ERBB-3 BINDING PROTEIN 1 acts downstream of TARGET OF RAPAMYCIN regulating multiple aspects of protein synthesis in the meristem and counteracts RETINOBLASTOMA RELATED to sustain cell growth.
Abstract
The ErbB-3 BINDING PROTEIN 1 (EBP1) drives growth, but the mechanism of how it acts in plants is little understood. Here, we show that EBP1 expression and protein abundance in Arabidopsis (Arabidopsis thaliana) are predominantly confined to meristematic cells and are induced by sucrose and partially dependent on TARGET OF RAPAMYCIN (TOR) kinase activity. Consistent with being downstream of TOR, silencing of EBP1 restrains, while overexpression promotes, root growth, mostly under sucrose-limiting conditions. Inducible overexpression of RETINOBLASTOMA RELATED (RBR), a sugar-dependent transcriptional repressor of cell proliferation, depletes meristematic activity and causes precocious differentiation, which is attenuated by EBP1. To understand the molecular mechanism, we searched for EBP1- and RBR-interacting proteins by affinity purification and mass spectrometry. In line with the double-stranded RNA-binding activity of EBP1 in human (Homo sapiens) cells, the overwhelming majority of EBP1 interactors are part of ribonucleoprotein complexes regulating many aspects of protein synthesis, including ribosome biogenesis and mRNA translation. We confirmed that EBP1 associates with ribosomes and that EBP1 silencing hinders ribosomal RNA processing. We revealed that RBR also interacts with a set of EBP1-associated nucleolar proteins as well as factors that function in protein translation. This suggests EBP1 and RBR act antagonistically on common processes that determine the capacity for translation to tune meristematic activity in relation to available resources.
Cell growth is generally required for cell proliferation, and although these two processes are separately regulated, they are closely connected and coordinated (Fox et al., 2018). The central conserved regulator of the cell cycle is the cyclin-dependent kinase (CDK), the activity of which (1) sets the unidirectional alternation of cell cycle phases, (2) enables cells to tune the cell cycle to match with developmental and environmental cues, or (3) gives a signal to exit from the cycle to allow cellular differentiation (De Veylder et al., 2007). A pivotal target of CDK that balances cell proliferation with differentiation is the RETINOBLASTOMA RELATED (RBR) protein, whose phosphorylation lifts the repression on the cell-cycle regulatory E2F transcription factors (Magyar et al., 2016). In plants, modulation in the levels of these cell cycle regulators, such as the overexpression of Cyclin D3;1 (Dewitte et al., 2003), silencing of the RBR protein (Gutzat et al., 2011), or overexpression of the E2FB transcription factor (Magyar et al., 2005), results in more but smaller cells and stunted and developmentally arrested plants. This led to the realization that acceleration of cell proliferation does not necessarily lead to increased plant growth.
The TARGET OF RAPAMYCIN (TOR) kinase pathway is regarded as the central regulator of cell growth in connection with light and sugars by generally boosting anabolic and repressing catabolic processes (Dobrenel et al., 2016a; González and Hall, 2017). Protein synthesis, being an energy and resource demanding cellular process, is one of the major cellular functions controlled by TOR signaling (González and Hall, 2017). S6 kinase (S6K) is a key downstream component of TOR signaling that regulates translation and ribosome biogenesis (Deprost et al., 2007; Schepetilnikov et al., 2013). Thus growth-stimulatory factors such as light, sugar, and auxin activate TOR and affect the translation apparatus in different ways (Liu et al., 2013; Dobrenel et al., 2016b; Chen et al., 2018; Enganti et al., 2018; Schepetilnikov and Ryabova, 2018). Correspondingly, elevated TOR expression boosts plant growth in Arabidopsis (Arabidopsis thaliana; Deprost et al., 2007) and increases yield in crop plants, specifically in limiting conditions (Bakshi et al., 2019). On the other hand, TOR silencing or chemical inhibition of TOR activity results in reduced growth (Deprost et al., 2007; Montané and Menand, 2013, 2019).
The growth regulatory TOR signaling drives cell proliferation by controlling the selective translation of key cell-cycle regulatory proteins, such as CLN3 G1 cyclin in yeast (Saccharomyces cerevisiae; Barbet et al., 1996) or Cyclin E in human (Homo sapiens) cells (Dowling et al., 2010). In plants, however, there appears to be an even more direct connection between growth and cell cycle control, as TOR phosphorylates and thus stimulates E2FA and E2FB activities (Xiong et al., 2013; Wu et al., 2019). More recent research shows that the YET ANOTHER KINASE1 (AtYAK1) gene, encoding a member of the dual-specificity Tyr phosphorylation-regulated kinase family, inhibits growth and cell proliferation by up-regulating the SIAMESE-RELATED (SMR) proteins when TOR activity is reduced (Barrada et al., 2019; Forzani et al., 2019). Moreover, the principal downstream effector of TOR, AtS6K1, interacts with RBR and facilitates its nuclear localization, which is important to impose repression on cell proliferation in sucrose limiting conditions (Henriques et al., 2013). Finally, S6K1, E2FB, and RBR are interconnected through negative feedback loops that might be important to toggle between cell proliferation and quiescence (Henriques et al., 2010).
The ErbB-3 BINDING PROTEIN 1 (EBP1) came to the forefront of interest in plants when it was found that it is rapidly induced during early stages of tuber formation, stimulated by sugar in potato (Solanum tuberosum; Horváth et al., 2006). Overexpression or silencing of EBP1 in potato could dose-dependently tune leaf and tuber growth and thus potato yield without any obvious developmental abnormalities (Horváth et al., 2006). Studying the cellular basis how EBP1 affects organ growth showed that it boosts cell proliferation in meristematic cells, resulting in more but smaller cells, which correlated with elevated expression of cell cycle regulators. In postmitotic cells, EBP1 enhances cell growth leading to a larger final cell size. On the molecular level EBP1 represses RBR protein abundance, whereas RBR negatively influences the EBP1 level. This antagonism suggested that EBP1 provides an important link between a growth driver and cell cycle regulation (Horváth et al., 2006). The initial finding that EBP1 enhances growth was verified for a number of EBP1 orthologs identified in different species (Cao et al., 2009; Cheng et al., 2016; Wang et al., 2016); however, another report described an opposite effect (Li et al., 2016). Additionally, EBP1 expression is induced upon abiotic stress, and when EPB1 levels are elevated, it can confer stress tolerance (Cao et al., 2009; Cheng et al., 2016). In line with being a positive regulator of growth, in a maize hybrid displaying hybrid vigor, EBP1 is expressed in an over dominant fashion (Wang et al., 2016). In a recent study, EBP1 was identified as an interactor with the FERONIA receptor-like kinase with a major function in regulating cell expansion. FERONIA directly phosphorylates EBP1, leading to its nuclear localization to regulate genes responsive to FERONIA signaling (Li et al., 2018).
A clue to understand EBP1’s molecular function in plants was its remarkable coexpression with genes involved in the regulation of protein synthesis in proliferating cells, the so-called ribi regulon (Horváth et al., 2006). In line with this, several additional supporting lines of evidence have been published. First, EBP1 expression is rapidly induced when growth is stimulated in the shoot meristem of dark-grown etiolated seedlings upon light exposure, correlating with the induction of genes involved in protein synthesis (López-Juez et al., 2008; Mohammed et al., 2018). Second, EBP1 is present in nucleoli in both animals (Squatrito et al., 2004; Liu et al., 2006) and in plants (Pendle et al., 2005), as evidenced by a carboxy-terminal Lys-rich motif (Karlsson et al., 2016) and an N-terminal nucleolar localization signal (Squatrito et al., 2004; Ko et al., 2016). Third, human EBP1 (HsEBP1) has a double-stranded RNA binding activity (Squatrito et al., 2004). It binds to rRNAs and ribosomes (Squatrito et al., 2006) and also supports ribosome biogenesis by interacting with the Pol I transcription factor TIF-IA (Nguyen le et al., 2015; Nguyen et al., 2019) and possibly with nucleolar nucleophosmin (Okada et al., 2007). Fourth, HsEbp1 also stabilizes several mRNAs by binding their 3′UTRs (Bose et al., 2006; Zhou et al., 2010; Pisapia et al., 2015). Finally, EBP1 has been implicated in translational control in Leishmania major (Norris-Mullins et al., 2014) and, repeatedly, in gene specific translation in cap-independent translation initiation of RNA viruses (Pilipenko et al., 2000) on the androgen receptor mRNA (Zhou et al., 2010; Zhou et al., 2011) by affecting eIF2α phosphorylation (Squatrito et al., 2006).
To gain insight into the growth-promoting function of EBP1, it is paramount to identify the molecular partners. To this end, we performed mass spectrometry to identify EBP1 interactors, which uncovered proteins with a broad range of RNA binding and, to a lesser extent, of DNA binding activities, and also proteins involved in ribosome biogenesis, splicing, translation, and transcription. Here, we confirm that EBP1 can associate with ribosomes and is distributed between cytoplasm, nucleus, and nucleolus, in line with its suggested role in translation and ribosome biogenesis. We demonstrate that the inhibition of EBP1 expression causes a defect in rRNA processing, which can be rescued by sucrose. Sugar could also rescue the growth defects during root development in the EBP1 silencing lines, suggesting EBP1 is specifically required to support protein translation in growth limiting conditions. Strikingly, overexpression of EBP1 can suppress both the loss of meristem activity and precocious differentiation that occurs upon induction of elevated RBR expression. In addition, we also found that RBR shares interacting partners with EBP1, and besides its canonical transcriptional repressor function, it potentially acts on common biological processes with EBP1 in ribosome biogenesis and protein translation. Thus, EBP1 and RBR antagonistically regulate translational capacity to determine the balance between meristematic activity and cellular differentiation.
