Misregulation of either MADS78 or MADS79 results in seed abnormalities, due to a mistimed developmental transition of the endosperm and downstream effects on grain quality.
Abstract
MADS box transcription factors (TFs) are subdivided into type I and II based on phylogenetic analysis. The type II TFs regulate floral organ identity and flowering time, but type I TFs are relatively less characterized. Here, we report the functional characterization of two type I MADS box TFs in rice (Oryza sativa), MADS78 and MADS79. Transcript abundance of both these genes in developing seed peaked at 48 h after fertilization and was suppressed by 96 h after fertilization, corresponding to syncytial and cellularized stages of endosperm development, respectively. Seeds overexpressing MADS78 and MADS79 exhibited delayed endosperm cellularization, while CRISPR-Cas9-mediated single knockout mutants showed precocious endosperm cellularization. MADS78 and MADS79 were indispensable for seed development, as a double knockout mutant failed to make viable seeds. Both MADS78 and 79 interacted with MADS89, another type I MADS box, which enhances nuclear localization. The expression analysis of Fie1, a rice FERTILIZATION-INDEPENDENT SEED-POLYCOMB REPRESSOR COMPLEX2 component, in MADS78 and 79 mutants and vice versa established an antithetical relation, suggesting that Fie1 could be involved in negative regulation of MADS78 and MADS79. Misregulation of MADS78 and MADS79 perturbed auxin homeostasis and carbon metabolism, as evident by misregulation of genes involved in auxin transport and signaling as well as starch biosynthesis genes causing structural abnormalities in starch granules at maturity. Collectively, we show that MADS78 and MADS79 are essential regulators of early seed developmental transition and impact both seed size and quality in rice.
Double fertilization is a characteristic feature of angiosperm reproduction. During double fertilization, two haploid sperm cells reach the embryo sac via the pollen tube. One sperm cell fuses with a haploid egg cell to form a diploid embryo and the other fuses with a diploid central cell to form a triploid endosperm cell composed of two maternal genomes and one paternal genome (Olsen, 2004). The embryo acts as a precursor to the next generation of the plant life cycle, and the endosperm is the terminal nutritive tissue. In eudicots, the embryo represents the majority of the tissue in mature seeds and the endosperm is ephemeral (Chaudhury et al., 2001). However, in monocots such as rice (Oryza sativa), wheat (Triticum aestivum), and maize (Zea mays), the endosperm persists and becomes the dominant tissue that stores starch and proteins, thus determining the mature seed size (Chaudhury et al., 2001; Gao et al., 2013). Endosperm is the primary source of calories for human (Homo sapiens) and livestock nutrition, thereby making it a valuable biological and economic entity (Sabelli and Larkins, 2009).
Endosperm development undergoes several distinct and overlapping phases categorized as follows: (1) early, the triploid nuclei undergo multiple rounds of mitotic divisions without cytokinesis (syncytium) before initiating cell wall formation (cellularization); (2) mid, differentiation of cells, grain filling that is primarily contributed by synthesis of starch and proteins, and endoreduplication; and (3) late, maturation and dehydration (Sabelli and Larkins, 2009). The rate and duration of endosperm proliferation during the syncytial phase is an important determinant of final seed size (Garcia et al., 2005). Normal endosperm development requires a timely transition from syncytium to cellularization (Brown et al., 1996; Olsen, 2004). Genomic imbalance between the parental genomes disrupts the timely initiation of cellularization (Brown et al., 1996; Haig, 2013; Wang et al., 2018). Excessive paternal dosage delays cellularization and causes endosperm overproliferation, resulting in bigger seeds (Scott et al., 1998; Stoute et al., 2012). Conversely, increased maternal dosage leads to precocious cellularization and smaller seeds. Either case may result in seed abortion as well (Scott et al., 1998; Stoute et al., 2012).
The transition of endosperm from the syncytial to cellularized state is regulated at both the genetic and epigenetic levels and is sensitive to environmental conditions (Folsom et al., 2014). Epigenetic regulation is mediated either by DNA methylation or FERTILIZATION-INDEPENDENT SEED-POLYCOMB REPRESSIVE COMPLEX2 (FIS-PRC2; Gehring, 2013; Pires, 2014). METHYLTRANSFERASE1 (MET1) predominantly maintains CG DNA methylation, and a cross between met1 as pollen donor and the wild type activates otherwise repressed maternal-specific genes (Finnegan et al., 1996; Genger et al., 1999; Köhler et al., 2012). The FIS-PRC2 complex regulates endosperm development by installing a repressive chromatin mark, trimethylation of histone H3 at Lys-27 (H3K27me3), on several MADS box and other seed development-related genes (Huh et al., 2007; Folsom et al., 2014; Zhang et al., 2018). MADS box transcription factors (TFs), divided into type I and type II (or MIKC type), are known for their role in floral organ identity and flowering time control. The type II genes have been extensively studied as floral homeotic genes, while only a few type I genes have been functionally characterized (Bemer et al., 2010; Masiero et al., 2011; Smaczniak et al., 2012; Chen et al., 2016). In Arabidopsis (Arabidopsis thaliana), a well-studied type I MADS box TF, AGAMOUS LIKE62 (AGL62) shows specific syncytial phase expression that declines with the progression of cellularization; the agl62 mutant undergoes precocious endosperm cellularization (Kang et al., 2008). Several type I MADS box genes in Arabidopsis are implicated in the fis-prc2 mutants, which fail to undergo endosperm cellularization (Chaudhury et al., 1997; Grossniklaus et al., 1998; Köhler et al., 2003; Walia et al., 2009; Zhang et al., 2018). Interestingly, genes encoding the components of the FIS-PRC2 complex are also imprinted and prone to misregulation due to paternal excess (Luo et al., 2000, 2009; Jullien and Berger, 2010). Thus, normal seed development requires a multifaceted interplay between parental genome dosage, the imprinting of FIS-PRC2 components, and the role of FIS-PRC2 in the imprinting of downstream factors (Pires, 2014).
Here, we report the functional characterization of two rice type I MADS box genes, MADS78 and MADS79, that are active during early seed development. We report that MADS78 and MADS79 are essential for normal seed development. Misregulation of either gene results in seed abnormalities due to mistimed developmental transition of the endosperm and downstream impacts on starch/grain quality. At least one of the two MADS genes is required for rice seed viability, suggesting partial functional redundancy. Overall, our work shows that MADS78 and MADS79 regulate agronomically important yield traits, including grain size, fertility, and grain quality, in rice.
RESULTS
Expression Analysis of MADS78 and 79
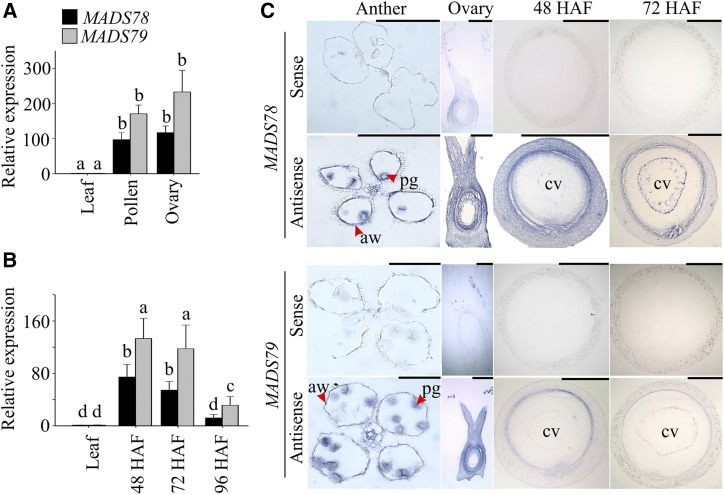
Based on phylogenetic analysis, type I MADS box genes are divided into three classes: Mα, Mβ, and Mγ (Parenicová et al., 2003). The rice genome has 32 type I MADS box proteins composed of 13 Mα, nine Mβ, and 10 Mγ proteins (Arora et al., 2007). Both MADS78 and MADS79 correspond to the Mα subclade and exhibit high sequence similarity (DNA, 83% and protein, 69%; Supplemental Fig. S1). The expression profile of these two genes in public databases was limited, as they were not represented on the microarray platforms. The RNA sequencing (RNA-seq)-based expression analysis also presents a challenge due to high sequence similarity, as one member is biased for mapping of most reads (Supplemental Table S1). Therefore, we determined the spatial and temporal expression patterns of MADS78 and MADS79 using reverse transcription quantitative PCR (RT-qPCR) assays and in situ hybridizations (Fig. 1). Both genes were preferentially expressed in pollen and ovary (Fig. 1A). Furthermore, tissue-specific expression patterns determined by in situ hybridization assays indicated that both genes were expressed in pollen grains as well as in anther walls (Fig. 1C). In developing seeds, the transcript abundance of both genes peaked at 48 h after fertilization (HAF), coinciding with the syncytium stage, where endosperm undergoes rapid nuclear division without forming cell walls (Fig. 1B). The transcript levels of both genes declined by 72 HAF and were significantly reduced at 96 HAF, which coincided with the onset and completion of endosperm cellularization, respectively (Fig. 1B). This decline in transcript levels was also confirmed via in situ hybridization assays where sense/antisense probes for both genes were used during each developmental stage (Fig. 1C). Furthermore, the preferential expression pattern of both genes during endosperm development was supported by the presence of regulatory motifs in their promoters (Supplemental Fig. S2). RY-element, Skn-1, and GCN4 motifs are known to establish seed- or endosperm-specific expression (Dickinson et al., 1988; Blackwell et al., 1994; Yoshihara et al., 1996).
Figure 1.
Expression analysis of MADS78 and MADS79. A and B, RT-qPCR analysis of MADS78 and MADS79 in mature pollen and unfertilized ovary (A) and early seed development (48, 72, and 96 HAF; B) relative to leaf tissue from wild-type plants. For statistical analysis, Student’s t test was used, and different lowercase letters indicate significant differences (P < 0.05). Error bars indicate sd (n = 6; two biological and three technical replicates). C, In situ hybridization of MADS78 and MADS79 in mature anthers, unfertilized ovary, and seeds at 48 and 72 HAF. aw, Anther wall; cv, central vacuole; pg, pollen grain. Bars = 0.2 mm.
