The genus Pseudovibrio is commonly associated with marine invertebrates, which are essential for ocean health and marine nutrient cycling. Traditionally, the phylogeny of the genus has been based on 16S rRNA gene analysis. The use of the 16S rRNA gene or any other single marker gene for robust phylogenetic placement has recently been questioned. We used a large set of marker genes from all available Pseudovibrio genomes for in-depth phylogenomic analyses. We identified divergent monophyletic basal lineages within the Pseudovibrio genus, including two strains isolated from polyclad flatworms. These strains showed reduced sulfur metabolism and biosynthesis capacities. The phylogenomic analyses revealed distinct evolutionary trajectories and ecological adaptations that differentiate the divergent strains from the other Pseudovibrio members and suggest that they fall into a novel genus. Our data show the importance of widening the use of phylogenomics for better understanding bacterial physiology, phylogeny, and evolution.
KEYWORDS: Pseudovibrio, evolution, symbiont, secondary metabolites, comparative genomics, natural selection
ABSTRACT
Bacteria belonging to the Pseudovibrio genus are widespread, metabolically versatile, and able to thrive as both free-living and host-associated organisms. Although more than 50 genomes are available, a comprehensive comparative genomics study to resolve taxonomic inconsistencies is currently missing. We analyzed all available genomes and used 552 core genes to perform a robust phylogenomic reconstruction. This in-depth analysis revealed the divergence of two monophyletic basal lineages of strains isolated from polyclad flatworm hosts, namely, Pseudovibrio hongkongensis and Pseudovibrio stylochi. These strains have reduced genomes and lack sulfur-related metabolisms and major biosynthetic gene clusters, and their environmental distribution appears to be tightly associated with invertebrate hosts. We showed experimentally that the divergent strains are unable to utilize various sulfur compounds that, in contrast, can be utilized by the type strain Pseudovibrio denitrificans. Our analyses suggest that the lineage leading to these two strains has been subject to relaxed purifying selection resulting in great gene loss. Overall genome relatedness indices (OGRI) indicate substantial differences between the divergent strains and the rest of the genus. While 16S rRNA gene analyses do not support the establishment of a different genus for the divergent strains, their substantial genomic, phylogenomic, and physiological differences strongly suggest a divergent evolutionary trajectory and the need for their reclassification. Therefore, we propose the novel genus Polycladidibacter gen. nov.
IMPORTANCE The genus Pseudovibrio is commonly associated with marine invertebrates, which are essential for ocean health and marine nutrient cycling. Traditionally, the phylogeny of the genus has been based on 16S rRNA gene analysis. The use of the 16S rRNA gene or any other single marker gene for robust phylogenetic placement has recently been questioned. We used a large set of marker genes from all available Pseudovibrio genomes for in-depth phylogenomic analyses. We identified divergent monophyletic basal lineages within the Pseudovibrio genus, including two strains isolated from polyclad flatworms. These strains showed reduced sulfur metabolism and biosynthesis capacities. The phylogenomic analyses revealed distinct evolutionary trajectories and ecological adaptations that differentiate the divergent strains from the other Pseudovibrio members and suggest that they fall into a novel genus. Our data show the importance of widening the use of phylogenomics for better understanding bacterial physiology, phylogeny, and evolution.
INTRODUCTION
The genus Pseudovibrio belongs to the alphaproteobacterial family Rhodobacteraceae. These bacteria have been isolated from various marine habitats worldwide, from polar to tropical regions (1). In recent years, this genus has attracted increasing attention due to its recurrent association with marine invertebrates and its ability to produce bioactive secondary metabolites (1, 2). Their great metabolic versatility allows members of the genus Pseudovibrio to thrive under conditions of various nutrient regimes by using diverse organic and inorganic substrates (3–6). Additionally, comparative genomic analyses shed light on the mechanisms potentially used by these bacteria to interact with their invertebrate hosts and microbiomes. Numerous secretion systems, effectors, toxins, and biosynthetic gene clusters (BGCs) likely producing bioactive compounds have been identified in almost all available genomes (7–10). Six type strains have been described so far, and an additional species was recently proposed based on genomic analyses (1, 11). Considering the descriptions of the type strains, the genus appears homogeneous both physiologically and phylogenetically. However, comparative genomics revealed the presence of clearly divergent strains (7, 10), but these discrepancies have not been resolved to date.
