The Arabidopsis acetate or polyketide pathway operates at the interface of central and lipid metabolism as well as supporting the much better studied phenylpropanoid pathway of flavonoid biosynthesis.
Abstract
The phenylpropanoid pathway of flavonoid biosynthesis has been the subject of considerable research attention. By contrast, the proposed polyketide pathway, also known as the acetate pathway, which provides malonyl-CoA moieties for the C2 elongation reaction catalyzed by chalcone synthase, is less well studied. Here, we identified four genes as candidates for involvement in the supply of cytosolic malonyl-CoA from the catabolism of acyl-CoA, based on coexpression analysis with other flavonoid-related genes. Two of these genes, ACC and KAT5, have been previously characterized with respect to their involvement in lipid metabolism, but no information concerning their relationship to flavonoid biosynthesis is available. To assess the occurrence and importance of the acetate pathway, we characterized the metabolomes of two mutant or transgenic Arabidopsis lines for each of the four enzymes of this putative pathway using a hierarchical approach covering primary and secondary metabolites as well as lipids. Intriguingly, not only flavonoid content but also glucosinolate content was altered in lines deficient in the acetate pathway, as were levels of lipids and most primary metabolites. We discuss these data in the context of our current understanding of flavonoids and lipid metabolism as well as with regard to improving human nutrition.
Flavonoids are secondary metabolites involved in several physiological responses to the environment, such as defense against herbivores, UV radiation, and pathogens (Peters and Constabel, 2002; Foster-Hartnett et al., 2007; Agati and Tattini, 2010; Schulz et al., 2015). They are ubiquitously distributed among plant species and act at the intersection between primary and secondary metabolism (Alseekh et al., 2015; Alseekh and Fernie, 2018). Flavonoid uptake in the diet is also associated with health benefits for humans (Homo sapiens), mostly due to their activity as antioxidants (Zhang et al., 2015; Zhao et al., 2016; Martin and Li, 2017; Tohge et al., 2017), and this class of molecules has recently been compellingly demonstrated to play an important function in normal plant root development (Silva-Navas et al., 2016; Tohge and Fernie, 2016). Taken together these observations render the comprehensive understanding of flavonoid biosynthesis an important fundamental research aim.
The basic backbone structure of flavonoids is synthesized by a combination of the phenylpropanoid and polyketide (or acetate) pathways, the former providing p-coumaroyl-CoA from Phe while the latter provides malonyl-CoA for C2 chain elongation by chalcone synthase (Winkel-Shirley, 2001, 2002; Tohge et al., 2018). Flavonoids are one of the major secondary products in the model plant Arabidopsis (Arabidopsis thaliana), and research in this species has considerably improved our understanding of this pathway (Saito et al., 2013). As a consequence of this research, most of the structural and regulatory genes of the shikimate/phenylpropanoid part of the pathway involved in their biosynthesis have been well described (for details see Saito et al., 2013).
Investigation of the transparent testa (tt) mutants of Arabidopsis demonstrated that the regulation of flavonoid biosynthesis is strongly controlled at the transcriptional level (Borevitz et al., 2000; Xu et al., 2015). Indeed three main classes of transcription factors (TFs) were identified that regulate anthocyanin and proanthocyanidin biosynthesis: MYB, basic helix-loop-helix motifs (bHLHs), and WD40 repeats, although so far only MYBs have been described as involved in flavonol biosynthesis (Hichri et al., 2011). Additionally, the different branches of flavonoid metabolism are described as being regulated by specific TFs. In Arabidopsis, PAP1 (MYB75) and PAP2 (MYB90) are regulators of anthocyanin structural genes (Borevitz et al., 2000; Zimmermann et al., 2004), MYB123 (tt2) is a seed-specific regulator of proanthocyanidin biosynthetic genes (Nesi et al., 2001), and MYB11, MYB12, and MY B111 are regulators of flavonol structural genes (Stracke et al., 2007).
Despite these advances one of the biggest gaps still existent in this metabolic network is the interplay of these pathways with primary metabolism. In particular the mechanisms involved in the mobilization of primary resources toward flavonoid biosynthesis remain relatively poorly investigated. That said, some studies suggest that its regulation is modulated by the very same TFs described in the previous paragraph. Interesting work on tomatoes (Solanum lycopersicum) overexpressing Arabidopsis MYB12 under the control of a fruit-specific E8 promoter (Luo et al., 2008) showed that MYB12 is capable of binding to the promoters and inducing genes from primary metabolism, including those encoding 3-deoxy-d-arabinoheptulosonate 7-phosphate synthase, a rate-limiting step in the shikimate pathway for Phe biosynthesis, and the plastidial enolase (Zhang et al., 2015). The constantly improving resources for coexpression analysis could assist the identification of new candidate genes involved in supplying precursors, energy, and reducing power for secondary metabolism pathways (Yonekura-Sakakibara et al., 2008). Aside from these avenues, an interesting target for exploring the communication of central carbon metabolism with flavonoid biosynthesis, also indicated by gene coexpression analysis, is the ketoacyl-CoA thiolase KAT5. KATs are essential enzymes for the conversion of lipids to acetyl-CoA in the peroxisomes through β-oxidation (Li-Beisson et al., 2013). Interestingly, the expression of the cytosolic KAT5 was recently shown to be highly correlated with genes involved in flavonol biosynthesis in several studies (Germain et al., 2001; Carrie et al., 2007; Yonekura-Sakakibara et al., 2008; Wiszniewski et al., 2012). It was therefore postulated that KAT5 may play a role in the mobilization of carbon resources acting as part of a regulatory network directing flux toward flavonoid biosynthesis. The production of malonyl-CoA in the cytosol through carboxylation of acetyl-CoA by acetyl-CoA carboxylase (ACC) is additionally postulated to be one of the essential substrates for flavonoid biosynthesis (Fig. 1). However, the regulation of its supply remains poorly investigated. In order to better understand the role of the so-called acetate pathway, here we analyze the metabolic phenotypes of a range of genotypes, covering the reactions linking acyl-CoA to malonyl-CoA that were identified within the coexpression network of KAT5 and characterized flavonoid genes. Mutant lines were either isolated from knockout collections or generated using the artificial microRNA (amiRNA) strategy. Given the importance of this pathway to both flavonoid and lipid metabolism we evaluated the levels of primary, secondary, and lipid metabolites in these lines, as well as the expression levels of key structural genes of the flavonoid pathway. The collective results are discussed in the context of our current understanding of both flavonoid metabolism and the interplay of plant primary and specialized metabolism.
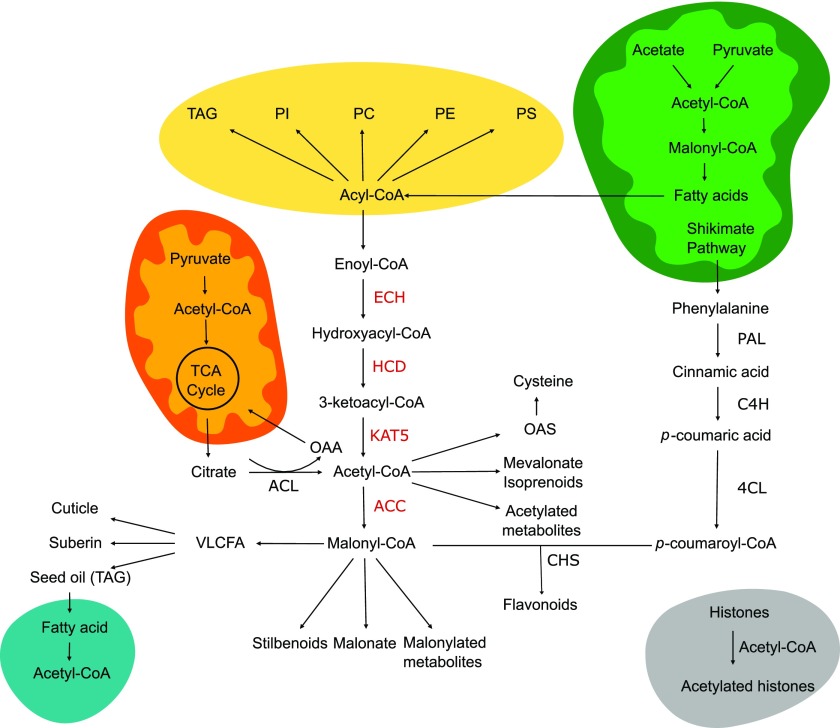
Figure 1.