Both MADS78 and 79 Heterodimerize with MADS87 and MADS89
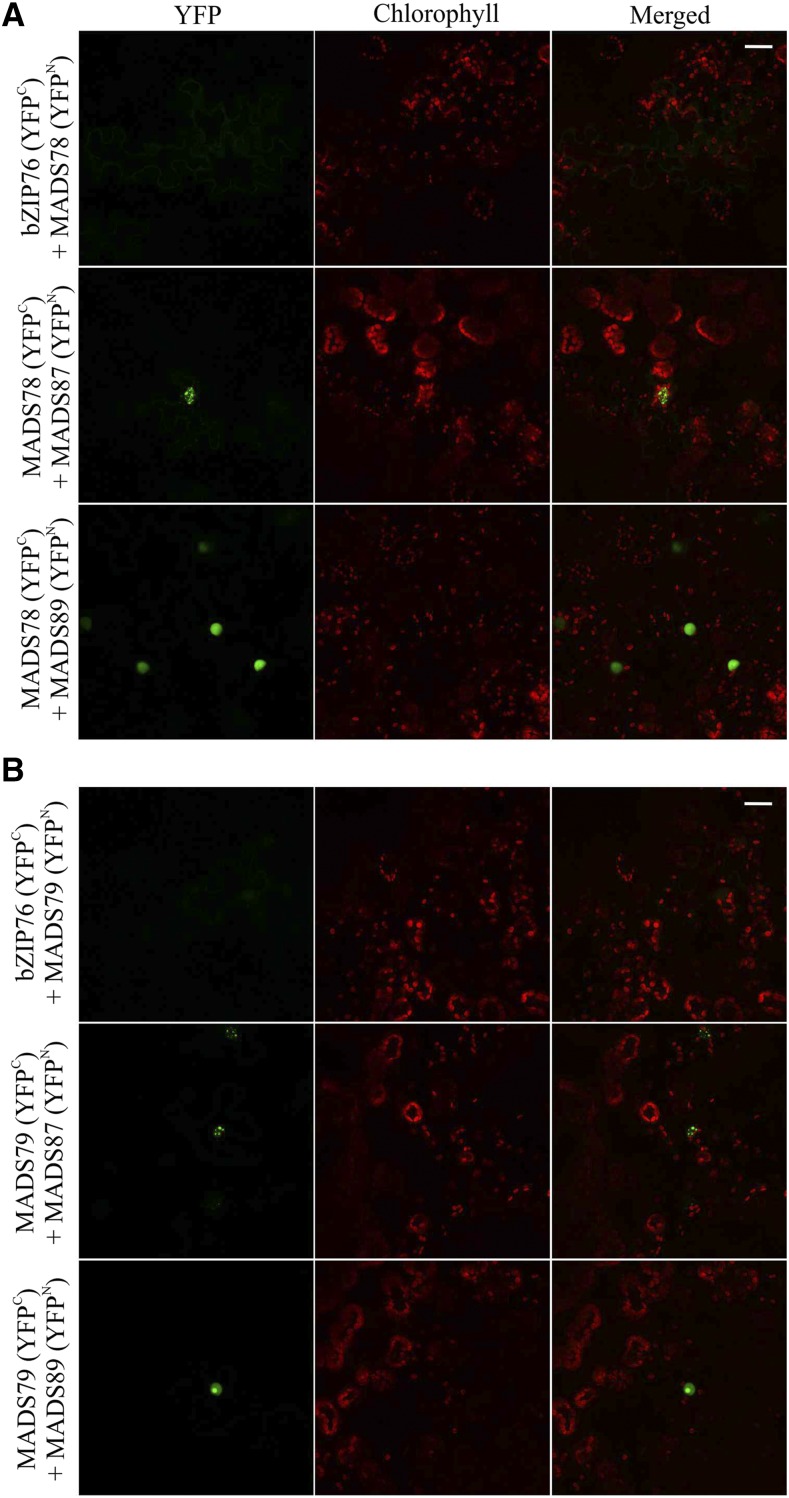
Type I MADS box proteins in Arabidopsis have a well-characterized interactome exhibiting extensive heterodimerization (de Folter et al., 2005). We next tested the nature and extent of interactions of MADS78 and MADS79 in rice. For this, we cloned the coding sequences (CDS) of 13 rice type I MADS box proteins from all three subclades: four Mα (MADS70, MADS77, MADS78, and MADS79), three Mβ (MADS90, MADS96, and MADS98), and six Mγ (MADS81, MADS83, MADS84, MADS87, MADS88, and MADS89). The resulting prey clones were cotransformed with MADS78 or MADS79 as bait in a bimolecular fluorescence complementation (BiFC) interaction assay. For controls, separate cotransformation of MADS78 and 79 was performed with an unrelated protein, bZIP76, as well as with an empty vector (Supplemental Fig. S3). Two Mγ member proteins, MADS87 and MADS89, showed positive interactions with both MADS78 and MADS79 (Fig. 2). Positive interactions were also observed when we cotransformed MADS87 or MADS89 with MADS78 or MADS79 by swapping the respective vectors to test the veracity of these interactions (Supplemental Fig. S4). Neither MADS78 nor MADS79 formed homodimers.
Figure 2.
Both MADS78 and MADS79 heterodimerize with MADS87 and MADS89. BiFC assay shows positive interaction of MADS78 (A) and MADS79 (B) with MADS87 and MADS89. For a control, we cotransformed both constructs with an unrelated gene (bZIP76). Bars = 25 µm.
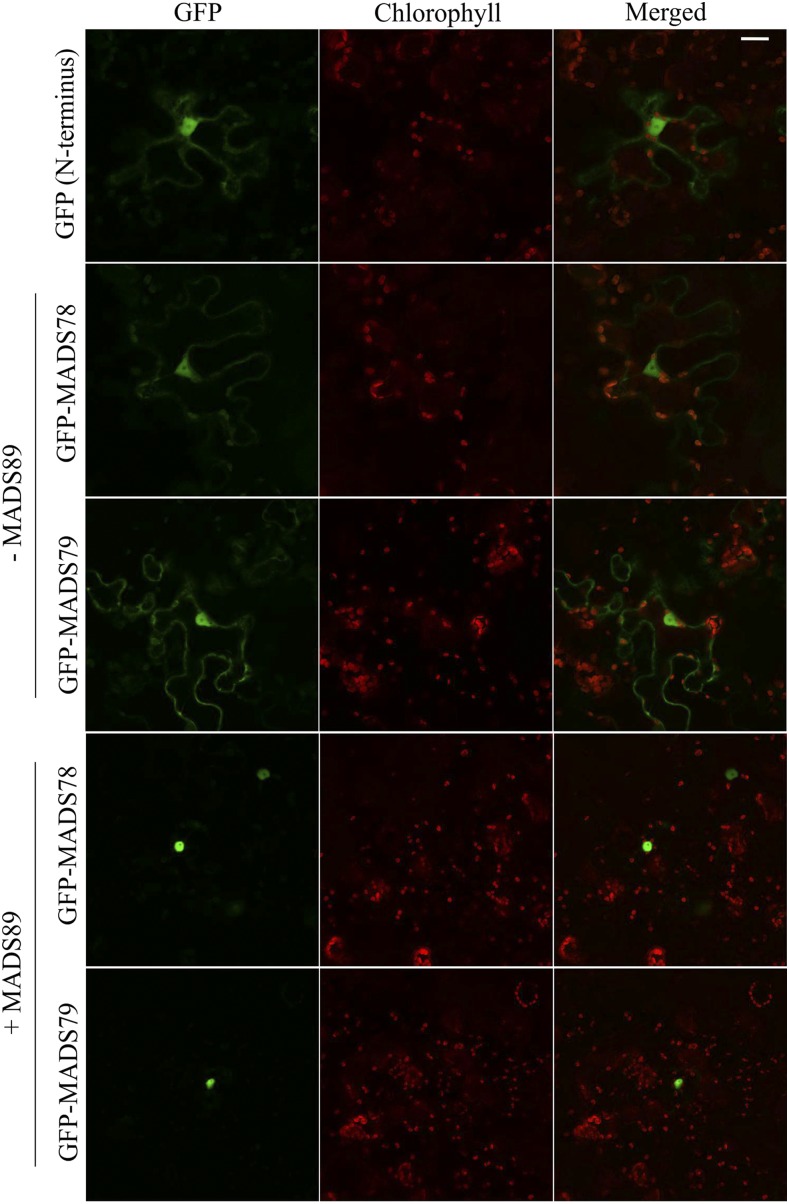
Since both MADS78 and MADS79 are TFs, they are expected to localize to the nucleus. To test this possibility, we fused GFP to the N or C terminus of the CDS for both genes. The N-terminal GFP fusion constructs of both genes localized to the nucleus as well as to the cytosol (Fig. 3). The C-terminal GFP fusion constructs for MADS78 localized exclusively to the nucleus, while MADS79 localized around chloroplasts (Supplemental Fig. S5A). In silico analysis revealed that neither gene carries a nuclear localization signal (NLS), suggesting that both genes likely require a recruiting factor or an anchor for nuclear transportation (Supplemental Table S2). Since both genes interact with the same Mγ proteins (MADS87 and MADS89; Fig. 2), we co-transformed the nonfluorescent BiFC construct (YFN43) encoding a positive interaction partner (MADS87 or MADS89) with MADS78 and MADS79 GFP fusions. When MADS78 (fused with GFP on its N terminus) was cotransformed with MADS89, the GFP-MADS78 fusion protein specifically localized to the nucleus (Fig. 3). Similarly, MADS79 fused with GFP on its N or C terminus and cotransformed with MADS89 specifically localized to the nucleus (Fig. 3; Supplemental Fig. S5A). However, cotransformation of MADS78 or MADS79 with the other positive interactor, MADS87, failed to alter their localization (Supplemental Fig. S5, B–D).
Figure 3.
MADS89 enhances the specificity of nuclear localization for MADS78 and MADS79. In the absence of MADS89, N-terminal GFP fusion constructs of MADS78 and MADS79 localize to nucleus and cytosol. Cotransformation of the respective constructs with MADS89 enhances their localization specifically to the nucleus. Empty vector was used as a control. Bar = 25 µm.
Overexpression of MADS78 and 79 Leads to High Spikelet Sterility and Delayed Cellularization
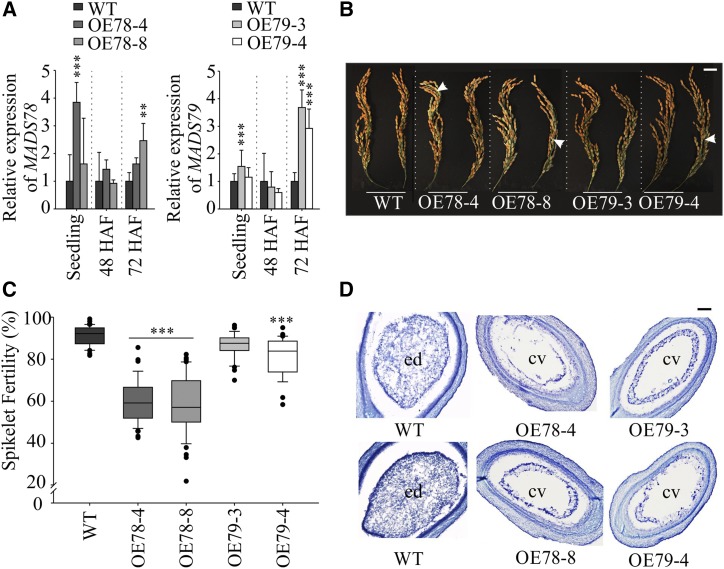
To functionally characterize MADS78 and MADS79, we generated ubiquitin-promoter-driven overexpression (OE) plant lines (OE78-4/OE78-8 and OE79-3/OE79-4 for MADS78 and MADS79, respectively; Fig. 4). We did not detect any phenotypic differences between OE and wild-type plants during the vegetative phase (Supplemental Fig. S6). However, we observed significant spikelet sterility in most of the OE lines for both genes compared with the wild type (Fig. 4, B and C). Seed sterility was more severe for OE78 compared with OE79 plants, where sterility was significantly higher only for one plant line (OE79-4) relative to the wild type (Fig. 4C). To determine the basis of seed sterility, we assayed the viability of mature pollen derived from OE plants. We did not find significant differences in pollen viability between OE and wild-type plant lines (Supplemental Fig. S7). Furthermore, careful inspection of the dehusked sterile seeds at maturity revealed flattened seed mass instead of aborted ovaries, suggesting postzygotic failure as the cause of the seed sterility phenotype (Supplemental Fig. S8). This observation prompted us to investigate postzygotic seed development during early stages (48, 72, and 96 HAF) when both MADS78 and MADS79 were preferentially expressed (Fig. 1B). We observed that developing seeds from OE lines could be categorized into two types: abnormal and normal. The abnormally developing seeds from the OE lines were distinguished by their smaller size relative to the normal-looking seeds, which were indistinguishable from the developing seeds of wild-type plants (Supplemental Fig. S9A). The percentage of normally developing seeds in the OE lines was significantly lower than in the wild type (Supplemental Fig. S9B). Consistent with the spikelet sterility phenotype, the lower percentage of normal developing seeds in the OE78 lines was much more severe than in the OE79 lines (Fig. 4C; Supplemental Fig. S9B). This suggested that the severity of sterility observed at maturity is determined during early seed development when both MADS78 and MADS79 are transcriptionally active. We also observed that fully developed mature seeds derived from the OE plant lines had significantly higher length-to-width ratios relative to wild-type seeds at maturity (Table 1). We observed decreases in 1,000-grain weight in both OE lines; however, the decrease was only significant for OE78 lines (Table 1). To further investigate the developmental defects caused by misregulation of these two genes, we examined cross sections of developing seeds from OE and wild-type plants. Seeds from wild-type plants completed endosperm cellularization by 96 HAF (Fig. 4D). However, seeds from OE MADS78 and MADS79 lines did not completely cellularize by 96 HAF. This suggests that the higher abundance of MADS78 or MADS79 and/or their ubiquitous expression causes delayed endosperm cellularization (Fig. 4D).
