The chloroplast proteins depress H2O2 accumulation in guard cells and drought resistance through OST1-dependent and OST1-independent pathways.
Abstract
Drought is one of the most deleterious environmental conditions affecting crop growth and productivity. Here we report the important roles of a nuclear-encoded chloroplast protein, PsbP Domain Protein 5 (PPD5), in drought resistance in Arabidopsis (Arabidopsis thaliana). From a forward genetic screen, a drought-resistant mutant named ppd5-2 was identified, which has a knockout mutation in PPD5. The ppd5 mutants showed increased H2O2 accumulation in guard cells and enhanced stomatal closure in response to drought stress. Further analysis revealed that the chloroplast-localized PPD5 protein interacts with and is phosphorylated by OST1, and phosphorylation of PPD5 increases its protein stability. Double mutant ppd5-2ost1-3 exhibited phenotypes resembling the ost1-3 single mutant with decreased stomatal closure, increased water loss, reduced H2O2 accumulation in guard cells, and hypersensitivity to drought stress. These results indicate that the chloroplast protein PPD5 negatively regulates drought resistance by modulating guard cell H2O2 accumulation via an OST1-dependent pathway. Interestingly, the thf1-1 mutant defective in the chloroplast protein THF1 displayed drought-resistance and H2O2 accumulation similar to the ppd5 mutants, but the thf1-1ost1-3 double mutant resembled the phenotypes of the thf1-1 single mutant. These results indicate that both OST1-dependent and OST1-independent pathways exist in the regulation of H2O2 production in chloroplasts of guard cells under drought stress conditions. Additionally, our findings suggest a strategy to improve plant drought resistance through manipulation of chloroplast proteins.
Drought is a frequent stress condition for crops and causes enormous losses in plant agriculture (Fedoroff et al., 2010). Plants have developed complex signaling pathways to cope with water deficit due to their sessile lifestyle. Under water deficit conditions, plants reduce water loss by promoting stomatal closure and limiting shoot growth, while they increase water uptake by promoting root growth (Maggio et al., 2006). Stress responses occur in various cell compartments such as the cell surface, cytoplasm, endoplasmic reticulum (ER), chloroplast, mitochondrion, peroxisome, and nucleus (Zhu, 2016). Although increasing evidence has suggested that cell organelles such as mitochondria and chloroplasts are important sites for stress responses, the stress response pathways in these organelles are not fully understood.
Chloroplast is the site for photosynthesis and energy production, and its function is crucial for plant growth and development (Foyer et al., 2003). The PSII Subunit P (PsbP) protein is a component of the oxygen-evolving complex (OEC) of PSII. The OEC of higher plants consists of three extrinsic proteins named as PsbO (OEC33), PsbP (OEC23), and PsbQ (OEC16; De Las Rivas et al., 2007; Sato, 2010), which shows similarity as well as distinct features compared with the OEC in cyanobacteria (Bricker et al., 2013). The PsbP protein is essential for the normal function and high efficiency of PSII by stabilizing the Mn cluster, and knock-out mutant of PsbP is lethal in Arabidopsis (Arabidopsis thaliana; Ifuku et al., 2005; Yi et al., 2007). Arabidopsis possesses two PsbP-like proteins (PPL1 and PPL2) and seven PsbP-domain proteins (PPD1-PPD7; Ifuku et al., 2008; Sato, 2010; Bricker et al., 2013). These proteins, together with PsbP, comprise the PsbP protein family with relatively low sequence identities among the family members. The PsbP family proteins show distinct functions in plants. For example, PPL1 functions in efficient repair of photo-damaged PSII under high-intensity light, whereas PPL2 is required for the accumulation of the NDH complex (Ishihara et al., 2007; Ifuku et al., 2008). PPD1 functions in assisting the assembly of PSI (Liu et al., 2012). Nonetheless, the functions of most PPD proteins in the PsbP family are largely unknown. By the gene expression profiles being analyzed, several genes belonging to the PsbP family, including PPD5, were found to be responsive to different stresses such as high light, and PPD5 was shown to be coexpressed with genes encoding cytosolic ribosomal proteins (Ifuku et al., 2010).
As the photosynthetic organelle, chloroplast produces significant amount of reactive oxygen species (ROS) and has been considered as an important intracellular site for abiotic stress responses in plants (Mignolet-Spruyt et al., 2016). Under drought and other stress conditions, stomatal closure can be mediated by signaling molecules such as abscisic acid (ABA), Ca2+, and ROS (Murata et al., 2015). The ABA signaling pathway in Arabidopsis consists of three major components: ABA receptor (RCAR/PYR1/PYLs), type-2C protein phosphatase (PP2C), and SNF1-related protein kinase 2 (SnRK2; Melcher et al., 2009; Park et al., 2009). ABA binds to the PYR/PYL family of receptors, resulting in inhibition of type-2C phosphatases (PP2C) and de-repression of the SnRK2s to activate downstream effector proteins, which mediates stomatal closure and other ABA responses (Fujii et al., 2009; Munemasa et al., 2015). OPEN STOMATA 1 (OST1), also known as SnRK2.6, is one of the SnRK2s that is activated by ABA (Mustilli et al., 2002). OST1 controls stomatal movement by phosphorylating various substrates. The anion channel SLAC1 is one of the substrates of OST1, and phosphorylation of SLAC1 promotes anion efflux and stomatal closure (Vahisalu et al., 2008). OST1 also phosphorylates RbohF, a plasma membrane-bound NADPH oxidase, leading to the production of apoplastic O2.- and H2O2 (Acharya et al., 2013).
In our previous study, we found that the chloroplast protein THF1 forms a complex with another chloroplast protein HCF106, and mutation in either gene causes elevated accumulation of ROS in guard cells, enhanced stomatal closure, and increased drought resistance (Wang et al., 2016). THF1 is essential for chloroplast development and plays a crucial role in regulating the PSII-LHCII complex during leaf senescence and in response to excess light (Huang et al., 2013). THF1 interacts with the plasma membrane–located G-protein GPA1 and functions in sugar signaling (Huang et al., 2006). In this study, we identified a drought-resistant mutant, ppd5-2, which is defective in the chloroplast protein PPD5, by screening a pool of mutants with Agrobacterium transferred DNA (T-DNA) insertions in nuclear genes encoding chloroplast proteins. We found that OST1 physically interacts with PPD5 and phosphorylates PPD5 at the Thr-283 residue. The double mutant ppd5-2ost1-3 displayed reduced stomatal closure, increased water loss, and reduced H2O2 accumulation in guard cells under drought stress, which resembles the drought-sensitive ost1-3 mutant. Interestingly, the drought-resistant mutant thf1-1 defective in the nuclear-encoded chloroplast protein THF1 showed phenotypes similar with ppd5, but the double mutant thf1-1ost1-3 behaved like thf1-1 with elevated levels of H2O2 in guard cells, enhanced stomatal closure, and reduced water loss under drought stress. These results indicate that PPD5 and THF1 are involved in OST1-dependent and OST1-independent pathways, respectively, to negatively regulate plant drought resistance.
RESULTS
PPD5 Negatively Regulates Plant Drought Resistance
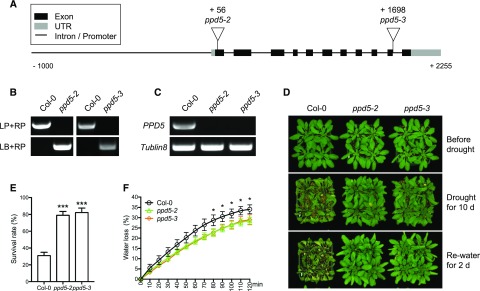
The ppd5-2 (SALK_039106C) mutant was identified from a genetic screen for drought-resistant mutants from a pool of T-DNA insertion mutants with mutations in nuclear genes encoding chloroplast proteins, as described in our previous study (Wang et al., 2016). The ppd5-2 mutant has a T-DNA insertion in the first exon of the nuclear gene At5g11450, which encodes a chloroplast protein named PsbP-domain protein 5 (PPD5; Fig. 1A). We obtained from the Arabidopsis Biological Resource Center (ABRC) another mutant line, ppd5-3 (SAIL_511_G10), which has a T-DNA insertion in the last exon of At5g11450 (Fig. 1A). Real time PCR results showed that both ppd5-2 and ppd5-3 were null mutant alleles of PPD5 (Fig. 1, B and C). Both the ppd5-2 and ppd5-3 mutants were more resistant to drought than the Col-0 wild type as indicated by their survival rates after drought treatment for 10 d (Fig. 1, D and E). The water loss rates of ppd5-2 and ppd5-3 were significantly lower than that of Col-0 wild type (Fig. 1F). The stomatal conductance and leaf transpiration of ppd5-2 and ppd5-3 were significantly lower than that of Col-0 wild type under drought stress (Supplemental Fig. S1, A–D), which suggested that the drought resistance of ppd5 mutants was likely due to altered stomatal regulation. The drought resistance function of PPD5 was further tested by a molecular complementation assay. The genomic sequence of PPD5 gene with its native promoter was amplified and fused with an YFP tag to generate the PPD5pro::PPD5-YFP construct, which was then introduced into the ppd5-2 mutant. Two transgenic lines, designated as COM #1 and COM #2, were selected for analysis (Supplemental Fig. S2, A and B). Immunoblotting assays confirmed the expression of PPD5-YFP fusion protein in the transgenic lines (Supplemental Fig. S2C). Plants (21 d old) of Col-0, ppd5-2, COM#1, and COM#2 grown in soil were subjected to drought stress treatment for 12 d. After rewatering for 3 d, the ppd5-2 mutant but not the two complementation lines COM #1 and COM #2 showed increased drought resistance compared with Col-0 wild type (Supplemental Fig. S2, D and E). These results show that the mutations in PPD5 are responsible for the drought-resistant phenotype of the mutants, suggesting that PPD5 is a negative regulator of drought resistance in Arabidopsis.
