Comparisons between root system architecture-associated genes identified from maize and sorghum via GWAS enabled by high-throughput phenotyping reveal conserved functional roles of syntenic orthologs.
Abstract
Determining the genetic control of root system architecture (RSA) in plants via large-scale genome-wide association study (GWAS) requires high-throughput pipelines for root phenotyping. We developed Core Root Excavation using Compressed-air (CREAMD), a high-throughput pipeline for the cleaning of field-grown roots, and Core Root Feature Extraction (COFE), a semiautomated pipeline for the extraction of RSA traits from images. CREAMD-COFE was applied to diversity panels of maize (Zea mays) and sorghum (Sorghum bicolor), which consisted of 369 and 294 genotypes, respectively. Six RSA-traits were extracted from images collected from >3,300 maize roots and >1,470 sorghum roots. Single nucleotide polymorphism (SNP)-based GWAS identified 87 TAS (trait-associated SNPs) in maize, representing 77 genes and 115 TAS in sorghum. An additional 62 RSA-associated maize genes were identified via expression read depth GWAS. Among the 139 maize RSA-associated genes (or their homologs), 22 (16%) are known to affect RSA in maize or other species. In addition, 26 RSA-associated genes are coregulated with genes previously shown to affect RSA and 51 (37% of RSA-associated genes) are themselves transe–quantitative trait locus for another RSA-associated gene. Finally, the finding that RSA-associated genes from maize and sorghum included seven pairs of syntenic genes demonstrates the conservation of regulation of morphology across taxa.
The spatial arrangements of root systems, i.e. root system architecture (RSA; Lynch, 1995), play a critical role in plant productivity and tolerance to environmental stresses. In maize (Zea mays), the majority of the root mass is found in the top 0.3 m of soil (Amos and Walters, 2006). This mass of roots has been referred to as the “root crown” (Trachsel et al., 2011), the “core root” (Grift et al., 2011), or the “core root system” (Hauck et al., 2015). The term core root system is used hereafter for two reasons. First, the term root crown originally referred only to the above-ground portion of the root system (Bray et al., 1959; Schwarz, 1972). Only more recently has this term been used to describe roots within the top 0.3 m of soil (Trachsel et al., 2011). Second, this term is easily confused with the term “crown roots,” which refers to postembryonic shoot-borne roots (Kiesselbach, 1999).
The genetic regulation of root development has been extensively studied in Arabidopsis (Arabidopsis thaliana; Dolan et al., 1993; Birnbaum et al., 2003; Petricka et al., 2012a, 2012b) resulting in an in-depth understanding of the relevant genes and pathways. Despite similarities in embryonic root systems and some shared mechanisms of genetic regulation, there are major anatomical differences between the root systems of Arabidopsis and cereal crops. The adult Arabidopsis root system comprises a tap root, a basal root, hypocotyl roots, internodal shoot-borne roots, and lateral roots (Zobel, 2016). By contrast, the maize root system is composed of the embryonic primary root and variable numbers of seminal roots, as well as postembryonic shoot-borne and lateral roots (Hochholdinger and Tuberosa, 2009). Similar to maize, sorghum (Sorghum bicolor) develops shoot-borne roots; however, sorghum lacks seminal roots (Singh et al., 2010). Based on these fundamental morphological differences, it is unlikely that a complete understanding of the genetic regulation of RSA in these species can be elucidated from Arabidopsis.
Whereas functional studies on qualitative mutants of genes with large effect sizes have deepened our understanding of the genetic control and developmental processes of the root systems of cereals, a comprehensive understanding of the genes underlying quantitative variation in RSA has not been achieved (Hochholdinger et al., 2018).
Genome-wide associated study (GWAS) offers the opportunity to identify genes affecting natural variation of quantitative traits via the association of markers across the genome with phenotypic variation within diversity panels (Xiao et al., 2017). With the ready availability of large numbers of genetic markers, phenotyping has become the bottleneck for GWAS. Several root phenotyping pipelines have been developed for genetic mapping (Topp et al., 2013; Zurek et al., 2015). Most of these studies were conducted on young plants grown in microcosms and mesocosms (Topp, 2016). However, it has been observed in multiple species that RSA varies across development and environments (Rauh et al., 2002; Magalhaes et al., 2004; Trachsel et al., 2013) and that roots grown under controlled conditions do not match those grown under field conditions (Poorter et al., 2016). Hence, if we wish to understand the genetic control of RSA as it relates to crop growth in target environments, it is necessary to phenotype roots grown under agronomically relevant field conditions. However, the throughput of current pipelines for analyzing RSA is insufficient to satisfy the phenotyping needs of large-scale GWAS. Hence, thus far, efforts to characterize RSA have mainly focused on quantitative trait locus (QTL) mapping using biparental populations with limited genetic diversity, population size, and mapping resolution (Thomson et al., 2003; Giuliani et al., 2005; Li et al., 2005; Liang et al., 2010; Cai et al., 2012; Atkinson et al., 2015; Richard et al., 2015; Guo et al., 2018). To date, few of the genes underlying RSA QTL in cereal crops have been cloned (Mai et al., 2014; Hochholdinger et al., 2018).
Maize and sorghum are both important crops, ranked first and fifth, respectively, in global cereal production (http://faostat.fao.org/). Maize and sorghum diverged from a common ancestor ∼12 mya (Swigoňová et al., 2004). Approximately 60% of annotated genes are syntenically conserved between these two species, and this syntenically conserved set of genes accounts for >90% of all genes characterized by forward genetics in maize (Schnable and Freeling, 2011; Schnable, 2015). Syntenic orthologs are more likely to retain consistent patterns of gene regulation and expression across related species (Davidson et al., 2012), and may be more likely to retain ancestral functional roles than nonsyntenic gene copies (Dewey, 2011). However, to date, the conservation of functional roles for syntenic orthologous gene pairs in related species has not been widely tested.
We report the development of Core Root Excavation using Compressed-air (CREAMD), a high-throughput pipeline suitable for the excavation and cleaning of field-grown roots, and Core Root Feature Extraction (COFE), a semiautomated pipeline to extract features from images of roots. CREAMD-COFE was used to phenotype roots from maize and sorghum diversity panels. Comparative analyses of maize and sorghum GWAS results provided strong evidence for shared genetic control of RSA in these two species and the conservation of functional roles for syntenic orthologous gene pairs.
RESULTS
CREAMD-COFE Enables the Efficient Excavation, Cleaning, and Phenotyping of Core Root Systems
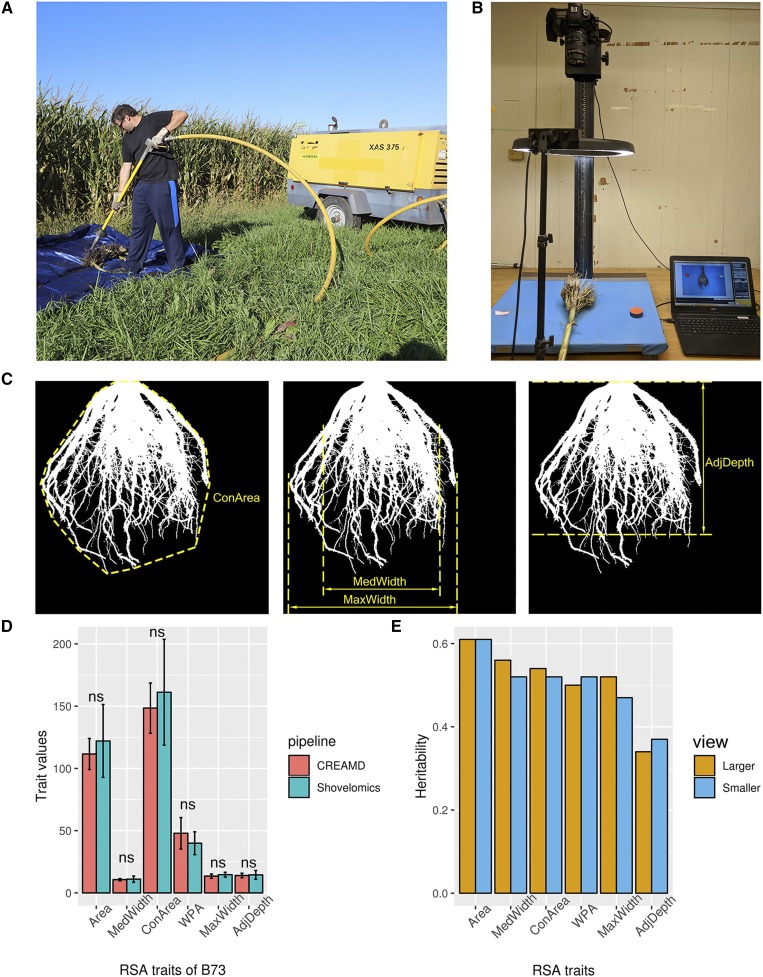
Manual excavation and cleaning of field-grown roots is labor intensive (Trachsel et al., 2011; Colombi et al., 2015). To simplify the phenotyping of RSA from field-grown plants and thereby enable large-scale genetic studies under agronomic conditions, we developed CREAMD, a pipeline for the rapid excavation and cleaning of roots. CREAMD uses compressed air to remove soil from core root systems (see “Materials and Methods”; Fig. 1; Supplemental Text S1).
Figure 1.
Extraction of RSA traits from binary images of core root systems using COFE. Illustration of root cleaning (A) and phenotyping of CREAMD pipeline (B) are shown. C, Illustration of four out of six traits extracted via COFE. D, Comparison of RSA trait values from the inbred line B73 extracted by COFE from roots collected using CREAMD or water-based root cleaning. Data are means ± sd; ns, not significant, Student’s t test; n = 15. E, Heritabilities of RSA trait values obtained from the SAM Diversity Panel via CREAMD-COFE; n = 3,196 roots per view. See Table 1 for abbreviations.
Following excavation and cleaning, core root systems were photographed (see “Materials and Methods”; Fig. 1). COFE, a semiautomated pipeline, was used to extract traits from the resulting images (Supplemental Text S1). COFE is an adaptation of the ARIA software (Pace et al., 2014), which had been developed for lab-based phenotyping of immature root systems.
