Cyanobacterial Rubisco large subunits form functional hybrids with tobacco small subunits and procarboxysome microcompartments via the linker protein CcmM35 in the absence of cognate small subunits.
Abstract
Much of the research aimed at improving photosynthesis and crop productivity attempts to overcome shortcomings of the primary CO2-fixing enzyme Rubisco. Cyanobacteria utilize a CO2-concentrating mechanism (CCM), which encapsulates Rubisco with poor specificity but a relatively fast catalytic rate within a carboxysome microcompartment. Alongside the active transport of bicarbonate into the cell and localization of carbonic anhydrase within the carboxysome shell with Rubisco, cyanobacteria are able to overcome the limitations of Rubisco via localization within a high-CO2 environment. As part of ongoing efforts to engineer a β-cyanobacterial CCM into land plants, we investigated the potential for Rubisco large subunits (LSU) from the β-cyanobacterium Synechococcus elongatus (Se) to form aggregated Rubisco complexes with the carboxysome linker protein CcmM35 within tobacco (Nicotiana tabacum) chloroplasts. Transplastomic plants were produced that lacked cognate Se Rubisco small subunits (SSU) and expressed the Se LSU in place of tobacco LSU, with and without CcmM35. Plants were able to form a hybrid enzyme utilizing tobacco SSU and the Se LSU, allowing slow autotrophic growth in high CO2. CcmM35 was able to form large Rubisco aggregates with the Se LSU, and these incorporated small amounts of native tobacco SSU. Plants lacking the Se SSU showed delayed growth, poor photosynthetic capacity, and significantly reduced Rubisco activity compared with both wild-type tobacco and lines expressing the Se SSU. These results demonstrate the ability of the Se LSU and CcmM35 to form large aggregates without the cognate Se SSU in planta, harboring active Rubisco that enables plant growth, albeit at a much slower pace than plants expressing the cognate Se SSU.
The need to produce sufficient food for a growing population requires increasing the productivity and efficiency of agriculture in order to increase yields by the estimated 70% that will be needed by 2050 (Lobell et al., 2009; Ray et al., 2012). Given its central role in crop growth and productivity, improving photosynthesis is one approach that has the potential to generate step-change improvements in crop yields and resource use efficiency (Long et al., 2006; Ort et al., 2015). One of the primary limitations to photosynthesis is the relative inefficiency of the central carbon-fixing enzyme Rubisco, in particular its lack of specificity for CO2 versus oxygen, which leads to the energetically costly photorespiratory cycle (Whitney et al., 2011; Carmo-Silva et al., 2015; Sharwood et al., 2016; Flamholz et al., 2019). Exemplifying this, at current atmospheric levels of CO2 and oxygen, Rubisco’s tendency to oxygenate rather than carboxylate its substrate ribulose 1,5-bisphosphate (RuBP) is estimated to reduce yields by as much as 36% and 20% in United States-grown soybean (Glycine max) and wheat (Triticum aestivum), respectively (Walker et al., 2016). Recent work has shown that limiting the costs of photorespiration by increasing its efficiency can provide dramatic benefits to plant growth (South et al., 2019).
Synthetic biology approaches hold promise for improving a number of facets of photosynthetic efficiency in crop plants (Maurino and Weber, 2013; Erb and Zarzycki, 2016; Orr et al., 2017). One example is the introduction of CO2-concentrating mechanisms (CCMs) into C3 crops to increase CO2 concentrations at the site of Rubisco, a strategy that is likely to dramatically reduce the propensity of Rubisco to carry out oxygenation reactions by creating an environment that favors the beneficial carboxylation reaction (Price et al., 2011; McGrath and Long, 2014; Hanson et al., 2016; Long et al., 2016). Significant research efforts are being invested in this area, with varying sources for the CCMs being engineered, such as C4 (Hibberd et al., 2008; Langdale, 2011) and Crassulacean acid metabolism (Borland et al., 2014; Yang et al., 2015) systems from plants and the pyrenoid- and carboxysome-based systems of algae and cyanobacteria, respectively (Rae et al., 2017; Mackinder, 2018).
The CCM employed by cyanobacteria uses a combination of factors to create a high-CO2 environment localized around Rubisco (Price et al., 2008; Hanson et al., 2016). Aggregation and encapsulation of Rubisco within a highly ordered icosahedral protein microcompartment, or carboxysome, allows colocalization of Rubisco and carbonic anhydrase to convert HCO3− to CO2 where it is needed and permits the movement of key molecules while limiting CO2 escape. Generating a high-CO2 environment is also facilitated by a complex system of inorganic carbon transporters on the cyanobacterial outer membrane that move either HCO3− or CO2 into the cytoplasm through active and passive mechanisms (Price, 2011). Modeling the incorporation of the various components of the CCM into plants suggests that once a fully functioning system is established within a land plant chloroplast, photosynthetic rates could be improved by as much as 60% (McGrath and Long, 2014). The resulting subsequent improvements in yield could facilitate a major change in crop productivity and resource use efficiency (Ort et al., 2015; Hanson et al., 2016).
Substantial progress has been made during recent years to unravel the molecular mechanisms of CCMs involving either carboxysomes or pyrenoids. In Synechococcus elongatus (Se) PCC7942, which produces β-carboxysomes, the ccmM gene gives rise to two proteins, CcmM58 and CcmM35, the latter arising from an internal ribosomal entry site (Long et al., 2007, 2010). CcmM35 possesses three tandem repeats of Rubisco small subunit-like domains and was initially thought to interact with Rubisco by replacing small subunits (Long et al., 2011). However, recent experiments suggest that CcmM35 binds Rubisco without releasing the small subunits (Ryan et al., 2019). A recent structural study revealed that the interaction between CcmM35 and Rubisco leads to dramatic phase separation (Wang et al., 2019). This nucleation of Rubisco holoenzymes by CcmM35 represents a critical first step in the assembly of β-carboxysomes (Cameron et al., 2013). In the pyrenoid of Chlamydomonas spp., similar phase separation was also observed when Rubisco and a repeat protein called Essential Pyrenoid Component1 (EPYC1) interact (Wunder et al., 2018). Likewise, in α-carboxysomes, Rubisco holoenzymes interact with a highly disordered repeat protein called Carboxysome operon S2 (CsoS2; Cai et al., 2015; Liu et al., 2018). In a recent breakthrough, Long et al. (2018) were able to assemble α-carboxysomes in tobacco (Nicotiana tabacum) chloroplasts by coexpressing Rubisco large and small subunit genes along with CsoS2 and a shell protein called CsoS1A from Cyanobium marinum PCC7001. In another study, the shell proteins of β-carboxysome transiently expressed in the chloroplasts of Nicotiana benthamiana were able to assemble structures similar to microcompartments (Lin et al., 2014a).
Our previous work demonstrated that replacing the Rubisco large subunit (LSU) gene in tobacco with the Rubisco large and small subunit genes from Se resulted in plants that can support photosynthetic growth under elevated CO2 conditions (Lin et al., 2014b; Occhialini et al., 2016). When CcmM35 was coexpressed in tobacco chloroplasts, the heterologous Rubisco was observed in a large aggregate with an appearance resembling a separate liquid phase (Lin et al., 2014b). In a previous study performed by another group, when the tobacco Rubisco LSU gene (rbcL) was replaced with that from S. elongatus PCC6301, no Rubisco LSU was detected in the transformed plant (Kanevski et al., 1999), and it was thought that the cyanobacterial LSU could not assemble with the plant small subunit (SSU) to form a functional enzyme.
Here, we investigated the assembly and functioning of cyanobacterial Rubisco within land plant chloroplasts when the Se LSU is expressed either with or without CcmM35 in the absence of the cognate cyanobacterial SSU. Analysis of transplastomic tobacco lines incorporating some cyanobacterial components but lacking the cognate SSU revealed that the Se LSU and CcmM35 are able to form large aggregates of Rubisco within tobacco chloroplasts. Although only low amounts of tobacco SSUs were present, the transplastomic lines characterized differed significantly in physiology and biochemistry from comparable lines that also coexpressed the cognate cyanobacterial SSU. Remarkably, albeit at slow rates, in the absence of the cognate SSUs, the hybrid cyanobacterial LSU-tobacco SSU expressed in tobacco chloroplasts with and without CcmM35 was active and supported plant growth.