Figure 1.
The ppd5 mutants show drought resistance phenotype. A, The schematic structure of PPD5 gene and the positions of T-DNA insertions in the ppd5 mutant alleles. B, PCR-based genotyping of the ppd5-2 and ppd5-3 mutants. LP, left primer; RP, right primer; LB, primer of the T-DNA left border. C, Real time PCR analysis of PPD5 expression in ppd5-2, ppd5-3, and Col-0 plants. D, Drought resistance assay of ppd5-2, ppd5-3, and Col-0. The 21-d-old plants were drought-treated for 10 d and re-watered for 2 d. E, Survival rates of ppd5-2, ppd5-3, and Col-0 after 2 d recovery from a 10-d drought stress treatment. Values are means ± sd (n = 3). F, Water loss in ppd5-2, ppd5-3, and Col-0 rosette leaves (n = 3, each contained three fully expanded leaves from 21-d-old plants with drought stress treatment for additional 5 d). Data represent means ± sd (n = 3). *P < 0.05 and ***P < 0.001, Student’s t test.
The ppd5 Mutations Enhance H2O2 Accumulation in Guard Cells
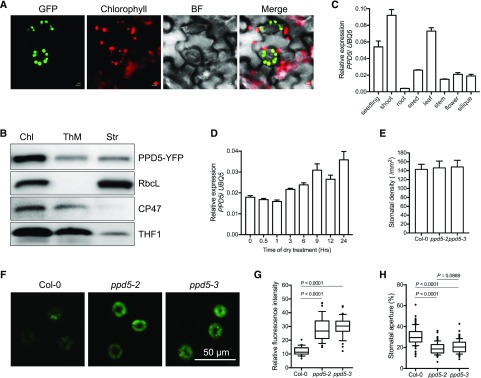
PPD5 belongs to the PsbP family of proteins with ten members: PsbP, PPL1, PPL2, and PPD1 to PPD7 (Supplemental Fig. S3). To determine the subcellular localization of PPD5, the complementation line COM #1 harboring the PPD5pro:PPD5-YFP was used. The YFP signals were detected only in the chloroplasts under a confocal microscope (Fig. 2A), indicating that PPD5 is a chloroplast protein. To pinpoint the localization of PPD5 in the chloroplast, immunoblot assays were carried out using fractionated proteins from plastids. The component of the core complex of PSII CP47 (PsbB) located in the inner membrane and the large subunit of Rubisco RbcL located in the stroma were used as maker proteins in this assay. The result showed that PPD5 existed in both the thylakoid membrane and stromal fractions (Fig. 2B). To analyze the expression pattern of PPD5, different tissues were collected for quantitative real-time PCR analysis. The expression of PPD5 could be detected in all tested tissues, with higher expression in leaves than in other tissues (Fig. 2C). The expression of PPD5 was slightly up-regulated by air dry treatment (Fig. 2D).
Figure 2.
The mutations in PPD5 enhance stomatal closure via increasing H2O2 accumulation in guard cells. A, Subcellular localization of PPD5-YFP fusion protein in the guard cells of transgenic Arabidopsis seedlings harboring the PPD5pro:PPD5-YFP construct. Chlorophyll, autofluorescence of chlorophyll; BF, bright field. Bars = 5 μm. B, Immunoblot analysis of RbcL, CP47, THF1, and PPD5 in fractionated chloroplast proteins. Chl, whole chloroplast; ThM, thylakoid membrane; Str, stromal fraction. C, The expression pattern of PPD5 in Arabidopsis. Values are means ± sd (n = 3). D, The expression levels of PPD5 in 21-d-old plant leaves treated with air dry. Values are means ± sd (n = 3). E, Stomatal density in the middle leaves of 4-week-old ppd5-2, ppd5-3, and Col-0. F, Representative images of the CM-H2DCFDA staining of H2O2 in guard cells of ppd5-2, ppd5-3, and Col-0 leaves of 21-d-old plants after a 5-d drought stress treatment. G, Quantitative measurement of H2O2 levels in guard cells of ppd5-2, ppd5-3, and Col-0 (n = 90, stomata from 21-d-old plants after drought stress for 5 d). H, Stomatal apertures of the middle leaves of 3-week-old ppd5-2, ppd5-3, and Col-0 plants after drought treatment for 5 d (n ≥ 80). Stomatal aperture was calculated by the percentage of width/length ratio. P-value was determined by Student’s t test.
To determine whether the ppd5 mutations may affect ROS production in guard cells, we measured the ROS levels in guard cells of ppd5-2, ppd5-3, and Col-0 using the fluorescent probe CM-H2DCFDA. After water was withheld for 5 d, the relative fluorescence intensities detected in ppd5-2 and ppd5-3 guard cells were significantly higher than that in Col-0 (Fig. 2, F and G), which indicated that ppd5-2 and ppd5-3 mutants accumulated more ROS in guard cells than Col-0 wild type under drought stress. The stomatal apertures of the leaves of ppd5-2, ppd5-3, and Col-0 after a 5-d drought stress treatment were also measured. The ppd5-2 and ppd5-3 mutants had significantly smaller stomatal apertures than the Col-0 wild type, although their stomatal densities were similar to that of the wild type (Fig. 2, E and H). These results suggested that there is increased ROS accumulation in guard cells and enhanced stomatal closure in the ppd5 mutants compared with the Col-0 wild type under drought stress.
PPD5 Physically Interacts with OST1
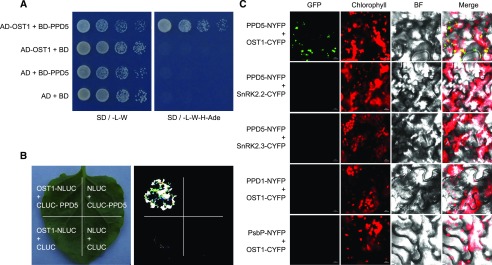
According to the Arabidopsis interactome database, OST1 may interact with PPD5 in the yeast two-hybrid assays (Arabidopsis Interactome Mapping Constortium, 2011). To confirm this interaction, we carried out a yeast two-hybrid assay and found that PPD5 could interact with OST1 (Fig. 3A). We further tested the interactions between PPD5 with other SnRK2 kinases, and the result showed that PPD5 could not interact with other SnRK2s in the yeast two-hybrid assays (Supplemental Fig. S4A). The interactions between OST1 and two PPD5 homologs, PsbP and PPD1, were also analyzed, and no interactions were detected in the yeast two-hybrid system (Supplemental Fig. S4B). These results indicate that PPD5 specifically interacts with OST1 (SnRK2.6). The interaction between PPD5 and OST1 was further verified using three additional assays. In the firefly luciferase complementation imaging assay (LCI), strong LUC signals were detected in the tobacco leaves only when both PPD5-CLUC and OST1-NLUC were expressed (Fig. 3B). The PPD5-OST1 interaction was also tested in a bimolecular fluorescence complementation (BiFC) assay using split YFP. Yellow fluorescence signals were only observed in the tobacco leaves carrying both the PPD5-NYFP and OST1-CYFP constructs (Fig. 3C; Supplemental Fig. S5). Neither SnRK2.2 nor SnRK2.3 interacted with PPD5 in tobacco leaves, and no interaction could be detected between OST1 and PPD1 or PsbP in the BiFC assay (Fig. 3C). In addition, we purified the recombinant biotin-MBP-PPD5 protein and 6×His-SUMO-OST1 protein and examined their interaction using the AlphaScreen assay (Melcher et al., 2009; Park et al., 2009; Cao et al., 2013). The result suggests a strong interaction between PPD5 and OST1 in vitro (Supplemental Fig. S6). These results support that PPD5 physically interacts with OST1.
Figure 3.
PPD5 physically interacts with OST1. A, Yeast two-hybrid assay showing an interaction between PPD5 and OST1. B, Split-LUC assay indicating an interaction between PPD5 and OST1 in Nicotiana benthamiana leaves. C, BiFC assay verifying the interaction between PPD5 and OST1 in N. benthamiana. BiFC assay analyzing the interaction between PPD5 and SnRK2s (OST1, SnRK2.2 and SnRK2.3), OST1 and PPD5 homologs, PPD1 and PsbP, in leaves of N. benthamiana. Chlorophyll, autofluorescence of chlorophyll; BF, bright field. Scale bars = 10 μm.