Overview of flavonoid, lipid, and acetyl-CoA metabolism in Arabidopsis. Cell compartments represented include mitochondria (orange), chloroplast (green), endoplasmic reticulum (yellow), peroxisome (blue), the nucleus (gray), and the cytosol (white). Enzymes in the pathway proposed in this work are highlighted in red. OAA, Oxaloacetate; ECH, enoyl-CoA hydratase; HCD, hydroxyacyl-CoA dehydrogenase; PAL, Phe ammonia lyase; C4H, cinnamate 4-hydroxylase; 4CL, 4-coumarate:coenzyme A ligase; CHS, chalcone synthase.
RESULTS
Selection of Candidate Genes Putatively Involved in Malonyl-CoA Supply for Flavonoid Biosynthesis
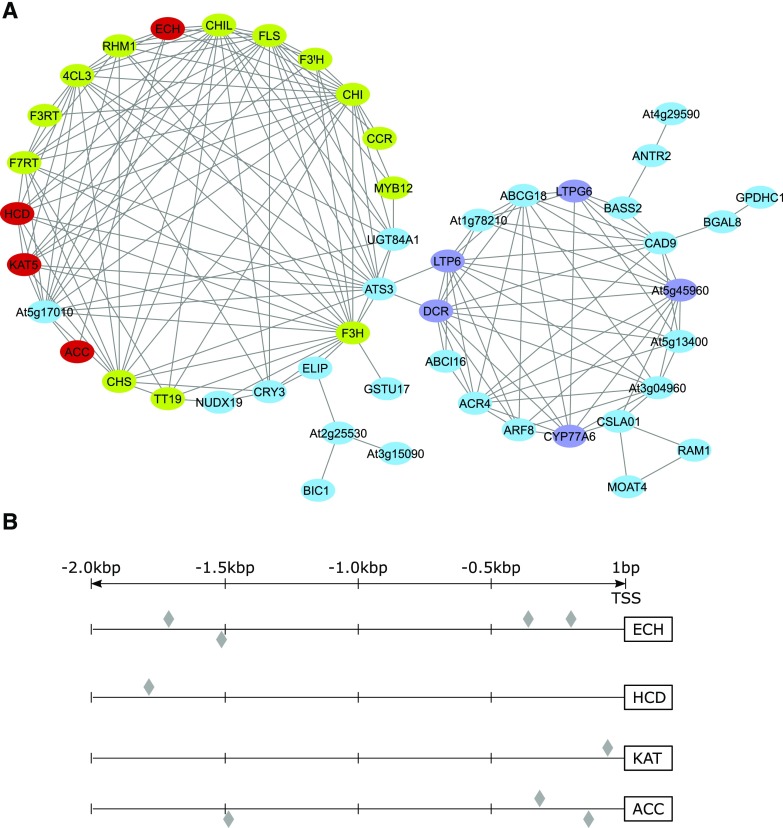
Coexpression network analyses previously performed by multiple groups have highlighted the coexpression of a 3-ketoacyl-CoA thiolase isoform (KAT5) with genes involved in flavonoid biosynthesis (Carrie et al., 2007; Yonekura-Sakakibara et al., 2008). KAT is one of the core enzymes in peroxisomal β-oxidation, catalyzing the breakdown of 3-ketoacyl-CoA to produce acetyl-CoA and a shorter acyl-CoA that can reenter the cycle (Li-Beisson et al., 2013). The Arabidopsis genome contains three loci that encode KAT enzymes, annotated as KAT1, KAT2, and KAT5, with KAT2 the only one expressed at significant levels during seed germination in Arabidopsis (Germain et al., 2001; Carrie et al., 2007; Li-Beisson et al., 2013). Both KAT2 and KAT5 are conserved in higher plants, and while the three isoforms from Arabidopsis are localized to the peroxisomes, KAT5 is the only one exhibiting an alternative transcript the product of which is targeted to the cytosol (Wiszniewski et al., 2012). To evaluate the possible role of this enzyme in flavonoid metabolism, we used the ATTED-II web platform (Obayashi et al., 2018) to select the top 50 genes of highest correlation with KAT5. The pairwise correlation for all selected genes was retrieved from ATTED-II and used for constructing the network depicted in Figure 2A (Supplemental Datasets S1 and S2). Genes were classified based on their annotations on The Arabidopsis Information Resource (Berardini et al., 2015), and we could observe a clear enrichment for genes functionally involved in flavonol and lipid metabolism. Interestingly, a group of three genes among the coexpression network (ECH [At1g06550], HCD [At1g65560], and ACC [At1g36160]) together with KAT5 included all necessary enzymatic steps for the production of acetyl-CoA from activated Acyl-CoA and subsequent carboxylation to malonyl-CoA (Fig. 1). Furthermore, putative orthologs of all these genes exhibited increased expression levels in Del/Ros1 transgenic tomato lines in which ectopic fruit‐specific expression of the snapdragon (Antirrhinum majus) TFs Delila (Del, bHLH) and Rosea1 (Ros1, Myb) led to the accumulation of flavonols and anthocyanins within the fruit (Tohge et al., 2015). In order to support our hypothesis, we also performed promoter analysis including a 2.0-kb region upstream to the transcription start site of each candidate gene using the web resource PlantPAN 3.0 (Chow et al., 2019). This analysis indicated the presence of a putative flavonoid-related MYB111 (PFG3) binding motif for all candidate genes (Fig. 2B; Supplemental Table S1). Taken together, the structure of this network and candidate gene promoters suggest a regulatory mechanism in which transcriptional activation of flavonoid biosynthesis is coregulated with mobilization of the cytosolic acyl-CoA pool via its degradation into the building block malonyl-CoA, a substrate for the production of the core flavonoid aglycone narigenin by chalcone synthase. Additionally, the acetate pathway may be regulated by the same transcriptional mechanism that activates downstream flavonoid specific genes, as well as the shikimate pathway involved in the production of Phe upstream to the phenylpropanoid branch of flavonoid biosynthesis (Zhang et al., 2015).
Figure 2.
Selection and analysis of candidate genes putatively involved in malonyl-CoA supply for flavonoid biosynthesis. A, KAT5 coexpression network. The top 50 genes coexpressing with KAT5 were selected based on the highest Spearman rank correlation coefficients obtained from the web platform ATTED-II (Obayashi et al., 2018). A pairwise correlation threshold of 0.55 was applied as a cutoff to establish the edges between pairs of nodes (genes). Genes that were not connected to any neighbor within the network were excluded from further analysis. Green circles represent the characterized flavonoid-related genes, purple circles the lipid-related genes, and red circles the candidate genes putatively involved in fatty acid degradation and production of malonyl-CoA selected for functional characterization. B, Promoter analysis of candidate genes. A 2.0-kb region upstream to the transcription start site (TSS) of each candidate gene was analyzed using the web resource PlantPAN 3.0 (Chow et al., 2019). Gray diamonds indicate predicted flavonoid-related MYB111 (PFG3) binding motifs.
Expression Analysis of Transgenic Lines
In order to investigate the metabolic roles of these genes, we obtained transfer DNA (T-DNA) mutants from the stock center (see “Materials and Methods”) and screened for homozygous lines exhibiting reduced expression of the target genes. T-DNA lines were obtained for ECH (ech1 and ech2), HCD (hcd1 and hcd2), and KAT5 (kat5-1). Mutant lines disrupting ACC activity were previously shown to exhibit very strong phenotypes including defective cotyledons, deformed leaves, short primary root, and impaired embryo morphogenesis and biosynthesis of the very-long-chain fatty acids (VLCFAs) associated with cuticular waxes and triacylglycerols (Baud et al., 2003; Lü et al., 2011; Chen et al., 2017). These led to the conclusion that our difficulties in acquiring homozygous T-DNA lines for this gene in particular were most probably due to an embryo-lethal phenotype. There are, however, mutants containing reductions in ACC activity that were only severe enough to cause a differential response under stress conditions associated with a reduced capacity of providing malonyl-CoA, but leading to no visible phenotype at normal growth conditions (Amid et al., 2012). Therefore, we proceeded by generating multiple amiRNA lines in order to obtain ACC genotypes exhibiting a less severe phenotype (acc1 and acc2). We also included the generation of an additional amiRNA KAT5 (kat5-2) in order to proceed with the further experiments on at least two independent lines for each candidate gene.