Figure 4.
Overexpression of MADS78 and MADS79 leads to partial spikelet sterility and delayed endosperm cellularization. A, RT-qPCR analysis of MADS78 and MADS79 expression in 11-d-old seedlings and developing seeds (48 and 72 HAF) in the wild-type (WT) and OE mutants for the respective tissue. Wild-type expression was used as a baseline for the corresponding tissue and gene. B, Representative mature panicle images of the OE mutants compared with the wild type. White arrows indicate sterile seeds. Images shown were digitally extracted and scaled for comparison. Bar = 2 cm. C, Quantification of spikelet fertility (%) in the wild type and OE mutants; n = 25 to 40 plants. D, Representative cross sections of developing seeds at 96 HAF from the wild type and OE mutants. cv, Central vacuole; ed, endosperm. Bar = 0.2 µm. For statistical analysis in A and C, Tukey’s test was used: ***P < 0.001 and **P < 0.01. Error bars indicate sd (n = 6; two biological and three technical replicates).
Table 1. Physiological and morphometric measurements of OE lines and single knockout mutants relative to the wild type.
Spikelet fertility, 1,000-grain weight, total tiller count, and total seeds per plant were measured at the whole-plant level (n = 25–50 plants). Area, perimeter, length, width, and length-to-width ratio were measured for individual seeds (n = 700–1,000). Tukey’s test was used for comparing the wild type and mutant lines: *, P < 0.05 and **, P < 0.001.
| Plant Line | 1,000-Grain Weight (g) | Tiller Count | Area (mm2) | Perimeter (mm) | Length (mm) | Width (mm) | Length-to-Width Ratio |
|---|---|---|---|---|---|---|---|
| Wild type | 21.60 ± 0.8 | 8.11 ± 1.0 | 12.13 ± 0.8 | 13.81 ± 0.5 | 5.05 ± 0.2 | 3.04 ± 0.1 | 1.65 ± 0.1 |
| OE78-4 | 19.09 ± 2.0* | 7.09 ± 0.7 | 11.57 ± 0.9** | 13.58 ± 0.6** | 5.01 ± 0.2* | 2.90 ± 0.1** | 1.72 ± 0.1** |
| OE78-8 | 19.22 ± 1.4* | 6.44 ± 1.0* | 13.00 ± 1.1** | 14.42 ± 0.6** | 5.36 ± 0.2** | 3.05 ± 0.1* | 1.75 ± 0.1** |
| OE79-3 | 20.18 ± 1.5 | 7.44 ± 1.0 | 11.34 ± 1.1** | 13.44 ± 1.0** | 4.97 ± 0.3** | 2.87 ± 0.2** | 1.73 ± 0.1** |
| OE79-4 | 19.65 ± 1.3 | 7.10 ± 1.2 | 11.97 ± 1.0 | 14.11 ± 0.6** | 5.38 ± 0.2** | 2.79 ± 0.1** | 1.93 ± 0.1** |
| mads78_17 | 19.17 ± 1.7** | 6.63 ± 1.0 | 12.00 ± 1.0* | 13.66 ± 0.7* | 4.97 ± 0.3* | 3.05 ± 0.01 | 1.62 ± 0.1 |
| mads78_19 | 21.95 ± 1.2 | 6.90 ± 1.3 | 12.38 ± 0.8 | 13.99 ± 0.5* | 5.14 ± 0.2** | 3.04 ± 0.01 | 1.69 ± 0.1** |
| mads79_23 | 21.74 ± 0.8 | 7.90 ± 1.2 | 12.45 ± 1.0* | 14.01 ± 0.6* | 5.17 ± 0.2** | 3.06 ± 0.01 | 1.68 ± 0.1** |
| mads79_24 | 19.40 ± 0.5** | 7.80 ± 0.8 | 10.92 ± 1.1** | 13.05 ± 0.7** | 4.79 ± 0.3** | 2.90 ± 0.01** | 1.65 ± 0.1 |
Single Knockout Mutants of MADS78 or MADS79 Show Precocious Endosperm Cellularization
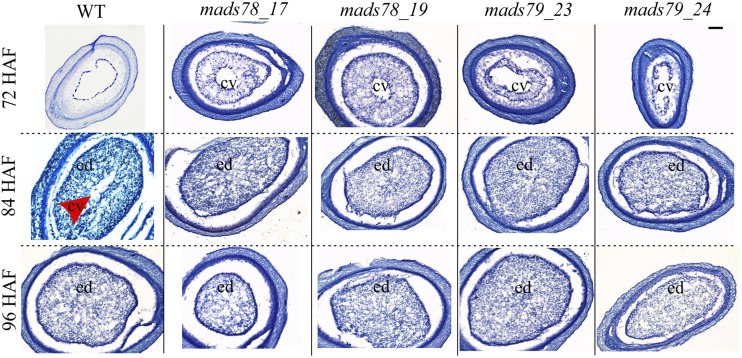
For each of the genes, we selected two homozygous single mutants (MADS78, mads78_17 and mads78_19; MADS79, mads79_23 and mads79_24). Both mutant lines corresponding to MADS78 had deletions in the targeted regions and resulted in truncated transcripts due to premature stop codons (Supplemental Fig. S10A). MADS79 mutants, mads79_23 and mads79_24, carried an insertion of adenine and a 4-bp deletion, respectively (Supplemental Fig. S10A). Neither of the MADS79 mutations led to premature stop codons. RT-qPCR analysis showed a decline in transcript abundance of MADS78 and MADS79 in the mutants relative to the wild type at 48 HAF (Supplemental Fig. S10, B and C). At reproductive maturity, we did not observe severe spikelet sterility in the mutants compared with the wild type (Supplemental Fig. S10, D and E). Like the OE lines, one single knockout mutant line of both genes (mads78_19 and mads79_23) showed a significant increase in length-to-width ratio of mature seeds relative to the wild type (Table 1). We detected significant decreases in 1,000-grain weight in single knockout lines of both MADS78 and MADS79 (Table 1). Next, we performed histochemical analysis of developing seeds to check the timing of endosperm cellularization. The knockout mutants exhibited a faster rate of endosperm cellularization relative to the wild type (Fig. 5). As a result, knockout mutants underwent complete endosperm cellularization by 84 HAF, whereas the wild type completed it by 96 HAF (Fig. 5). This suggests that seeds deficient in either of the two genes exhibit precocious endosperm cellularization.
Figure 5.
Single knockout mutants of MADS78 and MADS79 show precocious cellularization. Representative cross sections of developing seeds are shown at 72, 84, and 96 HAF in single knockout mutants of MADS78 and MADS79 compared with the wild type (WT). The red arrowhead points to the central vacuole region (cv). ed, Endosperm. Bar = 0.2 µm.
The Double Knockout Mutation for MADS78 and 79 Is Lethal
Given the high percentage of sequence similarity and overlap in MADS78 and MADS79 gene activity during rice reproductive and seed development, we next examined the combined role of MADS78 and MADS79. For this, we generated CRISPR-Cas9-mediated double knockout mutants. We obtained two homozygous T0 mutants corresponding to each of the single guide RNAs (sgRNAs): sgRNA-1 (mads78-79_5 and mads78-79_9) and sgRNA-2 (mads78-79_3 and mads78-79_4; Supplemental Fig. S11A). Both mads78-79_5 and mads78-79_9 double mutants (sgRNA-1) had a homozygous point mutation within both genes (insertion of adenine). In addition, mads78-79_9 possessed a substitution (guanine to thymine) in both alleles of MADS78 (Supplemental Fig. S11A). The mads78-79_3 (sgRNA-2) double knockout mutants had a homozygous insertion of an adenine in both genes, while mads78-79_4 had an insertion of thymine and guanine in MADS78 and MADS79, respectively (Supplemental Fig. S11A). All double knockout mutant lines were completely sterile, as they failed to develop mature seeds (Fig. 6; Supplemental Fig. S11B). On the other hand, we obtained seed from double knockout mutants that were heterozygous for one or both genes (Supplemental Fig. S11C). These data indicated that at least one functional gene (MADS78 or MADS79) is required for seed viability in rice.
Figure 6.
Double knockout mutation is lethal. A, Representative mature panicle images of T0 homozygous double knockout mutants compared with the wild type (WT). mads78-79_5 and mads78-79_9 are derived from sgRNA-1, while mads78-79_3 and mads78-79_4 correspond to sgRNA-2 (Supplemental Fig. S11). Bar = 2 cm. B, Representative mature seed images of the T0 homozygous double knockout mutants with husk. Bar = 1 cm. Images shown in A and B were digitally extracted and scaled for comparison.
Expression of MADS78 and MADS79 Is Negatively Associated with Fie1
Several type I MADS box genes in Arabidopsis (Walia et al., 2009; Zhang et al., 2018; Bjerkan et al., 2019) and rice (Folsom et al., 2014; Chen et al., 2018; Wang et al., 2018) are imprinted. Therefore, we examined the DNA methylation status of MADS78 and MADS79. McrBC (an endonuclease that cleaves DNA containing methylcytosine) assay indicated that promoter regions of MADS78 and 79 are methylated in the leaf tissue and developing seed (48 HAF; Supplemental Fig. S12, A and B). Furthermore, bisulfite sequencing confirmed methylation in the promoter region for MADS78 and the gene body for MADS79 in developing seeds (24, 48, 72, and 96 HAF; Supplemental Fig. S12C). No significant change in CG, CHG, or CHH methylation levels for either gene were detected in developing seeds (Supplemental Fig. S12C). Thus, the methylation status did not associate with changes in the gene expression profile of MADS78 and MADS79 (Fig. 1), suggesting that DNA methylation at the sites assayed is unlikely to play a direct regulatory role for these two genes.
