Two large deletions upstream from FLOWERING LOCUS T occurred independently in Eurasian and East-Asian cucumber populations and are associated with higher expression of this gene and earlier flowering.
Abstract
Flowering time plays a crucial role in the geographical adaptation of most crops during domestication. Cucumber (Cucumis sativus) is a major vegetable crop worldwide. From its tropical origin on the southern Asian continent, cucumber has spread over a wide latitudinal cline, but the molecular mechanisms underlying this latitudinal adaptation and the expansion of domesticated cucumber are largely unclear. Here, we report the cloning of two flowering time loci from two distinct cucumber populations and show that two large deletions upstream from FLOWERING LOCUS T (FT) are associated with higher expression of FT and earlier flowering. We determined that the two large deletions are pervasive and occurred independently in Eurasian and East-Asian populations. Nucleotide diversity analysis further revealed that the FT locus region of the cucumber genome contains a signature for a selective sweep during domestication. Our results suggest that large genetic structural variations upstream from FT were selected for and have been important in the geographic spread of cucumber from its tropical origin to higher latitudes.
Crop domestication is the outcome of selecting plants for increased adaptability to cultivation (Gepts, 2004; Gaudin et al., 2011; Chen et al., 2015) and has been a fundamental feature of human society since the origins of agriculture. The domestication of crop plants is accompanied by genetic changes, described as the domestication syndrome, which differentiate domesticated plants from their wild progenitors (Doebley et al., 2006; Jones et al., 2008; Wang et al., 2018a). The spread of agriculture from the areas of domestication has involved the dispersal of crop plants well beyond the native range of their progenitors and would often have required adaptation to new environments (Jones et al., 2008). In this respect, flowering time is a key phenomenon in the local adaptation of crops (Xue et al., 2008; Hung et al., 2012; Liu et al., 2014; Lu et al., 2017; Soyk et al., 2017; Zhang et al., 2018). A long suitable growth season facilitates late flowering, which leads to a longer vegetative growth period and the accumulation of more storage reserves, whereas a short growth season is associated with early flowering (Roux et al., 2006).
Cucumber (Cucumis sativus) is an economically important vegetable crop worldwide as well as a model system for sex determination studies and plant vascular biology (Huang et al., 2009; Li et al., 2019). Cucumber originated from India (Sebastian et al., 2010; Lv et al., 2012), where its wild form C. sativus var hardwickii grows. As a result of natural and artificial selection, cucumber has been adapted to a wide range of environments from the tropical areas to the temperate areas, spreading over 90° of latitude from Oceania to northern Europe. Phylogenetic analysis (Qi et al., 2013) revealed that cucumber collections worldwide are composed of four intercrossable botanical subgroups: the wild Indian group, the semiwild Xishuangbanna group, and two cultivated cucumber groups (Eurasian and East Asian). However, the molecular processes that contributed to cucumber domestication and dispersal are still not well understood.
In this study, we report that FLOWERING LOCUS T (CsFT) is presumably an important genetic determinant of flowering time variation in cucumber. Using near-isogenic lines (NILs), we studied two distinct populations to fine-map the late-flowering locus 1.1 (Lf1.1) and the early-flowering locus 1.1 (Ef1.1). We identified two independent large deletions, located a considerable distance upstream from CsFT, as candidate causal mutations associated with CsFT expression levels and flowering time. We show that the two large deletions are pervasive and originated independently in natural cucumber populations. Our results reveal that the CsFT locus was targeted by artificial selection and has been important in cucumber adaptation to higher latitudes.
RESULTS
A Large Diversity of Flowering Time in Four Cucumber Populations
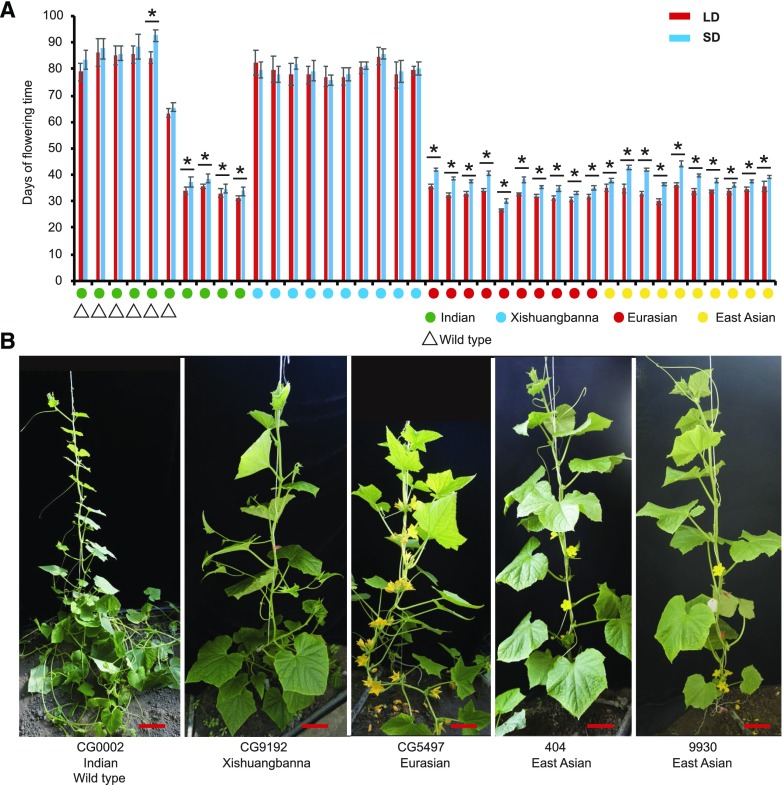
To investigate differences in flowering time between the four cucumber subgroups (Indian, Xishuangbanna, Eurasian, and East Asian), we measured the time to first male flower of 10 Indian accessions, consisting of six wild accessions (C. sativus var hardwickii) and four cultivated accessions, 10 Xishuangbanna accessions, 10 Eurasian accessions, and 10 East-Asian accessions (Supplemental Data Set S1). The plants were grown under long-day (LD; 16 h of light/8 h of dark) or short-day (SD; 8 h of light/16 h of dark) conditions. All of the 40 selected accessions were monoecious and their first flowers were male, regardless of whether they were grown under LD or SD conditions; thus, the flowering time was recorded for male flowers, representing the transition from vegetative growth to reproductive growth.
Under both LD and SD conditions, the wild and Xishuangbanna accessions flowered much later than the other cultivated cucumbers, after approximately 80 d (Fig. 1) compared with 30 to 40 d for the other accessions. There was no significant difference between Xishuangbanna flowering time under LD and SD conditions, which similarly happens in most wild cucumber accessions, indicating that the male flowering time of wild and semiwild Xishuangbanna cucumbers is insensitive to daylength. Compared with the SD condition, the LD condition could significantly accelerate the first male flowering time of cultivated cucumbers, especially Eurasian and East-Asian cucumbers (Fig. 1A).
Figure 1.