RESULTS
Cyanobacterial Rubisco LSU Can Support Carbon Fixation in Tobacco Chloroplasts in the Absence of Cognate SSU
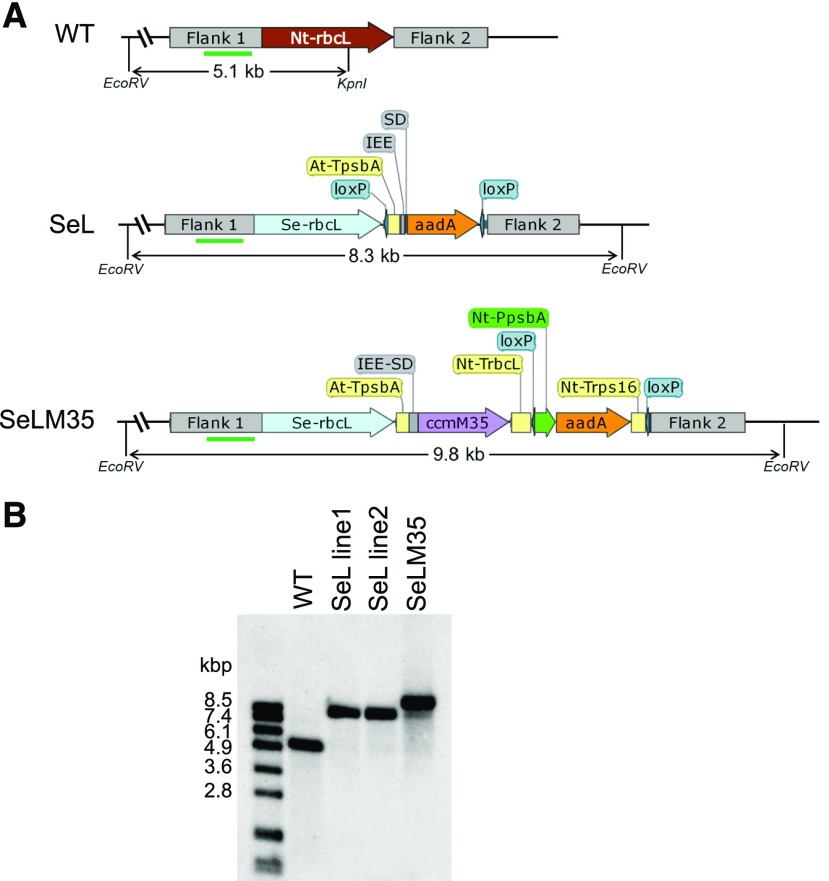
We generated two transplastomic tobacco lines named SeL and SeLM35 by replacing in frame the entire tobacco Rubisco LSU gene with that from Se. In the SeLM35 line, the ccmM35 gene was introduced downstream of the SerbcL gene to be coexpressed from the same chloroplast genome locus (Fig. 1A). We used the same regulatory elements at intergenic regions as described in our previous work, namely, a terminator, an intercistronic expression element (IEE), and a Shine-Dalgarno (SD) or ribosome-binding site (Lin et al., 2014b; Occhialini et al., 2016). In contrast to our previous work, the new transplastomic lines do not possess a corresponding cyanobacterial Rubisco SSU gene. The aminoglycoside-3″-adenyltransferase (aadA) selectable marker gene was incorporated into the same operon as the SerbcL gene in the SeL construct instead of a separate operon, as in the SeLM35 construct. We obtained homoplasmic transformed shoots after two rounds of selection and were able to transfer them to soil for growth under elevated CO2 (9,000 µL L−1). We collected seeds from two independent SeL lines and one SeLM35 line. Both DNA and RNA blots confirmed complete removal of the NtrbcL gene and its corresponding transcript in these plants (Fig. 1B; Supplemental Fig. S1). We also analyzed the transcripts containing SerbcL and ccmM35 genes in these lines together with SeLS and SeLSM35 lines generated in our previous study (Supplemental Fig. S1). The RNA blots showed bands arising from incomplete processing of IEE as well as read-through transcription of the downstream aadA operon, consistent with our previous observations (Occhialini et al., 2016).
Figure 1.
Replacement of the Rubisco LSU gene (rbcL) in tobacco chloroplasts with SerbcL with or without the ccmM35 gene. A, Gene arrangements of wild-type (WT), SeL, and SeLM35 tobacco lines along with the locations of the EcoRV and KpnI restriction sites used on the DNA blot. The binding site for the digoxigenin (DIG)-labeled DNA probe is shown in green bars. Seeds were obtained from two independent SeL lines and one SeLM35 line. B, DNA-blot analysis of wild-type, SeL, and SeLM35 samples digested with EcoRV and KpnI. All samples produced the expected band on the DNA blot.
Cyanobacterial Rubisco LSU and CcmM35 Aggregate in Procarboxysome Microcompartments in Tobacco Chloroplasts
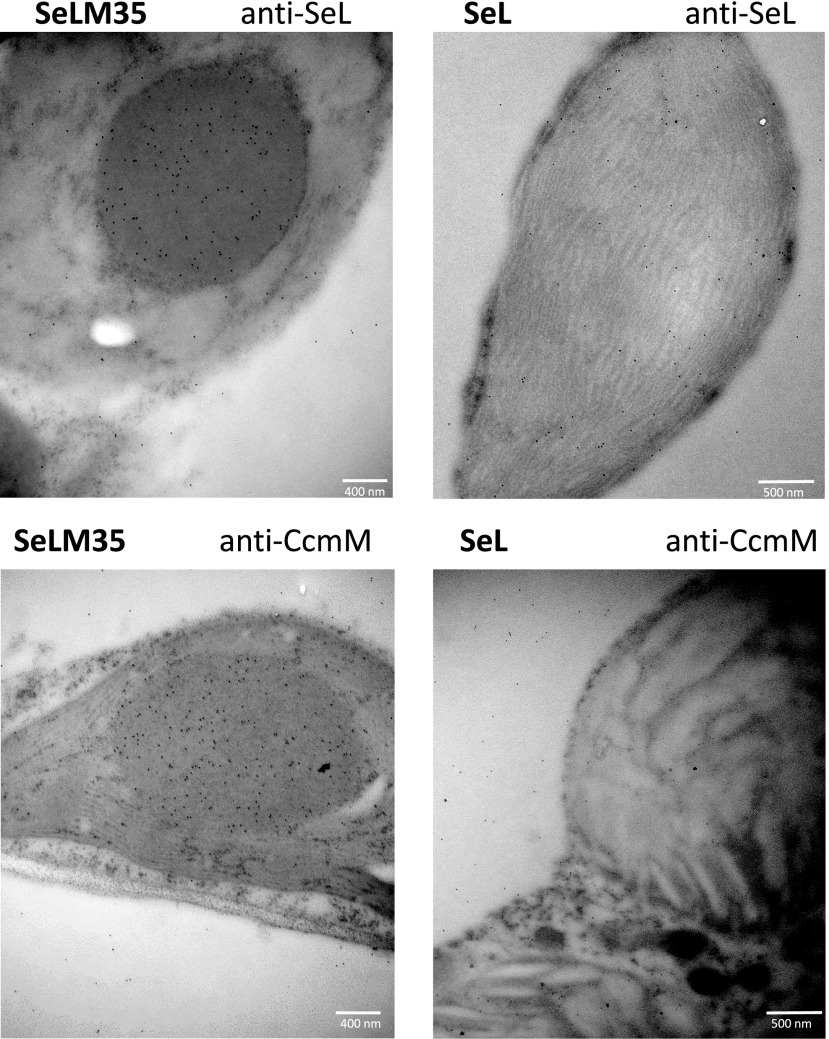
Expression of SeCcmM35 together with the cyanobacterial LSU in the SeLM35 transformant resulted in the formation of aggregates, or procarboxysome microcompartments, in tobacco chloroplasts (Fig. 2). These aggregates were similar in size and shape to those observed in plants containing both the large and small subunits of Rubisco and CcmM35 (SeLSM35; Supplemental Fig. S2) but were absent from tobacco plants expressing the Se LSU in the absence of CcmM35. Immunogold labeling confirmed the presence of the Se LSU and CcmM35 proteins within the SeLM35 procarboxysome compartments (Fig. 2; Supplemental Figs. S3 and S4). In comparison, in SeL plants, the SeLSU protein could be detected throughout the chloroplast and, as expected, the anti-CcmM antibody gave only background-level signal.
Figure 2.
Tobacco plants expressing cyanobacterial Rubisco LSUs and CcmM35 contain a procarboxysome compartment in the chloroplast. Immunolocalization is shown for Se proteins in the chloroplasts of transplastomic tobacco lines expressing the Rubisco LSU and CcmM35 (SeLM35) or the LSU alone (SeL). Electron micrographs show ultrathin sections of mesophyll cells probed with the indicated primary antibody and a secondary antibody conjugated to 10-nm gold particles. Additional images are presented in Supplemental Figures S3 and S4. Bars show size as indicated.
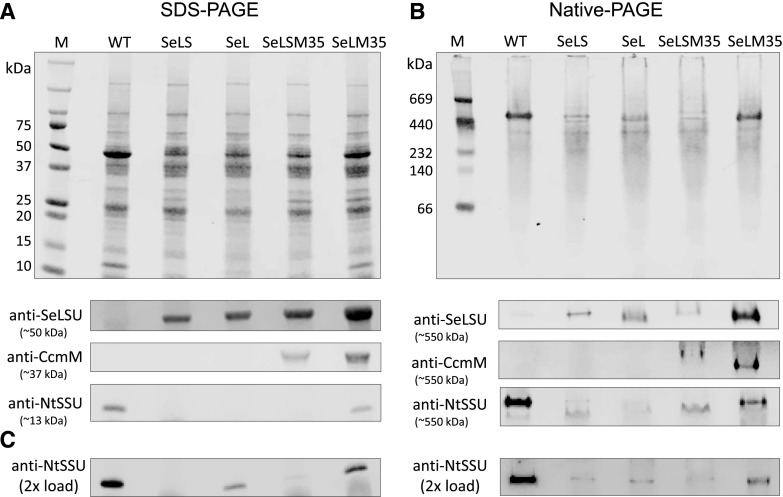
Gel electrophoresis and immunoblotting of leaf extracts demonstrated the presence of cyanobacterial LSU and CcmM35 in SeLM35 transplastomic plants (Fig. 3; Supplemental Fig. S5). Visually, the two proteins appear to be more abundant on a total soluble protein basis in these plants compared with SeLSM35. As expected, both proteins were absent from wild-type leaf extracts, and in SeLS and SeL plants, the Se LSU was present but CcmM35 was not observed. The tobacco SSU was detected in wild-type, SeL, and SeLM35 leaf extracts, although its abundance in SeL was very low, and visualization of the ∼13-kD SSU required a higher total soluble protein load to detect clearly using immunoblotting (Fig. 3C). Nondenaturing native PAGE suggested that CcmM35 is present in functional complexes with Rubisco in the tobacco transplastomic lines SeLSM35 and SeLM35 (Fig. 3B).