RESULTS
EBP1 Is Expressed in Root Meristematic Tissues
When ectopically overexpressed or silenced, EBP1 enhances or reduces the meristematic activity and correspondingly organ growth. However, when and where EBP1 is naturally expressed is not well documented (Horváth et al., 2006). To study the regulation of EBP1 expression, we followed the EBP1 promoter activity driving GUS and cyan-fluorescent protein (CFP) expression. For this, we cloned the entire intergenic region (1193bp) between EBP1 (At3g51800) and the nearest upstream gene (At3g51810). Around 25 Arabidopsis transgenic lines were generated for pEBP1:GUS and for pEBP1:CFP. For both, five lines were examined in detail, all showing the same expression profile in root tissues. We detected strong GUS activity in the primary meristem (Fig. 1A), in the region of the rapidly dividing transit amplifying cells and in the emerging lateral root meristematic cells (Fig. 1B). In the elongation and differentiation zones, we observed expression only at a low level, mainly in the vasculature. Similarly, the expression pattern of pEBP1:CFP was largely confined to the meristematic cells (Fig. 1C). In this aspect, the expression pattern overlaps with the expression pattern published in Arabidopsis.org or Plant eFP at https://bar.utoronto.ca/eplant. To see whether the entire intergenic region is required to regulate the transcription, we studied stepwise-shortened regions of the full promoter. As Supplemental Figure S1 illustrates, the region (-337-0) contains the essential regulatory sequences to induce transcription.
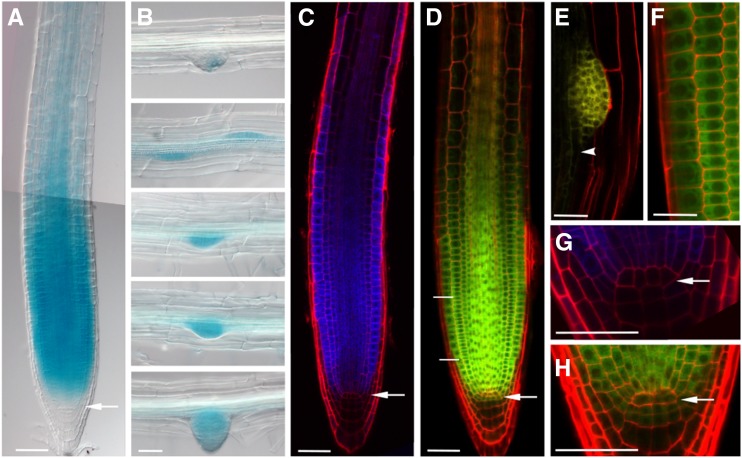
Figure 1.
Arabidopsis EBP1 is expressed and localized in actively dividing tissues during root development. Representative differential interference contrast images of EBP1 expression in the primary root (A) and during lateral root development (B) in the transgenic pEBP1:GUS lines after 3h GUS staining. The images of the different stages of lateral root development belong to the same root. C, Confocal microscopy images in the propidium-iodide–stained root sample of the pEBP1:CFP lines. D to F, Confocal microscopy images represent the localization of the EBP1-YFP protein in the pgEBP1:YFP transgenic line in the primary (D) and secondary (E) meristems. The enlarged sections of the meristematic region in (F) and of a lateral root initial in (E) are showing cytoplasmic localization of the fusion protein (scale bar = 25 μm), whereas (G) and (H) illustrate the lower level of expression in the slowly dividing stem cells and QC. In each case, 5- to 6-d-old seedling were analyzed. White arrow points toward the QC, and the white arrowhead in (E) labels the occasional nuclear localization of the EBP1-YFP protein in the differentiation zone. The merged image in (A) is a composite of two consecutive overlapping images of the same root, taken with the same median focal plane. The enlarged image in (F) comes from the meristematic region, labeled by two white lines in (D). Scale bars = 50 μm, unless stated otherwise.
To investigate whether EBP1 promoter activity and protein accumulation correspond, we fused the genomic coding region of EBP1 with the coding region of the yellow fluorescent protein (YFP) and expressed this 3′-terminal fusion under its native promoter (pgEBP1-YFP). For this, we used the entire intergenic region (1193bp) containing the 5′-upstream region, which also comprises the 5′UTR. Of over 30 regenerated transgenic lines, 5 lines were analyzed in detail and showed the same pattern of EBP1-YFP protein accumulation, which overlapped with the EBP1 promoter activity. EBP1-YFP protein was most abundant in root meristematic cells (Fig. 1D), specifically in the region of the transit amplifying cells and emerging lateral root primordia (Fig. 1E). Lower levels of EBP1-YFP signal were detected in the slowly dividing stem cells and even less in the quiescent center (QC; Fig. 1H). This pattern at the QC region was also closely mirrored in the pEBP1:CFP line (Fig. 1G). The abundance of EBP1-YFP protein also corresponds well with translatomics data, which demonstrate EBP1 mRNA is actively translated in the shoot apex (Tian et al., 2019).
In the transgenic lines, EBP1-YFP signal was most prominent in the cytoplasm of meristematic cells surrounding the centrally located nuclei (Fig. 1F). Similar cellular localization patterns were shown independently by Palm et al. (2019). The amino acid sequence of EBP1 shows high conservation to its human counterpart, which contains a motif for nuclear and nucleolar localization near to its C terminus (Squatrito et al., 2004; Karlsson et al., 2016). This motif may be more obscured when the fluorescent protein is fused to the C terminus of EBP1. Therefore, we labeled EBP1 both N-and C-terminally using the Cerulean fluorescent protein and transiently expressed it under the control of the 35S promoter. In the transiently transformed Nicotiana benthamiana leaves (Supplemental Fig. S2, A and B), both fusion proteins were found in the nucleoli, the nucleus, and cytosol. Similar localization was observed in onion (Allium cepa) epidermal cells where about 10% of Cerulean-EBP1 localized in the nucleoli with the remainder distributed between the nucleus and cytosol (Supplemental Fig. S2, C and D). Taken together, these data show that both the expression and protein accumulation of EBP1 are concentrated in the region of rapidly dividing root meristematic cells. Most of the protein is confined to the cytoplasm in meristematic cells, but it has the ability to enter into the nucleus and nucleolus in different tissue environments.
EBP1 Supports Root Meristem Activity in Sucrose Limiting Conditions
Cell cycle activity in the meristem is reliant on the availability of sugar and regulated by the TOR pathway (Ahmad et al., 2019). Therefore, we examined how the external addition of sucrose and chemical inhibition of TOR activity by AZD-8055 influence EBP1 transcription, EBP1 promoter activity, and EBP1-YFP protein levels. Endogenous EBP1 mRNA levels were determined in Col-0 seedlings grown on medium without sucrose and then transferred to fresh media with or without sucrose (2% and 0%, respectively) and with or without AZD-8055. EBP1 transcript levels increased upon sucrose addition but were suppressed by AZD-8055, irrespective of whether sucrose was present in the media (Fig. 2A). To be able to compare the effect of TOR kinase inhibitor in the presence of sucrose on EBP1 promoter activity and protein accumulation, we used our pEBP1:CFP and pgEBP1-YFP transgenic lines and followed the amounts of CFP and EBP1-YFP on protein blots. Both EBP1 promoter activity and EBP1-YFP protein levels were induced by sucrose and were affected by the TOR inhibitor (Fig. 2, B and C).
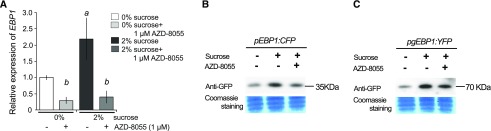
Figure 2.
Both the gene expression and the protein abundance of EBP1 depend on sugar availability and TOR kinase activity. A, Endogenous EBP1 transcript level was analyzed in wild-type seedlings (Col0) grown in sucrose free media (0%) for 6 d in continuous light. Subsequently, seedlings were moved to media with or without sucrose and/or the specific TOR inhibitor, AZD-8055 (0% sucrose, 0% sucrose + 1 µm AZD-8055, 2% sucrose, 2% sucrose + AZD-8055). Seedlings were harvested after 3-h treatment. EBP1 expression was set arbitrarily to 1 in the nontreated sample (0% Suc). Significance was determined using the one-way ANOVA and Tukey’s test; significant difference a: P-value < 0.05 between 2% versus 0% sucrose treatment, while b: P-value < 0.05 between AZD8055 treated versus nontreated samples. Values represent mean of relative expression of three biological repeats (n = 3) with sample size: n > 100, error bars: sd (StDv). The transgenic lines pEBP1:CFP (B) and pgEBP1:YFP (C) were germinated and grown on 0% sucrose, then transferred at 6 d after germination to 2% sucrose containing media or 2% sucrose supplemented with 1 µm AZD-8055 for 3-h treatment. Protein gel blot analysis was performed using a GFP-specific antibody to detect in EBP1 promoter activity (B) and in EBP1-YFP protein level (C).
To study how EBP1 level influences root growth, we generated inducible EBP1 silencing lines and screened them for growth retardation using 17-β-estradiol on a medium without sucrose. Two independent transformants with the EBP1 RNAi construct (iEBP1 line 1 and line 2) showed strongly reduced growth upon 17-β-estradiol treatment as silencing was induced and the EBP1 mRNA level diminished (Supplemental Fig. S3, A and B). However, from the T3 generation onward the retardation in root growth on sucrose-free media was no longer dependent on 17-β-estradiol, and the EBP1 level was already reduced without chemical induction, indicating that silencing became constitutive (Supplemental Fig. S4A). Therefore, in subsequent experiments we did not apply the inducer. To ascertain that EBP1 silencing is not influenced by sucrose, we determined the EBP1 level both in the presence and absence of sucrose. Although the level of EBP1 was induced by sucrose as shown above, the silencing remained effective in both iEBP1 lines (Supplemental Fig. S4A).
