Gram-negative bacteria contain lipopolysaccharide (LPS) coating their thin peptidoglycan cell wall. The presence of LPS has been suggested to be associated with a metabolic disorder of cattle—ruminal acidosis—through affecting ruminal bacteria. Ruminal acidosis could reduce feed intake and milk production and increase the incidence of diarrhea, milk fat depression, liver abscesses, and laminitis. However, how LPS affects bacteria associated with ruminal acidosis has not been studied. In this study, we investigated how LPS affects the growth of ruminal bacteria by pure cultures, including those that contribute to acidosis, and the functional genes of ruminal bacteria. Thus, this work serves to further our understanding of the roles of LPS in the pathogenesis of ruminal acidosis, as well as providing information that may be useful for the prevention of ruminal acidosis and reducetion of economic losses for farmers.
KEYWORDS: LPS, dairy cow, Gram-negative bacteria, ruminal bacteria
ABSTRACT
Lipopolysaccharide (LPS) has been reported to contribute to a ruminal acidosis of cattle by affecting ruminal bacteria. The goal of this study was to determine how LPS affects the growth of pure cultures of ruminal bacteria, including those that contribute to ruminal acidosis. We found that dosing LPS (200,000 EU) increased the maximum specific growth rates of four ruminal bacterial species (Streptococcus bovis JB1, Succinivibrio dextrinosolvens 24, Lactobacillus ruminis RF1, and Selenomonas ruminantium HD4). Interestingly, all the species ferment sugars and produce lactate, contributing to acidosis. Species that consume lactate or ferment fiber were not affected by LPS. We found that S. bovis JB1 failed to grow in LPS as the carbon source in the media; growth of S. bovis JB1 was increased by LPS when glucose was present. Growth of Megasphaera elsdenii T81, which consumes lactate, was not different between the detoxified (lipid A delipidated) and regular LPS. However, the maximum specific growth rate of S. bovis JB1 was greater in regular LPS than detoxified LPS. Mixed bacteria from a dual-flow continuous culture system were collected to determine changes of metabolic capabilities of bacteria by LPS, and genes associated with LPS biosynthesis were increased by LPS. In summary, LPS was not toxic to bacteria, and lipid A of LPS stimulated the growth of lactate-producing bacteria. Our results indicate that LPS not only is increased during acidosis but also may contribute to ruminal acidosis development by increasing the growth of lactic acid-producing bacteria.
IMPORTANCE Gram-negative bacteria contain lipopolysaccharide (LPS) coating their thin peptidoglycan cell wall. The presence of LPS has been suggested to be associated with a metabolic disorder of cattle—ruminal acidosis—through affecting ruminal bacteria. Ruminal acidosis could reduce feed intake and milk production and increase the incidence of diarrhea, milk fat depression, liver abscesses, and laminitis. However, how LPS affects bacteria associated with ruminal acidosis has not been studied. In this study, we investigated how LPS affects the growth of ruminal bacteria by pure cultures, including those that contribute to acidosis, and the functional genes of ruminal bacteria. Thus, this work serves to further our understanding of the roles of LPS in the pathogenesis of ruminal acidosis, as well as providing information that may be useful for the prevention of ruminal acidosis and reducetion of economic losses for farmers.
INTRODUCTION
Gram-negative bacteria contain lipopolysaccharide (LPS) coating their thin peptidoglycan cell wall, which is important for the structural and functional integrity of the bacteria (1). Lipopolysaccharides from all Gram-negative bacteria share essentially the same structure: an O-polysaccharide chain, an R core, and a lipid A moiety; the lipid A moiety, covalently linked to the inner core, carries the endotoxic activity of LPS (2, 3). The presence of LPS in the plasma has been correlated to multiple diseases, including sepsis and septic shock but also more recently to obesity and its associated metabolic disorders, in human beings (1). The presence of LPS also has been reported to contribute to a significant metabolic disorders in dairy cattle: subacute ruminal acidosis (SARA) (4). Subacute ruminal acidosis has been associated with reduced feed intake and milk production, diarrhea, milk fat depression, liver abscesses, and laminitis (5–7). The adverse effects of SARA are very costly and can affect animal health, especially for high-producing dairy cows (5).
In general, ruminal bacteria are predominantly Gram negative (8), and these bacteria are the major source of LPS in the rumen. Bacteria death and lysis are normal processes; LPS is thus normally present in ruminal fluid. However, the LPS concentration is much higher in grain-fed than in forage-fed cattle (9, 10), especially in cows with SARA, in which the LPS concentration ranges from 4- to 16-fold higher than in cows without SARA (11–13). Therefore, LPS has been suspected to contribute to the pathogenesis of SARA (4). Meanwhile, our previous study has reported that LPS dosing (200,000 endotoxin units [EU]) changed the ruminal bacterial population and stimulated the Gram-negative bacteria related to starch digestion, which may, in turn, develop into SARA (14).
However, how LPS alters the struture of ruminal bacteria and stimulates bacteria associated with ruminal acidosis has not been well studied. Therefore, the goal of this study was first to determine how LPS affects the growth of pure cultures of ruminal bacteria, including those that contribute to SARA, and then to evaluate how LPS affects the functional genes of mixed ruminal bacteria by shotgun sequencing. We expected that the current study would facilitate our understanding of the role of LPS on pathogenesis of SARA, as well as provide information that may be useful for the prevention of ruminal acidosis. We hypothesized that (i) dosing of LPS would affect Gram-negative bacteria that use starch as the substrate, (ii) LPS would replace glucose as a carbon source for growth of ruminal lactate-producing bacteria, (iii) LPS would be a growth factor for lactate-producing bacteria and toxic to non-starch-utilizing bacteria, and (iv) LPS would change functional genes of ruminal bacteria.
RESULTS
Effects of LPS on bacterial growth and fermentation end products.
In order to evaluate if LPS could only stimulate Gram-negative bacteria that utilize starch, we applied 200,000 EU of regular LPS to eight different ruminal bacterial species in pure culture. We found that the maximum specific growth rates of lactate-producing bacteria (Succinivibrio dextrinosolvens 24, Selenomonas ruminantium HD4, Lactobacillus ruminis RF1, and Streptococcus bovis JB1) were increased by regular LPS (R-LPS) dosing (200,000 EU) regardless of Gram reaction (Table 1). The concentration of LPS was chosen based on previous studies in which the ruminal concentration of LPS was measured in cows with SARA (11–13). The maximum specific growth rate of S. bovis JB1 was increased by 5.88%, that of Se. ruminantium HD4 by 7.83%, that of Su. dextrinosolvens 24 by 8.21%, and that of L. ruminis RF1 by 29.1% by LPS dosing (Table 1). The lag phases of Se. ruminantium HD4 and Su. dextrinosolvens 24 were reduced by LPS dosing (Table 1). However, dosing of R-LPS had no effects on the maximum specific growth rate or lag phase of fiber-utilizing bacteria (R. albus 7, Ruminococcus flavefaciens FD-1, and Fibrobacter succinogenes S85 (Table 1) or lactate-utilizing bacteria (Megasphaera elsdenii T81) (Table 1). Therefore, the results showed that LPS stimulated the growth of bacteria that ferment starch and produce lactate regardless of Gram reaction; however, species that consume lactate or ferment fiber were not affected by LPS.
TABLE 1.
Effects of regular lipopolysaccharide on lag phase and the maximum specific growth rate of bacteria based on the prediction from logistic functiona
| Bacterium and parameter | Value with treatmentb |
SEM | P value | |
|---|---|---|---|---|
| Control | R-LPS | |||
| Starch-utilizing bacteria | ||||
| Selenomonas ruminantium HD4 | ||||
| Lag, min | 243 | 234 | 5.23 | 0.02 |
| μmax, h−1 | 0.35 | 0.38 | 0.06 | <0.01 |
| Succinivibrio dextrinosolvens 24 | ||||
| Lag, min | 220 | 212 | 5.17 | 0.03 |
| μmax, h−1 | 0.11 | 0.12 | 0.00 | <0.01 |
| Lactobacillus ruminis RF1 | ||||
| Lag, min | 158 | 173 | 35.7 | 0.23 |
| μmax, h−1 | 0.07 | 0.09 | 0.02 | <0.01 |
| Streptococcus bovis JB1 | ||||
| Lag, min | 165 | 152 | 14.3 | 0.24 |
| μmax, h−1 | 1.02 | 1.08 | 0.06 | 0.04 |
| Fiber-utilizing bacteria | ||||
| Ruminococcus albus 7 | ||||
| Lag, min | 402 | 396 | 26.2 | 0.54 |
| μmax, h−1 | 0.40 | 0.41 | 0.14 | 0.21 |
| Ruminococcus flavefaciens FD-1 | ||||
| Lag, min | 567 | 563 | 58.9 | 0.78 |
| μmax, h−1 | 0.18 | 0.18 | 0.01 | 0.98 |
| Fibrobacter succinogenes S85 | ||||
| Lag, min | 446 | 449 | 54.3 | 0.43 |
| μmax, h−1 | 0.20 | 0.20 | 0.01 | 1.00 |
| Lactate-utilizing bacterium | ||||
| Megasphaera elsdenii T81 | ||||
| Lag, min | 253 | 238 | 42.3 | 0.54 |
| μmax, h−1 | 0.14 | 0.14 | 0.00 | 0.76 |
Lag, lag phase; μmax, maximum specific growth rate.
Control, control group (LPS-free anaerobic water); R-LPS, regular LPS (200,000 EU).
