PTC2, an AT-hook DNA binding protein, is required for tapetal programmed cell death and pollen wall patterning in rice.
Abstract
The timely programmed cell death (PCD) of the tapetum, the innermost somatic anther cell layer in flowering plants, is critical for pollen development, including the deposition and patterning of the pollen wall. Although several genes involved in tapetal PCD and pollen wall development have been characterized, the underlying regulatory mechanism remains elusive. Here we report that PERSISTENT TAPETAL CELL2 (PTC2), which encodes an AT-hook nuclear localized protein in rice (Oryza sativa), is required for normal tapetal PCD and pollen wall development. The mutant ptc2 showed persistent tapetal cells and abnormal pollen wall patterning including absent nexine, collapsed bacula, and disordered tectum. The defective tapetal PCD phenotype of ptc2 was similar to that of a PCD delayed mutant, ptc1, in rice, while the abnormal pollen wall patterning resembled that of a pollen wall defective mutant, Transposable Element Silencing Via AT-Hook, in Arabidopsis (Arabidopsis thaliana). Levels of anther cutin monomers in ptc2 anthers were significantly reduced, as was expression of a series of lipid biosynthetic genes. PTC2 transcript and protein were shown to be present in the anther after meiosis, consistent with the observed phenotype. Based on these data, we propose a model explaining how PTC2 affects anther and pollen development. The characterization of PTC2 in tapetal PCD and pollen wall patterning expands our understanding of the regulatory network of male reproductive development in rice and will aid future breeding approaches.
Pollen grains, the male gametophytes in flowering plants, form in anthers. Their development requires cell division and differentiation, programmed cell death (PCD), and support from sporophytic tissue development for lipid synthesis, modification, and transport. Mature pollen grains are protected by the exine, a two-layer structure including the inner nexine and outer sexine. The sexine is further divided into the tectum and the bacula. The main exine biopolymer, sporopollenin, is composed predominantly of lipidic and phenolic compounds and their derivatives (Ariizumi and Toriyama, 2011; Shi et al., 2015).
Genetic and biochemical studies have revealed the importance of tapetal cell death and the metabolism of lipids and phenolics in pollen grain development (Ariizumi and Toriyama, 2011; Shi et al., 2015). Tapetal cell death in flowering plants is a typical type of PCD, with characteristic cell shrinkage, DNA degradation, and the caspase-like proteolytic activity (Li et al., 2006; van Doorn et al., 2011; Niu et al., 2013; Daneva et al., 2016). Previous studies indicated that disruption of tapetal PCD, either delay or promotion, often adversely affect pollen development resulting in male sterility. The atmyb103 (Higginson et al., 2003) and CEP1 overexpressor (Zhang et al., 2014a) in Arabidopsis (Arabidopsis thaliana), PET1-CMS mutation in Helianthus annuus (Balk and Leaver, 2001) and osdex1 (Yu et al., 2016) in rice (Oryza sativa) displayed premature tapetal PCD. However, the mechanisms behind these observations were poorly understood. In rice, several basic Helix–Loop–Helix transcription factors including Undeveloped Tapetum1 (UDT1; Jung et al., 2005), Tapetum Degeneration Retardation (TDR; Li et al., 2006) Eternal Tapetum1 (EAT1; Ji et al., 2013; Niu et al., 2013), and TDR Interacting Protein2 (TIP2; Fu et al., 2014; Ko et al., 2014), one MYB transcription factor GAMYB (Aya et al., 2009), and one PHD finger DNA binding protein Persistent Tapetal Cell1 (PTC1; Li et al., 2011) have been reported to be required for tapetal PCD.
In rice, lipid and phenolic metabolism are indispensable for sporopollenin synthesis and pollen wall formation. Genes associated with lipid metabolism include Glycerol-3-Phosphate Acyltransferase3 (GPAT3; Men et al., 2017), CYP704B2 (Li et al., 2010), CYP703A3 (Yang et al., 2014), Defective Pollen Wall (DPW; Shi et al., 2011), Acyl-CoA Synthetase12 (ACOS12; Yang et al., 2017), Oryza sativa ATP Binding Cassette G26 (OsABCG26; Zhao et al., 2015), OsABCG15 (Qin et al., 2013), OsABCG3 (Chang et al., 2018), and OsC6 (Zhang et al., 2010). Loss of function of these genes often leads to defective pollen wall formation, indicating the important role of lipid metabolism on pollen wall formation. Reported genes involved in phenolic metabolism include Oryza sativa Polyketide Synthase1 (OsPKS1; Wang et al., 2013; Zou et al., 2017), OsPKS2 (Wang et al., 2013; Zhu et al., 2017), No Pollen1 (NP1; Chang et al., 2016; Liu et al., 2017), and DPW2 (Xu et al., 2017). Mutants of these genes also display abnormal pollen wall patterning, suggesting that cross linking between aromatic and aliphatic monomers is also important for pollen wall formation. The divergent functions of reported sporopollenin synthesis genes on different processes of pollen wall formation suggest that pollen wall formation is a well-coordinated process, manipulated by a complicated network of transcription factors, enzymes, and their substrates and products. However, our knowledge of this regulatory network is still limited.
Known tapetal PCD mutants show different types of pollen wall patterning. The mutants osdex1 (Yu et al., 2016), udt1 (Jung et al., 2005), tip2 (Fu et al., 2014; Ko et al., 2014), tdr (Li et al., 2006), and gamyb (Aya et al., 2009) are defective in pollen wall formation. The mutants ptc1 (Li et al., 2011) and eat1 (Ji et al., 2013; Niu et al., 2013) show abnormal pollen wall development, all of which contain a two-layer exine. It has been reported that Transposable Element Silencing Via AT-Hook (TEK), an AT-hook nuclear localized (AHL) family protein, is required for nexine formation in Arabidopsis. TEK regulates a series of arabinogalactan protein related genes (Lou et al., 2014; Jia et al., 2015), but genes involved in nexine formation have not been reported in rice to date.
Here we describe the functional characterization of an AHL family protein PTC2, expressed in roots and specifically in the tapetum after meiosis, which is required for tapetal PCD and pollen wall patterning in rice. Our results describe a new regulator controlling male reproduction in rice, which is involved in nexine formation in pollen.
RESULTS
Isolation and Genetic Analysis of ptc2
A male sterile mutant was identified from an existing rice mutant library (O. sativa ‘japonica’ ssp. 9522, generated with 60Co γ-ray irradiation; Chen et al., 2006), and was named ptc2 due to its phenotypic similarity to ptc1 (Li et al., 2011). The mutant ptc2-1 showed normal vegetative development (Fig. 1, A–C) but its anthers were smaller and paler than wild-type anthers (Fig. 1, D and E). Iodide staining showed that ptc2-1 anthers harbored no viable pollen grains (Fig. 1, F and G). All F1 progeny from the backcross of ptc2-1 with wild-type were fertile, and F2 plants showed a segregation of 96 fertile and 42 sterile plants (3:1, χ2 = 2.17), suggesting that the ptc2 phenotype is controlled by a single recessive mutation.
Figure 1.
Phenotypic comparison between the wild type (WT) and the ptc2-1 mutant. A, Wild-type plant (left) and the ptc2-1 mutant plant (right) after flowering. B, Wild-type panicle (left) and the ptc2-1 panicle (right) at the heading stage. C, Wild-type spikelet (left) and the ptc2-1 spikelet (right). D, Wild-type flower (left) and ptc2-1 flower (right) after removing lemma and palea. Pi, Pistil; St, stamen. E, Wild-type yellow anther (left) and ptc2-1 white anther (right). F, Wild-type pollen grains stained with I2-KI solution at stage 13. G, ptc2-1 pollen grains stained with I2-KI solution at stage 13. Scale bars = 10 cm (A), 2 cm (B), 2 mm (C), 1 mm (D), 500 μm (E), and 100 μm (F and G).
Phenotypic Analysis of ptc2
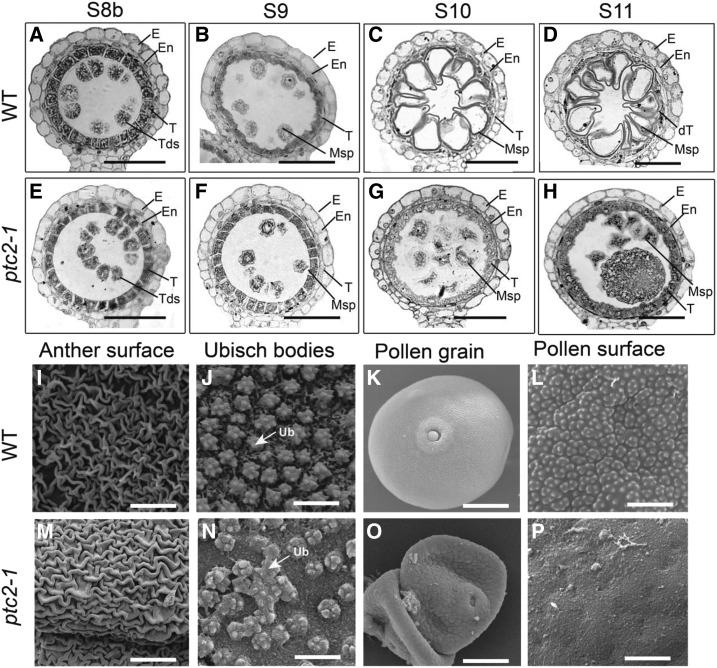
To investigate the cytological effect of ptc2, comparative semithin section analysis of wild-type and ptc2-1 anthers was performed. Anther developmental stages are as described by Zhang et al. (2011). No obvious morphological changes were observed in ptc2-1 anthers until after stage 8b, marked by tetra development after meiosis II, the formation of microspores enclosed by the callose wall, and the condensation and vacuolation of tapetal cells (Fig. 2, A and E). At stage 9, the wild-type anther formed free haploid microspores, and tapetal cells condense and begin to degenerate, with the production of characteristic orbicules/Ubisch bodies on the inner surface linking the microspores (Fig. 2B), while the ptc2-1 anther exhibited delayed tapetal condensation and disintegration (Fig. 2F). By stages 10 and 11, the wild-type tapetal cells have decayed into a thinner, discontinuous layer (Fig. 2, C and D), while the ptc2-1 tapetal cells have degenerated, and indeed, overproliferate (Fig. 2, G and H), which was similar to the reported phenotype of ptc1 (Li et al., 2011). Furthermore, the microspore wall was much thicker in ptc2-1 compared with wild type (Fig. 2, D and H). These data suggested that tapetal PCD and pollen wall formation are affected in the ptc2 mutant.
Figure 2.