Figure 3.
Protein composition of wild-type (WT) tobacco and transplastomic lines expressing β-cyanobacterial carboxysome components. A and B, Polypeptides in leaf extracts prepared from plants of each line were separated by denaturing SDS-PAGE (A) and nondenaturing native PAGE (B) and either stained with Coomassie Blue (top gels) or used for immunoblotting with antibodies against cyanobacterial Rubisco LSU (SeLSU) and CcmM35 and against the tobacco Rubisco small subunit (NtSSU; bottom gels). Images showing blotting of PAGE gels are slices from blots (Supplemental Fig. S5) and show the indicated size regions where the respective antibodies detect proteins of interest. For SDS-PAGE and native PAGE, 10 and 20 µg of total soluble protein was loaded per lane, respectively. Lanes marked M indicate protein markers containing proteins of a range of sizes as indicated at left of each gel. C, SDS-PAGE and native PAGE gels immunoblotted with antibody against NtSSU, loaded with 20 and 40 µg of total soluble protein, respectively.
Cyanobacterial Rubisco Activity Is Impaired by the Lack of a Cognate SSU within Tobacco Chloroplasts
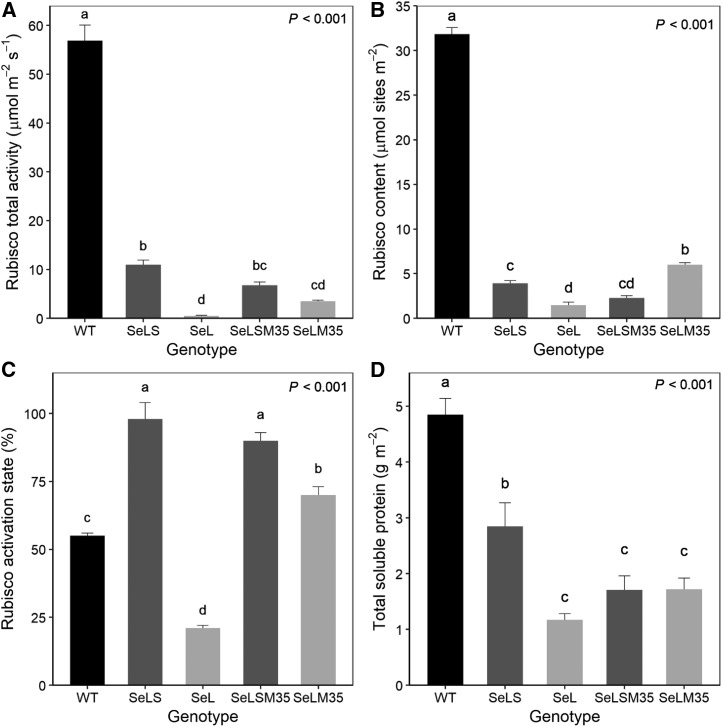
Consistent with previous efforts expressing Se Rubisco within tobacco chloroplasts, Rubisco content and activity on a leaf area basis were significantly lower in leaf extracts of all the transplastomic lines, representing less than 20% of the values in wild-type plants (Fig. 4). SeL plants in particular displayed minimal amounts of Rubisco. While Rubisco active sites in SeL were approximately 20% of SeLS plants expressing both Se Rubisco subunits (Fig. 4B), total activity in SeL was less than 5% of SeLS and approximately 1% of wild-type tobacco, consistent with the extremely slow growth of these plants (see below). SeLM35 plants had significantly more Rubisco active sites than other transplastomic lines, including SeLSM35, which also expresses the CcmM35 linker protein (Fig. 4B; P < 0.001), although Rubisco total activity was not significantly different between the two lines (Fig. 4A; P > 0.001).
Figure 4.
Rubisco and total soluble protein. Rubisco total activity (A), activation state (B), and content (C), and total soluble protein (D), are shown for wild-type (WT) tobacco and transplastomic lines expressing β-cyanobacterial carboxysome components from Se: Rubisco LSU (L), Rubisco SSU (S), and CcmM35 (M35). Values represent means ± se (n = 3–4 biological replicates). Letters denote significant differences (P < 0.05) as determined by Tukey’s honestly significant difference mean-separation test following ANOVA (P values are as indicated).
To ascertain the ability of tobacco chloroplasts to maintain active cyanobacterial Rubisco, we determined Rubisco activation states from wild-type and transplastomic plants under steady-state conditions. As anticipated, wild-type plants were observed to have a comparatively low activation state in high-CO2 conditions (Fig. 4C). Lines expressing both Se Rubisco subunits, with or without CcmM35, showed essentially fully active Rubisco. In contrast, in SeLM35 Rubisco, activation was approximately 70%, and in SeL, expressing just the cyanobacterial LSU, it was only approximately 20%. These data indicate that these complexes, although able to function, did not become fully active in these growth conditions.
All transplastomic lines displayed significantly lower total soluble protein compared with wild-type tobacco (Fig. 4D; P < 0.001), and this decrease was largely consistent with the decreased amount of Rubisco on an area basis (Supplemental Fig. S6). Alongside reduced total soluble protein and Rubisco content and in agreement with visual observations of these transplastomic plants, levels of chlorophyll a and b, and thus total chlorophyll, were significantly reduced (Supplemental Fig. S7). Chlorophyll a was more severely reduced, and with the exception of SeLS, all lines had a significantly reduced chlorophyll a/b ratio compared with wild-type tobacco.
Cyanobacterial Rubisco has been characterized to have a very high catalytic rate but also a poor affinity for CO2 (high KC value). In SeLS and SeLSM35 plants, values obtained for carboxylation rate (VC) and the Michaelis-Menten constant for CO2 (KC) were consistent with previous work (Table 1; Occhialini et al., 2016). Rubiscos from SeLM35 and SeL, which contain the cyanobacterial LSU but lack a cognate SSU, were able to carboxylate RuBP at significant rates. Immunoblotting suggested the presence of tobacco SSU in the Rubisco complex, but this was likely at a stoichiometric ratio lower than 1:1 in relation to the cyanobacterial LSU (Fig. 3). These two Rubisco enzymes had affinities for CO2 comparable to the enzyme from the transplastomic lines containing both the cyanobacterial LSU and SSU (Table 1).
Table 1. Rubisco catalytic properties.
Rubisco maximum VC and KC are shown for wild-type tobacco and transplastomic lines expressing β-cyanobacterial carboxysome components from Se: Rubisco LSU (L), Rubisco SSU (S), and CcmM35 (M35). Values represent means ± se (n = 3–5 biological replicates). *, Wild-type values are from Occhialini et al. (2016). Letters denote significant differences (P < 0.05) between transplastomic lines as determined by Tukey’s pairwise comparisons following ANOVA.
| Line | VC | KC |
|---|---|---|
| µmol mg−1 min−1 | µm | |
| Wild type* | 3.9 ± 0.2 | 9.0 ± 0.3 |
| SeLS | 15.0 ± 0.9 a | 168 ± 59 a |
| SeL | 0.6 ± 0.2 b | 105 ± 9 a |
| SeLSM35 | 10.9 ± 0.8 c | 133 ± 12 a |
| SeLM35 | 2.0 ± 0.3 b | 110 ± 22 a |
The Lack of a Cognate Rubisco SSU Also Impairs Photosynthetic Gas Exchange
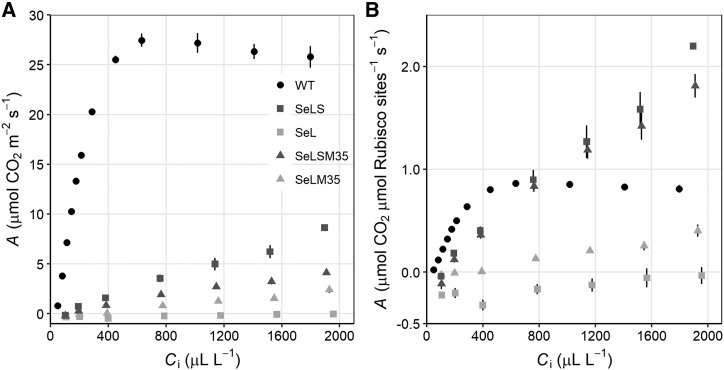
To evaluate the impact of the unusual Rubisco composition in the leaves of these transplastomic lines, gas-exchange measurements were carried out. At the levels present in these transplastomic plants and in the absence of a functional CCM, the faster catalytic rate of Se Rubisco does not confer an advantage in the photosynthetic rate per leaf area even at 2,000 µL L−1 CO2 (Fig. 5A). Consistent with a previous work, aggregating cyanobacterial Rubisco through the expression of CcmM35 in SeLSM35 plants slightly reduced photosynthetic rates on an area basis (Fig. 5A; Occhialini et al., 2016). SeLM35 photosynthetic rates show that the lack of the cognate Se SSU decreases photosynthetic rates even further (Fig. 5A). Most transplastomic lines showed a noticeable increase of photosynthesis under low-oxygen conditions (Supplemental Fig. S8). However, even at the highest CO2 concentration measured combined with 2% (v/v) oxygen, SeL plants displayed net photosynthetic rates that were barely above zero (Supplemental Fig. S8C).