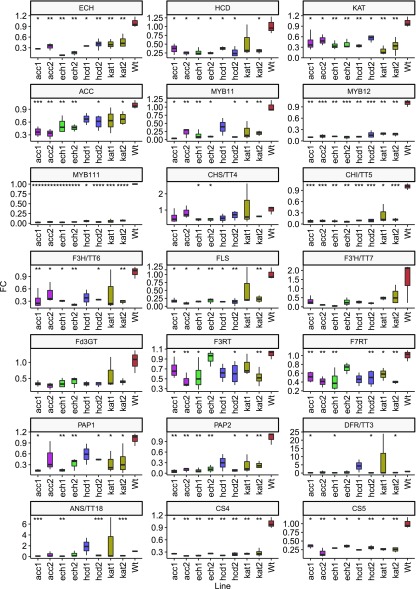
Gene expression analysis showed a decrease in expression levels not only of targeted genes for each line but also of all other candidates and most flavonoid related genes (Fig. 3; Supplemental Dataset S3). Interestingly, while these results are consistent with the coexpression between candidate genes and flavonoid related genes, it also suggests that acetyl-CoA and/or malonyl-CoA levels within the cytosol have a significant impact in the regulation of the whole pathway. Publicly available expression data (GEO Series accession number GSE 51215) for flavonoid mutants tt4 and tt4xMYB12OX support this hypothesis (Nakabayashi et al., 2014). Both genotypes exhibited reduced expression of several characterized flavonoid genes including MYB111, MYB12, pap-1d, FLS, Fd3GT, F3RT, and F7RT, as well as KAT5.
Figure 3.
Transcript levels of candidate genes and characterized flavonoid genes in wild type and mutant lines. Transcript levels were determined by reverse transcription quantitative PCR (RT-qPCR) and normalized to ACT2, UBQ10, and GAPDH3′. Boxplots represent at least three biological replicates, each with two technical replicates. Statistical significance of each line relative to the wild type is coded as ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05 by Student's two-tailed t test.
Metabolite Profiling of Transgenic Lines
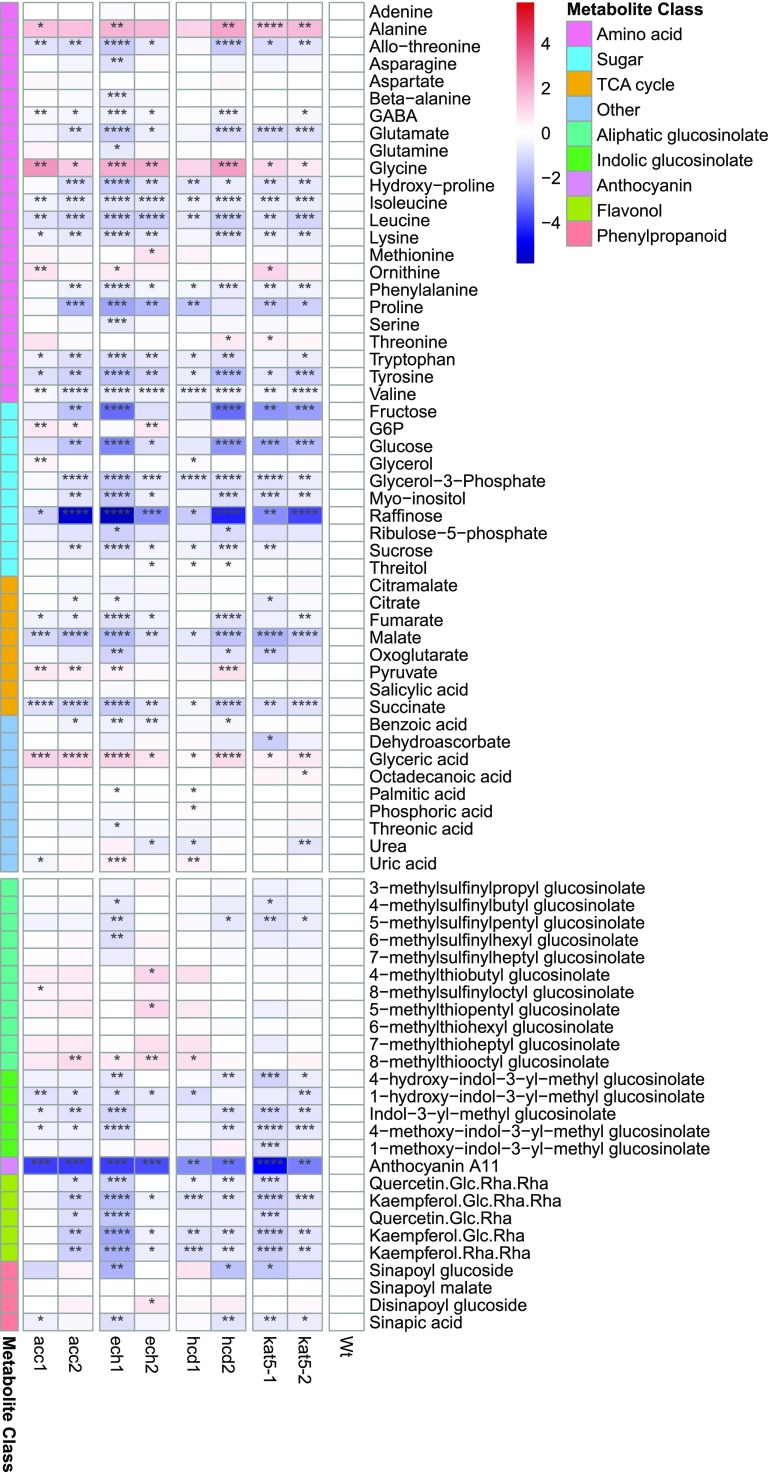
Under normal conditions, Arabidopsis ecotype Columbia (Col-0) leaves constitutively accumulate a number of flavonols, mainly kaempferol glycosides, but also, albeit at lower levels, quercetin glycosides (Saito et al., 2013). Several anthocyanins are also produced, but are often detected exclusively in response to certain stress conditions. The major anthocyanin detected under normal conditions is a cyanidin glycoside conjugated to p-coumaroyl, sinapoyl, and malonyl moieties called anthocyanin A11 (Saito et al., 2013). As expected, metabolite profiling of mutant lines (Fig. 4; Supplemental Datasets S4–S6; Fernie et al., 2011) showed a clear reduction for most flavonols as well as for the major anthocyanin A11. Despite the fact that flavonol and anthocyanin biosynthesis are regulated by different TFs, and that we observed mostly flavonol-specific genes within the coexpression network described here, such results are not surprising when one considers that these two metabolic pathways still share exactly the same precursors from central metabolism. More intriguing was the reduction observed in the Trp-derived indolic glucosinolates. This class of metabolites has been shown to interact with phenylpropanoid metabolism possibly via the interaction of indole-3-acetaldoxime, a precursor in indole glucosinolate biosynthesis, and MED5, a member of the mediator complex and thereby a repressor of phenylpropanoid metabolism (Kim et al., 2015). However, in the mutants studied here, it seems more likely that down-regulation of the shikimate pathway for production of both Phe and Trp is directly affecting the levels of the precursors of both pathways.
Figure 4.
Primary and secondary metabolite profiling of candidate gene mutant lines and wild type. Scales represent the log2 fold change relative to the wild type. Statistical significance for each line relative to the wild type is coded as ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05 by Student's two-tailed t test. The heatmap was produced using the function pheatmap in R.