Comparison of anther development in the wild-type (WT) and ptc2-1 mutant. A to H, Semithin sections of wild-type anthers (A–D) and ptc2-1 anthers (E–H). Stage 8b (A and E); Stage 9 (B and F); Stage 10 (C and G); and Stage 11 (D and H). I to P, SEM analysis of wild-type and the ptc2-1 mutant at stage 13. Anther surface of wild-type (I) and the ptc2-1 mutant (M). Inner surface of the anther walls layers of the wild-type (J) and the ptc2-1 mutant (N), showing Ubisch bodies. Pollen grains of the wild type (K) and ptc2-1 (O). Pollen surface of wild type (L) and the ptc2-1 mutant (P). dT, degraded tapetum; E, Epidermis; En, endothecium; Msp, microspores; Tds, tetrads; T, tapetum; Ub, Ubisch body. Scale bars = 50 µm (A–H), 10 µm (I and M), 2 µm (J and N), 20 μm (K and O), and 3 μm (L and P).
Scanning electron microscopy (SEM) was used to further investigate the morphological defects, focusing on late stage anthers and pollen grains (at stage 13). A marginal difference in the patterning of nano-ridges on the outer surface of anthers (Fig. 2, I and M) and a more obvious difference in the distribution of Ubisch bodies on the inner surface of anthers (Fig. 2, J and N) was observed between wild-type and the ptc2-1 mutant. Wild-type Ubisch bodies with sharp protrusions were evenly distributed (Fig. 2J), whereas ptc2-1 anthers displayed abnormally clustered and randomly distributed Ubisch bodies with relatively blunt protrusions (Fig. 2N). During pollen maturation, wild-type pollen grains were full, round, and with regular protrusions on the surface (Fig. 2, K and L), while ptc2-1 pollen grains were shrunken, with no regular protrusions on the surface (Fig. 2, O and P). These findings indicated abnormal anther surface development and pollen wall patterning in the ptc2 mutant.
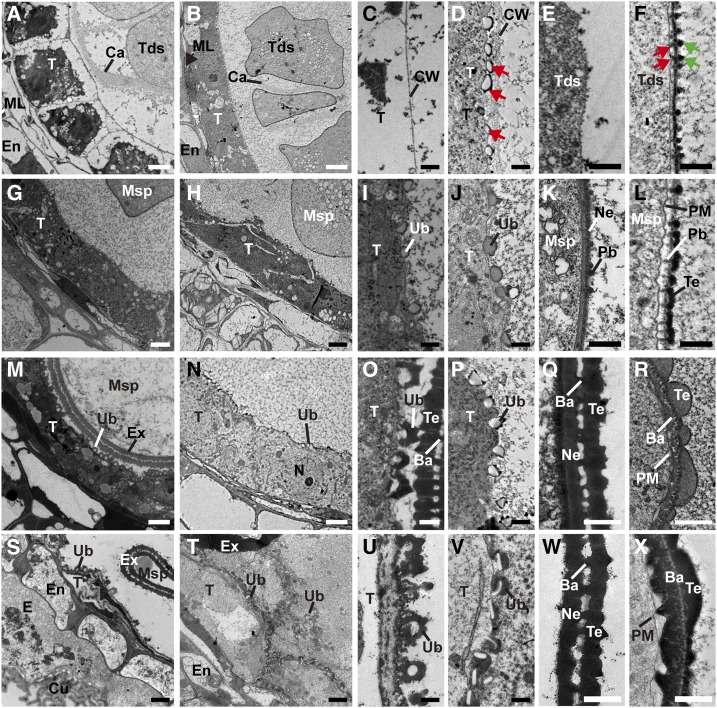
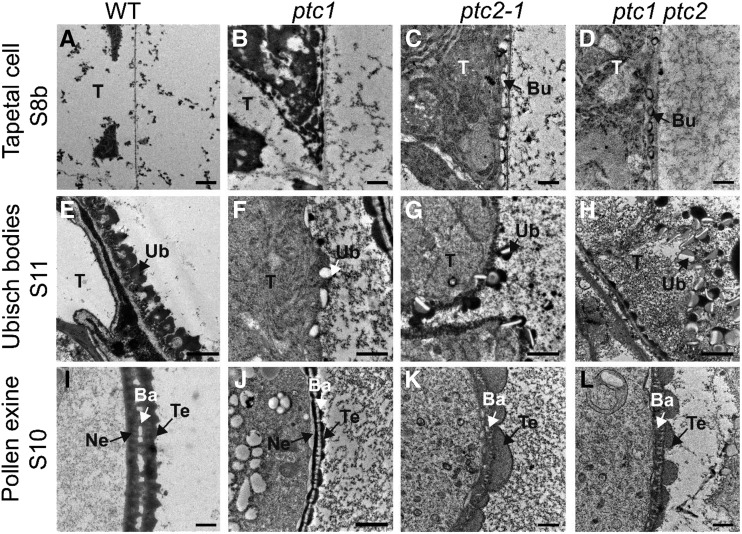
Transmission electron microscopy (TEM) was performed to gain more detailed insights into the developmental defects of ptc2-1 anthers. No obvious difference could be observed until after stage 8a (Supplemental Fig. S1, A and B). At stage 8b, wild-type tapetal cells became largely vacuolated and were covered by a thin layer of the cell wall (Fig. 3, A and C; Supplemental Fig. S1C). No exine was formed on the tetrad surface (Fig. 3E). By contrast, in ptc2-1 tapetal cells, tapetal cytoplasm was dense with many organelles, such as endoplastic recticulum and mitochondria (Fig. 3B; Supplemental Fig. S1D). In addition, some bubble-like structures were observed beneath the tapetal cell wall (Fig. 3D; Supplemental Fig. S1D). A thick layer with darkly stained dots (black arrows in Fig. 3F) was observed on the tetrad surface at this stage. Moreover, some thin and rod-shaped structures (Fig. 3F, red arrows) were observed between the thick layer and plasma membrane (PM; Fig. 3F). At stage 9, in wild-type anthers, Ubisch bodies emerged on the outer surface of tapetal cells (Fig. 3, G and I), and the nexine and probacula were observed on the microspore surface (Fig. 3K). Although ptc2-1 anthers appeared to form normal Ubisch bodies (Fig. 3, H and J), the mutant formed microspores with no nexine (Fig. 3L), Furthermore, the darkly stained dots (black arrows in Fig. 3F) on the microspore surface joined to form the tectum (Fig. 3L). Weakly stained probacula could be observed between the tectum and the PM (Fig. 3L). At stage 10, the wild-type anther showed dramatic degeneration of the tapetal layer with the formation of U-shaped Ubisch bodies (Fig. 3, M and O) and normal pollen exine (Fig. 3Q). However, as in ptc1 mutants, ptc2-1 anthers showed no obvious degeneration of tapetal cell layer with intact nuclei in tapetal cells (Fig. 3N; Supplemental Fig. S1, G and H). Furthermore, the ptc2-1 Ubisch bodies were relatively unchanged from stage 9 (Fig. 3P). On the ptc2-1 microspore surface, the nexine layer was indistinct, and the bacula seemed collapsed, covered by excessive but coarse tectum (Fig. 3R). At stage 11, the wild-type tapetal cell layer was completely degenerated and the exine was well developed (Fig. 3, S, U, and W). However, in ptc2-1 anthers, tapetal cells with abnormal Ubisch bodies were persistent (Fig. 3, T and V). On the microspore surface, the nexine was absent, while both tectum and bacula were disordered (Fig. 3X). These results supported the semithin section and SEM results that pollen wall patterning and tapetal PCD were abnormal in ptc2 mutants.
Figure 3.
TEM images of the anthers from the wild-type (WT) a and ptc2-1 mutant. Transverse sections showing tapetal cells (A and B), tapetal surface (C and D), and tetrad surface (E and F) of wild-type (A, C, and E) and ptc2-1 (B, D, and F) at stage 8b. Tapetal cells (G and H), tapetal surface (I and J), and exine (K and L) of wild-type (G, I, and K) and ptc2-1 (H, J, and L) at stage 9. Tapetal cells (M and N), tapetal surface (O and P), and exine (Q and R) of wild-type (M, O, and Q), and ptc2-1 (N, P, and R) at stage 10. Tapetal cells (S and T), tapetal surface (U and V), and exine (W and X) of wild-type (S, U, and W) and ptc2-1 (T, V, and X) at stage 11. Ba, bacula; Bu, bubble; Ca, callose; Cu, cuticle; CW, cell wall; E, epidermis; En, Endothecium; Ex, exine; ML, middle layer; Msp, microspore; N, nucleus; Ne, nexine; Pb, probacula; T, tapetum; Tds, tetrads; Te, tectum; Ub, Ubisch body. Red arrows in (D) show the bubble structure; green arrows in (F) show dot-like structure on tetrad surface and the red arrows in (F) shows a thin rod between PM and the thick layer. Scale bars = 2 μm (A, B, G, H, M, N, S, and T), 0.5 μm (C–F, I–L, O, P, U, and V), and 1 μm (Q, R, W, and X).
DNA Fragmentation in ptc2 Tapetal Cells Are Delayed
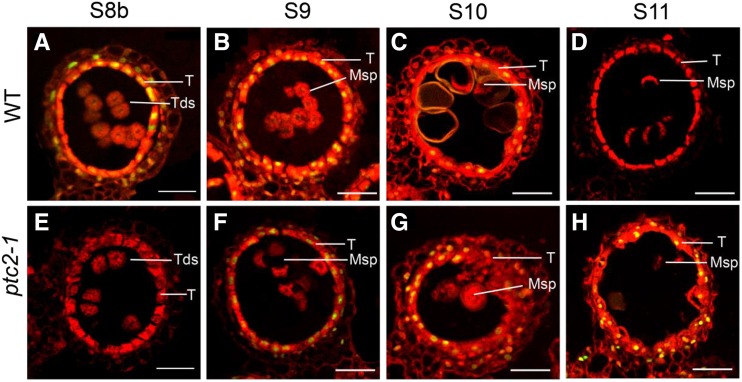
As TEM revealed the persistence of tapetal cells in ptc2 mutants in late anther development (stage 11), the terminal deoxynucleotidyl transferase-mediated dUTP nick-end labeling (TUNEL) assay was used to verify delayed PCD. In wild-type anthers, DNA fragmentation signals were clearly visualized at stage 8b (Fig. 4A) and peaked at stage 9 (Fig. 4B), before declining and disappearing by stage 11 (Fig. 4, C and D). In ptc2-1 anthers, DNA fragmentation signals were undetectable until stage 9 (Fig. 4, E and F), and strong by stages 10 and 11 (Fig. 4, G and H). These results further supported the delayed tapetal PCD and explained the phenotype of ptcs in the ptc2 mutant.
Figure 4.