OST1 Phosphorylates PPD5
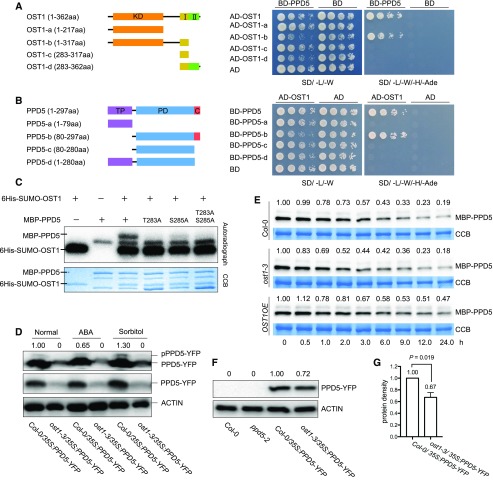
OST1 is an important kinase that regulates stress responses by phosphorylating various protein substrates in Arabidopsis. OST1 contains a kinase domain (KD) and two other domains (Fig. 4A), and its activity can be activated by osmotic stress and ABA. PPD5 possesses an N-terminal transit peptide domain (TP) for plastid localization, a PsbP domain (PD), and a C-terminal domain of unknown function (Fig. 4B). In order to determine the regions for OST1-PPD5 protein interaction, we constructed truncated PPD5 and OST1 and tested their interactions using yeast two-hybrid assays. The KD and osmotic stress-activated domain I (Domain I) of OST1 and the PD domain and the C-terminal domain of PPD5 were found to be sufficient for the interaction of these two proteins in the yeast two-hybrid system (Fig. 4, A and B). We further analyzed the subcellular localization of the truncated PPD5-b (80-287 amino acid, loss of the transit peptide of PPD5) and PPD5-d (1-280 amino acid, the C-terminal domain truncation) in N. benthamiana leaves. The yellow fluorescence of PPD5-b-YFP was detected in the cytoplasm and nucleus but not in the chloroplast, which indicates that the TP of PPD5 is required for its chloroplast localization, while PPD5-d was still localized in the chloroplast (Supplemental Fig. S7A). The interactions between OST1 and PPD5-b and PPD5-d were analyzed using BiFC assays, and the results show that PPD5-b lacking TP could still interact with OST1, but PPD5-d failed to interact with OST1 (Supplemental Fig. S7B), which confirms that the C-terminal domain of PPD5 is essential for its interaction with OST1.
Figure 4.
OST1 phosphorylates and stabilizes PPD5. A, Yeast two-hybrid assay for the interaction between the full-length PPD5 and OST1 deletion variants. I, domain I; II, domain II. B, The interaction of OST1 and PPD5 deletion variants analyzed by yeast two-hybrid assay. PD, PsbP domain; C, C-terminal domain. C, In vitro phosphorylation assay. Recombinant proteins PPD5, PPD5T283A, PPD5S285A, PPD5T283AS285A, and OST1 were combined for the in vitro phosphorylation assay. D, In vivo phosphorylation of PPD5 by OST1 under normal, ABA, and sorbitol treatment conditions. Proteins extracted from 10-d-old seedlings after treatment were separated in a Phos-tag containing SDS-PAGE gel and immuno-detected by an anti-GFP antibody. Normal SDS-PAGE gel was used as a control. The intensity of the pPPD5-YFP bands shown above the images was quantified by using Image Lab software (version 5.2.1). E, Western blotting showing the degradation of MBP-PPD5 proteins in different genotypes using cell-free assay. Quantitation of the MBP-PPD5 band intensities was done by using the Image Lab software (version 5.2.1). F, Western blotting showing the levels of PPD5-YFP fusion proteins in transgenic plants. ACTIN was used as a loading control. The intensity of the PPD5 bands relative to the ACTIN bands was shown above the images, and the intensity was quantified by using Image Lab software (version 5.2.1). G, Statistical analysis of the result shown in (F) revealing significant differences between Col-0 and ost1-3 (n = 3). P value was determined by Student’s t test.
To determine whether PPD5 is a substrate of OST1, purified recombinant PPD5 and OST1 proteins were subjected to in vitro phosphorylation assays and mass spectrometry (MS). Based on the MS analysis, the Thr-283 and Ser-285 residues at the C-terminal region of PPD5 are the likely phosphorylation sites. Wild-type PPD5 protein and three mutated PPD5 proteins—PPD5T283A, PPD5S285A, and PPD5T283AS285A—with changes in these two residues were expressed and used for further in vitro phosphorylation assays. The result showed that the Thr-283 mutation completely abolished the phosphorylation, whereas the Ser-285 mutation caused reduced phosphorylation (Fig. 4C). AlphaScreen assays revealed that the Thr-283 mutation in PPD5 significantly reduced its interaction with OST1, whereas the Ser-285 mutation had no obvious effect on the interaction (Supplemental Fig. S6). These results suggest that Thr-283 in PPD5 is the major site of phosphorylation by OST1. Phosphorylation of PPD5 was further verified using ost1-3 and Col-0 wild-type plants (Fig. 4D). Because the protein interaction assays showed that PPD5 only interacts with the KD and Domain I (osmotic stress-activated domain) but not the Domain II (ABA-responsive domain) of OST1, we tested the phosphorylation of PPD5 after both ABA and sorbitol (osmotic stress) treatments. A clear mobility shift of pPPD5-YFP protein was detected in Col-0 wild-type plants using SDS-PAGE with Phos-tag containing gels. The phosphorylation level of PPD5 increased after sorbitol treatment but decreased after ABA treatment. The band indicative of phosphorylated PPD5 was not detectable in the ost1 mutant. These results indicate that PPD5 is phosphorylated by OST1 in vivo and the phosphorylation is responsive to osmotic stress.
PPD5 Phosphorylation by OST1 Promotes Its Stability
Protein phosphorylation is a posttranslational modification that may modulate protein activity, stability, and/or subcellular localization. To determine whether the phosphorylation of PPD5 affects its stability, a cell-free protein degradation assay was performed. The recombinant MBP-PPD5 proteins were incubated with the total proteins from 10-d-old seedlings of Col-0, ost1-3, or OST1OE-#5. The degradation rate of MBP-PPD5 proteins did not show much difference after incubation for 6 h with the total proteins from Col-0 and ost1-3, whereas the MBP-PPD5 proteins incubated with the total proteins of OST1OE-#5 exhibited a slower degradation rate (Fig. 4E). The results indicate an important role of OST1-mediated PPD5 phosphorylation in regulating PPD5 stability. We crossed the ppd5-2/35S:PPD5-YFP transgenic plants with ost1-3 mutant plants and generated 35S:PPD5-YFP transgenic lines in ost1-3 and wild-type backgrounds. Immunoblotting assays showed that the protein level of PPD5-YFP was substantially higher in Col-0 than that in ost1-3 (Fig. 4. F and G). The protein levels of PPD5-YFP in independent transgenic lines with a similar transcript level of the transgene in ppd5-2 or ppd5-2ost1-3 mutants were analyzed. The result indicated that the PPD5-YFP level was lower in ppd5-2ost1-3 mutant plants compared with that in ppd5-2 mutant plants (Supplemental Fig. S8). These results support that phosphorylation of PPD5 by OST1 increases PPD5 stability.
Whether OST1-mediated PPD5 phosphorylation may affect the subcellular localization of PPD5 was determined by analyzing the PPD5-YFP protein in Col-0 and ost1-3. The results showed that the ost1-3 mutation did not affect the chloroplast localization of PPD5-YFP protein (Supplemental Fig. S9). Furthermore, we generated YFP fusion proteins with the PPD5 variants PPD5T283A, PPD5S285A, and PPD5T283AS285A, and compared the subcellular localization of these variants with the wild-type PPD5 in a transient assay in tobacco leaves. PPD5T283A, PPD5S285A, and PPD5T283AS285A were found to be localized in chloroplasts, which resembles the wild-type PPD5 (Supplemental Fig. S10). These results indicate that PPD5 phosphorylation mediated by OST1 is not required for the subcellular localization of PPD5.
The Drought Resistance of ppd5-2 Mutant Plants Requires OST1
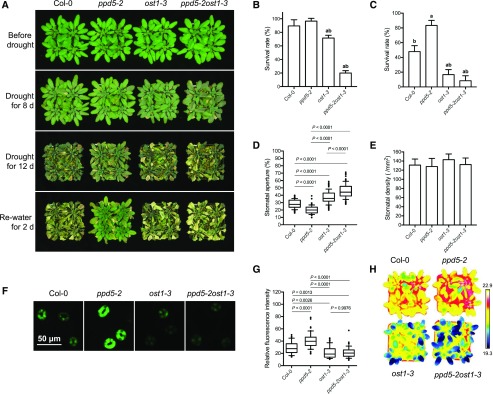
OST1 can be activated by ABA and plays a crucial role in controlling stomatal movement under drought stress. To determine whether PPD5 may genetically interact with OST1, we crossed ppd5-2 and ost1-3 mutant plants and generated homozygous ppd5-2ost1-3 double mutant plants (Supplemental Fig. S11). The ppd5-2ost1-3 double mutant exhibited a hypersensitivity to drought stress resembling the ost1 single mutant, whereas the ppd5-2 single mutant was more tolerant than Col-0 as expected (Fig. 5, A–C). The leaf temperature, which reflects the stomatal response and transpirational water loss, was measured using infrared thermal imaging. The leaf temperatures of ppd5-2ost1-3 and ost1-3 were substantially lower than those of ppd5-2 and Col-0 under drought stress (Fig. 5H). The water loss rates, stomatal conductance, and leaf transpiration in the leaves of ppd5-2ost1-3 were similar to ost1-3, but significantly higher than ppd5-2 and Col-0 (Supplemental Fig. S11). Consistent with the water loss rate result, the values of stomatal aperture in ppd5-2ost1-3 and ost1-3 were similar but significantly larger than that in ppd5-2 and Col-0 (Fig. 5, D and E). Because ppd5 mutants accumulated more ROS in guard cells under drought stress, we examined the ROS levels in the guard cells of ppd5-2ost1-3, Col-0, and the single mutants. After a 5-d drought stress treatment, the ROS level in the guard cells of ppd5-2ost1-3 was comparable with that in ost1-3, but significantly lower than that in ppd5-2 and Col-0, as indicated by the fluorescent intensity (Fig. 5, F and G). Collectively, these results show that the ppd5-2ost1-3 double mutant phenotypically resembles ost1-3 single mutant in response to drought stress and the ost1 mutation suppresses the drought resistance of ppd5-2.
