The physical interaction between naturally occurring mutant variants of Arabidopsis circadian clock components contributes to temperature compensation in the Cape Verde Islands.
Abstract
Circadian systems share the three properties of entrainment, free-running period, and temperature compensation (TC). TC ensures nearly the same period over a broad range of physiologically relevant temperatures; however, the mechanisms behind TC remain poorly understood. Here, we identify single point mutations in two key elements of the Arabidopsis circadian clock, GIGANTEA (GI) and ZEITLUPE (ZTL), which likely act as compensatory substitutions to establish a remarkably constant free-running period over a wide range of temperatures. Using near-isogenic lines generated from the introgression of the Cape Verde Islands (Cvi) alleles of GI and ZTL into the Landsberg erecta (Ler) background, we show how longer periods in the Cvi background at higher temperatures correlate with a difference in strength of the GI/ZTL interaction. Pairwise interaction testing of all GI/ZTL allelic combinations shows similar affinities for isogenic alleles at 22°C, but very poor interaction between GI (Cvi) and ZTL (Cvi) at higher temperature. In vivo, this would result in lower ZTL levels at high temperatures leading to longer periods in the Cvi background. Mismatched allelic combinations result in extremely strong or weak GI/ZTL interactions, indicating how the corresponding natural variants likely became fixed through epistatic selection. Additionally, molecular characterization of GI (Cvi) reveals a novel functional motif that can modulate the GI/ZTL interaction as well as nucleocytoplasmic partitioning. Taken together, these results identify a plausible temperature-dependent molecular mechanism, which contributes to the robustness of TC through natural variation in GI and ZTL alleles found on the Cape Verde Islands.
Circadian oscillators in eukaryotes are comprised of multiply interlocked transcriptional-translational circuits that create and sustain rhythms of ∼24 h (Hurley et al., 2016). The Arabidopsis circadian system complex incorporates at least 20 genes in the maintenance of a robust circadian period (Hsu and Harmer, 2014; Nohales and Kay, 2016). Most current models are largely based on transcriptional relationships among clock components, but post-translational processes, such as regulated proteolysis and splicing, are increasingly being viewed as equally important (Fujiwara et al., 2008; Kim et al., 2007, 2011; Seo and Mas, 2014).
Temperature compensation (TC) is a defining feature of all known circadian systems (Edwards et al., 2005; Narasimamurthy and Virshup, 2017). This mechanism maintains the pace, or period, of the clock stably over a broad range of physiologically relevant temperatures, establishing an accurate timing mechanism independent of ambient temperature. The molecular basis of TC is unclear. Numerous studies in a variety of circadian systems have implicated different genes and mechanisms as central to its maintenance, suggesting that more than one approach to solving this problem has evolved. The strongest common element across species is the role of protein phosphorylation. In Neurospora crassa, phosphorylation of the frequency protein by casein kinase2 (CK2) alters protein turnover, especially at higher temperatures, contributing to normal TC (Mehra et al., 2009). In mammals, CK1 phosphorylates PER2 and contributes to the action of a two-site phosphoswitch that controls PER2 stability in a temperature-dependent way, contributing to TC (Isojima et al., 2009; Narasimamurthy and Virshup, 2017).
In Arabidopsis, CK2 phosphorylates CCA1, reducing its binding to oscillator genes (Portolés and Más, 2010). Altering CK2 abundance disrupts TC, in part by altering CCA1 chromatin presence (Portolés and Más, 2010). Additional elements implicated in TC in Arabidopsis include the importance of the expression levels of certain clock proteins (e.g. LHY, CCA1, PRR7, and PRR9; Gould et al., 2006; Salomé et al., 2010), factors involved in alternative splicing (James et al., 2012; Wang et al., 2012; Schlaen et al., 2015; Marshall et al., 2016), and the photoreceptor cryptochrome (Gould et al., 2013).
Recent reports on the role of GIGANTEA (GI) have clarified its central role as a post-translational stabilizer of the F-box proteins ZEITLUPE (ZTL) and FKF1, in the control of circadian period and flowering time, respectively (Kim et al., 2007). In the absence of GI or in the presence of mutations that disrupt the GI–ZTL interaction, peak ZTL levels are reduced 4- to 5-fold (Kim et al., 2007). GI acts as a cochaperone of the ubiquitous protein chaperone, HSP90, to specify the maturation of ZTL into a functional component of the SCFZTL E3 ligase that targets the transcriptional repressors TOC1 and PRR5 for ubiquitylation and degradation (Cha et al., 2017). GI protein oscillates with a late evening phase, and a blue light-enhanced GI–ZTL interaction helps create and sustain an evening-phased post-translational rhythm of ZTL abundance (Kim et al., 2007). This contributes to the maintenance of high-amplitude oscillations of TOC1 and PRR5, which is essential for normal circadian period in Arabidopsis (Kim et al., 2007; Somers and Fujiwara, 2009).
TC in Arabidopsis has been previously examined by exploiting the natural variation between two accessions of Arabidopsis adapted to different thermal environments. One population was obtained from a warm climate, the Cape Verde Islands (Cvi; 15° N to 17° N), and the second from a temperate European location (Landsberg erecta, Ler; 53° N; Edwards et al., 2005). Recombinant inbred lines (Alonso-Blanco et al., 1998) from the two accessions were used to identify several QTLs for TC. Two strong candidates identified were the core clock components GI and ZTL, which were further resolved using near-isogenic lines (NILs; Edwards et al., 2005; Keurentjes et al., 2007). ZTLCvi was implicated to promote period lengthening at high temperatures whereas GICvi was associated with shortened period, at least at 22°C (Edwards et al., 2005). These results suggested opposing effects of these alleles on period at different temperatures, though no mechanism was suggested.
Here, we identify a likely molecular mechanism for TC that arises through the cooperative action of GICvi and ZTLCvi alleles. We propose that each variant alone drives opposing period effects but ZTLCvi compensates for the overall period shortening of GICvi at higher temperatures, resulting in strong TC. This period compensation effect is mediated by changes in ZTL abundance through a thermo-dependent GI-ZTL interaction. Additionally, the GICvi allele resides in a novel functional motif that modulates the GI/ZTL interaction as well as GI nuclear trafficking. Taken together, these results identify a probable molecular mechanism that increases the robustness of Arabidopsis TC capability through the natural variation of GI and ZTL at the high thermal environments of Cape Verde Island.
RESULTS
Natural Variants of GI and ZTL Mediate Temperature-Dependent Period Variations
A previous report demonstrated temperature-specific period effects associated with the Cvi alleles of GI and ZTL (GICvi and ZTLCvi) through leaf movement analysis using NILs, suggesting GI and ZTL are candidate loci underlying the temperature effects (Edwards et al., 2005). We obtained a more direct and molecular assessment of clock activity by crossing the appropriate NILs to a CCA1:LUC (Ler) reporter line and selecting for NILs expressing CCA1:LUC and harboring either GICvi only (GCZL), ZTLCvi only (GLZC), or both GICvi and ZTLCvi (GCZC), which were compared to the Ler (GLZL) control (Supplemental Fig. S1). We then characterized the temperature effect of these genotypes on circadian period.
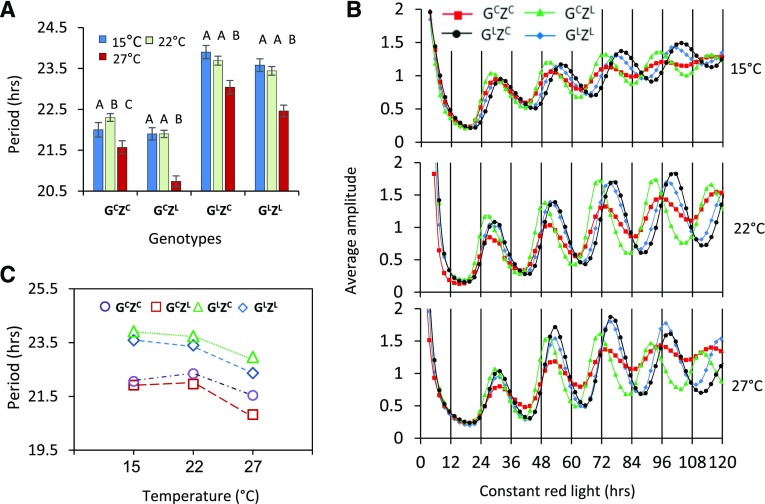
We first observed circadian rhythmicity for the NILs and wild-type Ler (GLZL) under continuous light at 15°C, 22°C, and 27°C (Fig. 1). The periods of each genotype were not significantly different within a genotype at 15°C and 22°C (Fig. 1A). However, for most genotypes periods shortened significantly at 27°C (Fig. 1A), although the magnitudes of the period changes were different among the four genotypes tested (Fig. 1, A and C; Supplemental Fig. S2, A and B). Most notably, GCZC displayed the least shortened period among the four lines, showing strong TC over the full 12°C temperature range. The two other NILs and Ler (GLZL) were extremely similar, with a strong tendency toward period shortening at higher ambient temperatures (Fig. 1C).