DNA fragmentation is delayed and persistent in the ptc2-1 mutant. A to D, TUNEL signal at stage 8b (A), stage 9 (B), stage 10 (C), and stage 11 (D) in wild-type (WT). E to H, TUNEL signal at stage 8b (E), stage 9 (F), stage 10 (G), and stage 11 (H) in the ptc2-1 mutant. The red fluorescence shows the propidium iodide staining of anther cells using confocal microscopy; yellow fluorescence shows the TUNEL and PI signal overlap. Msp, microspores; T, tapetum; Tds, tetrads. Scale bars = 20 μm.
Chemical Analysis of Anther Cuticular Lipids
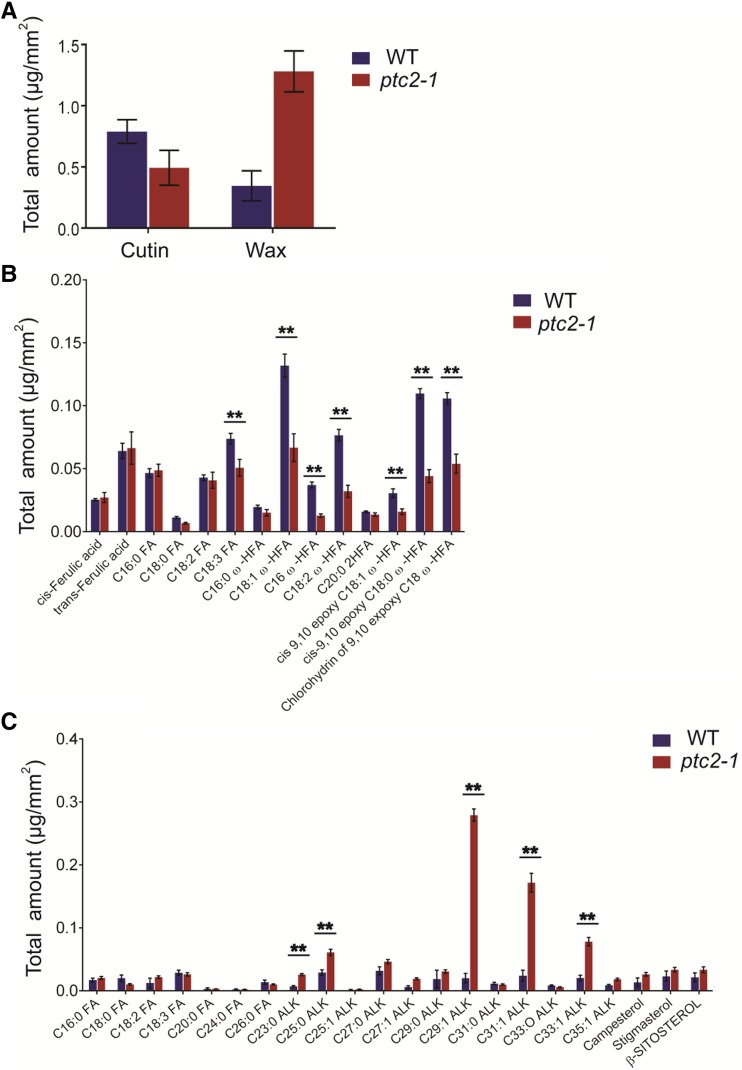
TEM results indicated that the ptc2 pollen wall was abnormally thick at late stages (stage 10 onwards, Fig. 3), while SEM had revealed a subtle difference in anther surface patterning between ptc2 and wild type (Fig. 2, I and M), prompting a detailed analysis of anther cuticular lipids. The cuticular wax constituents and cutin monomers from wild-type and ptc2-1 anthers were measured by gas chromatography–mass spectrometry and gas chromatography–flame ionization detection. The total amount of cutin on the wild-type anther surface was 0.79 μg/mm2 compared with 0.49 μg/mm2 on the ptc2-1 anther surface (Fig. 5A). The ω-hydroxy fatty acids, such as C18:1 ω-HFA, C18:2 ω-HFA, cis-9,10 epoxy C18:0 ω-HFA, and chlorohydrin of 9,10 epoxy C18 ω-HFA, were the dominant cutin monomers, and were present at significantly lower levels in ptc2-1 anthers (Fig. 5B). Most of the unhydroxylated fatty acids, such as C16:0 FA, C18:0 FA, and C18:2 FA, and ferulic acid were not present in significantly different amounts in wild-type and mutant anthers (Fig. 5B; Supplemental Table S1). The total amount of cuticular wax on the wild-type anther surface was 0.35 μg/mm2 compared with the much higher 1.28 μg/mm2 on ptc2-1 anther surfaces (Fig. 5A). The increase of cuticular wax was predominantly due to the significant increase in unsaturated alkanes, such as C29:1 ALK, C31:1 ALK, and C33:1 ALK (Fig. 5C; Supplemental Table S1). These results indicated that PTC2 may be involved in regulating the biosynthesis of lipidic compounds for anther cuticle formation, and the thicker pollen wall at late stages might be more related to the increase of wax content.
Figure 5.
Analysis of anther cutin and wax in the wild-type(WT) and ptc2-1 mutant. A, Total cutin and wax amounts per unit surface area in wild-type and ptc2-1 anthers. Error bars indicate ± sd (n = 5). B, Cutin monomers, amount per unit surface area in wild-type and ptc2-1 anthers. C, Wax constituents, amount per unit surface area in wild-type and ptc2-1 anthers. Error bars indicate ± sd (n = 5). **P < 0.01 by Student’s t-test. ALK, alkane; C16:0 FA, hexadecanoic acid; C18:0 FA, octadecanoic acid; C18:2 FA, linoleic acid; C18:3 FA, linolenic acid; C16:0 ω-HFA, 16-hydroxy-hexadecanoic acid; C18:1 ω-HFA, 18-hydroxy-octadecanoic acid; C16 DHFA, 16-dihydroxy-palmitic acid; C18:2 ω-HFA, 18-hydroxy-linoleic acid; C20:0 2HFA, 2‐hydroxyeicosanoic acid; cis‐9,10‐epoxy C18:1 ω-HFA, cis‐9,10‐epoxy‐18‐hydroxy‐oleic acid; cis‐9,10‐epoxy C18:0 ω-HFA,cis‐9,10‐epoxy 18‐hydroxy‐stearic acid; Chlorohydrin of 9,10-expoxy C18 ω-HFA, chlorohydrin of 9,10-epoxy-18-hydroxy-octadecanoic acid. Acids were analyzed as methyl esters, and hydroxyl groups were analyzed as trimethylsilyl esters.
Map-Based Cloning and Expression Analysis of PTC2
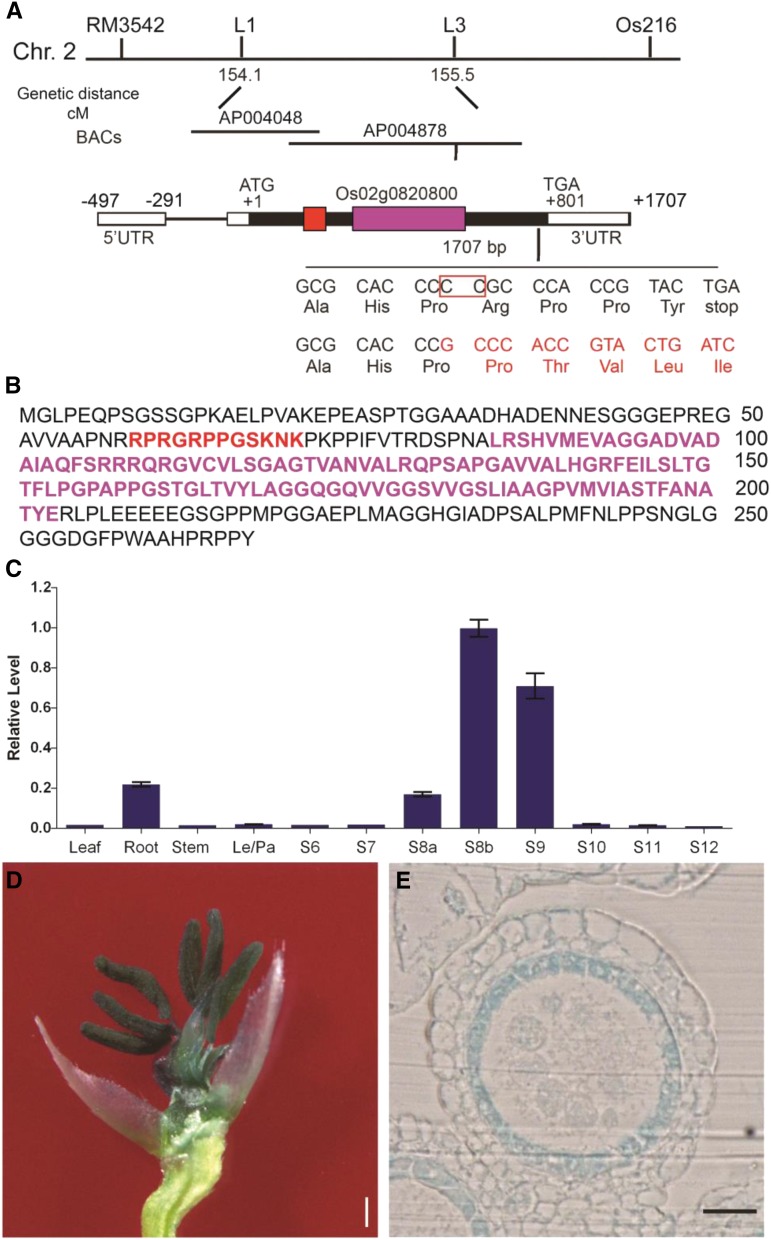
A map-based cloning approach was used to map PTC2 to chromosome 2 between the L1 and L3 markers, spanning 47 kb of genomic DNA. After sequencing potential candidate genes within this region, a two-bp deletion was identified just upstream of the stop codon of the gene corresponding to LOC_Os02g57520 (http://rice.plantbiology.msu.edu/), also annotated as Os02g0820800 (http://rapdb.dna.affrc.go.jp/). The frameshift caused by the mutation extended the predicted translated protein by 32 amino acids (Fig. 6A).
Figure 6.
Map-based cloning and the expression analysis of PTC2. A, Fine mapping of PTC2 on chromosome 2, showing Indel molecular marker positions. Schematic representation of the exon and intron organization of PTC2; the red box represents an AT-hook motif and the purple box represents PPC (DUF296) domain. UTR, untranslated region. B, Protein sequence highlighted with AT-hook motif and PPC (DUF296) domain; the red characters represents an AT-hook motif and the purple characters represents PPC (DUF296) domain. C, PTC2 relative expression in different rice tissues and tapetal developmental stages, analyzed by RT-qPCR. Error bars are ± sd of the three means from three technical repeats in each biological repeat, inferring the se for inferential statistics. D, GUS-stained flower after removal of palea and lemma from PTC2pro:GUS transgenic lines. E, GUS expression driven by PTC2 promoter in tapetum cell layer at early stage 9. Scale bars = 2 mm (D) and 50 μm (E).