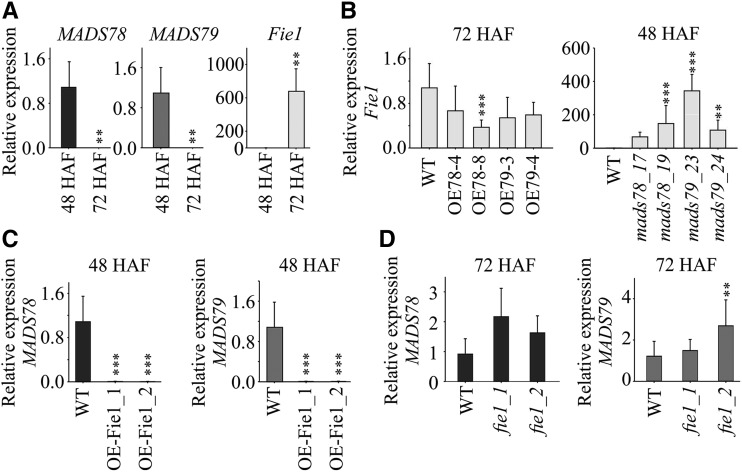
The FIS-PRC2 complex regulates imprinting by installing histone methylation marks on the target genes (Folsom et al., 2014). In this framework, we asked if Fie1, an essential component of the FIS-PRC2 complex, is a potential regulator of MADS78 and MADS79. The transcript abundance of Fie1 was negligible at 48 HAF but accumulated to significant levels by 72 HAF (Fig. 7A). Conversely, the transcript levels of MADS78 and MADS79 were higher at 48 HAF and declined considerably by 72 HAF in normally developing seeds (Fig. 7A). We next examined the expression of Fie1 in MADS78 and MADS79 mutant lines. Fie1 exhibited lower transcript abundance at 72 HAF in the OE lines compared with the wild type (Fig. 7B). The single knockout mutants of MADS78 and MADS79 accumulated more Fie1 transcripts at 48 HAF relative to the wild type (Fig. 7B). Moreover, in OE Fie1 mutants, the transcripts of both MADS78 and MADS79 were not detected at 48 HAF, when otherwise they were abundantly expressed in wild-type seeds (Fig. 7C). Also, expression of MADS78 and MADS79 was up-regulated at 72 HAF in knockout mutants of Fie1 (Fig. 7D). The expression analysis of Fie1 in MADS78 and MADS79 mutants and expression of MADS78 and MADS79 in Fie1 mutants establishes an inverse transcriptional association between MADS78 and MADS79 and Fie1. Collectively, our data suggest that MADS78 and MADS79 regulation is likely linked to the abundance of Fie1 and potentially mediated by the FIS-PRC2 complex.
Figure 7.
FIS-PRC2 potentially regulates MADS78 and MADS79. A, RT-qPCR analysis of MADS78, MADS79, and Fie1 at 48 and 72 HAF in the wild type (WT). Expression levels at 48 HAF were considered as baseline for each gene. B, RT-qPCR analysis of Fie1 in OE and single knockout mutants for MADS78 and MADS79 at 72 and 48 HAF, respectively. C and D, RT-qPCR analysis of MADS78 and MADS79 in OE and single knockout mutants for Fie1 at 48 and 72 HAF, respectively. For B to D, wild type expression was used as a baseline. For statistical analysis, the Holm-Sidak method was used: ***, P < 0.001 and **, P < 0.01. Error bars indicate sd (n = 6; two biological and three technical replicates).
Misregulation of MADS78 and/or 79 Disturbs Auxin Homeostasis
To further explore the basis of the observed phenotypes with respect to misregulated endosperm cellularization, we performed transcriptome analysis of developing seed from mutants (48 and 72 HAF; Supplemental Table S3). We searched the transcriptome data set for genes that were (1) preferentially activated during the syncytial stage (48 HAF) and down-regulated with the onset of cellularization (72 HAF; syncytial-associated genes) and (2) suppressed during the syncytial stage but activated with the progression of endosperm cellularization, referred to here as cellularization-associated genes (Folsom et al., 2014; Chen et al., 2016). As expected, OE mutants exhibiting delayed cellularization had higher transcript abundance for syncytium-associated genes at 48 and/or 72 HAF (Supplemental Table S3). Conversely, single knockout mutants with precocious cellularization exhibited increased transcript abundance of cellularization-associated genes at 72 HAF relative to the wild type (Supplemental Table S4).
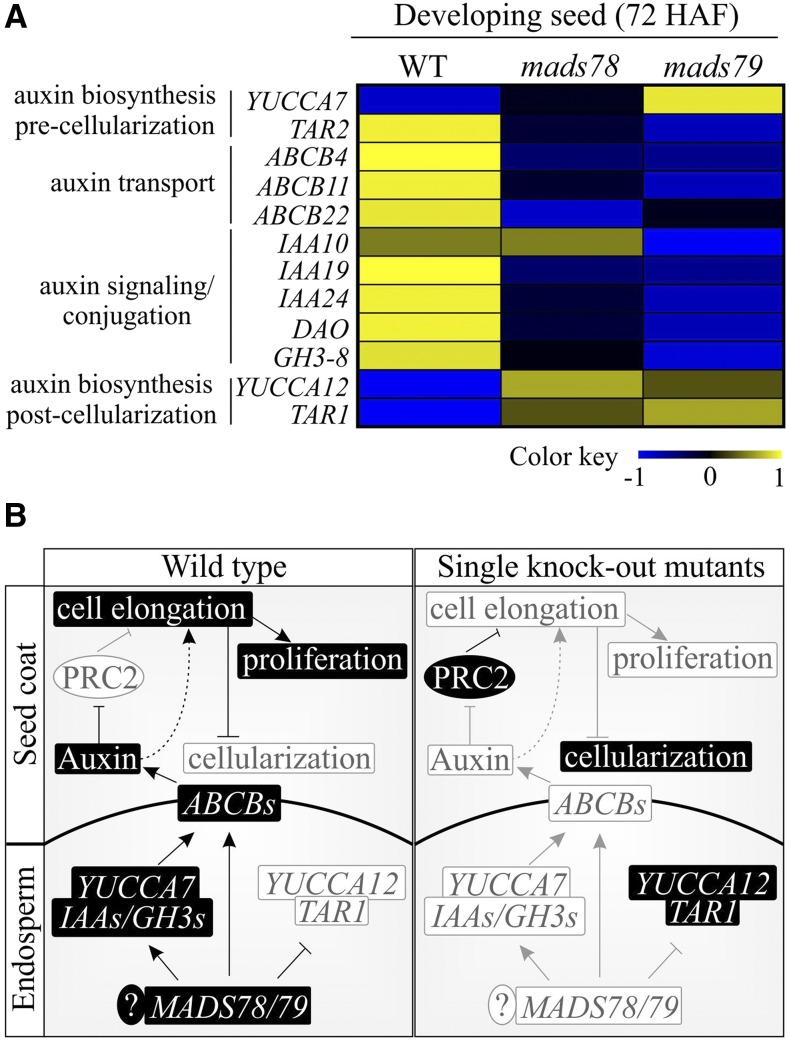
Auxin (indole-3-acetic acid [IAA]) plays an important role in the initiation of endosperm development and its maintenance during the proliferative phase in Arabidopsis (Figueiredo et al., 2015; Batista, 2019). Auxin synthesized in endosperm is exported to integuments via the ABCB-type transporter, P-GLYCOPROTEIN10 (PGP10; Figueiredo et al., 2016). Auxin blocks the PRC2 complex and thus facilitates cell elongation and differentiation. Seeds deficient in AGL62 have reduced auxin export due to suppressed PGP10, which releases PRC2 suppression. The activated PRC2 in turn inhibits cell elongation and differentiation leading to precocious cellularization (Figueiredo et al., 2016; Batista et al., 2019a). Since MADS79 is one of the orthologs of AGL62 and knocking out MADS78 or MADS79 accelerates cellularization, we explored the link between auxin homeostasis and MADS78 and MADS79 in our transcriptome data. For this, we combined the transcriptome analysis with a search for potential target genes of MADS78 and/or MADS79 (Supplemental Table S5). The CArG box motif is a signature binding motif for MADS box TFs (both type I and II TFs; Batista et al., 2019b). The rice ABCB-type transporter, ABCB4, which carries a CArG box-like DNA-binding motif in its promoter, showed lower transcript abundance in the absence of MADS78 or MADS79 compared with the wild type at 72 HAF (Fig. 8A). Likewise, other ABCB-type transporters, such as ABCB11, ABCB14, and ABCB22 (orthologs of Arabidopsis PGP10), also had lower transcript levels in single knockout mutants, while expression in OE lines was unaltered (Fig. 8A; Supplemental Table S5). Genes responsible for maintaining auxin levels either by oxidation (DIOXYGENASE FOR AUXIN OXIDATION [DAO]; Zhao et al., 2013) or amino acid conjugation (GRETCHEN HAGEN3 [GH3]; Jain and Khurana, 2009) also showed lower transcript abundance in single knockouts at 72 HAF (Fig. 8A; Supplemental Table S5). Furthermore, auxin signaling genes (IAA10, IAA19, and IAA24) exhibited lower transcript abundance in single knockout mutants at 72 HAF (Fig. 8A; Supplemental Table S5). Previous work has shown that ABCB4/11/14/22, DAO, GH3s, and IAAs are up-regulated in developing rice seeds at 24 HAF (Jain and Khurana, 2009; Uchiumi and Okamoto, 2010; French et al., 2014; Basunia and Nonhebel, 2019). TRYPTOPHAN AMINOTRANSFERASE-RELATED PROTEIN2 (TAR2), the auxin biosynthesis gene that is preferentially expressed prior to cellularization (Abu-Zaitoon et al., 2012), exhibited lower transcript levels in MADS78 and MADS79 mutants, while YUCCA7 was unaltered in the MADS78 mutant and showed higher transcript abundance in the MADS79 mutant (Fig. 8A). On the other hand, auxin biosynthesis genes (YUCCA12 and TAR1) preferentially expressed in postcellularization seeds (Abu-Zaitoon et al., 2012; Basunia and Nonhebel, 2019) exhibited higher transcript abundance in single knockouts at 72 HAF (Fig. 8A; Supplemental Table S5). YUCCA12 and TAR1 promoters have CArG box-binding motifs (Supplemental Table S5). These changes in transcript abundance and the presence of CArG box-binding motifs in promoters of auxin biosynthesis, transporters, and signaling-related genes (Supplemental Table S5) suggest a regulatory role for MADS78 and MADS79 in maintaining auxin levels in developing rice seeds (Fig. 8).
Figure 8.
Misregulation of MADS78 and/or MADS79 disturbs auxin homeostasis. A, Heat map based on normalized read counts for genes involved in auxin homeostasis in the wild type (WT) and single knockout mutants (mads78 and mads79) in developing seeds (72 HAF). The color key represents a range of normalized values. B, MADS78 and MADS79, auxin, and FIS-PRC2 collectively regulate the timely initiation of endosperm cellularization. MADS78 and MADS79 regulate some of the auxin biosynthesis (YUCCAs), signaling (IAAs and GH3s), and transport (ABCB-type transporters) related genes in developing endosperm. In normally developing syncytial stage seeds, auxin is exported from developing endosperm to the seed coat and blocks FIS-PRC2, thus ensuring normal endosperm proliferation and seed coat cell elongation. Seeds deficient in MADS78 and MADS79 have reduced auxin export, thus releasing auxin-mediated FIS-PRC2 suppression. The activated FIS-PRC2 blocks cell elongation and differentiation, which causes cellularization. The accelerated cellularization triggers the activation of auxin biosynthesis genes YUCCA12 and TAR1 that may promote endoreduplication, differentiation, and starch deposition during later stages of seed development. ? indicates additional, uncharacterized factors that might participate in this model.
Misregulation of MADS78 and/or 79 Alters Carbon Metabolism
We observed a chalky grain appearance for mature seeds of OE lines and knockout mutants (Fig. 9A). Further investigation via scanning electron microscopy (SEM) revealed structural abnormalities in the starch granules of the mutants. In contrast to densely packed polygonal starch granules in the wild type, mutants possessed loosely packed spherical starch granules resulting in larger openings between granules (Fig. 9A). Quantification of total starch content from mature seed as well as developing seed (10 DAF) did not show a significant difference between the mutants and the wild type (Supplemental Fig. S13A). However, we observed higher total protein content for developing seed in OE and single knockout mutants relative to wild-type seeds (Supplemental Fig. S13B). Abnormal starch granule packaging and higher total protein content are likely related to altered carbon and nitrogen metabolism. To investigate the metabolic status in the developing seeds (10 DAF), we performed metabolite profiling by gas chromatography (GC) coupled to mass spectrometry. Alterations in sugar metabolism were observed as evidenced by significant elevations in Glc, Fru, and myoinositol in OE lines and knockout mutants relative to the wild type (Supplemental Table S6). Taken together, our results suggest impaired metabolism of carbohydrates in the developing seeds of the mutant lines.