Figure 1.
Circadian phenotypes mediated by interactions between GI and ZTL natural alleles. A, Mean circadian period (CCA1pro:LUC) in Ler (GLZL) and three NILs harboring GICvi alone (GCZL), ZTLCvi alone (GLZC), or both GICvi and ZTLCvi (GCZC) at 15°C, 22°C, and 27°C under constant red light (35 μmol m−2 s−1) after entrainment in 12-h light/12-h dark cycles (LD). Error bars indicates ±se (n = 2 for 15°C, n = 3 for 22°C and 27°C; see Supplemental Fig. S2 for details). Different letters indicate significant differences at a given temperature within each line according to Tukey’s multiple comparisons. B, Normalized circadian traces. C, TC profiles determined by the mean period values of Ler and the NILs.
In addition, lines with GICvi (GCZC and GCZL) showed a significantly shorter period at all temperatures regardless of the ZTL allele (Fig. 1, A and C; Supplemental Fig. S2B). The periods of GCZC and GCZL were significantly shortened by ∼1.1 and 1.5 h at 22°C relative to GLZL, respectively (Supplemental Fig. S2B). The period of GCZL was similar to GCZC at 15°C and 22°C but was further shortened at 27°C. This indicates that the shortening effect of GICvi is most acute at high temperatures.
In contrast, ZTLCvi modulates the temperature-dependent period shortening effect of GICvi (Fig. 1). The two GLer lines (GLZC and GLZL) were similar in period at 15°C but at higher temperature the period of GLZC remained longer, relative to the Ler control (Supplemental Fig. S2B). A high temperature-dependent period lengthening effect of ZTLCvi is consistent with this difference, and when paired together with GICvi (GCZC) at 27°C, it can explain the strong TC seen in this line (Fig. 1, A and C). Overall, the NIL data support the notion that at higher temperatures GICvi has a period shortening effect that is counterbalanced by ZTLCvi, supporting a nearly unchanging period over this range of temperatures when the two alleles are together (GCZC; Fig. 1C). The findings are similar under white light, with the trend of GCZC less acute than for the other two NILs and the Ler parent line (Supplemental Fig. S3).
These conclusions are bolstered by statistical modeling analysis (Supplemental Methods). We analyzed the red and white-light data sets using a linear model with the period as an outcome and the loci GI, ZTL, and temperature (15°C, 22°C, and 27°C) as categorical covariates. This approach allowed consideration of which of these three factors, and their potential interactions, contributed to period variation. Based on this analysis, we found that GICvi has a significant period-shortening effect under red light, compared to GILer. In contrast, ZTLCvi has, to a lesser degree, significant period-lengthening effect, compared to ZTLLer (see Supplemental Methods; Supplemental Table S1). High temperature (27°C) also contributes to period shortening, as visually apparent in Figure 1. Under white light, the independent effects of the alleles are the same, but additionally there is a strong period lengthening effect contributed by the joint effect of GICvi and ZTLCvi at high temperature (see Supplemental Methods; Supplemental Table S2).
These period effects mediated by GICvi and ZTLCvi are unlikely to be due to background differences at other loci because independent segregants derived from the original NILs x Ler (CCA1-LUC) crosses (Supplemental Fig. S1) showed similar period effects among them (Supplemental Fig. S4), and additional segregants derived from the GCZC x Ler (CCA1-LUC) crosses (Supplemental Figs. S5 and S6) exhibited the expected period phenotypes.
GI-ZTL Allelic Interactions Can Account for High Temperature-Enhanced ZTL Accumulation
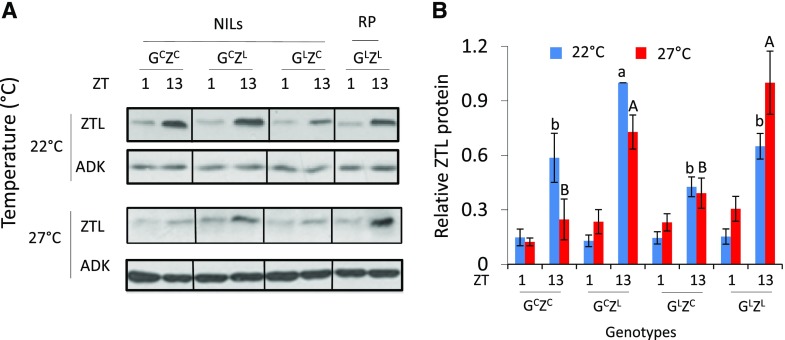
Circadian period and ZTL protein level are inversely correlated in Arabidopsis (Somers et al., 2004) and GI positively regulates ZTL protein levels through direct protein interactions (Kim et al., 2007). Therefore, we next tested whether the opposing effects GICvi and ZTLCvi have on period might arise from altered ZTL levels. We monitored ZTL levels at Zeitgeber time (ZT) 1 and 13 in seedlings of the NILs (GCZC, GLZC, and GCZL) and Ler (GLZL) grown at 22°C and 27°C under light/dark (LD) cycles (Fig. 2). We observed differences in steady-state levels of ZTL among the lines and at the different temperatures.
Figure 2.
Temperature effects on endogenous ZTL abundance. A, Endogenous ZTL levels in NILs and the recurrent parent (RP). Seedlings were grown for 7 d in 12-h light/12-h dark cycles (LD; 22°C), then continued at 22°C or 27°C for five additional days under LD cycles and sampled at ZT1 and ZT13. Immunoblot representative of at least three trials. B, Quantitation of ZTL levels. Error bars represent ±se (n = 3). Different letters indicate significant differences at a given temperature according to Tukey’s multiple comparisons, followed by ANOVA. ADK was used as loading control. Quantitation relative to ADK band and normalized to highest protein level at a given temperature.
Figure 2 shows that the shorter periods consistently associated with GICvi are not due to marked differences in ZTL levels. All genotypes have nearly identical ZTL levels at ZT1 at 22°C. At ZT13 (22°C), ZTL levels in both GLZC and GLZL are very similar to those in the shorter period GCZC line (Fig. 2B). Similarly, at 27°C, both GILer lines show longer periods than GICvi lines (Fig. 1, A and C), despite similar ZT13 ZTL levels in, for example, GCZL and GLZL at both time points (Fig. 2). This indicates that the consistently shorter periods of GICvi genotypes arise from effects of GI on other aspects of the circadian system.
Within a GI genotype, however, the period reflects the expected effects of ZTL and GI interactions inferred from the genetics, particularly at 27°C. The longer period of GLZC relative to GLZL at 27°C (Fig. 1) correlates well with the higher ZTL levels in GLZL (at ZT13) and its shorter period (Fig. 2). ZTLCvi is less abundant in the presence of GILer, especially at 27°C (Fig. 2), correlating with the longer period (Fig. 1).
For GICvi, ZTL protein abundance also correlates strongly with period in a ZTL allele-specific way. At 22°C, the ZTL level of GCZL is significantly higher relative to GCZC at ZT13, supporting a positive role of GICvi on ZTLLer stability and a shorter period (Fig. 2; Fig. Supplemental S2B). This effect is further accentuated at 27°C, where ZTL levels in GCZL are nearly 3-fold higher than those found in GCZC at ZT13 (Fig. 2B), correlating with the much shorter period of GCZL (Fig. 1; Supplemental Fig. S2B). Taken together, these results support the notion that GI controls ZTL accumulation depending on the particular combination of alleles. Hence, to account for the better TC of GCZC, we hypothesize that the longer period at 27°C arises from an inherently strong interaction of GICvi with ZTL being counterbalanced by an inherently weaker ZTLCvi interaction. This would result in lower accumulation of ZTL and the longer period.
Examination of the mRNA expression patterns of the GI and ZTL loci under the same temperatures did not show any significant differences among them, indicating these effects occur post-transcriptionally (Supplemental Fig. S7).
Temperature-Dependent ZTL Accumulation Results from Temperature Effects on GI-ZTL Interactions
We tested the above hypothesis by addressing the molecular basis of the different GI and ZTL allelic combinations on ZTL levels. Amino acid sequence alignments of the GI and ZTL alleles of Col, Ler, and Cvi accessions revealed that GICvi and ZTLCvi differ from GILer and ZTLLer by single amino acid substitutions of a Leu-to-Phe (L718F) and a Pro-to-Thr (P35T), respectively (Supplemental Fig. S8). Additionally, GICol is nearly identical to GILer except for an I-to-V substitution at residue 113 (Supplemental Fig. S8).