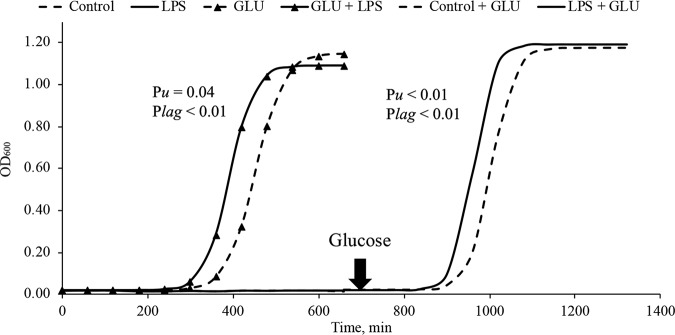
In order to determine the mechanism by which LPS stimulates growth of amylolytic lactic acid bacteria, we grew S. bovis JB1 in defined mediaum without a carbon source and added regular LPS as the only carbon source. We found S. bovis JB1 did not grow on it (Fig. 1), indicating that LPS cannot be utilized as the main carbon source for bacterial growth of S. bovis JB1. However, when glucose was present, Streptococcus bovis JB1 grew on it, and the maximum specific growth rate of S. bovis JB1 was increased by 15% compared to that of the control (Table 2), which indicates that the stimulating effects of LPS on ruminal bacteria occurs if other carbon sources are available. To validate this, glucose was then added to the Balch tubes in which S. bovis JB1 did not grow. We found that S. bovis JB1 regrew and that the maximum specific growth rate of S. bovis JB1 was increased by 15% (Fig. 1; Table 2) and the lag phase was reduced by 40 min by LPS dosing compared to those of the control (Table 2). Therefore, this confirmed that the stimulating effects of LPS on ruminal bacteria required other carbon sources to be present.
FIG 1.
Effects of replacing glucose with lipopolysaccharide as a carbon source on the growth of Streptococcus bovis JB1. Control, control group (LPS-free anaerobic water); LPS, regular LPS (200,000 EU) as a carbon source; GLU, glucose as a carbon source; GLU + LPS, glucose plus LPS (200,000 EU); control + GLU, control group plus glucose; LPS + GLU, regular LPS (200,000 EU) plus glucose.
TABLE 2.
Effects of regular lipopolysaccharide as a carbon source on the lag phase and the maximum specific growth rate of Streptococcus bovis JB1 based on the logistic function prediction
| Parameter | Value with treatmenta |
SEM | P value | |||
|---|---|---|---|---|---|---|
| Controlb | R-LPSb | GLU + controlc | GLU + R-LPSc | |||
| Without glucose added | ||||||
| Lag, min | — | — | 387 | 318 | 6.83 | <0.01 |
| μmax, h−1 | — | — | 0.50 | 0.58 | 0.10 | 0.04 |
| With glucose added | ||||||
| Lag, min | 292 | 256 | — | — | 5.72 | <0.01 |
| μmax, h−1 | 0.66 | 0.76 | — | — | 0.18 | <0.01 |
Control, control group (LPS-free anaerobic water); R-LPS, regular LPS (200,000 EU) as a carbon source; GLU, glucose as a carbon source; GLU + R-LPS, glucose plus R-LPS (200,000 EU).
Results were obtained after adding glucose. —, not applicable (no growth).
—, data not shown.
The LPS mainly consists of two components: polysaccharide and fatty acid. In order to evaluate which part would be the stimulator, the same concentrations of regular and detoxified LPS (D-LPS; lipid A delipidated) were applied to evaluate their effects on the growth of S. bovis JB1. We found that the maximum specific growth rate of S. bovis was increased by 7.5% in D-LPS (lipid A delipidated) and increased by 15% in R-LPS compared to that of the control (Table 3). The lag phase of S. bovis JB1 was not affected by D-LPS dosing, while it was reduced by 42 min by R-LPS dosing compared to that of the control (Table 3), indicating that lipid A may be the stimulator.
TABLE 3.
Effects of detoxified and regular lipopolysaccharide on lag phases and the maximum specific growth rates of Megasphaera elsdenii T81 and Streptococcus bovis JB1 based on the prediction from logistic function
| Bacterium and parameter | Value with treatmenta |
SEM | P value | ||
|---|---|---|---|---|---|
| Control | D-LPS | R-LPS | |||
| Megasphaera elsdenii T81 | |||||
| Lag, min | 181 | 172 | 158 | 24.1 | 0.15 |
| μmax, h−1 | 0.12 | 0.12 | 0.12 | 0.00 | 0.21 |
| Streptococcus bovis JB1 | |||||
| Lag, min | 293 A | 285 A | 251 B | 6.07 | <0.01 |
| μmax, h−1 | 0.67 C | 0.72 B | 0.77 A | 0.03 | <0.01 |
Control, control group (LPS-free anaerobic water); D-LPS, detoxified LPS (200,000 EU); R-LPS, regular LPS (200,000 EU). Least-squares means within the same row with different letters are significantly different (P < 0.05).
In order to evaluate if LPS was toxic to ruminal bacteria, the same concentration of regular and detoxified LPS (D-LPS; lipid A delipidated) were applied to evaluate their effects on the growth of M. elsdenii T81. We found that the maximum specific growth rate and lag phase of M. elsdenii T81 were not different among the control, detoxified, and regular LPS dosing (Table 3). Meanwhile, the growth rate of S. bovis JB1was greater in R-LPS and D-LPS. Therefore, we can conclude that LPS is not toxic to S. bovis JB1, the main lactic acid-producing bacteria during SARA.
However, we found that concentrations of fermentation end products (volatile fatty acids [VFA] and NH3-N) were not affected by treatments for all the tested bacteria, and concentrations of l-lactate, d-lactate, and total lactate were not affected by treatment for all tested bacteria as well (Tables 4 to 7).
TABLE 4.
Effects of regular lipopolysaccharide on ammonia nitrogen, VFA, and lactate concentrations in lactate-producing bacteria
| Bacterium and parameter | Value (mM) with treatmenta |
SEM | P value | |
|---|---|---|---|---|
| Control | R-LPS | |||
| Selenomonas ruminantium HD4 | ||||
| NH3-N | 3.45 | 3.47 | 0.35 | 0.90 |
| Acetate | 10.9 | 11.6 | 3.73 | 0.60 |
| Propionate | 4.85 | 5.08 | 0.49 | 0.23 |
| Valerate | 2.94 | 2.90 | 0.56 | 0.79 |
| d-Lactate | 5.07 | 4.96 | 0.09 | 0.42 |
| l-Lactate | 0.33 | 0.34 | 0.04 | 0.86 |
| dl-Lactate | 5.40 | 5.30 | 0.08 | 0.41 |
| Succinivibrio dextrinosolvens 24 | ||||
| NH3-N | 3.94 | 4.07 | 0.34 | 0.51 |
| Acetate | 27.9 | 28.3 | 0.04 | 0.22 |
| Propionate | 5.99 | 6.01 | 0.47 | 0.60 |
| Butyrate | 3.32 | 3.34 | 0.02 | 0.65 |
| d-Lactate | 4.50 | 4.43 | 0.13 | 0.44 |
| l-Lactate | — | — | — | — |
| dl-Lactate | 4.50 | 4.43 | 0.13 | 0.44 |
| Lactobacillus ruminis RF1 | ||||
| NH3-N | 4.08 | 4.27 | 0.50 | 0.37 |
| Acetate | 41.3 | 32.5 | 3.19 | 0.09 |
| Propionate | 13.3 | 10.6 | 0.92 | 0.17 |
| Butyrate | 6.99 | 5.63 | 0.54 | 0.19 |
| d-Lactate | 0.99 | 1.00 | 0.02 | 0.42 |
| l-Lactate | 7.50 | 7.20 | 0.51 | 0.35 |
| dl-Lactate | 8.50 | 8.22 | 0.56 | 0.41 |
| Streptococcus bovis JB1 with PC+VFA | ||||
| NH3-N | 4.05 | 3.83 | 0.14 | 0.30 |
| Acetate | 59.7 | 65.2 | 4.23 | 0.35 |
| Valerate | 0.91 | 0.85 | 0.05 | 0.34 |
| d-Lactate | 0.14 | 0.15 | 0.06 | 0.85 |
| l-Lactate | 6.81 | 7.13 | 0.68 | 0.47 |
| dl-Lactate | 6.96 | 7.28 | 0.64 | 0.45 |
Control, control group (LPS-free anaerobic water); LPS, regular LPS (200,000 EU). Lactate concentrations are millimolar. —, not applicable.
TABLE 5.
Effects of regular lipopolysaccharide on ammonia nitrogen, VFA, and lactate concentrations in fiber- and lactate-utilizing bacteria
| Bacterium and parameter | Value with treatmenta |
SEM | P value | |
|---|---|---|---|---|
| Control | R-LPS | |||
| Ruminococcus albus 7 | ||||
| NH3-N | 2.28 | 2.04 | 0.35 | 0.26 |
| Acetate | 26.5 | 25.8 | 0.35 | 0.24 |
| Propionate | 5.47 | 5.18 | 0.60 | 0.75 |
| Butyrate | 2.88 | 2.66 | 0.91 | 0.69 |
| Ruminococcus flavefaciens FD-1 | ||||
| NH3-N | 1.97 | 1.80 | 0.13 | 0.13 |
| Acetate | 31.6 | 31.3 | 1.08 | 0.64 |
| Propionate | 6.12 | 6.11 | 0.21 | 0.96 |
| Butyrate | 3.33 | 3.33 | 0.12 | 1.00 |
| Fibrobacter succinogenes S85 | ||||
| NH3-N | 1.69 | 1.79 | 0.31 | 0.59 |
| Acetate | 26.6 | 27.3 | 1.23 | 0.50 |
| Propionate | 5.91 | 6.08 | 0.26 | 0.51 |
| Butyrate | 3.28 | 3.34 | 0.14 | 0.54 |
| Megasphaera elsdenii T81 | ||||
| NH3-N | 6.18 | 6.24 | 0.97 | 0.92 |
| Acetate | 9.16 | 10.0 | 1.47 | 0.69 |
| Butyrate | 6.25 | 6.57 | 0.71 | 0.73 |
| Valerate | 3.02 | 2.80 | 0.39 | 0.71 |
| d-Lactate | 0.10 | 0.16 | 0.03 | 0.28 |
| l-Lactate | 0.07 | 0.15 | 0.18 | 0.30 |
| dl-Lactate | 0.15 | 0.29 | 0.08 | 0.28 |
Control, control group (LPS-free anaerobic water); LPS, regular LPS (200,000 EU). Lactate concentrations are millimolar.