Figure 5.
The ost1 mutation represses the drought-resistant phenotype of ppd5-2 mutant. A, Drought resistance assay of ppd5-2, ost1-3, ppd5-2ost1-3, and Col-0. 21-d-old plants was treated with drought stress for 8 d or 12 d and then rewatered for 2 d. B and C, The survival rate of ppd5-2, ost1-3, ppd5-2ost1-3, and Col-0 after 2 d recovery from drought stress treatment for 8 d (B) or 12 d (C). Values are means ± sd (n = 3 replicates, each replicate performing with 16 plants per line). The letters a and b above columns indicate significant difference relative to Col-0 and ppd5-2 mutant, respectively (P < 0.05, Student’s t test). D, Stomatal aperture of the middle leaves of 3-week-old ppd5-2, ost1-3, ppd5-2ost1-3, and Col-0 after drought stress treatment for 5 d (n ≥ 80). Stomatal aperture was calculated by the percentage of width/length ratio. E, Stomatal density of the middle leaves of 4-week-old ppd5-2, ost1-3, ppd5-2ost1-3, and Col-0. F, Representative images of CM-H2DCFDA staining of H2O2 in guard cell of ppd5-2, ost1-3, ppd5-2ost1-3, and Col-0 (n = 3 leaves, 30 stomata per leaf from 21-d-old plants after a 5-d drought stress treatment). G, Quantitative analysis of H2O2 levels in guard cells of ppd5-2, ost1-3, ppd5-2ost1-3, and Col-0 (n = 90). H, Leaf temperature of 21-d-old plants of ppd5-2, ost1-3, ppd5-2ost1-3, and Col-0 after drought stress treatment for 5 d.
The Drought Resistance of the thf1 Mutant Is Not Dependent on OST1
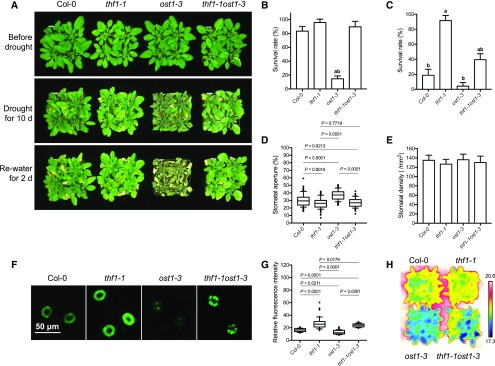
In our previous study, we showed that the drought-resistant mutant thf1-1 exhibited reduced stomatal aperture, reduced water loss, and increased H2O2 accumulation in guard cells compared with Col-0 under drought stress (Wang et al., 2016). Because the thf1 phenotypes resemble the phenotypes of ppd5, we tested whether the functions of THF1 and PPD5 in regulating drought resistance may be related. The thf1-1ost1-3 double mutant was generated, and drought resistance assays of thf1-1ost1-3, thf1-1, ost1-3 and Col-0 were carried out. We found that the thf1-1ost1-3 double mutant displayed phenotypes similar with thf1-1 but distinct from ost1-3. Under drought stress, thf1-1ost1-3 exhibited a higher survival rate than ost1-3, whereas thf1-1 showed the highest survival rate (Fig. 6, A–C). The leaf temperature of thf1-1ost1-3 was higher than that of ost1-3, with thf1-1 showing the highest leaf temperature (Fig. 6H). The stomatal aperture in thf1-1ost1-3 was also smaller than that in ost1-3, but was comparable with that in thf1-1 (Fig. 6, D and E). The ROS level in the guard cells of thf1-1ost1-3 double mutant was similar with that in thf1-1 but higher than that in ost1-3 under drought stress (Fig. 6, F and G). These results show that the ost1 mutation cannot suppress the drought resistance phenotype of thf1-1, suggesting that THF1 regulates plant drought response in an OST1-independent manner.
Figure 6.
Genetic interaction between thf1 and ost1 mutants. A, Drought resistant assay of thf1-1, ost1-3, thf1-1ost1-3, and Col-0. 21-d-old plants were treated with drought stress for 10 d and then re-watered for 2 d. B and C, The survival rate of thf1-1, ost1-3, thf1-1ost1-3, and Col-0 after 2 d recovery from drought stress treatment for 8 d (B) or 12 d (C). Values are means ± sd (n = 3 replicates, each replicate contained 16 plants per line). The letters a and b above columns indicate significant difference relative to Col-0 and thf1-1 mutant, respectively (P < 0.05, Student’s t test). D, Stomatal aperture in the middle leaves of 21-d-old plants of thf1-1, ost1-3, thf1-1ost1-3, and Col-0 after drought stress treatment for 5 d (n ≥ 80). Stomatal aperture was calculated by the percentage of width/length ratio. E, Stomatal density of the middle leaves of 4-week-old plants of thf1-1, ost1-3, thf1-1ost1-3, and Col-0. F, Representative images of CM-H2DCFDA staining of H2O2 in guard cells of thf1-1, ost1-3, thf1-1ost1-3, and Col-0. G, Quantitative analysis of H2O2 levels in guard cells of thf1-1, ost1-3, thf1-1ost1-3, and Col-0 (n = 90). H, Leaf temperature of 21-d-old plants of thf1-1, ost1-3, thf1-1ost1-3, and Col-0 after drought stress treatment for 5 d.
DISCUSSION
PsbP Family Proteins Function in Regulating Photosynthesis and Drought Stress Resistance
In plants, chloroplast is the organelle for photosynthesis and energy production, which is essential for plant growth and development. Chloroplast is also the compartment accommodating various metabolic pathways including ABA biosynthesis and ROS production. The regulation of these metabolic pathways is crucial for stomatal movement in response to environmental stresses (Zhu, 2016). An opened stoma allows CO2 to diffuse into leaf cells for photosynthesis. However, stomatal opening results in rapid water loss from leaves to the atmosphere, thus stomatal closing to reduce water loss is a critical response of plants to drought stress (Steudle, 2001; Maggio et al., 2006). Therefore, plants must balance photosynthesis and stress response by regulating stomatal apertures for appropriate growth under stress conditions.
Plants have evolved a series of mechanisms to regulate stomatal movement, one of which may involve the modulation of the photosystems in chloroplasts of guard cells. PsbP family proteins are important components of the oxygen-evolving complex (OEC) in chloroplasts and play crucial roles in photosynthesis. Some of the members in the PsbP family, including PPL1, PPL2 and PPD1, have been implicated in stress responses (Ishihara et al., 2007; Liu et al., 2012). In this study, we found that one of the PsbP members, PPD5, plays an important role in regulating stomatal movement under drought stress conditions. The ppd5 mutants accumulate more H2O2 in guard cells under drought stress, which leads to reduced stomatal aperture, decreased water loss, and thus improved drought stress resistance. It is possible that PPD5 is a component that can be modulated in the chloroplast to balance photosynthesis and stress response by regulating H2O2 production in guard cells. Chlorophyll fluorescence analyses in ppd5, ost1-3, ppd5-2ost1-3, and Col-0 wild type indicate that the mutations in PPD5 had no effect on the photosynthetic efficiency and capacity (Supplemental Fig. S1, E–H). A significant decrease in NPQ was observed in the ost1-3 and ppd5-2ost1-3 mutants (Supplemental Fig. S11), which suggests that the PPD5-OST1 interaction may serve as a photoprotective regulator via dissipating excess energy through heat emission.
Chloroplast-Affected ROS Accumulation Regulates Plant Drought Response
In our previous study, we found that the mutations in the HCF106 gene promote ROS accumulation in guard cells and improve drought resistance. HCF106 interacts with THF1, and this complex functions in thylakoid formation and chloroplast development. The thf1-1 resembles hcf106-1 under drought stress conditions (Wang et al., 2016). In this study, we identified another drought-resistant mutant, ppd5-2, that exhibits increased ROS accumulation, reduced stomatal aperture, and reduced water loss compared with the wild type under drought stress conditions. Unlike thf1-1 and hcf106-1, which show obvious developmental phenotypes (Huang et al., 2006; Wang et al., 2016), the ppd5 mutants grow normally like Col-0, suggesting that PPD5 is not important for plant development under normal growth conditions. These three mutants all exhibit increased ROS in guard cells and show drought resistance. This suggests that chloroplast-controlled ROS production positively regulate stomatal closure and drought resistance. However, PPD5 seems to function differently from HCF106 and THF1 in the control of ROS production, since ROS accumulation in the ppd5 but not thf1 mutant is dependent on OST1 (Fig. 7). HCF106 and THF1 are essential for chloroplast development, and mutations in THF1 or HCF106 result in malfunction of the photosystems in chloroplasts, causing increased accumulation of ROS in guard cells. This excess ROS at the cost of dysfunction of chloroplasts contribute to drought resistance, but it also causes growth defects. In contrast, ppd5 mutations result in ROS accumulation but do not cause growth defects. It appears that PPD5 may be a chloroplastic component that can be actively modulated to control ROS production in guard cells in response to drought stress. This notion is supported by our observation that PPD5 can be phosphorylated by OST1, which is a key kinase in the ABA signaling pathway.