In central metabolism, we observed a marked reduction of several amino acids, sugars, and organic acids involved in the tricarboxylic acid cycle. Interestingly, we initially expected to observe an increased flux through the portion of the tricarboxylic acid cycle providing most of the cytosolic pool of acetyl-CoA, namely the export of citrate from the mitochondria and subsequent conversion by cytosolic ATP citrate lyase (ACL; Fatland et al., 2005), in order to compensate for the reduction in acetyl-CoA. The results observed here, however, are more similar to what would be expected from oxidative stress, with an overall reduction of photosynthesis, glycolysis, and the tricarboxylic acid cycle-related metabolites and an increase of photorespiratory related metabolites Gly and glycerate, pyruvate, and Ala (Dumont and Rivoal, 2019). Moreover, it has been shown that plants deficient in ACL activity unexpectedly exhibit a 4-fold increase in anthocyanin accumulation despite the considerable reduction in cytosolic acetyl-CoA (Fatland et al., 2005), suggesting that the regulation of acetyl-CoA production for flavonoid biosynthesis may occur via a pathway other than the one the ACL belongs to. Indeed, KAT5 appears among differentially expressed genes in antisense ACL mutants (2.7 fold change [FC]), together with tt4 (5.0 FC) and F3H (3.2 FC; Fatland et al., 2005). Given these results, we also measured the expression of citrate synthase in order to compare the status of the tricarboxylic acid cycle in mutants versus the wild type. As we anticipated, we observed a reduced expression of genes encoding both mitochondrial isoforms of this enzyme, CSY4 and CSY5 (Fig. 3).
In summary, we observed a consistent decrease of flavonol accumulation across the mutant lines, together with a reduction in other secondary metabolites sharing precursors with the pathway. We could additionally observe extensive changes in central metabolism, particularly in glycolysis and the tricarboxylic acid cycle intermediates, suggesting increased oxidative damage in mutant lines.
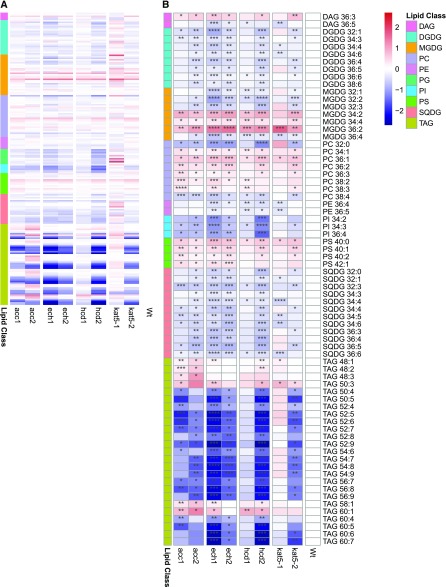
Lipid Profiling
Considering the obvious link between the candidate genes and acyl-CoA metabolism, differences were expected from lipidomic analysis of these mutant lines. Indeed, we could observe consistent changes in most classes of lipids detected (Fig. 5; Supplemental Datasets S7 and S8; Fernie et al., 2011). For a more detailed description of the lipid metabolism of Arabidopsis we refer to the literature (Chapman et al., 2013; Li-Beisson et al., 2013). In brief, fatty acids are synthesized in the chloroplasts, where they may traffic between the plastid stroma and the endoplasmatic reticulum where they are desaturated, converted to acyl-CoA, and released in the cytosol. This cytosolic pool of acyl-CoA, suggested here to serve as a substrate for malonyl-CoA production, can also be incorporated into membrane phospholipids and triacylglycerols (TAGs; Chapman et al., 2013). The phospholipids detected in our experiments include phosphatidylcholine (PC), phosphatidylethanolamine (PE), phosphatidylinositol (PI), phosphatidyl-Ser (PS), and phosphatidylglycerol. PEs and PGs exhibited few significant changes. PIs were reduced for most mutants, while both PCs and PSs were increased. Despite the reduction of PIs, it is tempting to speculate that the increase in PCs and PSs may be due to a lower consumption of the acyl-CoA leading to an increase in its available cytosolic pool. Intriguingly, the levels of TAGs which also rely upon the cytosolic pool of acyl-CoA, were drastically reduced. It is important to note that despite its essential role in flavonoid biosynthesis, one of the most vital functions of cytosolic malonyl-CoA is to provide the C2 building blocks for VLCFA elongation (Baud et al., 2003). These acyl chains are further incorporated into TAG molecules, cuticle, waxes or sphingolipids (Roesler et al., 1994; Baud et al., 2003). We do observe a significant reduction in the levels of several of those TAGs that incorporate VLCFAs, in our mutant lines. However, we also observe a reduction of those TAGs that do not incorporate such VLCFAs. Possible explanations for this observation include that TAG reserves may be reduced due to a reduction in carbon reserves or their mobilization to produce citrate via peroxisomal β-oxidation (Pracharoenwattana et al., 2005). It is also relevant to note here that we could not observe differences in one of the main products of fatty acid synthesis, palmitic acid (Ohlrogge and Browse, 1995; Burgos et al., 2011), suggesting that these phenotypes are not associated with an impairment in chloroplast fatty acid production but are rather related to some downstream factor, such as changes in lipid turnover or increased lipid peroxidation. Finally, a behavior similar to that previously described in stress situations associated with heat (Higashi et al., 2015) and dark (Burgos et al., 2011) could be observed for the main galactolipids constituting the chloroplast membrane, namely an overall decrease in digalactosyldiacylglycerols (DGDGs) and increase in monogalactosyldiacylglycerols. Again, we speculate that this may be due to a mild oxidative stress affecting the mutant lines. Additionally, DGDGs and sulfoquinovosyldiacylglycerols accumulate on membrane remodeling following Pi deficiency. This is known to be affected by changes in auxin signaling (Narise et al., 2010), a process in which flavonols are also known to be involved (Silva-Navas et al., 2016), establishing a possible link between flavonol deficiency and a change in the membrane remodeling capacity.
Figure 5.
Lipid profiling of candidate gene mutant lines and wild type. A, Profile of all lipids belonging to the different classes. B, Selected lipids differentially accumulating in mutant lines. Scales represent the log2 fold change relative to the wild type. Statistical significance for each line relative to the wild type is coded as ****P ≤ 0.0001, ***P ≤ 0.001, **P ≤ 0.01, and *P ≤ 0.05 by Student's two-tailed t test. The heatmap was produced using the function pheatmap. DAG, diacylglycerol; PG, phosphatidylglycerol; MGDG, monogalactosyldiacylglycerol, SQDG, sulfoquinovosyldiacylglycerol. The numbers in a lipid name describe the fatty acid chains on the lipid in the format (number of carbons in fatty acid chain):(number of double bonds in fatty acid chain), with unique numbering used to distinguish isomers.
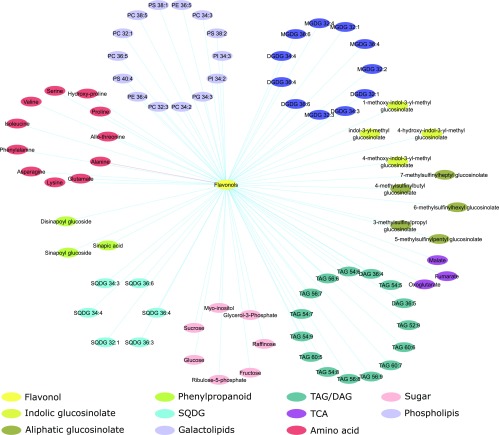
Metabolic Correlation Network
In order to aid visualization and summarize all the relevant metabolic results within the context of flavonol accumulation, the correlation network in Figure 6 (Supplemental Dataset S9) was computed. This network is based on Spearman rank correlation using an absolute threshold of 0.5 to establish edges between all metabolites and detected flavonols. The resulting network shows significant correlation between flavonoids and most of the metabolite classes previously described. Interestingly, we observe not only the strong correlation with indolic glucosinolates, previously described in "Metabolite Profiling of Transgenic Lines" and "Lipid Profiling", but also with other secondary metabolites that were not showing consistent significant changes across multiple mutant lines. These include sinapic acid derivatives, sharing the phenylpropanoid branch with flavonoid biosynthesis, and methylsulfinyl glucosinolates derived from Met. With regard to primary metabolism, the correlation of flavonols is particularly evident with sugars, but also with many amino acids, including those associated with the shikimate pathway as well as the tricarboxylic acid cycle-derived Glu. The only strong negative correlation observed was with Ala, which was increased in the mutant lines. Interestingly, among the tricarboxylic acid cycle compounds, we did not observe a strong correlation with citrate but only with oxoglutarate, malate, and fumarate. Among the lipids we could observe a strong correlation with several of the phospholipids derived from cytosolic acyl-CoAs, as well as with many of the TAGs, particularly those including VLCFAs, and a considerable number of the galactolipids MDGD and DGDG. This analysis underscores the role of the acetate pathway at the interface of primary and flavonoid metabolism, as well as being of importance, albeit lesser, in lipid biosynthesis.