Figure 9.
Misregulation of MADS78 and MADS79 causes impairment of starch granules. A, Representative images of mature seeds from MADS78 and MADS79 mutants (left column; bar = 1 cm) and their cross sections (right column; bar = 20 µm) using SEM. Images shown were digitally extracted and scaled for comparison. B, RT-qPCR analysis of selected genes involved in carbon metabolism at 72 HAF in OE and knockout mutants relative to the wild type (WT). The genes were selected based on RNA-seq analysis. For statistical analysis, Student’s t test was used: ***, P < 0.001; **, P < 0.01; and *, P < 0.05. Error bars indicate sd (n = 6; two biological and three technical replicates).
Genes involved in starch biosynthesis exhibited differential transcript abundance between the genotypes (Supplemental Table S7). We validated the expression of a selected subset of these genes by RT-qPCR (Fig. 9B). In this context, the seed-specific isoform of Suc synthase (Susy3), the two subunits of ADP-Glc pyrophosphorylase (AGPS2b and AGPL2), and its transporter BRITTLE1 showed higher transcript abundance in knockout mutants at 72 HAF, while the transcript levels were unaltered in OE lines (Fig. 9B; Supplemental Table S7). The granule-bound starch synthase (GBSSI), which is involved in amylose biosynthesis (Waxy), and its upstream regulators, NF-YB1, exhibited higher transcript levels in knockout mutants at 72 HAF (Fig. 9B; Supplemental Table S7). One of the amylopectin biosynthesis genes (starch synthase [SSIIa]), cytosolic pyruvate orthophosphate dikinase (PPDK), and the known regulators for seed storage proteins, rice basic Leu zipper (RISBZ1/bZIP58) and rice prolamin box binding factor (RPBF), were also altered. These genes showed higher transcript abundance in the developing seeds (72 HAF) of OE lines and knockout mutants (Fig. 9B; Supplemental Table S7). Collectively, our results indicate that misregulation of MADS78 and MADS79 during early seed development results in compromised seed quality.
DISCUSSION
Functional characterization of MADS78 and MADS79 revealed six key findings: (1) misregulation of the genes by either OE or knockout leads to delayed or precocious endosperm cellularization, respectively; (2) the double knockout mutant is not viable, thereby suggesting an indispensable role for MADS78 and MADS79 in rice seed development; (3) MADS78 and MADS79 physically interact with MADS89, which enhances their specificity to localize in the nucleus; (4) MADS78 and MADS79 are potentially regulated by the FIS-PRC2 complex; (5) MADS78 and MADS79, auxin, and FIS-PRC2 are important regulators of the timely transition of endosperm from syncytium to cellularization; and (6) misregulation of MADS78 and/or MADS79 alters grain quality and metabolism.
Functional characterization of MADS78 and MADS79 indicates that both genes are involved in regulating the rate of endosperm cellularization. Seeds deficient in either of the two genes exhibit slightly precocious cellularization (Fig. 5), and overabundance of either gene results in delayed cellularization (Fig. 4D). Knockout and OE lines of both genes do not arrest the initiation of cellularization but likely determine the duration required to complete cellularization. Seeds that carry mutations in both MADS78 and MADS79 are nonviable, indicating that at least one of these two genes is required for producing viable rice seeds (Fig. 6). This also suggests that MADS78 and MADS79 have partial functional redundancy. OE of MADS78 has a greater penalty on spikelet fertility compared with MADS79 (Fig. 4C). Although MADS78 and MADS79 do not dimerize with each other, their nuclear localization is enhanced when they heterodimerize with MADS89 (Fig. 3). We found gene expression of Fie1, a rice FIS-PRC2 member, to be negatively associated with the abundance of MADS78 and MADS79 transcripts in knockout and OE lines, suggesting a possible link between MADS78, MADS79, and Fie1 in regulating the rate of endosperm cellularization (Fig. 7).
MADS89 Enhances Nuclear Localization of MADS78 and MADS79
Both MADS78 and MADS79 belong to the Mα subclade and heterodimerized with two Mγ proteins, MADS87 and MADS89. Furthermore, RNA-seq analysis revealed that both MADS87 and MADS89 coexpressed with MADS78 and MADS79 (Supplemental Table S2). No homodimers were observed for either MADS78 or MADS79. In Arabidopsis, MADS box partnerships are limited to subclass interactions, as interclass interactions (e.g. between type I and type II MADS) have not been reported (de Folter et al., 2005). Furthermore, interactions within a specific subclade of type I proteins (Mα, Mβ, and Mγ) are rare, and members of a subclade mainly interact with a protein corresponding to another subclade. Mα proteins dimerize preferentially with members of Mβ and Mγ, and only a few interactions exist between Mβ and Mγ. Thus, it is proposed that Mα proteins are central components stabilizing higher order protein complexes (Immink et al., 2009). The necessity of at least one of MADS78 and MADS79 being functional is indicated by the complete sterility of double knockout mutants. Physical interactions of both MADS78 and MADS79 with MADS89 suggest partial functional redundancy. One possible role for this interaction could be to enhance the nuclear localization of MADS78 and MADS79 in the presence of MADS89. Both MADS78 and MADS79 lack a clear NLS (Supplemental Table S2). The retargeting experiment suggests that MADS78 and MADS79 require MADS89 and not MADS87 to increase the specificity of their localization to the nucleus (Fig. 6; Supplemental Fig. S4). The in silico analysis indicates that MADS89 carries an NLS on its N terminus, whereas MADS87 lacks an NLS (Supplemental Table S2). Likewise, in Arabidopsis, AGL62 (an ortholog of MADS79) heterodimerizes with AGL80 and AGL61, which are orthologs of MADS89 and MADS78, respectively (de Folter et al., 2005). Both AGL80 and AGL61 play crucial roles in the initiation of endosperm development (de Folter et al., 2005; Portereiko et al., 2006; Steffen et al., 2008).
Misregulation of MADS78 and MADS79 Affects Early Seed Development
MADS78 and MADS79 are preferentially expressed during the syncytial stage of endosperm development, and transcript levels decline as the endosperm cellularizes. Overexpressing either gene prevents the decline in their transcript pools at 72 HAF (Fig. 4A). Higher abundance of MADS78 or MADS79 in OE lines is concomitant with delayed cellularization in seeds that reach maturity. Seeds from wild-type plants are cellularized by 96 HAF. It is noteworthy that MADS78 and MADS79 OE seeds have a range of phenotypes. While some seeds reach maturity, others can be scored as abnormal as early as 48 HAF (before cellularization; Supplemental Fig. S9). These abnormal seeds are randomly distributed on the panicle. The development of individual seeds is partially independent of other seeds. Therefore, we hypothesize that certain developing seeds experience relatively higher syncytial activity than others and thus fail to cope with this abnormal developmental pace, eventually resulting in seed abortion. Seeds that abort during the early developmental stage contribute to the increased sterility of mature panicles. On the other hand, some seeds endure the abnormal developmental progression but get penalized with respect to delayed endosperm cellularization. Delayed cellularization in a subset of OE seeds could be due to (1) delayed transition from syncytium to cellularization of the endosperm and/or (2) reduced rate of cellularization because of overabundance of MADS78 and MADS79. Knockout mutants of MADS78 or MADS79 show precocious cellularization but do not impact seed viability (Fig. 5; Supplemental Fig. S10, D and E). These observations are consistent with the reported role of AGL62 (an ortholog of MADS79 in Arabidopsis) in endosperm cellularization (Kang et al., 2008; Bemer et al., 2010). agl62 mutant seeds show precocious cellularization, thus establishing its role in suppressing endosperm cellularization (Kang et al., 2008). Plants with mutations in both genes do not produce viable seed, indicating that at least one of these two genes should have a functional allele to produce viable seeds in rice (Fig. 6; Supplemental Fig. S11).
FIS-PRC2 Potentially Regulates MADS78 and MADS79
Normal endosperm development is tightly controlled by genome dosage, as paternal excess leads to endosperm overproliferation, while increase in maternal dosage results in precocious endosperm cellularization (Scott et al., 1998; Stoute et al., 2012). FIS-PRC2 is one of the key components that aids in maintaining genome dosage by regulating seed-specific type I MADS box genes (Folsom et al., 2014). In this context, Fie1, which itself is regulated by repressive histone methylation (H3K9me2), regulates type I MADS box genes (MADS82, MADS87, and AGL36) by H3K27me3 histone methylation in rice (Folsom et al., 2014). The expression of type I MADS box genes MADS78 and MADS79 is negatively associated with Fie1 (Fig. 7). Overexpressing either MADS78 or MADS79 prolongs the syncytium stage, thereby delaying the induction of Fie1 expression (Fig. 7B). On the other hand, in single knockout mutants of MADS78 and MADS79, Fie1 transcripts accumulate at 48 HAF while remaining absent in the wild type (Fig. 7B). Furthermore, expression of MADS78 and MADS79 is repressed at 48 HAF in the mutant lines overexpressing Fie1, whereas their expression is temporally extended in Fie1 knockout mutants (Fig. 7, C and D). Our results are consistent with previous work that reported an up-regulation of AGL62 (an ortholog of MADS79) in PcG mutants of Arabidopsis (Kang et al., 2008).
Interplay between Auxin, Type I MADS Box TFs, and FIS-PRC2 Ensures Timely Endosperm Cellularization
We detected CArG box-binding motifs in the promoter of an auxin transporter, ABCB4 (Supplemental Table S5), which exhibits lower transcript abundance in knockout mutants (Fig. 8A). Moreover, auxin biosynthesis (TAR2) and signaling (IAAs and GH3s) genes, which are preferentially expressed before cellularization (Abu-Zaitoon et al., 2012), also showed lower transcript abundance in knockout mutants, resulting in altered auxin homeostasis (Fig. 8). This could lead to decreased auxin export, which relieves FIS-PRC2 suppression in the seed coat (Figueiredo et al., 2016), as evidenced by increased transcript abundance of Fie1 in knockout mutants (Fig. 7B). The activated FIS-PRC2 likely blocks cell elongation and differentiation, which induces precocious cellularization (Batista et al., 2019a). Early cellularization in the absence of MADS78 or MADS79 triggers activation of the CArG box-containing auxin biosynthesis genes YUCCA12 and TAR1 (Fig. 8A) that may have a role in subsequent endoreduplication, differentiation, and starch deposition during later stages of seed development (Abu-Zaitoon et al., 2012; Basunia and Nonhebel, 2019). Detection of CArG box-binding motifs in promoters of auxin biosynthesis enzyme-, transporter-, and signaling-related genes further supports the regulatory role of type I MADS box TFs in maintaining auxin levels and/or distribution (Supplemental Table S5). Collectively, our findings present evidence that type I MADS box TFs (MADS78 and MADS79 and potentially other type I genes), in conjunction with auxin homeostasis and Fie1, regulate the timely transition of endosperm from syncytial stage to cellularization, which is necessary for normal seed development (Fig. 8B).