Figure 7.
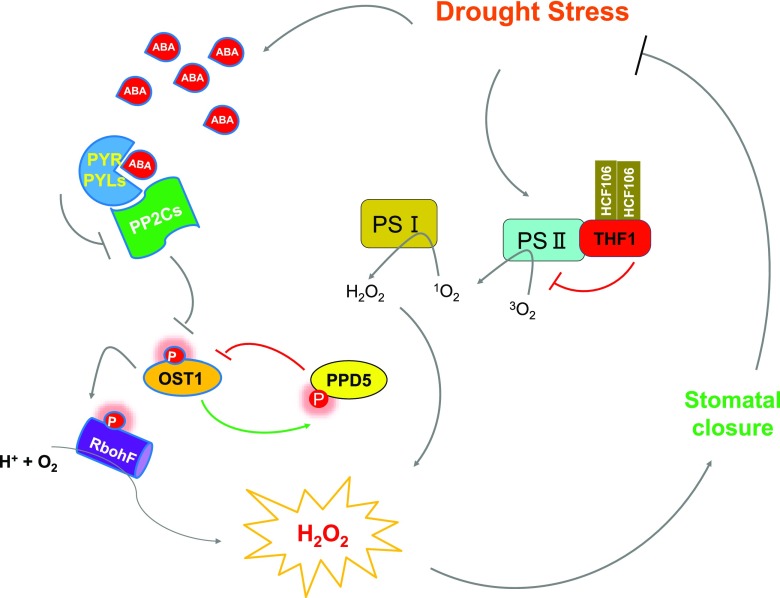
A working model depicting the function of the two chloroplast proteins in plant drought resistance. Drought stress promotes the biosynthesis of ABA and activates the ABA signaling pathway. In addition to phosphorylating the plasma membrane-bound NADPH oxidase RbohF and promoting the production of H2O2, OST1 can also interact and phosphorylate the chloroplast protein PPD5, which negatively regulates ROS and ABA accumulation under drought stress conditions. This negative regulation may involve a competition for OST1 between PPD5 and other OST1 substrate proteins. Another chloroplast protein, THF1, modulates H2O2 accumulation and plant drought resistance in an OST1-independent manner.
OST1 Stabilizes PPD5 by Phosphorylation
Different types of posttranslational modifications (PTMs), including acetylation, methylation, S-nitrosylation, sumoylation, glycosylation, and phosphorylation, have been identified in chloroplast proteins, and are thought to be important for chloroplast function (Lehtimäki et al., 2015; Grabsztunowicz et al., 2017). Among these PTMs, protein phosphorylation has been recognized as a major PTM to modulate photosynthetic light reactions, carbon assimilation, and gene expression in chloroplasts (Baginsky, 2016; Grieco et al., 2016). Phosphorylation of proteins in chloroplasts was mostly carried out by chloroplast-localized kinases, including the well-characterized photosynthetic machinery protein kinases STN7 and STN8 (White-Gloria et al., 2018). In this study, we discovered phosphorylation of the chloroplast protein PPD5 by the protein kinase OST1, which is not a chloroplast protein (Supplemental Fig. S7). Interestingly, PPD5 and OST1 appear to interact near or around the chloroplast (Fig. 3C; Supplemental Fig. S5), suggesting that the phosphorylation may occur in the cytoplasmic side of the chloroplast membrane. PPD5 was detected in both the thylakoid membrane and stroma of the chloroplast (Fig. 2B). Alternatively, PPD5 may be phosphorylated before being imported into the chloroplast. Protein phosphorylation has been implicated in regulating the import of chloroplast proteins (Zufferey et al., 2017) and the stability of the chloroplast F1 ATP synthase (Schmidt et al., 2017). Our results indicate that phosphorylation of PPD5 by OST1 stabilizes PPD5 (Fig. 4, E–G) but does not affect its import to and localization in chloroplasts (Supplemental Fig. S, 9 and 10). Indeed, the fluorescent intensity of PPD5-YFP in the chloroplasts of Col-0 wild type was clearly higher than that in the ost1 mutant (Supplemental Fig. S9), which further supports the role of OST1 in modulating PPD5 stability. In addition to the regulation of photosynthetic light reactions, protein phosphorylation in chloroplasts was also suggested to be a strategy for coping with environmental stresses such as heat and drought (Chen et al., 2013; Chen and Hoehenwarter, 2015). In this study, we found that PPD5 is a chloroplast factor affecting stomatal movement and drought resistance (Figs. 1 and 2). Therefore, it is possible that the OST1-mediated PPD5 phosphorylation is a stress acclimation mechanism transducing drought stress signals into the action of chloroplasts to control stomatal movement.
PPD5 Modulates Plant Drought Resistance Through an OST1-Dependent Pathway
Drought stress activates OST1 (aka SnRK2.6), which mediates the phosphorylation of various protein substrates. Phosphorylation of RbohF by OST1 results in ROS accumulation in the cytoplasm (Sirichandra et al., 2009); and phosphorylation of the anion channel SLAC1 promotes anion efflux (Vahisalu et al., 2008), whereas phosphorylation of KAT1 inhibits K+ influx (Islam et al., 2015). These phosphorylation events mediated by OST1 contribute to promoting stomatal closure under drought stress conditions. In this study, we found that PPD5 is another substrate of OST1 and phosphorylation of PPD5 increases its stability (Fig. 4, C–G). However, unlike other OST1-mediated phosphorylation events, phosphorylation of PPD5 may attenuate stomatal closure (Fig. 7). The ppd5 mutations cause ROS accumulation in guard cells and enhance stomatal closure and drought resistance. Therefore, under drought stress conditions, phosphorylation of PPD5 by activated OST1 stabilizes PPD5, limiting ROS accumulation and thus inhibiting stomatal closure. This could be important for fine-tuning stomatal aperture in response to drought stress. The ppd5 and ost1 mutants display opposite phenotypes in ROS accumulation and drought resistance, whereas the ost1ppd5 double mutant resembles the phenotypes of ost1 (Fig. 5). This suggests that the ost1 mutation is genetically epistatic to the ppd5 mutation and accumulation of ROS in the guard cells of ppd5 mutant might need functional OST1. Because phosphorylated PPD5 showed stronger interaction with OST1 than nonphosphorylatable variants (Supplemental Fig. S6), the binding of phosphorylated PPD5 with OST1 may sequestrate OST1 in the chloroplast membrane and thus may attenuate the function of OST1 in phosphorylating other substrates such as the plasma membrane-localized RbohF (Fig. 7). Therefore, the interaction between PPD5 and OST1 could be a mechanism to regulate the availability of OST1 to interact and phosphorylate other substrate proteins such as RbohF.
The communication between the chloroplast and cytoplasm or nucleus is achieved via anterograde and retrograde signaling pathways. The retrograde signaling pathways play important roles in regulating plant drought resistance by controlling stomatal movement (Chan et al., 2016). Several molecules have been identified as the chloroplast retrograde signals, including methylerythritol cyclodiphosphate (MEcPP) and 3′-phosphoadenosine 5′-phosphate (PAP; Estavillo et al., 2011; Xiao et al., 2012). The chloroplast-generated H2O2 is also regarded as a retrograde signal owing to its small size, lower toxicity, and longer half-life compared with other ROS (Møller and Sweetlove, 2010). Enhanced ROS levels have been proposed to elevate ABA biosynthesis or inhibit ABA degradation, and the cross-talk between ROS and ABA signaling regulates guard cell movement under drought stress (Galvez-Valdivieso et al., 2009; Suzuki et al., 2013; Mittler and Blumwald, 2015). Because the ppd5 mutants accumulate more H2O2 in guard cells under drought stress (Fig. 2, F and G) and PPD5 is a substrate of the ABA signaling component OST1 (Fig. 4), we asked whether the increased ROS affects ABA biosynthesis. The ABA contents in leaves of wild type and ppd5 mutants under both normal and drought stress conditions were determined, but no difference in the ABA content was detected when plants were grown under normal conditions and after a 3-d drought treatment. However, the ABA content in ppd5 mutants accumulated to a higher level than that in wild type plants after 6-d drought treatment (Supplemental Fig. S12), which suggests that PPD5 may be involved in regulating ABA metabolism. It is also possible that the ppd5 mutation alters the environment in the chloroplast and thus favors ABA biosynthesis in this organelle.
MATERIALS AND METHODS
Plant Materials and Growth Conditions
The Arabidopsis (Arabidopsis thaliana) genetic materials used in this study are in the Columbia-0 background. The T-DNA insertion mutants, ppd5-2 (SALK_039106C), ppd5-3 (SAIL_511_G10), and ost1-3 (SALK_008068C), were obtained from the Arabidopsis Biological Resource Center. The thf1-1 was kindly provided by Dr. Jirong Huang. After genetic crossing, the ppd5-2ost1-3 and thf1-1ost1-3 homozygous double mutants were identified by using PCR genotyping. For seedlings growing in petri dish, seeds were surface-sterilized and stored in sterile water at 4°C for 2 d, and the seeds were then sown on 0.5× Murashige and Skoog medium (pH 5.8) containing 1% (w/v) Suc and 0.6% (w/v) agar. For plants growing in soil, the 7-d-old seedlings grown in agar plates were transplanted in soil and grow in a growth room at 22°C with 16-h light/8-h dark (long-day) condition.