Figure 6.
Metabolic correlation network. The correlation matrix was computed using the R package Hmisc and represented using cytoscape v3.6.1 (Shannon et al., 2003). Nodes represent metabolites and edges represent pairs of metabolites with a Spearman rank correlation above an absolute threshold of 0.5. The node color code indicates the different metabolite classes in the figure. Edge colors represent positive (blue) and negative (red) correlation coefficients between the pairs of metabolites.
DISCUSSION
Flavonoid biosynthesis is arguably one of the most well characterized plant secondary metabolic pathways. Flavonoids are known to possess high nutritional value due to their antioxidant activity and are conserved across higher plants (Tohge et al., 2013; Martin and Li, 2017), attracting great attention from breeding and metabolic engineering perspectives. While intensive research in the last decades generated vast knowledge regarding flavonoid biosynthesis and regulation, few advances were made regarding its integration with primary metabolism. However, flavonoid biosynthesis, together with the rest of phenylpropanoid metabolism, represents a significant sink of carbon resources for the plant (Saito et al., 2013), and must be tightly connected to central metabolism. That said, recent work strongly suggests that flavonoid-related TFs do play a role in directing resources from central metabolism toward flavonoid biosynthesis (Zhang et al., 2015).
Strategies for gene annotation based on coexpression network analysis following a guilt-by-association approach (Usadel et al., 2009) tend to work particularly well for genes and pathways that are transcriptionally regulated, hence the success of this approach in identifying many secondary metabolism structural and regulatory genes (Achnine et al., 2005; Tohge et al., 2005; Yonekura-Sakakibara et al., 2008; Alejandro et al., 2012; Wisecaver et al., 2017). In the current study, an investigation of flavonoid coexpression network led to an interesting gene, KAT5, that is strongly correlated to many flavonol-specific genes. By expanding the KAT5 coexpression network, it became evident that the enrichment of flavonol and fatty acid-related genes strongly suggests a link between fatty acid degradation and flavonol biosynthesis. The roles associated with KAT5, ECH, HCD, and ACC identified by this approach fit perfectly with the production of malonyl-CoA, one of the building blocks for the backbone of flavonoids (Saito et al., 2013), from activated acyl CoAs via a process similar to β-oxidation. Of all of these genes, ACC is the only one with a clear connection to flavonoid biosynthesis. ACC is described as the main source of cytosolic malonyl-CoA, essential for many processes, including long-chain fatty acid biosynthesis and flavonoid production (Baud et al., 2003; Amid et al., 2012; Chen et al., 2017). It is interesting to note that the proposed pathway for malonyl-CoA production seems to be specifically related to the flavonols and not to other flavonoids, such as anthocyanins, suggesting the regulation of this pathway by a flavonol-specific MYB TF (Tohge et al., 2005; Stracke et al., 2007). Considering the pattern of accumulation in the different branches of the pathway, it is observed that flavonols are constitutively accumulated in the leaves, while anthocyanin accumulation is usually induced by stress. These considerations allow us to speculate that this alternative malonyl-CoA supply and its specificity toward flavonols may be related to the different behaviors of each branch of the pathway and their costs to central metabolism.
The results obtained for the mutants show a clear effect on flavonoid accumulation with a consistent reduction of flavonol accumulation in all mutant lines. Changes in other classes of secondary metabolites, namely anthocyanins and indolic glucosinolates, were attributed to a reduction in the availability of substrates shared by these pathways, including malonyl-CoA and the precursors of both Phe and Trp in the shikimate pathway.
Previous work overexpressing Arabidopsis MYB12 in tomatoes under the control of a fruit-specific promoter demonstrated clear changes in primary metabolism. This included higher flux of carbon through glycolysis, the pentose phosphate pathway, and the tricarboxylic acid cycle, with a significant reduction in the content of major sugars and increase in aromatic amino acids other than Phe (Zhang et al., 2015). Their data provide solid evidence of an important role of the reprogramming of central metabolism following the up-regulation of flavonoid biosynthesis. The most marked differences in primary metabolism observed in this work were the reductions in amino acids, sugars, and tricarboxylic acid cycle intermediates. Interestingly, the phenotype of these mutants is far less severe than that observed following deficiency in expression of ACL (Fatland et al., 2005), suggesting what was previously anticipated, that this route of acetyl-CoA production is less crucial than that of ACL, which recent studies have suggested is closely associated to the activity of the tricarboxylic acid cycle via a thioredox-intermediated approach (Daloso et al., 2015). Indeed, our results combined with previous observations on ACL unexpected overaccumulation of anthocyanins (Fatland et al., 2005), as well as the behavior of central metabolism under oxidative stress, exhibiting reduction on glycolysis and tricarboxylic acid cycle activity (Dumont and Rivoal, 2019), suggest that this may be an alternative route to sustain the synthesis of these antioxidant compounds even under conditions of stress in which the flux of acetyl-CoA from the tricarboxylic acid cycle is reduced.
This alternative supply of acetyl-CoA via acyl-CoA degradation implies a significant role of lipid metabolism; hence, the alterations observed in lipid profiling were expected. The accumulation of a cytosolic pool of acyl-CoAs is suggested to be a limiting step in the biosynthesis of many lipids (Li-Beisson et al., 2013), and we anticipated that a reduction in the mobilization of these substrates for acetyl-CoA production would lead to an increase in its derived lipids. We did observe a significant increase of PSs and PCs, which are some of the main phospholipids produced from cytosolic acyl-CoA, despite the reduction in TAGs and PIs. It is important to notice, however, that many of the TAGs reduced are indeed products of VLCFAs that need cytosolic malonyl-CoA for the acyl chain extension. Additionally, while TAGs are a well described source of energy for germinating seeds via β-oxidation (Germain et al., 2001), their role is not so well understood in leaves, where some evidence suggests their action as intermediates of membrane turnover during senescence and in mechanisms of excess acyl-CoA scavenging followed by peroxisome degradation (Chapman et al., 2013).
Finally, the fact that no visible phenotype is discernible to the naked eye was somewhat surprising for us, because we had anticipated that the reason the acetate pathway had not been identified as important for flavonoid metabolism in the mutant screens that unraveled the phenylpropanoid pathway (Winkel-Shirley, 2001) was that they resulted in lethality due to pleotropic effects on lipid or primary metabolism. This is not the case; however, it is important to note that there were no obvious alterations in seed or leaf pigmentation. Still, it remains to be seen how they respond under stress conditions, where such changes may be more easily observed.