Misregulation of Early Seed Development Alters Grain Quality Traits
Timely transition of endosperm from syncytial to cellularization ensures optimal nutritional quality of the seed, which is also dependent upon successful import of photoassimilates from source tissues (Aguirre et al., 2018). Misregulation of MADS78 and/or MADS79, associated with mistimed endosperm cellularization, also causes structural abnormalities in kernel starch (Fig. 9A). The starch biosynthesis genes were either unchanged or down-regulated in OE lines and up-regulated in single knockout mutants (Fig. 9B; Supplemental Table S7). This alteration implied that OE mutants exhibiting delayed cellularization postpone the transition of developing seed from the prestorage phase to the storage phase, while single knockout mutants with precocious cellularization reach the storage phase earlier. The transcript level changes for starch biosynthesis genes did not corroborate with starch quantity; however, abnormal starch granule structures were observed (Fig. 9A). This is not entirely unexpected, because the starch granule structure is determined not only by starch content but also by protein content, protein-starch interactions, amylose-lipid complexes, and short and long chains of amylopectin, which collectively define cooking and milling qualities of rice (Singh et al., 2007; Chen et al., 2012). The discrepancy between starch quantity and related gene expression is linked to the cytosolic (cy) isoform of PPDK, which is preferentially expressed in endosperm, especially during syncytial/endosperm cellularization, and down-regulated during subsequent stages by posttranslational mechanisms (Kang et al., 2005; Chastain et al., 2006). cyPPDK catalyzes a reversible reaction between pyruvate and phosphoenolpyruvate, thereby modulating carbon metabolism in favor of amino acid or lipid biosynthesis (Aoyagi and Bassham, 1984; Kang et al., 2005). In our study, cyPPDK showed higher transcript abundance in single knockout mutants (Fig. 9B; Supplemental Table S7). These results indicate that carbon influx is possibly directed toward either amino acid or lipid biosynthesis. Higher transcript abundance for SSIIa, RISBZ1/bZIP58, and RPBF was detected for both OE lines and knockout mutants (Fig. 9B; Supplemental Table S7). Since normal starch deposition and endosperm development are impaired in both OE lines and knockout mutants, it is plausible that SSIIa and potentially other genes are misregulated (at the transcript level) as a consequence of abnormal seed development triggered by overabundance or deficiency of MADS78 and MADS79. It is important to note that in normally developing seeds, MADS78 and MADS79 transcript abundance declines as endosperm initiates starch biosynthesis in nuclei that have cellularized.
CONCLUSION
MADS78 and MADS79 play an important role in regulating rice seed size and shape. Their misregulation is associated with temporal defects in early seed development and abnormal starch deposition. Since rice yield and milling quality are significantly impacted by unfavorable environments, such as drought and temperature stress, it will be useful to explore if MADS78 and MADS79 could be mediating the stress sensitivity of rice yield and quality traits in future research.
MATERIALS AND METHODS
Protein Sequence Alignment and Promoter Analysis
Protein sequences for MADS78 (LOC_Os09g02830) and MADS79 (LOC_Os01g74440) were downloaded from RGAP7. The SMART database (Letunic et al., 2012) was used to predict domains. The T-COFFEE program was used for aligning protein sequences of MADS78 and MADS79 (Di Tommaso et al., 2011). For promoter analysis, the regions 2 kb upstream of the transcription start sites of both MADS78 and MADS79 were downloaded from RGAP7 and analyzed using the PlantCARE database (Lescot et al., 2002).
Generation of Transgenic Plants
For generating overexpression constructs, full-length CDS corresponding to MADS78 and MADS79 were amplified. OE mutants of Fie1 (LOC_Os08g04290) were considered from Folsom et al. (2014). The amplified products were cloned into pENTR/D-TOPO entry vector (Invitrogen). PCR products were moved from entry to destination vector (Ubi-NC1300) using LR clonase (Invitrogen). The final constructs were transformed into Agrobacterium tumefaciens strain EHA105. Calli from rice (Oryza sativa ‘Kitaake’) were transformed as described by Cheng et al. (1998). Hygromycin was used as a selective agent to screen the transformed calli. To generate CRISPR-Cas9 mutants, sgRNAs were designed using CRISPR-P 1.0 (http://crispr.hzau.edu.cn/CRISPR/) to specifically target either MADS78 or MADS79 and Fie1 (Lei et al., 2014). Considering the high percentage of sequence similarity between MADS78 and MADS79, sgRNAs were designed to target nonconserved genic regions. For targeting MADS78, an sgRNA that targeted a region downstream of the coiled-coil domain was selected. An sgRNA targeting a region between the MADS and coiled-coil domains was designed for MADS79 (Supplemental Fig. S10A). A list of potential off-targets corresponding to genic regions (exonic or intronic) for each sgRNA is provided in Supplemental Table S8. To create double knockout constructs, we designed two sgRNAs (sgRNA-1 and -2) targeting both MADS78 and MADS79 in their conserved MADS and coiled-coil domains (Supplemental Fig. S11A). With the aim of introducing point mutations in both genes, separate transformations for both sgRNAs were performed. The primer sequences used in the study are listed in Supplemental Table S9.
CRISPR/Cas9 constructs were developed following the protocols described by Lowder et al. (2015). pYPQ141C was used to clone all sgRNAs, which were then transferred to a destination vector (pANIC6B) along with a vector containing Cas9 (pYPQ167) using LR clonase. The destination vector was transformed into A. tumefaciens, which was further used to infect calli. For single knockout mutants, T1 segregates carrying mutations and lacking Cas9 (confirmed by GUS screening assay) were considered for downstream analysis (Lowder et al., 2015). T0, T1, and T2 plants were screened for mutations by Sanger sequencing. T2 or later generations of mutant plant lines were used in the study. For experiments, mature rice (‘Kitaake’) seeds were manually dehusked and sterilized with bleach (40%, v/v) and water. The seeds were germinated in the dark on one-half-strength Murashige and Skoog medium. The germinated seedlings were transplanted in soil amid controlled greenhouse conditions: 16 h of light and 8 h of dark at 28°C ± 1°C and 23°C ± 1°C, respectively, and a relative humidity of 55% to 60% (Sandhu et al., 2019).
Genomic DNA and RNA Extraction, RT-qPCR
To screen for mutations in the single and double knockout mutants, genomic DNA was isolated from leaves using the Suc method (Berendzen et al., 2005). The region of interest was amplified using Kapa3G Plant PCR Kits (Kapa Biosystems) according to the manufacturer’s protocol. The amplicon was then sequenced for genotyping. Total RNA was extracted from the following tissues: seedling, leaves, mature pollen, unfertilized ovary, and developing seed (24, 48, 72, and 96 HAF), using the RNeasy Plant mini kit (Qiagen). RNA was subsequently DNase treated. Early seed developmental stages corresponding to 24, 48, 72, and 96 HAF (normally developing seed tissues: category B; Supplemental Fig. S9), as well as unfertilized ovaries, were collected as described (Folsom et al., 2014). Spikelets were marked at the time of fertilization, which is evident by the opening of a floret, to track the precise developmental stage. Mature pollen grains were collected as described (Paul et al., 2016, 2017) with minor adjustments. After anthesis, the protruded anthers were collected in 500 µL of germination solution (2 mm boric acid, 2 mm calcium nitrate, 2 mm magnesium sulfate, and 1 mm potassium nitrate). The collected anthers were vortexed, filtered through muslin cloth, and centrifuged for 1 min at 1,000g. The pelleted pollen grains were washed multiple times in 200 µL of germination solution to remove unwanted material. The collected pollen grains were immersed in liquid nitrogen and stored at −80°C.
One microgram of total RNA was used for cDNA synthesis using the SuperScript VILO cDNA synthesis kit (Invitrogen). The RT-qPCR (10 µL) comprising gene-specific primers, SYBR Green Master Mix (Bio-Rad), and template was conducted using the Lightcycler 480 Real-Time PCR System (Roche). Thermal cycling conditions were used as described by Fragkostefanakis et al. (2015). As an endogenous control, ubiquitin (UBQ5) genes were used as reference genes (Jain et al., 2006). Data were analyzed using standard methods (Livak and Schmittgen, 2001). For RT-qPCR assays, a minimum of two independent biological replicates and three technical replicates were conducted.
Pollen Viability Assay, Agronomic and Morphometric Measurements
For pollen viability assays, mature pollen was collected and subjected to staining with Alexander dye (Alexander, 1980) and Lugol’s solution (I2/KI). Pollen number was determined using a Fuchs-Rosenthal cell counter (Fragkostefanakis et al., 2016). Three independent biological replicates were performed for the pollen viability assay. For determining grain morphology, seeds were harvested at maturity, and only fully developed seeds were scored to calculate percentage of spikelet fertility. The fully developed seeds were dehusked using a rice husker (Kett TR-130). The dehusked seeds were scanned using an Epson Expression 12000 XL scanner and were analyzed by SmartGrain software to obtain seed size measurements (Tanabata et al., 2012). For developing seed analysis, florets marked at the time of fertilization were tracked for 4 d after fertilization (24, 48, 72, and 96 HAF). Each marked floret was opened and evaluated for seed abortion at the designated time. Images of the normal and aborted seeds were representative across all three replicates.
DNA Methylation Assay
Genomic DNA was extracted from two biological replicates of seeds at 24, 48, 72, and 96 HAF using the Qiagen DNeasy Plant Mini Kit. Genomic DNA was then treated with sodium bisulfite (EZ DNA Methylation Kit; Zymo Research). Whole-genome bisulfite sequencing was performed using an Illumina high-throughput sequencing platform to generate 126-bp paired end reads. After filtering the low-quality regions using Trimmomatic, v 0.32 (Bolger et al., 2014), reads were mapped to the reference genome (rice ssp. japonica MSU v7.0), and methylated sites were called using the Bismark v0.16.3 pipeline (Krueger and Andrews, 2011). Methylation levels of sites corresponding to the genomic regions of MADS78 and MADS79 were plotted using R-studio. McrBC assay was performed as mentioned by Folsom et al. (2014).
Histochemical Analysis and RNA in Situ Hybridization Assay
Normally developing seed tissue (category B; Supplemental Fig. S9) corresponding to 72, 84, and 96 HAF was harvested and fixed in 1 mL of formaldehyde (2%, v/v), acetic acid (5%, v/v), and ethanol (60% [v/v]; FAE solution) for 16 h, followed by a 70% (v/v) ethanol wash and stored overnight at 4°C. The dehydration procedure was performed with an ethanol series (85%, 95%, and 100% [v/v]) for 1 h each at room temperature. Samples were incubated in ethanol:xylene (1:1) for 1 h, followed by xylene (100%) for 2 h, and then transferred to 500 µL of xylene and Paraplast tablets at 60°C. Samples were then transferred and embedded in Paraplast. Ten-micrometer sections were made using a rotary microtome (Leica RM2125 RTS). RNA in situ hybridization was performed following the protocol described by Chen et al. (2016). Gene-specific fragments were amplified for MADS78 and MADS79. The respective amplicons were cloned into pGEM-T Easy vector (Promega). RNA probes were generated by in vitro transcription using the DIG RNA labeling kit (Roche). The RNA transcripts by T7 and SP6 polymerase were applied to slides as sense and antisense probes, respectively. Images from cross sections and in situ hybridization were observed using a bright-field microscope (Leica DM-2500) and represent six independent biological replicates.
Localization and BiFC
For localization, the CDS regions of MADS78 and MADS79 were cloned into pGWB5 and pGWB6 vectors (obtained from TAIR), generating protein fusions between GFP and their C and N termini, respectively. Empty vector was used as a control. For BiFC, full-length CDS of 11 rice type I MADS box genes (MADS70, MADS77, MADS81, MADS83, MADS84, MADS87, MADS88, MADS89, MADS90, MADS96, and MADS98) were amplified and cloned into either YFC43 or YFN43 containing the C- or N-terminal half of yellow fluorescent protein (YFP), respectively. Gene selection for the interaction study was based on their spatiotemporal coexpression with MADS78 and MADS79 (Chen et al., 2016). To rule out any false positives, we swapped the vectors for MADS78 and MADS79 with positive interactors (MADS87 and MADS89). Empty vector and bZIP76, an unrelated TF that is expressed during early seed development, were used for controls. Also, we cotransformed the MADS78 and MADS79 GFP fusions (cloned in pGWB5 and pGWB6) with a nonfluorescent BiFC construct (YFN43), encoding a positive interaction partner (MADS87 or MADS89), to test the translocation efficacy of each gene into the nucleus. For both localization and BiFC experiments, Nicotiana benthamiana plants were hand infiltrated using needleless syringes containing suspensions of A. tumefaciens (EHA105 strain) transformed with each vector corresponding to a gene of interest. Two days later, fluorescence signals were observed using a confocal laser scanning microscope (Nikon A1). The following conditions were used for imaging: GFP, excitation at 488 nm and emission at 500 to 550 nm; YFP, excitation at 514.5 nm and emission at 525 to 555 nm (pseudocolored green); chlorophyll, excitation at 640.9 nm and emission at 663 to 738 nm. The images shown for localization and BiFC experiments are representative of four independent experiments with three to four biological replicates in each experiment. For in silico NLS prediction, we used cNLS mapper (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi; Kosugi et al., 2009).