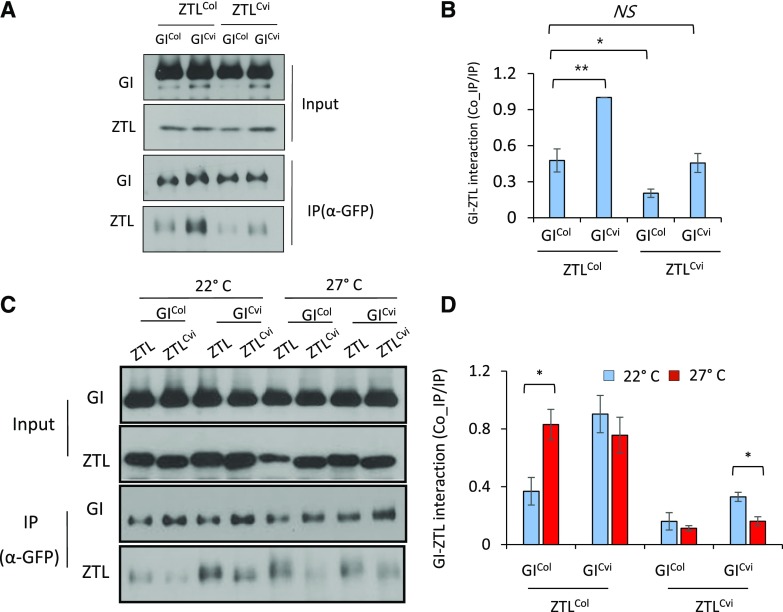
To determine whether these differences alter the GI-ZTL interaction, we performed quantitative coimmunoprecipitation (co-IP) assays analyzing all combinations of GI and ZTL natural alleles through transient expression in Nicotiana benthamiana (Fig. 3). In these interaction experiments, we compared the more commonly used Columbia (Col-0) alleles to Cvi alleles, so we first established that the interactibility of GICol and GILer with ZTLCol/Ler are not significantly different (P = 0.210, two-sample Student’s t test). In vivo, ZTL protein levels in the Col and Ler accessions are very similar, suggesting that ZTL interacts equally well with both GI alleles (Supplemental Fig. S9).
Figure 3.
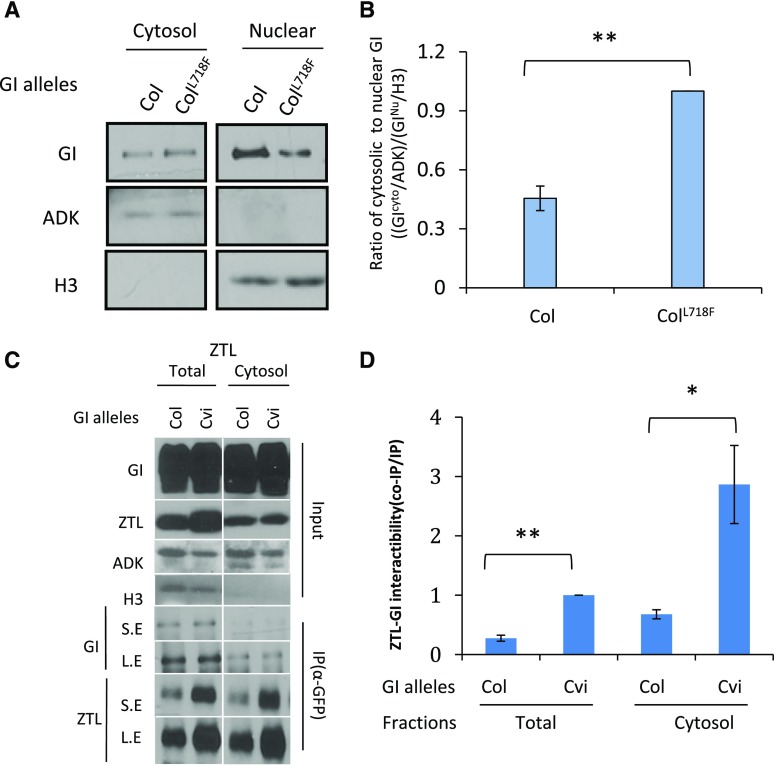
GI and ZTL temperature-dependent interactions. A and B, N. benthamiana plants were grown at 22°C under 16-h light/8-h dark cycles (LD) and protein extracts from leaves transiently expressing GICol-GFP and GICvi –GFP (GICvi) coexpressed with ZTLCol or ZTLCvi were harvested at ZT12. Anti-GFP immunoprecipitates (IP) were probed with anti-ZTL polyclonal antibody (co-IP). Quantitation based on the ratio of co-IP/IP signals, normalized to strongest interaction. Image is representative of at least three trials; error bars represent ±se (n = 3). C and D, As in (A) and (B), but for plants maintained in 22°C or 27°C before extraction and processing. Statistical difference between pairs (Student’s t test) is shown as *P < 0.05, **P < 0.01. NS, not significant.
ZTLCol or ZTLCvi were coinfiltrated with tagged GICol-GFP or GICvi-GFP into N. benthamiana and the association of ZTL with GI-GFP in the immunoprecipitates was measured at 22°C (Fig. 3). When a ZTL allele was paired with the complementary GI variant naturally found in that accession, the extent of the interaction was very similar: GICol/ZTLCol and GICvi/ZTLCvi interactions were nearly identical (Fig. 3B). In contrast, the test of the GICvi /ZTLCol interaction resulted in significantly larger amounts of coimmunoprecipitated ZTL, whereas the GICol /ZTLCvi combination showed the weakest interactions (Fig. 3B). These results strongly suggest an epistatic selection for similar ZTL-GI interactiveness within an accession under 22°C.
We extended these tests in N. benthamiana that were maintained at 27°C for 3 d after infiltration. Here, we found that the paired Col-0 alleles of GI and ZTL interacted >2-fold better at 27°C relative to that seen at 22°C (Fig. 3, C and D). Conversely, ZTL-GI Cvi interactions at 27°C were reduced by 2-fold (Fig. 3). These results are consistent with the long period (Fig. 1, A and C) and lower amounts (Fig. 2) of ZTLCvi observed in the GCZC NIL at 27°C.
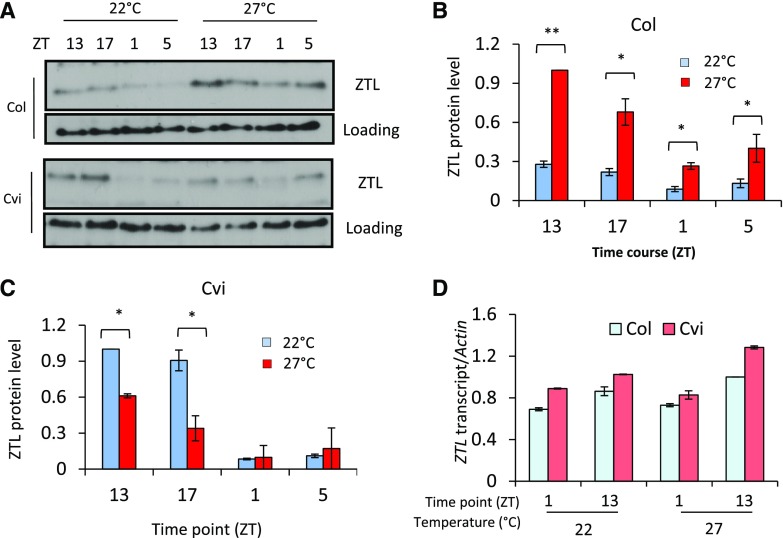
We conducted a more extensive time series and compared endogenous ZTL protein accumulation within two Cvi accessions and Col at 22°C and 27°C. ZTL levels in Col were consistently higher at all-time points at 27°C, consistent with a shorter period at that temperature (Fig. 4, A and B). In contrast, peak ZTLCvi levels were lower at 27°C over the time series (Fig. 4, A and C). When samples were compared on the same immunoblot with the same exposure, similar trends were observed (Supplemental Fig. S10). ZTL transcript levels are not markedly different under these conditions, indicating that the temperature-dependent effects on ZTL protein levels are determined post-translationally (Fig. 4D). Taken together, results from both the transient coexpression data and in vivo ZTL levels demonstrate that a temperature-dependent interaction between GI and ZTL results in changes in the steady state of ZTL over a diurnal time course.
Figure 4.