Figure 5.
Response of net CO2 assimilation (A) to intercellular CO2 concentrations (Ci). Rates are expressed on an area basis (A) and on a Rubisco active site basis (B) for leaves of wild-type (WT) tobacco and transplastomic lines expressing β-cyanobacterial carboxysome components from Se: Rubisco LSU (L), Rubisco SSU (S), and CcmM35 (M35). Values represent means ± se (n = 3–4 biological replicates).
As a fully functional cyanobacterial CCM within tobacco will ideally require less Rubisco than wild-type plants, we also determined Rubisco content in the leaves used for gas-exchange analyses. When CO2 assimilation was normalized by Rubisco active site concentration, neither SeLM35 nor SeL outperformed wild-type plants even at 2,000 µL L−1 CO2 (Fig. 5B). Consistent with an earlier work, at CO2 levels well above ambient SeLS and SeLSM35, plants showed higher photosynthesis per Rubisco active site (Fig. 5B; Occhialini et al., 2016). Even accounting for very low Rubisco content, SeL plants show null normalized rates even at intercellular CO2 concentration of 2,000 µL L−1 CO2 (Fig. 5B). This is consistent with the observation that even a short exposure of several hours in ambient CO2 conditions leads to tissue damage, and that even in growth conditions of 4,000 µL L−1 CO2, SeL plants are extremely slow to develop (see below).
Replacement of Tobacco Rubisco LSUs with Cyanobacterial LSUs Impairs Growth Irrespective of Other Components
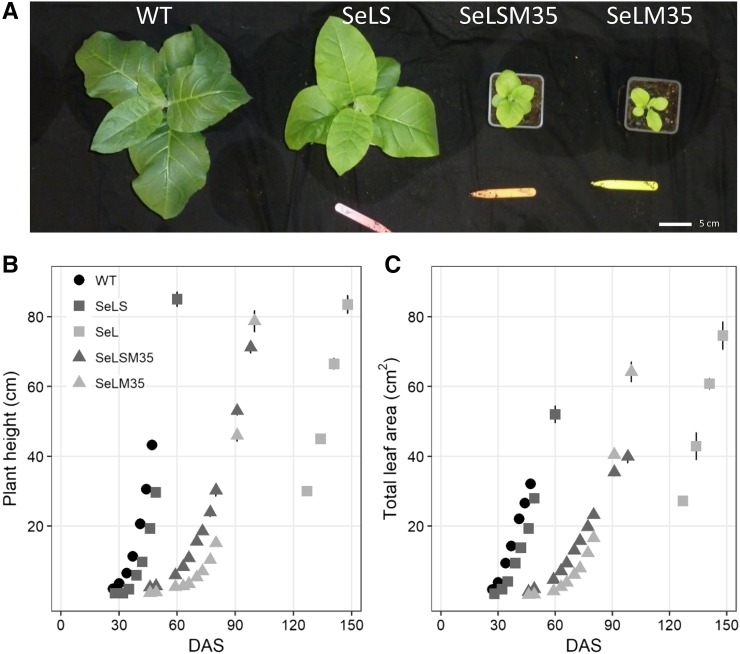
Transplastomic plants where the native tobacco Rubisco LSU was replaced with the Se LSU with or without the carboxysome linker protein CcmM35 (SeLSM35 and SeLM35) grew slowly even at 4,000 µL L−1 CO2 when compared with both the wild type and lines expressing both Se Rubisco subunits (SeLS; Fig. 6A; Supplemental Fig. S6; Supplemental Table S1). Germination time was similar between all lines (∼7 d). Plant height and total leaf area of SeLSM35 and SeLM35 plants started to visibly increase 60 d after sowing, and the growth rate for the subsequent 15 d was significantly slower in SeLM35 plants lacking the Se SSU compared with SeLSM35 (P < 0.05; Fig. 6, B and C; Supplemental Table S2). SeL plants expressing only the Se LSU were dramatically slower in growth (P < 0.001), which necessitated germination in tissue culture for establishment before transferring to soil. These plants took approximately three times as long as SeLS plants to reach a plant height of ∼80 cm (Fig. 6B). SeL and SeLM35 plants produced numerous smaller leaves, consistent with the other line expressing CcmM35, SeLSM35 (Supplemental Fig. S9). Both SeL and SeLM35 were noticeably paler than wild-type controls and transplastomic lines expressing the Se SSU (Supplemental Figs. S7, S9, and S10).
Figure 6.
Plant development and growth traits. Shown are a photograph of 33-d-old plants grown in parallel in 4,000 µL L−1 CO2 (A), plant height (B), and leaf area (C) development during the growth cycle of wild-type (WT) tobacco and transplastomic lines expressing β-cyanobacterial carboxysome components from Se: Rubisco LSU (L), Rubisco SSU (S), and CcmM35 (M35). Values represent means ± se (n = 3–5 biological replicates). DAS, Days after sowing.
DISCUSSION
This study describes two new transplastomic tobacco lines, SeL and SeLM35, where the native rbcL gene has been replaced with its cyanobacterial counterpart without the SerbcS gene. Previous work had shown the ability of Rubisco LSU and SSU from S. elongatus to assemble and function within tobacco chloroplasts and to form large aggregates of linked Rubisco complexes in the presence of CcmM35 (Lin et al., 2014a; Occhialini et al., 2016). Our results here show that cyanobacterial LSU interacts with the carboxysome linker protein CcmM35 in the absence of a cognate cyanobacterial SSU and forms procarboxysome-like aggregates in tobacco chloroplasts. In contrast to a previous study where no cyanobacterial LSU was detected in a similar tobacco transformant (Kanevski et al., 1999), we were able to detect the cyanobacterial LSU as well as catalytic activity of Rubisco in both SeL and SeLM35 lines (Table 1). It should be noted that the cyanobacterial LSU expressed in the previous study had the first eight residues at its N terminus replaced by the first 11 residues of the tobacco LSU, possibly leading to lower stability of the modified LSU or inhibition of its assembly with the tobacco SSU (Kanevski et al., 1999).
Relative to comparable lines expressing Se SSU, both SeLM35 and SeL plants showed delayed growth (Fig. 6) and developed more numerous but smaller leaves (Supplemental Figs. S9 and S10). SeL was not able to grow autotrophically from seeds even in high CO2 levels and required establishment on tissue culture medium. Similar effects have been seen when engineering Rubisco in tobacco, where either the introduction of a foreign LSU (Whitney and Andrews, 2001; Sharwood et al., 2008) or mutation of the native tobacco LSU (Whitney et al., 1999) leads to very low Rubisco amount and/or very poor activity.
Rubisco from both SeLM35 and SeL had dramatically slower maximum catalytic rates compared with the native Se enzyme (SeLS; Table 1), consistent with the slower growth of these plants. Combined with the significantly lower Rubisco active sites, this led to much lower Rubisco activity on a leaf area basis (Fig. 4). In both lines containing CcmM35, the Rubisco catalytic rate was worse than that of β-cyanobacterial Rubisco extracted from SeLS, where no aggregation occurs, which would suggest a putative negative impact of CcmM35 on Rubisco activity in the Se plants and agrees with previous work with the SeLSM35 line (Occhialini et al., 2016). This is consistent with previous observations from plants expressing α-cyanobacterial Rubisco within a minimal α-carboxysome from Cyanobium marinum PCC7001 (Long et al., 2018). The authors found that the Rubisco catalytic rate was approximately halved when determined for Rubisco from tobacco chloroplasts; however, after high-speed centrifugation to remove insoluble carboxysomes, rates were consistent with those obtained from either the native cyanobacterium or expressed without linker proteins within tobacco. The movement of metabolites such as RuBP may be similarly inhibited by the formation of large β-procarboxysomes of LSU-CcmM35, as observed via KMRuBP measurements made on tobacco derived minimal α-carboxysomes (Long et al., 2018). The large size of the observed procarboxysomes in SeLSM35 and SeLM35 plants, relative to native cyanobacterial carboxysomes, appears likely to have influenced metabolite movement. This highlights that an important part of balancing expression of the various components is not only to ensure correct formation of a functional carboxysome but also to achieve a suitably sized microcompartment. However, Rubisco extracted from SeLM35 was significantly more active than the enzyme extracted from SeL plants, showing that in the absence of Se SSU, CcmM35 helps sequester more tobacco SSU, possibly by increasing the stability of the hybrid L8S8 enzyme or facilitating its assembly (Fig. 3).
The very low activity observed for SeL Rubisco that lacked the cognate SSU from cyanobacteria agrees with in vitro findings from a number of previous studies investigating the ability of LSU-only Rubisco to perform catalysis (Andrews and Ballment, 1984; Jordan and Chollet, 1985; Andrews, 1988). In studies including cyanobacterial Rubisco, in vitro preparations containing only L8 octameric cores typically had detectable activity corresponding to only ∼1% of the cyanobacterial holoenzyme, and even the addition of heterologous SSU from spinach (Spinacia oleracea) led to dramatic increases in activity (Andrews, 1988). The cyanobacterial L8 core binds the spinach SSU with an affinity 1 order of magnitude lower than its native SSU, and the activity of the hybrid enzyme was only half that of the enzyme with homologous subunits (Andrews and Lorimer, 1985). This suggests that the minimal activity observed for SeL Rubisco, ∼5% of SeLS (Fig. 4), may in part result from a substoichiometric amount of tobacco SSU’s binding to cyanobacterial L8 cores.