SEM
For SEM, mature rice grains were transversely cut, placed on conductive tape attached to aluminum SEM stubs, and vacuum dried in a sample drying oven at 40°C for 1 week. Afterward, samples were sputter coated with chromium using a Denton Vacuum Desk V sputter coater and imaged with the Hitachi-S4700 Field-Emission SEM device at 50×, 200×, and 2,000× (Dhatt et al., 2019). Three independent biological replicates were used for the observations.
Metabolite Profiling and Quantification of Total Starch and Protein Content
Polar metabolites were extracted from 25 mg of developing seeds (10 DAF), and 50 µL of extract was dried and derivatized as described (Lisec et al., 2006). Five biological replicates were considered for metabolite profiling, which was performed using a 7200 GC-QTOF system (Agilent) with an HP-5MS UI column (30 m, 0.25 mm, and 0.25 µm; Agilent). GC and ionization parameters followed those described by Lisec et al. (2006). Mass spectrometry detection was performed in TOF mode with a 50-Hz scan rate. Peaks in the reference sample containing equal volumes of all samples were annotated with the metabolites in the Fiehn GC/MS Metabolomics RTL Library (Agilent) by MassHunter Unknown Analysis (Agilent) with manual curation. Peak areas of each representative ion were quantified by MassHunter Quantitative Analysis (Agilent). Following background subtraction, peak area was normalized to that of the internal standard (ribitol) and sample fresh weight to calculate relative metabolite levels with orbital unit. The parameters used for peak annotation and peak area values are listed in Supplemental Table S10. Starch and total protein contents were determined in the pellet of metabolite extraction as described (Gibon et al., 2009).
RNA-Seq Analysis
Isolation of total RNA from developing seed (48 and 72 HAF) of the wild type, OE lines, and single knockout mutants was performed as described above. The NuGen Universal Plus mRNA-Seq kit was used for preparing RNA-seq libraries. The libraries were sequenced on a single-read, 75-bp high-output flow cell on the NextSeq 550 device. For the analysis, each RNA-seq read was trimmed using Trimmomatic to ensure that it had a minimum length of 70 bp and an average quality score greater than 30 (Bolger et al., 2014). All trimmed short reads were mapped to the rice genome (RGAP v7.0) using TopHat, allowing up to two base mismatches per read (Trapnell et al., 2009). Reads mapped to multiple locations were not considered. Numbers of reads in genes were counted by the software tool HTSeq-count using corresponding rice gene annotations and the union resolution mode (Supplemental Table S4; Anders et al., 2015).
Target Gene Analysis
FIMO (http://meme-suite.org/tools/fimo) was used to search for MADS box-binding sites (Grant et al., 2011). Promoter sequences (−2,000 bp upstream) downloaded from the MSU7 annotation (Kawahara et al., 2013) were scanned for matches with the CArG box-binding sequence (Batista et al., 2019b) obtained from JASPAR 2018 (http://jaspar.genereg.net/; Khan et al., 2018).
Accession Numbers
Gene accession numbers are displayed in Supplemental Table S2.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Protein sequence alignment for MADS78 and MADS79.
Supplemental Figure S2. In silico promoter analysis.
Supplemental Figure S3. Empty vector used as an additional control for BiFC assays.
Supplemental Figure S4. Confirmation of positive protein-protein interactions.
Supplemental Figure S5. Localization of MADS78 and 79 in the absence and presence of the positive interactors, MADS89 and 87.
Supplemental Figure S6. Representative pictures for overexpression mutants at the vegetative stage.
Supplemental Figure S7. Pollen viability assay.
Supplemental Figure S8. Mature sterile seeds from overexpression mutants of MADS78 and MADS79.
Supplemental Figure S9. Seed abortion analysis during early seed development.
Supplemental Figure S10. Single knockout mutants for MADS78 and MADS79.
Supplemental Figure S11. Double knockout mutations are lethal.
Supplemental Figure S12. DNA methylation analysis.
Supplemental Figure S13. Quantification of total starch and total protein content from developing and mature seeds.
Supplemental Table S1. RNA-seq-based normalized read counts for MADS78 and MADS79.
Supplemental Table S2. Normalized read counts for type I MADS box genes in developing rice seeds (48 and 72 HAF).
Supplemental Table S3. Summary of RNA-seq libraries.
Supplemental Table S4. Normalized read count for syncytial- and cellularization-associated genes.
Supplemental Table S5. Potential target genes for MADS78 and MADS79.
Supplemental Table S6. Levels of metabolites in the developing grains (10 DAF) of MADS78 and MADS79 mutants.
Supplemental Table S7. Normalized read count for carbon metabolism-related genes in mutants and the wild type at 72 HAF.
Supplemental Table S8. List of potential off-targets (as per CRISPR-P 1.0) corresponding to genic region (exonic or intronic) for sgRNAs and their normalized read counts in single knockout mutants.
Supplemental Table S9. List of primers used in the study.
Supplemental Table S10. Parameters used for peak annotation and row peak area data for metabolite profiling.
Acknowledgments
We thank Christian Elowsky, Jules Russ, and You Zhou from the Morrison Core Research Facility, Center for Biotechnology, University of Nebraska-Lincoln, for help with microscopy work. RNA-seq data analysis was completed utilizing the Holland Computing Center of the University of Nebraska. We thank Martha Rowe (Department of Agronomy and Horticulture, University of Nebraska-Lincoln) for help with the histochemical analysis.
Footnotes
This work was supported by the National Science Foundation (grant no. 1736192 to H.W.).
Articles can be viewed without a subscription.
References
- Abu-Zaitoon YM, Bennett K, Normanly J, Nonhebel HM (2012) A large increase in IAA during development of rice grains correlates with the expression of tryptophan aminotransferase OsTAR1 and a grain-specific YUCCA. Physiol Plant 146: 487–499 [DOI] [PubMed] [Google Scholar]
- Aguirre M, Kiegle E, Leo G, Ezquer I (2018) Carbohydrate reserves and seed development: An overview. Plant Reprod 31: 263–290 [DOI] [PubMed] [Google Scholar]
- Alexander MP. (1980) A versatile stain for pollen fungi, yeast and bacteria. Stain Technol 55: 13–18 [DOI] [PubMed] [Google Scholar]
- Anders S, Pyl PT, Huber W (2015) HTSeq: A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyagi K, Bassham JA (1984) Pyruvate orthophosphate dikinase of c(3) seeds and leaves as compared to the enzyme from maize. Plant Physiol 75: 387–392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arora R, Agarwal P, Ray S, Singh AK, Singh VP, Tyagi AK, Kapoor S (2007) MADS-box gene family in rice: Genome-wide identification, organization and expression profiling during reproductive development and stress. BMC Genomics 8: 242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basunia MA, Nonhebel HM (2019) Hormonal regulation of cereal endosperm development with a focus on rice (Oryza sativa). Funct Plant Biol 46: 493–506 [DOI] [PubMed] [Google Scholar]
- Batista RA. (2019) Signalling mechanisms and epigenetic regulation of seed development in Arabidopsis thaliana. PhD thesis. Swedish University of Agricultural Sciences, Uppsala, Switzerland [Google Scholar]
- Batista RA, Figueiredo DD, Santos-González J, Köhler C (2019a) Auxin regulates endosperm cellularization in Arabidopsis. Genes Dev 33: 466–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batista RA, Moreno-Romero J, Qiu Y, van Boven J, Santos-González J, Figueiredo DD, Köhler C (2019b) The MADS-box transcription factor PHERES1 controls imprinting in the endosperm by binding to domesticated transposons. eLife 8: 616698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemer M, Heijmans K, Airoldi C, Davies B, Angenent GC (2010) An atlas of type I MADS box gene expression during female gametophyte and seed development in Arabidopsis. Plant Physiol 154: 287–300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berendzen K, Searle I, Ravenscroft D, Koncz C, Batschauer A, Coupland G, Somssich IE, Ülker B (2005) A rapid and versatile combined DNA/RNA extraction protocol and its application to the analysis of a novel DNA marker set polymorphic between Arabidopsis thaliana ecotypes Col-0 and Landsberg erecta. Plant Methods 1: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjerkan KN, Hornslien KS, Johannessen IM, Krabberød AK, van Ekelenburg YS, Kalantarian M, Shirzadi R, Comai L, Brysting AK, Bramsiepe J, et al. (2019) Genetic variation and temperature affects hybrid barriers during interspecific hybridization. Plant J doi:10.1111/tpj.14523 [DOI] [PubMed] [Google Scholar]
- Blackwell TK, Bowerman B, Preiss JR, Weintraub H (1994) Formation of a monomeric DNA binding domain by Skn-1 bZIP and homeodomain elements. Science 266: 621–628 [DOI] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Olsen OA (1996) Development of the endosperm in rice (Oryza sativa L.): Cellularization. J Plant Res 109: 301–313 [Google Scholar]
- Chastain CJ, Heck JW, Colquhoun TA, Voge DG, Gu XY (2006) Posttranslational regulation of pyruvate, orthophosphate dikinase in developing rice (Oryza sativa) seeds. Planta 224: 924–934 [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Koltunow A, Payne T, Luo M, Tucker MR, Dennis ES, Peacock WJ (2001) Control of early seed development. Annu Rev Cell Dev Biol 17: 677–699 [DOI] [PubMed] [Google Scholar]
- Chaudhury AM, Ming L, Miller C, Craig S, Dennis ES, Peacock WJ (1997) Fertilization-independent seed development in Arabidopsis thaliana. Proc Natl Acad Sci USA 94: 4223–4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Begcy K, Liu K, Folsom JJ, Wang Z, Zhang C, Walia H (2016) Heat stress yields a unique MADS box transcription factor in determining seed size and thermal sensitivity. Plant Physiol 171: 606–622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Li T, Zhu S, Liu Z, Shi Z, Zheng X, Chen R, Huang J, Shen Y, Luo S, et al. (2018) Characterization of imprinted genes in rice reveals conservation of regulation and imprinting with other plant species. Plant Physiol 177: 1754–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang M, Ouwerkerk PBF (2012) Molecular and environmental factors determining grain quality in rice. Food Energy Secur 1: 111–132 [Google Scholar]
- Cheng X, Sardana RK, Altosaar I (1998) Rice transformation by Agrobacterium infection In Cunningham C, and Porter AJR, eds, Recombinant Proteins from Plants Methods in Biotechnology, Vol Vol 3 Humana Press, Totawa, NJ, pp 1–9 [Google Scholar]
- de Folter S, Immink RG, Kieffer M, Parenicová L, Henz SR, Weigel D, Busscher M, Kooiker M, Colombo L, Kater MM, et al. (2005) Comprehensive interaction map of the Arabidopsis MADS box transcription factors. Plant Cell 17: 1424–1433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhatt BK, Abshire N, Paul P, Hasanthika K, Sandhu J, Zhang Q, Obata T, Walia H (2019) Metabolic dynamics of developing rice seeds under high night-time temperature stress. Front Plant Sci 10: 1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickinson CD, Evans RP, Nielsen NC (1988) RY repeats are conserved in the 5′-flanking regions of legume seed-protein genes. Nucleic Acids Res 16: 371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Tommaso P, Moretti S, Xenarios I, Orobitg M, Montanyola A, Chang JM, Taly JF, Notredame C (2011) T-Coffee: A web server for the multiple sequence alignment of protein and RNA sequences using structural information and homology extension. Nucleic Acids Res 39: W13–W17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Hennig L, Köhler C (2016) Auxin production in the endosperm drives seed coat development in Arabidopsis. eLife 5: 1–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueiredo DD, Batista RA, Roszak PJ, Köhler C (2015) Auxin production couples endosperm development to fertilization. Nat Plants 1: 15184. [DOI] [PubMed] [Google Scholar]