The field of microbial taxonomy has changed considerably during the last decade (12, 13), since the advent of phylogenomics greatly expanded the phylogeneticists’ toolbox to classify both cultured and uncultured species (12, 14). Currently, the 16S rRNA gene is the most widely used taxonomic marker and is considered a gold standard for the classification of both Bacteria and Archaea. However, the use of the 16S rRNA gene alone has failed to provide robust classification on multiple occasions (15–18). In contrast, phylogenomic analyses consider a set of several hundred phylogenetic marker genes/proteins, ensuring more-robust tree topologies and increased phylogenetic resolution (12, 14, 16–21). Moreover, the development of overall genome relatedness indices (OGRI) has significantly improved our ability to classify both cultured and uncultured microorganisms (12, 14, 17, 22, 23). Previously, standard polyphasic approaches have placed strains with pronounced genome reduction, namely, Pseudovibrio stylochi and Pseudovibrio hongkongensis, in the Pseudovibrio genus (24, 25), despite recent questioning of such a classification (10). The genome reduction raises the issue of whether these Pseudovibrio strains have evolved under different selection regimes. In fact, evolutionary models state that larger bacterial genome sizes are usually the result of effective natural selection that would maintain genes in a genome, thus counteracting their loss due to neutral processes (26).
Here, we used all 51 currently available Pseudovibrio genomes to perform an in-depth phylogenomic reconstruction of the evolutionary history of this genus and to resolve the taxonomic discrepancy concerning P. stylochi and P. hongkongensis. We use this extensive data set to infer the evolutionary trajectories that led to the divergent lineages, and we place the data in an ecological perspective. Using phylogenomics, comparative genomics, and phenotyping, we show that P. stylochi and P. hongkongensis form a basal monophyletic group within the genus that is genomically and metabolically divergent from the other Pseudovibrio strains. For these reasons, we propose the new genus Polycladidibacter gen. nov. to describe the previously named Pseudovibrio stylochi and Pseudovibrio hongkongensis species.
RESULTS
Phylogenomics and genome composition.
Some of the 51 publicly available Pseudovibrio genomes have been sequenced multiple times. Hence, we selected only the ones with the fewest contigs, more than 90% completeness, and less than 2% contamination, obtaining a total of 45 high-quality genomes (Table 1). Their lengths differed considerably among the strains, with the type strains P. stylochi and P. hongkongensis having the smallest genomes (3.6 and 3.7 Mbp) and strains Tun.PHSC04-5.I4 (here PHSC04) and POLY-S9 (here PolyS9) having the largest (6.5 Mbp; Table 1). GC content levels were very similar among the strains, ranging from 47% in P. stylochi to 52.7% in P. japonicus.
TABLE 1.