There are two major potential sources of error between auto-extracted trait values and ground truth: (1) errors introduced via the projection of three-dimensional (3D) traits onto a two-dimensional (2D) image; and (2) errors in the extraction of trait values from 2D images. To distinguish between these two potential sources of error, we compared COFE-extracted trait values to trait values obtained by manually measuring 3D core root systems (ground truth) and to trait values manually extracted (using ImageJ) from 2D photos of the same core root systems. These comparisons were performed for ∼5% of all collected maize and sorghum core root systems (“Materials and Methods”). The coefficient of determination (r2) between COFE’s auto-extraction trait values and manual measurements of maximum width and depth from 3D core root systems are 0.54 and 0.46, respectively. By contrast, the r2 for the same two traits between COFE’s auto-extracted trait values and measurements obtained using ImageJ from photos are 0.88 and 0.87, respectively (see “Materials and Methods”; Supplemental Fig. S1). These results demonstrate that COFE can accurately extract trait values from 2D images of core root systems (Fig. 1) and that much of the difference between COFE-extracted trait values and ground truth is due to the challenge of representing 3D core root systems in 2D images.
The air-based root cleaning pipeline, CREAMD, increases the speed of root cleaning 6.5-fold as compared with a previously described water-based root cleaning pipeline previously described by Trachsel et al. (2011; Supplemental Table S1), while yielding comparably intact core root systems; trait values obtained from 15 plants of each of four maize genotypes via CREAMD-COFE (“Materials and Methods”) are similar to those obtained via the water-based root cleaning pipeline (Fig. 1; Supplemental Fig. S2). In addition to being substantially faster than the water-based root cleaning pipeline without comprising root quality, CREAMD can be conducted at remote field sites that lack access to water.
Phenotypic Variation of RSA in Maize
Three biological replications of 369 inbred lines from the SAM Diversity Panel (Leiboff et al., 2015) were grown (“Materials and Methods”). Core root systems from up to three competitive plants (“Materials and Methods”) from each of the three replications were excavated and cleaned using CREAMD. Each core root system was first photographed using a camera angle selected to obtain a view from a neighboring plant in the row in which the plant under analysis was grown (view 1) and then again after rotating the core root system by 90° (clockwise when viewing from above), resulting in view 2 (“Materials and Methods”). Trait values of core root systems of maize from the two views did not exhibit statistically significant differences (Supplemental Table S2), suggesting maize plants do not substantially alter their RSA in response to neighbors, at least at the planting densities used here (“Materials and Methods”). Even so, when viewed from above core root systems do not exhibit radial symmetry (see “Materials and Methods”; Supplemental Fig. S3). Consequently, for subsequent analyses, we classified the two images of each core root system as the larger and smaller on a per trait basis (see “Materials and Methods”; Supplemental Fig. S4; Supplemental Table S3).
COFE was used to extract the following six types of traits from both images of each core root system (Fig. 1; Table 1; Supplemental Text S2; Supplemental Figs. S4–S6). Because we extracted traits from both images of each root, a total of twelve traits were extracted. Maximum and median widths (designated smMaxWidth, lgMaxWidth, smMedWidth, and lgMedWidth) served as measures of the horizontal expansion of core root systems. The Adjusted Depth (smAdjDepth and lgAdjDepth), which is the root depth at which the ratio of root pixels to total pixels exhibits the highest heritability (Supplemental Fig. S5), was used as a measure of the depth of the core root system. Convex hull (smConArea and lgConArea), the minimum set of points that define a polygon containing all the pixels of a core root in an image, was used to describe the overall expansion of a core root system. The penultimate trait was total root area (smArea and lgArea), which is the total number of pixels of roots in a photograph.
Table 1. Abbreviations of RSA Traits.
| Abbreviation | Trait |
|---|---|
| Area | Root area |
| ConArea | Convex hull area |
| MedWidth | Median width |
| MaxWidth | Maximum width |
| WPA | Width-profile angle |
| AdjDepth | Adjusted depth |
The final extracted trait was root angle. The scientific literature does not offer a consistent definition of root angle, particularly among, but even within, species (Vitha et al., 2000; Li et al., 2005; Hargreaves et al., 2009; Singh et al., 2010; Courtois et al., 2013; Richard et al., 2015), among developmental stages (Omori and Mano, 2007; Fang et al., 2009; Trachsel et al., 2011; Pace et al., 2014; Zurek et al., 2015), and across environments (Topp et al., 2013; Uga et al., 2013; Huang et al., 2018). Due to the low heritabilities (<0.2) of two previously defined measures of root angle (CA) and top angle (IAngRt; Trachsel et al., 2011; Colombi et al., 2015), we defined a root angle trait based on width profiles (smWPA and lgWPA). High values of WPA are associated with steep roots. WPA exhibits higher heritabilities (0.50 for lgWPA and 0.52 for smWPA) than the two previously described root angle traits (Fig. 1; Supplemental Fig. S6; Supplemental Text S2).
The heritabilities of the twelve traits ranged from 0.47 for smMaxWidth to 0.61 for smArea and lgArea, with the exception of smAdjDepth and lgAdjDepth, which had the lowest heritabilities (0.33 and 0.37; Fig. 1). For five of the six types of root traits (Area, ConArea, MedWdith, MaxWidth, Adjusted Depth) the two views (large and small) were positively correlated. Correlations between larger and smaller views of the collected RSA traits range from 0.92 for MaxWidth to 0.98 for Area (Supplemental Table S4). The pairwise Pearson correlation coefficients ranged from 0.45 (between smAdjustedDepth and smMedWidth) to 0.97 (between smArea and smConArea). Both views of WPA exhibited negative correlations with all other RSA traits (Supplemental Table S5).
To determine correlations between RSA and above-ground traits, we compared the 12 RSA traits with four above-ground traits: plant height, plant ear height, flowering time (days to anthesis), and node number data from Leiboff et al. (2015). Even though the root and above-ground traits were collected in different environments, both views of five of the six types of root traits (Area, ConArea, MedWdith, MaxWidth, Adjusted Depth) were positively correlated with all four above-ground traits. Pairwise Pearson correlation coefficients ranged from 0.36 (between smMaxWidth and node number) to 0.59 (between lgArea and plant ear height). Similarly, both views of WPA exhibited negative correlations with all four above-ground traits (Supplemental Table S5). These correlations between RSA and above-ground traits support the hypothesis that by selecting for the latter breeders may have inadvertently selected for the former.
GWAS for RSA Traits
FarmCPU accounts for kinship and population structure in GWAS (Liu et al., 2016). An efficient implementation of FarmCPU termed FarmCPUpp (Kusmec and Schnable, 2018) was used to perform GWAS on the SAM Diversity Panel, which was previously genotyped with ∼1.2 m SNPs (Leiboff et al., 2015). RSA trait values were adjusted to account for field-based spatial variation (“Materials and Methods”). The 107 significant SNPs were associated with six types of RSA traits (each of which has two views, resulting in a total of 12 traits) using a false discovery rate (FDR) cutoff of <0.05 (Supplemental Table S6; Benjamini and Hochberg, 1995). Only 20% (20/107) of these trait-associated SNPs (TASs) were associated with both views of the same trait (i.e. 10 pairs of TASs), a result that is consistent with our finding that roots do not exhibit radial symmetry (Supplemental Fig. S3). In addition, ∼6% (7/107) of the TASs were associated with two or more traits, a result consistent with the high correlations among traits (Supplemental Table S4). For 77/87 of the TASs (88%) it was possible to identify a candidate gene (“SNP-genes”), which was defined as the gene nearest a TAS within a 20-kb window centered on that TAS (Supplemental Table S6).
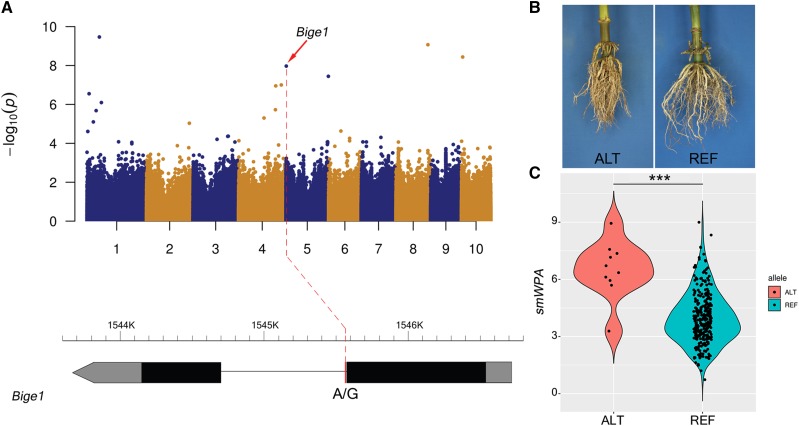
A SNP located within GRMZM2G148937, Big embryo 1 (Bige1; Fig. 2), was associated with the trait smWPA. Bige1, which encodes a MATE transporter, is one of only eight cloned maize genes with a known function in root development (Hochholdinger et al., 2018). A loss-of-function mutant of Bige1 displays increased number of seminal roots and lateral organs during vegetative development as compared with wild-type controls (Suzuki et al., 2015). Within the SAM Diversity Panel, inbred lines that carry the ALT (i.e. the non-B73) allele of Bige1 have significantly higher mean values of smWPA (P = 0.01) than those that carry the REF (i.e. the B73) allele (Fig. 2). Based on published trait data (Leiboff et al., 2015), inbred lines homozygous for the ALT allele of bige1 were shorter and flowered earlier than those homozygous for the REF allele, a result consistent with those of Suzuki et al. (2015). Inbred lines homozygous for the ALT allele of bige1 also exhibit reduced ear height and plant height (Supplemental Fig. S7). In addition, based on published shoot apical meristem (SAM) phenotypic data (Leiboff et al., 2015), inbred lines that carry the ALT allele of Bige1 have SAMs with larger radii. This is consistent with a previous report that SAM radius is correlated with flowering time, ear height, and plant height (Leiboff et al., 2015).