Temperature-dependent ZTL accumulation depends on genotype. A to C, Seedlings (Col, Cvi) were grown for 7 d in 12-h light/12-h dark cycles (LD) at 22°C, continued at 22°C or 27°C for an additional 5 d under LD cycles, then sampled at the indicated ZT. Endogenous ZTL protein was probed with anti-ZTL polyclonal antibody. ZTL abundance was quantitated relative to a nonspecific band above the target protein and normalized to the most abundant level within each genotype for both temperatures. D, ZTL transcript abundance determined by reverse transcription quantitative PCR and normalized to ACTIN2. Cvi data are from four independent trials; two with Cvi-0 and two with Cvi-1 together with the corresponding Col accession. Statistical difference between pairs (Student’s t test) is shown as *P < 0.05, **P < 0.01. Error bars (B–D) represent ±se (n = 4).
GICvi Enhances GI Cytosolic Localization and Strengthens the GI-ZTL Interaction
GI is present in both the nucleus and cytosol and the relative partitioning of GI can affect ZTL levels (Kim et al., 2013a). Previous work indicated residues 543–783 are sufficient for nuclear targeting, within which the Cvi/Ler polymorphisms (GI718) reside (Huq et al., 2000). Therefore, we next tested whether the differences in ZTLCvi and ZTLCol/Ler accumulation might also arise, in part, from differences in the nucleocytoplasmic partitioning of the different GI alleles. We tested the effect of GICvi on nucleocytoplasmic partitioning through transient expression. GICvi displayed 2-fold enriched presence in the cytosol relative to GICol (P < 0.001, one-way ANOVA; Fig. 5, A and B).
Figure 5.
GICvi enhances cytosolic partitioning and ZTL interaction. A and B, GICol-GFP and GICvi-GFP transiently expressed in N. benthamiana and separated into cytosolic and nuclear fractions. Immunoblot is representative of three trials. Levels of cytosolic and nuclear GI protein were normalized to ADK and H3. Error bars represent ±se of three independent trials. C and D, N. benthamiana leaves transiently and ectopically expressing GICol –GFP or GICvi –GFP coexpressed with ZTLCol were harvested at ZT12. Anti-GFP immunoprecipitates (IP) were probed with anti-ZTL polyclonal antibody (CoIP). Quantitation based on the ratio of co-IP/IP signals and normalized to the GICvi-ZTL total interaction. Image representative of at least three trials; error bars represent ±se (n = 3). S.E, short exposure; L.E, long exposure. Statistical differences between pairs (Student’s t test) is shown as *P < 0.05, **P < 0.01.
As ZTL is localized in the cytosol, we tested whether the allele-specific differential interactions that we observed among the different GI and ZTL alleles were due to the higher affinity of GIL718F for ZTL or its predominant cytosolic localization. We compared the interaction of GICol and GIL718F with ZTL and found that in both total and cytosolic extracts GIL718F interacts 3-fold more strongly with ZTL relative to GICol, indicating that the enhanced protein accessibility of GIL718F to ZTL cannot explain the strong interaction (Fig. 5, C and D). We confirmed in vitro that the GICvi allele has intrinsically higher affinity for ZTL using GST-tagged ZTL (P < 0.05, two-sample Student’s t test; Supplemental Fig. S11).
GI718L Is within a Functional Unit that Modulates GI/ZTL Interaction and GI Nucleocytoplasmic Localization
Closer inspection of the sequence shows that the L718F substitution occurs within a highly conserved Leu-rich region at residues 712–721 (LXXLXXLXXL; Supplemental Fig. S12) that is found across many taxa. Hence, it is plausible that the L-to-F difference in GICvi at GI(718) interrupts nuclear targeting either by modifying this motif or a neighboring nuclear localization sequence.
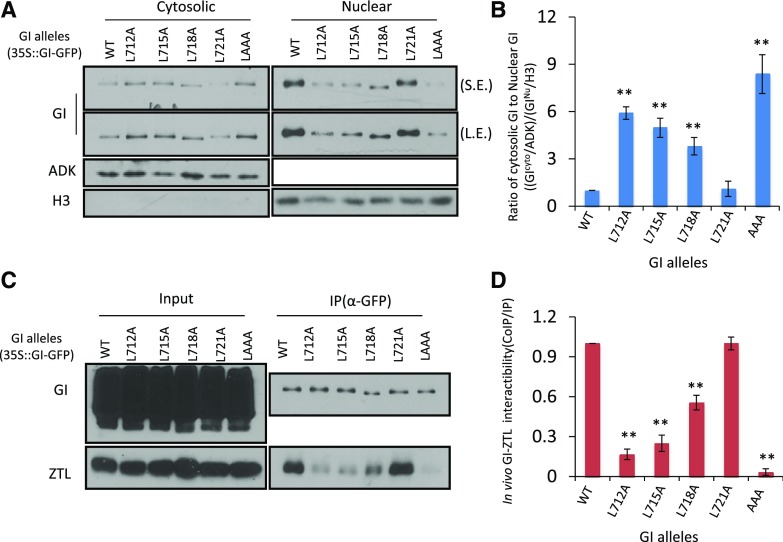
To determine whether this motif is required for either the GI/ZTL interaction or nuclear import, we created serial L-to-A mutations in the LXXLXXLXXL region (Fig. 6). We found that single L-to-A mutations (L712A, L715A, and L718A) within this region diminished the relative nuclear abundance, with substitutions closer to the N terminus being the most effective. Moreover, the triple mutant (L715A, L718A, and L721A) markedly suppressed the nuclear presence of GI relative to that of the single mutants (Fig. 6, A and B). Notably, the nuclear accumulation of GIL721A is comparable to the wild type (GICol), demonstrating the necessity of only the L712XXL715XXL718 motif for nuclear accumulation (Fig. 6, A and B).
Figure 6.
The L712XXL715XXL718 motif determines GI nucleocytoplasmic partitioning and ZTL interaction. A and B, N. benthamiana leaves transiently and ectopically expressing GICol-GFP, GIL712A-GFP, GIL715A-GFP, GIL718A-GFP, GIL721A-GFP or GILAAA-GFP were separated into cytosolic and nuclear protein fractions and detected with anti-GFP antibody. Immunoblots quantitated as the ratios of cytosolic to nuclear GI. Levels of cytosolic and nuclear GI protein were normalized to ADK and H3, respectively. Error bars represent ±se of three independent trials. C and D, N. benthamiana leaves transiently and ectopically expressing GI alleles in (A) and (B) coexpressed with ZTLCol were harvested at ZT12. Anti-GFP immunoprecipitates (IP) were probed with anti-ZTL polyclonal antibody (Co-IP). Interactions were quantitated by the ratio of co-IP/IP signals, normalized to the strongest interaction. Gel image is representative of at least three trials; error bars represent ±se (n = 3). Statistical difference between pairs (Student’s t test) is shown as *P < 0.05 and **P < 0.01. WT, wild type.
We next determined if this same motif affects the GI-ZTL interaction. Quantitative co-IPs with these GI mutant alleles and ZTL showed that mutations in the motif weaken the GI-ZTL interaction (Fig. 6, C and D). GIL718A diminished the interaction slightly (<2-fold) relative to the wild type, but GI L712A and GIL715A strongly decreased the interaction further. Furthermore, the triple GI mutant allele completely abrogated the interaction (Fig. 6, C and D). Together, these data indicate that the LXXLXXL motif is a necessary determinant for both the GI-ZTL protein interaction as well as nucleocytoplasmic localization. Both functions are most strongly affected by loss of the more N-terminal Leu residues (Fig. 6, B and D).
Natural Variants of GIL718F and ZTLP35T Are Unique
To investigate how frequently the GIL718F and ZTLP35T alleles might emerge in nature, we used Polymorph 1001 to examine the sequences of the GI and ZTL loci in 1135 Arabidopsis accessions (https://tools.1001genomes.org/polymorph/), in addition to ecotypes adapted to hot areas, including Tanzania (Tanz-1 and -2) and Marrakesh, Morocco (Aitba-2 and Toufl-1; Supplemental Fig. S13). With the exception of Cvi, none of these Arabidopsis accessions, including hot-adapted ecotypes, harbored either GIL718F or ZTLP35T. Considering the functional impacts of the natural alleles for TC, the co-occurrence of these alleles in Cvi may be an evolutionary outcome unique to Cvi, where the average daily temperature is much higher than in many other Arabidopsis habitats.
DISCUSSION
Temperature-Dependent GI-ZTL Interactions Alter Period through Effects on ZTL Abundance
In this study, we have exploited the natural genetic variation between two accessions of Arabidopsis to identify a role for ZTL and GI in the TC of the circadian clock. We propose that the mechanism behind this process hinges on a differential, temperature-dependent interaction between the two proteins based on two single amino acid changes within each of them, specifically L718F in GI and P35T in ZTL, which are present in the Cvi accession and absent in the Col and Ler alleles. We show that pairings between mismatched alleles result in a much reduced interaction (GICol/ZTLCvi) or an enhanced interaction (GICvi/ZTLCol) relative to like allelic pairings (GICol/ZTLCol and (GICvi/ZTLCvi; Fig. 3) at 22°C, which are balanced to result in very similar interactivity. However, at higher temperatures (27°C) this balance is lost, with a much reduced interaction seen between GICvi/ZTLCvi and more enhanced interaction apparent between GICol/ZTLCol (Fig. 3D). We propose that these interaction differences are sufficient to account for difference in the net accumulation of ZTL in the two backgrounds: a much higher level of ZTL in Col-0 at 27°C versus 22°C compared to in the Cvi accession under the same conditions (Fig. 4). This finding is consistent with the role of GI in the maturation of functional ZTL (Kim et al., 2007; Cha et al., 2017).