Cucumber populations show great flowering time diversity and insensitivity to daylength. A, Time to first male flower of 10 Indian accessions, 10 Xishuangbanna accessions, 10 Eurasian accessions, and 10 East Asian accessions under LD (red bars) and SD (blue bars) conditions. The green circles, blue circles, red circles, and yellow circles indicate the Indian, Xishuangbanna, Eurasian, and East Asian cucumbers, respectively. The bars and lines indicate means and sd of eight individual plants. Triangles represent the wild-type accessions. Asterisks represent significant differences of flowering time (*, P < 0.05, Student’s t test). B, Representative 45-d-old accessions from four cucumber populations exhibiting major flowering time diversity. The cucumber accessions CG9192, CG5479, 404, and 9930 are also the parents of the genetic populations used in this study. Bars = 15 cm.
Positional Cloning of Lf1.1 of Xishuangbanna Cucumber
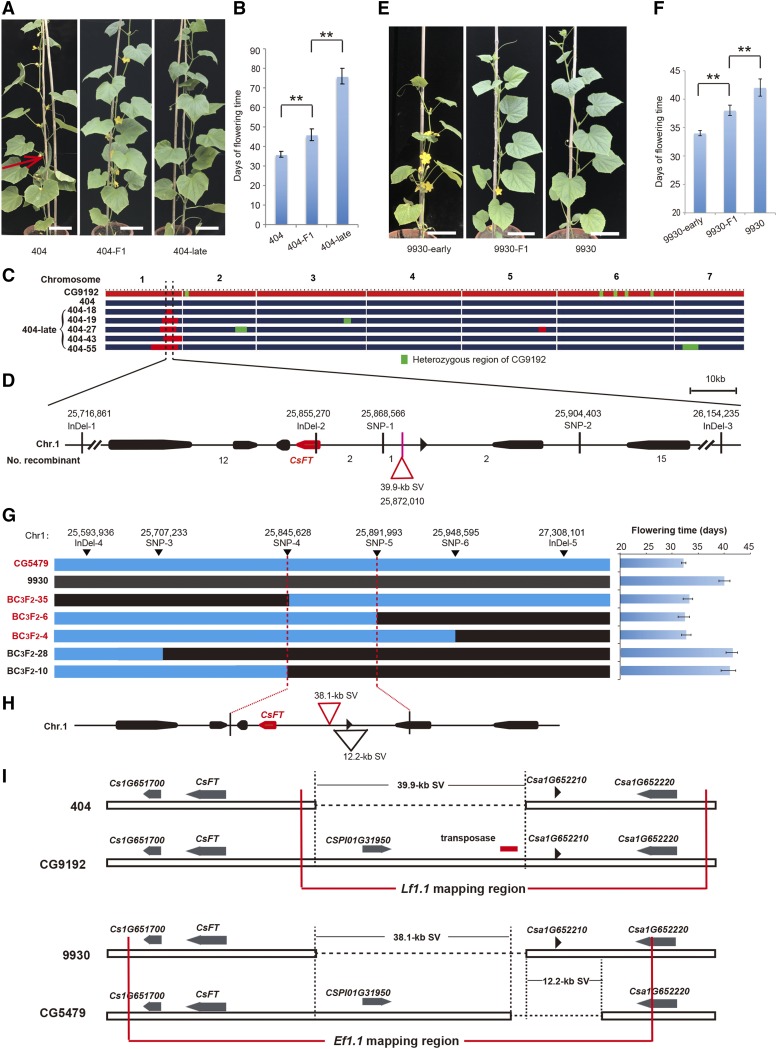
In a previous study, we generated NILs using the accession CG9192 (Xishuangbanna) as the donor parent and the accession 404 (East-Asian type) as the recurrent parent (Figs. 1B and 2, A and B; Wang et al., 2015). We observed that accession CG9192 flowered approximately 40 d later than accession 404. We identified a series of lines displaying later flowering characteristics that contained a common region (25.2–28.1 Mb) on the end of chromosome 1 (Fig. 2C), indicating that there was a genetic factor conferring the late-flowering trait in that region.
Figure 2.
Map-based cloning of Lf1.1 and Ef1.1 in two distinct F2 populations. A and B, Phenotypes and first male flowering times of NILs 404, 404-late, and the heterozygous 404-F1. C, Construction of NILs 404 and 404-late. The red color represents the homozygous region of accession CG9192, and the dark blue color shows homozygous regions of accession 404. The green color indicates heterozygous regions of CG9192. The region with dotted lines represents the position of the Lf1.1 locus on chromosome 1 of cucumber. D, High-resolution mapping of Lf1.1. The numbers above the markers indicate the physical positions on chromosome 1. Numbers below the map indicate the numbers of recombinants detected between the corresponding markers. The red triangle represents a fragment of 39.9 kb absent in accession 404 compared with CG9192. E and F, Phenotypes and first male flowering times of NILs 9930-early, 9930, and the heterozygous 9930-F1. G, Linkage analysis of Ef1.1 in the BC3F2 population from the CG5479 × 9930 cross. The numbers above the markers indicate the physical positions. CG5479, BC3F2-35, BC3F2-6, and BC3F2-4 were all early flowering (red text); 9930, BC3F2-28, and BC3F2-10 were late flowering (black text). The blue boxes represent the homozygous region of accession CG5479, and the black boxes show homozygous regions of accession 9930. The graph at right shows the flowering time of F3 families of recombinant plants. H, Genes from the fine-mapping region of Ef1.1 in the 9930 reference genome. The numbers above the markers indicate the physical positions on chromosome 1. The red triangle represents a fragment of 38.1 kb absent in 9930 compared with CG5479, and the black triangle represents a fragment of 12.2 kb absent in CG5479 compared with 9930. I, Schematic illustration of the mapping regions of the Lf1.1 and Ef1.1 loci. SVs are shown in the mapping regions of the Lf1.1 and Ef1.1 loci. Horizontal dashed lines represent large deletions, and the red lines represent the Lf1.1 and Ef1.1 mapping regions. The gray arrow shapes represent the annotated genes, and the transposase is labeled with a red rectangle. The bars and lines indicate means and sd of eight individual plants in B, F, and G. **, P < 0.01 (Student’s t test) for flowering time. The black shapes along the chromosome represent the annotated genes and the red shape represents gene CsFT in D and H. Bars in A and E = 15 cm.
To identify the causal genetic factor underlying Lf1.1, we analyzed a segregating population of 3,104 BC3F2 plants derived from the NILs, using two flanking insertion/deletion (InDel) markers (InDel-1 and InDel-3), and identified 32 recombinants. The late-flowering Lf1.1 was ultimately limited to a 35,837-bp interval (in the 9930 reference genome) between two single-nucleotide polymorphism (SNP) markers (SNP1 at nucleotide 25,868,566 and SNP2 at nucleotide 25,904,403 on chromosome 1). The recombination events between SNP1 and SNP2 were confirmed in an analysis of the corresponding F3 families (Fig. 2D).