Figure 2.
Association of Bige1 (GRMZM2G148937) with maize smWPA. A, Manhattan plot of SNP-based GWAS for smWPA; gene model with the position indicated of the RSA-associated SNP within the intron. B, Representative root images of inbred lines homozygous for the ALT (non-B73) and REF (B73) alleles of the RSA-assocciated SNP within Bige1. Illustrated inbred lines are LH52 (ALT allele) and LH57 (REF allele). C, Distribution of trait values of inbred lines homozygous for the ALT and REF alleles. Student’s t test; ***P < 0.001.
Homologs from other species for 10 of the remaining 76 SNP-genes (13%) are known to influence RSA (Supplemental Table S7). For example, GRMZM2G143756, a maize homolog of an Arabidopsis ABCG transporter, was associated with lgArea. Members of a clade of five Arabidopsis ABCG transporters are required for the synthesis of an effective suberin barrier in roots, and seedlings of the abcg2 abcg6 and abcg20 triple mutant of Arabidopsis exhibit fewer lateral root primordia and fewer lateral roots than wild-type controls (Yadav et al., 2014). In potato (Solanum tuberosum), ABCG1-RNA interference plants exhibit reduced suberin content in root exodermis cells and tuber periderm cells. The lower suberin content leads to reduced root volume (Landgraf et al., 2014). GRMZM2G013128, a maize homolog of the Arabidopsis SMXL3 gene, was associated with variation in both the smMaxWidth and lgMedWidth traits. In Arabidopsis, SMXL3 is highly expressed in root vasculature; double mutants of smxl3;smxl4 and smxl3;smxl5 exhibit reduced primary root lengths as compared with wild-type controls (Wallner et al., 2017). GRMZM2G013324, a maize homolog of Arabidopsis SHV3, was associated with variation in the trait lgMaxWidth. SHV3 encodes a glycerophosphoryl diester phosphodiesterase–like protein. A mutant of shv3 exhibits a defective root hair phenotype (Jones et al., 2006). GRMZM2G400907, a maize homolog of GTE4 in Arabidopsis, was associated with variation in smMedWidth. GTE4 is a Bromodomain and Extra Terminal domain factor, which functions in the maintenance of the mitotic cell cycle. An Arabidopsis mutant of gte4 exhibits significant shorter primary roots and defective lateral roots (Airoldi et al., 2010).
eRD-GWAS of Maize RSA
Conventional GWAS uses SNPs as the explanatory variable. By contrast, eRD-GWAS uses gene expression levels as the explanatory variables to associate genes with phenotypic variation (Lin et al., 2017). Because eRD-GWAS has been shown to identify gene/trait associations that are complementary to those identified via SNP-based GWAS (Lin et al., 2017), we also conducted eRD-GWAS.
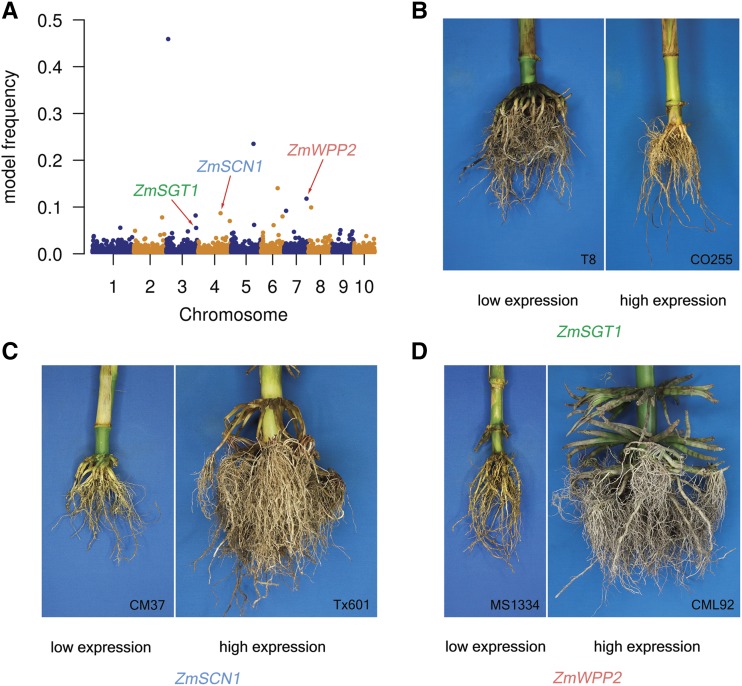
RNA sequencing (RNA-seq) data from 2-cm tips of germinating seedling roots are available for a subset (n = 246) of the SAM Diversity Panel (Kremling et al., 2018). eRD-GWAS was conducted on this subset of the SAM Diversity Panel, resulting in the identification of 62 gene-trait associations (Supplemental Table S8). Thirty-four percent (21/62) of “eRD-genes” are associated with more than two RSA traits, whereas 42% (26/62) are associated with the two views of the same RSA traits. For example, GRMZM2G021410, which encodes a putative α/β-hydrolase superfamily protein, is associated with all six root traits. Twelve of the 62 unique eRD-genes (19%) have homologs in Arabidopsis or Medicago truncatula with known functions in root development (Supplemental Table S9). For example, Arabidopsis homologs of four eRD-genes associated with variation in the smaller view of root area (smArea) of maize (Fig. 3) have been associated with root development in Arabidopsis. An Arabidopsis mutant, sgt1b (a homolog of GRMZM2G105019), exhibits auxin-resistant root growth under low concentrations of auxin (Gray et al., 2003). An RNA interference mutant of wpp2 (a homolog of GRMZM2G309970) exhibits delayed root development, reduced root length, and fewer lateral roots as compared with wild-type controls (Patel et al., 2004). SCN1 (a homolog of GRMZM2G012814) encodes a RhoGTPase GDP dissociation inhibitor (RhoGDI) that restricts the initiation of root hairs to trichoblasts (Carol et al., 2005).
Figure 3.
Expression levels of three maize homologs of Arabidopsis root-related genes were associated with smArea via eRD-GWAS. A, Manhattan plot of eRD-GWAS for smArea. Three homologs of Arabidopsis root-related genes— ZmSGT1 (GRMZM2G105019), ZmSCN1 (GRMZM2G012814), and zmWPP2 (GRMZM2G309970) —were detected. Correlation coeffecients (r) of expression levels and trait values of smArea for the three genes are −0.23, 0.25, and 0.22, respectively. P < 0.01 for all correlations. B to D, Representative root images of inbred lines having extremely low and extremely high expression levels of the three candidate genes.
Network Analyses of RSA-Associated Genes
Expression quantitative trait loci (eQTL) mapping is used to identify DNA polymorphisms associated with variation in gene regulation (Gilad et al., 2008). 110 of the RSA-candidate genes were expressed in at least half of the 246 genotypes used for eRD-GWAS. eQTL analyses were conducted for each of the 66/77 qualified SNP-genes and each of the 44/62 eRD-genes that passed this expression profile criterion (“Materials and Methods”). At an FDR cutoff of <0.05, 601 eQTL were identified for 58/66 (88%) of the SNP-genes and 39/44 (89%) of the eRD-genes (Supplemental Table S10). In cis and trans, 69/601 (11.5%) and 447/511 (88.5%) of these eQTL acted, respectively (Supplemental Table S11; “Materials and Methods”). For at least one of the other 61 genes, 36 of the 97 (= 58 +3 9; 37%) SNP-genes and eRD-genes are themselves transeQTL. This level of enrichment is statistically significant (P = 2.2e-16, “Materials and Methods”), and suggests the existence of a regulatory network involving both SNP-genes and eRD-genes.
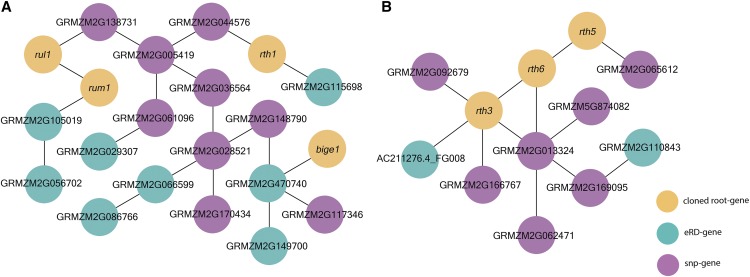
To further explore the existence of a regulatory network, a Gaussian Graphical model (GGM) was used to construct a GGM-based coexpression network for the 246 genotypes using the RNA-seq data from root tips that had been used in the eQTL analyses, and thereby identify putative regulatory relationships among the 139 RSA-associated genes (77 SNP-genes and 62 eRD-genes) and the nine root-related genes (eight cloned maize root-related genes, plus rum1-like1, a homeolog of rum1; “Materials and Methods”; Hochholdinger et al., 2018). In total, 26 unique RSA-associated genes (16/77 SNP-genes and 10/62 eRD-genes) are coexpressed with one or more cloned maize root-related genes. For example, 17 root candidate genes (nine SNP-genes and eight eRD-genes) were included in the GGM-based coexpression network that contains rth1, rum1, rul1, and bige1 (Fig. 4). The rth1 gene encodes the SEC3 subunit (Wen et al., 2005) of the exocyst complex (Hála et al., 2008) that controls the exocytotic growth of root hair tip. The rum1 gene encodes an AUX/IAA protein and plays key roles in lateral and seminal root formation (Woll et al., 2005; Zhang et al., 2016), whereas rul1 is a homeolog of rum1 that exhibits 92% sequence identity and shares the canonical features of AUX/IAA protein (von Behrens et al., 2011). In another module of the GGM-based coexpression network, nine root candidate genes (seven SNP-genes and two eRD-genes) were coexpressed with rth3, rth5, and rth6 (Fig. 4). Rth5 and rth6 play important roles in cellulose biosynthesis and are involved in root hair elongation (Nestler et al., 2014; Li et al., 2016), whereas rth3 is a member of COBRA gene family that is required for root hair elongation and contributes to grain yield (Wen and Schnable, 1994; Hochholdinger et al., 2008).