In turn, the lower ZTL levels correlate with the longer period in the Cvi accession at higher temperatures (Fig. 4; Somers et al., 2004). The period-shortening effect of the GICvi allele is more acute at higher temperatures and it is the weaker interaction between GICvi and ZTLCvi that results in the longer free-running period. Thus, the evidence suggests that temperature-dependent interactions between different ZTL and GI alleles underlie TC in this environmental setting.
Interestingly, an analogous relationship exists in the mouse (Mus musculus) clock, also as a mechanism of TC. Here, a balance between temperature-sensitive substrate binding and temperature-sensitive product binding underlies how CKIδ-dependent phosphorylation of its substrate (PER) is subject to TC (Shinohara et al., 2017). A single amino acid change at the enzyme′phosphopeptide binding site of CKIδ markedly alters the temperature dependency of CKIδ-dependent phosphorylation of PER by impairing ADP-dependent product binding. This mutant shows much greater degradation activity of PER2 protein at a higher temperature compared to wild-type CKIδ, resulting in shorter periods. Thus, a temperature dependency in substrate binding and product release is inherent in CKIδ structure, akin to how the temperature-dependent interaction between GI and ZTL turns on two specific, complementary residues within the two proteins.
GICvi Shortens Period Independent of the ZTL Allele
The single L718F transition of GICvi results in a consistently shorter period than the GILer allele regardless of the ZTL allele with which it is paired and under all the growth temperatures tested (Fig. 1; Supplemental Fig. S2). This could indicate that GICvi is expressed at higher levels than GILer, hence raising ZTL levels, resulting in shorter periods. However, GCZL and GLZL have similar ZTL levels at 27°C (Fig. 2B), yet differ in the period by >2 h (Fig. 1; Supplemental Fig. S2). Similarly, at 22°C ZTL levels in GCZC and GLZC are about the same (Fig. 2B), yet GCZC period is >1 h shorter than GLZC (Supplemental Fig. S2). Thus, it appears that GI can also control circadian period through other mechanisms, separate from its effects on ZTL accumulation. This notion is supported by the fact that many GI mutants display both short periods and reduced ZTL levels (Kim et al., 2007; Somers et al., 2007). Interestingly, this strong period shortening effect of GICvi is diminished under white light (Supplemental Fig. S3), although the tendencies seen under red light for each NIL are still present. This finding suggests that the added presence of blue light may differentially affect one variant of GI over the other.
Two known GI interactors, ELF3 and ELF4, are core components of the tripartite Evening Complex (EC; ELF3-ELF4-LUX), a key element of the circadian clock that also helps integrate environmental signals with endogenous processes (Nusinow et al., 2011; Herrero et al., 2012; Ezer et al., 2017). ELF4 interacts with and sequesters GI in the nucleus, restricting its chromatin access (Kim et al., 2013b). Further, ELF3, together with COP1, controls GI turnover in the dark (Yu et al., 2008). Hence, allelic differences among the Cvi and Ler/Col accessions at one or more of these loci could be partially responsible for the ZTL-independent period differences observed in the NILs that are linked to GICvi. For example, natural variation in ELF3 nuclear abundance has been associated with shortened free-running period, though the molecular basis of the difference in its nucleocytoplasmic partitioning is unknown (Anwer et al., 2014). Similarly, known variation in the poly-Gln (polyQ) tract length in ELF3 (7–26 polyQs) can correlate with ELF3-dependent phenotypes (Undurraga et al., 2012; Press and Queitsch, 2017). ELF3Cvi (nine polyQs) and ELF3Ler (17 polyQs) may interact differently with GICvi, leading to period differences.
Although nuclear levels of GI have only been linked to effects on flowering time, through interaction with FKF1, natural variation in ELF4 nuclear sequestration of GI may indirectly affect period through GI effects on clock gene expression. Similarly, GI expression levels are affected by temperature, which is mediated through direct binding of the EC to the GI promoter (Mizuno et al., 2014). Natural variation in the relative affinity of the EC components for each other, and its chromatin residence, may subsequently alter GI levels in a temperature-specific manner. Further testing of the expression levels and interaction features of the Cvi and Ler/Col alleles of GI, ELF3, and ELF4 may resolve the ZTL-independent effects of GI on period.
The GI Leu-Rich Motif (LXXLXXL) Controls the GI-ZTL Interaction and GI Partitioning
We found that the L718F substitution in GI increases its affinity to ZTL but L718A strongly diminishes the interaction. Furthermore, serial changes at L712, 715, and 718 to A but not at L721 additively compromise both the GI-ZTL interaction and GI nuclear presence. Taken together, these results clearly support the notion that the L718 residue of GI is a part of Leu-rich unit motif (L712xxL715xxL718) that facilitates both the GI/ZTL interaction and nucleocytoplasmic partitioning. This sequence is superficially similar to the Leu-rich repeat sequence motifs (LxxLxLxxNxL or LxxLxLxxCxxL) that form the core of this well-characterized protein interaction domain (Matsushima and Miyashita, 2012). However, this stand-alone motif in GI is not part of a larger, repeating sequence (typically 4–30 repeats) and is likely unrelated to the structural and signaling functions found in plant receptor kinases (Leu-rich repeat receptor kinases).
Using the nuclear export sequence (NES) predictor, LocNES, the L712xxL715xxL718 motif was shown to reside within the second most likely NES of GI (Xu et al., 2015). Replacing L718 with either A or F greatly diminishes the predicted likelihood of this motif as a functional NES. If these changes disrupted an NES, we would predict a greater nuclear accumulation of GI. However, we observe a stronger cytosolic GI accumulation with both mutations (Figs. 5 and 6), suggesting that this region affects GI nucleocytoplasmic partitioning through a different mechanism. The Leu-rich motif also lies within a region (residues 543–783) previously identified as controlling nucleocytoplasmic partitioning (Huq et al., 2000). Strongly predicted nuclear localization sequence (SGSKRPRSEY) and NES (CAGVELASRLLFVV) motifs also reside within this stretch. Hence, our findings corroborate this previous report as defining an essential nucleocytoplasmic partitioning region of GI.
Our previous mapping of the GI-ZTL interaction domains identified the ZTL N terminus as essential for GI interaction but did not resolve the necessary GI elements (Kim et al., 2007). The Leu-rich motif now identifies a unique region within GI that is clearly essential for ZTL binding. The very large size of GI (1,173 residues) and many binding partners (Mishra and Panigrahi, 2015; Cha et al., 2017) suggests that it can fold to present multiple interaction surfaces. GI, ZTL, and HSP90 form a tripartite complex in which the GI N terminus (amino acids 1–391) interacts with the middle domain of HSP90 (Cha et al., 2017), and now a central region (amino acids 712–718) is essential for the GI-ZTL interaction. Taken together, these findings begin to establish functional domains of GI essential to its role in the circadian clock.
Potential Epistatic Selection of GICvi and ZTLCvi
Epistatic interactions have been well-documented as underlying autoimmune responses and pathogen resistance in Arabidopsis (Bomblies et al., 2007; Alcázar et al., 2009; Chae et al., 2014; Zhu et al., 2018). Genetic analyses in these studies have implicated certain gene clusters or individual loci as important, but the biochemical or physical mechanism underlying the resultant phenotypes (e.g. poor growth, necrosis) has rarely been identified. Fixation of allelic pairs through the simultaneous selection of combinations of genotypes at two loci may arise through epistatic selection (Schlosser and Wagner, 2008; Nasrallah, 2013). Compensatory mutations, or amino acid substitutions, in interacting proteins that, together, offset their individual effects, are one type of epistatic interaction.
Compensatory intramolecular substitutions that affect RNA secondary structure have been described for HIV and the pre-mRNA of alcohol dehydrogenase (Kirby et al., 1995; Assis, 2014). In these examples, mutations that disrupt base pairing in an element of RNA secondary structure are considered deleterious if they destabilize a functionally important structure. If a second compensatory mutation arises, the original structure may be restabilized and fitness restored (Kirby et al., 1995). Similarly, intraprotein compensatory mutations can be recovered, for example, as second-site suppressors of temperature-sensitive folding mutations, which can restore protein activity or overall fitness (Mitraki et al., 1991, 1993; DePristo et al., 2005).