A common theme in the organization of Rubisco enzymes within both carboxysomes of photosynthetic bacteria and pyrenoids from green algae appears to be through interactions with a disordered repeat protein such as CcmM35 in β-carboxysomes, CsoS2 in α-carboxysomes, and EPYC1 in pyrenoids (Long et al., 2011; Cai et al., 2015; Mackinder et al., 2016). In the case of β-carboxysomes and pyrenoids, the Rubisco enzymes were sequestered into a separate liquid phase by these linker proteins (Freeman Rosenzweig et al., 2017; Wunder et al., 2018; Wang et al., 2019). EPYC1 and CsoS2 were shown to interact only with the SSU (Liu et al., 2018; Atkinson et al., 2019), whereas both the LSU and SSU are involved in binding CcmM35 based on a cryo-electron microscopy structural model, and the L8 core alone was insufficient to form a separate liquid phase with CcmM35 (Wang et al., 2019). Thus, the tobacco SSUs are likely involved in the formation of CcmM35-Rubisco aggregates in SeLM35 plants, although the stoichiometry between the Se LSU and tobacco SSUs was not determined. Indeed, the residues in Se SSU critical for interaction with CcmM35 are well conserved in tobacco SSU (Supplemental Fig. S11; Wang et al., 2019).
The poor photosynthetic performance of these transplastomic lines in the absence of a functional CCM with all the necessary components is unsurprising. However, the ability of some lines to outperform wild-type plants on a per Rubisco basis at higher CO2 levels suggests that, provided with high CO2 concentrations such as those within a fully formed β-carboxysome shell in a complete CCM, the Rubisco levels within these plants may be sufficient to support improved rates of carbon assimilation. Consistent with this, Long and colleagues (2018) observed that leaf discs from plants expressing α-cyanobacterial Rubisco produced similar photosynthetic rates to wild-type tobacco plants in 2% (v/v) CO2 conditions within a membrane inlet mass spectrometry system. Thus, and even considering the associated nitrogen costs of producing the shell components, reducing the typically very large investment into Rubisco by C3 plants may represent an overall nitrogen saving (McGrath and Long, 2014). An issue that is highly likely to be encountered when dealing with the numerous other components of the carboxysome shell is to optimize expression levels, and this may also be necessary for Rubisco. An increasing understanding of the role of chaperones for Rubisco assembly (Feiz et al., 2014; Salesse-Smith et al., 2018; Wilson and Hayer-Hartl, 2018; Conlan et al., 2019) may provide avenues to increase Se Rubisco amounts, should this become necessary to support the desired number of carboxysomes per chloroplast, in order to drive higher photosynthetic rates within a fully formed CCM. It is also possible that adjusting the chloroplast regulatory sequences used to express Se Rubisco subunits may be sufficient to increase the Rubisco amount.
The ability of CcmM35 to link Se LSU in planta without a cognate SSU shows that tobacco SSUs can not only substitute Se SSU to form functional hybrid Rubisco but can also result in an enzyme to which CcmM35 can bind. While the Se SSU does not appear to be essential for the formation of a procarboxysome, the differences shown here based on its presence in a procarboxysome highlight its importance for full Rubisco functionality and carboxysome structural organization. These results support the likely necessity of coengineering cognate subunits from a distant foreign Rubisco, as part of efforts to engineer a foreign Rubisco into crop plants (Whitney and Andrews, 2001; Sharwood et al., 2008), and for more complex engineering of CCMs such as carboxysomes and pyrenoids from cyanobacteria and algae, respectively (Atkinson et al., 2016; Rae et al., 2017).
The carboxysome alone will be insufficient to attain higher rates of photosynthesis without the removal of existing stromal carbonic anhydrase and the addition of transporters to pump high levels of HCO3− into the chloroplast (Hanson et al., 2016; Long et al., 2018; Desmarais et al., 2019). There have been recent improvements in approaches to tackle the issue of localizing these inorganic carbon pumps (Rolland et al., 2016; Uehara et al., 2016) alongside advances in understanding the role of the various carbonic anhydrases (Hu et al., 2015; DiMario et al., 2016). Furthermore, there is now a better understanding of the actual ratios of components in β-carboxysomes (Sun et al., 2019), engineering of β-carboxysome shells to obtain cryo-electron microscopy structural models (Cai et al., 2016; Sutter et al., 2019), an assembly of full β-carboxysomes in Escherichia coli (Fang et al., 2018), and recent successes with α-carboxysomes (Long et al., 2018). These advances provide encouragement that ongoing research is steadily moving toward the ability to assemble these complex, powerful CCMs within plants to improve photosynthesis with the ultimate goal of improving global food security.
MATERIALS AND METHODS
Construction of Chloroplast Transformation Vectors
All primers used were obtained from Integrated DNA Technologies and are listed in Supplemental Table S2. Phusion high-fidelity DNA polymerase, FastDigest restriction enzymes, and T4 DNA ligase from Thermo Scientific were used to generate amplicons, restriction digests, and ligation products, respectively. The ligation products were transformed into chemically competent DH5α Escherichia coli and selected on Luria-Bertani agar medium with 100 μg mL−1 ampicillin. A template vector to hold each DNA piece was first constructed as follows. The aadA operon from the BJF-070 vector (Hanson et al., 2013) was removed by self-ligation of the NsiI digest. An amplicon was generated from the resulting vector using NsiI-BJF3 and BamHI-BJF5 primers and ligated into the BamHI and NsiI sites of the vector to introduce SbfI and NotI sites upstream of the NsiI locus. The resulting vector, BJFE-BB, was used as a vector to hold each DNA element between the SbfI and NotI sites using BB-XXX-f and BB-XXX-r primers, where XXX stands for the name of each DNA element. Once ligated into the BJFE-BB vector, each DNA element was flanked by SbfI-MluI upstream and MauBI-NotI downstream. Since MluI and MauBI restriction sites have compatible cohesive ends, these DNA parts can be assembled in any desired order using an approach similar to the BioBrick method (Shetty et al., 2008). Specifically, we assembled an aadA module comprising loxP-At_TpsbA-IEE-SD-RBS-aadA-loxP. We then modified the pGEM-F1-rbcL-F2 vector described previously (Lin et al., 2014b) by introducing an SbfI site immediately downstream of the SerbcL gene. It was accomplished by ligating the amplicon generated with HindIII-LSUE5 and T1L-IEE3 primers into the HindIII and XbaI sites to obtain the pCT-rbcL-BB2 vector. Next, an XbaI+AscI digest of the amplicon from TrbcL5 and AscI-LSUFl2r primers was ligated into XbaI and MluI sites of the pCT-rbcL-BB2 vector to obtain the pCT-rbcL-BB vector. Finally, we introduced the aadA module between the SbfI and NotI sites of the pCT-rbcL-BB vector to obtain the pCT-rbcL-BB-aadA vector used to generate the SeL chloroplast transformant tobacco (Nicotiana tabacum) line. pCT-rbcL-ccmM35, described previously (Lin et al., 2014b), was used in the generation of SeLM35 tobacco chloroplast transformant.
Generation of Transplastomic Tobacco Plants
We introduced transformation vectors into 2-week-old tobacco (‘Samsun’) seedlings with the Biolistic PDS-1000/He Particle Delivery System (Bio-Rad Laboratories) and a tissue culture-based selection method as described previously (Occhialini et al., 2016). Briefly, about 10 μg of DNA was mixed with 100 μL of 50 mg mL−1 0.6-μm gold nanoparticles, 100 μL of 2.5 m CaCl2, and 40 μL of 0.1 m spermidine free base by vortexing for about 1 min. The gold particles were then pelleted in a microcentrifuge at 1,000 rpm for 8 s and resuspended in 180 μL of 70% (v/v) ethanol. After washing of the gold particles was repeated one more time, the pellet was resuspended in about 60 μL of 100% ethanol and then spread on 10 microcarrier discs used for bombardment. Two days later, the leaves from the bombarded seedlings were cut into halves and placed on RMOP agar shoot regeneration medium with 500 μg mL−1 spectinomycin for 4 to 6 weeks at 23°C under 14 h of light per day. The shoots arising were cut into 5-mm2 pieces and subjected to a second round of selection on the same medium for another 4 to 6 weeks. The regenerated shoots were then transferred to Murashige and Skoog agar medium for rooting and subsequently transferred to soil for growth in a chamber with elevated CO2 (∼9,000 µL L−1) until the seeds were collected. Total DNA was extracted from leaf tissues using cetyl-trimethyl-ammonium bromide buffer, digested with EcoRV+KpnI restriction enzymes, separated on a 1% (w/v) agarose gel, transferred to a nylon membrane, and detected with a DIG-labeled DNA probe as described previously (Lin et al., 2014b).