TABLE 6.
Effects of regular lipopolysaccharide as the carbon source on ammonia nitrogen, VFA, and lactate concentrations in Streptococcus bovis JB1 with defined media
| Parameter | Value with treatmenta |
SEM | P value | |||
|---|---|---|---|---|---|---|
| Controlb | R-LPSb | GLU + Controlc | GLU + R-LPSc | |||
| Without glucose added | ||||||
| NH3-N | — | — | 2.10 | 2.25 | 0.14 | 0.40 |
| Acetate | — | — | 37.4 | 38.1 | 2.16 | 0.76 |
| Valerate | — | — | 1.51 | 1.46 | 0.23 | 0.18 |
| d-Lactate | — | — | 0.04 | 0.05 | 0.01 | 0.77 |
| l-Lactate | — | — | 8.83 | 8.88 | 0.08 | 0.79 |
| dl-Lactate | — | — | 8.85 | 8.90 | 0.09 | 0.77 |
| With glucose added | ||||||
| NH3-N | 2.07 | 2.40 | — | — | 0.18 | 0.26 |
| Acetate | 32.4 | 33.2 | — | — | 1.35 | 0.66 |
| Valerate | 1.97 | 1.53 | — | — | 0.34 | 0.21 |
| d-Lactate | 0.04 | 0.05 | — | — | 0.01 | 0.69 |
| l-Lactate | 8.77 | 8.83 | — | — | 0.11 | 0.71 |
| dl-Lactate | 8.78 | 8.85 | — | — | 0.12 | 0.71 |
Control, control group (LPS-free anaerobic water); R-LPS, regular LPS (200,000 EU) as a carbon source; GLU, glucose as a carbon source; GLU + R-LPS, glucose plus R-LPS (200,000 EU). Lactate concentrations are millimolar.
Results were obtained after adding glucose. —, not applicable (no growth).
—, data not shown.
TABLE 7.
Effects of detoxified and regular lipopolysaccharide on ammonia nitrogen, VFA, and lactate concentrations in Megasphaera elsdenii T81 and Streptococcus bovis JB1 with defined media
| Bacterium and parameter | Value with treatmenta |
SEM | P value | ||
|---|---|---|---|---|---|
| Control | D-LPS | R-LPS | |||
| Megasphaera elsdenii T81 | |||||
| NH3-N | 5.82 | 5.75 | 5.48 | 0.29 | 0.63 |
| Acetate | 12.5 | 13.3 | 12.2 | 1.46 | 0.83 |
| Butyrate | 7.87 | 8.98 | 7.67 | 0.88 | 0.56 |
| Valerate | 2.37 | 2.24 | 2.35 | 0.19 | 0.84 |
| d-Lactate | 0.12 | 0.14 | 0.13 | 0.01 | 0.55 |
| l-Lactate | 0.06 | 0.06 | 0.05 | 0.01 | 0.53 |
| dl-Lactate | 0.18 | 0.20 | 0.18 | 0.02 | 0.55 |
| Streptococcus bovis JB1 | |||||
| NH3-N | 2.75 | 2.54 | 2.48 | 0.11 | 0.13 |
| Acetate | 44.8 | 47.2 | 46.7 | 4.83 | 0.41 |
| Valerate | 1.32 | 1.25 | 1.34 | 0.15 | 0.41 |
| d-Lactate | 0.00 | 0.01 | 0.01 | 0.01 | 0.57 |
| l-Lactate | 8.80 | 8.72 | 8.74 | 0.08 | 0.75 |
| dl-Lactate | 8.82 | 8.74 | 8.76 | 0.08 | 0.79 |
Control, control group (LPS-free anaerobic water); D-LPS, detoxified LPS (200,000 EU); R-LPS, regular LPS (200,000 EU). Lactate cconcentrations are millimolar.
Ruminal microbiome functional changes by LPS dosing.
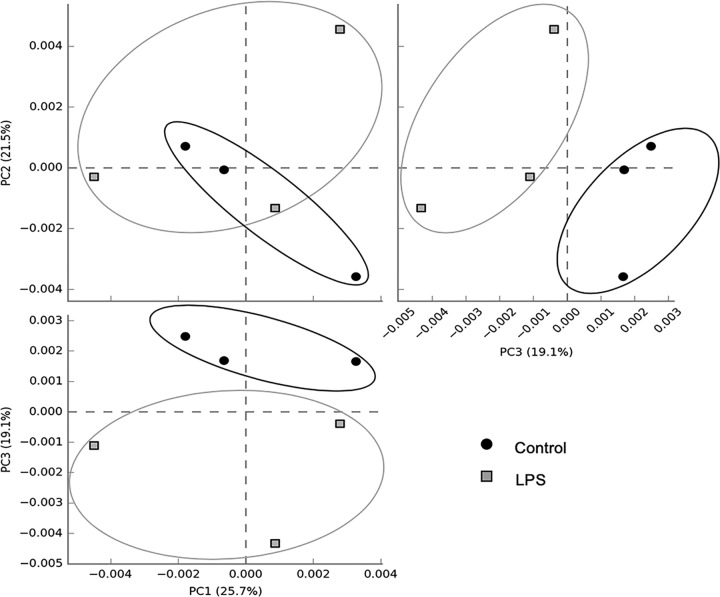
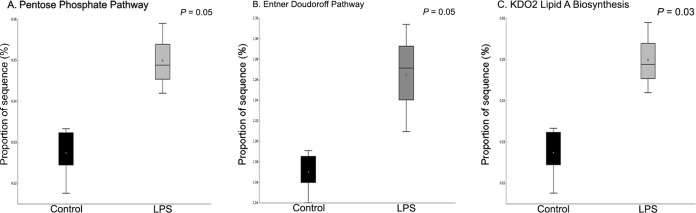
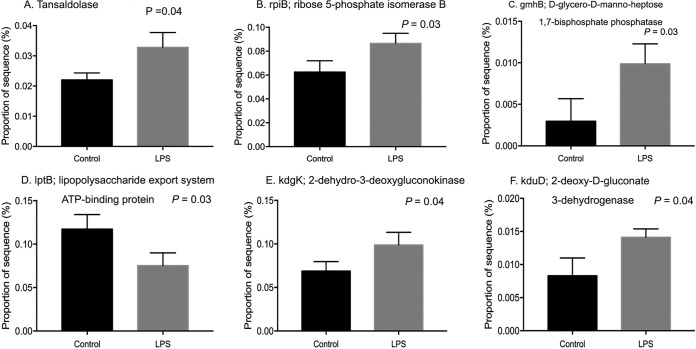
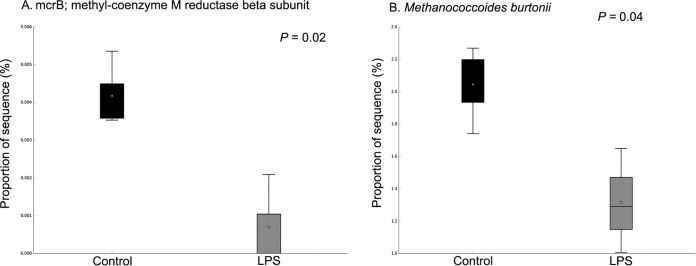
In order to evaluate the effects of LPS on metabolic capabilities of ruminal bacteria, we dosed 200,000 EU of R-LPS in a dual-flow continuous culture system and then mixed ruminal bacteria which were collected to perform shotgun sequencing. We found that ruminal microbiome functions were different between the control and LPS dosing (Fig. 2). The relative abundances of sequence reads of 35 functional genes and 86 functional genes were affected by LPS dosing based on KEGG data set and SEED data set, respectively (see Fig. S1 and S2 in the supplemental material). LPS dosing showed a greater abundance of sequences of functional genes related to the pentose phosphate pathway (PPP) (Fig. 3A), Entner-Doudoroff pathway (EDP) (Fig. 3B), and 3-deoxy-d-manno-octulosonic acid (2-keto-3-deoxyoctonate [KDO2])–lipid A biosynthesis (Fig. 3C) than the control. The proportions of sequences associated with transaldolase (Fig. 4A), ribose 5-phosphate isomerase B (rpiB) (Fig. 4B), and d-glycero-d-manno-heptose 1,7-bisphosphate phosphatase (gmhB) (Fig. 4C) were increased by LPS dosing, while the proportion of sequences associated with LPS export system ATP-binding protein (lptB) (Fig. 4D) was decreased by LPS dosing compared to that of the control. Meanwhile, the abundances of sequence of functional genes related to 2-dehydro-3-deoxygluconokinase (kdgK) (Fig. 4E) and deoxy-d-gluconate 3-dehydrogenase (kduD) (Fig. 4F) were increased by LPS dosing as well. Sequence abundances of functional genes associated with methyl-coenzyme M reductase beta subunit (mcrB) (Fig. 5A) and sequence abundance of Methanococcoides burtonii (Fig. 5B) were decreased by LPS dosing.