LOC_Os02g57520 is predicted to encode an AT-hook DNA binding protein (Zhao et al., 2014). PTC2 contains an AT-hook DNA binding domain at the N terminus, and a Plant and a Prokaryote Conserved domain (PPC), also known as the Domain of Unknown Function #296 (DUF296), at the C terminus (Fig. 6, A and B). The frameshift mutation in ptc2-1 occurs 3′ to the sequence encoding the PPC/DUF296 domain (Fig. 6A). The PTC2 protein sequence shows high similarity to type I AT-hook motifs from the AHL family from land plants (Supplemental Fig. S2).
To confirm that LOC_Os02g57520 is the mutated gene causing phenotypic defects, a binary plasmid containing the promoter and wild-type genomic PTC2 DNA was introduced into homozygous ptc2-1 plants. The male sterile phenotype of ptc2-1 was fully complemented in the transgenic plants (Supplemental Fig. S3A). Furthermore, an additional allele, named ptc2-2, with a one-bp insertion early in the coding sequence that leads to a frameshift in the coding sequence, was generated by the CRISPR/Cas9 system (Supplemental Fig. S3B). The mutant ptc2-2 showed identical defects in male fertility and tapetum degradation to ptc2-1 (Supplemental Fig. S3, C–F). These findings indicated that the PTC2 mutation was responsible for observed defects in male fertility in ptc2 mutants.
The spatial-temporal expression patterns of PTC2 were assessed by reverse transcription quantitative PCR (RT-qPCR). PTC2 expression was detectable in roots but not in other vegetative tissues, with a high and otherwise specific expression in anthers, starting at stage 8a and peaking at stage 8b, before disappearing at stage 10 (Fig. 6C). Consistent with these results, transgenic plants expressing the PTC2pro:GUS construct showed GUS activity mainly in tapetal cell layers in anthers (Fig. 6, D and E). These expression data confirmed that PTC2 was closely associated with anther and pollen development in rice.
Subcellular Localization of PTC2 in Anthers
AHL family proteins are predicted to localize in the nucleus. To validate this prediction, the PTC2 coding sequence fused with GFP driven by the Cauliflower mosaic virus 35S promoter was transiently expressed in epidermal cells of tobacco leaves. Compared with the ubiquitous localization of GFP not fused to PTC2 (Supplemental Fig. S4, A–C), PTC2-GFP fluorescence was specifically detected in the nucleus (Supplemental Fig. 4, D–F), suggesting that PTC2 was likely a nucleus localized protein.
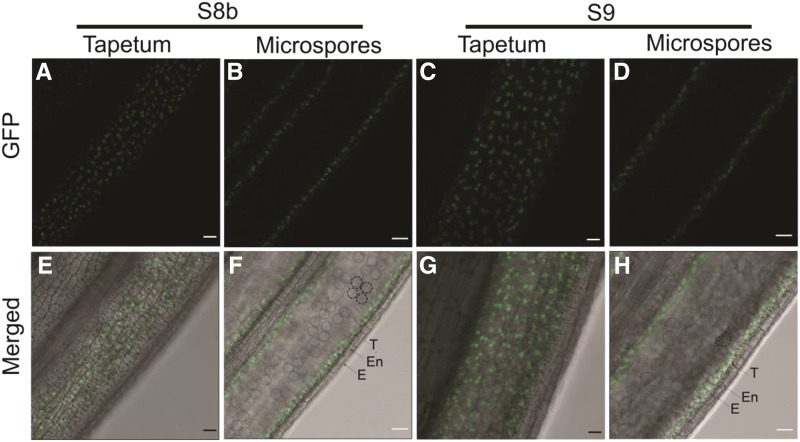
PTC2 localization in rice cells was analyzed after transforming a construct of PTC2 genomic DNA fused with GFP driven by the PTC2 native promoter into ptc2 mutant plants. The transgenic lines were fertile, indicating the correct localization and function of PTC2-GFP protein (Supplemental Fig. S5). The GFP signal was detectable as dots in the tapetal cell layer at stages 8b and 9 (Fig. 7), consistent with the gene expression data and GUS staining assay (Fig. 6).
Figure 7.
Wild-type PTC2 protein localization in ptc2-1 anther. Images were acquired through a GFP filter (A–D) and merged with bright field imaging (E–H). A, B, E, and F, Anther view focusing on tapetum (A and E) and microspores (B and F) at stage 8b. C, D, G, and H, Anther view focusing on tapetum (C and G) and microspores (D and H) at stage 9. Dotted circle (F and H) showing tetrads and microspores respectively. E, Epidermis; En, endothecium; T, tapetum. Scale bars = 20 μm.
Phenotypic Analysis of ptc1 ptc2
Because TEM data indicated that ptc2-1 showed similar developmental defects to ptc1 (Figs. 2 and 3; Li et al., 2011), the ptc1 ptc2 double mutant was generated to study the relationship between these two genes. Compared with the wild-type, the ptc1 ptc2 double mutant exhibited ptcs with many cellular organelles, like that of single mutant ptc1 and ptc2 anthers (Fig. 8, A–D). The double mutant exhibited formation of bubble-like structures at stage 8b, identical to ptc2-1 (Fig. 8, C and D). The Ubisch bodies at stage 9 were abnormal in ptc1 ptc2, more similar in morphology to those of ptc2-1 than ptc1 (Fig. 8, E–H). In addition, ptc1 ptc2 displayed an abnormal exine, with a collapsed bacula, disordered tectum, and no nexine, almost identical to that of ptc2-1 rather than ptc1 (Fig. 8, I–L). These data indicated that PTC2 and PTC1 have a similar function in the regulation of tapetal PCD, while PTC2 was epistatic to PTC1 for pollen wall development.
Figure 8.
TEM images of the anthers from the wild-type (WT), ptc1, ptc2-1, and ptc1 ptc2 mutants. A to D, Transverse sections showing tapetal cells at stage 8b. E to H, Transverse sections showing Ubisch bodies at stage 11. I to L, Transverse sections showing exine at stage 10. Bu, bubble-like structure; Ba, bacula; Ne, nexine; T, tapetum; Te, tectum; Ub, Ubisch body. Scale bars = 0.5 μm (A–H and J) and 1 μm (I, K, and L).
Transcriptome Analysis in Anthers
As PTC2 encodes an AHL protein family transcription factor involved in tapetal PCD and pollen wall patterning (Figs. 3, 4, and 6), PTC2 may regulate a series of tapetal PCD and lipids synthetic or transport genes. We performed transcript profiling of ptc2-1 anthers at stages 9 and 10. The principal component analysis indicates distinct patterns of gene expression between wild-type and ptc2 mutant at both stages (Supplemental Fig. S6A). Analysis of the false discovery rate (Log10 FDR) and fold change (Log2FC; Supplemental Fig. S6, B and C) revealed 3,129 genes with significantly changed values compared with wild-type genes (FDR < 0.05 and |Log2FC| > 2). Of these, 330 genes were upregulated, and 368 genes were downregulated at both stages. Fifty-three genes were upregulated and 781 genes downregulated only at stage 9, while 1,283 genes were upregulated and 304 genes downregulated only at stage 10. Ten genes were downregulated at stage 9 and upregulated at stage 10 (Supplemental Fig. S6D; Supplemental Tables S2 and S3).
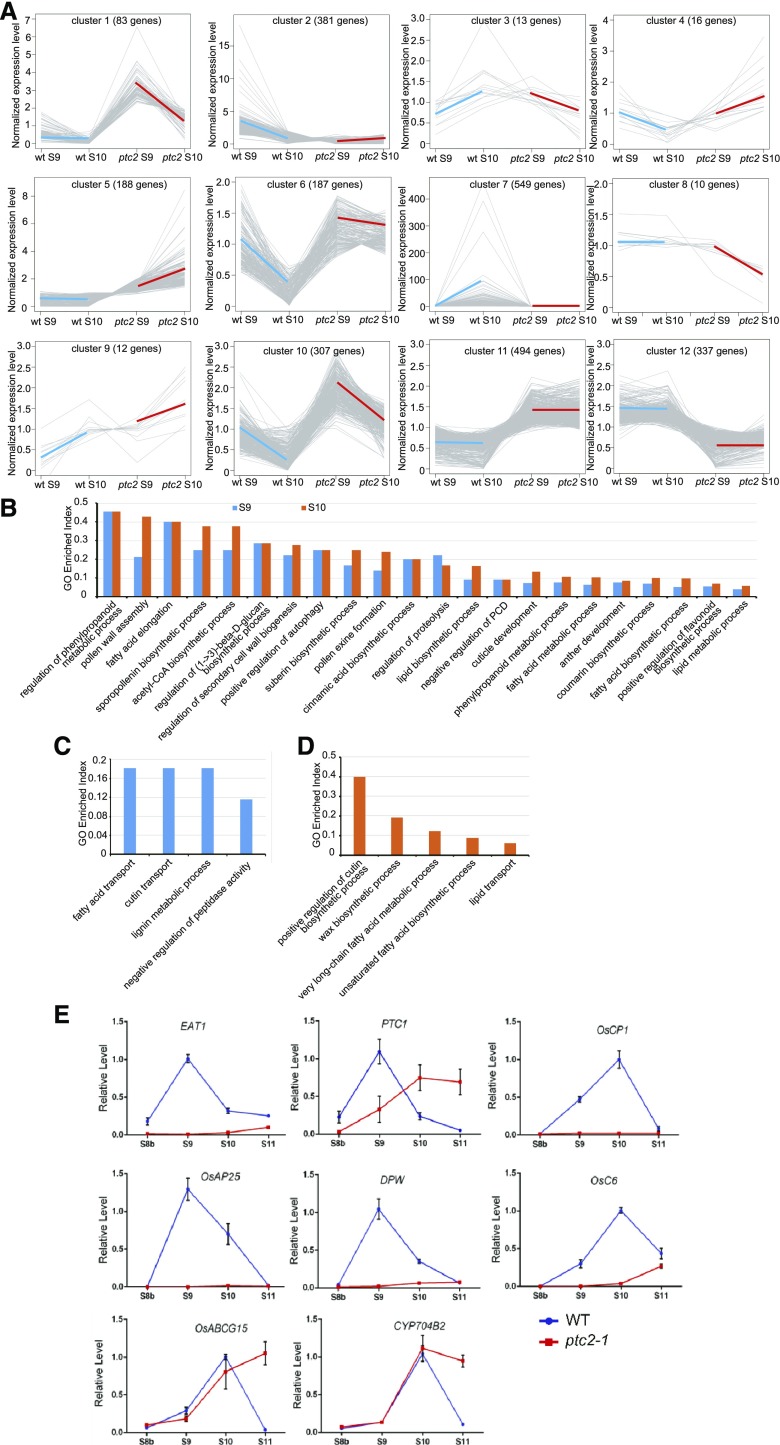
Among the 3,129 differentially expressed genes (DEGs), 2,577 highly expressed genes (fragments > 200) were selected for coexpression clustering analysis. Based on their expression in wild-type and ptc2 mutants, these genes formed 12 clusters (Supplemental Table S4). Clusters 1, 6, and 10 contained genes more highly expressed in mutant anthers at both stages, with higher expression at stage 9 than in stage 10. Genes in clusters 11 and 12 were expressed similarly at both stages, though more highly in mutant anthers in cluster 11, and more highly in wild-type anthers in cluster 12. The expression level of genes in cluster 7 was substantially induced at stage 10 in wild type; however, the induction was absent in ptc2 (Fig. 9A).