To obtain the PPD5pro:PPD5-YFP fusion construct, the genomic region containing the PPD5 gene with 1500-bp native promoter was amplified and cloned into the pCambia1300-N1-YFP vector. The 35S:PPD5-YFP fusion was constructed by inserting the full-length coding sequence (CDS) of PPD5 into a modified pCambia1300-N1-YFP vector with the 35S promoter. These constructs were then transformed into Agrobacterium stain GV3101 and introduced to the ppd5-2 mutant using the floral dip method. The T3 homozygous transgenic seedlings were selected and used for further analysis. For the analysis of PPD5 in different genotypes, a single homozygous T3 of the PPD5 overexpression line PPD5OE-#3 was selected to cross with ost1-3, and the homozygous lines were selected from the F2 generation and used for immunoblotting and subcellular localization analysis.
Plastid Fractionation
The isolation of chloroplast and thylakoid membrane and stromal proteins from 21-d-old seedlings was performed as previously described (Huang et al., 2006). The CP47 (AS04 038), RbcL (AS03 037), and THF1 (AS07 240) antibodies used for immunoblot analysis were purchased from Agrisera.
Yeast Two-Hybrid Assay
Yeast two-hybrid assay was carried out as described previously (Ren et al., 2017). The open reading frames of SnRK2.1 - SnRK2.10, and PPD5, PsbP, PPD1 were amplified by PCR from the complementary DNA (cDNA) of Arabidopsis Col-0 plants and cloned into pGAD-T7 vector and pGBK-T7 vector, respectively. To generate truncated OST1 and PPD5, the corresponding CDS of OST1 and PPD5 were amplified and cloned into pGAD-T7 vector and pGBK-T7 vector, forming OST1-a (1-217 amino acid), OST1-b (1-317 amino acid), OST1-c (283-317 amino acid), OST1-d (283-362 amino acid), and PPD5-a (1-79 amino acid), PPD5-b (80-297 amino acid), PPD5-c (80-280 amino acid), PPD5-d (1-280 amino acid) constructs. After the constructs were confirmed by sequencing, each pair of the combinations of the AD and BD constructs were co-transformed into the yeast strain AH109 (Saccharomyces cerevisiae), and empty vectors were cotransformed as negative controls. The transformed yeast cells were plated on SD/-Leu/-Trp agar medium for 2 d at 30°C; the well-grown clones were then used to test the interaction. The transformants were spotted on SD/-Leu/-Trp/-His/-Ade agar medium with a series of dilutions (10−1, 10−2, 10−3) and incubated at 30°C for 3 d before photographing. The primers used were listed in the Supplemental Table S1.
BiFC Assay
The full-length CDS of OST1, SnRK2.2, and SnRK2.3 were amplified and cloned into the p2YC vector to generate an OST1-CYFP, SnRK2.2-CYFP, and SnRK2.3-CYFP constructs, and the full-length CDS of PPD5, PsbP, PPD1 and the truncated PPD5-b (80-297 amino acid), PPD5-d (1-280 amino acid) were cloned into the p2YN vector to create a PPD5-NYFP, PsbP-NYFP, PPD1-NYFP, PPD5-b-NYFP, and PPD-d-NYFP constructs. The plasmids were transformed into Agrobacterium stain GV3101, and the combination of plasmids were infiltrated into tobacco (Nicotiana benthamiana) leaves using the method as described by Waadt and Kudla (2008). The leaves were incubated at 22°C in a greenhouse for 36–48 h, and the fluorescence signals were observed and photographed using a confocal laser-scanning microscope (Zeiss LSM880). The fluorescent signals in guard cells were detected by using the confocal microscope Leica TSC SP8 STED 3X.
Split-LUC Complementation Assay
The full-length CDS of OST1 gene was amplified and cloned into pCambia-NLUC vector to generate an OST1-NLUC construct, and the full-length CDS of PPD5 gene was cloned into pCambia-CLUC to create a PPD5-CLUC construct. The constructs were transformed into Agrobacterium stain GV3101, and the combination of Agrobacteria were cotransferred into tobacco leaves. After incubation at 22°C in a greenhouse for 48 h, the leaves were infiltrated with 1 μm luciferin and then put in dark for 10 min. The luminescence signals were detected using a low-light cooled CCD imaging system (Lumazone; Roper Scientific).
Gene Expression Analysis
The seedlings (12-d-old), shoot and root (from 21-d-old seedlings), leaves, stems, flowers, siliques (from 6-week-old plants), and seeds of Col-0 wild type were used for tissue-specific expression analysis. For gene expression under stress conditions, 12-d-old seedlings were treated with air dry for the indicated hours showing in the figures. Total RNA was extracted with TRIzol reagent (Invitrogen), and cDNAs were synthesized by reverse transcription using the iScript cDNA synthesis kit (Bio-Rad). All quantitative real-time PCR analyses in this study were performed in a CFX96 Real-time system (Bio-Rad) using the ChamQ SYBR qPCR Master Mix (Vazyme Biotech) following the manufacturer’s protocol. Each analysis included three biological replicates. AtUBQ5 was used as an internal control. The primers used were listed in the Supplemental Table S1.
Subcellular Localization
To determine the subcellular localization of PPD5, 7-d-old transgenic seedlings harboring PPD5pro:PPD5-YFP were collected for detecting the YFP signals by a confocal laser-scanning microscope (Zeiss LSM880). The sequences of OST1, PPD5, and its truncated forms PPD5-b (80-297 amino acid), PPD5-d (1-280 amino acid) were cloned from the constructs used in Y2H to generate 35S:OST1-YFP, 35S:PPD5-YFP, 35S:PPD5-b-YFP, and 35S:PPD5-d-YFP constructs. For analysis of the subcellular localization of PPD5′s variants PPD5T283A, PPD5S285A and PPD5T283AS285A, 35S:PPD5T283A-YFP, 35S:PPD5S285A-YFP, and 35S:PPD5T283AS285A-YFP constructs were generated from the normal 35S:PPD5-YFP construct using site mutagenesis. The constructs were transformed into Agrobacterium stain GV3101 and transferred into tobacco leaves. After incubation at 22°C in a greenhouse for 48 h, the YFP signals were detected by using a confocal laser-scanning microscope (Zeiss LSM880).
Physiological Assays
For drought resistance assay, 7-d-old seedlings in agar plates were transferred to soil and grow in a growth room at 22°C with 10 h light/14 h dark (short day) cycle for 2 weeks, and the plants were then subjected to drought treatment by withholding water for the indicated days showing in the figures. The plants were rewatered for 2 or 3 d, and the survival rates were counted. The experiments were performed three times, and each experiment contained 12 plants per line.
For the measurement of stomatal aperture, 3-week-old plants grown at 22°C with 10-h light/14-h dark (short day) cycles were withheld water for 5 d and the stomatal apertures were measured by using a leaf surface imprint method as described previously (Yu et al., 2008). Five leaves per line and ten fields per leaf were analyzed by taking images using the Olympus DP72 imaging system for statistical analysis of the stomatal aperture and stomatal density.
For the measurement of water loss of detached leaves, plants were grown under short-day conditions for 21 d with additional 5-d’s withhold-watering, and the fully expanded detached leaves (three or four individual leaves per genotype) were harvested and weighed using an electronic balance at the indicated times showing in the figures. A percentage of the decreased fresh weight was calculated as the water loss rate. The experiments were performed three times.
For the measurement of leaf temperature, 3-week-old plants grown at 22°C under short-day conditions were withheld water for 5 d and photographed with an IR imager (FLIR) system. The experiments were performed three times, and each experiment contained three replicates.
The stomatal conductance and leaf transpiration were measured using the sixth leaf with the LI-6400XT Portable Photosynthesis System (LI-COR Biosciences) as described previously (Wang et al., 2018). Leaves were placed in the chamber set at an ambient CO2 concentration of 400 μmol mol−1 and a temperature of 22°C, and irradiance was set to 300 μmol m−2 s−1. A reading was recorded every 2 min after the IRGA (infrared gas analyzer) conditions were stabilized.
Analysis of Chlorophyll Fluorescence
Chlorophyll fluorescence were analyzed using a portable fluorometer PAM-2500 (Walz) as described in the manufacturer’s instructions. The 21-d-old plants grown in soil under short-day condition were subjected to withholding water for 0 or 5 d, and then the mature rosette leaves of plants were collected for detecting the chlorophyll fluorescence after a 30-min darkness-adaption. A slow kinetics program regime of AL exposure times, saturating light pulses, and real-time recording of parameters Fv/Fm, ΦY(II), qP, and NPQ were performed by the PAM-2500 and the associated software PamWin-3 (http://www.walz.com/).
H2O2 Staining in Guard Cells
The CM-H2DCFDA staining assay to detect H2O2 content in guard cells was performed as described previously (Wang et al., 2016). Briefly, the abaxial epidermal strips of the leaves were harvested from 23-d-old plants grown under short-day conditions after withhold-watering for additional 5 d and floated in 0.1 m potassium phosphate buffer (pH 7.0). The samples were transferred to 2 μm (final concentration) CM-H2DCFDA in 0.1 m potassium phosphate buffer (pH 7.0) and incubated for 20 min at 22°C in dark. To remove the excess staining buffer, the samples were washed three times with a wash buffer (0.1 mm KCl, 0.1 mm MgCl2) for 10 min each time. The fluorescent signals in the guard cells of the strips were observed using an Olympus DP72 microscope.