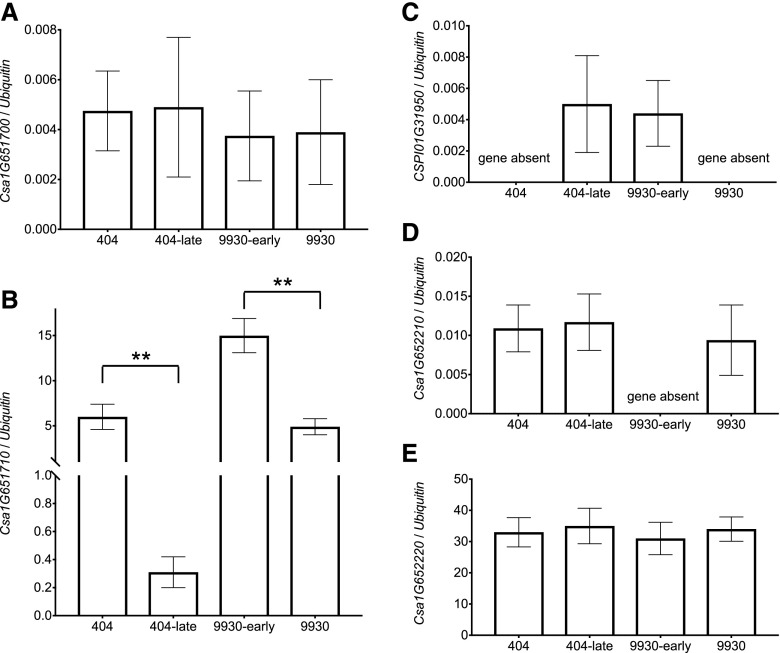
The 35,837-bp mapping region contained no SNP or InDel variants between cucumber accessions 404 and CG9192, except for a 39.9-kb structural variant (SV), a fragment of 39.9-kb sequences present in accession CG9192 but absent from accession 404 (Fig. 2D). We investigated the expression of the genes adjacent to the 39.9-kb SV in the 404 and 404-late NILs (Csa1G651700, Csa1G651710, Csa1G652210, and Csa1G652220) and found that only Csa1G651710 exhibited a significant difference (Fig. 3). A pectin lyase-like superfamily protein, CSPI01G31950, was predicted by gene structure to be located in the 39.9-kb fragment; however, CSPI01G31950 also exists in many other early-flowering accessions (such as the 10 Eurasian accessions in Fig. 1A), so we concluded that CSPI01G31950 is not likely to be responsible for the reduced flowering time. Csa1G651710 is annotated to encode a phosphatidylethanolamine-binding protein (PEBP), and a phylogenetic tree was constructed using protein sequences of the seven cucumber PEBP members and six Arabidopsis (Arabidopsis thaliana) PEBP members (Supplemental Fig. S1). Based on a phylogenetic analysis, Csa1G651710 was predicted to be an ortholog of Arabidopsis FLOWERING LOCUS T (FT; Supplemental Fig. S1), and the 39.9-kb SV is located 16.5 kb upstream of Csa1G651710 (named CsFT here), suggesting that sequences within the 39.9-kb fragment of CG9192 may be contributing to repressed expression of CsFT and later flowering time.
Figure 3.
Expression analysis of the genes adjacent to the 39.9-kb SV and the 16.2-kb SV in two pairs of NILs (404/404-late and 9930/9930-early). The mRNA levels of genes Csa1G651700 (A), Csa1G651710 (B), CSPI01G31950 (C), Csa1G652210 (D), and Csa1G652220 (E) were measured in mature leaves of cucumber plants by reverse transcription quantitative PCR (RT-qPCR). Data shown are relative to the control Ubiquitin (Csa3G778350) gene and represent means ± se of three biological replicates. **, P < 0.01 (Student’s t test).
Positional Cloning of Ef1.1 of Eurasian Cucumber
In a previous study, we found that accession CG5479 (Eurasian type) flowered about 7 d earlier than accession 9930 (East-Asian type; Fig. 1B) and performed quantitative trait loci mapping to delimit the major early-flowering locus Ef1.1 to an 890-kb genomic region on chromosome 1 (Lu et al., 2014). To clone the Ef1.1 locus, we generated NILs (Fig. 2, E and F) in which the Ef1.1 locus of the CG5479 genotype was introgressed into 9930 by backcrossing three times, to construct a large BC3F2 population (n = 4,320). A total of 56 recombinants were identified with two InDel markers (InDel-4 and InDel-5) in the BC3F2 population. Following a recombinant-derived progeny testing strategy, Ef1.1 was narrowed down to an interval of 46,365 bp on chromosome 1, between markers SNP25845628 and SNP25891993 (corresponding to positions 25,845,628 to 25,891,993 of chromosome 1), according to the 9930 reference genome (Fig. 2, G and H). The recombination events between the two markers were also confirmed in the corresponding F3 families. We compared the sequences of the 46,365-bp mapping region between 9930 and CG5479 and identified two SVs located 16.5 kb upstream from CsFT (Fig. 2, H and I). We examined the expression levels of the genes adjacent to the two SVs (Csa1G651700, Csa1G651710, and Csa1G652220) in the NILs (9930 and 9930-early), and only Csa1G651710 exhibited significantly different expression (Fig. 3), suggesting that the two SVs affect CsFT expression.
Intriguingly, the Lf1.1 and Ef1.1 mapping regions overlapped between the two distinct populations (Fig. 2I), and the two candidate causal SVs for Lf1.1 and Ef1.1 were found to be situated at the same location (16.5 kb upstream of CsFT in the 9930 reference genome), raising the possibility that common allelic variations upstream from CsFT lead to flowering time diversity. Notably, the CsFT gene, a critical regulator of the transition from vegetative to reproductive growth (Corbesier et al., 2007; Huijser and Schmid, 2011; Andrés and Coupland, 2012), is located within the overlapping mapping interval of the two populations, and there are no other flowering time-associated genes located in, or adjacent to, the mapping interval. Given that CsFT showed significantly different expression between the two NILs (404/404-late and 9930/9930-early), we inferred that CsFT is most likely to be the gene underlying the flowering time variation observed in both F2 populations.