- Finnegan EJ, Peacock WJ, Dennis ES (1996) Reduced DNA methylation in Arabidopsis thaliana results in abnormal plant development. Proc Natl Acad Sci USA 93: 8449–8454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folsom JJ, Begcy K, Hao X, Wang D, Walia H (2014) Rice fertilization-Independent Endosperm1 regulates seed size under heat stress by controlling early endosperm development. Plant Physiol 165: 238–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkostefanakis S, Mesihovic A, Simm S, Paupière MJ, Hu Y, Paul P, Mishra SK, Tschiersch B, Theres K, Bovy A, et al. (2016) HsfA2 controls the activity of developmentally and stress-regulated heat stress protection mechanisms in tomato male reproductive tissues. Plant Physiol 170: 2461–2477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fragkostefanakis S, Simm S, Paul P, Bublak D, Scharf KD, Schleiff E (2015) Chaperone network composition in Solanum lycopersicum explored by transcriptome profiling and microarray meta-analysis. Plant Cell Environ 38: 693–709 [DOI] [PubMed] [Google Scholar]
- French SR, Abu-Zaitoon Y, Uddin MM, Bennett K, Nonhebel HM (2014) Auxin and cell wall invertase related signaling during rice grain development. Plants (Basel) 3: 95–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Xu H, Shen Y, Wang J (2013) Transcriptomic analysis of rice (Oryza sativa) endosperm using the RNA-Seq technique. Plant Mol Biol 81: 363–378 [DOI] [PubMed] [Google Scholar]
- Garcia D, Fitz Gerald JN, Berger F (2005) Maternal control of integument cell elongation and zygotic control of endosperm growth are coordinated to determine seed size in Arabidopsis. Plant Cell 17: 52–60 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehring M. (2013) Genomic imprinting: Insights from plants. Annu Rev Genet 47: 187–208 [DOI] [PubMed] [Google Scholar]
- Genger RK, Kovac KA, Dennis ES, Peacock WJ, Finnegan EJ (1999) Multiple DNA methyltransferase genes in Arabidopsis thaliana. Plant Mol Biol 41: 269–278 [DOI] [PubMed] [Google Scholar]
- Gibon Y, Pyl ET, Sulpice R, Lunn JE, Höhne M, Günther M, Stitt M (2009) Adjustment of growth, starch turnover, protein content and central metabolism to a decrease of the carbon supply when Arabidopsis is grown in very short photoperiods. Plant Cell Environ 32: 859–874 [DOI] [PubMed] [Google Scholar]
- Grant CE, Bailey TL, Noble WS (2011) FIMO: Scanning for occurrences of a given motif. Bioinformatics 27: 1017–1018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus U, Vielle-Calzada J, Hoeppner M, Gagliano W (1998) Maternal control of embryogenesis by MEDEA, a polycomb group gene in Arabidopsis. Science 280: 446–450 [DOI] [PubMed] [Google Scholar]
- Haig D. (2013) Kin conflict in seed development: An interdependent but fractious collective. Annu Rev Cell Dev Biol 29: 189–211 [DOI] [PubMed] [Google Scholar]
- Huh JH, Bauer MJ, Hsieh TF, Fischer R (2007) Endosperm gene imprinting and seed development. Curr Opin Genet Dev 17: 480–485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Immink RG, Tonaco IA, de Folter S, Shchennikova A, van Dijk AD, Busscher-Lange J, Borst JW, Angenent GC (2009) SEPALLATA3: The ‘glue’ for MADS box transcription factor complex formation. Genome Biol 10: R24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Khurana JP (2009) Transcript profiling reveals diverse roles of auxin-responsive genes during reproductive development and abiotic stress in rice. FEBS J 276: 3148–3162 [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP (2006) Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochem Biophys Res Commun 345: 646–651 [DOI] [PubMed] [Google Scholar]
- Jullien PE, Berger F (2010) Parental genome dosage imbalance deregulates imprinting in Arabidopsis. PLoS Genet 6: e1000885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HG, Park S, Matsuoka M, An G (2005) White-core endosperm floury endosperm-4 in rice is generated by knockout mutations in the C-type pyruvate orthophosphate dikinase gene (OsPPDKB). Plant J 42: 901–911 [DOI] [PubMed] [Google Scholar]
- Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20: 635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawahara Y, De La Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. (2013) Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (N Y) 6: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan A, Fornes O, Stigliani A, Gheorghe M, Castro-Mondragon JA, van der Lee R, Bessy A, Chèneby J, Kulkarni SR, Tan G, et al. (2018) JASPAR 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res 46: D260–D266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Hennig L, Spillane C, Pien S, Gruissem W, Grossniklaus U (2003) The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev 17: 1540–1553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Köhler C, Wolff P, Spillane C (2012) Epigenetic mechanisms underlying genomic imprinting in plants. Annu Rev Plant Biol 63: 331–352 [DOI] [PubMed] [Google Scholar]
- Kosugi S, Hasebe M, Tomita M, Yanagawa H (2009) Systematic identification of cell cycle-dependent yeast nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci USA 106: 10171–10176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger F, Andrews SR (2011) Bismark: A flexible aligner and methylation caller for Bisulfite-Seq applications. Bioinformatics 27: 1571–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei Y, Lu L, Liu HY, Li S, Xing F, Chen LL (2014) CRISPR-P: A web tool for synthetic single-guide RNA design of CRISPR-system in plants. Mol Plant 7: 1494–1496 [DOI] [PubMed] [Google Scholar]
- Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, Van de Peer Y, Rouzé P, Rombauts S (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res 30: 325–327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P (2012) SMART 7: Recent updates to the protein domain annotation resource. Nucleic Acids Res 40: D302–D305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisec J, Schauer N, Kopka J, Willmitzer L, Fernie AR (2006) Gas chromatography mass spectrometry-based metabolite profiling in plants. Nat Protoc 1: 387–396 [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−Δ Δ CT method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lowder LG, Zhang D, Baltes NJ, Paul JW III, Tang X, Zheng X, Voytas DF, Hsieh TF, Zhang Y, Qi Y (2015) A CRISPR/Cas9 toolbox for multiplexed plant genome editing and transcriptional regulation. Plant Physiol 169: 971–985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Bilodeau P, Dennis ES, Peacock WJ, Chaudhury A (2000) Expression and parent-of-origin effects for FIS2, MEA, and FIE in the endosperm and embryo of developing Arabidopsis seeds. Proc Natl Acad Sci USA 97: 10637–10642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo M, Platten D, Chaudhury A, Peacock WJ, Dennis ES (2009) Expression, imprinting, and evolution of rice homologs of the polycomb group genes. Mol Plant 2: 711–723 [DOI] [PubMed] [Google Scholar]
- Masiero S, Colombo L, Grini PE, Schnittger A, Kater MM (2011) The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 23: 865–872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA. (2004) Nuclear endosperm development in cereals and Arabidopsis thaliana. Plant Cell 16(Suppl): S214–S227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, et al. (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: New openings to the MADS world. Plant Cell 15: 1538–1551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P, Chaturvedi P, Mesihovic A, Ghatak A, Weckwerth W, Schleiff E (2017) Protocol for enrichment of the membrane proteome of mature tomato pollen. Bio Protoc 7: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul P, Chaturvedi P, Selymesi M, Ghatak A, Mesihovic A, Scharf KD, Weckwerth W, Simm S, Schleiff E (2016) The membrane proteome of male gametophyte in Solanum lycopersicum. J Proteomics 131: 48–60 [DOI] [PubMed] [Google Scholar]
- Pires ND. (2014) Seed evolution: Parental conflicts in a multi-generational household. Biomol Concepts 5: 71–86 [DOI] [PubMed] [Google Scholar]
- Portereiko MF, Lloyd A, Steffen JG, Punwani JA, Otsuga D, Drews GN (2006) AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell 18: 1862–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA (2009) The development of endosperm in grasses. Plant Physiol 149: 14–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandhu J, Zhu F, Paul P, Gao T, Dhatt BK, Ge Y, Staswick P, Yu H, Walia H (2019) PI-Plat: A high-resolution image-based 3D reconstruction method to estimate growth dynamics of rice inflorescence traits. bioRxiv 835306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott RJ, Spielman M, Bailey J, Dickinson HG (1998) Parent-of-origin effects on seed development in Arabidopsis thaliana. Development 125: 3329–3341 [DOI] [PubMed] [Google Scholar]
- Singh N, Nakaura Y, Inouchi N, Nishinari K (2007) Fine structure, thermal and viscoelastic properties of starches separated from indica rice cultivars. Starke 59: 10–20 [Google Scholar]
- Smaczniak C, Immink RGH, Angenent GC, Kaufmann K (2012) Developmental and evolutionary diversity of plant MADS-domain factors: Insights from recent studies. Development 139: 3081–3098 [DOI] [PubMed] [Google Scholar]
- Steffen JG, Kang IH, Portereiko MF, Lloyd A, Drews GN (2008) AGL61 interacts with AGL80 and is required for central cell development in Arabidopsis. Plant Physiol 148: 259–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoute AI, Varenko V, King GJ, Scott RJ, Kurup S (2012) Parental genome imbalance in Brassica oleracea causes asymmetric triploid block. Plant J 71: 503–516 [DOI] [PubMed] [Google Scholar]
- Tanabata T, Shibaya T, Hori K, Ebana K, Yano M (2012) SmartGrain: High-throughput phenotyping software for measuring seed shape through image analysis. Plant Physiol 160: 1871–1880 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL (2009) TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchiumi T, Okamoto T (2010) Rice fruit development is associated with an increased IAA content in pollinated ovaries. Planta 232: 579–592 [DOI] [PubMed] [Google Scholar]
- Walia H, Josefsson C, Dilkes B, Kirkbride R, Harada J, Comai L (2009) Dosage-dependent deregulation of an AGAMOUS-LIKE gene cluster contributes to interspecific incompatibility. Curr Biol 19: 1128–1132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Yuan J, Ma Y, Jiao W, Ye W, Yang DL, Yi C, Chen ZJ (2018) Rice interploidy crosses disrupt epigenetic regulation, gene expression, and seed development. Mol Plant 11: 300–314 [DOI] [PubMed] [Google Scholar]
- Yoshihara T, Washida H, Takaiwa F (1996) A 45-bp proximal region containing AACA and GCN4 motif is sufficient to confer endosperm-specific expression of the rice storage protein glutelin gene, GluA-3. FEBS Lett 383: 213–218 [DOI] [PubMed] [Google Scholar]
- Zhang S, Wang D, Zhang H, Skaggs MI, Lloyd A, Ran D, An L, Schumaker KS, Drews GN, Yadegari R (2018) FERTILIZATION-INDEPENDENT SEED-Polycomb Repressive Complex 2 plays a dual role in regulating type I MADS-box genes in early endosperm development. Plant Physiol 177: 285–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Z, Zhang Y, Liu X, Zhang X, Liu S, Yu X, Ren Y, Zheng X, Zhou K, Jiang L, et al. (2013) A role for a dioxygenase in auxin metabolism and reproductive development in rice. Dev Cell 27: 113–122 [DOI] [PubMed] [Google Scholar]