CONCLUSION
The results presented here support the hypothesis that the flavonoid regulatory network is not only involved in activating flavonoid-specific genes and mobilizing primary substrates from the shikimate and phenylpropanoid pathways but is also able to regulate carbon flux through the much less studied acetate pathway. Our data here experimentally confirm the operation of this pathway and validate the identity of the genes which encode it in Arabidopsis. They additionally allow the assessment of its importance in flavonoid and lipid biosynthesis. In both cases, it would seem to be an important supporting pathway rather than the dominant pathway; however, deficiency in this pathway does result in clear deficiencies of both flavonol and lipid levels. In the case of lipids, the mechanism described is independent of the main source of acetyl-CoA via the export of citrate from the tricarboxylic acid cycle to the cytosol and the activity of ACL. Hence, this pathway allows Arabidopsis to mobilize alternative carbon resources from its acyl-CoA pool, and to modulate flavonoid concentration without compromising the low concentration and rapid turnover pool of cytosolic acetyl-CoA, central for many other vital processes such as terpenoid biosynthesis and VLCFA biosynthesis.
MATERIALS AND METHODS
Coexpression Network Analysis
Coexpression information for a network consisting of the top 50 genes of highest correlation with KAT5 was obtained from ATTED-II (Obayashi et al., 2018). The pairwise correlation for all selected genes was used for constructing the network depicted in Figure 2A (Supplemental Dataset S1), where edges represent Spearman rank correlation coefficients above a threshold of 0.55. Genes that were not connected to any other node were omitted from the network. The results were imported and manipulated in the software Cytoscape v 3.6.1 (Shannon et al., 2003).
amiRNA Silencing
Oligonucleotides for multiple amiRNA silencing of KAT5 and ACC genes (Supplemental Table S2) were designed using the web-tool WMD3 (http://wmd3.weigelworld.org/cgi-bin/webapp.cgi; Ossowski et al., 2008). The amiRNA precursors were introduced by overlapping PCR into the plasmid pRS300 according to the protocol available in the WMD3 webpage (http://wmd3.weigelworld.org/downloads/Cloning_of_artificial_microRNAs.pdf; Schwab et al., 2006), with minor changes to include the sequences necessary for further Gateway cloning. The final expression vectors were obtained by transferring the multiple amiRNA precursor fragments into the destination vector pB7WG2 for constitutive expression under the 35S promoter (Karimi et al., 2007). All constructs were confirmed by DNA sequencing, transformed into Agrobacterium tumefaciens-competent cells GV3101 by electroporation (800 Ω, 1.8 kV, capacitance 25 μF), and subsequently used for Arabidopsis (Arabidopsis thaliana) transformation via the floral dip method (Clough and Bent, 1998). Transformed plants were selected in media with hygromycin, and T3 plants were used for all further analysis.
Plant Material and Growth Conditions
Arabidopsis Col-0 plants were used as the wild-type plant in this study. T-DNA mutants were purchased from the Nottingham Arabidopsis Stock Center (Alonso et al., 2003). At least two independent lines (T-DNA and/or amiRNA) were obtained for each of the candidate genes. Plants were cultured on soil in a growth chamber (CU-36L4, Percival Scientific) for 35 d at 21°C in 8 h light/16 h dark cycles with light intensity of 250 μE. After this period, 10 biological replicates of each line, constituted by the whole rosette, were harvested, immediately frozen in liquid nitrogen and powdered in a ball mill.
RNA Isolation and RT-qPCR
Total RNA was isolated from four biological replicates for each line using the extraction kit “Nucleospin RNA Plant” (Macherey-Nagel; https://www.mn-net.com/) and treated with Turbo DNase (Invitrogen; www.thermofisher.com) to remove contaminating DNA. RNA quantity was determined with a NanoDrop spectrophotometer (Thermo Fisher; www.thermofisher.com), and complementary DNA (cDNA) was synthesized from 300 ng RNA using a Maxima First Strand cDNA Synthesis Kit (Thermo Fisher; www.thermofisher.com). For RT-qPCR, cDNA equivalent to 5 ng total RNA was used as a template, and PCR was performed with PowerSYBER Green PCR Master Mix (Applied Biosystems; www.thermofisher.com) and 250 nm of each primer (Supplemental Table S3) in a total volume of 5 μL. Two technical replicates for each sample were analyzed in an ABI Prism 7900 HT sequence detection system (Applied Biosystems; www.thermofisher.com) with the following cycling program: 2 min at 50°C, 15 min at 95°C, 45 cycles of 15 s at 95°C, and 1 min at 60°C. Primer pair efficiencies were calculated by analyzing amplification curves from a standard cDNA dilution range. Expression levels were calculated using the 2–∆∆Ct method normalized to the mean transcript level of ACT2, UBQ10, and GAPDH3′.
Metabolite Profiling
Metabolites were extracted for each one of the 10 biological replicates of leaf tissues using a modified version of the protocol previously described by (Hummel et al., 2011). Fifty milligrams of the frozen material was extracted with 1 mL of precooled (−20°C) extraction buffer (methanol/methyl-tert-butyl-ether [1:3, v/v] mixture, spiked with 0.1 μg/mL of PE 34:0 [17:0, 17:0], and PC 34:0 [17:0, 17:0] as internal standards). After 10 min incubation in 4°C and sonication for 10 min in a sonic bath, 500 μL of water/methanol mixture spiked with 0.2 μg/mL of Ribitol and Isovitexin as internal standards was added. Samples were centrifuged (5 min, 14 000g) to separate the lipophilic and polar phases. Aliquots of 150 μL from the lipophilic phase and 150 μL and 300 μL from the aqueous phase were collected and dried under vacuum. The dried lipophilic phase was suspended in 200 μL of Acetonitrile/isopropanol (7:3 [v/v]) and used for lipid analysis according to Hummel et al. (2011). The first aliquot of the polar phase was derivatized and analyzed by gas chromatography time-of-flight mass spectrometry following the protocol described by Lisec et al. (2006). The second aliquot was directly suspended in 80% (v/v) methanol:water and analyzed according to the protocol described by Tohge and Fernie (2010). Secondary metabolism and lipids were analyzed exactly as described in Garbowicz et al. (2018)
Statistical Analysis
All statistical analysis and plots were performed in R (R Development Core Team, 2017) using log-transformed data and the packages ggplot2 (Wickham, 2016), ggpubr (https://CRAN.R-project.org/package=ggpubr), and pheatmap (https://CRAN.R-project.org/package=pheatmap). Data normality was assessed by the Shapiro-Wilk test and by visual inspection of quantile-quantile plots. Outliers were removed using the interquartile ranges for each metabolite and line. A Student's two-tailed t test was performed to compare the differences between metabolites for each line and the wild type.
Correlation Network Analysis
Metabolites correlation matrix was computed using the R package Hmisc (https://CRAN.R-project.org/package=Hmisc). Spearman rank correlation was used for calculating the correlations, and an absolute value of 0.5 was set as a threshold to establish an edge between two metabolites. The resulting network (Fig. 6) was represented using cytoscape v3.6.1 (Shannon et al., 2003).
Accession Numbers
The AGI locus numbers for the genes discussed in this article are as follows: At1g06550 (ECH), At1g65560 (HCD), At5g48880 (KAT5), At1g36160 (ACC), At1g56650 (PAP1/MYB75), At1g66390 (PAP2/MYB90), At3g62610 (MYB11), At2g47460 (MYB12), At5g49330 (MYB111), At5g13930 (CHS/TT4), At3g55120 (CHI/TT5), At3g51240 (F3H/TT6), At5g07990 (F3′H/TT7), At5g08640 (FLS), At5g17050 (UGT78D2/Fd3GT), At1g30530 (UGT78D1/F3RT), At1g06000 (UGT89C1/F7RT), At5g42800 (DFR/TT3), and At4g22880 (ANS/TT18).
Supplemental Data
The following supplemental materials are available.
Supplemental Table S1. Promoter analysis.
Supplemental Table S2. amiRNA oligo sequences.
Supplemental Table S3. RT-qPCR primer sequences.
Supplemental Dataset S1. Coexpression network edges.
Supplemental Dataset S2. Coexpression network annotations.
Supplemental Dataset S3. RT-qPCR raw data.
Supplemental Dataset S4. Liquid chromatography-mass spectrometry metabolite reporting list.
Supplemental Dataset S5. Gas chromatography-mass spectrometry metabolite reporting list.
Supplemental Dataset S6. Metabolite data.
Supplemental Dataset S7. Liquid chromatography-mass spectrometry lipid reporting list.
Supplemental Dataset S8. Lipid data.