Two Independent Deletion Events Are Present Upstream from CsFT in the Eurasian and East-Asian Populations
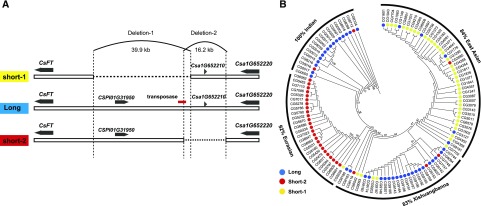
To assess whether there were any structural polymorphisms in the upstream region (UR) of CsFT, we analyzed the depth of sequencing reads at the CsFT locus by comparing a 104.5-kb region (a 100-kb region upstream from the start codon and the 4.5-kb gene body region) in high-quality sequence data derived from 115 cucumber accessions (Qi et al., 2013). Given that parts of the genome sequence may have been lost in cultivated populations during domestication, we used the assembled wild cucumber genome CG0002 as the reference (Qi et al., 2013). This analysis revealed the three types of URs of CsFT that varied in size, termed long, short-1, and short-2 (Fig. 4A; Supplemental Fig. S2). Through combined analysis of the deletion positions and the sequences around the deletion sites, we inferred that the long type of UR was ancestral (Supplemental Figs. S3 and S4). A series of primers was also designed, based on the deletion break points, to validate the three types of URs by PCR and Sanger sequencing (Supplemental Fig. S5; Supplemental Data Set S2). To generate further supporting evidence, we performed a phylogenetic analysis of the 115 cucumber lines in our collection, using all SNPs within a 21-kb region around the CsFT locus (a 16.5-kb region upstream from the start codon and a 4.5-kb gene body region; Fig. 4B). This suggested that the long type of UR from Indian cucumber was ancestral and that the structural variation originated from two independent events: a 39.9-kb deletion (designated as deletion-1) and a 16.2-kb deletion (designated as deletion-2), giving rise to the short-1 and short-2 URs, respectively (Fig. 4B; Supplemental Figs. S3 and S4).
Figure 4.
Large SVs upstream from CsFT in different cucumber populations. A, Schematic illustration of SVs giving rise to the short-1, short-2, and long types URs of CsFT. The 39.9- and 16.2-kb deletions are designated as deletion-1 and deletion-2, respectively. The gray arrow shapes represent the annotated genes, and the transposase is labeled with a red rectangle. B, Maximum-likelihood phylogeny of the 4.5-kb gene body regions and 16.5-kb URs of CsFT from 115 cucumber accessions. Blue, red, and yellow circles denote long, short-2, and short-1 type URs of CsFT, respectively. The bootstrap values are shown at the nodes (%). The numbers on the circular lines indicate the proportions of cucumber accessions from different subgroups in each branch.
We observed that the deletion-1 fragment harbored a transposase (with similarity to transposase Tnp2), which carried some frameshift mutations. Furthermore, a five-nucleotide (GTTTT) duplication was identified on both sides of the break points in deletion-1 (Supplemental Fig. S3), consistent with a transposase-mediated origin of the SV, although the duplicated sequences are not related to any known transposable element excision footprint/insertion point.
Three Types of URs Are Associated with Differences in CsFT Expression
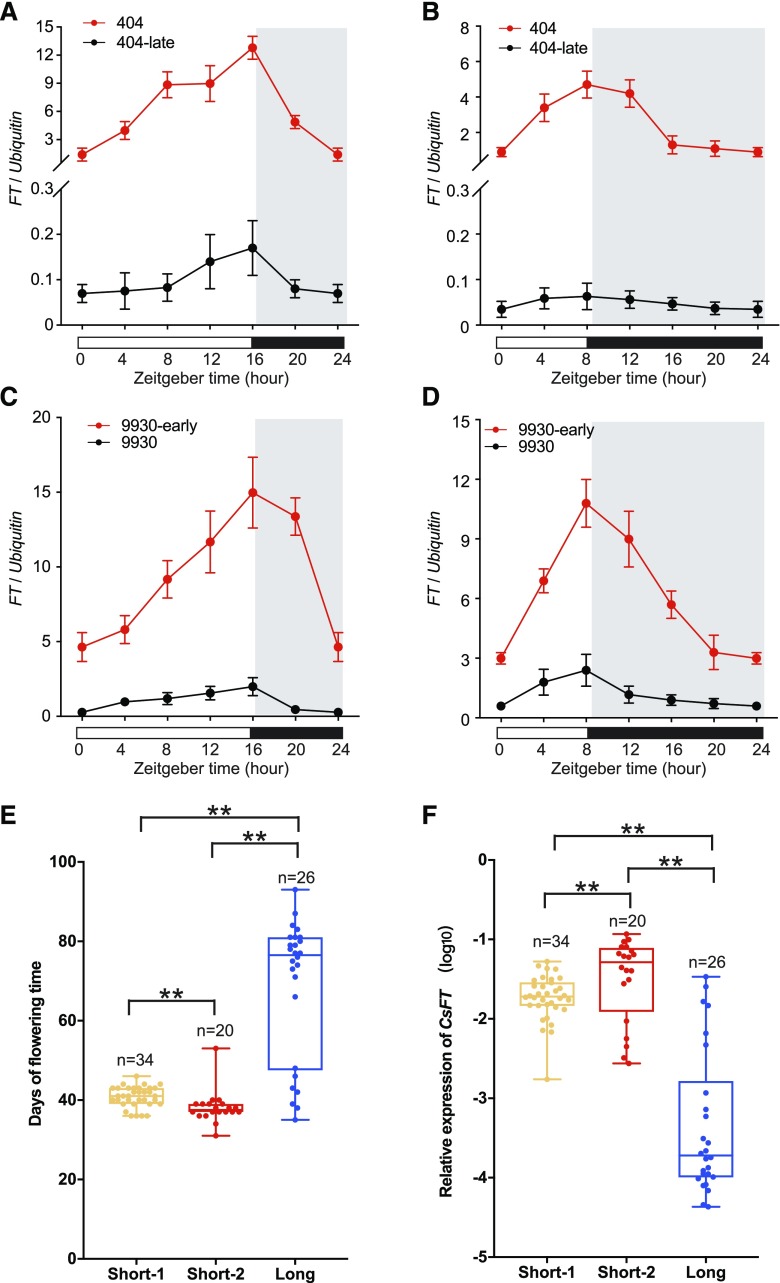
In order to detect how the CsFT transcript level is affected by the three types of URs under different daylength conditions, we therefore examined the diurnal expression patterns of CsFT in mature NIL leaves (404/404-late and 9930/9930-early) under artificial LD and SD conditions. Under both conditions, CsFT expression in 404 was higher, with a clear diurnal rhythm, than CsFT in 404-late, in which expression was always extremely low (Fig. 5, A and B). CsFT expression in both 9930-early and 9930 showed a clear diurnal rhythm, with a peak at dusk under both LD and SD conditions, and the level of CsFT mRNA in 9930-early was higher than in 9930 at all time points during the day (Fig. 5, C and D). Consistent with the CsFT expression levels, the 404-late flowering time was about 40 d later than that of 404 under both LD and SD conditions, and 9930-early flowered approximately 5 d earlier than 9930 under both LD and SD conditions (Supplemental Fig. S6). Thus, the short-1 and short-2 URs are both associated with higher CsFT transcript levels in the NILs than the long UR type, independent of the photoperiod.
Figure 5.