Figure 4.
Gaussian Graphical model–based coexpression networks. Two clusters illustrating putative regulatory relationships among RSA-associated genes (A) and cloned root genes (B) are shown. Yellow dots indicate cloned root-related genes, green dots indicate genes identified via eRD GWAS, and purple dots indicate genes identified via SNP-based GWAS.
Comparative GWAS for RSA of Maize and Sorghum
Core root systems of up to five competitive plants were also collected and phenotyped using the CREAMD-COFE pipeline for a subset (n = 294) of the Sorghum Association Panel (SAP; Casa et al., 2008), which will be designated the SAP-RSA (Supplemental Table S12). The SAP-RSA was grown in Mead, NE (“Materials and Methods”), and phenotyped using CREAMD-COFE for the same RSA traits as was done for maize. The heritabilities and pairwise correlations of these traits in sorghum were similar to maize (Supplemental Fig. S8). GWAS for the SAP-RSA was conducted using 205k SNPs from published Genotyping by Sequencing (GBS) data (Morris et al., 2013). In total, 132 TASs (comprising 115 unique TASs) were detected for the RSA traits with FDR <0.05 (Supplemental Table S13). Among the 132 sorghum TASs, 9% (12/132) were associated with multiple RSA traits or two views of the same RSA trait. Whereas the minor allele frequencies of sorghum TASs are similar to those of the maize, the effect sizes of TASs, which is an estimate of the contribution of each SNP to the total genetic variance (Park et al., 2011), from sorghum are significantly larger than those from maize (P < 0.01; Supplemental Fig. S9), presumably reflecting the greater statistical power of the maize GWAS, resulting in a greater ability to detect smaller effect loci.
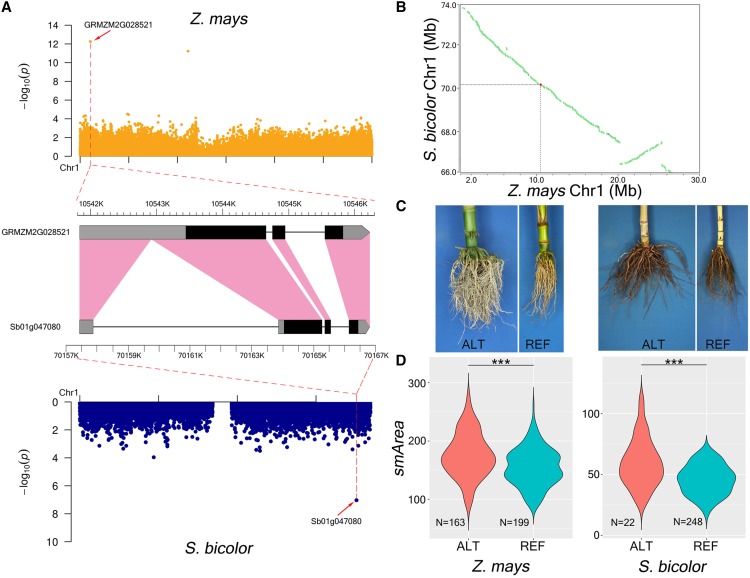
The similarities of maize and sorghum RSAs (Yamauchi et al., 1987), in addition to the syntenic relationship of their genomes (Schnable et al., 2011, 2012), led us to hypothesize that these species have conserved genetic control for RSA. To test this hypothesis, a comparison was conducted between the unique TASs from GWAS for RSA for maize and sorghum. Syntenic genes were identified within 20-kb windows centered on maize TASs and 500-kb windows centered on sorghum TASs (“Materials and Methods”). These window sizes were selected based on average linkage disequilibrium (LD) values of 10 kb and 250 kb for the SAM diversity and SAP-RSA panels, respectively (“Materials and Methods”). With use of an FDR cutoff of <0.05 for both species, seven pairs of syntenic genes were identified (Supplemental Table S14). Based on a permutation test, this is more overlap than would be expected by chance (P = 1e-04, “Materials and Methods”). For example, GRMZM2G028521, annotated as maize citrate transporter 1 (citt1), was identified via SNP-based GWAS for smArea and lgMaxWidth. Its sorghum homolog Sb01g047080 was 138 kb away from the sorghum TAS associated with both smArea and lgMaxWidth (Fig. 5). Although some syntenic gene pairs were not associated with the same RSA traits in maize and sorghum, the associated traits exhibited high correlations.
Figure 5.
Comparative GWAS between maize and sorghum for smArea. A, Manhattan plots of Chromosome 1 from SNP-based GWAS for smArea of maize (top) and sorghum (botttom) identified a pair of RSA-associated syntentic genes; homologous sequences are indicated in pink. B, Genomic positions of the syntenic gene pair from (A). C, Inbred lines of maize (left pair; LH150 and A188) and sorghum (right pair; White Kafir and D940Y) fixed for ALT and REF alleles of the SNPs associated with smArea. D, Distribution of trait values of maize (left) and sorghum (right) inbred lines homozygous for the ALT and REF alleles of the SNPs associated with smArea. Student’s t test; ***P < 0.001.
This is also more overlap than we detected in two pairs of intraspecific GWAS for above-ground traits conducted as controls. First, we conducted GWAS for multiple traits using mostly previously published data from two genetically distinct maize diversity panels grown in separate environments. The Yan panel, which consists of 368 inbred lines, was grown in China (Li et al., 2013; Yang et al., 2014), whereas the SAM Diversity Panel (Leiboff et al., 2015) was grown in the United States (“Materials and Methods”). These panels do not include any shared inbred lines. Both panels were phenotyped for four traits: plant height, plant ear height, flowering time, and ear length. Data for the Yan and SAM Diversity Panels were obtained from Yang et al. (2014) and Leiboff et al. (2015), respectively (except for EL of the SAM Diversity Panel, which is previously unpublished data, see “Materials and Methods”). Through GWAS conducted using an FDR cutoff of <0.05 for both panels, 24 and 18 TASs were detected from the Yan and SAM Diversity Panels, respectively (Supplemental Table S15). With use of methodology similar to that described for the comparative interspecific GWAS for RSA (“Materials and Methods”), no overlapping TASs were identified between the two maize panels, even using window sizes as large as 100 kb. Next, we conducted another pair of intraspecific GWAS on two diversity panels that consisted of the same inbred lines and that were genotyped with the same set of SNPs, but that were grown in different environments and phenotyped by different groups. Ninety-seven percent (273/282) of the members of the Maize 282 association panel (Peiffer et al., 2014) are a subset of the SAM panel. This subset of 273 inbred lines will be referred to as the “Maize273” and “SAM273” panels. Both panels were phenotyped for four traits: plant height, plant ear height, flowering time, and ear length (“Materials and Methods”). Detected from the Maize273 and SAM273 panels were 15 and 13 TASs, respectively (Supplemental Table S16). Even though presumably genetically identical inbred lines were analyzed with the same genotyping data, only two overlapping TAS were identified. The number of shared candidates did not increase even when using window sizes up to 100 kb. The absence of shared signals identified via GWAS conducted within a single species and the very small number of overlapping signals within a single diversity panel provides further evidence that the multiple pairs of RSA-associated syntenic genes detected between the two species is significant.
DISCUSSION
Accurate phenotyping is an essential component of GWAS. Phenotyping RSA, i.e. the topology and distribution of roots (Lynch, 1995), is challenging due to tradeoffs between throughput and intactness (Topp et al., 2016). To enable high-throughput excavation and cleaning of core root systems, thereby making feasible GWAS for RSA, we developed the CREAMD pipeline, which offers a 6X speed advantage in root cleaning as compared with conventional water-based methods (Trachsel et al., 2011; Colombi et al., 2015), while yielding comparably intact core root systems. In addition, the towable air-compressor, which is the key component of the CREAMD pipeline, simplifies the phenotyping of RSA in multiple environments, even when a nearby water source is not available. This promises to make the study of genotype-environment interactions of RSA feasible.
Another phenotyping challenge is the complicated topology and structure of RSA, particularly of adult plants. Like others (Trachsel et al., 2011; Topp et al., 2013; Pace et al., 2014), we used multiple 2D images in an effort to capture more of the 3D complexity of RSA. To convert these images to trait values we developed the COFE software, which offers several advantages relative to alternative software packages such as GiA Roots (Galkovskyi et al., 2012) and DIRT (Das et al., 2015). The accuracy and flexibility of the CREAMD-COFE pipeline is supported both by comparisons to ground truth data and the relatively high heritabilities observed across highly diverse germplasm that exhibits highly divergent RSA phenotypes. Because the density cutoffs used for the AdjDepth traits were selected to maximize heritabilities (Supplemental Fig. S5) and these cutoffs are likely to be affected by factors such as soil type, crop management, excavation date, and weather, we recommend determining the optimal cutoffs for each independent project.
The availability of the CREAMD-COFE pipeline enabled us to conduct high-throughput phenotyping of RSA traits in diversity panels of adult, field-grown maize, and sorghum plants. After collecting phenotypic data, we used two complementary GWAS approaches to identify RSA-associated maize genes. Conventional SNP-based GWAS associate variation in SNP genotypes across a diversity panel with phenotypic variation. By contrast, eRD-GWAS uses expression levels of genes as the explanatory variable for GWAS (Lin et al., 2017). The robustness of eRD-GWAS is demonstrated by the fact that even though read counts obtained from RNA-seq data from root tips excised from germinating seedlings were used as the explanatory variable for the RSA of adult field-grown plants, it was still possible to identify strongly supported candidate genes. Consistent with Lin et al. (2017), few RSA-associated genes were detected by both GWAS approaches, providing further evidence that the two approaches are complementary.
The ability of our pipeline to detect true positives is supported by the finding that homologs of 16% (22/139) of the RSA-associated maize genes are known to affect RSA in other species, one of the highest confirmation rates reported in crops (Xiao et al., 2017). In addition, 26 RSA-associated genes are coregulated with genes previously shown to affect RSA and 37% of RSA-associated genes are themselves transeQTL for at least one other qualified RSA-associated gene (again, significantly more than would be expected by chance). Finally, we detected substantially more pairs of RSA-associated syntenic genes in maize and sorghum than would be expected by chance. In combination, these results provide strong support for the accuracies of our gene/trait associations and demonstrate that the CREAMD-COFE pipeline is sufficiently accurate for use in GWAS.