FIG 2.
Principal-coordinate analysis plots (PCA) performed on microbial functional similarities between control and lipopolysaccharide dosing in a dual-flow continuous culture system based on the KEGG database.
FIG 3.
Effects of lipopolysaccharide dosing on the proportion of sequence ssociated with the pentose phosphate pathway (A; KEGG), Entner-Doudoroff pathway (B; SEED), and KDO2-lipid A biosynthesis (C; SEED) level 3.
FIG 4.
Effects of lipopolysaccharide dosing on the proportion of sequence of functional genes associated with the transaldolase gene (A), rpiB (B), gmhB (C), lptB (D), kdgK (E), and kduD (F) based on SEED (B) and KEGG (A and C to F) data sets.
FIG 5.
Effects of lipopolysaccharide dosing on the proportions of sequences associated with mcrB (A) and Methanococcoides burtonii (B).
DISCUSSION
Based on our previous study, LPS dosing (200,000 EU) stimulated the Gram-negative bacteria related to starch digestion (Succinimonas, Anaeroplasma, Succinivibrio, Succiniclasticum, and Ruminobacter) (14). Therefore, we hypothesized that LPS dosing would only affect Gram-negative bacteria that mainly used starch or another soluble carbohydrate as their main substrates. From the current study, we found that LPS dosing increased the maximum specific growth rates while decreasing the lag phases of Su. dextrinosolvens 24 and Se. ruminantium HD4, Gram-negative bacteria that mainly use starch as a substrate (15, 16). However, dosing LPS also increased the maximum specific growth rates of S. bovis JB1 and L. ruminis RF1, which are abundant species in the rumens of animals adapted to high-grain diets (17) and in the rumens of animals with acidosis (18). Even though they are also starch utilizers and lactate producers, both are Gram-positive bacteria. Therefore, the current results were against our first hypothesis that LPS dosing would only affect Gram-negative bacteria that mainly used starch or another soluble carbohydrate as a substrate. Actually, dosing of LPS could stimulate lactate-producing bacteria regardless of their Gram reactions.
Ruminal bacteria that rapidly ferment starch or soluble sugars and contribute to rapid accumulations of dl-lactic acid and VFA include S. dextrinosolvens, Se. ruminantium, S. bovis, and Lactobacillus spp., which are considered to play important roles in ruminal acidosis (18). Therefore, the stimulating effect of LPS on the growth of these lactate-producing bacteria indicates that LPS not only is produced during acidosis but also may contribute to ruminal acidosis development.
Due to its central role in the prevention of acid accumulation in the rumen, Megasphaera elsdenii (Gram negative) is also predominant in the rumens of animals with ruminal acidosis (19). However, dosing of LPS had no effects on the maximum specific growth rate or lag phase of M. elsdenii T81, as well as its fermentation end products. Meanwhile, we sought to evaluate if LPS dosing would affect other bacteria; therefore, three main fiber-utilizing bacteria (R. albus 7, R. flavefaciens FD-1, and F. succinogenes S85) that use cellobiose as their substrates were tested, and we found that LPS dosing had no effects on their maximum specific growth rates, lag phases, or fermentation end products. Therefore, dosing of LPS mainly stimulated lactate-producing bacteria and not the fiber-utilizing (R. albus 7, R. flavefaciens FD-1, and F. succinogenes S85) or lactate-utilizing (M. elsdenii T81) bacteria based on the current study.
Next, we wished to investigate the potential mechanism of how LPS stimulates the growth of these lactate-producing bacteria. LPS is an amphipathic molecule that contains fatty acyl chains attached to a polysaccharide containing up to 200 sugars (20). Therefore, we thought that LPS may be catabolized by the enzymes secreted by bacteria and used as the carbon source for their growth. Streptococcus bovis JB1 with the defined medium was used to test this hypothesis. We found that when LPS replaced glucose as the carbon source in the defined medium, S. bovis JB1 did not grow on it. However, when the glucose was added into the LPS dosing medium, Streptococcus bovis JB1 started to grow and the maximum specific growth rate was increased. Meanwhile, the stimulating effect of LPS on the growth of S. bovis JB1 was present regardless of time of glucose inclusion. LPS dosing increased the maximum specific growth rate about 15% before and after glucose addition. Therefore, LPS could not replace glucose as the carbon source for S. bovis JB1 growth, and the stimulating effect of LPS on the growth of lactate-producing bacteria can be observed when there is another soluble carbon source (e.g., starch or glucose) present. The fermentation end products of all the tested bacteria were not affected by LPS dosing. Therefore, the hypothesis that LPS is used as the carbon source for lactate-producing bacteria and thus stimulates their growth was rejected; otherwise, due to the additional carbon source (LPS) in the medium, the fermentation end products may be increased by LPS dosing.
We thought that the presence of LPS may work as a factor to stimulate the growth of lactate-producing bacteria. It has been reported that LPS is essential for all tested ruminal strains of Anaeroplasma abactoclasticum (a starch-producing ruminal bacterium) in pure culture and 0.25 mg/liter of LPS stimulated growth of strain 6-1 (21). Unfortunately, the mechanism of how LPS stimulated Anaeroplasma abactoclasticum 6-1 was not elucidated from the study. LPS is composed of a lipid A moiety, inner and outer core oligosaccharides, and the O antigens, where lipid A consists of fatty acyl chains and the rest are polysaccharides (20). We questioned which part of LPS (fatty acyl chain or polysaccharide) could potentially act to stimulate the growth of lactate-producing bacteria. To evaluate this, the same concentrations of delipidated LPS (lipid A delipidated) and intact (regular) LPS were tested for their effects on the growth of S. bovis JB1. We found that when LPS was mainly present as polysaccharide (lipid A delipidated), the maximum specific growth rate of S. bovis JB1 was increased by only 7.5% and the lag phase was not affected. However, the maximum specific growth rate was increased by 15% and the lag phase was decreased by 42 min by the regular/intact LPS (lipid A present). Therefore, the stimulating effects of LPS on the growth of S. bovis JB1 could mainly contribute to the presence of lipid A of LPS. Future studies could perhaps focus on evaluating the effects of pure lipid A of LPS on the growth of lactate-producing bacteria to validate this.
Meanwhile, lipid A, covalently linked to the inner core, carries the endotoxic activity of LPS (1). We thought that LPS may be toxic to other non-lactate-producing ruminal bacteria, increasing the competition of lactate-producing bacteria and thus stimulating their growth. Megasphaera elsdenii T81 was chosen because it is the main lactate-utilizing ruminal bacterium and is dominantly present in ruminal acidosis. Megasphaera elsdenii T81 treated with the same concentrations of detoxified (lipid A delipidated) and regular LPS was used to test this hypothesis. Removal of the fatty acid portions of lipid A results in a detoxified LPS with an endotoxin level about 10,000 times lower than that of the regular LPS based on the Sigma-Aldrich protocol. However, there were no difference on the maximum specific growth rate and lag phase of M. elsdenii T81 among control, detoxified LPS, and regular LPS dosing. Meanwhile, S. bovis JB1 had a lower maximum specific growth rate with detoxified LPS than with regular LPS. Therefore, LPS may have no toxic effects on ruminal bacteria. In turn, the endotoxin carrier—lipid A—may act as a factor to stimulate the growth of lactate-producing bacteria.
Shotgun sequencing was performed to evaluate the effects of LPS dosing on the metabolic capabilities of ruminal bacteria. It was observed that dosing of LPS increased the sequence abundances of microbial functional genes associated with LPS biosynthesis pathway. KDO2-lipid A and ADP-d-glycero-β-d-manno-heptose are two necessary precursors for LPS biosynthesis (22). The sequence abundances of functional genes associated with KDO2-lipid A biosynthesis were increased by LPS dosing; the sequence abundance of gmhB involved in ADP-d-glycero-β-d-manno-heptose biosynthesis was also increased by LPS dosing. Meanwhile, the sequence abundances of microbial functional genes associated with rpiB and the transaldolase gene, which are involved in d-ribulose-5-phosphate (d-ribulose-5P) and d-sedoheptulose-7-phosphate synthesis, respectively, were increased by LPS dosing. d-Ribulose-5-phosphate and d-sedoheptulose-7-phosphate could initiate LPS biosynthesis (23), which could be converted into KDO2-lipid A biosynthesis (24) and ADP-l-glycero-β-d-manno-heptose (25) to synthesis, respectively (23, 24, 26). Sequence abundances of microbial functional genes associated with kdgK and kduD, which are involved in the synthesis of 2-dehydro-3-deoxy-6-phospho-d-gluconate (the precursor of d-ribulose-5P) based on the PPP pathway and LPS biosynthesis pathway, were increased by LPS dosing. Therefore, dosing of LPS mainly increased the microbial functional genes associated with the LPS biosynthesis pathway, and microbial functional genes associated with the PPP and EDP were increased by LPS dosing to increase the precursors for LPS biosynthesis.
However, the sequence abundance associated with LPS export system ATP-binding protein (lptB) was decreased by LPS dosing. The energy responsible for driving LPS transport is derived from ATP hydrolysis catalyzed by lptB (20, 27, 28). This energy must be coupled to move across the periplasm and outer membrane (OM) in Gram-negative bacteria, as there is no ATP in these compartments (20). In vivo studies demonstrated that all Lpt proteins are required for the transport of LPS; depletion of any of the Lpt proteins leads to the accumulation of LPS in the periplasmic leaflet of the inner membrane (IM) (29, 30). Therefore, sequence abundance of lptB was decreased by LPS dosing, which may lead to the accumulation of LPS in the periplasmic leaflet of IM of bacteria. LPS accumulation in the IM of bacteria may increase the osmotic pressure and then may increase LPS release from bacteria into the rumen.