CsFT expression is associated with three types of URs. A to D, Diurnal expression patterns of CsFT in mature leaves of NILs 404/404-late (A and B) and 9930/9930-early (C and D) over 24 h under artificial LD (16 h of light/8 h of dark) and SD (8 h of light/16 h of dark) conditions. White regions represent light periods, and gray regions represent dark periods. Data shown are relative to the control gene CsUbiquitin and represent means ± se of three biological replicates (n = 10 plants). Zeitgeber time is a standardized 24-h notation of the phase in an entrained circadian cycle, and 0 indicates the beginning of day. E and F, Flowering time and CsFT expression levels in different cucumber accessions. Cucumber accessions were classified into three types of URs of CsFT. Details regarding the cucumber accessions used to generate data for this figure are listed in Supplemental Data Set S3. Data are plotted for n accessions per type of CsFT UR, and all of the data points are presented with box plots showing the median, upper and lower quartiles, and minimum and maximum range values for each haplotype. For quantitative analyses of flowering time and CsFT transcript levels of theses 80 accessions, 10 individual plants were analyzed per accession. **, P < 0.01 (Student’s t test).
To further assess the contributions of the three types of URs, we analyzed their effects on gene expression and flowering time in the collected cucumber accessions (Fig. 5, E and F; Supplemental Data Set S3). Through statistical analysis, we found that cucumber accessions carrying the short-1 and short-2 URs flowered significantly earlier than the long accessions (Fig. 5E), and short-2 accessions flowered somewhat earlier than short-1 accessions. The CsFT gene was expressed at the highest level in short-2 accessions, at an intermediate level in short-1 accessions, and at the lowest level in long accessions (Fig. 5F), consistent with flowering time. We noted that some long accessions exhibited similar CsFT expression levels and flowering time to those of short-1 and short-2 accessions, suggesting that these accessions might contain other genetic loci that accelerate flowering.
Domestication Features of a Long-Distance SV Upstream from CsFT
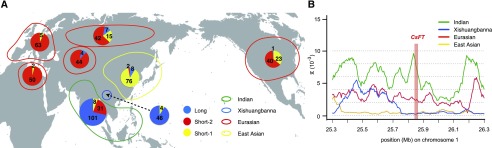
We genotyped the three types of URs of CsFT in 286 cucumber accessions worldwide (a core collection of 115 cucumber accessions and another 171 cucumber accessions we collected) by PCR (Supplemental Fig. S5; Supplemental Data Set S4) and associated the different types of URs with geographic coordinates (Fig. 6A). These results revealed that the three types of URs are highly linked to the geographic distribution of the four cucumber subgroups. The long type UR variant is associated with lower latitudes, which makes up the largest proportion of Indian (72.1%) and Xishuangbanna (92%) populations (Fig. 6A; Supplemental Fig. S7). Compared with the lower frequency distribution of short-1 (5.7%) and short-2 (22.2%) type URs in the Indian population, the short-1 and short-2 types of URs tend to accumulate significantly in higher latitudes (88.4% in East Asian and 80.7% in Eurasian, respectively; Fig. 6A; Supplemental Fig. S7). Furthermore, the geographical distribution of the three types of URs is consistent with genome-wide phylogenetic studies (Lv et al., 2012; Qi et al., 2013; Wang et al., 2018b) that distinguished Eurasian and East-Asian populations, indicating that CsFT UR variation associated with flowering time was one of the selective signatures during domestication of Eurasian and East-Asian cucumbers.
Figure 6.
Footprints of CsFT locus selection. A, Geographical distribution of three types of CsFT URs. A total of 286 cucumber accessions (Supplemental Data Set S4) were used, consisting of 26 North-African accessions, 43 East-Asian accessions, 70 Southeast-Asian accessions, 25 Xishuangbanna accessions, 32 Russian accessions, 34 European accessions, 24 Central-Asian accessions, and 32 North-American accessions. Distributions of long, short-2, and short-1 URs across geographic areas are shown as pie charts, colored in blue, red, and yellow, respectively, with the allele numbers indicated. The colored lines represent the four cucumber populations: Indian (green), Xishuangbanna (blue), Eurasian (red), and East Asian (yellow). B, The genomic region around the CsFT locus shows evidence of a domestication sweep. The distribution of π is shown for the Indian (green), Xishuangbanna (blue), Eurasian (red), and East-Asian (yellow) populations. The CsFT locus is located in the pink region.
To further assess whether the altered flowering time conferred an adaptive advantage and was subject to selection during cucumber breeding, we screened the published genetic diversity data set of four cucumber groups among 115 cucumber accessions (Qi et al., 2013) for potential signatures of selection surrounding the CsFT locus. We found that the nucleotide diversity (π) of the region (a 16.5-kb region upstream from the start codon and a 4.5-kb gene body region) around CsFT was much lower in cultivated populations (Eurasian and East Asian) compared with the Indian population (Fig. 6B).
Collectively, allele expansion and the significant reduction of π implied a possible domesticated trace at the region around the CsFT locus in East-Asian and Eurasian populations. The results above suggest that CsFT short-1 and short-2 URs have undergone positive selection during domestication of Eurasian and East-Asian cucumbers and that these two types of URs may bring CsFT under the influence of stronger promotive effects, thereby decreasing flowering time, which facilitates the improvement of cucumber adaptation to temperate and cold geographic areas.
DISCUSSION
Cucumber is a warm-season plant, with an optimal growth temperature of 20°C to 30°C, and is widely distributed from low latitudes to high latitudes, where there are significant differences in photoperiod and temperature. In this study, we found that most of the Indian and Xishuangbanna cucumber accessions growing at low latitudes have the long UR of CsFT, giving rise to late flowering times. In contrast, most cultivated cucumbers (Eurasian and East Asian) growing at higher latitudes have short-1 and short-2 URs of CsFT, resulting in earlier flowering times. We propose here a model for cucumber adaptation to higher latitudes. Cucumber originated in low-latitude tropical areas with long-term high-temperature environments throughout the year, and the wild progenitor of cucumber in these areas evolved into late-flowering plants. In the process of differentiation of cultivated cucumber subgroups at higher latitudes, there is a gradual decrease in temperature and thus a shortened period suitable for cucumber growth, and consequently, the cucumber genotypes in these areas were domesticated as early-flowering crops. In this study, we found that the transition from vegetative to reproductive growth in cucumber was not sensitive to photoperiod. We speculate that after long-term artificial selection, cucumbers were domesticated as crops with a shorter growth period and higher expression of CsFT that were adapted to lower temperature changes in higher latitude areas.
Despite the association between flowering time variation and the three types of CsFT URs, the flowering times of some accessions could not be explained by these SVs. For example, four accessions (CG1601, CG1149, CG1778, and CG3127) from the East-Asian group showed an early-flowering trait despite carrying a late-flowering long type UR allele. We propose that there are loci other than CsFT that determine flowering time in these accessions. This is consistent with previous studies reporting cucumber flowering time quantitative trait loci on chromosome 1, chromosome 5, and chromosome 6 (Dijkhuizen and Staub, 2002; Fazio et al., 2003; Weng et al., 2010; Bo et al., 2015; Pan et al., 2017).