We photographed each core root system from two directions. Initially, we were surprised that there was little overlap between the SNPs or genes associated with a given trait from the two views. However, in contrast with published reports (Colombi et al., 2015), we showed that core root systems are not radially symmetrical. As a consequence of this asymmetry, the two 2D images we captured of a given core root system typically exhibited different trait values. It is therefore not surprising that we often identified different genes as being associated with the same nominal “trait” from the smaller and larger views of the same core root system.
Seven pairs of syntenic maize and sorghum genes are associated with RSA traits, which is significantly more overlap than would be expected by chance. Chen et al. (2016) used a comparative GWAS approach to identify shared genetic control among maize and rice homologs for biochemical composition of grain and leaves. Their analysis relied upon conservation of biochemical pathways across taxa. It was not obvious that the regulation of morphology would be shared across taxa as has been demonstrated by this study.
There is substantial overlap among the RSA-associated genes detected in maize and sorghum that were grown in different environments but phenotyped by the same group using the same methodology. This is in line with the observation that overall gene expression profiles of maize roots are substantially more influenced by genotype than by environmental stress factors such as drought (Marcon et al., 2017). By contrast, we found no overlap among trait-associated genes from the same panel of maize inbred lines that had been genotyped with the same markers, but were grown in different environments and phenotyped by different groups. Although these two groups were nominally measuring the same traits, the lack of overlap among trait-associated genes suggests that differences in phenotyping methodologies and hence trait values may be a major contributor to differences in GWAS results among experiments.
Given its fast rate of LD decay, GWAS in maize results in single-gene or near single-gene resolution. By contrast, as a consequence of its slower rate LD decay as compared with maize (Morris et al., 2013), GWAS in sorghum does not (Li et al., 2015). However, due to the syntenic relationship between maize and sorghum (Schnable et al., 2011, 2012), our data indicate that GWAS in maize has the potential to identify candidate genes in that control quantitative traits within large chromosomal windows of sorghum.
More generally, our results suggest that comparative multispecies GWAS has the potential to enhance our understanding of within-species genetic architecture. Indeed, some RSA-associated genes detected in maize but not sorghum may be false-negative associations (and vice-versa). This is because within a given species it may not be possible to detect an association between a gene and a relevant trait as a consequence of (among other factors) low minor allele frequencies, small effect sizes and/or evolutionary histories (Lai et al., 2018). Hence, just as phenotypes of qualitative mutants identified in one species can inform our understanding of gene function in related species (Lin et al., 2012; Huang et al., 2017; Wang et al., 2018), GWAS results from one species have the potential to identify candidate genes in related species that are not detectable via single-species GWAS.
CONCLUSION
We report on a high-throughput phenotyping pipeline that uses compressed air to harvest and clean roots, thereby overcoming current throughput limitations. We used this approach to phenotype RSA in both maize and sorghum diversity panels and then conducted GWAS. The finding that homologs of 16% (22/139) of the detected RSA-associated maize genes are known to affect RSA in other species (one of the highest confirmation rates reported in any crop) demonstrates the accuracy of our phenotyping and analysis pipeline and suggests that the RSA-associated genes detected in this study are worthy of further investigation and exploration for use in crop improvement. Comparisons between high-confidence, RSA-associated genes identified from maize and sorghum via GWAS revealed conserved functional roles of syntenic orthologs in regulating quantitative variation. Our findings suggest that GWAS results from one species have the potential to identify candidate genes in related species that are not detectable in that second species as a consequence of, for example, low minor allele frequencies, small effect sizes, and/or differing evolutionary histories.
MATERIALS AND METHODS
Germplasm for GWAS
Three fully randomized replications of 380 maize (Zea mays) inbred lines were grown at the Iowa State University’s Curtiss Research Farm in Ames, IA with a planting date of May 9, 2017 and an interrow spacing of 88.9 cm and an average within row plant-to-plant spacing of 25.4 cm. Only phenotypic data from the 369 lines in the SAM Diversity Panel (Leiboff et al., 2015) were used for GWAS.
For sorghum (Sorghum bicolor), a subset of the SAP (Casa et al., 2008) was grown at the Agronomy Farm of University of Nebraska-Lincoln (UNL), Mead, NE with a planting date of May 15, 2017 and a planting density an interrow spacing of 72 cm and an average within row plant-to-plant spacing of 7.7 cm. Phenotypic data from 294 accessions of the SAP, referred to as the SAP-RSA, were used for GWAS.
CREAMD - Collection and Phenotyping of Core Root Systems
Core root systems, each with ∼0.3-m3 volume (Hauck et al., 2015), of typically three competitive plants (i.e. plants that are not the terminal plant at the beginning or end of a row nor adjacent to a missing plant within a row) within a row were excavated and cleaned on site using a towable commercial air compressor and an AirSpade device (Supplemental Text S2). Up to three maize roots were collected from up to three biological replications (i.e. plants grow in three different one-row plots) for a total of up to nine roots per genotype. Roots of each genotype were collected within 1 week of the end of flowering for that genotype. For the SAP-RSA, typically five competitive sorghum plants within each row were excavated and cleaned between October 12 and 18 (2017) following the same protocol. In most cases for both species it was possible to harvest competitive plants. Harvested plants that were noncompetitive (i.e. adjacent to a missing plant, or that were a border plant) or that had lodged were recorded for subsequent statistical modeling (see “Materials and Methods”).
Cleaned core root systems were imaged on a customized board (40.6 cm × 50.8 cm) covered with blue fabric. Core root systems were positioned on the center of the imaging board and a dimmable 45.7 cm-diameter light ring (Neewer Technology Co.) was placed directly beneath the camera lens to provide evenly distributed lighting to reduce shadows. A round orange marker (ø = 5.1 cm) and a tag containing an ID number for each plant were placed on the imaging platform next to the corresponding core root system. All images were captured using an EOS 5D Mark III camera with an EF 24-105 mm f/4L IS USM lens (Canon), positioned 125 cm above the imaging board surface using an adjustable mount. The camera was controlled using a laptop computer (Latitude 3550, Dell) running EOS Utility 3 software (Version 3.6.30.0) to capture images. Two images from two orthogonal views (North and West) of the core root system were taken based on the spray-painted identifier. Images were stored using JPEG file format.
For both the maize and sorghum diversity panels, a random 5% of all collected core root systems (149 maize roots and 56 sorghum roots) were chosen for ground truth measurement. The maximum width and depth of core root system were manually measured for both of the two orthogonal views (Supplemental Fig. S2). In addition, ImageJ (Schneider et al., 2012) was used to measure maximum width and depth from the images of the same sets of roots.
To determine whether core root systems exhibit radial symmetry, we collected four to six plants of the inbred lines B73 and Mo17 from three locations near Ames, Iowa on September 23, 2018: Curtiss Farm (GPS: GPS: 42°00’N, 93°39’W, planting date: May 31, 2018, planting density: ∼36 cm within row, 3 m between rows), Marsden Farm (GPS: 42°00’N, 93°47’W, planting date: May 23, 2018, planting density: ∼36 cm within row, 3 m between rows), and South Woodruff Farm (GPS: 41°58’N, 93°41’W, planting date: June 15, 2018; planting density: ∼25 cm within row, 75 cm between rows; Supplemental Fig. S3).
COFE - Image Analysis and Feature Extraction
For image analysis, we used MATLAB (The Mathworks) to develop an interactive software, Core Root Feature Extraction (COFE). Captured images were analyzed via a two-phase process: preprocessing and trait extraction (Supplemental Text S1). During preprocessing, the first visible node above the soil line of a core root system is identified by the user. Then, the software automatically generates a binary image of the root according to user-defined settings. During automated trait extraction the software uses a blurring and thresholding algorithm to prune roots that aberrantly stick out from the core root system and then extracts traits from the core root system.
Comparison of CREAMD vs. Water-Based Root Cleaning
The inbred lines B73, LH185, and PHN46 and the commercial hybrid Hoegemeyer 7089, grown during the summer of 2017 at the Curtiss Farm, were used to compare CREAMD versus a water-based root cleaning pipeline (Trachsel et al., 2011). For each method, 15 competitive plants of each genotype were processed at the time of grain harvest on October 24, 2017. The cleaning of roots with pressurized air is described in the CREAMD protocol (“Materials and Methods”; Supplemental Text S1). For the water-based root cleaning, the excavated core root system was soaked in water for ∼1 h and then water washed as described (Trachsel et al., 2011; Colombi et al., 2015). Traits were extracted using COFE from images of core root systems excavated and cleaned by both methods.
Comparative GWAS between Maize and Sorghum
For the analysis of maize RSA phenotypes, the best linear unbiased prediction of traits extracted from COFE were calculated by treating genotype and planting row as random effects, and lodging and border status as fixed effects using R package ‘lme4’ v1.1-21 (Bates et al., 2015; Supplemental Table S17). Broad sense heritability was calculated for all RSA traits for both maize and sorghum (Cai et al., 2012). For the analysis of sorghum RSA phenotypes, means of extracted trait values of all plants having the same genotype were calculated, after removing extreme values, i.e. those that were 1.5× larger than the 3rd quartile (Supplemental Table S18).