This concept can be expanded to consider the interaction between two or more proteins (Schlosser and Wagner, 2008). For example, the strong interactions between two key sex determination proteins, FEM-3 and TRA-2, which occur within three different species of Caenorhabditis, fail when tested across the species (Haag et al., 2002). These results strongly suggest a compensatory coevolution of binding partners. For GI and ZTL, we find that cross-accession alleles still interact, but with altered affinities; the divergence of interaction has not progressed to the extent found in Caenorhabditis species. Further interaction testing of GI and ZTL alleles from other Arabidopsis species and relatives could shed insight into how important their affinities are for period maintenance in general.
Compensatory substitutions may arise in different ways, depending on the strength of natural selection. If selection is very weak and the population size is small, a deleterious mutation may become fixed in the population before a second (compensatory) mutation arises, and then subsequently the pair becomes fixed. Under stronger selection, the first mutation may appear deleterious and compensatory substitution can only occur if the second mutation arises before the first is selected against (Nasrallah, 2013). However, without knowing more about the evolutionary history of the origins of GICvi and ZTLCvi or the conditions under which their interaction was selected, it is not possible to know how they became fixed in the Cvi population.
The pairing of these specific GI and ZTL alleles in Cvi is unique among known Arabidopsis accessions. Our data supports the notion that both alleles were selected to help establish strong TC at high temperatures. Whereas it might be expected that other populations indigenous to high-temperature environments would show the same or similar compensatory mutations in GI and ZTL, no common or universal molecular mechanism of TC has been identified in either plants or animals. Hence, it is likely different solutions have independently evolved to attain period stability at high ambient temperatures. In this case, field tests could, in principle, be performed in which all four combinations of the Cvi and Ler/Col-0 GI and ZTL alleles were created (possibly via CRISPR-Cas mediated genome editing) in the Cvi background and tested in a high-temperature field environment for reproductive success. Reduced seed set or reduced survival of populations with Col/Ler alleles in place of the Cvi alleles would support the notion of a selective advantage conferred by the endogenous Cvi alleles. This outcome would be strong support for our hypothesis that the GI and ZTL Cvi alleles were selected for high temperature stabilization of circadian period.
Recent genomic analysis of Arabidopsis accessions sampled from within Africa and Asia suggests a history of the species that includes a more widespread historical distribution across Africa than previously considered (Durvasula et al., 2017; Zou et al., 2017; Fulgione and Hancock, 2018). Earlier assessments, based on 1,135 Arabidopsis accessions, concluded that Arabidopsis is comprised largely of a single majority group (1,109 accessions) out of six diverged groups, all survivors from multiple glacial refugia. From this analysis, Cvi-0 is in an outlier group, with recent evidence indicating an African origin based on S-haplotype analysis (Nordborg et al., 2005; The 1001 Genomes Consortium, 2016; Durvasula et al., 2017). The average daily temperature of the Cape Verde Islands tropical bioclimatic zone is higher over the year, with only mild variation, ranging from 25°C (77°F) in January to 29°C (84.2°F) in September (Abarca et al., 2001). Thus, the colonizing Arabidopsis populations on the Cape Verde Islands were likely exposed to thermal selective pressures very different from continental accessions, particularly at the high end of the temperature range.
The Cape Verde Islands environment is geographically remote and genetically isolated, yet GI and ZTL are remarkably conserved in their peptide similarity compared to alleles found elsewhere, except for these two substitutions. The evidence we provide here strongly suggests these are compensatory mutations, possibly arising through epistatic selection, which create an appropriately balanced GI/ZTL interaction at high temperatures to attain an adaptively temperature-compensated circadian clock.
MATERIALS AND METHODS
Plant Material
To obtain NILs covering GICvi and ZTLCvi, we screened candidate NILs through SNPs in GI (C to T at nucleotide 3,061, AT1G22770) and ZTL (C to A at 103, AT5G57360) from NILs in which the Cvi accession had been introgressed into the Landsberg erecta (Ler) background (Keurentjes et al., 2007). CCA1: luciferase (Ler; Wang et al., 2013) were crossed to the relevant NIL lines and luminescent progeny homozygous for GICvi or ZTLCvi or both were selected for subsequent rhythmic assay and protein analysis.
CCA1:luciferase was transformed into the Cvi-0 background via standard techniques. Two independent transformants were selected for subsequent study.
The NILs (CS717143–CS717147), Arabidopsis (Arabidopsis thaliana) ecotypes Cvi-0 (CS28198), Cvi-1 (CS28199), Tanz-1 (CS75719), Tanz-2 (CS75720), Aitba-2 (CS76347), and Toufl-1 (CS76348)) were obtained from the Arabidopsis Biological Resource Center (The Ohio State University).
Growth Condition and Temperature Treatments
All plant materials were grown under 12-h white fluorescent light (50–60 μmol m−2 s−1)/12-h dark cycles (LD) for 8–12 d on Murashige and Skoog (MS; GIBCO BRL) medium with 3% (w/v) Suc and 1% (w/v) agar as described in Somers et al. (2004). For all period analyses, seedlings were grown on MS agar under LD cycles for 5–7 d at 22°C and then transferred to constant red light (35 µmol m−2 s−1) at 15°C, 22°C, or 27°C. For protein expression experiments, 7-d–old LD-grown (12-h light/12-h dark) seedlings (MS agar media) at 22°C were then incubated at 15°C, 22°C, or 27°C for five additional days under the LD cycles. Seedlings were harvested at the indicated ZT times.
Rhythmic Assay
Seedlings for luminescence rhythm analysis were grown and period estimates determined as described in Somers et al. (2004). Constant white light (5,700 K) during circadian imaging experiments was provided by LED RX30 lamps (Heliospectra). Red light was provided by a 660-nm LED array.
Vector Construction
The 35S::GI-GFP, GI complementary DNA (cDNA) clone was PCR-amplified using the full-length GI cDNA derived from the Columbia ecotype (Col-0; Kim et al., 2007) using the primers 5′-GAA TTC ATG GCT AGT TCA TCT TCA T-3′ and 5′-CGC GGA TCC ATG GCT AGT TCA TCT TCA TCT GAG-3′. The amplified GI cDNA digested with EcoRI and BamHI was ligated into pENTR2B and transferred to pMDC85 via Gateway cloning. The 35S::ZTL was prepared by recombination of an entry clone that contained the ZTL cDNA (Kim et al., 2013a) to pMDC32. GI alleles, including GIL718F, GIL712A, GIL715A, GIL718A, and GIL721A, and the ZTL allele ZTLP35T were created through the site-directed mutagenesis with a QuikChange system (Stratagene) using GI and ZTL cDNA sequences in the entry clone.
Protein Analysis
Detection of ZTL and GI-GFP, adenosine kinase (ADK), and Histone H3 from Arabidopsis protein extracts was performed as described in Kim et al. (2007). For immunoprecipitation of GI-GFP and Co-IP of ZTL, anti-GFP polyclonal antibody was preincubated with protein A agarose (Invitrogen) at 4°C. After incubation for 1 h with gentle rotation, immune complexes were washed three times, then resuspended in SDS-PAGE sample buffer, briefly heated, and subjected to SDS-PAGE followed by immunoblotting. To quantify the protein interactions, the signal intensity of coimmunoprecipitated protein was normalized to that of primary-immunoprecipitate. For ZTL immunoblotting, protein was transferred to a PVDF membrane (Bio-Rad) and incubated with affinity-purified anti-ZTL antibody (1:500) for 4 h at room temperature or overnight at 4°C. The signal was visualized using peroxidase-conjugated secondary antibody (1:3,000–5,000; GE or Sigma-Aldrich) by enhanced chemiluminescence (ECL SuperSignal West Pico Chemiluminescent substrate; Pierce Biotechnology).
Isolation of nuclear and cytosolic proteins from tobacco tissue was performed with a CelLytic PN Isolation/Extraction Kit (Sigma-Aldrich) as described in Wang et al. (2010). Purification of each compartment was validated by immunodetection using cytosolic (ADK) and nuclear (Histone H3) marker proteins.
For the GST binding assay, GST and GST-ZTL were expressed in Escherichia coli BL21(DE3) and purified with glutathione-agarose (Sigma-Aldrich). Concentration of each protein bound to the glutathione-agarose was determined by Coomassie staining. Equal amounts of glutathione beads of GST and GST-ZTL were used for the in vitro binding assay. Protein extracts from Nicotiana benthamiana leaves transiently expressing GI-GFP were incubated with ZTL-GST resin. ZTL-bound GI alleles-GFP was detected with an anti-GFP antibody (cat. no. ab6556; Abcam).