The ratio of π between Indian and East-Asian cucumbers (πIndian/πEast-Asian) at the region around CsFT was 163.4 (at genome-wide top 1.1%), and therefore suggestive of a domestication event in the East-Asian population. Although the πIndian/πEurasian value at the same region was 4 (at genome-wide top 36.7%), the frequency of short-2 type UR (22.2%) was much higher than that of short-1 type UR (5.7%) in the Indian population (Supplemental Fig. S7), suggesting the possibility that the genetic diversity of the germplasms containing short-2 type UR that migrated from the Indian to the Eurasian population was much higher; thus, the πEurasian value at this region was higher. These findings further suggest that the CsFT locus was positively selected during the domestication of Eurasian and East-Asian cucumbers.
Our study showed that there are three types of URs of CsFT (long, short-1, and short-2) associated with flowering time whose geographic distribution is related to cucumber population structure: the long type is most frequent in Indian and Xishuangbanna populations, the short-1 in East Asian, and the short-2 in Eurasian cucumbers. Considering that flowering time is one of the important traits for cucumber domestication and breeding concerns, our results further enhance the previous notion that there are two distinct domestication directions of cucumber from India, one to East Asia and the other to the West (including Central Asia, Europe, North America, and Africa; Lv et al., 2012; Qi et al., 2013; Wang et al., 2018b).
The Xishuangbanna population is semiwild and is supposed to be a single dispersal that occurred from the Indian population (Lv et al., 2012). The Xishuangbanna population has a very narrow genetic base, and the genome-wide π of Xishuangbanna (1.06 × 10−3) is similar to that of East Asian (1.03 × 10−3) but much lower than that of Eurasian (1.85 × 10−3) and Indian (4.48 × 10−3; Lv et al., 2012; Qi et al., 2013). We thus hypothesized that the ancestor of Xishuangbanna cucumbers might only be derived from some specific late-flowering germplasms carrying the long type UR in the Indian population. In the process of the formation of the Xishuangbanna population, the long type UR associated with the late-flowering trait has not been altered obviously by local ethnic residents, probably due to the suitable temperature for cucumber growth throughout the year. These factors may lead to the lower π around the CsFT locus in the Xishuangbanna population.
Our findings that the short-1 and short-2 type UR alleles were associated with higher expression of CsFT suggested that both types of alleles would result in faster-flowering plants. These plants could have important agronomic value, particularly for the early yield of cucumber fruit. We investigated the yield of NILs at 35 d after seedlings were transplanted in the field (Supplemental Fig. S8). Notably, accession 404 produced ∼0.6 kg of fruit per plant, and the heterozygous F1 produced only 58% (∼0.35 kg per plant) of that value, while the 404-late line did not flower within that time. Similarly, the fruit yield of 9930-early was about 20% higher than that of 9930 at the early stage (Supplemental Fig. S8). Thus, the two deletions upstream from CsFT promote an earlier flowering and so provide a potentially valuable target for tuning cucumber productivity.
In summary, the CsFT locus is a major source of cucumber adaptation to higher-latitude regions. Our findings provide an important perspective on cucumber flowering time control and latitudinal adaptation, which may help drive improvements in cucumber breeding for temperate and cold zones.
MATERIALS AND METHODS
Plant Growth
For flowering time assessment in different daylengths, 40 cucumber (Cucumis sativus) accessions (listed in Supplemental Data Set S1) from a cucumber core collection (Qi et al., 2013) were grown in phytotrons under LD (16 h of light/8 h of dark) and SD (8 h of light/16 h of dark) conditions at a temperature ranging from 18°C to 25°C for 28 d, then the cucumber plants were transplanted in a greenhouse in Beijing under natural light conditions. To further assess the contributions of the three types of URs in more cucumber accessions, 80 cucumber accessions that are all monoecious in sex expression were selected (listed in Supplemental Data Set S3), and the 80 cucumber accessions were grown in a greenhouse in Beijing under natural light conditions. At least eight individual plants of each accession were used for flowering time assessment. Male flowering time was calculated on the days from sowing to anthesis of the first male flower.
Construction of NILs
In order to identify the Lf1.1 locus, accessions CG9192 and 404 were used to construct NILs and segregating populations. CG9192 is a Xishuangbanna type cucumber from tropical southwest China, and 404 is an East-Asian type cucumber from the north of China. Both accessions are monoecious and have been resequenced (Wang et al., 2015). Accession CG9192 usually flowered about 40 d later compared with accession 404. A series of BC3S2 lines with a late-flowering trait were developed using accession CG9192 as donor and accession 404 as recurrent parent, and these late-flowering lines were screened by whole cucumber genome chip, which contains 181 loci for an SNP test (Supplemental Data Set S5). We chose one BC3S2 late-flowering line, 404-18, whose background was most similar to that of line 404 for the construction of the BC3F2 segregation population.
Two cucumber accessions, CG5479 and 9930, were used as parental lines to develop segregating populations for mapping the Ef1.1 locus. CG5479 (also named Muromskij) from Russia is a Eurasian type cucumber whose genome has been resequenced (Qi et al., 2013), and 9930 is an East-Asian type cucumber whose draft genome assembly is available (Huang et al., 2009). Both lines are monoecious in sex expression, and accession CG5479 usually flowered about 7 d earlier than accession 404. We used accession CG5479 as donor and accession 9930 as recurrent parent; nine BC3S2 lines with an early-flowering trait were screened by whole cucumber genome chip, which contains 193 loci for an SNP test (Supplemental Data Set S5). The early-flowering lines 9930-17 and 9930 were crossed for construction of the BC3F2 segregation population.
Positional Cloning of Lf1.1 and Ef1.1
For fine-mapping of cucumber Lf1.1 and Ef1.1, we developed InDel and SNP markers (Supplemental Data Set S2) on chromosome 1 based on the polymorphic sequences. PCR products corresponding to InDel markers were subjected to electrophoresis in 6% (w/v) polyacrylamide, and the presence of SNP markers was validated by PCR and Sanger sequencing.
We identified 32 recombinants for Lf1.1 and 56 recombinants for Ef1.1 using flanking markers and observed the time to the first male flower of all the recombinants. The F3 families of these recombinants were tested to confirm the recombination events, with 20 individuals per family.
Phylogenetic Analysis of CsFT Protein
PEBPs were obtained from The Arabidopsis Information Resource (http://www.arabidopsis.org) and the Cucurbit Genomics Database (http://cucurbitgenomics.org). These protein sequences were aligned by ClustalW and manually edited when necessary. A phylogenetic tree was constructed using a maximum-likelihood method with bootstrapping based on 1,000 pseudoreplicates with MEGA7 software (Kumar et al., 2016).