Overview of features of publicly available Pseudovibrio genomesa
| Species | Strain | Length (bp) |
% GC |
No. of contigs |
GenBank accession no. |
Completeness (%) |
Contamination (%) |
% core |
|---|---|---|---|---|---|---|---|---|
| Pseudovibrio hongkongensis | MCCC 1K00451 (UST20140214-015B) | 3,746,600 | 53.3 | 39 | LLWC00000000.1 | 98.44 | 0.1 | 44.27 |
| Pseudovibrio stylochi | MCCC 1K00452 (UST20140214-052) | 3,682,052 | 47 | 43 | LLWE00000000.1 | 98.38 | 0.93 | 46.75 |
| Pseudovibrio ascidiaceicola | DSM 16392 | 5,878,784 | 51.2 | 39 | PRJEB20602b | 99.35 | 0.29 | 28.89 |
| Pseudovibrio axinellae | DSM 24994 | 5,204,899 | 50.3 | 78 | PRJEB20602b | 99.27 | 1.47 | 32.73 |
| Pseudovibrio denitrificans | JCM 12308 | 6,117,547 | 52.3 | 27 | PRJEB20602b | 99.71 | 0.73 | 28.44 |
| Pseudovibrio japonicus | NCIMB 14279 | 4,972,544 | 52.7 | 17 | PRJEB20602b | 100 | 1.61 | 33.96 |
| Pseudovibrio sp. | Tun.PHSC04-5.I4 | 6,549,844 | 50.3 | 8 | FNLB00000000.1 | 100 | 0.73 | 25.42 |
| Pseudovibrio sp. | 1D03 | 5,774,703 | 51.4 | 22 | PRJEB20602b | 99.65 | 0.93 | 29.53 |
| Pseudovibrio sp. | 1F12 | 5,843,738 | 51.4 | 28 | PRJEB20602b | 99.41 | 0.93 | 29.31 |
| Pseudovibrio sp. | 2G02 | 5,870,506 | 51.4 | 25 | PRJEB20602b | 99.35 | 0.29 | 28.87 |
| Pseudovibrio sp. | 2G05 | 5,759,198 | 51.4 | 20 | PRJEB20602b | 99.35 | 0.59 | 29.82 |
| Pseudovibrio sp. | 2H03 | 5,999,042 | 51.5 | 39 | PRJEB20602b | 99.65 | 0.93 | 28.11 |
| Pseudovibrio sp. | 2H05 | 5,943,579 | 51.3 | 28 | PRJEB20602b | 99.06 | 0.29 | 29.82 |
| Pseudovibrio sp. | 2H06 | 6,090,655 | 51.3 | 21 | PRJEB20602b | 99.65 | 0.29 | 28.23 |
| Pseudovibrio sp. | 3C10 | 6,048,288 | 51.3 | 36 | PRJEB20602b | 99.06 | 0.29 | 27.93 |
| Pseudovibrio sp. | 4B07 | 5,121,938 | 52.8 | 51 | PRJEB20602b | 99.71 | 1.03 | 33.15 |
| Pseudovibrio sp. | 4H03 | 5,788,437 | 51.4 | 26 | PRJEB20602b | 99.35 | 0.29 | 29.24 |
| Pseudovibrio sp. | 4H10 | 5,776,139 | 51.4 | 14 | PRJEB20602b | 99.65 | 0.29 | 29.60 |
| Pseudovibrio sp. | 7A08 | 5,758,403 | 51.4 | 22 | PRJEB20602b | 99.35 | 0.59 | 29.85 |
| Pseudovibrio sp. | 7F08 | 5,782,811 | 51.4 | 27 | PRJEB20602b | 99.35 | 0.29 | 29.50 |
| Pseudovibrio sp. | 7G12 | 5,845,319 | 51.4 | 29 | PRJEB20602b | 99.41 | 0.93 | 29.33 |
| Pseudovibrio sp. | 7H02 | 5,844,273 | 51.4 | 28 | PRJEB20602b | 99.41 | 0.93 | 29.29 |
| Pseudovibrio sp. | 7H04 | 5,589,230 | 51.3 | 23 | PRJEB20602b | 99.65 | 1.22 | 30.65 |
| Pseudovibrio sp. | 8G08 | 5,940,829 | 51.4 | 36 | PRJEB20602b | 99.65 | 0.44 | 28.87 |
