2019. An inhibitor of fatty acid synthase thioesterase domain with improved cytotoxicity against breast cancer cells and stability in plasma. J. Pharmacol. Exp. Ther. 371: 171–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.