Next, we studied how seedling growth was influenced by the addition of external sucrose when EBP1 was silenced. Although a large portion of the iEBP1 seedlings (both line 1 and line 2) showed growth arrest and retarded growth on sucrose-free media, this inhibition was largely suppressed when seeds were germinated on 1% sucrose (Fig. 3, A and B). In an insertional mutant, ebp1-3 (CS854731), we observed an even more pronounced growth retardation in the absence of sucrose and recovery on sucrose-free media (Fig. 3, A and B). Leaf development showed similar effects in the absence and presence of sucrose (Supplemental Fig. S4, B and C). The level of growth retardation closely matched the decline in EBP1 expression in the silencing lines (Supplemental Fig. S4A). To test whether the arrested growth upon EBP1 silencing can be rescued, we transferred seedlings from sucrose-free onto sucrose-containing media and found that root growth indeed recovered (Supplemental Fig. S4, D and E).
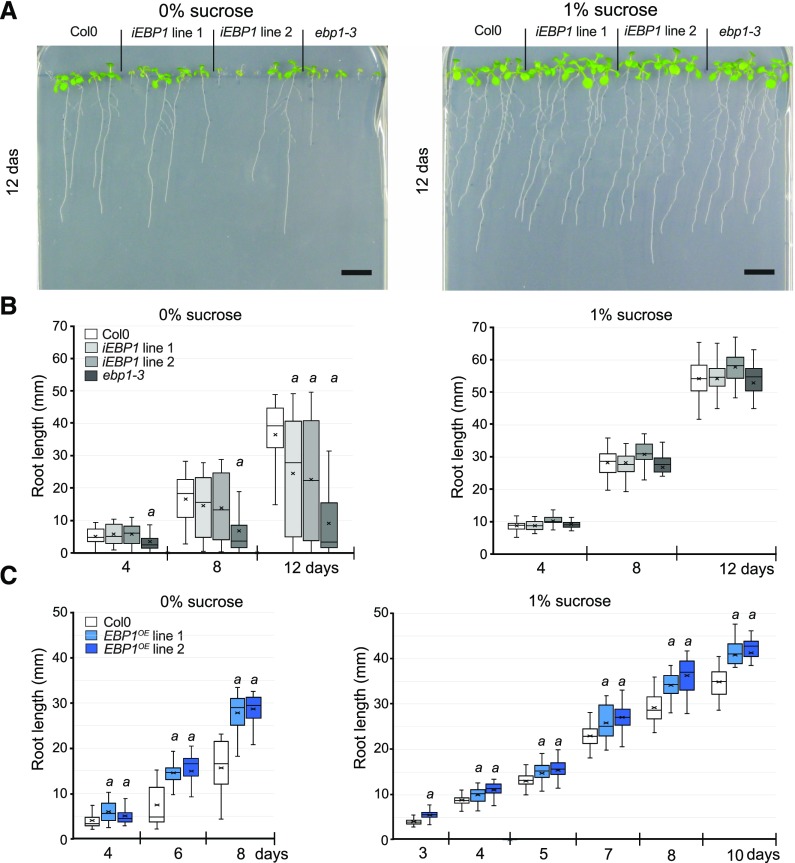
Figure 3.
Sugar can compensate for the lack of EBP1 expression and an elevated level of EBP1 can suppress sugar-dependent root growth deficiencies. A, Root growth of two independent EBP1 RNAi (iEBP1) lines and the insertional mutant, ebp1-3 compared with Col0, germinated and grown (12 d) in the absence (0%) and presence of sucrose (1%). Scale bars = 1 cm. B, Boxplot analyses show the quantification and distribution of the root length (in millimeters) of seedlings grown on 0% and 1% sucrose media at the given time points after sowing. The boxplot gives the mean line and the mean marker (cross); the quartile calculation was done exclusive of the median on root-length measurements from n = 3, n > 35 in each repeat. Significance was determined by Student t test a: P-value < 0.01. C, Quantitative analysis of root growth of the transgenic EBP1OE lines (referred as lines 19.2 and 43.2, respectively in Horváth et al., 2006) compared with Col0, germinated and grown on media with or without sucrose (0%, 1%). Boxplot analysis was carried out as in (B); n = 3, n > 35 in each repeat; significance was determined by Student t test a: P-value < 0.01. The level of EBP1 expression upon silencing and overexpression is shown in Supplemental Figure S4A and Figure 4C, respectively.
Ectopic EBP1 overexpression can dose-dependently promote growth in potato and Arabidopsis (Horváth et al., 2006). We asked whether this promotion depends on sucrose availability. On sucrose-free medium, we found that roots of the two independent EBP1 overexpression lines 1 and 2 (Horváth et al., 2006) grew significantly better than Col-0, but the difference in root length between the transgenic lines and the control was less pronounced on sucrose-containing media (Fig. 3C). Taken together, the facts that sucrose rescued the growth retardation caused by EBP1 silencing and, vice versa, that EBP1 overexpression overcame the sucrose limitation, suggest that EBP1 has a role in coordinating root growth with sugar availability.
EBP1 Supports Maintenance of the Root Meristem and Counteracts Cell Differentiation Induced by RBR
Sugar availability controls cell proliferation through the RBR pathway (Magyar et al., 2016). Silencing of RBR results in excessive cell proliferation, while elevating the RBR level leads to premature differentiation, exhausting the stem-cell pool, and arresting division of the amplifying cells. As a result, the root meristem is gradually reduced as cells become differentiated (Wildwater et al., 2005). EBP1 and RBR levels are counter-inhibited in cultured cells (Horváth et al., 2006). To follow how the EBP1 level responds to RBR-induced differentiation in the root, we introgressed pEBP1:CFP and pgEBP1-YFP into the dexamethasone-inducible RBR overexpression line (RBROE). Upon induction of RBR expression, both the EBP1 promoter activity and EBP1-YFP protein level progressively diminished, together with the meristematic zone, as differentiation progressed (Fig. 4, A and B).
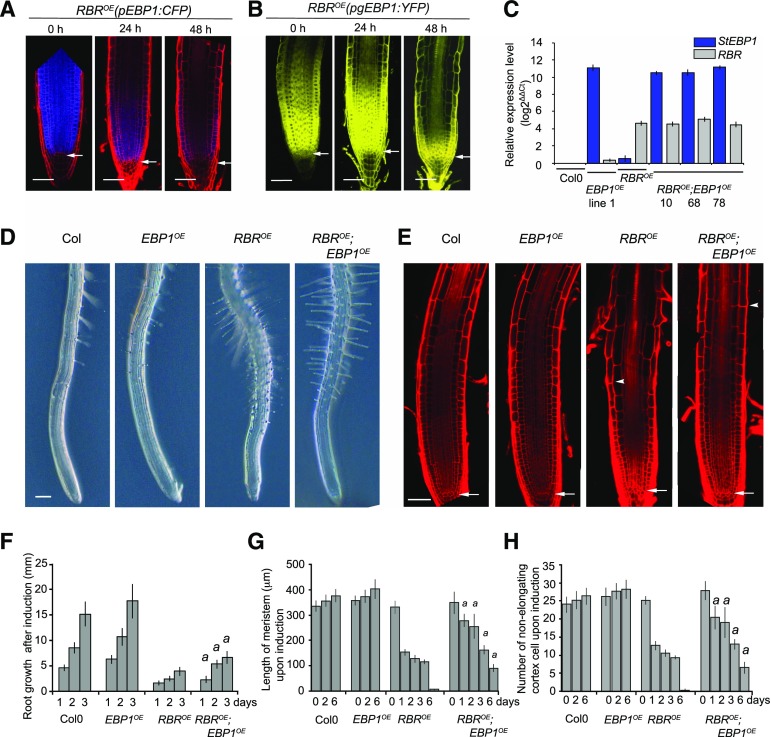
Figure 4.
Elevated level of EBP1 delays differentiation upon induction of RBR expression. A, Confocal microscopy images of the transgenic lines, RBROE(pEBP1:CFP) and RBROE(pgEBP1:YFP). B, Upon induction of RBR with dexamethasone (dex) treatment (1 μm), at the given time points. Note, the loss of meristem structure and progressive differentiation due to RBR induction and the concomitant drop in EBP1 transcription and reduction of EBP1-YFP abundance. C, To generate the RBROE;EBP1OE lines, the transgenic lines RBROE (Wildwater et al., 2005) and EBP1OE (line 1; Horváth et al., 2006) constitutively overexpressing the Solanum tuberosum EBP1 (StEBP1) were introgressed. The expression levels for RBR and EBP1 were analyzed by quantitative reverse transcription-PCR in three independent introgressed lines (RBROE;EBP1OE line 10, 68, 78; F3 generation) using RBR- and StEBP1-specific primers. As a control, the parental lines (RBROE and EBP1OE) and Col0 were tested. For the introgressed line 10, the transgenic line EBP1OE line 1 was used as a parent, while for the lines 68 and 78, EBP1OE line 2. D, Representative root samples of Col0, EBP1OE, RBROE and RBROE;EBP1OE lines upon 24 h dex (1 μm) induction. See also Supplemental Figure S5A. E, Confocal images of propidium iodide–stained root samples from Col0, EBP1OE, RBROE and RBROE;EBP1OE lines upon 72 h dex (1 μm) treatment. In (D) and (E), the arrowhead shows the position of the first differentiating epidermal cell, while in (E) the arrow points to the QC. Scale bars =1 mm (D) and 50 μm (E). F, Root growth (millimeters) after 24, 48, and 72 h dex (1 μm) treatment. Induction with dex started on 6-d-old seedlings. G, Meristem length and number of non-elongating cortical cells (H) of Col0, EBP1OE, RBROE and RBROE;EBP1OE lines after 0, 24, 48, 72, and 144 h dex (1 μm) induction. The region of the transit amplifying cells in the RBROE root meristem is fully differentiated around 96 h dex treatment; n > 3, n > 15 seedlings at each repeat for each genotype. In (F), (G), and (H), values represent means with StDv. a: P-value < 0.01 shows the significance of the measured values of RBROE;EBP1OE line compared with RBROE at the given time point.