Figure 9.
RNA-seq and RT-qPCR analysis of wild-type (WT) and ptc2 mutant plants. A, Coexpression clustering of genes expressed at stages 9 and 10 in wild-type anthers and ptc2 anthers. B, GEI of DEGs in ptc2 enriched at both stages 9 and 10. C, GEI of DEGs enriched only at stage 9. D, GEI of DEGs enriched only at stage 10. E, RT-qPCR analysis of genes related to sporopollenin metabolism and tapetal PCD in the wild-type and ptc2-1 mutant from stages 8b to 11. Error bars are ± sd of the three means from three technical repeats in each biological repeat, inferring the se for inferential statistics. OsC6, encoding lipid transfer protein; CYP704B2, CYTOCHROME P450 family member
DEGs from clusters 1 to 8, which exhibited different patterns of expression from stage 9 to stage 10 in the wild-type and mutant anthers, were selected for gene ontology (GO) analysis. We used a GO Enrichment Index (GEI), which compares the frequency of DEGs with a given GO term compared with the frequency of genes in this GO term (Supplemental Table S5). Genes associated with tissue and wall development, lipid biosynthesis and metabolism, and cell death, were highly enriched at both stages (Fig. 9B; bold in Supplemental Table S5). Genes associated with fatty acid and cutin transport, and lignin metabolism were enriched at stage 9 only (Fig. 9C; bold in Supplemental Table S5), and genes associated with cutin, wax and fatty acid biosynthesis, fatty acid metabolism, and lipid transport were enriched at stage 10 (Fig. 9D; bold in Supplemental Table S5). These DEGs may be responsible for the pollen wall defects in ptc2 anthers.
Known genes involved in tapetal PCD were selected for RT-qPCR analysis across four developmental stages to explore the tapetal PCD defect in ptc2 anthers. EAT1 and PTC1 are tapetal PCD key regulators expressed after meiosis (Li et al., 2011; Ji et al., 2013; Niu et al., 2013). The expression level of EAT1 was relatively low at all stages (Fig. 9E), consistent with its inclusion in cluster 2 (Supplemental Table S4). The expression of PTC1 (cluster 6) was downregulated at stage 9 but upregulated at stages 10 and 11 (Fig. 9E). Oryza sativa Cys Protease1 (OsCP1) and Oryza sativa Aspartic Protease25 (OsAP25) are the reported downstream targets of TDR and EAT1, respectively (Li et al., 2006; Niu et al., 2013). Expression of these genes was low in ptc2-1 anthers at each stage (Fig. 9E), consistent with their inclusion in cluster 2. The observed downregulation of PCD-associated genes matched with the delayed tapetal PCD phenotype in ptc2 anthers.
We also examined the expression of some genes related to pollen wall formation. The sporopollenin synthesis gene, DPW (Shi et al., 2011), and the sporopollenin transport gene, OsC6 (Zhang et al., 2010), both belonging to cluster 2, showed no induction during anther development in ptc2-1 anthers (Fig. 9E; Supplemental Table S4). CYP704B2 (Li et al., 2010), and OsABCG15 (Qin et al., 2013; Zhao et al., 2015) were not differently expressed in ptc2 anthers (Supplemental Table S4), consistent with the RT-qPCR results. Furthermore, the expression level of both genes was higher at stage 11 in the mutant compared with the wild type (Fig. 9E). These diverse patterns of changes in pollen wall development-related gene expression suggests a complex regulatory network regulated by PTC2 for pollen wall development.
DISCUSSION
Rice is an ideal model monocot plant, with many resources that facilitate the discovery of mechanisms underlying male gametophyte development (Zhang and Liang, 2016). In this study, we have cloned and functionally characterized a male-sterile associated gene, PTC2, which encodes an AHL family protein in rice. The ptc2 mutant displayed tapetal cells that did not degrade during anther development, abnormal pollen wall and anther surface patterning with altered cuticle composition, and different expression patterns of several genes associated with sporopollenin biosynthesis, modification, transport, and regulation of tapetal PCD. Our results provided new insights into plant male reproductive biology.
PTC2 Is Required for Tapetal PCD
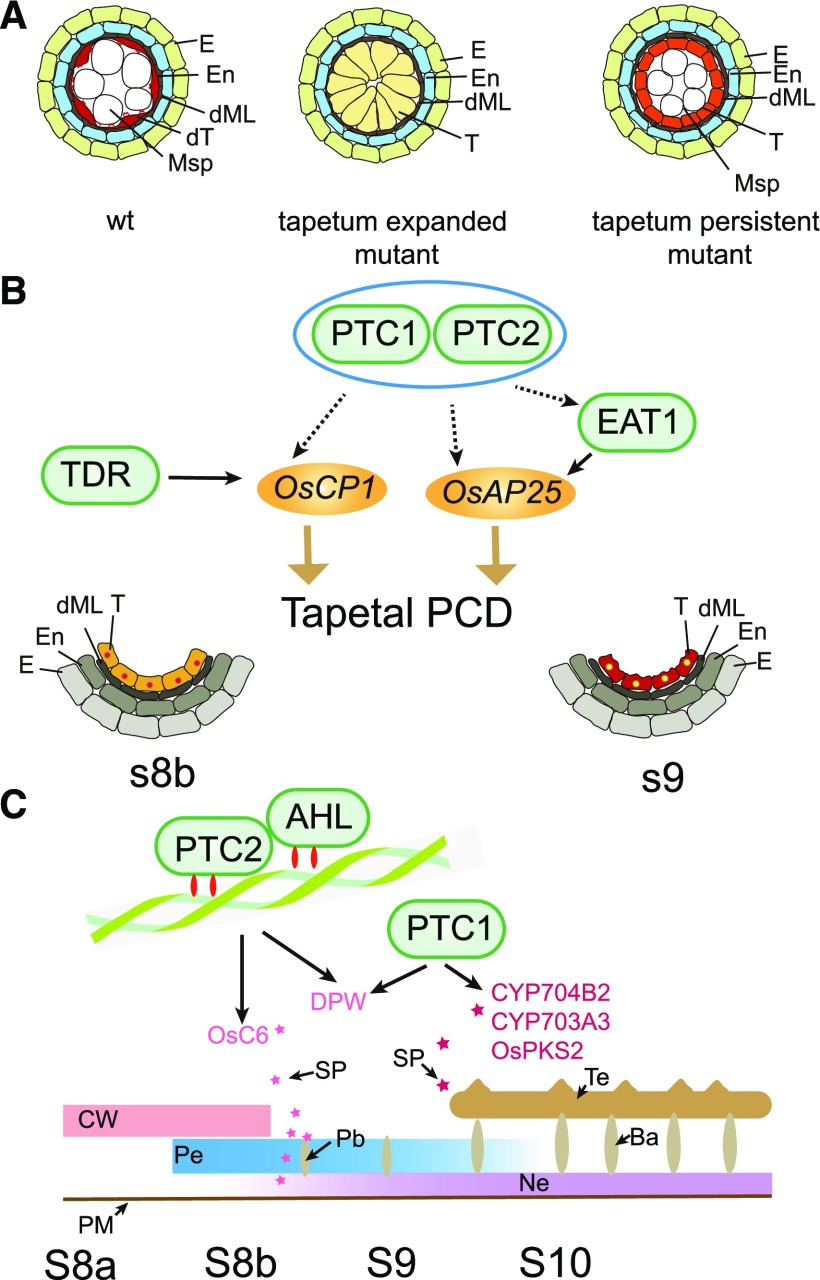
In rice, a series of tapetal PCD delayed mutants have indicated the importance of tapetal PCD for pollen development. These mutants can be classified into two groups. Tapetum expanded mutants, such as tip2 (Fu et al., 2014; Ko et al., 2014), udt1(Jung et al., 2005), gamyb (Aya et al., 2009), and tdr (Li et al., 2006), exhibit resistance to tapetum degradation, with highly expanded tapetal cells lacking organelles present at late developmental stage (Fig. 10A). Interestingly, all these transcription factors are expressed at premeiotic stages, with the exception of the ubiquitously expressed GAMYB, which has a posttranscription regulatory effect (Jung et al., 2005; Li et al., 2006; Tsuji et al., 2006; Aya et al., 2009; Fu et al., 2014; Ko et al., 2014; Lin et al., 2017). Tapetum persistent mutants, such as ptc1 (Li et al., 2011) and eat1 (Ji et al., 2013; Niu et al., 2013), also have a tapetum resistant to degradation. These tapetal cells retain their normal size, but rather than displaying a large central vacuole, contain many organelles such as mitochondria, ER and others, indicating metabolic activity (Fig. 10A). In contrast, these genes are highly expressed at postmeiotic stages (Li et al., 2011; Ji et al., 2013; Niu et al., 2013).
Figure 10.
Model for tapetal PCD mutants and PTC2 effect on pollen wall formation. A, Model for tapetal PCD mutants. B, Model for PTC2 on tapetum development. Blue oval indicates the same pathway. Different stars indicate the different sporopollenin for bacula and tectum formation. C, Model for PTC2 on pollen wall formation. Ba, bacula; CW, cell wall; dML, degraded middle layer; dT, degraded tapetum; E, Epidermis; En, endothecium; Msp, microspores; Ne, nexine; Pb, probacula; Pe, primexine; SP, sporopollenin; T, tapetal layer; Te, tectum; WT, wild type.