Analyses of the Transcripts of Transgenes on RNA Blots
The transcripts were analyzed on RNA blots using the procedure described previously with the same DIG-labeled RNA probes (Occhialini et al., 2016). Briefly, RNA samples were prepared from leaf tissues with a PureLink RNA mini kit (Life Technologies), and their concentrations were estimated with a Qubit RNA BR assay kit. About 1 μg of each RNA sample was mixed with northernMax formaldehyde load dye (Life Technologies) with 50 μg mL−1 ethidium bromide and incubated at 65°C for 15 min before they were loaded onto a 1.3% (w/v) agarose gel with 2% (v/v) formaldehyde. After separation at 7 V cm−1 for about 2 h, the gel was washed three times in diethylpyrocarbonate-treated water for 10 min each and incubated in 20× SSC for 45 min before the RNAs were transferred to a positively charged nylon membrane under capillary action. The membrane was then exposed to UV radiation with a Stratalinker UV Crosslinker, hybridized with 200 ng of each DIG-labeled RNA probe in ∼4 mL of DIG EasyHyb buffer (Roche) at 68°C overnight, and detected with anti-DIG-AP antibody and CDP-Star chemiluminescent substrate (Roche).
Plant Material
Seeds of wild-type (cv Samsun) and transplastomic tobacco were sown into trays of a commercial potting mix (Petersfield Products) with a slow-release fertilizer (Osmocote; Scotts UK Professional). Seedlings were thinned out after approximately 2 weeks, with individual plants transferred to 1-L pots after 3 weeks. Seeds of SeL were sown into tissue culture pots containing agar-solidified Murashige and Skoog medium containing 1% (w/v) Suc before transferring to soil after 8 weeks. Plants were grown in a controlled-environment chamber (Microclima 1750; Snijders Scientific). The chamber was set at day/night temperatures of 24°C/22°C with a 16-h photoperiod at 60% humidity. The ambient CO2 concentration within the chamber was maintained at 4,000 ± 400 µL L−1 using the integrated CO2 controller. CO2 levels were also monitored in the chamber with a Vaisala hand-held GM70 m device. Plants were kept well watered. Space limitations within growth chambers necessitated growing plants in batches for growth analysis.
Fixation and Embedding of Plant Tissue, Immunogold Labeling, and Transmission Electron Microscopy
Small pieces (1 × 1.5 mm) of tissue from fully expanded leaves of plants equivalent in size to 33-DAS wild-type plants were incubated in fixative (4% [v/v] paraformaldehyde and 2.5% [v/v] glutaraldehyde in 0.05 m sodium phosphate buffer, pH 7.2) for 2 h at room temperature with rotation. A vacuum was used to aid infiltration. After washing three times for 10 min each in 0.05 m sodium phosphate buffer, pH 7.2, the tissue was dehydrated in an ethanol series (50%, 70%, 80%, and 90% [v/v]) at room temperature for 30 min at each step and finally 100% ethanol for 1 h. Tissue was infiltrated with LR white resin (Agar Scientific), first by incubating for 1 h in 100% ethanol:LR white (1:1, v/v), then for 2 h in 100% LR white, and finally overnight in 100% LR white. Specimens were transferred to microtubes overfilled with fresh 100% LR white resin. The tubes were sealed with plastic film, and the resin was polymerized at 50°C for 16 h.
Ultrathin sections (∼90 nm) of embedded leaf material were captured on gold gilded grids (Agar Scientific) and used for immunogold labeling. Samples were blocked for 30 min in 1% (w/v) BSA in phosphate-buffered saline (PBS) and then incubated in primary antibody solution (antibody diluted 1:100 in 1% BSA in PBS) for 1.5 h. Grids were washed three times for 10 min each with 1% BSA in PBS before incubation for 1 h with secondary goat anti-rabbit antibody conjugated to 10-nm gold particles (Agar Scientific; 1:100 antibody dilution prepared in 1% BSA in PBS). Grids were washed three times for 10 min each in 1% BSA in PBS and three times for 5 min each in distilled water before air drying. Images were obtained at 80 kV using a JEOL 1010 (JEOL) microscope equipped with a digital AMT NanoSprint500 camera (Deben).
Gel Electrophoresis and Immunoblotting
Soluble protein extracts were analyzed for the presence of proteins via both denaturing SDS-PAGE and nondenaturing (native PAGE) gel electrophoresis. SDS-PAGE and immunoblotting were carried out as described by Perdomo et al. (2018) using Mini-Protean TGX gels (Bio-Rad). Nondenaturing gels were run using a Tris-Gly buffering system at 4°C according to the manufacturer’s instructions. For both types of electrophoresis, immunoblotting was conducted as described by Perdomo et al. (2018) using the Se LSU and CcmM antibodies described previously (Lin et al., 2014b) and a plant SSU antibody (AS07 259; Agri-Sera).
Rubisco Biochemistry
Rubisco activities and activation state in leaf extracts were determined as described by Carmo-Silva et al. (2017), except that homogenate centrifugation was done at a reduced 300g for 1 min. Chlorophyll content in the homogenates was determined by the method of Wintermans and de Mots (1965) using ethanol and measuring absorbance in a microplate reader (SPECTROstar Nano; BMG LabTech). Total soluble protein in the same supernatant as used for Rubisco activity assays was determined via Bradford assay (Bradford, 1976). The amount of Rubisco was also quantified in the same supernatant by a [14C]carboxyarabinitol-1,5-bisphosphate-binding assay (Whitney et al., 1999).
Rubisco catalytic properties were determined essentially as described previously (Prins et al., 2016; Orr and Carmo-Silva, 2018) with the following changes: leaf discs were ground in extraction buffer, followed by centrifugation at 300g and 4°C for 1 min. Supernatants were immediately used for assays, which was previously found to be suitable with similar cyanobacterial Rubisco complexes (Lin et al., 2014b). Additional higher CO2 concentrations (180, 280, and 410 µm) were also used for catalysis assays to enable the determination of KC.
Photosynthesis Measurements
Photosynthetic gas exchange was measured in healthy leaves that had recently reached full expansion, typically leaf 4 or 5 on plants of approximately 45 cm in height. An LI-6800F portable gas-exchange system (LI-COR) was used to enclose a 6-cm2 portion of leaf, with constant irradiance of 600 µmol photons m−2 s−1 supplied by the cuvette head light-emitting diodes, a vapor pressure deficit of 1.2 ± 0.03 kPa, and a flow rate of 300 µmol m−2 s−1. Leaf temperature was maintained at 24°C ± 1°C. For all measurements, the entire gas-exchange system was positioned inside the plant growth chamber and controlled remotely via an Ethernet connection. After the cuvette was clamped onto a leaf, the chamber door was kept closed to minimize fluctuations in CO2 levels and the plant was allowed to stabilize for at least 15 min at 3,000 µL L−1 CO2 prior to commencing measurements. For transplastomic tobacco lines, the ambient CO2 concentration was subsequently decreased to 100 µL L−1, followed by increases to 200, 400, 800, 1,200, 1,600, 2,000, and 2,500 µL L−1 CO2. For wild-type tobacco, additional concentrations were used such that increases in CO2 went from 50 to 100, 150, 200, 250, 300, 400, 600, 800, 1,200, 1,600, 2,000, and 2,500 µL L−1. For all leaves measured, a separate CO2 response curve was determined under 2% (v/v) oxygen conditions using a balanced air gas cylinder for input, using otherwise identical settings.
Plant Biomass
Leaf numbers and leaf measurements were taken every 3 to 7 d from four or five individuals for each line (two in the SeL line). Plant height was measured from the soil level to the growing point. Measurements were initiated at 28 DAS for the wild type and SeLS, 46 DAS for SeLSM35 and SeLm35, and 127 DAS for SeL, due to the differing growth rates between lines, and continued until the initiation of flowering. At the end of the growth period, final leaf measurements were taken and leaf area was measured using an LI-3100C leaf area machine (LI-COR). Leaf areas were then derived for all time points.
Statistical Analysis
Statistical differences between trait means were tested via one-way ANOVA. In cases where an effect of genotype was observed (P < 0.05), a posthoc Tukey’s test was used to conduct multiple pairwise comparisons. Statistical analyses were performed using RStudio (version 1.1.453; R Studio Team, 2019) and R (version 3.5.0; R Core Development Team, 2013). Plots were prepared with ggplot2 (Wickham, 2016). Plant height and leaf area data analyses involved fitting curves to the exponential phase of growth and comparing means of the curve coefficients using ANOVA. Where a significant difference was observed between lines, a posthoc Holm-Sidak test was used for multiple pairwise comparisons. Analyses were performed in SigmaPlot (version 13; Systat Software).
Accession Numbers
Sequence data for cyanobacterial RbcL and CcmM35 can be found in the GenBank data library under accession numbers AIM40198.1 and AIM40200.1, respectively.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. RNA blots of wild-type and transplastomic tobacco lines.
Supplemental Figure S2. Presence of procarboxysome compartments in tobacco transplastomic plants containing cyanobacterial Rubisco LSUs and CcmM35, with and without Rubisco SSUs.
Supplemental Figure S3. Electron micrographs of tobacco plants expressing cyanobacterial Rubisco LSUs and CcmM35 containing a procarboxysome compartment in the chloroplast.
Supplemental Figure S4. Additional examples of electron micrographs of tobacco plants expressing cyanobacterial Rubisco LSUs and CcmM35 with a procarboxysome compartment in the chloroplast.
Supplemental Figure S5. Western blots of SDS-PAGE and native PAGE gels used to examine protein composition of wild-type tobacco and transplastomic lines expressing β-cyanobacterial carboxysome components.