Protein Purification and In Vitro Phosphorylation Assay
The purification of recombinant proteins was performed as described previously (Ren et al., 2017). The full-length CDS of PPD5 gene was amplified from Arabidopsis Col-0 cDNA and inserted into the pMAL-c5x vector. The construct was transformed into Escherichia coli BL21 (DE3) to express a recombinant MBP-PPD5 fusion protein. The point mutated forms PPD5T283A, PPD5S285A, PPD5T283AS285A were generated from the normal PPD5-pMal-c5x construct using site mutagenesis. The OST1 CDS was cloned and expressed in BL21 cells for expression of the 6×His-SUMO-OST1 recombinant protein. The in vitro phosphorylation assay was performed as described previously (Klimczak and Cashmore, 1993) with some modifications. Briefly, the purified protein of MBP-PPD5 or its variants MBP-PPD5T283A, MBP-PPD5S285A, MBP-PPD5T283AS285A was mixed with 6×His-SUMO-OST1 recombinant protein and incubated in the reaction buffer (50 mm Tris-HCl [pH 8.0], 10 mm MgCl2, 1 mm dithiothreitol, 10 mm ATP) containing 5 μCi [ 𝛾-32P ] ATP for 1 h at 37°C. The reaction mixtures were boiled at 98°C for 10 min after adding the 6× sample buffer, and the proteins were then separated by 10% SDS-PAGE. The gel was washed with a wash buffer containing 10% (v/v) ethanol and 10% (v/v) HAc for three times and autoradiographed using Fujifilm FLA 9000 plus DAGE according to the manufacturer instruction. For the AlphaScreen assay, the truncated PPD5 (33 aa-end) and its variants were generated and expressed as biotin-MBP-PPD5 recombinant proteins. Interactions between 6×His-SUMO-OST1 and biotin-MBP-PPD5s were assessed by luminescence-based AlphaScreen technology (Perkin Elmer) as previously described (Melcher et al., 2009).
Cell-Free Protein Degradation Assay
The cell-free protein degradation assay was conducted as described previously (Chen et al., 2018) with some modifications. Briefly, 10-d-old seedlings of Col-0, ost1-3, OST1OE-#5 plants grown in 0.5× MS plates were harvested and the total proteins were extracted in buffer (50 mm Tris-HCl, pH 8.0; 400 mm NaCl; 0.5% [v/v] Nonidet P-40; 10% [v/v] glycerol; 1 mm EDTA; 1 mm dithiothreitol; and 1 mm phenylmethylsulfonyl fluoride). Protein concentration was determined by using Qubit 2 Fluorometer (Invitrogen, Thermo Fisher Scientific). The purified recombinant MBP-PPD5 proteins were incubated with the same amounts of total proteins extracted from Col-0, ost1-3 and OST1OE-#5. The mixtures were incubated at 22°C for 0, 0.5, 1, 2, 3, 6, 9, 12, or 24 h. The reaction mixtures were then stopped by incubation in ice for 5 min followed by boiling at 98°C for 10 min after adding the 6× sample buffer. Protein abundances of MBP-PPD5 were determined by immunoblot assay using anti-MBP antibody (No. A00190-100, GenScript). The intensity of the bands was quantified by Image Lab software (version 5.2.1; Bio-Rad).
Detection of Phosphorylated Proteins by Phos-Tag
The 10-d-old seedlings were collected and subjected to 50 μm ABA treatment for 1 h, 0.8 m sorbitol for 1 h, or without treatment. The total proteins were extracted and separated on a 10% SDS-PAGE gel containing 100 μm MnCl2 and 50 μm Phos-tag (Phos-tag acrylamide AAL-107). A normal 10% SDS-PAGE gel without the Phos-tag was used as a control. The separated proteins were then transferred to a nitrocellulose filter membrane and immune-detected by an anti-GFP antibody (No. 11814460001; Roche). The intensity of the bands was quantified by Image Lab software (version 5.2.1; Bio-Rad).
Accession Numbers
Sequence data from this article can be found in The Arabidopsis Information Resource (http://www.arabidopsis.org/) under the following accession numbers: PPD5 (AT5G11450), PPD1 (AT4G15510), PPD2 (AT2G28605), PPD3 (AT1G76450), PPD4 (AT1G77090), PPD6 (AT3G56650), PPD7 (AT3G05410), PPL1 (AT3G55330), PPL2 (AT2G39470), PsbP (AT1G06680), OST1/SnRK2.6 (AT4G33950), SnRK2.1 (AT5G08590), SnRK2.2 (AT3G50500), SnRK2.3 (AT5G66880), SnRK2.4 (AT1G10940), SnRK2.5 (AT5G63650), SnRK2.7 (AT4G40010), SnRK2.8 (AT1G78290), SnRK2.9 (AT2G23030), SnRK2.10 (AT1G60940), THF1 (AT2G20890), ACT2 (AT3G18780), UBQ5 (AT3G62250).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. The stomatal conductance and transpiration rate were decreased in ppd5 mutant plants under drought stress.
Supplemental Figure S2. Molecular complementation of the drought-resistant phenotype of ppd5 mutant.
Supplemental Figure S3. Phylogenetic analysis of the PsbP family proteins in Arabidopsis.
Supplemental Figure S4. The interaction of PPDs and SnRK2s in yeast.
Supplemental Figure S5. PPD5 physically interacts with OST1 in guard cell.
Supplemental Figure S6. Alpha-Screen assay for PPD5-OST1 interactions.
Supplemental Figure S7. Analysis of the subcellular localization of PPD5 deletion variants and their interactions with OST1 by BiFC assay.
Supplemental Figure S8. The transcript and protein levels of PPD5-YFP in ppd5-2ost1-3 double mutant plants.
Supplemental Figure S9. Subcellular localization of PPD5-YFP in Col-0, ost1-3, ppd5-2, and ppd5-2ost1-3.
Supplemental Figure S10. Subcellular localization of PPD5 and its variants.
Supplemental Figure S11. The stomatal conductance and transpiration rate were increased in ppd5-2 mutant by ost1-3 mutation under drought stress.
Supplemental Figure S12. ABA content in rosette leaves during drought stress.
Supplemental Table S1. Primers used in this study.
Acknowledgments
We thank Dr. Minjie Cao for technical assistance and helpful suggestions.
Footnotes
This work was supported by the Chinese Academy of Sciences (CAS) (Strategic Priority Research Program; XDB27040108 to J.-K.Z.).