Interactions between GI and ZTL were performed with total and cytosolic proteins from transient expression in N. benthamiana of 35S::GI-GFP and 35::ZTL and immunoprecipitations were performed as reported in Wang et al. (2010).
Reverse Transcription Quantitative PCR
Reverse transcription quantitative PCR was performed using conditions and primers as described in Kim et al. (2013a).
Polymorphism Analysis
The tool Polymorph 1001 (https://tools.1001genomes.org/polymorph/) was used to scan 1,135 accessions for amino acid polymorphisms at ZTL and GI, with the settings: Variant type, SNP; Impact, moderate; and effect type, missense. There were 96 variants found for ZTL (relative to Col) and only Cvi (strain 6911) diverged from Col with a missense that resulted in a P35T change. Similarly, using the same criteria for GI, there were 783 variants and only Cvi diverged from Col with a missense that resulted in an L718F change.
Accession Numbers
Sequence data from this article can be found in the European Molecular Biology Laboratory databases under accession numbers: At1g22770 (GI;https://www.ncbi.nlm.nih.gov/gene/838883) and At5g57360 (ZTL;https://www.ncbi.nlm.nih.gov/gene/835842).
Supplemental Data
The following supplemental materials are available:
Supplemental Figure S1. Creation and identification of CCA1:LUC expressing NILs.
Supplemental Figure S2. Tabular and graphic summary of the temperature-dependent period effects of Ler and the NILs.
Supplemental Figure S3. Circadian phenotypes under white light mediated by interactions between GI and ZTL natural alleles.
Supplemental Figure S4. Allele-dependent period effects of the NILs supported by additional segregants.
Supplemental Figure S5. Period estimates of independent GCZL lines derived from GCZC.
Supplemental Figure S6. Period estimates of independent GLZC lines derived from GCZC.
Supplemental Figure S7. The period phenotypes of NILs are not based on transcript abundance differences.
Supplemental Figure S8. Amino acid sequence profile of GI and ZTL alleles in Col, Ler, and Cvi accessions.
Supplemental Figure S9. GILer and GICol interact similarly with ZTL.
Supplemental Figure S10. Endogenous ZTL levels in Col-0 and Cvi.
Supplemental Figure S11. GI Cvi strongly interacts with ZTL in vitro.
Supplemental Figure S12. L718F occurs at a highly conserved LxxLxxLxxL region.
Supplemental Figure S13. GIL718F and ZTLP35T alleles are unique.
Supplemental Table S1. Table of regression coefficients from a linear model (lm in R) with the periods as outcomes and the genetic loci GI and ZTL and temperature (15°C, 22°C, and 27°C) as categorical covariates under red light.
Supplemental Table S2. Table of regression coefficients from a linear model with the periods as outcomes and the genetic loci GI and ZTL and temperature (15°C, 22°C, and 27°C) as categorical covariates under white light.
Supplemental Methods. Linear model analysis.
Acknowledgments
We thank Dr. Maarten Koornneef (Max Planck Institute for Plant Breeding Research) and Dr. Joost Keurentjes (Wageningen University) for sharing unpublished data, Dr. Micheal Sovic (Center for Applied Plant Sciences, Ohio State University) for advice on using the program R, and Dr. Janet Best (Ohio State University) for advice on statistical modeling efforts.
Footnotes
This work was supported by the National Institutes of Health (R01GM093285 to D.E.S.), the Next-Generation BioGreen21 Program (PJ01327305), the Rural Development Administration, Republic of Korea (to D.E.S.), and the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture, Forestry and Fisheries through Export Promotion Technology Development Program, Ministry of Agriculture, Food and Rural Affairs (117033-03-1-HD030 to T.-S.K.).
Articles can be viewed without a subscription.
References
- Abarca D, Roldán M, Martín M, Sabater B (2001) Arabidopsis thaliana ecotype Cvi shows an increased tolerance to photo-oxidative stress and contains a new chloroplastic copper/zinc superoxide dismutase isoenzyme. J Exp Bot 52: 1417–1425 [DOI] [PubMed] [Google Scholar]
- Alcázar R, García AV, Parker JE, Reymond M (2009) Incremental steps toward incompatibility revealed by Arabidopsis epistatic interactions modulating salicylic acid pathway activation. Proc Natl Acad Sci USA 106: 334–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso-Blanco C, Peeters AJ, Koornneef M, Lister C, Dean C, van den Bosch N, Pot J, Kuiper MT (1998) Development of an AFLP based linkage map of Ler, Col and Cvi Arabidopsis thaliana ecotypes and construction of a Ler/Cvi recombinant inbred line population. Plant J 14: 259–271 [DOI] [PubMed] [Google Scholar]
- Anwer MU, Boikoglou E, Herrero E, Hallstein M, Davis AM, Velikkakam James G, Nagy F, Davis SJ (2014) Natural variation reveals that intracellular distribution of ELF3 protein is associated with function in the circadian clock. eLife 3: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assis R. (2014) Strong epistatic selection on the RNA secondary structure of HIV. PLoS Pathog 10: e1004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bomblies K, Lempe J, Epple P, Warthmann N, Lanz C, Dangl JL, Weigel D (2007) Autoimmune response as a mechanism for a Dobzhansky–Muller-type incompatibility syndrome in plants. PLoS Biol 5: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cha JY, Kim J, Kim TS, Zeng Q, Wang L, Lee SY, Kim WY, Somers DE (2017) GIGANTEA is a co-chaperone which facilitates maturation of ZEITLUPE in the Arabidopsis circadian clock. Nat Commun 8: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae E, Bomblies K, Kim ST, Karelina D, Zaidem M, Ossowski S, Martín-Pizarro C, Laitinen RA, Rowan BA, Tenenboim H, et al. (2014) Species-wide genetic incompatibility analysis identifies immune genes as hot spots of deleterious epistasis. Cell 159: 1341–1351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Weinreich DM, Hartl DL (2005) Missense meanderings in sequence space: A biophysical view of protein evolution. Nat Rev Genet 6: 678–687 [DOI] [PubMed] [Google Scholar]
- Durvasula A, Fulgione A, Gutaker RM, Alacakaptan SI, Flood PJ, Neto C, Tsuchimatsu T, Burbano HA, Picó FX, Alonso-Blanco C, et al. (2017) African genomes illuminate the early history and transition to selfing in Arabidopsis thaliana. Proc Natl Acad Sci USA 114: 5213–5218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards KD, Lynn JR, Gyula P, Nagy F, Millar AJ (2005) Natural allelic variation in the temperature-compensation mechanisms of the Arabidopsis thaliana circadian clock. Genetics 170: 387–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezer D, Jung JH, Lan H, Biswas S, Gregoire L, Box MS, Charoensawan V, Cortijo S, Lai X, Stöckle D, et al. (2017) The Evening Complex coordinates environmental and endogenous signals in Arabidopsis. Nat Plants 3: 17087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara S, Wang L, Han L, Suh SS, Salomé PA, McClung CR, Somers DE (2008) Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283: 23073–23083 [DOI] [PubMed] [Google Scholar]
- Fulgione A, Hancock AM (2018) Archaic lineages broaden our view on the history of Arabidopsis thaliana. New Phytol 219: 1194–1198 [DOI] [PubMed] [Google Scholar]
- Gould PD, Locke JC, Larue C, Southern MM, Davis SJ, Hanano S, Moyle R, Milich R, Putterill J, Millar AJ, et al. (2006) The molecular basis of temperature compensation in the Arabidopsis circadian clock. Plant Cell 18: 1177–1187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould PD, Ugarte N, Domijan M, Costa M, Foreman J, Macgregor D, Rose K, Griffiths J, Millar AJ, Finkenstädt B, et al. (2013) Network balance via CRY signalling controls the Arabidopsis circadian clock over ambient temperatures. Mol Syst Biol 9: 650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haag ES, Wang S, Kimble J (2002) Rapid coevolution of the nematode sex-determining genes fem-3 and tra-2. Curr Biol 12: 2035–2041 [DOI] [PubMed] [Google Scholar]