The increase of the sequence abundance associated with LPS biosynthesis may be due to the increase in the sequence abundance of Gram-negative bacteria by LPS dosing. With the increase of Gram-negative bacteria by LPS dosing, the demands of LPS biosynthesis for outer membrane assembly were increased. Therefore, the sequence abundances of functional genes associated with the LPS biosynthesis pathway were increased. Our previous study also reported that LPS dosing mainly stimulated the Gram-negative bacteria related to starch digestion (the genera Succinimonas, Anaeroplasma, Succinivibrio, Succiniclasticum, and Ruminobacter) (14). In turn, LPS dosing-increased growth of Gram-negative bacteria may be due to stimulation of functional genes involved in the LPS biosynthesis pathway. However, in order to validate how exactly LPS affects the LPS biosynthesis pathway, collecting RNA samples from the single strains of Gram-negative bacteria and analyzing the genes involved in LPS biosynthesis by quantitative PCR (qPCR) or transcriptome sequencing (RNA-seq) would be suggested.
Sequence abundances of functional genes associated with the methyl-coenzyme M reductase beta subunit (mcrB) involved in methane metabolism were decreased by LPS dosing. The enzyme encoded by the mcrB gene catalyzes the reductive cleavage of methyl-coenzyme M [CoM-S-CH3; 2-(methylthio)ethanesulfonate] using coenzyme B (CoB; 7-mercaptoheptanoylthreonine phosphate) as reductant and produce methane and the mixed heterodisulfide of CoB and CoM (CoM-S-S-CoB) (31). This is the final step in methanogenesis. At the same time, the sequence abundance of Methanococcoides burtonii was decreased by LPS dosing as well. Methanococcoides burtonii is known as a methanogen; methanogens produce methane by decomposition of organic carbons under anaerobic conditions (32). Therefore, dosing of LPS might decrease methane production by directly affecting methanogen and the functional genes involved in methane formation. In the future, studies could focus on increased ruminal LPS concentration as a function of the growth or lysis of Gram-negative bacteria and thus help to understand and possibly develop practical strategies to control ruminal methane emissions.
Conclusion.
Dosing of LPS mainly stimulated the growth of lactate-producing bacteria (S. bovis JB1, Se. ruminantium HD4, Su. dextrinosolvens 24, and L. ruminis RF1), regardless of Gram reaction, and not fiber-utilizing bacteria (R. albus 7, R. flavefaciens FD-1, and F. succinogenes S85) or a lactate-utilizing bacterium (M. elsdenii T81). Lipopolysaccharide could not replace glucose as a carbon source for lactate-producing bacteria (S. bovis JB1), and stimulating effects of LPS on the growth of lactate-producing bacteria were observed only when there was another soluble carbon source (e.g., starch or glucose) present. Dosing of LPS has no toxic effects on tested bacteria, and the presence of lipid A or intact LPS was the main contributor to the stimulating effects of LPS on the growth of lactate-producing bacteria. The sequence abundances of functional genes associated with the PPP, EDP, and LPS biosynthesis pathway were increased by LPS dosing, while the sequence abundance of functional genes involved in methanogenesis and sequence abundance of Methanococcoides burtonii were decreased by LPS dosing. In summary, LPS stimulated the growth of ruminal bacteria that use starch and produce lactate. Our results indicate that LPS not only is increased during acidosis but also may contribute to ruminal acidosis development by increasing the growth of lactic acid-producing bacteria; dosing of LPS may stimulate the LPS biosynthesis of ruminal bacteria.
MATERIALS AND METHODS
Organisms.
Fiber-utilizing bacteria (Ruminococcus albus 7 (Gram negative), Ruminococcus flavefaciens FD-1 (Gram negative), Fibrobacter succinogenes S85 (Gram negative), lactate-producing bacteria (Streptococcus bovis JB1 [Gram positive], Succinivibrio dextrinosolvens 24 [Gram negative], Lactobacillus ruminis RF1 [Gram positive], and Selenomonas ruminantium HD4 [Gram negative]), and a lactate-utilizing bacterium (Megasphaera elsdenii T81 [Gram negative]) were chosen for the current study. The Gram characteristic was based on the Gram reaction. Ruminococcus albus 7, R. flavefaciens FD-1, F. succinogenes S85, and M. elsdenii T81 were obtained from P. Weimer (USDA-ARS, Madison, WI). Streptococcus bovis JB1 and Su. dextrinosolvens 24 were obtained from the ATCC. Lactobacillus. ruminis RF1 was obtained from the DSMZ. Selenomonas ruminantium HD4 was obtained from Michael Flythe (USDA-ARS, Lexington, KY). The bacteria were preserved in anaerobic dilution solution with 50% glycerol in Balch tubes with butyl rubber stoppers and aluminum crimp seals and stored at −20°C.
Maintenance and growth of strains.
For each experimental run, each strain was revived from the Balch tube that was stored at −20°C and transferred twice in the same fresh media as the ones used before experiments started. The stored bacteria can be freeze-thawed multiple times without effect on viability of bacteria (33). During the experiment, 0.1 ml of homogeneous stock culture was transferred from the previous stock culture by a 1-ml syringe with a 22-gauge needle into fresh medium vials that were sealed with a flanged butyl rubber stopper and aluminum crimp seal. Anaerobic water and regular and detoxified LPS were also transferred into the Balch tubes by a 1-ml syringe with a 22-gauge needle after transfer of bacteria. The glucose stock was added to Balch tubes of defined medium right before transfer of bacteria. All of the procedures were done on a disinfected bench with 75% ethanol and under anaerobic conditions.
Unless otherwise stated, strains were grown anaerobically under O2-free CO2 in Balch tubes with butyl rubber stoppers and aluminum crimp seals and incubated at 39°C without shaking. The optical density (OD) before (ODb) and after (OD0) bacterium inclusion were measured at 600 nm on spectrophotometer (Genesys 20; Thermo Scientific, Waltham, MA) for each experimental run and for each strain. Measurements of OD at 600 nm (ODt) were collected every hour except in the case of S. bovis JB1, for which measurement s were collected every 30 min until bacterial growth reached a plateau. The real OD of strains was calculated as ODt − ODb. Once bacterial growth reached a plateau, the culture vials were decapped and stock culture medium was collected and stored in −20°C for later determination of the fermentation end products (NH3-N, VFA, and lactate).
Media.
Phosphate carbonate minus VFA (PC–VFA) medium contained 4 g of glucose, 292 mg of K2HPO4, 292 mg of KH2PO4, 480 mg of (NH4)2SO4, 480 mg of NaCl, 49 mg of MgSO4·7H2O, 64 mg of CaCl2·2H2O, 4 g of Na2CO3, 1 g of Trypticase peptone (product 212750; BD), and 0.5 g of yeast extract (product 212750; BD) per liter (34, 35). Resazurin was added as a redox indicator. Strains grown on PC–VFA included S. bovis JB1, M. elsdenii T81, and Se. ruminantium HD4.
In order to stimulate growth, short-chain fatty acids were added to the above-described medium, giving PC+VFA. Short-chain fatty acids included 1.7 ml of acetic acid, 0.6 ml of propionic acid, 0.4 ml of n-butyric acid, 0.1 ml of n-valeric acid, 0.1 ml of isovaleric acid, 0.1 ml of isobutyric acid, and 0.1 ml of 2-methylbutyric acid per liter (15, 34, 35). Strains grown on PC+VFA included L. ruminis RF1 and Su. dextrinosolvens 24. In order to grow fiber-utilizing bacteria (F. succinogenes S85, R. albus 7, and R. flavefaciens FD1), 4 g of glucose in PC+VFA medium was replaced with 3 g of cellobiose per liter of medium (34).
To determine if LPS would replace glucose as carbon source for the growth of S. bovis JB1, a defined medium was modified from PC-VFA medium (36, 37) in which, per liter of the medium, 4 g of glucose, 1 g of Trypticase peptone, and 0.5 g of yeast extract were removed and 5 ml of Pfennings heavy metal solution, 1 ml of hemin solution, and 10 ml of Russell vitamin solution were added to the medium. Pfennings heavy metal solution included 0.5 g of EDTA disodium salt, 0.2 g of FeSO4·7H2O, 0.2 g of MnCl2·4H2O, 0.01 g of ZnSO4·7H2O, 0.03 g of H3BO4, 0.02 g of CoCl2·6H2O, 0.001 g of CuCl2·2H2O, 0.002 g of NiCl2·6H2O, and 0.003 g of NaMoO4·2H2O per liter (38). Hemin solution included 2.8 g of KOH, 250 ml of 95% ethanol, and 1 g of hemin per liter (39). Russell vitamin solution included 0.1 g of pyridoxamine dihydrochloride, 0.1 g of pyridoxal hydrochloride, 0.1 g of pyridoxine, 0.2 g of riboflavin, 0.2 g of thiamine HCl, 0.2 g of nicotinamide, 0.2 g of calcium pantothenate, 0.1 g of lipoic acid, 0.01 g of 4-aminobenzoic acid, 0.005 g of folic acid, 0.005 g of biotin, and 0.005 U of coenzyme B12 in 1 liter of 0.1 M KH2PO4 (36; M. Flythe, personal communication).
Anaerobic water, LPS, and glucose stock preparation. (i) Anaerobic water.
Nonpyrogenic water (catalog no. W50-500; Lonza Group Ltd., Basel, Switzerland), was boiled for 5 to 7 min and dispensed into culture tubes while flushing with N2, and cultured tubes were capped with butyl rubber stoppers and aluminum crimp seals for autoclaving.