References
- Acharya BR, Jeon BW, Zhang W, Assmann SM (2013) Open Stomata 1 (OST1) is limiting in abscisic acid responses of Arabidopsis guard cells. New Phytol 200: 1049–1063 [DOI] [PubMed] [Google Scholar]
- Arabidopsis Interactome Mapping Consortium (2011) Evidence for network evolution in an Arabidopsis interactome map. Science 333: 601–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baginsky S. (2016) Protein phosphorylation in chloroplasts - a survey of phosphorylation targets. J Exp Bot 67: 3873–3882 [DOI] [PubMed] [Google Scholar]
- Bricker TM, Roose JL, Zhang P, Frankel LK (2013) The PsbP family of proteins. Photosynth Res 116: 235–250 [DOI] [PubMed] [Google Scholar]
- Cao M, Liu X, Zhang Y, Xue X, Zhou XE, Melcher K, Gao P, Wang F, Zeng L, Zhao Y, et al. (2013) An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants. Cell Res 23: 1043–1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan KX, Phua SY, Crisp P, McQuinn R, Pogson BJ (2016) Learning the languages of the chloroplast: Retrograde signaling and beyond. Annu Rev Plant Biol 67: 25–53 [DOI] [PubMed] [Google Scholar]
- Chen HH, Qu L, Xu ZH, Zhu JK, Xue HW (2018) EL1-like casein kinases suppress ABA signaling and responses by phosphorylating and destabilizing the ABA receptors PYR/PYLs in Arabidopsis. Mol Plant 11: 706–719 [DOI] [PubMed] [Google Scholar]
- Chen Y, Hoehenwarter W (2015) Changes in the phosphoproteome and metabolome link early signaling events to rearrangement of photosynthesis and central metabolism in salinity and oxidative stress response in Arabidopsis. Plant Physiol 169: 3021–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YE, Zhao ZY, Zhang HY, Zeng XY, Yuan S (2013) The significance of CP29 reversible phosphorylation in thylakoids of higher plants under environmental stresses. J Exp Bot 64: 1167–1178 [DOI] [PubMed] [Google Scholar]
- De Las Rivas J, Heredia P, Roman A (2007) Oxygen-evolving extrinsic proteins (PsbO,P,Q,R): Bioinformatic and functional analysis. Biochim Biophys Acta 1767: 575–582 [DOI] [PubMed] [Google Scholar]
- Estavillo GM, Crisp PA, Pornsiriwong W, Wirtz M, Collinge D, Carrie C, Giraud E, Whelan J, David P, Javot H, et al. (2011) Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23: 3992–4012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedoroff NV, Battisti DS, Beachy RN, Cooper PJ, Fischhoff DA, Hodges CN, Knauf VC, Lobell D, Mazur BJ, Molden D, et al. (2010) Radically rethinking agriculture for the 21st century. Science 327: 833–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foyer CH, Parry M, Noctor G (2003) Markers and signals associated with nitrogen assimilation in higher plants. J Exp Bot 54: 585–593 [DOI] [PubMed] [Google Scholar]
- Fujii H, Chinnusamy V, Rodrigues A, Rubio S, Antoni R, Park SY, Cutler SR, Sheen J, Rodriguez PL, Zhu JK (2009) In vitro reconstitution of an abscisic acid signalling pathway. Nature 462: 660–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvez-Valdivieso G, Fryer MJ, Lawson T, Slattery K, Truman W, Smirnoff N, Asami T, Davies WJ, Jones AM, Baker NR, Mullineaux PM (2009) The high light response in Arabidopsis involves ABA signaling between vascular and bundle sheath cells. Plant Cell 21: 2143–2162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabsztunowicz M, Koskela MM, Mulo P (2017) Post-translational modifications in regulation of chloroplast function: Recent advances. Front Plant Sci 8: 240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grieco M, Jain A, Ebersberger I, Teige M (2016) An evolutionary view on thylakoid protein phosphorylation uncovers novel phosphorylation hotspots with potential functional implications. J Exp Bot 67: 3883–3896 [DOI] [PubMed] [Google Scholar]
- Huang J, Taylor JP, Chen JG, Uhrig JF, Schnell DJ, Nakagawa T, Korth KL, Jones AM (2006) The plastid protein THYLAKOID FORMATION1 and the plasma membrane G-protein GPA1 interact in a novel sugar-signaling mechanism in Arabidopsis. Plant Cell 18: 1226–1238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W, Chen Q, Zhu Y, Hu F, Zhang L, Ma Z, He Z, Huang J (2013) Arabidopsis thylakoid formation 1 is a critical regulator for dynamics of PSII-LHCII complexes in leaf senescence and excess light. Mol Plant 6: 1673–1691 [DOI] [PubMed] [Google Scholar]
- Ifuku K, Ishihara S, Sato F (2010) Molecular functions of oxygen-evolving complex family proteins in photosynthetic electron flow. J Integr Plant Biol 52: 723–734 [DOI] [PubMed] [Google Scholar]
- Ifuku K, Ishihara S, Shimamoto R, Ido K, Sato F (2008) Structure, function, and evolution of the PsbP protein family in higher plants. Photosynth Res 98: 427–437 [DOI] [PubMed] [Google Scholar]
- Ifuku K, Yamamoto Y, Ono TA, Ishihara S, Sato F (2005) PsbP protein, but not PsbQ protein, is essential for the regulation and stabilization of photosystem II in higher plants. Plant Physiol 139: 1175–1184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara S, Takabayashi A, Ido K, Endo T, Ifuku K, Sato F (2007) Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis. Plant Physiol 145: 668–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Islam MM, Ye W, Matsushima D, Khokon MA, Munemasa S, Nakamura Y, Murata Y (2015) Inhibition by acrolein of light-induced stomatal opening through inhibition of inward-rectifying potassium channels in Arabidopsis thaliana. Biosci Biotechnol Biochem 79: 59–62 [DOI] [PubMed] [Google Scholar]
- Klimczak LJ, Cashmore AR (1993) Purification and characterization of casein kinase I from broccoli. Biochem J 293: 283–288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehtimäki N, Koskela MM, Mulo P (2015) Posttranslational modifications of chloroplast proteins: An emerging field. Plant Physiol 168: 768–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Yang H, Lu Q, Wen X, Chen F, Peng L, Zhang L, Lu C (2012) PsbP-domain protein1, a nuclear-encoded thylakoid lumenal protein, is essential for photosystem I assembly in Arabidopsis. Plant Cell 24: 4992–5006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maggio A, Zhu JK, Hasegawa PM, Bressan RA (2006) Osmogenetics: Aristotle to Arabidopsis. Plant Cell 18: 1542–1557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melcher K, Ng LM, Zhou XE, Soon FF, Xu Y, Suino-Powell KM, Park SY, Weiner JJ, Fujii H, Chinnusamy V, et al. (2009) A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature 462: 602–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignolet-Spruyt L, Xu E, Idänheimo N, Hoeberichts FA, Mühlenbock P, Brosché M, Van Breusegem F, Kangasjärvi J (2016) Spreading the news: Subcellular and organellar reactive oxygen species production and signalling. J Exp Bot 67: 3831–3844 [DOI] [PubMed] [Google Scholar]
- Mittler R, Blumwald E (2015) The roles of ROS and ABA in systemic acquired acclimation. Plant Cell 27: 64–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Møller IM, Sweetlove LJ (2010) ROS signalling--specificity is required. Trends Plant Sci 15: 370–374 [DOI] [PubMed] [Google Scholar]
- Munemasa S, Hauser F, Park J, Waadt R, Brandt B, Schroeder JI (2015) Mechanisms of abscisic acid-mediated control of stomatal aperture. Curr Opin Plant Biol 28: 154–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata Y, Mori IC, Munemasa S (2015) Diverse stomatal signaling and the signal integration mechanism. Annu Rev Plant Biol 66: 369–392 [DOI] [PubMed] [Google Scholar]
- Mustilli AC, Merlot S, Vavasseur A, Fenzi F, Giraudat J (2002) Arabidopsis OST1 protein kinase mediates the regulation of stomatal aperture by abscisic acid and acts upstream of reactive oxygen species production. Plant Cell 14: 3089–3099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park SY, Fung P, Nishimura N, Jensen DR, Fujii H, Zhao Y, Lumba S, Santiago J, Rodrigues A, Chow TF, et al. (2009) Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324: 1068–1071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Z, Wang Z, Zhou XE, Shi H, Hong Y, Cao M, Chan Z, Liu X, Xu HE, Zhu JK (2017) Structure determination and activity manipulation of the turfgrass ABA receptor FePYR1. Sci Rep 7: 14022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato N. (2010) Phylogenomic and structural modeling analyses of the PsbP superfamily reveal multiple small segment additions in the evolution of photosystem II-associated PsbP protein in green plants. Mol Phylogenet Evol 56: 176–186 [DOI] [PubMed] [Google Scholar]
- Schmidt C, Beilsten-Edmands V, Mohammed S, Robinson CV (2017) Acetylation and phosphorylation control both local and global stability of the chloroplast F1 ATP synthase. Sci Rep 7: 44068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sirichandra C, Gu D, Hu HC, Davanture M, Lee S, Djaoui M, Valot B, Zivy M, Leung J, Merlot S, Kwak JM (2009) Phosphorylation of the Arabidopsis AtrbohF NADPH oxidase by OST1 protein kinase. FEBS Lett 583: 2982–2986 [DOI] [PubMed] [Google Scholar]
- Steudle E. (2001) The cohesion-tension mechanism and the acquisition of water by plant roots. Annu Rev Plant Physiol Plant Mol Biol 52: 847–875 [DOI] [PubMed] [Google Scholar]
- Suzuki N, Miller G, Salazar C, Mondal HA, Shulaev E, Cortes DF, Shuman JL, Luo X, Shah J, Schlauch K, et al. (2013) Temporal-spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell 25: 3553–3569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vahisalu T, Kollist H, Wang YF, Nishimura N, Chan WY, Valerio G, Lamminmäki A, Brosché M, Moldau H, Desikan R, et al. (2008) SLAC1 is required for plant guard cell S-type anion channel function in stomatal signalling. Nature 452: 487–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R, Kudla J (2008) In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). CSH Protoc 2008: pdb prot4995. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang F, Hong Y, Huang J, Shi H, Zhu JK (2016) Two chloroplast proteins suppress drought resistance by affecting ROS production in guard cells. Plant Physiol 172: 2491–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Wang F, Hong Y, Yao J, Ren Z, Shi H, Zhu J-K (2018) The flowering repressor SVP confers drought resistance in Arabidopsis by regulating abscisic acid catabolism. Mol Plant 11: 1184–1197 [Google Scholar]
- White-Gloria C, Johnson JJ, Marritt K, Kataya A, Vahab A, Moorhead GB (2018) Protein kinases and phosphatases of the plastid and their potential role in starch metabolism. Front Plant Sci 9: 1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Savchenko T, Baidoo EE, Chehab WE, Hayden DM, Tolstikov V, Corwin JA, Kliebenstein DJ, Keasling JD, Dehesh K (2012) Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149: 1525–1535 [DOI] [PubMed] [Google Scholar]
- Yi X, Hargett SR, Liu H, Frankel LK, Bricker TM (2007) The PsbP protein is required for photosystem II complex assembly/stability and photoautotrophy in Arabidopsis thaliana. J Biol Chem 282: 24833–24841 [DOI] [PubMed] [Google Scholar]
- Yu H, Chen X, Hong YY, Wang Y, Xu P, Ke SD, Liu HY, Zhu JK, Oliver DJ, Xiang CB (2008) Activated expression of an Arabidopsis HD-START protein confers drought tolerance with improved root system and reduced stomatal density. Plant Cell 20: 1134–1151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu JK. (2016) Abiotic stress signaling and responses in plants. Cell 167: 313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufferey M, Montandon C, Douet V, Demarsy E, Agne B, Baginsky S, Kessler F (2017) The novel chloroplast outer membrane kinase KOC1 is a required component of the plastid protein import machinery. J Biol Chem 292: 6952–6964 [DOI] [PMC free article] [PubMed] [Google Scholar]