To investigate whether EBP1 has a role in meristem maintenance when RBR is overexpressed, we crossed EBP1OE line 1 and 2 with RBROE. Homozygous offspring of three independent introgressed lines were analyzed and showed elevated levels of both RBR and EBP1 transcripts (Fig. 4C). As expected, induction of RBR overexpression reduced root growth and shortened the meristem, as measured by the distance from the tip to the first differentiating root hair. Strikingly, both effects of RBR overexpression were significantly attenuated by EBP1 overexpression (Fig. 4, D to H; Supplemental Fig. S5A). Five days after RBR induction the entire root meristem organization was lost and cells appeared vacuolated, but the elevated level of EBP1 prevented this process and supported meristematic function and root growth for a longer period (Supplemental Fig. S5, B and C). Taken together, the EBP1 level is important to maintain meristematic activity and to counteract the progression of differentiation imposed by RBR overexpression.
EBP1 Predominantly Interacts with Proteins Involved in Ribosome Biogenesis and Protein Translation
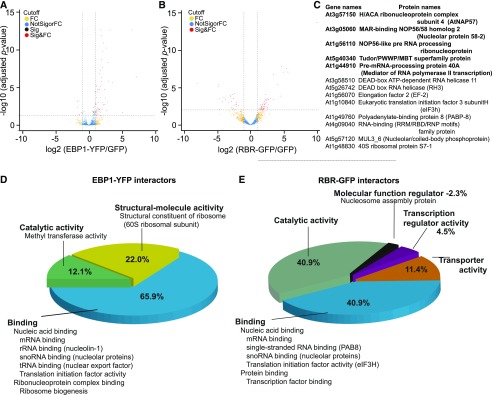
To gain insights into the molecular function of EBP1, we searched for EBP1 interacting proteins in a pull-down experiment and identified them by mass spectrometry. To do this we took advantage of the transgenic line pgEBP1-YFP and used p35S:GFP as a control. We identified and quantified EBP1-YFP associated proteins against proteins associated with the control GFP alone in six replicates each using label-free mass spectrometry (Hubner et al., 2010). To certify interactors with statistical confidence, we computed the false discovery rate and the amount ratio between proteins identified in EBP1-YFP vs GFP pull-downs and established thresholds as visualized in the volcano plot (Fig. 5A; Supplemental Methods). We identified 55 specific interactors with EBP1-YFP at high confidence (Supplemental Table S1). Gene ontology overrepresentation analysis based on molecular function placed 52 out of the 55 EBP1 interactors into different aspects of RNA binding (Fig. 5D; Supplemental Table S1). Classifying them according to biological processes, ribosome functions and regulation of protein translation, ribosome biogenesis, small nuclear RNA synthesis, and modifications and RNA splicing were the only prominent processes (Supplemental Table S1). Taken together, mass spectrometry identification of interactors suggested that EBP1 has the potential to regulate multiple aspects of ribosome biogenesis and protein translation.
Figure 5.
Functional classification of the EBP1 and RBR interacting proteins identified by mass-spectrometric analysis. Volcano plots showing the enrichment of EBP1-interacting (A) and RBR-interacting proteins (B) recovered from seedlings expressing the EBP1-YFP (A) and RBR-GFP (B) fusion proteins compared with the proteins identified from the transgenic lines Col0(p35S:GFP). The x axis shows the fold enrichment (FC) between the fusion protein and GFP alone (log2); whereas the y axis illustrates significance (Sig), the adjusted P-value (−log10) of the pulled-down proteins with anti-GFP antibody from EBP1-YFP (A) and RBR-GFP (B) seedlings compared with GFP-expressing Col0 transgenic plants. The vertical and horizontal stippled lines demarcate a 2-fold enrichment and an adjusted p-value of 0.05, respectively; proteins in the top right portion of the graph are considered to be the candidates for EBP1- (A) and RBR (B)-interactors. C, The table summarizes the list of RBR interactors involved in RNA binding (13) and highlights those that are common in both sets, the RBR and EBP1 interactors (5). D and E, The pie charts represent the major categories by their molecular functions using the gene ontology classification tool PANTHER version 14.1. Notable subcategories are also listed. For details on protein names and functions refer to Supplemental Tables S1 and S2.
Intrigued by the molecular and functional antagonism between EBP1 and RBR, we also searched for RBR-interacting proteins (Fig. 5, B and E; Supplemental Table S2) using the pgRBR-GFP line (Magyar et al., 2012). The list of RBR-interactors (55) included known partners, such as E2FB and DPB and additional, previously unidentified interactors. Among which 13 were annotated with the molecular function of RNA-binding, involved in ribosome biogenesis and protein translation (Fig. 5C). Five of these also interacted with EBP1, including members of the H/ACA ribonucleoprotein complex, known to catalyze the site-specific pseudo-uridylation and most of the methylation of rRNA (Watkins and Bohnsack, 2012); NOP56, which operates in trans to assist with the maturation of rRNAs (Lykke-Andersen et al., 2018); and finally the pre-mRNA-processing protein 40A, which binds the carboxyl-terminal domain of the largest subunit of RNA polymerase II and functions as a scaffold for RNA processing (Morris and Greenleaf, 2000). This set of common interactors points toward an as yet unknown aspect of RBR function that converges with EBP1 and explaining their opposing functions in the meristem.
EBP1 Is Associated with the Ribosome and Supports Global Translation in Sucrose Limiting Conditions
Given that EBP1 was found to interact with a number of ribosomal proteins by mass spectrometry, we asked whether EBP1 is preferentially associated with single ribosomes, their subunits, or with polysomes. In a high-density sucrose gradient (15% to 50%), EBP1 fusion protein cofractionated with polysomes and ribosomal subunits (Fig. 6A), in keeping with its association with 60S subunit proteins in the pull-down experiments (Fig. 5). After separating 40S, 60S, and 80S subunits in a low-sucrose gradient (7% to 20%), EBP1 also appeared in fractions containing only 40S, as well as in the nonribosomal fraction (Fig. 6A). Taken together, these data confirm that EBP1 has the potential to interact with the ribosome in polysomal and nonpolysomal contexts.
Figure 6.
EBP1 associates with cytosolic ribosomes and supports ribosome biogenesis. A, Absorbance profile of a sucrose density gradient after fractionation of whole tissue from Col0(pgEBP1-YFP) seedlings (12 d). The x axis represents the sedimentation distance in a 15% to 50% (red trace) or 7% to 20% (blue stippled trace) sucrose gradient, while the y axis represents A254 (A254). Peaks corresponding to 40S, 60S subunits, and 80S ribosomes are indicated on the individual profiles; polysomes are to the right of the 80S. Under the graph, the immunoblot analysis (EBP1-YFP) illustrates the distribution of EBP1-YFP in the total cell extract (CE) and in the eight fractions of the different gradients, while the ethidium bromide-stained agarose gels (EtBr gel) show the distribution of 18S (40S subunit) and 25S (60S subunit) rRNAs also in the eight fractions from the different gradients as shown in the graph. B and C, Polysome profiling of EBP1 RNAi lines (iEBP1 line 1 and line 2). Seedlings were grown for 12 d without sucrose (B) and with 1% sucrose (C). Total cell extracts from equal amounts of whole seedlings from Col0 and EBP1 RNAi transgenics were fractionated on 15% to 50% sucrose density gradients.
To test whether EBP1 supports translation, we obtained polysome profiles of 12-d-old EBP1 RNAi seedlings grown without sucrose supplementation and after phenotypic rescue using 1% sucrose. When polysomes were normalized by fresh weight, the EBP1 RNAi plants grown on 0% sucrose contained fewer ribosomes overall than the wild-type control, whereas at 1% sucrose, this difference disappeared (Fig. 6, B and C). However, the polysome-to-monosome (80S) ratio was not obviously affected by sucrose. Thus, the deficiency in ribosomes was rescued by sucrose, matching with the effect of sucrose on the growth kinetics shown earlier (Fig. 3; Supplemental Fig. S4).