(ii) LPS stock.
The concentrations of regular LPS (Escherichia coli O111:B4, L2630; Sigma-Aldrich Co., St. Louis, MO) and delipidated/detoxified LPS (lipid A delipidated, Escherichia coli O111:B4, L3023; Sigma-Aldrich Co.) were 200,000 EU, the same as in our previous study (14). The LPS was prepared under anaerobic conditions, where 100 mg of LPS was added to 250 ml of anaerobic nonpyrogenic water while flushing with N2, to generate 0.4 mg/ml of LPS. The LPS stock was then filtered through a 0.22-μm polyethersulfone (PES) membrane syringe filter (Celltreat, Pepperell, MA) into Balch tubes that were previously flushed with N2 and autoclaved. A 0.5-ml volume of 0.4-mg/ml regular or detoxified LPS stock was equal to 200,000 EU of LPS (1 ng/ml = 10 endotoxin units based on the Sigma-Aldrich protocol, the volume of culture media in Balch tubes of 10 ml).
(iii) Glucose stock.
Glucose stock was prepared under anaerobic conditions, where 4.2 g of glucose was added to 70 ml of anaerobic nonpyrogenic water while flushing with N2, to generate 0.06 g/ml of glucose. The glucose stock was then filtered through a 0.22-μm PES membrane syringe filter into 160-ml culture tubes with flanged butyl stoppers and aluminum crimp seals that were previously flushed with N2 and autoclaved. The glucose stock concentration was the same as in the PC−VFA and PC+VFA media.
Experimental design and measurement of bacterial growth. (i) Experiment 1.
The goal of experiment 1 was to evaluate effects of regular LPS (200,000 EU) on fiber-utilizing bacteria (R. albus 7, R. flavefaciens FD-1, and F. succinogenes S85), lactate-producing bacteria (S. bovis JB1, Su. dextrinosolvens 24, L. ruminis RF1, and Se. ruminantium HD4), and a lactate-utilizing bacterium (M. elsdenii T81). Treatments were (i) a control (0.5 ml of anaerobic water) and (ii) regular LPS (0.5 ml of 0.4-mg/ml regular LPS). There were at least three experimental runs per strain, and each treatment had three replications. The experimental procedure flowchart is presented in Fig. 6.
FIG 6.
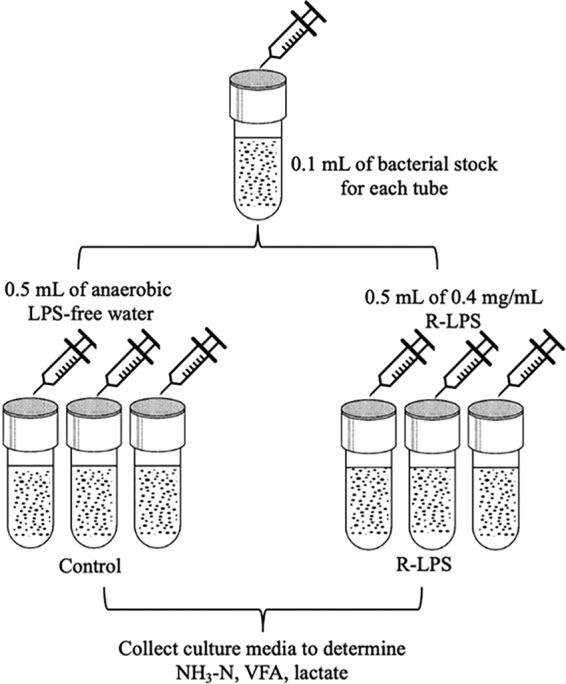
Experimental procedure flowchart, experiment 1. Control, control group (LPS-free anaerobic water); R-LPS, regular LPS (200,000 EU).
(ii) Experiment 2.
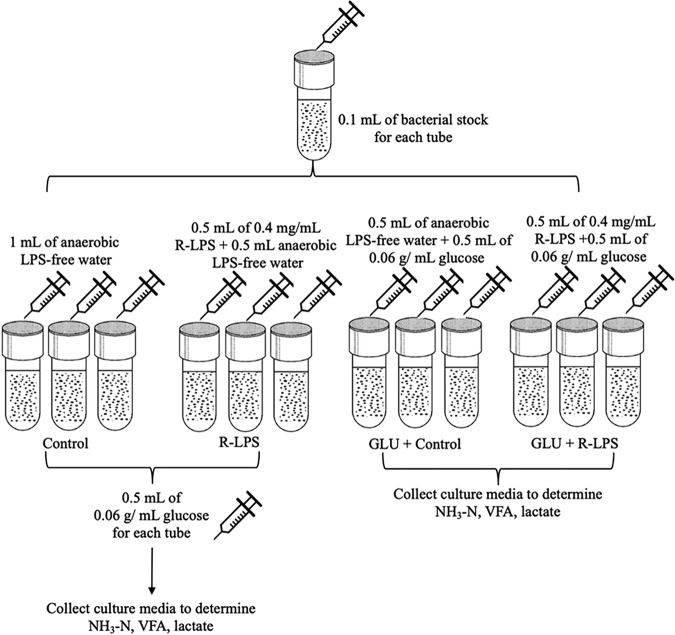
The goal of experiment 2 was to evaluate if LPS (200, 000 EU) could replace glucose as a carbon source for S. bovis JB1 growth with the defined medium. Treatments were (i) a control (1 ml of anaerobic water), (ii) LPS (0.5 ml of anaerobic water plus 0.5 ml of 0.4-mg/ml regular LPS), (iii) GLU (0.5 ml of anaerobic water plus 0.5 ml of 0.06 mg/ml glucose), and (iv) GLU plus LPS (0.5 ml of 0.4-mg/ml regular LPS plus 0.5 ml of 0.06-g/ml glucose). Once growth of S. bovis JB1 in GLU and GLU plus LPS reached a plateau, S. bovis JB1 did not grow in control and LPS media; 0.5 ml of 0.06-g/ml glucose was then added into control and LPS tubes to measure the growth of S. bovis JB1. There were five experimental runs, and each treatment had three replications. The experimental procedure flowchart is presented in Fig. 7.
FIG 7.
Experimental procedure flowchart, experiment 2. Control, control group (LPS-free anaerobic water); LPS, regular LPS (200,000 EU) as a carbon source; GLU, glucose as a carbon source; GLU + LPS, glucose plus LPS (200,000 EU).
(iii) Experiment 3.
The goal of experiment 3 was to evaluate the toxicity of LPS (200,000 EU) on S. bovis JB1 in a defined medium with glucose added as a carbon source and M. elsdenii T81 in PC−VFA medium. Treatments were (i) a control (0.5 ml of anaerobic water), (ii) R-LPS (0.5 ml of 0.4-mg/ml regular LPS), and (iii) D-LPS (0.5 ml of 0.4-mg/ml detoxified LPS). There were five experimental runs per strain, and each treatment had three replications. The experimental procedure flowchart is presented in Fig. 8.
FIG 8.
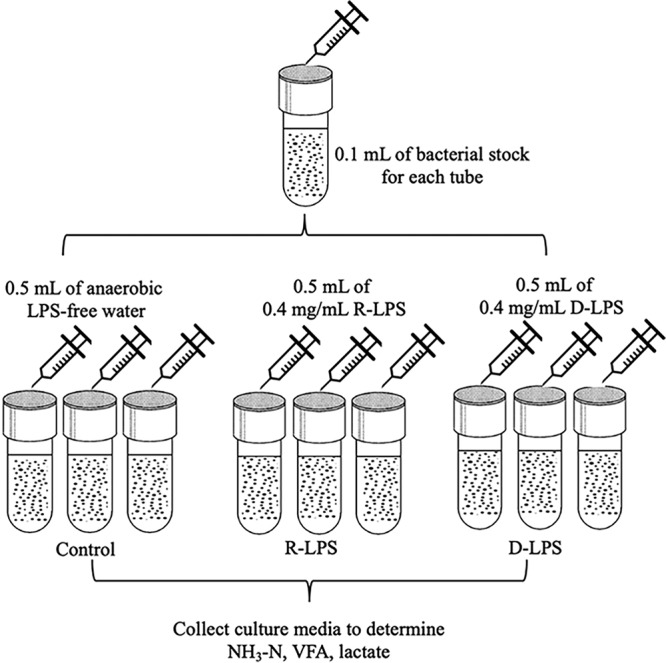
Experimental procedure flowchart, experiment 3. Control, control group (LPS-free anaerobic water); R-LPS, regular LPS (200,000 EU); D-LPS, detoxified LPS (200,000 EU).
Ammonia, VFA, and lactate analyses. (i) Sample preparation.
Ten milliliters of stock culture medium used for NH3 and VFA concentration analyses was preserved with 0.1 ml of 50% H2SO4. After that, samples were centrifuged at 10,000 × g for 15 min at 4°C. One milliliter of supernatant was filtered through a 0.22-μm syringe filter (Tisch Scientific, North Bend, OH) into 2-ml vials used for VFA analysis, and the rest of acidic samples were preserved for NH3-N analysis.
(ii) Ammonia nitrogen concentration analysis.
Ammonia nitrogen concentration analysis was adapted from Broderick and Kang (40). Briefly, 2 μl of samples, 100 μl of phenol, and 80 μl of hypochlorite were added to each well of a flat-bottom 96-well plate, mixed well, and then incubated at 95°C for 5 min. After cooling, the plate was read at 620 nm on a microplate spectrophotometer (Spectra Max 340 PC; Molecular Devices Corporation, Sunnyvale, CA).
(iii) Volatile fatty acid analysis.