To conduct GWAS on the maize SAM diversity and sorghum SAP-RSA panels, we used 1.2M (Leiboff et al., 2015) and 205k (Morris et al., 2013) SNPs, respectively, without filtering for minor allele frequencies (Bomba et al., 2017). GWAS for both species were conducted using a C++ implementation of FarmCPU (Liu et al., 2016), termed FarmCPUpp (Kusmec and Schnable, 2018). Based on simulation studies, for moderately complex traits, FarmCPU has been reported to have the best metrics for both the detection of gene-trait associations and false-positive metrics (Miao et al., 2018). The first three principle components calculated using TASSEL 5.0 were used as covariates to control for population structure (Bradbury et al., 2007). LD values of both panels were calculated using PLINK v1.90 (Purcell et al., 2007). Based on the average rates of LD in the diversity panels, 20- and 500-kb windows centered on TASs were used to identify candidate genes in maize and sorghum, respectively. Maize AGPv2 genes models (Schnable et al., 2009) and sorghum V1.14 genes models (Paterson et al., 2009) that overlapped with the defined windows for each species using the BEDtools software (V2.23.0; Quinlan and Hall, 2010) were considered to be candidate genes.
In addition to SNP-based GWAS, eRD-GWAS (Lin et al., 2017) was conducted on a subset of the SAM Diversity Panel (n = 246 inbred lines) for which RNA-seq data from seedling root tissue were available (Kremling et al., 2018). Genes with model frequencies over an arbitrary cutoff of 0.05 were designated as candidate genes (eRD-genes).
Maize and sorghum syntenic genes were identified following the methods of Zhang et al. (2017) using the reference genomes RefGen V2 for maize and Sbi1.4 for sorghum (Supplemental Table S19). The permutation test was conducted by shuffling the maize-sorghum table 10,000 times and counting the number of pairs of syntenic genes obtained from each trial (Supplemental Table S19).
eQTL and Coexpression Network
eQTL analyses were conducted on the same 246 maize inbred lines as were used for eRD-GWAS, and using the same GWAS method (i.e. FarmCPU) and SNPs as were used for the maize RSA GWAS (see above), with the gene expression values as phenotypes and the SNPs as explanatory variables. Only those maize RSA candidate genes expressed in at least 50% of the 246 lines were included in this analysis. An eQTL was defined as acting in cis if it was within a window that extends 500 kb upstream and 500 kb downstream of the gene it regulates; eQTL outside this 1-Mb window were defined as acting in trans. Ratios of cis- and transeQTL were relatively stable with window sizes ranging from 50 kb to 2 Mb (Supplemental Table S10). The eQTL with the smallest p-value within each 50-kb window was selected for further analyses. The enrichment test of RSA-associated genes and transeQTL was performed using the “fisher.test ()” function in R.
Graphical Gaussian model–based coexpression networks were constructed using the R package ‘bnlearn’ v4.4.1 (Scutari, 2010) with 5,000 bootstraps implemented with the constraint-based learning algorithm max-min parents and children (mmpc).
Comparative Intraspecific GWAS
Both phenotypic and genotypic data of the Yan panel were retrieved from MaizeGo (http://www.maizego.org/Resources.html). SNP data for the Yan panel were generated by Li et al. (2013) from RNA-seq and MaizeSNP50 BeadChip. Phenotypic data of the Maize 282 panel were retrieved from Panzea (http://cbsusrv04.tc.cornell.edu/users/panzea/filegateway.aspx?category=Phenotypes). Phenotypic data of plant height, plant ear height, and flowering time of the SAM Diversity Panel were from Leiboff et al. (2015). Ear length data were collected from two fully randomized replications of 369 maize inbred lines from the SAM Diversity Panel (Leiboff et al., 2015) in October 2016, at Iowa State University’s Curtiss Research Farm (42°00’N, 93°39’W) in Ames, Iowa (Supplemental Table S20). Genotypic data for both the Maize273 and SAM273 panels is a subset of the data used for the root-GWAS of the SAM Diversity Panel. GWAS was conducted with the same protocol as in comparative GWAS between maize and sorghum (see above section), except an arbitrarily relaxed window of 100 kb, centered on the TAS was used here.
COFE Software is available at https://bitbucket.org/baskargroup/cofe/src/master/.
Accession Numbers
The maize sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers SRP055871. The sorghum SNP data were downloaded from https://www.morrislab.org/data.
Supplemental Data
The following supplemental materials are available.
Supplemental Text S1. CREAMD-COFE protocols.
Supplemental Text S2. Definition of Width-Profile Angle (WPA).
Supplemental Figure S1. Ground truth validation for trait values extracted from COFE. 298 images from 149 maize plants were analyzed.
Supplemental Figure S2. Comparisons of trait values extracted using COFE from roots of three genotypes.
Supplemental Figure S3. Maize core root systems grown in three environments (Curtiss, Marsden, and South Woodruff farms) exhibit a lack of radial symmetry.
Supplemental Figure S4. Classification of images taken from two angles (North and West) into larger and smaller view on a per trait basis.
Supplemental Figure S5. Illustration of algorithm for determining root depth (AdjDepth) trait values.
Supplemental Figure S6. Width-Profile Angle (WPA) was used to measure root angle.
Supplemental Figure S7. Above-ground trait values of inbred lines homozygous for the ALT and REF alleles of bige1.
Supplemental Figure S8. Correlations among RSA traits for 294 sorghum inbred lines.
Supplemental Figure S9. Minor allele frequency (MAF) and the absolute value of effect sizes of maize and sorghum TAS.
Supplemental Table S1. Time required to process 60 core root systems via CREAMD and water-based root cleaning.
Supplemental Table S2. RSA traits do not exhibit statistically different values between two orthogonal views (North and West) of the maize SAM Diversity Panel.
Supplemental Table S3. Classification of trait values of root area (Area) from two angles (North and West) into larger and smaller views on a per trait basis.
Supplemental Table S4. Correlation coefficients between larger and smaller views of RSA traits in the maize SAM diversity and sorghum (SAP-RSA) panels.
Supplemental Table S5. Correlations among RSA traits and above-ground traits in maize.
Supplemental Table S6. Maize TAS and SNP-genes at FDR < 0.05.
Supplemental Table S7. Arabidopsis homologs with known root-related functions of maize SNP-genes
Supplemental Table S8. List of eRD-genes.
Supplemental Table S9. Arabidopsis and Medicago homologs with known root-related functions of maize eRD-genes.
Supplemental Table S10. List of cis- and transeQTL.
Supplemental Table S11. Percentage of cis- and transeQTL for qualified maize RSA-associated genes using different window sizes.
Supplemental Table S12. List of inbred lines in used in GWAS for maize (SAM Diversity Panel) and sorghum (SAP-RSA).
Supplemental Table S13. Sorghum TAS at FDR < 0.05.
Supplemental Table S14. Syntenic maize-sorghum gene pairs detected via comparative GWAS.
Supplemental Table S15. List of Yan panel and SAM Diversity Panel TAS for four traits (PH, PEH, DTA, EL).
Supplemental Table S16. List of TAS for four traits (PH, PEH, DTA, EL) identified via GWAS conducted on the maize273 and SAM273 panels.
Supplemental Table S17. RSA trait values (BLUP) of maize SAM Diversity Panel.
Supplemental Table S18. RSA trait values of sorghum SAP-RSA panel.
Supplemental Table S19. List of syntenic genes.
Supplemental Table S20. Ear length trait values (BLUP) of maize SAM Diversity Panel.
Acknowledgments
We thank Dr. Dan Nettleton (Iowa State University) for statistical consultation; Cheng-Ting “Eddy” Yeh (P. Schnable Lab) for bioinformatics support; Aaron Kusmec, Hung-Ying Lin, Qiang Liu, and Yan Zhou (graduate students in the P. Schnable Lab) for useful discussions; Colton McNinch (a former graduate student in the P. Schnable Lab) for early testing of the AirSpade and software; Dr. Melinda Yerka (University of Nevada, Reno) for designing the sorghum field layout; Maureen Booth, Daniel Russell, Leo Savage, Cameron Lahn, Trenton Houston (undergraduate research assistants in the P. Schnable Lab), and Tyler Greenwald (undergraduate research assistant at the University of Nebraska, Lincoln) for assisting with root collection and/or phenotyping; and Drs. Lakshmi Attigala and An-Ping Hsia (P. Schnable Lab) for assistance editing the article.
Footnotes
This work was supported in part by the DOE Advanced Research Projects Agency - Energy (ARPA-E: grant no. DE-AR0000826).
Articles can be viewed without a subscription.