EBP1 Supports Ribosome Biogenesis
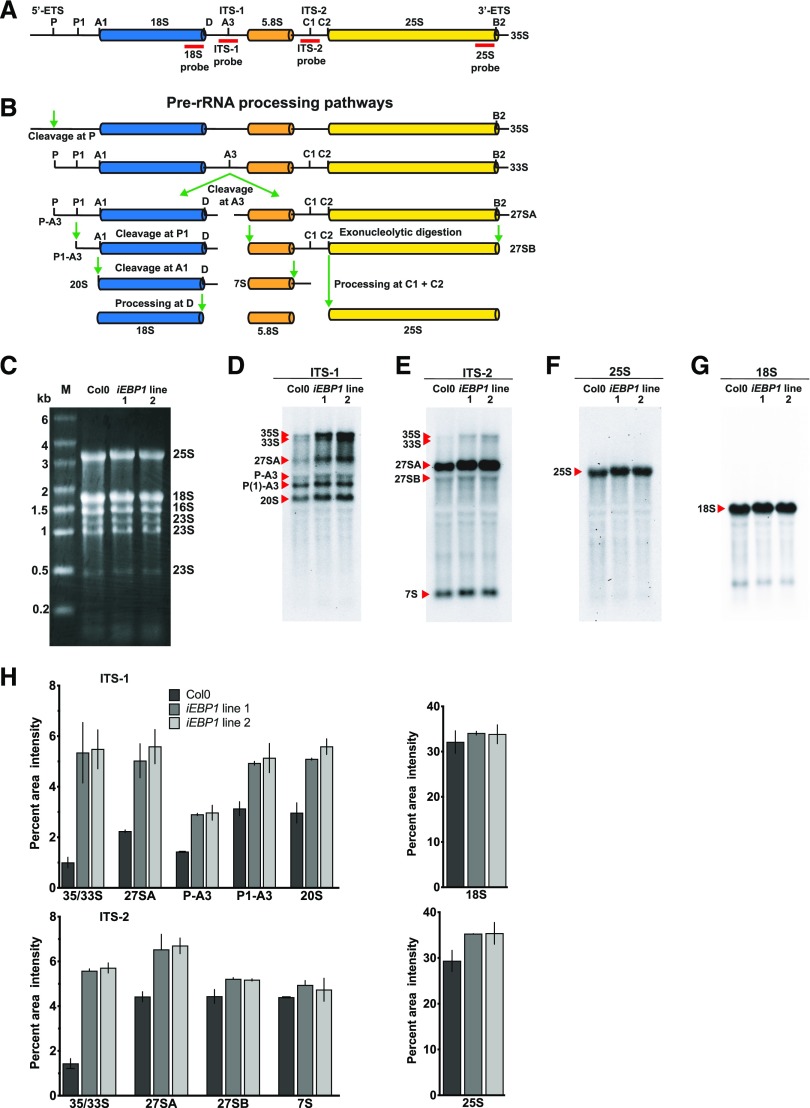
To further delineate the potential molecular functions of EBP1 in plants, especially in light of its association with proteins involved in ribosome biogenesis in Arabidopsis (Supplemental Table S1) and in human (Squatrito et al., 2004), we examined the iEBP1 lines for defects in rRNA processing. We conducted northern blot analyses of rRNA processing intermediates with probes against the internal transcribed spacers ITS-1 and ITS-2, as well as the mature 18S and 25S rRNA (Fig. 7, A and B). Despite equal loading of all three samples (Fig. 7C), the immature forms of the nuclear rRNAs were elevated in the two iEBP1 lines compared with the wild-type control (Fig. 7. D and E). In contrast, the abundance of the mature 18S and 25S rRNAs was indistinguishable between the Col-0 and RNAi lines (Fig. 7, F and G). These data suggest that the early processing events take place rapidly, as indicated by small pool sizes of the processing intermediates, whereas the later events (e.g. P[1]-A3 to 20S) appear to be slower, as shown by larger pool sizes (Fig. 7, D and E). Through quantification of the signals (Fig. 7H), we confirmed that the early processing events were particularly sensitive to the reduction in the EBP1 level, suggesting that EBP1 affects early processing steps in rRNA maturation. However, as the later processing intermediates (e.g. P[1]-A3 and 20S) were not depleted in the EBP1 RNAi lines, we suggest that EBP1 also supports the efficiency of processing steps at later stages. In conclusion, these data implicate EBP1 in ribosome biogenesis and, either directly or indirectly, in rRNA processing. The molecular defects in ribosome biogenesis may explain the slow growth of the EBP1 RNAi lines. In the rRNA processing experiments, we did not observe any imbalance in the ratios of specific rRNAs when samples containing equal amounts of total RNA were compared (Fig. 7C). The requirement for EBP1 for rRNA processing was rescued by growing the seedlings on sucrose-containing media (Supplemental Fig. S6, A to E, −S to +S). This result is in keeping with the conclusion that the slow growth of EBP1 RNAi plants is due, at least in part, to rRNA processing defects.
Figure 7.
EBP1 RNAi transgenic plants accumulate rRNA processing intermediates. Schematic representation of the Arabidopsis full length 35S pre-rRNA transcript indicating processing sites and the position of the probes used for northern blot analysis (red bars; A) and pre-rRNA processing pathway based on Zakrzewska-Placzek at al. (2010) (B). ETS, external transcribed spacer; ITS, internal transcribed spacer. C, Ethidium bromide-stained 1% (w/v) agarose gel showing equal loading of total RNA (4 µg) from Col0 and two independent EBP1 RNAi transgenic lines 1 and 2 grown without sucrose. Molecular weight marker (M) is shown on left, and the position of the rRNAs is indicated on the right. D to G, northern blot analysis of total RNA from Col0 and EBP1RNAi lines using digoxigenin-labeled specific probes against ITS-1 (D), ITS-2 (E), 25S (F), and 18S (G), visualized by chemiluminescence. Positions of the full length pre-rRNA transcript (35S) and the processed products are indicated on the left. H, Quantification of the signal intensity of the various rRNA transcript species shown in (D) to (G). The x axis indicates the precursors and processed rRNAs, while the y axis shows relative signal intensity. Values represent means and error bars stand for se from two northern blot experiments carried out with two independent biological materials.
DISCUSSION
In plants, EBP1 attracted attention as a growth driver both in Arabidopsis and in crop species, where it was shown to increase potato yield (Horváth et al., 2006) and to associate with hybrid vigor in maize (Wang et al., 2016). However, the molecular function and how EBP1 acts to increase plant growth and crop yield remained unknown. Here we show that EBP1 associates with proteins that bind RNA and are involved in a broad range of functions from ribosome biogenesis to transcriptional and translational regulation. Specifically, (1) plant EBP1 can localize to nucleoli, (2) EBP1 is found in cellular fractions containing ribosomal subunits as well as polysomes, and (3) silencing of EBP1 causes imbalances in the pattern of rRNA processing intermediates. In addition, we find that EBP1 antagonizes RBR action to maintain proliferation in the meristem, and sustains root growth, specifically in sucrose-limiting conditions.
Nutrient limiting conditions as well as abiotic stress constrain growth. It is of particular importance that plants maintain the proliferation potential in the meristem under these conditions (Julkowska and Testerink, 2015). For example, drought triggers jasmonate signaling, which counteracts the decline of EBP1 expression as leaf development progresses and cell proliferation gradually ceases (Kim et al., 2017). The same regulation is also observed for the large majority of genes coregulated with EBP1 in the ribi regulon (Noir et al., 2013). Thus, drought perception through jasmonate signaling has been suggested to establish a “ready-to-go” state that enables rapid recovery of meristematic functions after the stress subsides. Correspondingly, elevated EBP1 expression confers resistance to abiotic stresses such as cold (Cao et al., 2009) and drought (Cheng et al., 2016). Under stress, survival and growth are separate events, and it is likely that EBP1 is involved in the latter (Skirycz et al., 2011).
Here we show that EBP1 is important to maintain root meristem activity and growth in sucrose-limiting conditions. Thus, as during abiotic stress, EBP1 is also important to sustain growth when assimilates are limited. Consistent with previous data (Deprost et al., 2007), we show here that EBP1 mRNA and protein levels are stimulated by TOR kinase. sucrose stimulates cell proliferation in multiple ways, which include TOR-mediated (Xiong et al., 2013; Dobrenel et al., 2016a) and TOR-independent pathways. For example, sugar controls CDK-driven RBR phosphorylation, which lifts RBR-repression on cell proliferation (Magyar et al., 2012). This event may be TOR-mediated, because TOR, acting through YAK1 kinase, inhibits the expression of a class of CDK inhibitors, the SMRs, to promote root growth (Barrada et al., 2019). We now understand better how EBP1 assists in these events. Specifically, while RBR overexpression halts root growth by depleting stem cells and enhancing differentiation (Wildwater et al., 2005), EBP1 counteracts this RBR-driven repression and allows the maintenance of root meristem activity and growth. Moreover, overexpression of EBP1 bypasses the requirement for sugar to support root growth. Taken together, TOR activity is relayed to control growth by regulating both the level of EBP1 and the repressor activity of RBR. A remarkable result is that EBP1 silencing specifically leads to sensitivity under limited sugar availability, whereas addition of sucrose to the media largely compensates for the loss of EBP1. This result confirms that sucrose, while stimulating EBP1 expression, also supports growth in other EBP1-independent ways, such as the TOR-dependent pathways regulating E2F and RBR (Magyar et al., 2012; Xiong et al., 2013; Barrada et al., 2019).
Both EBP1 expression and EBP1 protein are largely confined to meristems, such as the root apical and lateral root meristems. Translatomic data also demonstrate that EBP1 mRNA is actively translated in the shoot apex (Tian et al., 2019). Furthermore, EBP1 expression progressively diminishes as the meristematic cells are exhausted when RBR overexpression is induced. This shows that EBP1 level is tightly linked with meristematic activity. Fluorescently tagged EBP1 could be detected throughout the cytosol, nucleus, and nucleolus, although the nuclear and nucleolar accumulation were largely masked when EBP1 was tagged at its C terminus. We conclude that EBP1 is likely found in all three compartments as suggested by the N-terminally tagged version, because (1) growth factor signaling promoted nuclear accumulation of EBP1-GFP (Li et al., 2018), (2) others have detected tagged EBP1 proteins in plant nuclei of various cell types (Zhang et al., 2005; Cao et al., 2009; Li et al., 2016; Palm et al., 2019), and (3) native Arabidopsis EBP1 was detected in a purified nucleolar preparation (Pendle et al., 2005).
In line with the localization of EBP1 in the cytoplasm, nucleus, and nucleolus, we identified EBP1-interacting proteins broadly distributed in the cell but remarkably almost exclusively involved in various aspects of protein synthesis. We identified EBP1 in association with ribosomal proteins of the 60S subunit, as well as the eukaryotic translation initiation factor eIF5B. EBP1 was detected in sucrose gradient fractions that contain polysomes, suggesting association of EBP1 with mature cytosolic ribosomes as well as ribosomal subunits, similar to the association pattern of human EBP1 (Squatrito et al., 2004; Squatrito et al., 2006). Related to the nucleolar localization of EBP1, we found EBP1 interaction with ribosome biogenesis factors, such as multiple subunits of the H/ACA ribonucleoprotein complex involved in pre-rRNA processing (Watkins and Bohnsack, 2012), with nucleolar S-adenosyl-l-Met-dependent methyltransferases, with other rRNA-processing proteins (Lykke-Andersen et al., 2018) and the multifunctional protein nucleolin (Jia et al., 2017). Similar to EBP1, nucleolin is also most abundant in meristematic tissues and, in parallel with d-type cyclin, it is rapidly induced when cell proliferation resumes after nutrient starvation (Bögre et al., 1996). Besides the proteins with nucleolar functions, EBP1 also interacted with proteins involved in transcriptional and posttranscriptional regulation. In conclusion, similarly to human EBP1 with RNA binding activity (Squatrito et al., 2004; Squatrito et al., 2006), the common feature of EBP1 interacting proteins is that they function as part of ribonucleoprotein complexes.