Supplemental Dataset S9. Metabolite correlation network.
Footnotes
This work was supported by the Brazilian National Council for Scientific and Technological Development (CNPq; scholarship to L.P.S.) and the Max Planck Society (International Max Planck Research School, 'Primary Metabolism and Plant Growth' program).
Articles can be viewed without a subscription.
References
- Achnine L, Huhman DV, Farag MA, Sumner LW, Blount JW, Dixon RA (2005) Genomics-based selection and functional characterization of triterpene glycosyltransferases from the model legume Medicago truncatula. Plant J 41: 875–887 [DOI] [PubMed] [Google Scholar]
- Agati G, Tattini M (2010) Multiple functional roles of flavonoids in photoprotection. New Phytol 186: 786–793 [DOI] [PubMed] [Google Scholar]
- Alejandro S, Lee Y, Tohge T, Sudre D, Osorio S, Park J, Bovet L, Lee Y, Geldner N, Fernie AR, et al. (2012) AtABCG29 is a monolignol transporter involved in lignin biosynthesis. Curr Biol 22: 1207–1212 [DOI] [PubMed] [Google Scholar]
- Alonso JM, Stepanova AN, Leisse TJ, Kim CJ, Chen H, Shinn P, Stevenson DK, Zimmerman J, Barajas P, Cheuk R, et al. (2003) Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Alseekh S, Fernie AR (2018) Metabolomics 20 years on: What have we learned and what hurdles remain? Plant J 94: 933–942 [DOI] [PubMed] [Google Scholar]
- Alseekh S, Tohge T, Wendenberg R, Scossa F, Omranian N, Li J, Kleessen S, Giavalisco P, Pleban T, Mueller-Roeber B, et al. (2015) Identification and mode of inheritance of quantitative trait loci for secondary metabolite abundance in tomato. Plant Cell 27: 485–512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amid A, Lytovchenko A, Fernie AR, Warren G, Thorlby GJ (2012) The sensitive to freezing3 mutation of Arabidopsis thaliana is a cold-sensitive allele of homomeric acetyl-CoA carboxylase that results in cold-induced cuticle deficiencies. J Exp Bot 63: 5289–5299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud S, Guyon V, Kronenberger J, Wuillème S, Miquel M, Caboche M, Lepiniec L, Rochat C (2003) Multifunctional acetyl-CoA carboxylase 1 is essential for very long chain fatty acid elongation and embryo development in Arabidopsis. Plant J 33: 75–86 [DOI] [PubMed] [Google Scholar]
- Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E (2015) The Arabidopsis Information Resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 53: 474–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borevitz JO, Xia Y, Blount J, Dixon RA, Lamb C (2000) Activation tagging identifies a conserved MYB regulator of phenylpropanoid biosynthesis. Plant Cell 12: 2383–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos A, Szymanski J, Seiwert B, Degenkolbe T, Hannah MA, Giavalisco P, Willmitzer L (2011) Analysis of short-term changes in the Arabidopsis thaliana glycerolipidome in response to temperature and light. Plant J 66: 656–668 [DOI] [PubMed] [Google Scholar]
- Carrie C, Murcha MW, Millar AH, Smith SM, Whelan J (2007) Nine 3-ketoacyl-CoA thiolases (KATs) and acetoacetyl-CoA thiolases (ACATs) encoded by five genes in Arabidopsis thaliana are targeted either to peroxisomes or cytosol but not to mitochondria. Plant Mol Biol 63: 97–108 [DOI] [PubMed] [Google Scholar]
- Chapman KD, Dyer JM, Mullen RT (2013) Commentary: Why don’t plant leaves get fat? Plant Sci 207: 128–134 [DOI] [PubMed] [Google Scholar]
- Chen C, Li C, Wang Y, Renaud J, Tian G, Kambhampati S, Saatian B, Nguyen V, Hannoufa A, Marsolais F, et al. (2017) Cytosolic acetyl-CoA promotes histone acetylation predominantly at H3K27 in Arabidopsis. Nat Plants 3: 814–824 [DOI] [PubMed] [Google Scholar]
- Chow C-N, Lee TY, Hung YC, Li G-Z, Tseng KC, Liu YH, Kuo PL, Zheng HQ, Chang WC (2019) PlantPAN3.0: A new and updated resource for reconstructing transcriptional regulatory networks from ChIP-seq experiments in plants. Nucleic Acids Res 47(D1): D1155–D1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Daloso DM, Müller K, Obata T, Florian A, Tohge T, Bottcher A, Riondet C, Bariat L, Carrari F, Nunes-Nesi A, et al. (2015) Thioredoxin, a master regulator of the tricarboxylic acid cycle in plant mitochondria. Proc Natl Acad Sci USA 112: E1392–E1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumont S, Rivoal J (2019) Consequences of oxidative stress on plant glycolytic and respiratory metabolism. Front Plant Sci 10: 166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fatland BL, Nikolau BJ, Wurtele ES (2005) Reverse genetic characterization of cytosolic acetyl-CoA generation by ATP-citrate lyase in Arabidopsis. Plant Cell 17: 182–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernie AR, Aharoni A, Willmitzer L, Stitt M, Tohge T, Kopka J, Carroll AJ, Saito K, Fraser PD, DeLuca V (2011) Recommendations for reporting metabolite data. Plant Cell 23: 2477–2482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster-Hartnett D, Danesh D, Peñuela S, Sharopova N, Endre G, Vandenbosch KA, Young ND, Samac DA (2007) Molecular and cytological responses of Medicago truncatula to Erysiphe pisi. Mol Plant Pathol 8: 307–319 [DOI] [PubMed] [Google Scholar]
- Garbowicz K, Liu Z, Alseekh S, Tieman D, Taylor M, Kuhalskaya A, Ofner I, Zamir D, Klee HJ, Fernie AR, et al. (2018) Quantitative trait loci analysis identifies a prominent gene involved in the production of fatty acid-derived flavor volatiles in tomato. Mol Plant 11: 1147–1165 [DOI] [PubMed] [Google Scholar]
- Germain V, Rylott EL, Larson TR, Sherson SM, Bechtold N, Carde J-P, Bryce JH, Graham IA, Smith SM (2001) Requirement for 3-ketoacyl-CoA thiolase-2 in peroxisome development, fatty acid β-oxidation and breakdown of triacylglycerol in lipid bodies of Arabidopsis seedlings. Plant J 28: 1–12 [DOI] [PubMed] [Google Scholar]
- Hichri I, Barrieu F, Bogs J, Kappel C, Delrot S, Lauvergeat V (2011) Recent advances in the transcriptional regulation of the flavonoid biosynthetic pathway. J Exp Bot 62: 2465–2483 [DOI] [PubMed] [Google Scholar]
- Higashi Y, Okazaki Y, Myouga F, Shinozaki K, Saito K (2015) Landscape of the lipidome and transcriptome under heat stress in Arabidopsis thaliana. Sci Rep 5: 10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel J, Segu S, Li Y, Irgang S, Jueppner J, Giavalisco P (2011) Ultra performance liquid chromatography and high resolution mass spectrometry for the analysis of plant lipids. Front Plant Sci 2: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M, Depicker A, Hilson P (2007) Recombinational cloning with plant gateway vectors. Plant Physiol 145: 1144–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JI, Dolan WL, Anderson NA, Chapple C (2015) Indole glucosinolate biosynthesis limits phenylpropanoid accumulation in Arabidopsis thaliana. Plant Cell 27: 1529–1546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li-Beisson Y, Shorrosh B, Beisson F, Andersson MX, Arondel V, Bates PD, Baud S, Bird D, Debono A, Durrett TP, et al. (2013) Acyl-lipid metabolism. The Arabidopsis Book 11: e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Lü S, Zhao H, Parsons EP, Xu C, Kosma DK, Xu X, Chao D, Lohrey G, Bangarusamy DK, Wang G, et al. (2011) The glossyhead1 allele of ACC1 reveals a principal role for multidomain acetyl-coenzyme A carboxylase in the biosynthesis of cuticular waxes by Arabidopsis. Plant Physiol 157: 1079–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo J, Butelli E, Hill L, Parr A, Niggeweg R, Bailey P, Weisshaar B, Martin C (2008) AtMYB12 regulates caffeoyl quinic acid and flavonol synthesis in tomato: Expression in fruit results in very high levels of both types of polyphenol. Plant J 56: 316–326 [DOI] [PubMed] [Google Scholar]