Concentrations of VFA in ruminal fluid samples were determined in a liquid-liquid solvent extraction using ethyl acetate (41). Ruminal fluid supernatant was mixed with a meta-phosphoric acid–crotonic acid (internal standard) solution at a 5:1 ratio, and samples were frozen overnight, thawed, and centrifuged for 15 min at 10,000 × g. Supernatant was transferred into glass tubes and mixed with ethyl acetate in a 2:1 ratio of ethyl acetate to supernatant. After the tubes were shaken vigorously and the fractions were allowed to separate, the ethyl acetate fraction (top layer) was transferred to vials. Samples were analyzed by gas chromatography (Agilent 7820A gas chromatograph; Agilent Technologies, Palo Alto, CA) using a flame ionization detector and a capillary column (CP-WAX 58 FFAP, 25 m by 0.53 mm, Varian CP7767; Varian Analytical Instruments, Walnut Creek, CA). Column temperature was maintained at 110°C, and injector and detector temperatures were 200 and 220°C, respectively.
(iv) Lactate analysis.
The stock culture medium used for lactate analysis was first boiled at 100°C for 10 min and then centrifuged at 10,000 × g for 10 min at 4°C. The supernatants were then used for lactate analysis. The UV method was used to determine d- and l-lactic acid concentrations by using a d-lactic acid/l-lactic acid kit from R-Biopharm AG (Darmstadt, Germany). Concentrations of d- and l-lactic acid were analyzed on a flat-bottom 96-well plate and read at 340 nm on a microplate spectrophotometer. The analysis procedure was described in detail in reference 14.
Shotgun sequencing and metagenomic data processing. (i) Library preparation and sequencing.
In order to evaluate the effects of LPS on metabolic capabilities of ruminal bacteria, we dosed 200,000 EU of R-LPS in a dual-flow continuous culture system that was designed to mimic the ruminal microbial fermentation. There were four fermentors randomly assigned within Latin square to receive either control or regular LPS (200,000 EU) over 3 periods with 7 days of diet adaptation followed by 4 days of bacterial sampling per period. Bacterial samples were collected at 2, 5, 7, and 9 h after morning feeding per sampling day. Then the same amount of bacterial sample from the same treatment and period were pooled for both control and LPS treatments. There were three replications per treatment, and a total of 6 bacterial samples were processed for the DNA isolation and shotgun sequencing. The experimental diets, sample collection, and DNA extraction processing were described in detail in reference 14.
A Qubit 2.0 fluorometer (Invitrogen, Carlsbad, CA) was used to measure the DNA concentration of each sample, and each sample was diluted to around 0.2 ng/μl for library preparation. Libraries were prepared for sequencing by using a Nextera XT DNA library prep kit (FC-131-1001; Illumina Inc., San Diego, CA). The libraries were constructed according to the Nextera XT DNA library prep kit reference guide (document number 15031942 v03; February 2018). The generated library plus 1% PhiX spike in control and were sequenced on Illumina Miseq using MiSeq reagent kit V2 (2 × 250 cycles; Illumina) according to the manufacturer’s protocol.
(ii) Shotgun metagenomic data processing.
The paired-end sequences obtained were merged by Sickle (42), and low-quality bases (quality score lower than 20) and sequences below 50 nucleotides were removed. Metagenome assembly was done by metaSPAdes with k-mers of 33, 55, and 77 (43). The assembly sequences were annotated with the Metagenomics Rapid Annotation (MG-RAST) pipeline, version 4.0.3 (44). Functional profiles were generated using the normalized abundance of sequence matches to the SEED (45) and KEGG (46) databases, respectively. A table of the frequency of hits to each individual function for each metagenome was generated and normalized. To identify hits, BLASTX was used (47) with a minimum alignment length of 50 bp, minimal identity of 60%, and an E value cutoff of <1 × 10−5. The matrices generated were exported and used for statistical analyses.
Statistical analysis.
Logistic function was used to predict the growth rate (μ) and lag phase (lag), as the R2 values were all greater than 99.5% and MSE were lower than 0.001 for each experimental run of tested strains. According to reference 48, the logistic function used in the study was
where Y is real ODt, y0 is initial OD0, μmax is maximum specific growth rate, and C is increase of OD from OD0 to ODt.
The logistic model was fit to the data by nonlinear regression using NLIN procedure of SAS software (version 9.4; SAS Institute Inc., Cary, NC). Several possible starting values were specified for each parameter, so that the NLIN procedure evaluated the model at each combination of initial values on the grid, using for the first iteration of the fitting process the combination yielding the smallest residual sum of squares. The initial values supplied were different per run per strain, and the selection of the starting values was based on visual inspection of the growth curve. The uniqueness of the final solution achieved in each case was checked by changing the initial parameter estimates within a reasonable range of each data set.
The predicted maximum specific growth rate and lag phase were used as input for SAS analysis. The effects of treatment on maximum specific growth rate and lag phase, and concentrations of ammonia, VFA, and lactate were subjected to least-square analysis of variance (ANOVA) using the MIXED procedure of SAS. All data were checked for the linear model assumption. The statistical model used was
where y is a dependent variable, μ is overall mean, is fixed effect of treatments, is experimental run, and is the random error. Experimental run was considered the random effect. Different covariance structures were tested, and the one with the lowest Akaike information criterion values were chosen.
The statistical differences between the sequencing data were determined using the Statistical Analysis of Metagenomic Profiles v2.1.3 (STAMP) software (49). For functional profiling, a table with hit frequency for each metagenome was generated from MG-RAST and used as input. P values were calculated using two-sided Welch’s t test (50). Corrected P values below 0.05 were considered significant. Data will be made fully available and without restriction.
Data availability.
All sequences from this project have been deposited at the National Center for Biotechnology Information Sequence Read Archive under accession number SRP216606.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge Patricia Spoto Correa (University of São Paulo) and Peixin Fan (University of Florida) for help with shotgun sequencing analysis and the NextGen DNA sequencing laboratory – Interdisciplinary Center for Biotechnology Research at the University of Florida for performing the MiSeq sequencing runs. We also acknowledge the labmates from the lab of Antonio P. Faciola for the discussion and support during the study and James Colee (University of Florida, IFAS Statistical Consulting) for help with the statistical analysis.
We declare no conflicts of interest.
This work was supported mainly by startup funds from APF and also by the Hatch Project (accession no. 1002754 and 1002352) from the USDA National Institute of Food and Agriculture to T.J.H.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Guerville M, Boudry G. 2016. Gastrointestinal and hepatic mechanisms limiting entry and dissemination of lipopolysaccharide into the systemic circulation. Am J Physiol Gastrointest Liver Physiol 311:G1–G15. doi: 10.1152/ajpgi.00098.2016. [DOI] [PubMed] [Google Scholar]
- 2.Caroff M, Karibian D. 2003. Structure of bacterial lipopolysaccharides. Carbohydr Res 338:2431–2447. doi: 10.1016/j.carres.2003.07.010. [DOI] [PubMed] [Google Scholar]
- 3.Erridge C, Bennett-Guerrero E, Poxton IR. 2002. Structure and function of lipopolysaccharides. Microbes Infect 4:837–851. doi: 10.1016/s1286-4579(02)01604-0. [DOI] [PubMed] [Google Scholar]
- 4.Huber TL. 1976. Physiological effects of acidosis on feedlot cattle. J Anim Sci 43:902–909. doi: 10.2527/jas1976.434902x. [DOI] [PubMed] [Google Scholar]
- 5.Plaizier JC, Krause DO, Gozho GN, McBride BW. 2008. Subacute ruminal acidosis in dairy cows: the physiological causes, incidence and consequences. Vet J 176:21–31. doi: 10.1016/j.tvjl.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 6.Plaizier JC, Khafipour E, Li S, Gozho GN, Krause DO. 2012. Subacute ruminal acidosis (SARA), endotoxins and health consequences. Anim Feed Sci Technol 172:9–21. doi: 10.1016/j.anifeedsci.2011.12.004. [DOI] [Google Scholar]
- 7.Kleen JL, Upgang L, Rehage J. 2013. Prevalence and consequences of subacute ruminal acidosis in German dairy herds. Acta Vet Scand 55:48. doi: 10.1186/1751-0147-55-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nagaraja TG, Lechtenberg KF. 2007. Acidosis in feedlot cattle. Vet Clin North Am Food Anim Pract 23:333–350. doi: 10.1016/j.cvfa.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 9.Nagaraja TG, Fina LR, Bartley EE, Anthony HD. 1978. Endotoxic activity of cell-free rumen fluid from cattle fed hay or grain. Can J Microbiol 24:1253–1261. doi: 10.1139/m78-201. [DOI] [PubMed] [Google Scholar]
- 10.Andersen PH, Bergelin B, Christensen KA. 1994. Effect of feeding regimen on concentration of free endotoxin in ruminal fluid of cattle. J Anim Sci 72:487–491. doi: 10.2527/1994.722487x. [DOI] [PubMed] [Google Scholar]