| Pseudovibrio sp. | 8H04 | 5,681,860 | 49 | 18 | PRJEB20602b | 100 | 0.29 | 29.84 |
| Pseudovibrio sp. | 8H06 | 6,056,598 | 51.4 | 40 | PRJEB20602b | 99.35 | 1.22 | 27.91 |
| Pseudovibrio sp. | AB108 | 5,910,732 | 51.4 | 49 | PRJEB20602b | 99.35 | 0.29 | 28.92 |
| Pseudovibrio sp. | AB111 | 5,922,117 | 51.3 | 36 | PRJEB20602b | 99.06 | 0 | 28.61 |
| Pseudovibrio sp. | AB113 | 5,372,543 | 51.4 | 13 | PRJEB20602b | 99.65 | 0.29 | 31.60 |
| Pseudovibrio sp. | AD13 | 6,001,312 | 51.3 | 36 | LMCC00000000.1 | 99.71 | 0.59 | 28.17 |
| Pseudovibrio sp. | AD14 | 6,201,736 | 51.3 | 57 | LMCD00000000.1 | 99.71 | 0.93 | 27.13 |
| Pseudovibrio sp. | AD15 (W74) | 6,190,724 | 51.2 | 64 | LMCJ00000000.1 | 99.56 | 0.93 | 27.11 |
| Pseudovibrio sp. | AD26 | 6,181,400 | 51.2 | 159 | LMCE00000000.1 | 99.41 | 1.17 | 27.13 |
| Pseudovibrio sp. | AD37 | 5,875,058 | 51.1 | 224 | LMCF00000000.1 | 99.41 | 0.88 | 28.31 |
| Pseudovibrio sp. | AD46 | 6,124,061 | 51.3 | 85 | LMCG00000000.1 | 99.71 | 0.59 | 27.98 |
| Pseudovibrio sp. | AD5 | 6,061,014 | 51.3 | 66 | LMCH00000000.1 | 99.41 | 0.29 | 28.03 |
| Pseudovibrio sp. | AD8 (W64) | 5,935,921 | 51.3 | 49 | LMCI00000000.1 | 99.56 | 0.29 | 28.52 |
| Pseudovibrio sp. | BC118 | 5,812,663 | 51.4 | 23 | PRJEB20602b | 99.65 | 0.29 | 29.12 |
| Pseudovibrio sp. | FO-BEG1 | 5,916,782 | 52.4 | 2 | CP003147.1; CP003148.1 | 99.71 | 0.59 | 28.99 |
| Pseudovibrio sp. | JCM 19062 | 4,607,025 | 52.3 | 286 | BAXV00000000.1 | 88.11 | 0.81 | NA |
| Pseudovibrio sp. | JE062 | 5,717,078 | 52.5 | 53 | ABXL00000000.1 | 99.02 | 0.29 | 30.24 |
| Pseudovibrio sp. | POLY-S9 (PPL9) | 6,559,398 | 50.8 | 163 | LCWX00000000.1 | 93.17 | 0.59 | 24.97 |
| Pseudovibrio sp. | AB134 | 5,975,631 | 52.1 | 39 | MIEL00000000.1 | 99.71 | 0.59 | 28.58 |
| Pseudovibrio sp. | AD30 (WM33) | 5,745,729 | 51.2 | 159 | LMCK00000000.1 | 99.41 | 0.29 | 28.84 |
| Pseudovibrio sp. | AU243 | 5,748,378 | 51.3 | 23 | NZOMPG00000000 | 99.41 | 0.93 | 29.39 |
| Pseudovibrio sp. | Frex01 (Alg231-02) | 5,963,068 | 51.3 | 26 | GCA_900143565.1 | 99.35 | 0.29 | 28.77 |
Genome completeness and contamination were assessed using CheckM v1.0.13 (69) after annotation of the assemblies with Prokka v1.12 (68). P. axinellae, P. denitrificans, and P. ascidiaceicola were independently sequenced by different institutions. NA, not analyzed. Type strains are indicated in bold type. Strain JCM19062 was excluded from further analysis because its completeness was <90%.