- Herrero E, Kolmos E, Bujdoso N, Yuan Y, Wang M, Berns MC, Uhlworm H, Coupland G, Saini R, Jaskolski M, et al. (2012) EARLY FLOWERING4 recruitment of EARLY FLOWERING3 in the nucleus sustains the Arabidopsis circadian clock. Plant Cell 24: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu PY, Harmer SL (2014) Wheels within wheels: The plant circadian system. Trends Plant Sci 19: 240–249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huq E, Tepperman JM, Quail PH (2000) GIGANTEA is a nuclear protein involved in phytochrome signaling in Arabidopsis. Proc Natl Acad Sci USA 97: 9789–9794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JM, Loros JJ, Dunlap JC (2016) Circadian oscillators: Around the transcription–translation feedback loop and on to output. Trends Biochem Sci 41: 834–846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isojima Y, Nakajima M, Ukai H, Fujishima H, Yamada RG, Masumoto KH, Kiuchi R, Ishida M, Ukai-Tadenuma M, Minami Y, et al. (2009) CKIε/δ-dependent phosphorylation is a temperature-insensitive, period-determining process in the mammalian circadian clock. Proc Natl Acad Sci USA 106: 15744–15749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- James AB, Syed NH, Bordage S, Marshall J, Nimmo GA, Jenkins GI, Herzyk P, Brown JW, Nimmo HG (2012) Alternative splicing mediates responses of the Arabidopsis circadian clock to temperature changes. Plant Cell 24: 961–981 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keurentjes JJ, Bentsink L, Alonso-Blanco C, Hanhart CJ, Blankestijn-De Vries H, Effgen S, Vreugdenhil D, Koornneef M (2007) Development of a near-isogenic line population of Arabidopsis thaliana and comparison of mapping power with a recombinant inbred line population. Genetics 175: 891–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Geng R, Gallenstein RA, Somers DE (2013a) The F-box protein ZEITLUPE controls stability and nucleocytoplasmic partitioning of GIGANTEA. Development 140: 4060–4069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TS, Kim WY, Fujiwara S, Kim J, Cha JY, Park JH, Lee SY, Somers DE (2011) HSP90 functions in the circadian clock through stabilization of the client F-box protein ZEITLUPE. Proc Natl Acad Sci USA 108: 16843–16848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim WY, Fujiwara S, Suh SS, Kim J, Kim Y, Han L, David K, Putterill J, Nam HG, Somers DE (2007) ZEITLUPE is a circadian photoreceptor stabilized by GIGANTEA in blue light. Nature 449: 356–360 [DOI] [PubMed] [Google Scholar]
- Kim Y, Lim J, Yeom M, Kim H, Kim J, Wang L, Kim WY, Somers DE, Nam HG (2013b) ELF4 regulates GIGANTEA chromatin access through subnuclear sequestration. Cell Reports 3: 671–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby DA, Muse SV, Stephan W (1995) Maintenance of pre-mRNA secondary structure by epistatic selection. Proc Natl Acad Sci USA 92: 9047–9051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall CM, Tartaglio V, Duarte M, Harmon FG (2016) The Arabidopsis sickle mutant exhibits altered circadian clock responses to cool temperatures and temperature-dependent alternative splicing. Plant Cell 28: 2560–2575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima N, Miyashita H (2012) Leucine-rich repeat (LRR) domains containing intervening motifs in plants. Biomolecules 2: 288–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehra A, Shi M, Baker CL, Colot HV, Loros JJ, Dunlap JC (2009) A role for casein kinase 2 in the mechanism underlying circadian temperature compensation. Cell 137: 749–760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra P, Panigrahi KC (2015) GIGANTEA—an emerging story. Front Plant Sci 6: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitraki A, Danner M, King J, Seckler R (1993) Temperature-sensitive mutations and second-site suppressor substitutions affect folding of the P22 tailspike protein in vitro. J Biol Chem 268: 20071–20075 [PubMed] [Google Scholar]
- Mitraki A, Fane B, Haase-Pettingell C, Sturtevant J, King J (1991) Global suppression of protein folding defects and inclusion body formation. Science 253: 54–58 [DOI] [PubMed] [Google Scholar]
- Mizuno T, Nomoto Y, Oka H, Kitayama M, Takeuchi A, Tsubouchi M, Yamashino T (2014) Ambient temperature signal feeds into the circadian clock transcriptional circuitry through the EC night-time repressor in Arabidopsis thaliana. Plant Cell Physiol 55: 958–976 [DOI] [PubMed] [Google Scholar]
- Narasimamurthy R, Virshup DM (2017) Molecular mechanisms regulating temperature compensation of the circadian clock. Front Neurol 8: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasrallah CA. (2013) The dynamics of alternative pathways to compensatory substitution. BMC Bioinformatics 14(Suppl 15): S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohales MA, Kay SA (2016) Molecular mechanisms at the core of the plant circadian oscillator. Nat Struct Mol Biol 23: 1061–1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordborg M, Hu TT, Ishino Y, Jhaveri J, Toomajian C, Zheng H, Bakker E, Calabrese P, Gladstone J, Goyal R, et al. (2005) The pattern of polymorphism in Arabidopsis thaliana. PLoS Biol 3: e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475: 398–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Portolés S, Más P (2010) The functional interplay between protein kinase CK2 and CCA1 transcriptional activity is essential for clock temperature compensation in Arabidopsis. PLoS Genet 6: e1001201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Press MO, Queitsch C (2017) Variability in a short tandem repeat mediates complex epistatic interactions in Arabidopsis thaliana. Genetics 205: 455–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salomé PA, Weigel D, McClung CR (2010) The role of the Arabidopsis morning loop components CCA1, LHY, PRR7, and PRR9 in temperature compensation. Plant Cell 22: 3650–3661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlaen RG, Mancini E, Sanchez SE, Perez-Santángelo S, Rugnone ML, Simpson CG, Brown JW, Zhang X, Chernomoretz A, Yanovsky MJ (2015) The spliceosome assembly factor GEMIN2 attenuates the effects of temperature on alternative splicing and circadian rhythms. Proc Natl Acad Sci USA 112: 9382–9387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlosser G, Wagner GP (2008) A simple model of co-evolutionary dynamics caused by epistatic selection. J Theor Biol 250: 48–65 [DOI] [PubMed] [Google Scholar]
- Seo PJ, Mas P (2014) Multiple layers of posttranslational regulation refine circadian clock activity in Arabidopsis. Plant Cell 26: 79–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara Y, Koyama YM, Ukai-Tadenuma M, Hirokawa T, Kikuchi M, Yamada RG, Ukai H, Fujishima H, Umehara T, Tainaka K, et al. (2017) Temperature-sensitive substrate and product binding underlie temperature-compensated phosphorylation in the clock. Mol Cell 67: 783–798.e20 [DOI] [PubMed] [Google Scholar]
- Somers DE, Fujiwara S (2009) Thinking outside the F-box: Novel ligands for novel receptors. Trends Plant Sci 14: 206–213 [DOI] [PubMed] [Google Scholar]
- Somers DE, Fujiwara S, Kim WY, Suh SS (2007) Posttranslational photomodulation of circadian amplitude. Cold Spring Harb Symp Quant Biol 72: 193–200 [DOI] [PubMed] [Google Scholar]
- Somers DE, Kim WY, Geng R (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16: 769–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The 1001 Genomes Consortium (2016) 1,135 Genomes reveal the global pattern of polymorphism in Arabidopsis thaliana. Cell 166: 481–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Undurraga SF, Press MO, Legendre M, Bujdoso N, Bale J, Wang H, Davis SJ, Verstrepen KJ, Queitsch C (2012) Background-dependent effects of polyglutamine variation in the Arabidopsis thaliana gene ELF3. Proc Natl Acad Sci USA 109: 19363–19367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Fujiwara S, Somers DE (2010) PRR5 regulates phosphorylation, nuclear import and subnuclear localization of TOC1 in the Arabidopsis circadian clock. EMBO J 29: 1903–1915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Kim J, Somers DE (2013) Transcriptional corepressor TOPLESS complexes with pseudoresponse regulator proteins and histone deacetylases to regulate circadian transcription. Proc Natl Acad Sci USA 110: 761–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Wu F, Xie Q, Wang H, Wang Y, Yue Y, Gahura O, Ma S, Liu L, Cao Y, et al. (2012) SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 24: 3278–3295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Marquis K, Pei J, Fu SC, Cağatay T, Grishin NV, Chook YM (2015) LocNES: A computational tool for locating classical NESs in CRM1 cargo proteins. Bioinformatics 31: 1357–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu JW, Rubio V, Lee NY, Bai S, Lee SY, Kim SS, Liu L, Zhang Y, Irigoyen ML, Sullivan JA, et al. (2008) COP1 and ELF3 control circadian function and photoperiodic flowering by regulating GI stability. Mol Cell 32: 617–630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu W, Zaidem M, Van de Weyer AL, Gutaker RM, Chae E, Kim ST, Bemm F, Li L, Todesco M, Schwab R, et al. (2018) Modulation of ACD6 dependent hyperimmunity by natural alleles of an Arabidopsis thaliana NLR resistance gene. PLoS Genet 14: e1007628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou YP, Hou XH, Wu Q, Chen JF, Li ZW, Han TS, Niu XM, Yang L, Xu YC, Zhang J, et al. (2017) Adaptation of Arabidopsis thaliana to the Yangtze River basin. Genome Biol 18: 239. [DOI] [PMC free article] [PubMed] [Google Scholar]