Although the precise role of EBP1 in ribosome biogenesis remains unclear, the related metallopeptidase-fold protein Arx1 is a canonical 60S biogenesis factor (Greber, 2016). Seedlings with reduced EBP1 level showed a defect in rRNA processing that affected specifically the early processing intermediates. Just as the growth phenotype was rescued by sucrose, so was the rRNA processing delay in the EBP1 silencing lines. These results are consistent with the assumption that EBP1 has a role in ribosome biogenesis. Nevertheless, we cannot rule out that Arabidopsis EBP1 also supports rRNA transcription, as was described in human cells (Nguyen le et al., 2015). However, our data point to a defect during rRNA processing, because a transcriptional defect upon EBP1 silencing would reduce rather than increase the abundance of rRNA processing intermediates.
In vertebrates EBP1 is expressed in two isoforms; the longer p48 isoform is generally associated with cell proliferation and tumorigenesis, specifically by inhibiting cell death and promoting Pol I-dependent rRNA expression. The shorter p42 isoform lacks the N terminus of p48 and is considered to have a tumor suppressor role. Together with Rb, E2F1, and chromatin modifying proteins, HsEBP1 p42 represses cell proliferation (Liu et al., 2006; Ko et al., 2016; Nguyen et al., 2018; Nguyen et al., 2019). The internal methionine that initiates translation of the p42 isoform is not conserved in plants. Thus, there is no evidence that the shorter suppressive form exists in plants. In line with these findings, the plant EBP1 promotes cell growth and division (Horváth et al., 2006; Cheng et al., 2016).
In addition, in this study we show that EBP1 overexpression can compensate for the pro-differentiation activity of RBR in the root stem cell niche. Inducible overexpression of RBR downregulated EBP1 expression and rapidly abolished proliferation competence. In contrast, overexpression of EBP1 countered RBR’s pro-differentiation activity and rescued the maintenance of cell proliferation around the stem cell niche. Thus, EBP1 expression is in an antagonistic relationship with the pro-differentiation factor, RBR. Our data illustrate how stem cell fate is maintained by a balance between pro-differentiation signals and the pro-growth agenda of the ribosome biogenesis machinery.
The striking finding that EBP1 and RBR share common interactors involved in ribosome biogenesis and that RBR associated with a number of proteins in the cytoplasmic translation machinery may shed light on how EBP1, in conjunction with RBR, regulates meristem activity. Future research will elaborate on our hypothesis that RBR and EBP1 counteract to regulate meristem function through acting on a common biological process to regulate the capacity for protein synthesis. In conclusion, our findings on the role of EBP1 in root meristem activity and growth could be important in breeding to improve yield and yield stability in crops.
MATERIALS AND METHODS
Plant Material and Growth Conditions
Arabidopsis (Arabidopsis thaliana) seeds were sterilized, incubated at 4°C for 2 d, then germinated and grown vertically on half-strength Murashige and Skoog salt plant media (Sigma, cat #2633024) with different concentration of Phytoagar under a long day cycle of 16 h light (80 ± 10 μmol m−2 s−1)/8 h dark at 22°C and 50% humidity. Sucrose was added at 1%, 2% (w/v), or omitted, as specified. Arabidopsis ecotype Columbia (Col-0) was used as the wild-type control as the genetic background to generate transgenic lines and as a source of DNA to clone either promoter or coding regions. Transgenic lines, EBPOE lines 1 and 2, overexpressing the potato (Solanum tuberosum) EBP1 (StEBP1, Horváth et al., 2006 referred as line 19 and 43), and RBROE (Wildwater et al., 2005) were described earlier. The insertional line, CS854731, named as ebp1-3 (Li et al., 2018), was obtained from the Salk Institute (http://signal.sal.edu).
Generating Transcriptional and Translational Fusion Proteins
Both transcriptional and translational fusions were constructed by the Multisite Gateway System (Invitrogen) by amplifying the putative regulatory and genomic coding sequences with gene-specific primers. To generate the p35S:Cerulean-EBP1 reporter construct and the EBP1 inducible silencing lines, EBP1 RNAi, Arabidopsis EBP1 cDNA was amplified and cloned in the Gateway compatible vectors. The plant binary vector system, pGREEN was used in the Agrobacterium tumefaciens C589(pMP90)-mediated plant transformation with the floral dip method. Details are described in Supplemental Data S1 and primer sequences summarized in Supplemental Table S3.
Root Measurement and Microscopy
Photographs of vertically or horizontally grown seedlings were taken with a digital camera (Canon). The primary root length was measured using ImageJ version 1.41 software (http://rsb.info.nih.gov/ij/index.html) and analyzed statistically with GraphPad Prism 7.0a (GraphPad Software). The root meristematic zone was determined by measuring the distance from the QC to the first elongating cortex cells; the same region was used to count the number of cortex cells with no signs of rapid elongation. For measurements and to follow tissue-specific expression, either light (Zeiss Axioscope with Nomarski optics) or confocal microscopy (Zeiss LSM710 and Leica SP8) was carried out. To take differential interference contrast images, chloral-hydrate treated roots were used; starch granules were visualized with 1% (w/v) lugol solution. Seedlings (T3 generation) were stained for GUS activity for 3–6 h at 37°C in 0.5 mg/mL X-gluc (Biosynth AG), dissolved according to the manufacturer’s recommendation. For confocal microscopy, to visualize cell walls, 5–10 μg/mL propidium iodide (Sigma-Aldrich) solution was applied. For analysis, longitudinal, single sections were taken with the same median focal plane. The final images were built and merged from partially overlapping sections with Adobe Illustrator CS6.
Chemical Treatments
The introgression lines, EBP1OE;RBROE (F3 generation) were germinated and grown on vertical plates (1% sucrose media). The 5- to 6-d-old seedlings were transferred to 1 μm dexamethasone-containing plates, and growth was scored over a time series (24 h–144 h period). RBROE, RBROE backcrossed to Col-0, EBP1OE, and Col-0 were used as a control to follow root growth, meristem size, and level of gene expression. To screen transformants of the silencing lines, EBP1 RNAi, seeds (T1 and T2) were germinated and grown on 5 μm 17-β-estradiol-containing medium supplied with 1% sucrose or no sucrose. Next, the candidate lines were grown in normal growth conditions and transferred to 17-β-estradiol medium with or without sucrose. The induction and level of silencing was measured by quantitative reverse transcription PCR.
Immunoblot and AZD Treatment
For immunoblot analysis, the transgenic lines Col-0(pgEBP1-YFP) and Col-0(pEBP1:CFP) were germinated and grown for 5 d on sucrose-free medium in 12-h light/12-h dark conditions. On the 5th day, at ZT0 timepoint, seedlings were transferred to medium containing either 2% sucrose, 2% sucrose + 1 µm AZD8055 or to fresh sucrose-free medium. Samples were harvested 3 h later by flash-freezing the whole seedlings. Total protein was extracted from samples using extraction buffer containing 75 mm NaCl, 25 mm Tris-HCl (pH = 7.5), 15 mm β-glycerolphosphate, 15 mm EGTA, 15 mm p-nitrophenylphosphate, 10 mm MgCl2, 1 mm dithiothreitol, 1 mm NaF, 0.5 mm Na3VO4, 0.5 mm phenylmethylsulfonyl fluoride, and 0.1% Tween 20 (Magyar et al., 2005). The total protein samples were separated in a 10% acrylamide gel using SDS-PAGE and transferred to polyvinylidene difluoride membrane. Antibodies used for detection: Anti-GFP mouse monoclonal antibody (ROCHE) in 1:1000 dilution and goat anti-mouse IgG-HRP Secondary Antibody (Sigma) in 1:10000 dilution.
Northern Blot Analysis, Polysome Profiling, and Protein Fractionation
Northern blot analyses were performed with 4 μg of total RNA from seedlings grown under the indicated conditions. Blots were probed with digoxigenin-labeled probes corresponding to the internal transcribed spacer 1 and 2, 18S and 25S rRNAs, and signal was detected with chemiluminescent reagent, imaged on a BioRad ChemiDoc instrument, and quantified with ImageJ version 1.41 software (http://rsb.info.nih.gov/ij/index.html). Polysome profiles were obtained from 12-d-old Col-0 or EBP1 RNAi whole seedlings grown without or with 2% sucrose essentially as described (Kim et al., 2007). Then 100 μl supernatant was layered on either a 2-mL 15-50% or a 2 mL 7% to 20% linear gradient prepared with a Hoefer gradient maker and centrifuged at 50,000 rpm (Beckmann TLS55 rotor) for 1 h 10 min at 4°C. After the UV A254 and fractionation into eight equal fractions were monitored, EBP1 protein was detected by immunoblotting with GFP-specific antibody (Lokdarshi et al., 2016). For detailed procedures see Supplemental Data S1.
Protein Complex Isolation, LC-MS/MS Identification, and Statistical Analysis
A detailed description and link to the raw datasets are given in the Supplemental Data S1.
Accession Numbers
Accession numbers are At3g51800, EBP1; At3g12280, RBR; the other accession numbers are listed in Supplemental Table S3.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. EBP1 is expressed in actively dividing tissues during root development.
Supplemental Figure S2. EBP1 localizes to nucleoli, nuclei and cytosol.
Supplemental Figure S3. Reduced level of EBP1 results in retarded growth in the absence of sucrose.
Supplemental Figure S4. Root and leaf growth deficiency in the absence of sugar and recovery of root growth defects upon sucrose supplementation in the EBP1 transgenic lines.
Supplemental Figure S5. Elevated level of EBP1 delays the differentiation induced by RBR overexpression.
Supplemental Figure S6. sucrose supplementation alleviates the requirement for EBP1 in rRNA processing.