Phylogenetic Analysis of 115 Cucumber Accessions Based on the SNPs within a 21-kb Region of the CsFT Locus
A total of 350 SNPs among the cucumber core collection (115 accessions) in the 4.5-kb gene body region and 16.5-kb UR of CsFT were extracted from the previously reported data set (Qi et al., 2013). Next, these variations were converted into phylip format and sent to PHYLIP version 3.6 (http://evolution.genetics.washington.edu/phylip.html) to generate a consensus tree using maximum-likelihood estimation with a bootstrap resampling time of 100. The final tree plot was modified using MEGA7 software (Kumar et al., 2016).
Mapping Resequencing Reads to the CG0002 Reference Genome
Previously generated resequencing reads from the core collection of 115 cucumber accessions (Qi et al., 2013) were mapped to the CG0002 reference genome (Qi et al., 2013) by BWA (Li and Durbin, 2009) with default parameters, and SAMtools (Li and Durbin, 2009) was used to perform sorting and duplicate marking of the resulting BAM-format files. These BAM alignment results were subsequently passed to the bamCoverage program within the deep Tools suite (Ramírez et al., 2016) to calculate the genome coverage of the 104.5-kb region (a 100-kb region upstream of the start codon and the 4.5-kb gene body of CsFT) on the CG0002 reference genome (parameter -r chr1:29,207,609-29,311,798) of the 115 accessions. The window size was set to 300 bp.
PCR Testing for the Three Types of URs
The PCR primers were designed as shown in Supplemental Figure S5, and the sequences of the primers are listed in Supplemental Data Set S2. DNA was isolated from leaves of cucumber accessions. Three pairs of primers (P1/P2, P3/P4, and P9/P10) were first used to detect all accessions. If the PCR fragments from the P9/P10 primers amplified the expected size in some accessions, these accessions were analyzed with another four pairs of primers (P5/P6, P7/P8, P11/P12, and P13/P14). PCR was performed using KOD-FX DNA polymerase (Toyobo), and PCR products were loaded on 1% agarose gels.
RNA Extraction and RT-qPCR Analysis
The third leaves from NILs (404/404-late and 9930/9930-early) and the collected cucumber accessions (Supplemental Data Set S3) at the four-leaf stage were collected, and total RNA was isolated using an EasyPure Plant RNA Kit (TransGen Biotech). First-strand cDNA was synthesized from 1 mg of total RNA using FastQuant RT Super Mix (Tiangen). PCR was performed on an ABI 7900 using SYBR Premix (Roche) using the following program: 3 min at 95°C followed by 40 cycles of 20 s at 95°C, 30 s at 60°C, and 20 s at 72°C, according to the manufacturer’s instructions. Ubiquitin (Csa3G778350) was used as the internal control for RT-qPCR. Three independent biological experiments were performed in all cases. The PCR primers used in these experiments are listed in Supplemental Data Set S2.
For the analysis of diurnal expression patterns, the two pairs of NILs (404/404-late and 9930/9930-early) were grown in a phytotron under either LD (16 h of light/8 h of dark) or SD (8 h of light/16 h of dark) conditions with 25°C constant temperature. Then we investigated CsFT expression on day 28 after sowing. The third leaf of each plant at the four-leaf stage was collected and stored in liquid nitrogen. Samples were collected every 4 h for 24 h. Three independent biological experiments were performed in all cases. Total RNA extraction, first-strand cDNA synthesis, and RT-qPCR analysis were performed as described above.
Identification of Selective Sweeps
To identify the selection region around CsFT, the SNPs near the CsFT locus in the cucumber genome were obtained (corresponding to Chr01: 25.3–26.3 Mb, 9930 genome V2). We measured the level of π sing a 100-kb window with a step size of 10 kb in the Indian population, Xishuangbanna population, Eurasian population, and East-Asian population.
Statistical Analyses
For quantitative analyses of flowering time, at least eight individual plants were analyzed per genotype or ecotype, and the numbers of individuals are given in the figure legends. For expression analyses using RT-qPCR, 10 individual plants were pooled per sample and three biological replicates were performed. Statistical calculations were conducted in Microsoft Excel 2016, and mean values for each measured parameter were compared using two-way ANOVA or two-tailed, two-sample Student’s t test.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL data libraries and the Cucurbit Genomics Database (cucurbitgenomics.org) under the following accession numbers: CsFT (Csa1G651710), AtATC (AT2G27550), AtBFT (AT5G62040), AtFT (AT1G65480), AtMFT (AT1G18100), AtTFL1 (AT5G03840), and AtTSF (AT4G20370). The sequences of cucumber genes (Csa1G651700, CSPI01G31950, Csa1G652210, and Csa1G652220 in Fig. 3 and Csa3G776350, Csa6G452100, Csa6G152360, Csa3G180440, Csa1G071830, and Csa3G807330 in Supplemental Fig. S1) can also be found in the Cucurbit Genomics Database.
Supplemental Data
The following supplemental materials are available.
Supplemental Figure S1. Phylogenetic analysis of PEBP proteins in cucumber and Arabidopsis.
Supplemental Figure S2. Schematic illustration of the mapped reads from different cucumber accessions on top of the wild-type CG0002 reference genome.
Supplemental Figure S3. Alignment of the polymorphic region between the long type and the short-1 type URs.
Supplemental Figure S4. Alignment of the polymorphic region between the long type and the short-2 type URs.
Supplemental Figure S5. PCR primers designed for testing the short-1, short-2, and long URs.
Supplemental Figure S6. Flowering time of the two pairs of NILs under LD and SD conditions.
Supplemental Figure S7. Frequency distribution of three types of URs in four cucumber populations.
Supplemental Figure S8. Yield test of two pairs of NILs at the early stage.
Supplemental Data Set S1. The 40 cucumber accessions listed for flowering time assessment.
Supplemental Data Set S2. Primers listed in this study.
Supplemental Data Set S3. The 80 cucumber accessions listed for assessing the contributions of the three types of URs
Supplemental Data Set S4. The 286 cucumber accessions used for geographical distribution of three types of URs.
Supplemental Data Set S5. The information of SNP chips used for genotyping in this study.
Acknowledgments
We thank Dr. Tongbing Su and Dr. Chunzhi Zhang for advice on article improvement.
Footnotes
This work was supported by the China Postdoctoral Science Foundation (2017M613227 and 2018T111109), the Fundamental Research Funds for the Central Universities (2452019048), and the National Natural Science Foundation of China (31701914).
Articles can be viewed without a subscription.