The male sterile mutant, ptc2, displayed delayed tapetal PCD confirmed by semithin section observation, TEM and TUNEL assays (Figs. 2–4). TEM observation also indicated that tapetal cells were persistent at late stages, with many organelles (Fig. 3), indicating that ptc2 is a tapetum persistent mutant. PTC2 transcripts were highly expressed at stages 8b and 9, i.e. in postmeiotic stages (Fig. 6), which supports its role as a tapetum persistent mutant.
The ptc1 ptc2 double mutant exhibited similar tapetum developmental defects compared with either ptc1 or ptc2 single mutants (Fig. 8). Similarly, the two genes exhibit similar spatial and temporal expression patterns, providing further evidence that PTC2 and PTC1 had similar functions in directing tapetal PCD.
PTC2 Is Required for Pollen Wall Patterning
Pollen wall formation starts during stage 8b in rice, during which initial primexine deposition occurs on the tetrad surface followed by callose degradation and microspore release into lobes (Zhang et al., 2010; Ariizumi and Toriyama, 2011; Shi et al., 2015). The nexine and sexine, which determine pollen wall patterning, are then synthesized (Scott, 1994; Ariizumi and Toriyama, 2011; Shi et al., 2015). The probacula is formed along the primexine at late stage 8b and early stage 9, and subsequently the tectum forms on the probacula at early stage 9. After the pollen wall is deposited, biosynthesis of sporopollenin begins, which is then laid down on the microspore surface to form the two-layer exine from late stage 9 to stage 11 (Li et al., 2010; Li and Zhang, 2010; Ariizumi and Toriyama, 2011; Shi et al., 2015; Yu et al., 2016; Liu et al., 2017; Zhu et al., 2017).
In ptc2 anthers, both the tectum and the probacula were formed earlier, at stage 8b, followed by further deposition of excess tectum, and abnormal development of the bacula (Fig. 3, F and L). In addition, no nexine could be observed at stage 9, and at later stages, excess material was observed where the bacula developed (Fig. 3, L and R). PTC2 protein was detectable at stages 8b and 9 in tapetal cells, consistent with the developmental defects observed in ptc2-1 mutant (Figs. 6 and 7).
The nexine in ptc1 was more uneven than in eat1, and completely absent in ptc2 (Li et al., 2011; Niu et al., 2013; Fig. 3), suggesting that the pollen wall patterning was divergent in tapetum persistent mutants. The ptc1 ptc2 double mutant displayed similar pollen wall patterning with ptc2 (Fig. 8), indicating that PTC2 is epistatic to PTC1 during pollen wall development, however, the epistasis could not be observed during tapetal PCD regulation. Thus, the tapetum persistent mutants, while having extremely similar phenotypes on the tapetal development, have quite different effects on pollen wall patterning.
The mutant tek in Arabidopsis displays a similar phenotype to ptc2 in rice, with an absent nexine and earlier, excess formation of tectum (Lou et al., 2014). TEK and PTC2 both belong to subfamily A of the AHL family (Zhao et al., 2014). These findings emphasize the importance of AHL family proteins on pollen wall patterning. However, TEK belongs to subfamily A2, while PTC2 belongs to subfamily A5 (Zhao et al., 2014), and the sequence similarity was comparatively low (Supplemental Fig. S2). Furthermore, the TEK subclade in subfamily A2 from AHL family is dicot-specific (Zhao et al., 2014). Rice members of subfamily A2, LOC_Os02g25020 and LOC_Os06g04540, are more closely related to AtAHL22, AtAHL24, and AtAHL26 than to TEK (AHL16; Zhao et al., 2014); AtAHL22 is involved in flowering (Yun et al., 2012), while the biological function of the other two proteins has not been reported. Conversely, the closest Arabidopsis putative orthologs of PTC2 in the A5 subfamily are AtAHL19 and AtAHL20 (Zhao et al., 2014), which have diverse roles in fungal resistance, PAMP-induced gene expression, and hypocotyl growth (Lu et al., 2010; Yadeta et al., 2011; Zhao et al., 2013). Thus, between rice and Arabidopsis at least, it was divergent in protein structure–function conservation through evolution. While the monocot putative ortholog of TEK may have been lost, its function has been highly conserved in PTC2. It will be interesting in the future to explore the more detailed biological function of AHL family members, especially their role in pollen development, as well as their evolutionary relationships.
The Putative Mechanisms of PTC2 Emphasizes Its Role in Male Sterility
The first hallmark of tapetal PCD, DNA fragmentation, occurs at stage 8b (Li et al., 2006; Yu et al., 2016), suggesting that gene expression during meiosis (stage 8) is critical for tapetal PCD initiation. Disturbing the gene expression profile through transcription factor mutation could therefore affect tapetum morphology at late stages. TIP2 modulates tapetal PCD directly via regulating expression levels of TDR and EAT1 (Fu et al., 2014; Ko et al., 2014). The direct regulation of OsCP1 by TDR, and of OsAP25 by EAT1, has been verified in rice (Li et al., 2006; Niu et al., 2013). Expression of EAT1, OsCP1, and OsAP25 peaks after PTC2, temporally consistent with their expression being directly or indirectly regulated by PTC2. The expression levels of these genes were largely downregulated in ptc2 (Fig. 9; Supplemental Table S4), as well as in ptc1 and tdr mutants (Zhang et al., 2008; Li et al., 2011), which matches with the tapetal PCD defect. Notably, some PCD-related genes showing altered expression level in tdr (Zhang et al., 2008) did not have altered expression patterns in ptc2 (Supplemental Tables S2 and S3), suggesting that PTC2 and TDR might modulate tapetal PCD through different downstream effectors (Fig. 10B). Our phenotypic analysis indicated that tapetal PCD of ptc2 was similar to that of ptc1 (Figs. 2–4), implying that PTC2 and PTC1 function in the same tapetal PCD pathway, likely by regulating overlapping downstream targets such as OsCP1 and OsAP25 (Fig. 10B). Nevertheless, further identification of downstream targets of PTC2 will lead to a better understanding of the regulatory mechanism of PTC2 in tapetal PCD.
In tapetum expanded mutants, the anther surface is smooth, with no exine deposited on the pollen surface and a large number of fatty acids dramatically downregulated (Jung et al., 2005; Li et al., 2006; Zhang et al., 2008; Fu et al., 2014). Conversely, in tapetum persistent mutants, the anther surface was very similar to the wild-type, and a pollen wall could be observed despite the abnormal morphology (Li et al., 2011; Niu et al., 2013; Figs. 2 and 3). Accordingly, unmodified fatty acids were not changed, and a moderate reduction of ω-hydroxy fatty acids was measured (Fig. 5). The anther cuticle and pollen wall share biosynthetic pathways in the tapetum (Zhao et al., 2015), which seem to be more widely affected in tapetum expanded mutants than in tapetum persistent mutants. We hypothesize that transcription factors involved in tapetum expansion might also affect biosynthesis of lipids and phenolics for anther cuticle and pollen wall development, while transcription regulators involved in tapetum persistence might modify the lipids and phenolics biosynthesis in the tapetum.
PTC2 and TEK belong to the same subfamily of AHL proteins in land plants (Zhao et al., 2014), and their mutant phenotypes exhibited similar pollen wall patterning (Fig. 3; Lou et al., 2014). AHL proteins contain AT-hook DNA binding motifs that interact with the minor groove of matrix attachment regions to mediate anchoring of specific DNA sequences to the nuclear matrix, affecting chromatin architecture as well as transcription of target genes (Reeves, 2001; Sgarra et al., 2006). AHL proteins can also interact with other AHL proteins to regulate gene expression (Zhao et al., 2013), so we speculate that PTC2 may regulate downstream targets through interaction with other AHL members and/or transcription factors. PTC2 and TEK may have conserved downstream targets that direct pollen wall patterning (Fig. 10C). Characterization of further AHL proteins in rice may reveal a regulatory network that works with PTC2 to modulate plant reproduction.
The expression of the sporopollenin biosynthetic gene, DPW, was downregulated in ptc2, ptc1, and tdr, all of which showed defects in nexine development (Fig. 9E; Supplemental Table S4; Li et al., 2006, 2011). No nexine formed in dpw anthers (Shi et al., 2011). The expression level of a sporopollenin transport gene, OsC6, was downregulated in ptc2 (Fig. 9E; Supplemental Table S4) and tdr, but not in ptc1, and accordingly, bacula development was disrupted in ptc2 and tdr, but not in ptc1 (Fig. 3; Li et al., 2006, 2011), as it was in the OsC6 RNAi mutant (Zhang et al., 2010). Therefore, DPW and OsC6 might be regulated by PTC2 involved in pollen wall patterning. The expression level of CYP704B2 was not changed in ptc2 anthers (Fig. 9); however, its expression level was downregulated in ptc1 anthers at stage 9 (Li et al., 2011). Thus PTC1 and PTC2 might regulate pollen wall patterning through an alternative regulatory network. More importantly, the pollen wall patterning is largely different between ptc2 and ptc1 (Fig. 8); therefore, we believe that PTC2 and PTC1 might regulate pollen wall patterning in an independent pathway (Fig. 10C). The induction of CYP704B2 and OsABCG15 at stages 10 and 11 (Fig. 9E; Supplemental Table S4) might be due to indirect regulation by PTC2. The downregulation of ω-hydroxylated fatty acids, the products of CYP704B2 at late stages (Fig. 5), might be contributed by alternative enzymes, for CYP704B2 plays a role at stage 9 (Li et al., 2010) during which the expression level of CYP704B2 was not changed in ptc2 anthers (Fig. 9). The wax content was overaccumulated in ptc2 anthers at late stages; however, none of the genes required for wax synthesis, including GL family (Islam et al., 2009), WDA1 (Jung et al., 2006), and DWA1 (Zhu and Xiong, 2013) were upregulated at stages 9 and 10 (Supplemental Table S4). We believe that the phenotype might be affected by the expression profile at late stages, which is not a direct result of PTC2 mutation.
In summary, we have cloned and characterized PTC2, an AHL family protein from rice, which is required for tapetal PCD and pollen wall patterning. PTC2 might function with different transcription regulators to play a role in tapetal PCD and pollen wall patterning.
MATERIALS AND METHODS
Plant Materials, Growth Conditions, and Gene Mapping of PTC2
All plants used in this study were cultivated in the paddy field of Shanghai Jiao Tong University during 2013 to 2019. The F2 population was obtained from the cross between ptc2-1 (Oryza sativa ‘japonica’ ssp. 9522) and wild-type GuangLuAi plants (O. sativa ‘indica’). Selection was based on the male sterile phenotype. Indel molecular markers for mapping were designed based on polymorphisms between the two parents. Mapping was performed as reported in Li et al. (2006) and Yu et al. (2016).