Supplemental Table S1. List of EBP1 interacting proteins.
Supplemental Table S2. List of RBR interacting proteins.
Supplemental Table S3. Primers used in this study.
Supplemental Data S1. https://doi.org/10.6084/m9.figshare.10069247.
Acknowledgments
We thank Dr. Patrick Giavalisco (Max Planck Institute Cologne) for initial discussions on EBP1, linking it with protein translation; Dr. Tessa Burch-Smith and Dr. Daniel M Roberts (University of Tennessee) for valuable advice with northern blot analysis and for providing anti-GFP antibody, respectively; Imma Perez-Salamo for valuable discussions (Royal Holloway, University of London); and Christian Bachem (Wageningen University and Research) for critical reading of the manuscript.
Footnotes
This work was supported by the National Science Foundation (NSF) (grants MCB-1546402 and IOS-1456988), National Institutes of Health (grant R15 GM129672), and the Donald L Akers Jr Faculty Enrichment Fellowship (to A.G.v.A); the European Commission (EC) (Marie-Curie IEF fellowship, FP7-PEOPLE-2012-IEF-330789 to B.M.H.; FP7-PEOPLE-2012-IEF.330713 to C.P.); the Netherlands Organization for Scientific Research (Spinoza grant to B.S.); Biotechnology and Biological Sciences Research Council (BBSRC) (BBSRC-NSF grant BB/M025047/1 to C.P. and L.B.); and Magyar Tudomanyos Akademia (Hungarian Academy of Sciences) (GINOP-2.3.2-15-2016-00032 to A.P.-S. and GINOP-2.3.2-15-2016-00001 to A.P.-S. and Z.M.).
[CC-BY]: Article free via Creative Commons CC-BY 4.0 license.
References
- Ahmad Z, Magyar Z, Bögre L, Papdi C (2019) Cell cycle control by the target of rapamycin signalling pathway in plants. J Exp Bot 70: 2275–2284 [DOI] [PubMed] [Google Scholar]
- Bakshi A, Moin M, Madhav MS, Kirti PB (2019) Target of rapamycin, a master regulator of multiple signalling pathways and a potential candidate gene for crop improvement. Plant Biol (Stuttg) 21: 190–205 [DOI] [PubMed] [Google Scholar]
- Barbet NC, Schneider U, Helliwell SB, Stansfield I, Tuite MF, Hall MN (1996) TOR controls translation initiation and early G1 progression in yeast. Mol Biol Cell 7: 25–42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrada A, Djendli M, Desnos T, Mercier R, Robaglia C, Montané MH, Menand B (2019) A TOR-YAK1 signaling axis controls cell cycle, meristem activity and plant growth in Arabidopsis. Development 146: dev171298. [DOI] [PubMed] [Google Scholar]
- Bögre L, Jonak C, Mink M, Meskiene I, Traas J, Ha DT, Swoboda I, Plank C, Wagner E, Heberle-Bors E, Hirt H (1996) Developmental and cell cycle regulation of alfalfa nucMs1, a plant homolog of the yeast Nsr1 and mammalian nucleolin. Plant Cell 8: 417–428 [PMC free article] [PubMed] [Google Scholar]
- Bose SK, Sengupta TK, Bandyopadhyay S, Spicer EK (2006) Identification of Ebp1 as a component of cytoplasmic bcl-2 mRNP (messenger ribonucleoprotein particle) complexes. Biochem J 396: 99–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao P, Song J, Zhou C, Weng M, Liu J, Wang F, Zhao F, Feng D, Wang B (2009) Characterization of multiple cold induced genes from Ammopiptanthus mongolicus and functional analyses of gene AmEBP1. Plant Mol Biol 69: 529–539 [DOI] [PubMed] [Google Scholar]
- Chen GH, Liu MJ, Xiong Y, Sheen J, Wu SH (2018) TOR and RPS6 transmit light signals to enhance protein translation in deetiolating Arabidopsis seedlings. Proc Natl Acad Sci USA 115: 12823–12828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H, Chen X, Zhu J, Huang H (2016) Overexpression of a Hevea brasiliensis ErbB-3 Binding protein 1 gene increases drought tolerance and organ size in Arabidopsis. Front Plant Sci 7: 1703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Inzé D (2007) The ins and outs of the plant cell cycle. Nat Rev Mol Cell Biol 8: 655–665 [DOI] [PubMed] [Google Scholar]
- Deprost D, Yao L, Sormani R, Moreau M, Leterreux G, Nicolaï M, Bedu M, Robaglia C, Meyer C (2007) The Arabidopsis TOR kinase links plant growth, yield, stress resistance and mRNA translation. EMBO Rep 8: 864–870 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewitte W, Riou-Khamlichi C, Scofield S, Healy JM, Jacqmard A, Kilby NJ, Murray JA (2003) Altered cell cycle distribution, hyperplasia, and inhibited differentiation in Arabidopsis caused by the D-type cyclin CYCD3. Plant Cell 15: 79–92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrenel T, Caldana C, Hanson J, Robaglia C, Vincentz M, Veit B, Meyer C (2016a) TOR signaling and nutrient sensing. Annu Rev Plant Biol 67: 261–285 [DOI] [PubMed] [Google Scholar]
- Dobrenel T, Mancera-Martínez E, Forzani C, Azzopardi M, Davanture M, Moreau M, Schepetilnikov M, Chicher J, Langella O, Zivy M, et al. (2016b) The Arabidopsis TOR kinase specifically regulates the expression of nuclear genes coding for plastidic ribosomal proteins and the phosphorylation of the cytosolic ribosomal protein S6. Front Plant Sci 7: 1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowling RJ, Topisirovic I, Alain T, Bidinosti M, Fonseca BD, Petroulakis E, Wang X, Larsson O, Selvaraj A, Liu Y, et al. (2010) mTORC1-mediated cell proliferation, but not cell growth, controlled by the 4E-BPs. Science 328: 1172–1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enganti R, Cho SK, Toperzer JD, Urquidi-Camacho RA, Cakir OS, Ray AP, Abraham PE, Hettich RL, von Arnim AG (2018) Phosphorylation of ribosomal protein RPS6 integrates light signals and circadian clock signals. Front Plant Sci 8: 2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forzani C, Duarte GT, Van Leene J, Clement G, Huguet S, Paysant-Le-Roux C, Mercier R, De Jaeger G, Leprince AS, Meyer C (2019) Mutations of the AtYAK1 Kinase Suppress TOR Deficiency in Arabidopsis. Cell Rep 27: 3696–3708 [DOI] [PubMed] [Google Scholar]
- Fox S, Southam P, Pantin F, Kennaway R, Robinson S, Castorina G, Sánchez-Corrales YE, Sablowski R, Chan J, Grieneisen V, et al. (2018) Spatiotemporal coordination of cell division and growth during organ morphogenesis. PLoS Biol 16: e2005952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- González A, Hall MN (2017) Nutrient sensing and TOR signaling in yeast and mammals. EMBO J 36: 397–408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greber BJ. (2016) Mechanistic insight into eukaryotic 60S ribosomal subunit biogenesis by cryo-electron microscopy. RNA 22: 1643–1662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutzat R, Borghi L, Fütterer J, Bischof S, Laizet Y, Hennig L, Feil R, Lunn J, Gruissem W (2011) RETINOBLASTOMA-RELATED PROTEIN controls the transition to autotrophic plant development. Development 138: 2977–2986 [DOI] [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Bögre L (2013) S6K1 and E2FB are in mutually antagonistic regulatory links controlling cell growth and proliferation in Arabidopsis. Plant Signal Behav 8: e24367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriques R, Magyar Z, Monardes A, Khan S, Zalejski C, Orellana J, Szabados L, de la Torre C, Koncz C, Bögre L (2010) Arabidopsis S6 kinase mutants display chromosome instability and altered RBR1-E2F pathway activity. EMBO J 29: 2979–2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horváth BM, Magyar Z, Zhang Y, Hamburger AW, Bakó L, Visser RG, Bachem CW, Bögre L (2006) EBP1 regulates organ size through cell growth and proliferation in plants. EMBO J 25: 4909–4920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubner NC, Bird AW, Cox J, Splettstoesser B, Bandilla P, Poser I, Hyman A, Mann M (2010) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol 189: 739–754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia W, Yao Z, Zhao J, Guan Q, Gao L (2017) New perspectives of physiological and pathological functions of nucleolin (NCL). Life Sci 186: 1–10 [DOI] [PubMed] [Google Scholar]
- Julkowska MM, Testerink C (2015) Tuning plant signaling and growth to survive salt. Trends Plant Sci 20: 586–594 [DOI] [PubMed] [Google Scholar]
- Karlsson T, Altankhuyag A, Dobrovolska O, Turcu DC, Lewis AE (2016) A polybasic motif in ErbB3-binding protein 1 (EBP1) has key functions in nucleolar localization and polyphosphoinositide interaction. Biochem J 473: 2033–2047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BH, Cai X, Vaughn JN, von Arnim AG (2007) On the functions of the h subunit of eukaryotic initiation factor 3 in late stages of translation initiation. Genome Biol 8: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JM, To TK, Matsui A, Tanoi K, Kobayashi NI, Matsuda F, Habu Y, Ogawa D, Sakamoto T, Matsunaga S, Bashir K, Rasheed S, et al. (2017) Acetate-mediated novel survival strategy against drought in plants. Nat Plants 3: 17097. [DOI] [PubMed] [Google Scholar]
- Ko HR, Chang YS, Park WS, Ahn JY (2016) Opposing roles of the two isoforms of ErbB3 binding protein 1 in human cancer cells. Int J Cancer 139: 1202–1208 [DOI] [PubMed] [Google Scholar]