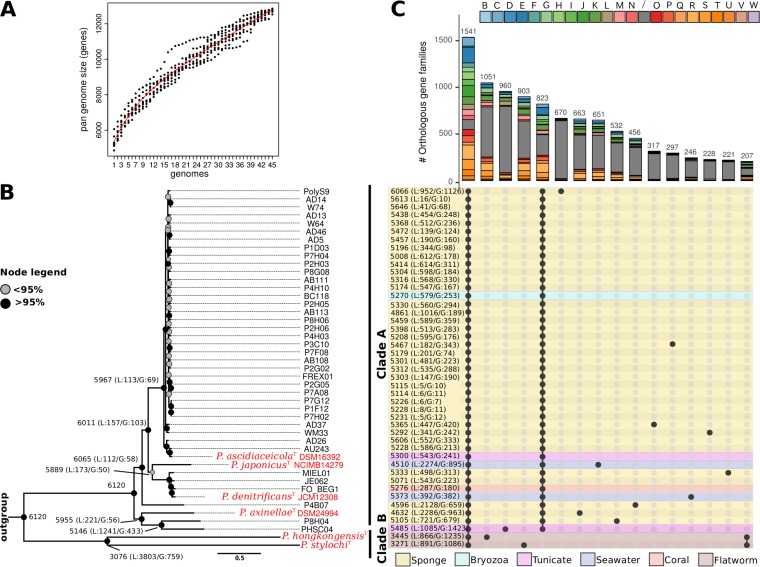
After identifying the homologous genes within the genus Pseudovibrio, we inferred the core and pan-genomes of the genus from the proteome of each strain. The core genome is defined here as the set of orthologous protein families shared among all strains of the genus Pseudovibrio, and the term pan-genome refers to the entire set of homologous families present in at least one strain. The core genome was composed of 1,541 orthologous families, representing 25% to 46% of all coding sequences in the strains’ genomes (Fig. 1; see also Table 1). The pan-genome contained 19,576 homologous families and was open, which means that considering genomes of additional strains will likely result in an increase in the pan-genome size (Fig. 1A).
FIG 1.
Genomic composition of the Pseudovibrio genus. (A) A resampling procedure was used to estimate the dimension of the pan-genome of the genus. (B) After inclusion of the outgroup Labrenzia alexandrii DFL-11 (GCA_000158095.2) in the homologous gene family identification, a phylogenomic tree was constructed selecting only those genes within the core genomes that could give a well-resolved tree topology and that did not show signs of recombination. Bootstrapping with 100 replicates was performed to obtain approximate Bayes branch support values. (C) Distribution of the homologous gene families among the Pseudovibrio strains, visualized using the UpSetR package (100). The histogram indicates the number of orthologous gene families, with their distribution profiles indicated by filled circles in the matrix at the bottom. Bars of the histogram are colored according to the COG categories to which the orthologous families have been assigned. The panel reporting the distribution profile of the orthologous families is colored according to the source of isolation of the strains. Only orthologous gene families containing > 200 genes are reported.
In order to perform a robust phylogenomic reconstruction, we included the genome of strain Labrenzia alexandrii DFL-11 (GCA_000158095.2) from the family Rhodobacteraceae as an outgroup in the calculation of the homologous gene families. The protein-encoding genes in the core genome of all Pseudovibrio strains and the outgroup were filtered to obtain 552 robust markers used in the phylogenomic reconstruction (Fig. 1B). We defined 9 lineages within the tree using average nucleotide identity (ANI) as the criterion. A lineage is defined here as a group of strains that have ≥90% ANI. This resulted in identification of nine lineages: lineage 1, containing the majority of the genomes available to date, including the type strain P. ascidiaceicola; lineage 2, containing the type strain P. japonicus; lineage 3, containing the type strain P. denitrificans and the newly proposed species P. brasiliensis (11); lineage 4, containing the type strain P. axinellae; lineages 5 and 6, containing strains P4B07 and P8H04, respectively, both isolated from a marine sponge (9); lineage 7, containing strain PHSC04 isolated from a marine tunicate (9); and lineages 8 and 9, containing the two type strains P. stylochi and P. hongkongensis, respectively (Fig. 1B; see also Fig. S1 in the supplemental material). Overall, the lineages fell into two distinct clades: clade A, containing lineages 1 to 7, and clade B, containing lineages 8 and 9 (Fig. 1B). In almost all cases, the nine lineages were characterized by strong branch support. Interestingly, clade B diverged strongly from the other lineages and formed a basal monophyletic group. Consistent with previous 16S rRNA gene phylogenies (3), the isolation sources did not follow the phylogenetic relatedness of the strains. The only exception was observed for the strains of clade B, which were both isolated from marine flatworms.