References
- Airoldi CA, Rovere FD, Falasca G, Marino G, Kooiker M, Altamura MM, Citterio S, Kater MM (2010) The Arabidopsis BET bromodomain factor GTE4 is involved in maintenance of the mitotic cell cycle during plant development. Plant Physiol 152: 1320–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos B, Walters DT (2006) Maize root biomass and net rhizodeposited carbon. Soil Sci Soc Am J 70: 1489–-1503 [Google Scholar]
- Atkinson JA, Wingen LU, Griffiths M, Pound MP, Gaju O, Foulkes MJ, Le Gouis J, Griffiths S, Bennett MJ, King J, Wells DM (2015) Phenotyping pipeline reveals major seedling root growth QTL in hexaploid wheat. J Exp Bot 66: 2283–2292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B, Walker S (2015) Fitting linear mixed-effects models using lme4. J Stat Softw 67: 1 [Google Scholar]
- Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: A practical and powerful approach to multiple testing. J R Stat Soc B 57: 289–300 [Google Scholar]
- Birnbaum K, Shasha DE, Wang JY, Jung JW, Lambert GM, Galbraith DW, Benfey PN (2003) A gene expression map of the Arabidopsis root. Science 302: 1956–1961 [DOI] [PubMed] [Google Scholar]
- Bomba L, Walter K, Soranzo N (2017) The impact of rare and low-frequency genetic variants in common disease. Genome Biol 18: 77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: Software for association mapping of complex traits in diverse samples. Bioinformatics 23: 2633–2635 [DOI] [PubMed] [Google Scholar]
- Bray JR, Lawrence DB, Pearson LC (1959) Primary production in some Minnesota terrestrial communities for 1957. Oikos 10: 38–49 [Google Scholar]
- Cai H, Chen F, Mi G, Zhang F, Maurer HP, Liu W, Reif JC, Yuan L (2012) Mapping QTLs for root system architecture of maize (Zea mays L.) in the field at different developmental stages. Theor Appl Genet 125: 1313–1324 [DOI] [PubMed] [Google Scholar]
- Carol RJ, Takeda S, Linstead P, Durrant MC, Kakesova H, Derbyshire P, Drea S, Zarsky V, Dolan L (2005) A RhoGDP dissociation inhibitor spatially regulates growth in root hair cells. Nature 438: 1013–1016 [DOI] [PubMed] [Google Scholar]
- Casa AM, Pressoir G, Brown PJ, Mitchell SE, Rooney WL, Tuinstra MR, Franks CD, Kresovich S (2008) Community resources and strategies for association mapping in Sorghum. Crop Sci 48: 30–40 [Google Scholar]
- Chen W, Wang W, Peng M, Gong L, Gao Y, Wan J, Wang S, Shi L, Zhou B, Li Z, et al. (2016) Comparative and parallel genome-wide association studies for metabolic and agronomic traits in cereals. Nat Commun 7: 12767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombi Kirchgessner TN, eLe Mari CA, York LM, Lynch JP, Hund A (2015) Next generation shovelomics: Set up a tent and REST. Plant Soil 388: 1–20 [Google Scholar]
- Courtois B, Audebert A, Dardou A, Roques S, Ghneim-Herrera T, Droc G, Frouin J, Rouan L, Gozé E, Kilian A, Ahmadi N, Dingkuhn M (2013) Genome-wide association mapping of root traits in a japonica rice panel. PLoS One 8: e78037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das A, Schneider H, Burridge J, Ascanio AKM, Wojciechowski T, Topp CN, Lynch JP, Weitz JS, Bucksch A (2015) Digital imaging of root traits (DIRT): A high-throughput computing and collaboration platform for field-based root phenomics. Plant Methods 11: 51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RM, Gowda M, Moghe G, Lin H, Vaillancourt B, Shiu SH, Jiang N, Robin Buell C (2012) Comparative transcriptomics of three Poaceae species reveals patterns of gene expression evolution. Plant J 71: 492–502 [DOI] [PubMed] [Google Scholar]
- Dewey CN. (2011) Positional orthology: Putting genomic evolutionary relationships into context. Brief Bioinform 12: 401–412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan L, Janmaat K, Willemsen V, Linstead P, Poethig S, Roberts K, Scheres B (1993) Cellular organisation of the Arabidopsis thaliana root. Development 119: 71–84 [DOI] [PubMed] [Google Scholar]
- Fang S, Yan X, Liao H (2009) 3D reconstruction and dynamic modeling of root architecture in situ and its application to crop phosphorus research. Plant J 60: 1096–1108 [DOI] [PubMed] [Google Scholar]
- Galkovskyi T, Mileyko Y, Bucksch A, Moore B, Symonova O, Price CA, Topp CN, Iyer-Pascuzzi AS, et al. (2012) GiA Roots: Software for the high throughput analysis of plant root system architecture. BMC Plant Biol 12: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Rifkin SA, Pritchard JK (2008) Revealing the architecture of gene regulation: The promise of eQTL studies. Trends Genet 24: 408–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giuliani S, Sanguineti MC, Tuberosa R, Bellotti M, Salvi S, Landi P (2005) Root-ABA1, a major constitutive QTL, affects maize root architecture and leaf ABA concentration at different water regimes. J Exp Bot 56: 3061–3070 [DOI] [PubMed] [Google Scholar]
- Gray WM, Muskett PR, Chuang H-W, Parker JE (2003) Arabidopsis SGT1b is required for SCF(TIR1)-mediated auxin response. Plant Cell 15: 1310–1319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grift TE, Novais J, Bohn M (2011) High-throughput phenotyping technology for maize roots. Biosyst Eng 110: 40–48 [Google Scholar]
- Guo J, Chen L, Li Y, Shi Y, Song Y, Zhang D, Li Y, Wang T, Yang D, Li C (2018) Meta-QTL analysis and identification of candidate genes related to root traits in maize. Euphytica 214: 223 [Google Scholar]
- Hála M, Cole R, Synek L, Drdová E, Pecenková T, Nordheim A, Lamkemeyer T, Madlung J, Hochholdinger F, Fowler JE, Zárský V (2008) An exocyst complex functions in plant cell growth in Arabidopsis and tobacco. Plant Cell 20: 1330–1345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hargreaves CE, Gregory PJ, Bengough AG (2009) Measuring root traits in barley (Hordeum vulgare ssp. vulgare and ssp. spontaneum) seedlings using gel chambers, soil sacs and X-ray microtomography. Plant Soil 316: 285–297 [Google Scholar]
- Hauck AL, Novais J, Grift TE, Bohn MO (2015) Characterization of mature maize (Zea mays L.) root system architecture and complexity in a diverse set of Ex-PVP inbreds and hybrids. Springerplus 4: 424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Tuberosa R (2009) Genetic and genomic dissection of maize root development and architecture. Curr Opin Plant Biol 12: 172–177 [DOI] [PubMed] [Google Scholar]
- Hochholdinger F, Wen TJ, Zimmermann R, Chimot-Marolle P, da Costa e Silva O, Bruce W, Lamkey KR, Wienand U, Schnable PS (2008) The maize (Zea mays L.) roothairless3 gene encodes a putative GPI-anchored, monocot-specific, COBRA-like protein that significantly affects grain yield. Plant J 54: 888–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochholdinger F, Yu P, Marcon C (2018) Genetic control of root system development in maize. Trends Plant Sci 23: 79–88 [DOI] [PubMed] [Google Scholar]
- Huang G, Liang W, Sturrock CJ, Pandey BK, Giri J, Mairhofer S, Wang D, Muller L, Tan H, York LM, et al. (2018) Rice actin binding protein RMD controls crown root angle in response to external phosphate. Nat Commun 9: 2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Jiang H, Zhu C, Barry K, Jenkins J, Sandor L, Schmutz J, Box MS, Kellogg EA, Brutnell TP (2017) Sparse panicle1 is required for inflorescence development in Setaria viridis and maize. Nat Plants 3: 17054. [DOI] [PubMed] [Google Scholar]
- Jones MA, Raymond MJ, Smirnoff N (2006) Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45: 83–100 [DOI] [PubMed] [Google Scholar]
- Kiesselbach TA. (1999) The Structure and Reproduction of Corn 50th Anniversary Edition. CSHL Press, Cold Spring Harbor, NY [Google Scholar]
- Kremling KAG, Chen SY, Su MH, Lepak NK, Romay MC, Swarts KL, Lu F, Lorant A, Bradbury PJ, Buckler ES (2018) Dysregulation of expression correlates with rare-allele burden and fitness loss in maize. Nature 555: 520–523 [DOI] [PubMed] [Google Scholar]
- Kusmec A, Schnable PS (2018) FarmCPUpp: Efficient large-scale genomewide association studies. Plant Direct; [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai X, Yan L, Lu Y, Schnable JC (2018) Largely unlinked gene sets targeted by selection for domestication syndrome phenotypes in maize and sorghum. Plant J 93: 843–855 [DOI] [PubMed] [Google Scholar]
- Landgraf R, Smolka U, Altmann S, Eschen-Lippold L, Senning M, Sonnewald S, Weigel B, Frolova N, Strehmel N, Hause G, et al. (2014) The ABC transporter ABCG1 is required for suberin formation in potato tuber periderm. Plant Cell 26: 3403–3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leiboff S, Li X, Hu HC, Todt N, Yang J, Li X, Yu X, Muehlbauer GJ, Timmermans MCP, Yu J, Schnable PS, Scanlon MJ (2015) Genetic control of morphometric diversity in the maize shoot apical meristem. Nat Commun 6: 8974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Peng Z, Yang X, Wang W, Fu J, Wang J, Han Y, Chai Y, Guo T, Yang N, et al. (2013) Genome-wide association study dissects the genetic architecture of oil biosynthesis in maize kernels. Nat Genet 45: 43–50 [DOI] [PubMed] [Google Scholar]
- Li L, Hey S, Liu S, Liu Q, McNinch C, Hu HC, Wen TJ, Marcon C, Paschold A, Bruce W, Schnable PS, Hochholdinger F (2016) Characterization of maize roothairless6 which encodes a D-type cellulose synthase and controls the switch from bulge formation to tip growth. Sci Rep 6: 34395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Li X, Fridman E, Tesso TT, Yu J (2015) Dissecting repulsion linkage in the dwarfing gene Dw3 region for sorghum plant height provides insights into heterosis. Proc Natl Acad Sci USA 112: 11823–11828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Mu P, Li C, Zhang H, Li Z, Gao Y, Wang X (2005) QTL mapping of root traits in a doubled haploid population from a cross between upland and lowland japonica rice in three environments. Theor Appl Genet 110: 1244–1252 [DOI] [PubMed] [Google Scholar]