Characterization of Mutant Plant Phenotypes
Photographs showing vegetative growth of whole plants and panicles were captured with a model no. DSLR E995 (Nikon). Anthers from different developmental stages were collected as described in Zhang et al. (2011). Spikelets, flowers, and anthers at anther developmental stage 12 were photographed with a model no. S8AP0 stereomicroscope (Leica). Semithin section, SEM and TEM microscopy, and TUNEL assays were performed as described in Li et al. (2006) and Yu et al. (2016).
Plasmid Construction and Plant Transformation
For complementation, a total 4,892-bp region was amplified from wild-type O. sativa ‘japonica’ spp. 9522 genomic DNA, using primers PTC2.comp-F and PTC2.comp-R, which spanned an 801-bp open reading frame region, a 3,674-bp upstream region, and a 417-bp downstream region. The fragment was cloned into pCAMBIA1301-PTC2pro:PTC2, and transformed into a ptc2-1 callus using Agrobacterium tumefaciens EHA105.
For protein localization in the anther, a 4,201-bp region without stop codon, which contained a 3,403-bp upstream region and 798-bp open reading frame minus stop codon, was amplified from wild-type 9522 genomic DNA, using the primers PTC2-GFP-F and PTC2-GFP-R. The fragments were ligated into a GFP-fusion vector pCAMBIA1301-GFP digested with SalI and SpeI using the In-Fusion HD cloning kit (Takara Bio) to generate the PTC2pro:PTC2-GFP construct, which was transformed into a ptc2-1 callus using A. tumefaciens EHA105.
For GUS staining, a 3,651-bp upstream region of PTC2 was amplified from wild-type 9522 genomic DNA, using the primers PTC2-GUS-F and PTC2-GUS-R. The fragment was ligated into GUS-expression vectors pCAMBIA1301:GUS digested with SalI and NcoI using the In-Fusion HD cloning kit (Takara Bio) to generate the PTC2pro:GUS construct, which was transformed into a wild-type rice callus with A. tumefaciens EHA105.
For protein localization in tobacco (Nicotiana tabacum) leaves, full-length 798-bp PTC2 CDS was amplified from wild-type complementary DNA (anther developmental S9) using PTC2-CDS-GFP-F and PTC2-CDS-GFP-R primers and introduced into pGreen-35S vector to obtain pGreen 35Spro:PTC2 CDS-GFP. The construct was transformed with A. tumefaciens GV3101 strain and infiltrated into 4-week–old N. tabacum leaves (Sparkes et al., 2006; Xu et al., 2017).
The sgRNA-Cas9 plant expression vectors were constructed as reported in Zhang et al. (2014b). The sgRNA-Cas9 plant expression vector was kindly provided by Jiankang Zhu’s Lab (Shanghai Center for Plant Stress Biology, Chinese Academy of Sciences). The primers used in constructing the sgRNA vector for PTC2 were designed as described in Zhang et al., (2014b). All primers used in this study are listed in Supplemental Table S6.
GUS Staining Assay
Spikelets from different anther developmental stages from transgenic lines were collected as described in Zhang et al. (2010). Samples were treated, stained, and photographed as described in Xu et al. (2017).
GFP Localization of PTC2 in Rice
Spikelets of transgenic lines from the same panicle at different developmental stages were observed under a model no. TCS SP5 confocal microscope (Leica). Anthers were observed using the 20× objective lens (cat. no. HC PL APO 20×/0.7 IMM; Leica Microsystems). GFP fluorescence signals were imaged at an excitation wavelength of 488 nm and an emission wavelength of 520–580 nm. The photos were taken with a model no. LAS AF camera (Leica Microsystems).
RT-qPCR Assay
Total RNA from rice leaves, roots, lemma, palea, and anthers at different developmental stages was extracted as described in Yu et al. (2016). A Primescript RT reagent kit with a genomic DNA eraser (Takara) was used to synthesize the cDNA from 1 μg of RNA for each sample. RT-qPCR was performed with a lightCycler system (Roche) using SuperReal PreMix Plus (SYBR Green; Tiangen Biotech), according to the manufacturer’s instructions. Expression levels of target genes were reported relative to the level of actin in the same tissue. RT-qPCR profiles were generated as described in Yu et al. (2016). Three technical replicates in three biological replicates were measured for each data point. Three F values were obtained from three technical repeats in each biological repeat, and the mean value from each biological repeat was calculated form the three F values of one biological repeat. Bars indicated the variation of ± sd of means from each of the biological repeats. Primers used for RT-qPCR are listed in Supplemental Table S6.
RNA Sequencing Analysis
Stages 9 and 10 anthers from wild type and ptc2-1 were collected for RNA sequencing (RNA-seq). Sequencing was performed by the Beijing Genomics Institution.
After data filtering and quality control, the sequence data were mapped to Nipponbare genomic sequence (http://rice.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/pseudomolecules/version_7.0/all.dir/) using the program hisat2 (http://ccb.jhu.edu/software/hisat2/index.shtml). The expression level was calculated by the program HTSeq (http://htseq.readthedocs.io/en/release_0.9.1/) and DEGs were screened by the program DESeq2 (http://bioconductor.org/packages/release/bioc/html/DESeq2.html). Venn diagrams were created at http://bioinfogp.cnb.csic.es/tools/venny/. The expression level of an interest gene was normalized by the fragments mean from three biological repeats of this gene divided by its median from the fragments. Coexpression clustering was performed using the software coseq (http://bioconductor.org). GO analysis was performed with the novelbio platform (www.novelbrain.com).
Analysis of Anther Wax and Cutin Monomers
Ten milligrams of anthers from wild-type and ptc2-1 plants were harvested into liquid nitrogen. The composition of cutin and wax, along with internal standards, was analyzed as described in Xu et al., 2017). Student’s t-test was used for the statistical analysis.
Accession Numbers
Sequence data from this article for the cDNA and genomic DNA of PTC2 can be found in the GenBank/EMBL/Gramene data libraries under accession numbers LOC_Os02g0820800 and LOC_Os02g57520, respectively. The accession numbers for genes for RT-qPCR are listed: EAT1 (Os04g0599300), PTC1 (Os09g0449000), OsCP1 (Os04g0670500), OsAP25 (Os03g0186900), DPW (Os03g0167600), OsC6 (Os11g0582500), OsABCG15 (Os06g0607700), CYP704B2 (Os03g0168600).
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. TEM images of the anthers from the wild-type (WT), ptc1 and ptc2-1 mutants.
Supplemental Figure S2. Sequence alignment of selected type-I AHL proteins.
Supplemental Figure S3. Complementation analysis of PTC2 and phenotype of ptc2-2 obtained by CRISPR/Cas9 system.
Supplemental Figure S4. Subcellular localization of PTC2 in tobacco leaf epidermal cells.
Supplemental Figure S5. PTC2-GFP transgenic line in ptc2-1 showing restoration of male fertility.
Supplemental Figure S6. RNA-seq analysis of wild-type and ptc2-1 anthers.
Supplemental Table S1. Cutin and wax measurement in anther (μg/mm2).
Supplemental Table S2. DEGs in RNA-seq results at anther developmental stage 9.
Supplemental Table S3. DEGs in RNA-seq results at anther developmental stage 10.
Supplemental Table S4. Coexpression clustering analysis of DEGs.
Supplemental Table S5. GO analysis of DEGs. Enriched GO categories for Figure 9.
Supplemental Table S6. Primers used in this study.
Acknowledgments
We thank Dr. Guorun Qu for his assistance in anther cutin wax analysis and Lu Zhu for TEM sections preparation. We thank Dr. Yanjie Zhang for her contribution to map-based cloning and Dr. Natalie Betts for helpful comments on the article.
Footnotes
This work was supported by the National Key Research and Development Program of China (grant nos. 2016YFD0100804 and 2016YFD0100903), the National Natural Science Foundation of China (grant nos. 31861163002 and 31700276), the Innovative Research Team, Ministry of Education, and 111 Project (grant no. B14016), and the China Scholarship Council (full PhD scholarship to M.U.).
Articles can be viewed without a subscription.