- Li C, Liu X, Qiang X, Li X, Li X, Zhu S, Wang L, Wang Y, Liao H, Luan S, Yu F (2018) EBP1 nuclear accumulation negatively feeds back on FERONIA-mediated RALF1 signaling. PLoS Biol 16: e2006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Yu G, Sun X, Zhang X, Liu J, Pan H (2016) AcEBP1, an ErbB3-binding protein (EBP1) from halophyte Atriplex canescens, negatively regulates cell growth and stress responses in Arabidopsis. Plant Sci 248: 64–74 [DOI] [PubMed] [Google Scholar]
- Liu MJ, Wu SH, Wu JF, Lin WD, Wu YC, Tsai TY, Tsai HL, Wu SH (2013) Translational landscape of photomorphogenic Arabidopsis. Plant Cell 25: 3699–3710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Z, Ahn JY, Liu X, Ye K (2006) Ebp1 isoforms distinctively regulate cell survival and differentiation. Proc Natl Acad Sci USA 103: 10917–10922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lokdarshi A, Conner WC, McClintock C, Li T, Roberts DM (2016) Arabidopsis CML38, a calcium sensor that localizes to ribonucleoprotein complexes under hypoxia stress. Plant Physiol 170: 1046–1059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Juez E, Dillon E, Magyar Z, Khan S, Hazeldine S, de Jager SM, Murray JA, Beemster GT, Bögre L, Shanahan H (2008) Distinct light-initiated gene expression and cell cycle programs in the shoot apex and cotyledons of Arabidopsis. Plant Cell 20: 947–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lykke-Andersen S, Ardal BK, Hollensen AK, Damgaard CK, Jensen TH (2018) Box C/D snoRNP Autoregulation by a cis-Acting snoRNA in the NOP56 Pre-mRNA. Mol Cell 72: 99–111 [DOI] [PubMed] [Google Scholar]
- Magyar Z, Bögre L, Ito M (2016) DREAMs make plant cells to cycle or to become quiescent. Curr Opin Plant Biol 34: 100–106 [DOI] [PubMed] [Google Scholar]
- Magyar Z, De Veylder L, Atanassova A, Bakó L, Inzé D, Bögre L (2005) The role of the Arabidopsis E2FB transcription factor in regulating auxin-dependent cell division. Plant Cell 17: 2527–2541 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magyar Z, Horváth B, Khan S, Mohammed B, Henriques R, De Veylder L, Bakó L, Scheres B, Bögre L (2012) Arabidopsis E2FA stimulates proliferation and endocycle separately through RBR-bound and RBR-free complexes. EMBO J 31: 1480–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed B, Bilooei SF, Dóczi R, Grove E, Railo S, Palme K, Ditengou FA, Bögre L, López-Juez E (2018) Converging light, energy and hormonal signaling control meristem activity, leaf initiation, and growth. Plant Physiol 176: 1365–1381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montané MH, Menand B (2013) ATP-competitive mTOR kinase inhibitors delay plant growth by triggering early differentiation of meristematic cells but no developmental patterning change. J Exp Bot 64: 4361–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montané MH, Menand B (2019) TOR inhibitors: From mammalian outcomes to pharmacogenetics in plants and algae. J Exp Bot 70: 2297–2312 [DOI] [PubMed] [Google Scholar]
- Morris DP, Greenleaf AL (2000) The splicing factor, Prp40, binds the phosphorylated carboxyl-terminal domain of RNA polymerase II. J Biol Chem 275: 39935–39943 [DOI] [PubMed] [Google Scholar]
- Nguyen DQ, Hoang DH, Nguyen TTV, Ho HD, Huynh V, Shin JH, Ly QT, Thi Nguyen DD, Ghoda L, Marcucci G, Nguyen LXT (2019) Ebp1 p48 promotes oncogenic activities in human colon cancer cells through regulation of TIF-90-mediated ribosomal RNA synthesis. J Cell Physiol 234: 17612–17621 [DOI] [PubMed] [Google Scholar]
- Nguyen DQ, Hoang DH, Nguyen Vo TT, Huynh V, Ghoda L, Marcucci G, Nguyen LXT (2018) The role of ErbB3 binding protein 1 in cancer: Friend or foe? J Cell Physiol 233: 9110–9120 [DOI] [PubMed] [Google Scholar]
- Nguyen XT, Lee Y, Urbani L, Utz PJ, Hamburger AW, Sunwoo JB, Mitchell BS (2015) Regulation of ribosomal RNA synthesis in T cells: Requirement for GTP and Ebp1. Blood 125: 2519–2529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noir S, Bömer M, Takahashi N, Ishida T, Tsui TL, Balbi V, Shanahan H, Sugimoto K, Devoto A (2013) Jasmonate controls leaf growth by repressing cell proliferation and the onset of endoreduplication while maintaining a potential stand-by mode. Plant Physiol 161: 1930–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norris-Mullins B, VanderKolk K, Vacchina P, Joyce MV, Morales MA (2014) LmaPA2G4, a homolog of human Ebp1, is an essential gene and inhibits cell proliferation in L. major. PLoS Negl Trop Dis 8: e2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada M, Jang SW, Ye K (2007) Ebp1 association with nucleophosmin/B23 is essential for regulating cell proliferation and suppressing apoptosis. J Biol Chem 282: 36744–36754 [DOI] [PubMed] [Google Scholar]
- Palm D, Streit D, Shanmugam T, Weis BL, Ruprecht M, Simm S, Schleiff E (2019) Plant-specific ribosome biogenesis factors in Arabidopsis thaliana with essential function in rRNA processing. Nucleic Acids Res 47: 1880–1895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pendle AF, Clark GP, Boon R, Lewandowska D, Lam YW, Andersen J, Mann M, Lamond AI, Brown JW, Shaw PJ (2005) Proteomic analysis of the Arabidopsis nucleolus suggests novel nucleolar functions. Mol Biol Cell 16: 260–269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU (2000) A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev 14: 2028–2045 [PMC free article] [PubMed] [Google Scholar]
- Pisapia L, Barba P, Cortese A, Cicatiello V, Morelli F, Del Pozzo G (2015) EBP1 protein modulates the expression of human MHC class II molecules in non-hematopoietic cancer cells. Int J Oncol 47: 481–489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Dimitrova M, Mancera-Martínez E, Geldreich A, Keller M, Ryabova LA (2013) TOR and S6K1 promote translation reinitiation of uORF-containing mRNAs via phosphorylation of eIF3h. EMBO J 32: 1087–1102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schepetilnikov M, Ryabova LA (2018) Recent discoveries on the role of TOR (Target of Rapamycin) signaling in translation in plants. Plant Physiol 176: 1095–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skirycz A, Vandenbroucke K, Clauw P, Maleux K, De Meyer B, Dhondt S, Pucci A, Gonzalez N, Hoeberichts F, Tognetti VB, et al. (2011) Survival and growth of Arabidopsis plants given limited water are not equal. Nat Biotechnol 29: 212–214 [DOI] [PubMed] [Google Scholar]
- Squatrito M, Mancino M, Donzelli M, Areces LB, Draetta GF (2004) EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene 23: 4454–4465 [DOI] [PubMed] [Google Scholar]
- Squatrito M, Mancino M, Sala L, Draetta GF (2006) Ebp1 is a dsRNA-binding protein associated with ribosomes that modulates eIF2α phosphorylation. Biochem Biophys Res Commun 344: 859–868 [DOI] [PubMed] [Google Scholar]
- Tian C, Wang Y, Yu H, He J, Wang J, Shi B, Du Q, Provart NJ, Meyerowitz EM, Jiao Y (2019) A gene expression map of shoot domains reveals regulatory mechanisms. Nat Commun 10: 141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T, Sui Z, Liu X, Li Y, Li H, Xing J, Song F, Zhang Y, Sun Q, Ni Z (2016) Ectopic expression of a maize hybrid up-regulated gene, ErbB-3 binding Protein 1 (ZmEBP1), increases organ size by promoting cell proliferation in Arabidopsis. Plant Sci 243: 23–34 [DOI] [PubMed] [Google Scholar]
- Watkins NJ, Bohnsack MT (2012) The box C/D and H/ACA snoRNPs: Key players in the modification, processing and the dynamic folding of ribosomal RNA. Wiley Interdiscip Rev RNA 3: 397–414 [DOI] [PubMed] [Google Scholar]
- Wildwater M, Campilho A, Perez-Perez JM, Heidstra R, Blilou I, Korthout H, Chatterjee J, Mariconti L, Gruissem W, Scheres B (2005) The RETINOBLASTOMA-RELATED gene regulates stem cell maintenance in Arabidopsis roots. Cell 123: 1337–1349 [DOI] [PubMed] [Google Scholar]
- Wu Y, Shi L, Li L, Fu L, Liu Y, Xiong Y, Sheen J (2019) Integration of nutrient, energy, light, and hormone signalling via TOR in plants. J Exp Bot 70: 2227–2238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiong Y, McCormack M, Li L, Hall Q, Xiang C, Sheen J (2013) Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496: 181–186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zakrzewska-Placzek M, Souret FF, Sobczyk GJ, Green PJ, Kufel J (2010) Arabidopsis thaliana XRN2 is required for primary cleavage in the pre-ribosomal RNA. Nucleic Acids Res 38: 4487–4502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang WK, Shen YG, He XJ, Du BX, Xie ZM, Luo GZ, Zhang JS, Chen SY (2005) Characterization of a novel cell cycle-related gene from Arabidopsis. J Exp Bot 56: 807–816 [DOI] [PubMed] [Google Scholar]
- Zhou H, Mazan-Mamczarz K, Martindale JL, Barker A, Liu Z, Gorospe M, Leedman PJ, Gartenhaus RB, Hamburger AW, Zhang Y (2010) Post-transcriptional regulation of androgen receptor mRNA by an ErbB3 binding protein 1 in prostate cancer. Nucleic Acids Res 38: 3619–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Zhang Y, Hamburger AW (2011) EBP1 inhibits translation of androgen receptor mRNA in castration resistant prostate cancer cells. Anticancer Res 31: 3129–3135 [PMC free article] [PubMed] [Google Scholar]