References
- Andrés F, Coupland G (2012) The genetic basis of flowering responses to seasonal cues. Nat Rev Genet 13: 627–639 [DOI] [PubMed] [Google Scholar]
- Bo K, Ma Z, Chen J, Weng Y (2015) Molecular mapping reveals structural rearrangements and quantitative trait loci underlying traits with local adaptation in semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis Qi et Yuan). Theor Appl Genet 128: 25–39 [DOI] [PubMed] [Google Scholar]
- Chen YH, Gols R, Benrey B (2015) Crop domestication and its impact on naturally selected trophic interactions. Annu Rev Entomol 60: 35–58 [DOI] [PubMed] [Google Scholar]
- Corbesier L, Vincent C, Jang S, Fornara F, Fan Q, Searle I, Giakountis A, Farrona S, Gissot L, Turnbull C, et al. (2007) FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science 316: 1030–1033 [DOI] [PubMed] [Google Scholar]
- Dijkhuizen A, Staub JE (2002) QTL conditioning yield and fruit quality traits in cucumber (Cucumis sativus L.) effects of environment and genetic background. J New Seeds 4: 1–30 [Google Scholar]
- Doebley JF, Gaut BS, Smith BD (2006) The molecular genetics of crop domestication. Cell 127: 1309–1321 [DOI] [PubMed] [Google Scholar]
- Fazio G, Staub JE, Stevens MR (2003) Genetic mapping and QTL analysis of horticultural traits in cucumber (Cucumis sativus L.) using recombinant inbred lines. Theor Appl Genet 107: 864–874 [DOI] [PubMed] [Google Scholar]
- Gaudin ACM, McClymont SA, Raizada MN (2011) The nitrogen adaptation strategy of the wild teosinte ancestor of modern maize, Zea mays subsp. parviglumis. Crop Sci 51: 2780 [Google Scholar]
- Gepts P. (2004) Domestication as a long-term selection experiment. Plant Breed Rev 24: 1–44 [Google Scholar]
- Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P, et al. (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41: 1275–1281 [DOI] [PubMed] [Google Scholar]
- Huijser P, Schmid M (2011) The control of developmental phase transitions in plants. Development 138: 4117–4129 [DOI] [PubMed] [Google Scholar]
- Hung HY, Shannon LM, Tian F, Bradbury PJ, Chen C, Flint-Garcia SA, McMullen MD, Ware D, Buckler ES, Doebley JF, et al. (2012) ZmCCT and the genetic basis of day-length adaptation underlying the postdomestication spread of maize. Proc Natl Acad Sci USA 109: E1913–E1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones H, Leigh FJ, Mackay I, Bower MA, Smith LMJ, Charles MP, Jones G, Jones MK, Brown TA, Powell W (2008) Population-based resequencing reveals that the flowering time adaptation of cultivated barley originated east of the Fertile Crescent. Mol Biol Evol 25: 2211–2219 [DOI] [PubMed] [Google Scholar]
- Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33: 1870–1874 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Q, Li H, Huang W, Xu Y, Zhou Q, Wang S, Ruan J, Huang S, Zhang Z (2019) A chromosome-scale genome assembly of cucumber (Cucumis sativus L.). Gigascience 8: giz072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Adrian J, Pankin A, Hu J, Dong X, von Korff M, Turck F (2014) Induced and natural variation of promoter length modulates the photoperiodic response of FLOWERING LOCUS T. Nat Commun 5: 4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Lin T, Klein J, Wang S, Qi J, Zhou Q, Sun J, Zhang Z, Weng Y, Huang S (2014) QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor Appl Genet 127: 1491–1499 [DOI] [PubMed] [Google Scholar]
- Lu S, Zhao X, Hu Y, Liu S, Nan H, Li X, Fang C, Cao D, Shi X, Kong L, et al. (2017) Natural variation at the soybean J locus improves adaptation to the tropics and enhances yield. Nat Genet 49: 773–779 [DOI] [PubMed] [Google Scholar]
- Lv J, Qi J, Shi Q, Shen D, Zhang S, Shao G, Li H, Sun Z, Weng Y, Shang Y, et al. (2012) Genetic diversity and population structure of cucumber (Cucumis sativus L.). PLoS ONE 7: e46919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Y, Qu S, Bo K, Gao M, Haider KR, Weng Y (2017) QTL mapping of domestication and diversifying selection related traits in round-fruited semi-wild Xishuangbanna cucumber (Cucumis sativus L. var. xishuangbannanesis). Theor Appl Genet 130: 1531–1548 [DOI] [PubMed] [Google Scholar]
- Qi J, Liu X, Shen D, Miao H, Xie B, Li X, Zeng P, Wang S, Shang Y, Gu X, et al. (2013) A genomic variation map provides insights into the genetic basis of cucumber domestication and diversity. Nat Genet 45: 1510–1515 [DOI] [PubMed] [Google Scholar]
- Ramírez F, Ryan DP, Grüning B, Bhardwaj V, Kilpert F, Richter AS, Heyne S, Dündar F, Manke T (2016) deepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44: W160–W165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux F, Touzet P, Cuguen J, Le Corre V (2006) How to be early flowering: An evolutionary perspective. Trends Plant Sci 11: 375–381 [DOI] [PubMed] [Google Scholar]
- Sebastian P, Schaefer H, Telford IRH, Renner SS (2010) Cucumber (Cucumis sativus) and melon (C. melo) have numerous wild relatives in Asia and Australia, and the sister species of melon is from Australia. Proc Natl Acad Sci USA 107: 14269–14273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soyk S, Müller NA, Park SJ, Schmalenbach I, Jiang K, Hayama R, Zhang L, Van Eck J, Jiménez-Gómez JM, Lippman ZB (2017) Variation in the flowering gene SELF PRUNING 5G promotes day-neutrality and early yield in tomato. Nat Genet 49: 162–168 [DOI] [PubMed] [Google Scholar]
- Wang M, Li W, Fang C, Xu F, Liu Y, Wang Z, Yang R, Zhang M, Liu S, Lu S, et al. (2018a) Parallel selection on a dormancy gene during domestication of crops from multiple families. Nat Genet 50: 1435–1441 [DOI] [PubMed] [Google Scholar]
- Wang S, Yang X, Xu M, Lin X, Lin T, Qi J, Shao G, Tian N, Yang Q, Zhang Z, et al. (2015) A rare SNP identified a TCP transcription factor essential for tendril development in cucumber. Mol Plant 8: 1795–1808 [DOI] [PubMed] [Google Scholar]
- Wang X, Bao K, Reddy UK, Bai Y, Hammar SA, Jiao C, Wehner TC, Ramírez-Madera AO, Weng Y, Grumet R, et al. (2018b) The USDA cucumber (Cucumis sativus L.) collection: Genetic diversity, population structure, genome-wide association studies, and core collection development. Hortic Res 5: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weng Y, Johnson S, Staub JE, Huang SW (2010) An extended microsatellite genetic map of cucumber, Cucumis sativus L. HortScience 45: 880–886 [Google Scholar]
- Xue W, Xing Y, Weng X, Zhao Y, Tang W, Wang L, Zhou H, Yu S, Xu C, Li X, et al. (2008) Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat Genet 40: 761–767 [DOI] [PubMed] [Google Scholar]
- Zhang S, Jiao Z, Liu L, Wang K, Zhong D, Li S, Zhao T, Xu X, Cui X (2018) Enhancer-promoter interaction of SELF PRUNING 5G shapes photoperiod adaptation. Plant Physiol 178: 1631–1642 [DOI] [PMC free article] [PubMed] [Google Scholar]