References
- Ariizumi T, Toriyama K (2011) Genetic regulation of sporopollenin synthesis and pollen exine development. Annu Rev Plant Biol 62: 437–460 [DOI] [PubMed] [Google Scholar]
- Aya K, Ueguchi-Tanaka M, Kondo M, Hamada K, Yano K, Nishimura M, Matsuoka M (2009) Gibberellin modulates anther development in rice via the transcriptional regulation of GAMYB. Plant Cell 21: 1453–1472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk J, Leaver CJ (2001) The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13: 1803–1818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, Chen Z, Wang N, Xie G, Lu J, Yan W, Zhou J, Tang X, Deng XW (2016) Construction of a male sterility system for hybrid rice breeding and seed production using a nuclear male sterility gene. Proc Natl Acad Sci USA 113: 14145–14150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Z, Jin M, Yan W, Chen H, Qiu S, Fu S, Xia J, Liu Y, Chen Z, Wu J, et al. (2018) The ATP-binding cassette (ABC) transporter OsABCG3 is essential for pollen development in rice. Rice (N Y) 11: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Chu HW, Yuan Z, Pan AH, Liang WQ, Huang H, Shen MS, Zhang DB (2006) Isolation and genetic analysis for rice mutants treated with 60Co γ-ray. J Xiamen Univ Nat Sci 45: 81–85 [Google Scholar]
- Daneva A, Gao Z, Van Durme M, Nowack MK (2016) Functions and regulation of programmed cell death in plant development. Annu Rev Cell Dev Biol 32: 441–468 [DOI] [PubMed] [Google Scholar]
- Fu Z, Yu J, Cheng X, Zong X, Xu J, Chen M, Li Z, Zhang D, Liang W (2014) The rice basic helix–loop–helix transcription factor TDR INTERACTING PROTEIN2 is a central switch in early anther development. Plant Cell 26: 1512–1524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higginson T, Li SF, Parish RW (2003) AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J 35: 177–192 [DOI] [PubMed] [Google Scholar]
- Islam MA, Du H, Ning J, Ye H, Xiong L (2009) Characterization of Glossy1-homologous genes in rice involved in leaf wax accumulation and drought resistance. Plant Mol Biol 70: 443–456 [DOI] [PubMed] [Google Scholar]
- Ji C, Li H, Chen L, Xie M, Wang F, Chen Y, Liu YG (2013) A novel rice bHLH transcription factor, DTD, acts coordinately with TDR in controlling tapetum function and pollen development. Mol Plant 6: 1715–1718 [DOI] [PubMed] [Google Scholar]
- Jia QS, Zhu J, Xu XF, Lou Y, Zhang ZL, Zhang ZP, Yang ZN (2015) Arabidopsis AT-hook protein TEK positively regulates the expression of arabinogalactan proteins for Nexine formation. Mol Plant 8: 251–260 [DOI] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee DY, Lee YS, Schreiber L, Franke R, Faust A, Yephremov A, Saedler H, Kim YW, et al. (2006) Wax-deficient anther1 is involved in cuticle and wax production in rice anther walls and is required for pollen development. Plant Cell 18: 3015–3032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung KH, Han MJ, Lee YS, Kim YW, Hwang I, Kim MJ, Kim YK, Nahm BH, An G (2005) Rice Undeveloped Tapetum1 is a major regulator of early tapetum development. Plant Cell 17: 2705–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ko SS, Li MJ, Sun-Ben Ku M, Ho YC, Lin YJ, Chuang MH, Hsing HX, Lien YC, Yang HT, Chang HC, et al. (2014) The bHLH142 transcription factor coordinates with TDR1 to modulate the expression of EAT1 and regulate pollen development in rice. Plant Cell 26: 2486–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Pinot F, Sauveplane V, Werck-Reichhart D, Diehl P, Schreiber L, Franke R, Zhang P, Chen L, Gao Y, et al. (2010) Cytochrome P450 family member CYP704B2 catalyzes the ω-hydroxylation of fatty acids and is required for anther cutin biosynthesis and pollen exine formation in rice. Plant Cell 22: 173–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Yuan Z, Vizcay-Barrena G, Yang C, Liang W, Zong J, Wilson ZA, Zhang D (2011) PERSISTENT TAPETAL CELL1 encodes a PHD-finger protein that is required for tapetal cell death and pollen development in rice. Plant Physiol 156: 615–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Zhang D (2010) Biosynthesis of anther cuticle and pollen exine in rice. Plant Signal Behav 5: 1121–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li N, Zhang DS, Liu HS, Yin CS, Li XX, Liang WQ, Yuan Z, Xu B, Chu HW, Wang J, et al. (2006) The rice tapetum degeneration retardation gene is required for tapetum degradation and anther development. Plant Cell 18: 2999–3014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Yu J, Pearce SP, Zhang D, Wilson AZ (2017) RiceAntherNet: A gene co-expression network for identifying anther and pollen development genes. Plant J 92: 1076–1091 [DOI] [PubMed] [Google Scholar]
- Liu Z, Lin S, Shi J, Yu J, Zhu L, Yang X, Zhang D, Liang W (2017) Rice No Pollen 1 (NP1) is required for anther cuticle formation and pollen exine patterning. Plant J 91: 263–277 [DOI] [PubMed] [Google Scholar]
- Lou Y, Xu XF, Zhu J, Gu JN, Blackmore S, Yang ZN (2014) The tapetal AHL family protein TEK determines nexine formation in the pollen wall. Nat Commun 5: 3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Zou Y, Feng N (2010) Overexpression of AHL20 negatively regulates defenses in Arabidopsis. J Integr Plant Biol 52: 801–808 [DOI] [PubMed] [Google Scholar]
- Men X, Shi J, Liang W, Zhang Q, Lian G, Quan S, Zhu L, Luo Z, Chen M, Zhang D (2017) Glycerol-3-Phosphate Acyltransferase 3 (OsGPAT3) is required for anther development and male fertility in rice. J Exp Bot 68: 513–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu N, Liang W, Yang X, Jin W, Wilson ZA, Hu J, Zhang D (2013) EAT1 promotes tapetal cell death by regulating aspartic proteases during male reproductive development in rice. Nat Commun 4: 1445. [DOI] [PubMed] [Google Scholar]
- Qin P, Tu B, Wang Y, Deng L, Quilichini TD, Li T, Wang H, Ma B, Li S (2013) ABCG15 encodes an ABC transporter protein, and is essential for post-meiotic anther and pollen exine development in rice. Plant Cell Physiol 54: 138–154 [DOI] [PubMed] [Google Scholar]
- Reeves R. (2001) Molecular biology of HMGA proteins: Hubs of nuclear function. Gene 277: 63–81 [DOI] [PubMed] [Google Scholar]
- Scott R. (1994) Pollen exine: The sporopollenin enigma and the physics of pattern In RJ Scott and MA Stead, eds, Molecular and Cellular Aspects of Plant Reproduction. University Press, Cambridge, UK, pp. 49–81 [Google Scholar]
- Sgarra R, Lee J, Tessari MA, Altamura S, Spolaore B, Giancotti V, Bedford MT, Manfioletti G (2006) The AT-hook of the chromatin architectural transcription factor high mobility group A1a is arginine-methylated by protein arginine methyltransferase 6. J Biol Chem 281: 3764–3772 [DOI] [PubMed] [Google Scholar]
- Shi J, Cui M, Yang L, Kim YJ, Zhang D (2015) Genetic and biochemical mechanisms of pollen wall development. Trends Plant Sci 20: 741–753 [DOI] [PubMed] [Google Scholar]
- Shi J, Tan H, Yu XH, Liu Y, Liang W, Ranathunge K, Franke RB, Schreiber L, Wang Y, Kai G, et al. (2011) Defective pollen wall is required for anther and microspore development in rice and encodes a fatty acyl carrier protein reductase. Plant Cell 23: 2225–2246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes IA, Runions J, Kearns A, Hawes C (2006) Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat Protoc 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Tsuji H, Aya K, Ueguchi-Tanaka M, Shimada Y, Nakazono M, Watanabe R, Nishizawa NK, Gomi K, Shimada A, Kitano H, et al. (2006) GAMYB controls different sets of genes and is differentially regulated by microRNA in aleurone cells and anthers. Plant J 47: 427–444 [DOI] [PubMed] [Google Scholar]
- van Doorn WG, Beers EP, Dangl JL, Franklin-Tong VE, Gallois P, Hara-Nishimura I, Jones AM, Kawai-Yamada M, Lam E, Mundy J, et al. (2011) Morphological classification of plant cell deaths. Cell Death Differ 18: 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Lin YC, So J, Du Y, Lo C (2013) Conserved metabolic steps for sporopollenin precursor formation in tobacco and rice. Physiol Plant 149: 13–24 [DOI] [PubMed] [Google Scholar]
- Xu D, Shi J, Rautengarten C, Yang L, Qian X, Uzair M, Zhu L, Luo Q, An G, Waßmann F, et al. (2017) Defective Pollen Wall 2 (DPW2) encodes an Acyl transferase required for rice pollen development. Plant Physiol 173: 240–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yadeta KA, Hanemian M, Smit P, Hiemstra JA, Pereira A, Marco Y, Thomma BP (2011) The Arabidopsis thaliana DNA-binding protein AHL19 mediates verticillium wilt resistance. Mol Plant Microbe Interact 24: 1582–1591 [DOI] [PubMed] [Google Scholar]
- Yang X, Liang W, Chen M, Zhang D, Zhao X, Shi J (2017) Rice fatty acyl-CoA synthetase OsACOS12 is required for tapetum programmed cell death and male fertility. Planta 246: 105–122 [DOI] [PubMed] [Google Scholar]
- Yang X, Wu D, Shi J, He Y, Pinot F, Grausem B, Yin C, Zhu L, Chen M, Luo Z, et al. (2014) Rice CYP703A3, a cytochrome P450 hydroxylase, is essential for development of anther cuticle and pollen exine. J Integr Plant Biol 56: 979–994 [DOI] [PubMed] [Google Scholar]
- Yu J, Meng Z, Liang W, Behera S, Kudla J, Tucker MR, Luo Z, Chen M, Xu D, Zhao G, et al. (2016) A rice Ca2+ binding protein is required for tapetum function and pollen formation. Plant Physiol 172: 1772–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun J, Kim YS, Jung JH, Seo PJ, Park CM (2012) The AT-hook motif-containing protein AHL22 regulates flowering initiation by modifying FLOWERING LOCUS T chromatin in Arabidopsis. J Biol Chem 287: 15307–15316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Liang W (2016) Improving food security: Using male fertility for hybrid breeding. In S Sanders, T Hicklin, eds, Pushing the Boundaries of Scientific Research: 120 Years of Addressing Global Issues. Science/AAAS, Washington, DC, pp. 45-48 [Google Scholar]
- Zhang D, Liang W, Yin C, Zong J, Gu F, Zhang D (2010) OsC6, encoding a lipid transfer protein, is required for postmeiotic anther development in rice. Plant Physiol 154: 149–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DS, Liang WQ, Yuan Z, Li N, Shi J, Wang J, Liu YM, Yu WJ, Zhang DB (2008) Tapetum degeneration retardation is critical for aliphatic metabolism and gene regulation during rice pollen development. Mol Plant 1: 599–610 [DOI] [PubMed] [Google Scholar]
- Zhang D, Liu D, Lv X, Wang Y, Xun Z, Liu Z, Li F, Lu H (2014a) The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 26: 2939–2961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang D, Luo X, Zhu L (2011) Cytological analysis and genetic control of rice anther development. J Genet Genomics 38: 379–390 [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, Mao Y, Yang L, Zhang H, Xu N, et al. (2014b) The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J 12: 797–807 [DOI] [PubMed] [Google Scholar]
- Zhao G, Shi J, Liang W, Xue F, Luo Q, Zhu L, Qu G, Chen M, Schreiber L, Zhang D (2015) Two ATP Binding Cassette G Transporters, rice ATP Binding Cassette G26 and ATP Binding Cassette G15, collaboratively regulate rice male reproduction. Plant Physiol 169: 2064–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Favero DS, Peng H, Neff MM (2013) Arabidopsis thaliana AHL family modulates hypocotyl growth redundantly by interacting with each other via the PPC/DUF296 domain. Proc Natl Acad Sci USA 110: E4688–E4697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J, Favero DS, Qiu J, Roalson EH, Neff MM (2014) Insights into the evolution and diversification of the AT-hook Motif Nuclear Localized gene family in land plants. BMC Plant Biol 14: 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Xiong L (2013) Putative megaenzyme DWA1 plays essential roles in drought resistance by regulating stress-induced wax deposition in rice. Proc Natl Acad Sci USA 110: 17790–17795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Yu J, Shi J, Tohge T, Fernie AR, Meir S, Aharoni A, Xu D, Zhang D, Liang W (2017) The polyketide synthase OsPKS2 is essential for pollen exine and Ubisch body patterning in rice. J Integr Plant Biol 59: 612–628 [DOI] [PubMed] [Google Scholar]
- Zou T, Xiao Q, Li W, Luo T, Yuan G, He Z, Liu M, Li Q, Xu P, Zhu J, et al. (2017) OsLAP6/OsPKS1, an orthologue of Arabidopsis PKSA/LAP6, is critical for proper pollen exine formation. Rice (N Y) 10: 53. [DOI] [PMC free article] [PubMed] [Google Scholar]