Supplemental Figure S6. Rubisco content expressed as grams per square meter.
Supplemental Figure S7. Chlorophyll contents of transplastomic lines.
Supplemental Figure S8. Response of leaf CO2 assimilation to intercellular CO2 concentrations under atmospheric levels and 2% oxygen.
Supplemental Figure S9. Comparison of leaf size in transplastomic plants.
Supplemental Figure S10. Plant photographs at a comparable growth stage.
Supplemental Figure S11. Multiple sequence alignment of cyanobacterial and tobacco Rubisco SSUs.
Supplemental Table S1. Plant growth data analyses.
Supplemental Table S2. Oligonucleotide sequences used in the construction of transformation vectors.
Acknowledgments
We thank Dr. Sam Taylor (Lancaster University) for support and advice with gas-exchange measurements and Nigel Fullwood (Lancaster University) for guidance with transmission electron microscopy. We also thank the reviewers for helpful comments on the article.
Footnotes
This work was supported by the U.K. Biotechnology and Biological Sciences Research Council (grant no. BB/I024488/1 to M.A.J.P.) and the U.S. National Science Foundation (grant no. EF-1105584 to M.R.H.).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Andrews TJ. (1988) Catalysis by cyanobacterial ribulose-bisphosphate carboxylase large subunits in the complete absence of small subunits. J Biol Chem 263: 12213–12219 [PubMed] [Google Scholar]
- Andrews TJ, Ballment B (1984) Active-site carbamate formation and reaction-intermediate-analog binding by ribulosebisphosphate carboxylase/oxygenase in the absence of its small subunits. Proc Natl Acad Sci USA 81: 3660–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews TJ, Lorimer GH (1985) Catalytic properties of a hybrid between cyanobacterial large subunits and higher plant small subunits of ribulose bisphosphate carboxylase-oxygenase. J Biol Chem 260: 4632–4636 [PubMed] [Google Scholar]
- Atkinson N, Feike D, Mackinder LCM, Meyer MT, Griffiths H, Jonikas MC, Smith AM, McCormick AJ (2016) Introducing an algal carbon-concentrating mechanism into higher plants: Location and incorporation of key components. Plant Biotechnol J 14: 1302–1315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson N, Velanis CN, Wunder T, Clarke DJ, Mueller-Cajar O, McCormick AJ (2019) The pyrenoidal linker protein EPYC1 phase separates with hybrid Arabidopsis-Chlamydomonas Rubisco through interactions with the algal Rubisco small subunit. J Exp Bot 70: 5271–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borland AM, Hartwell J, Weston DJ, Schlauch KA, Tschaplinski TJ, Tuskan GA, Yang X, Cushman JC (2014) Engineering Crassulacean acid metabolism to improve water-use efficiency. Trends Plant Sci 19: 327–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM. (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cai F, Bernstein SL, Wilson SC, Kerfeld CA (2016) Production and characterization of synthetic carboxysome shells with incorporated luminal proteins. Plant Physiol 170: 1868–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai F, Dou Z, Bernstein SL, Leverenz R, Williams EB, Heinhorst S, Shively J, Cannon GC, Kerfeld CA (2015) Advances in understanding carboxysome assembly in Prochlorococcus and Synechococcus implicate CsoS2 as a critical component. Life (Basel) 5: 1141–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cameron JCJ, Wilson SC, Bernstein SLS, Kerfeld CAC (2013) Biogenesis of a bacterial organelle: The carboxysome assembly pathway. Cell 155: 1131–1140 [DOI] [PubMed] [Google Scholar]
- Carmo-Silva E, Andralojc PJ, Scales JC, Driever SM, Mead A, Lawson T, Raines CA, Parry MAJ (2017) Phenotyping of field-grown wheat in the UK highlights contribution of light response of photosynthesis and flag leaf longevity to grain yield. J Exp Bot 68: 3473–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmo-Silva E, Scales JC, Madgwick PJ, Parry MAJ (2015) Optimizing Rubisco and its regulation for greater resource use efficiency. Plant Cell Environ 38: 1817–1832 [DOI] [PubMed] [Google Scholar]
- Conlan B, Birch R, Kelso C, Holland S, De Souza AP, Long SP, Beck JL, Whitney SM (2019) BSD2 is a Rubisco-specific assembly chaperone, forms intermediary hetero-oligomeric complexes, and is nonlimiting to growth in tobacco. Plant Cell Environ 42: 1287–1301 [DOI] [PubMed] [Google Scholar]
- Desmarais JJ, Flamholz AI, Blikstad C, Dugan EJ, Laughlin TG, Oltrogge LM, Chen AW, Wetmore K, Diamond S, Wang JY, et al. (2019) DABs are inorganic carbon pumps found throughout prokaryotic phyla. Nat Microbiol 4: 2204–2215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiMario RJ, Quebedeaux JC, Longstreth DJ, Dassanayake M, Hartman MM, Moroney JV (2016) The cytoplasmic carbonic anhydrases βCA2 and βCA4 are required for optimal plant growth at low CO2. Plant Physiol 171: 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erb TJ, Zarzycki J (2016) Biochemical and synthetic biology approaches to improve photosynthetic CO2-fixation. Curr Opin Chem Biol 34: 72–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang Y, Huang F, Faulkner M, Jiang Q, Dykes GF, Yang M, Liu LN (2018) Engineering and modulating functional cyanobacterial CO2-fixing organelles. Front Plant Sci 9: 739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feiz L, Williams-Carrier R, Belcher S, Montano M, Barkan A, Stern DB (2014) A protein with an inactive pterin-4a-carbinolamine dehydratase domain is required for Rubisco biogenesis in plants. Plant J 80: 862–869 [DOI] [PubMed] [Google Scholar]
- Flamholz AI, Prywes N, Moran U, Davidi D, Bar-On YM, Oltrogge LM, Alves R, Savage D, Milo R (2019) Revisiting trade-offs between Rubisco kinetic parameters. Biochemistry 58: 3365–3376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman Rosenzweig ES, Xu B, Kuhn Cuellar L, Martinez-Sanchez A, Schaffer M, Strauss M, Cartwright HN, Ronceray P, Plitzko JM, Förster F, et al. (2017) The eukaryotic CO2-concentrating organelle is liquid-like and exhibits dynamic reorganization. Cell 171: 148–162.e19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson MR, Gray BN, Ahner BA (2013) Chloroplast transformation for engineering of photosynthesis. J Exp Bot 64: 731–742 [DOI] [PubMed] [Google Scholar]
- Hanson MR, Lin MT, Carmo-Silva AE, Parry MAJ (2016) Towards engineering carboxysomes into C3 plants. Plant J 87: 38–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd JM, Sheehy JE, Langdale JA (2008) Using C4 photosynthesis to increase the yield of rice: Rationale and feasibility. Curr Opin Plant Biol 11: 228–231 [DOI] [PubMed] [Google Scholar]
- Hu H, Rappel WJ, Occhipinti R, Ries A, Böhmer M, You L, Xiao C, Engineer CB, Boron WF, Schroeder JI (2015) Distinct cellular locations of carbonic anhydrases mediate carbon dioxide control of stomatal movements. Plant Physiol 169: 1168–1178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan DB, Chollet R (1985) Subunit dissociation and reconstitution of ribulose-1,5-bisphosphate carboxylase from Chromatium vinosum. Arch Biochem Biophys 236: 487–496 [DOI] [PubMed] [Google Scholar]
- Kanevski I, Maliga P, Rhoades DF, Gutteridge S (1999) Plastome engineering of ribulose-1,5-bisphosphate carboxylase/oxygenase in tobacco to form a sunflower large subunit and tobacco small subunit hybrid. Plant Physiol 119: 133–142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langdale JA. (2011) C4 cycles: Past, present, and future research on C4 photosynthesis. Plant Cell 23: 3879–3892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Devonshire J, Hines KM, Parry MAJ, Hanson MR (2014a) β-Carboxysomal proteins assemble into highly organized structures in Nicotiana chloroplasts. Plant J 79: 1–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MT, Occhialini A, Andralojc PJ, Parry MAJ, Hanson MR (2014b) A faster Rubisco with potential to increase photosynthesis in crops. Nature 513: 547–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, He X, Lim W, Mueller J, Lawrie J, Kramer L, Guo J, Niu W (2018) Deciphering molecular details in the assembly of alpha-type carboxysome. Sci Rep 8: 15062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobell DB, Cassman KG, Field CB (2009) Crop yield gaps: Their importance, magnitudes, and causes. Annu Rev Environ Resour 34: 179–204 [Google Scholar]
- Long BM, Badger MR, Whitney SM, Price GD (2007) Analysis of carboxysomes from Synechococcus PCC7942 reveals multiple Rubisco complexes with carboxysomal proteins CcmM and CcaA. J Biol Chem 282: 29323–29335 [DOI] [PubMed] [Google Scholar]
- Long BM, Hee WY, Sharwood RE, Rae BD, Kaines S, Lim YL, Nguyen ND, Massey B, Bala S, von Caemmerer S, et al. (2018) Carboxysome encapsulation of the CO2-fixing enzyme Rubisco in tobacco chloroplasts. Nat Commun 9: 3570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long BM, Rae BD, Badger MR, Price GD (2011) Over-expression of the β-carboxysomal CcmM protein in Synechococcus PCC7942 reveals a tight co-regulation of carboxysomal carbonic anhydrase (CcaA) and M58 content. Photosynth Res 109: 33–45 [DOI] [PubMed] [Google Scholar]