- 11.Gozho GN, Krause DO, Plaizier JC. 2007. Ruminal lipopolysaccharide concentration and inflammatory response during grain-induced subacute ruminal acidosis in dairy cows. J Dairy Sci 90:856–866. doi: 10.3168/jds.S0022-0302(07)71569-2. [DOI] [PubMed] [Google Scholar]
- 12.Khafipour E, Krause DO, Plaizier JC. 2009. A grain-based subacute ruminal acidosis challenge causes translocation of lipopolysaccharide and triggers inflammation. J Dairy Sci 92:1060–1070. doi: 10.3168/jds.2008-1389. [DOI] [PubMed] [Google Scholar]
- 13.Khafipour E, Krause DO, Plaizier JC. 2009. Alfalfa pellet-induced subacute ruminal acidosis in dairy cows increases bacterial endotoxin in the rumen without causing inflammation. J Dairy Sci 92:1712–1724. doi: 10.3168/jds.2008-1656. [DOI] [PubMed] [Google Scholar]
- 14.Dai X, Paula EM, Lelis ALJ, Silva LG, Brandao VLN, Monteiro HF, Fan P, Poulson SR, Jeong KC, Faciola AP. 2019. Effects of lipopolysaccharide dosing on bacterial community composition and fermentation in a dual-flow continuous culture system. J Dairy Sci 102:334–350. doi: 10.3168/jds.2018-14807. [DOI] [PubMed] [Google Scholar]
- 15.Caldwell DR, Bryant MP. 1966. Medium without rumen fluid for nonselective enumeration and isolation of rumen bacteria. Appl Environ Microbiol 14:794–801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stewart CS, Hj F, Bryant MP. 1997. The rumen bacteria, p 45 In Hobson PN, Stewart CS (ed), The rumen microbial ecosystem, 2nd ed Blackie Academic and Professional, London, United Kingdom. [Google Scholar]
- 17.Ricke SC, Martin SA, Nisbet DJ. 1996. Ecology, metabolism, and genetics of ruminal Selenomonads. Crit Rev Microbiol 22:27–65. doi: 10.3109/10408419609106455. [DOI] [PubMed] [Google Scholar]
- 18.Nagaraja TG, Titgemeyer EC. 2007. Ruminal acidosis in beef cattle: the current microbiological and nutritional outlook. J Dairy Sci 90:E17–E38. doi: 10.3168/jds.2006-478. [DOI] [PubMed] [Google Scholar]
- 19.Counotte GHM, Prins RA, Janssen R, deBie MJA. 1981. Role of Megasphaera elsdenii in the fermentation of dl-[2-13C]lactate in the rumen of dairy cattle. Appl Environ Microbiol 42:649–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okuda S, Sherman DJ, Silhavy TJ, Ruiz N, Kahne D. 2016. Lipopolysaccharide transport and assembly at the outer membrane: the PEZ model. Nat Rev Microbiol 14:337–345. doi: 10.1038/nrmicro.2016.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robinson JP, Hungate RE. 1973. Acholeplasma bactoclasticum sp. n., an anaerobic Mycoplasma from the bovine rumen. Int J Syst Bacteriol 23:171–181. doi: 10.1099/00207713-23-2-171. [DOI] [Google Scholar]
- 22.Xu L, Huo M, Sun C, Cui X, Zhou D, Crittenden JC, Yang W. 2017. Bioresources inner-recycling between bioflocculation of Microcystis aeruginosa and its reutilization as a substrate for bioflocculant production. Sci Rep 7:43784. doi: 10.1038/srep43784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Taylor PL, Blakely KM, de Leon GP, Walker JR, McArthur F, Evdokimova E, Zhang K, Valvano MA, Wright GD, Junop MS. 2008. Structure and function of sedoheptulose-7-phosphate isomerase, a critical enzyme for lipopolysaccharide biosynthesis and a target for antibiotic adjuvants. J Biol Chem 283:2835–2845. doi: 10.1074/jbc.M706163200. [DOI] [PubMed] [Google Scholar]
- 24.Mosberg JA, Yep A, Meredith TC, Smith S, Wang P-F, Holler TP, Mobley HLT, Woodard RW. 2011. A unique arabinose 5-phosphate isomerase found within a genomic island associated with the uropathogenicity of Escherichia coli CFT073. J Bacteriol 193:2981–2988. doi: 10.1128/JB.00033-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kneidinger B, Graninger M, Puchberger M, Kosma P, Messner P. 2001. Biosynthesis of nucleotide-activated d-glycero-d-manno-heptose. J Biol Chem 276:20935–20944. doi: 10.1074/jbc.M100378200. [DOI] [PubMed] [Google Scholar]
- 26.Sarkar M, Maganti L, Ghoshal N, Dutta C. 2012. In silico quest for putative drug targets in Helicobacter pylori HPAG1: molecular modeling of candidate enzymes from lipopolysaccharide biosynthesis pathway. J Mol Model 18:1855–1866. doi: 10.1007/s00894-011-1204-3. [DOI] [PubMed] [Google Scholar]
- 27.Okuda S, Freinkman E, Kahne D. 2012. Cytoplasmic ATP hydrolysis powers transport of lipopolysaccharide across the periplasm in E. coli. Science 338:1214–1217. doi: 10.1126/science.1228984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman DJ, Lazarus MB, Murphy L, Liu C, Walker S, Ruiz N, Kahne D. 2014. Decoupling catalytic activity from biological function of the ATPase that powers lipopolysaccharide transport. Proc Natl Acad Sci U S A 111:4982–4987. doi: 10.1073/pnas.1323516111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ruiz N, Gronenberg LS, Kahne D, Silhavy TJ. 2008. Identification of two inner-membrane proteins required for the transport of lipopolysaccharide to the outer membrane of Escherichia coli. Proc Natl Acad Sci U S A 105:5537–5542. doi: 10.1073/pnas.0801196105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sperandeo P, Lau FK, Carpentieri A, De Castro C, Molinaro A, Deho G, Silhavy TJ, Polissi A. 2008. Functional analysis of the protein machinery required for transport of lipopolysaccharide to the outer membrane of Escherichia coli. J Bacteriol 190:4460–4469. doi: 10.1128/JB.00270-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hallam SJ, Girguis PR, Preston CM, Richardson PM, DeLong EF. 2003. Identification of methyl coenzyme M reductase A (mcrA) genes associated with methane-oxidizing archaea. Appl Environ Microbiol 69:5483–5491. doi: 10.1128/aem.69.9.5483-5491.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Franzmann PD, Springer N, Ludwig W, Conway De Macario E, Rohde M. 1992. A methanogenic archaeon from Ace Lake, Antarctica: Methanococcoides burtonii sp. nov. Syst Appl Microbiol 15:573–581. doi: 10.1016/S0723-2020(11)80117-7. [DOI] [Google Scholar]
- 33.Teather RM. 1982. Maintenance of laboratory strains of obligately anaerobic rumen bacteria. Appl Environ Microbiol 44:499–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chow JM, Van Kessel JAS, Russell JB. 1994. Binding of radiolabeled monensin and lasalocid to ruminal microorganisms and feed. J Anim Sci 72:1630–1635. doi: 10.2527/1994.7261630x. [DOI] [PubMed] [Google Scholar]
- 35.Tao J, Diaz RK, Teixeira CV, Hackmann TJ. 2016. Transport of a fluorescent analogue of glucose (2-NBDG) versus radiolabeled sugars by rumen bacteria and Escherichia coli. Biochemistry 55:2578–2589. doi: 10.1021/acs.biochem.5b01286. [DOI] [PubMed] [Google Scholar]
- 36.Cotta MA, Russell JB. 1982. Effect of peptides and amino acids on efficiency of rumen bacterial protein synthesis in continuous culture. J Dairy Sci 65:226–234. doi: 10.3168/jds.S0022-0302(82)82181-4. [DOI] [Google Scholar]
- 37.Bond DR, Russell JB. 2000. Protonmotive force regulates the membrane conductance of Streptococcus bovis in a non-ohmic fashion. Microbiology 146:687–694. doi: 10.1099/00221287-146-3-687. [DOI] [PubMed] [Google Scholar]
- 38.Schaefer DM, Davis CL, Bryant MP. 1980. Ammonia saturation constants for predominant species of rumen bacteria. J Dairy Sci 63:1248–1263. doi: 10.3168/jds.S0022-0302(80)83076-1. [DOI] [PubMed] [Google Scholar]
- 39.Holdeman L, Moore W. 1972. Anaerobe laboratory manual. Virginia Polytechnic Institute and State University, Blacksburg, VA. [Google Scholar]
- 40.Broderick GA, Kang JH. 1980. Automated simultaneous determination of ammonia and total amino acids in ruminal fluid and in vitro media. J Dairy Sci 63:64–75. doi: 10.3168/jds.S0022-0302(80)82888-8. [DOI] [PubMed] [Google Scholar]
- 41.Ruiz-Moreno M, Binversie E, Fessenden SW, Stern MD. 2015. Mitigation of in vitro hydrogen sulfide production using bismuth subsalicylate with and without monensin in beef feedlot diets. J Anim Sci 93:5346–5354. doi: 10.2527/jas.2015-9392. [DOI] [PubMed] [Google Scholar]
- 42.Joshi NA, Fass JN. 2011. Sickle: a sliding-window, adaptive, quality-based trimming tool for FastQ files (version 1.33). https://github.com/najoshi/sickle.
- 43.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meyer F, Paarmann D, D’Souza M, Olson R, Glass EM, Kubal M, Paczian T, Rodriguez A, Stevens R, Wilke A, Wilkening J, Edwards RA. 2008. The metagenomics RAST server—a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9:386. doi: 10.1186/1471-2105-9-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M, Goto S. 2000. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pla ML, Oltra S, Esteban MD, Andreu S, Palop A. 2015. Comparison of primary models to predict microbial growth by the plate count and absorbance methods. Biomed Res Int 2015:365025–365014. doi: 10.1155/2015/365025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Parks DH, Tyson GW, Hugenholtz P, Beiko RG. 2014. STAMP: statistical analysis of taxonomic and functional profiles. Bioinformatics 30:3123–3124. doi: 10.1093/bioinformatics/btu494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Welch BL. 1947. The generalization of ‘Student’s’ problem when several different population variances are involved. Biometrika 34:28. doi: 10.2307/2332510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences from this project have been deposited at the National Center for Biotechnology Information Sequence Read Archive under accession number SRP216606.