- Liang Q, Cheng X, Mei M, Yan X, Liao H (2010) QTL analysis of root traits as related to phosphorus efficiency in soybean. Ann Bot 106: 223–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin H, Liu Q, Li X, Yang J, Liu S, Huang Y, Scanlon MJ, Nettleton D, Schnable PS (2017) Substantial contribution of genetic variation in the expression of transcription factors to phenotypic variation revealed by eRD-GWAS. Genome Biol 18: 192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Li X, Shannon LM, Yeh CT, Wang ML, Bai G, Peng Z, Li J, Trick HN, Clemente TE, et al. (2012) Parallel domestication of the Shattering1 genes in cereals. Nat Genet 44: 720–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12: e1005767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch J. (1995) Root architecture and plant productivity. Plant Physiol 109: 7–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes JV, Garvin DF, Wang Y, Sorrells ME, Klein PE, Schaffert RE, Li L, Kochian LV (2004) Comparative mapping of a major aluminum tolerance gene in sorghum and other species in the poaceae. Genetics 167: 1905–1914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mai CD, Phung NTP, To HTM, Gonin M, Hoang GT, Nguyen KL, Do VN, Courtois B, Gantet P (2014) Genes controlling root development in rice. Rice (N Y) 7: 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon C, Paschold A, Malik WA, Lithio A, Baldauf JA, Altrogge L, Opitz N, Lanz C, Schoof H, Nettleton D, et al. (2017) Stability of single-parent gene expression complementation in maize hybrids upon water deficit stress. Plant Physiol 173: 1247–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao C, Yang J, Schnable JC (2018) Optimising the identification of causal variants across varying genetic architectures in crops. Plant Biotechnol J 17: 893–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GP, Ramu P, Deshpande SP, Hash CT, Shah T, Upadhyaya HD, Riera-Lizarazu O, Brown PJ, Acharya CB, Mitchell SE, et al. (2013) Population genomic and genome-wide association studies of agroclimatic traits in sorghum. Proc Natl Acad Sci USA 110: 453–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler J, Liu S, Wen TJ, Paschold A, Marcon C, Tang HM, Li D, Li L, Meeley RB, Sakai H, et al. (2014) Roothairless5, which functions in maize (Zea mays L.) root hair initiation and elongation encodes a monocot-specific NADPH oxidase. Plant J 79: 729–740 [DOI] [PubMed] [Google Scholar]
- Omori F, Mano Y (2007) QTL mapping of root angle in F2 populations from maize ‘B73’ × teosinte ‘Zea luxurians.’. Plant Root 1: 57–65 [Google Scholar]
- Pace J, Lee N, Naik HS, Ganapathysubramanian B, Lübberstedt T (2014) Analysis of maize (Zea mays L.) seedling roots with the high-throughput image analysis tool ARIA (Automatic Root Image Analysis). PLoS One 9: e108255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Gail MH, Weinberg CR, Carroll RJ, Chung CC, Wang Z, Chanock SJ, Fraumeni JF Jr., Chatterjee N (2011) Distribution of allele frequencies and effect sizes and their interrelationships for common genetic susceptibility variants. Proc Natl Acad Sci USA 108: 18026–18031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel S, Rose A, Meulia T, Dixit R, Cyr RJ, Meier I (2004) Arabidopsis WPP-domain proteins are developmentally associated with the nuclear envelope and promote cell division. Plant Cell 16: 3260–3273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. (2009) The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556 [DOI] [PubMed] [Google Scholar]
- Peiffer JA, Romay MC, Gore MA, Flint-Garcia SA, Zhang Z, Millard MJ, Gardner CAC, McMullen MD, Holland JB, Bradbury PJ, Buckler ES (2014) The genetic architecture of maize height. Genetics 196: 1337–1356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Schauer MA, Megraw M, Breakfield NW, Thompson JW, Georgiev S, Soderblom EJ, Ohler U, Moseley MA, Grossniklaus U, Benfey PN (2012a) The protein expression landscape of the Arabidopsis root. Proc Natl Acad Sci USA 109: 6811–6818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petricka JJ, Winter CM, Benfey PN (2012b) Control of Arabidopsis root development. Annu Rev Plant Biol 63: 563–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poorter H, Fiorani F, Pieruschka R, Wojciechowski T, van der Putten WH, Kleyer M, Schurr U, Postma J (2016) Pampered inside, pestered outside? Differences and similarities between plants growing in controlled conditions and in the field. New Phytol 212: 838–855 [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: A tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81: 559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM (2010) BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauh L, Basten C, Buckler S IV (2002) Quantitative trait loci analysis of growth response to varying nitrogen sources in Arabidopsis thaliana. Theor Appl Genet 104: 743–750 [DOI] [PubMed] [Google Scholar]
- Richard CAI, Hickey LT, Fletcher S, Jennings R, Chenu K, Christopher JT (2015) High-throughput phenotyping of seminal root traits in wheat. Plant Methods 11: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC. (2015) Genome evolution in maize: From genomes back to genes. Annu Rev Plant Biol 66: 329–343 [DOI] [PubMed] [Google Scholar]
- Schnable JC, Freeling M (2011) Genes identified by visible mutant phenotypes show increased bias toward one of two subgenomes of maize. PLoS One 6: e17855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC, Freeling M, Lyons E (2012) Genome-wide analysis of syntenic gene deletion in the grasses. Genome Biol Evol 4: 265–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable JC, Springer NM, Freeling M (2011) Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc Natl Acad Sci USA 108: 4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, et al. (2009) The B73 maize genome: Complexity, diversity, and dynamics. Science 326: 1112–15 [DOI] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 Years of image analysis. Nat Methods 9: 671–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarz M. (1972) Influence of root crown temperature on plant development. Plant Soil 37: 435–439 [Google Scholar]
- Scutari M. (2010) Learning Bayesian networks with the bnlearn R Package. J Stat Softw 35: 1–2221603108 [Google Scholar]
- Singh V, van Oosterom EJ, Jordan DR, Messina CD, Cooper M, Hammer GL (2010) Morphological and architectural development of root systems in sorghum and maize. Plant Soil 333: 287–299 [Google Scholar]
- Suzuki M, Sato Y, Wu S, Kang B-H, McCarty DR (2015) Conserved functions of the MATE transporter BIG EMBRYO1 in regulation of lateral organ size and initiation rate. Plant Cell 27: 2288–2300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swigoňová Z, Lai J, Ma J, Ramakrishna W, Llaca V, Bennetzen JL, Messing J (2004) Close split of sorghum and maize genome progenitors. Genome Res 14(10A): 1916–1923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson MJ, Tai TH, McClung AM, Lai XH, Hinga ME, Lobos KB, Xu Y, Martinez CP, McCouch SR (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107: 479–493 [DOI] [PubMed] [Google Scholar]
- Topp CN. (2016) Hope in change: The role of root plasticity in crop yield stability. Plant Physiol 172: 5–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp CN, Bray AL, Ellis NA, Liu Z (2016) How can we harness quantitative genetic variation in crop root systems for agricultural improvement? J Integr Plant Biol 58: 213–225 [DOI] [PubMed] [Google Scholar]
- Topp CN, Iyer-Pascuzzi AS, Anderson JT, Lee C-R, Zurek PR, Symonova O, Zheng Y, Bucksch A, Mileyko Y, Galkovskyi T, et al. (2013) 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci USA 110: E1695–E1704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP (2013) Maize root growth angles become steeper under low N conditions. F Crop Res 140: 18–31 [Google Scholar]
- Trachsel S, Kaeppler SM, Brown KM, Lynch JP (2011) Shovelomics: High throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil 341: 75–87 [Google Scholar]
- Uga Y, Sugimoto K, Ogawa S, Rane J, Ishitani M, Hara N, Kitomi Y, Inukai Y, Ono K, Kanno N, et al. (2013) Control of root system architecture by DEEPER ROOTING 1 increases rice yield under drought conditions. Nat Genet 45: 1097–1102 [DOI] [PubMed] [Google Scholar]
- Vitha S, Zhao L, Sack FD (2000) Interaction of root gravitropism and phototropism in Arabidopsis wild-type and starchless mutants. Plant Physiol 122: 453–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Behrens I, Komatsu M, Zhang Y, Berendzen KW, Niu X, Sakai H, Taramino G, Hochholdinger F (2011) Rootless with undetectable meristem 1 encodes a monocot-specific AUX/IAA protein that controls embryonic seminal and post-embryonic lateral root initiation in maize. Plant J 66: 341–353 [DOI] [PubMed] [Google Scholar]
- Wallner ES, López-Salmerón V, Belevich I, Poschet G, Jung I, Grünwald K, Sevilem I, Jokitalo E, Hell R, Helariutta Y, et al. (2017) Strigolactone- and karrikin-independent SMXL proteins are central regulators of phloem formation. Curr Biol 27: 1241–1247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Li W, Fang C, Xu F, Liu Y, Wang Z, Yang R, Zhang M, Liu S, Lu S, et al. (2018) Parallel selection on a dormancy gene during domestication of crops from multiple families. Nat Genet 50: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Wen T-J, Hochholdinger F, Sauer M, Bruce W, Schnable PS (2005) The roothairless1 gene of maize encodes a homolog of sec3, which is involved in polar exocytosis. Plant Physiol 138: 1637–1643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen T-J, Schnable PS (1994) Analysis of mutants of three genes that influence root hair development in Zea mays (Gramineae) suggest that root hairs are dispensable. Am J Bot 81: 833–842 [Google Scholar]
- Woll K, Borsuk LA, Stransky H, Nettleton D, Schnable PS, Hochholdinger F (2005) Isolation, characterization, and pericycle-specific transcriptome analyses of the novel maize lateral and seminal root initiation mutant rum1. Plant Physiol 139: 1255–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao Y, Liu H, Wu L, Warburton M, Yan J (2017) Genome-wide association studies in maize: Praise and stargaze. Mol Plant 10: 359–374 [DOI] [PubMed] [Google Scholar]
- Yadav V, Molina I, Ranathunge K, Castillo IQ, Rothstein SJ, Reed JW (2014) ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell 26: 3569–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi A, KoNo Y, Jiro T (1987) Comparison of root system structures of 13 species of cereals. Jpn J Crop Sci 56: 618–631 [Google Scholar]
- Yang N, Lu Y, Yang X, Huang J, Zhou Y, Ali F, Wen W, Liu J, Li J, Yan J (2014) Genome wide association studies using a new nonparametric model reveal the genetic architecture of 17 agronomic traits in an enlarged maize association panel. PLoS Genet 10: e1004573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Marcon C, Tai H, von Behrens I, Ludwig Y, Hey S, Berendzen KW, Hochholdinger F (2016) Conserved and unique features of the homeologous maize Aux/IAA proteins ROOTLESS WITH UNDETECTABLE MERISTEM 1 and RUM1-like 1. J Exp Bot 67: 1137–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Ngu DW, Carvalho D, Liang Z, Qiu Y, Roston RL, Schnable JC (2017) Differentially regulated orthologs in sorghum and the subgenomes of maize. Plant Cell 29: 1938–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobel RW. (2016) Arabidopsis: An adequate model for dicot root systems? Front Plant Sci 7: 58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurek PR, Topp CN, Benfey PN (2015) Quantitative trait locus mapping reveals regions of the maize genome controlling root system architecture. Plant Physiol 167: 1487–1496 [DOI] [PMC free article] [PubMed] [Google Scholar]