- Long BM, Rae BD, Rolland V, Förster B, Price GD (2016) Cyanobacterial CO2-concentrating mechanism components: Function and prospects for plant metabolic engineering. Curr Opin Plant Biol 31: 1–8 [DOI] [PubMed] [Google Scholar]
- Long BM, Tucker L, Badger MR, Price GD (2010) Functional cyanobacterial β-carboxysomes have an absolute requirement for both long and short forms of the CcmM protein. Plant Physiol 153: 285–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long SP, Zhu XG, Naidu SL, Ort DR (2006) Can improvement in photosynthesis increase crop yields? Plant Cell Environ 29: 315–330 [DOI] [PubMed] [Google Scholar]
- Mackinder LCM. (2018) The Chlamydomonas CO2-concentrating mechanism and its potential for engineering photosynthesis in plants. New Phytol 217: 54–61 [DOI] [PubMed] [Google Scholar]
- Mackinder LCM, Meyer MT, Mettler-Altmann T, Chen VK, Mitchell MC, Caspari O, Freeman Rosenzweig ES, Pallesen L, Reeves G, Itakura A, et al. (2016) A repeat protein links Rubisco to form the eukaryotic carbon-concentrating organelle. Proc Natl Acad Sci USA 113: 5958–5963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurino VG, Weber APM (2013) Engineering photosynthesis in plants and synthetic microorganisms. J Exp Bot 64: 743–751 [DOI] [PubMed] [Google Scholar]
- McGrath JM, Long SP (2014) Can the cyanobacterial carbon-concentrating mechanism increase photosynthesis in crop species? A theoretical analysis. Plant Physiol 164: 2247–2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Occhialini A, Lin MT, Andralojc PJ, Hanson MR, Parry MAJ (2016) Transgenic tobacco plants with improved cyanobacterial Rubisco expression but no extra assembly factors grow at near wild-type rates if provided with elevated CO2. Plant J 85: 148–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr DJ, Carmo-Silva E (2018) Extraction of Rubisco to determine catalytic constants In Covshoff S, ed, Photosynthesis. Humana Press, New York, pp 229–238 [DOI] [PubMed] [Google Scholar]
- Orr DJ, Pereira AM, da Fonseca Pereira P, Pereira-Lima Í, Zsögön A, Araújo WL (2017) Engineering photosynthesis: Progress and perspectives. F1000 Res 6: 1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ort DR, Merchant SS, Alric J, Barkan A, Blankenship RE, Bock R, Croce R, Hanson MR, Hibberd JM, Long SP, et al. (2015) Redesigning photosynthesis to sustainably meet global food and bioenergy demand. Proc Natl Acad Sci USA 112: 8529–8536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdomo JA, Sales CRG, Carmo-Silva E (2018) Quantification of photosynthetic enzymes in leaf extracts by immunoblotting In Covshoff S, ed, Photosynthesis. Humana Press, New York, pp 215–227 [DOI] [PubMed] [Google Scholar]
- Price GD. (2011) Inorganic carbon transporters of the cyanobacterial CO2 concentrating mechanism. Photosynth Res 109: 47–57 [DOI] [PubMed] [Google Scholar]
- Price GD, Badger MR, von Caemmerer S (2011) The prospect of using cyanobacterial bicarbonate transporters to improve leaf photosynthesis in C3 crop plants. Plant Physiol 155: 20–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price GD, Badger MR, Woodger FJ, Long BM (2008) Advances in understanding the cyanobacterial CO2-concentrating-mechanism (CCM): Functional components, Ci transporters, diversity, genetic regulation and prospects for engineering into plants. J Exp Bot 59: 1441–1461 [DOI] [PubMed] [Google Scholar]
- Prins A, Orr DJ, Andralojc PJ, Reynolds MP, Carmo-Silva E, Parry MAJ (2016) Rubisco catalytic properties of wild and domesticated relatives provide scope for improving wheat photosynthesis. J Exp Bot 67: 1827–1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Development Team (2013) A Language and Environment for Statistical Computing. http://www.r-project.org/
- R Studio Team (2019) RStudio Cloud: Integrated Development for R. https://www.rstudio.com/
- Rae BD, Long BM, Förster B, Nguyen ND, Velanis CN, Atkinson N, Hee WY, Mukherjee B, Price GD, McCormick AJ (2017) Progress and challenges of engineering a biophysical CO2-concentrating mechanism into higher plants. J Exp Bot 68: 3717–3737 [DOI] [PubMed] [Google Scholar]
- Ray DK, Ramankutty N, Mueller ND, West PC, Foley JA (2012) Recent patterns of crop yield growth and stagnation. Nat Commun 3: 1293. [DOI] [PubMed] [Google Scholar]
- Rolland V, Badger MR, Price GD (2016) Redirecting the cyanobacterial bicarbonate transporters bica and sbta to the chloroplast envelope: Soluble and membrane cargos need different chloroplast targeting signals in plants. Front Plant Sci 7: 185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan P, Forrester TJB, Wroblewski C, Kenney TMG, Kitova EN, Klassen JS, Kimber MS (2019) The small RbcS-like domains of the β-carboxysome structural protein CcmM bind RubisCO at a site distinct from that binding the RbcS subunit. J Biol Chem 294: 2593–2603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salesse-Smith CE, Sharwood RE, Busch FA, Kromdijk J, Bardal V, Stern DB (2018) Overexpression of Rubisco subunits with RAF1 increases Rubisco content in maize. Nat Plants 4: 802–810 [DOI] [PubMed] [Google Scholar]
- Sharwood RE, Ghannoum O, Whitney SM (2016) Prospects for improving CO2 fixation in C3-crops through understanding C4-Rubisco biogenesis and catalytic diversity. Curr Opin Plant Biol 31: 135–142 [DOI] [PubMed] [Google Scholar]
- Sharwood RE, von Caemmerer S, Maliga P, Whitney SM (2008) The catalytic properties of hybrid Rubisco comprising tobacco small and sunflower large subunits mirror the kinetically equivalent source Rubiscos and can support tobacco growth. Plant Physiol 146: 83–96 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shetty RP, Endy D, Knight TF Jr. (2008) Engineering BioBrick vectors from BioBrick parts. J Biol Eng 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- South PF, Cavanagh AP, Liu HW, Ort DR (2019) Synthetic glycolate metabolism pathways stimulate crop growth and productivity in the field. Science 363: eaat9077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Wollman AJM, Huang F, Leake MC, Liu LN (2019) Single-organelle quantification reveals stoichiometric and structural variability of carboxysomes dependent on the environment. Plant Cell 31: 1648–1664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter M, Laughlin TG, Sloan NB, Serwas D, Davies KM, Kerfeld CA (2019) Structure of a synthetic β-carboxysome shell. Plant Physiol 181: 1050–1058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara S, Adachi F, Ito-Inaba Y, Inaba T (2016) Specific and efficient targeting of cyanobacterial bicarbonate transporters to the inner envelope membrane of chloroplasts in Arabidopsis. Front Plant Sci 7: 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, VanLoocke A, Bernacchi CJ, Ort DR (2016) The costs of photorespiration to food production now and in the future. Annu Rev Plant Biol 67: 107–129 [DOI] [PubMed] [Google Scholar]
- Wang H, Yan X, Aigner H, Bracher A, Nguyen ND, Hee WY, Long BM, Price GD, Hartl FU, Hayer-Hartl M (2019) Rubisco condensate formation by CcmM in β-carboxysome biogenesis. Nature 566: 131–135 [DOI] [PubMed] [Google Scholar]
- Whitney SM, Andrews TJ (2001) Plastome-encoded bacterial ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) supports photosynthesis and growth in tobacco. Proc Natl Acad Sci USA 98: 14738–14743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, Houtz RL, Alonso H (2011) Advancing our understanding and capacity to engineer nature’s CO2-sequestering enzyme, Rubisco. Plant Physiol 155: 27–35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitney SM, von Caemmerer S, Hudson GS, Andrews TJ (1999) Directed mutation of the Rubisco large subunit of tobacco influences photorespiration and growth. Plant Physiol 121: 579–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H. (2016) ggplot2: Elegant Graphics for Data Analysis Using the Grammar of Graphics. Springer-Verlag, New York [Google Scholar]
- Wilson RH, Hayer-Hartl M (2018) Complex chaperone dependence of Rubisco biogenesis. Biochemistry 57: 3210–3216 [DOI] [PubMed] [Google Scholar]
- Wintermans JFGM, De Mots A (1965) Spectrophotometric characteristics of chlorophylls a and b and their phenophytins in ethanol. Biochim Biophys Acta 109: 448–453 [DOI] [PubMed] [Google Scholar]
- Wunder T, Cheng SLH, Lai SK, Li HY, Mueller-Cajar O (2018) The phase separation underlying the pyrenoid-based microalgal Rubisco supercharger. Nat Commun 9: 5076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Cushman JC, Borland AM, Edwards EJ, Wullschleger SD, Tuskan GA, Owen NA, Griffiths H, Smith JAC, De Paoli HC, et al. (2015) A roadmap for research on Crassulacean acid metabolism (CAM) to enhance sustainable food and bioenergy production in a hotter, drier world. New Phytol 207: 491–504 [DOI] [PubMed] [Google Scholar]