- Martin C, Li J (2017) Medicine is not health care, food is health care: Plant metabolic engineering, diet and human health. New Phytol 216: 699–719 [DOI] [PubMed] [Google Scholar]
- Nakabayashi R, Yonekura-Sakakibara K, Urano K, Suzuki M, Yamada Y, Nishizawa T, Matsuda F, Kojima M, Sakakibara H, Shinozaki K, et al. (2014) Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J 77: 367–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narise T, Kobayashi K, Baba S, Shimojima M, Masuda S, Fukaki H, Ohta H (2010) Involvement of auxin signaling mediated by IAA14 and ARF7/19 in membrane lipid remodeling during phosphate starvation. Plant Mol Biol 72: 533–544 [DOI] [PubMed] [Google Scholar]
- Nesi N, Jond C, Debeaujon I, Caboche M, Lepiniec L (2001) The Arabidopsis TT2 gene encodes an R2R3 MYB domain protein that acts as a key determinant for proanthocyanidin accumulation in developing seed. Plant Cell 13: 2099–2114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obayashi T, Aoki Y, Tadaka S, Kagaya Y, Kinoshita K (2018) ATTED-II in 2018: A plant coexpression database based on investigation of the statistical property of the mutual rank index. Plant Cell Physiol 59: e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohlrogge J, Browse J (1995) Lipid biosynthesis. Plant Cell 7: 957–970 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossowski S, Schwab R, Weigel D (2008) Gene silencing in plants using artificial microRNAs and other small RNAs. Plant J 53: 674–690 [DOI] [PubMed] [Google Scholar]
- Peters DJ, Constabel CP (2002) Molecular analysis of herbivore-induced condensed tannin synthesis: Cloning and expression of dihydroflavonol reductase from trembling aspen (Populus tremuloides). Plant J 32: 701–712 [DOI] [PubMed] [Google Scholar]
- Pracharoenwattana I, Cornah JE, Smith SM (2005) Arabidopsis peroxisomal citrate synthase is required for fatty acid respiration and seed germination. Plant Cell 17: 2037–2048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Development Core Team (2017). R: A Languange and Environment for Statistical Computing. R Foundation Statistical Computing, Vienna, Austria: http//www.R-project.org/. [Google Scholar]
- Roesler KR, Shorrosh BS, Ohlrogge JB (1994) Structure and expression of an Arabidopsis acetyl-coenzyme A carboxylase gene. Plant Physiol 105: 611–617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito K, Yonekura-Sakakibara K, Nakabayashi R, Higashi Y, Yamazaki M, Tohge T, Fernie AR (2013) The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant Physiol Biochem 72: 21–34 [DOI] [PubMed] [Google Scholar]
- Schulz E, Tohge T, Zuther E, Fernie AR, Hincha DK (2015) Natural variation in flavonol and anthocyanin metabolism during cold acclimation in Arabidopsis thaliana accessions. Plant Cell Environ 38: 1658–1672 [DOI] [PubMed] [Google Scholar]
- Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18: 1121–1133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 13: 2498–2504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva-Navas J, Moreno-Risueno MA, Manzano C, Téllez-Robledo B, Navarro-Neila S, Carrasco V, Pollmann S, Gallego FJ, Del Pozo JC (2016) Flavonols mediate root phototropism and growth through regulation of proliferation-to-differentiation transition. Plant Cell 28: 1372–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stracke R, Ishihara H, Huep G, Barsch A, Mehrtens F, Niehaus K, Weisshaar B (2007) Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J 50: 660–677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Fernie AR (2010) Combining genetic diversity, informatics and metabolomics to facilitate annotation of plant gene function. Nat Protoc 5: 1210–1227 [DOI] [PubMed] [Google Scholar]
- Tohge T, Fernie AR (2016) Specialized metabolites of the flavonol class mediate root phototropism and growth. Mol Plant 9: 1554–1555 [DOI] [PubMed] [Google Scholar]
- Tohge T, Nishiyama Y, Hirai MY, Yano M, Nakajima J, Awazuhara M, Inoue E, Takahashi H, Goodenowe DB, Kitayama M, Noji M, Yamazaki M, et al. (2005) Functional genomics by integrated analysis of metabolome and transcriptome of Arabidopsis plants over-expressing an MYB transcription factor. Plant J 42: 218–235 [DOI] [PubMed] [Google Scholar]
- Tohge T, Perez de Souza L, Fernie AR (2017) Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J Exp Bot 68: 4013–4028 [DOI] [PubMed] [Google Scholar]
- Tohge T, Perez de Souza L, Fernie AR (2018) On the natural diversity of phenylacylated-flavonoid and their in planta function under conditions of stress. Phytochem Rev 17: 279–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohge T, Watanabe M, Hoefgen R, Fernie AR (2013) The evolution of phenylpropanoid metabolism in the green lineage. Crit Rev Biochem Mol Biol 48: 123–152 [DOI] [PubMed] [Google Scholar]
- Tohge T, Zhang Y, Peterek S, Matros A, Rallapalli G, Tandrón YA, Butelli E, Kallam K, Hertkorn N, Mock H-P, et al. (2015) Ectopic expression of snapdragon transcription factors facilitates the identification of genes encoding enzymes of anthocyanin decoration in tomato. Plant J 83: 686–704 [DOI] [PubMed] [Google Scholar]
- Usadel B, Obayashi T, Mutwil M, Giorgi FM, Bassel GW, Tanimoto M, Chow A, Steinhauser D, Persson S, Provart NJ (2009) Co-expression tools for plant biology: Opportunities for hypothesis generation and caveats. Plant Cell Environ 32: 1633–1651 [DOI] [PubMed] [Google Scholar]
- Wickham H. (2016) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag, New York [Google Scholar]
- Winkel-Shirley B. (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126: 485–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkel-Shirley B. (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5: 218–223 [DOI] [PubMed] [Google Scholar]
- Wisecaver JH, Borowsky AT, Tzin V, Jander G, Kliebenstein DJ, Rokas A (2017) A global coexpression network approach for connecting genes to specialized metabolic pathways in plants. Plant Cell 29: 944–959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiszniewski AAG, Smith SM, Bussell JD (2012) Conservation of two lineages of peroxisomal (Type I) 3-ketoacyl-CoA thiolases in land plants, specialization of the genes in Brassicaceae, and characterization of their expression in Arabidopsis thaliana. J Exp Bot 63: 6093–6103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Dubos C, Lepiniec L (2015) Transcriptional control of flavonoid biosynthesis by MYB-bHLH-WDR complexes. Trends Plant Sci 20: 176–185 [DOI] [PubMed] [Google Scholar]
- Yonekura-Sakakibara K, Tohge T, Matsuda F, Nakabayashi R, Takayama H, Niida R, Watanabe-Takahashi A, Inoue E, Saito K (2008) Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene-metabolite correlations in Arabidopsis. Plant Cell 20: 2160–2176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Butelli E, Alseekh S, Tohge T, Rallapalli G, Luo J, Kawar PG, Hill L, Santino A, Fernie AR, et al. (2015) Multi-level engineering facilitates the production of phenylpropanoid compounds in tomato. Nat Commun 6: 8635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Q, Zhang Y, Wang G, Hill L, Weng J-K, Chen X-Y, Xue H, Martin C (2016) A specialized flavone biosynthetic pathway has evolved in the medicinal plant, Scutellaria baicalensis. Sci Adv 2: e1501780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann IM, Heim MA, Weisshaar B, Uhrig JF (2004) Comprehensive identification of Arabidopsis thaliana MYB transcription factors interacting with R/B-like BHLH proteins. Plant J 40: 22–34 [DOI] [PubMed] [Google Scholar]