In clade B, 1,396 homologous gene families were absent that were otherwise identified in 41 (≥95%) of 43 strains of the other lineages (Fig. 1C and data not shown). This underlines the strong divergence of clade B from the rest of the investigated Pseudovibrio strains. The clusters of orthologous groups (COG) functional annotation (27, 28) revealed that proteins involved in various metabolic processes were present within these sets (Fig. 1C) and that they were significantly enriched in proteins involved in lipid metabolism, transporters, and transcription regulators of the AcrR family (see Table S1 in the supplemental material). Surprisingly, only a small fraction of the genes missing in clade B showed signs of horizontal acquisition. On average, 2.8% of these genes were located on mobile genetic elements (MGEs) and 1.5% were identified as potentially horizontally acquired (Table S2). Therefore, it is likely that most genes defining the differences between clade A and clade B are not part of the mobilome. Notably, although many systems (e.g., molybdate transport and spermidine and putrescine transport) were encoded in all genomes, the corresponding genes were not always identified as orthologous between clade B and clade A strains, highlighting the strong divergence between these clades.
Interestingly, clade B and strain PHSC04 harbored >900 unique homologous protein families that were not detected in any other Pseudovibrio genome (Fig. 1C). While the unique proteins encoded by clade B fell into multiple COG categories, most unique proteins of PHSC04 could not be functionally classified (Fig. 1C). We hypothesize that this large number of unclassified unique proteins could represent an indication of ongoing pseudogenization resulting in genome rearrangements and eventually genome reduction. Consistent with this finding, a large number of short (200-to-300-bp) genes was detected (Fig. S2) in strains PHSC04 and PolyS9 (see reference 29 for a similar approach). Moreover, we found that the genome and, in particular, the unique proteins of strain PHSC04 were highly enriched in MGEs and related genes, such as those encoding transposases, insertion sequences, and methylases, supporting the hypothesis of increased genome rearrangements (Fig. S3; see also Table S1). This enrichment in mobile elements seemed to be unique for strain PHSC04 and did not correlate with the genome size.
P. stylochi and P. hongkongensis have a reduced sulfur metabolism.
Comparison of KEGG pathways (30, 31) revealed that several gene sets for sulfur utilization and for complete transport systems of sugars, amino acids, and micronutrients were present in most clade A strains but absent in clade B strains (Table S3). For example, the KEGG module for “thiosulfate oxidation by SOX complex” was complete in ≥95% of the clade A strains but was absent in both clade B strains (Table S3). To verify the genome-derived hypotheses positing differential utilization of sulfur compounds, we compared growth levels of P. denitrificans DN34 (32) (clade A) and of P. hongkongensis UST20140214-015BT (24) (clade B). When thiosulfate was added to sulfur-free glucose medium, proliferation was observed only in P. denitrificans (Table 2), suggesting that this strain oxidizes thiosulfate to sulfate via the SOX pathway. In contrast, no growth was observed in P. hongkongensis, suggesting that this strain cannot oxidize thiosulfate to sulfate, likely due to lack of the SOX pathway, thus resulting in the lack of growth.
TABLE 2.
Results of comparisons of P. hongkongensis and P. denitrificans grown in artificial seawater mediuma
| Substrate tested | P. denitrificans | P. hongkongensis |
|---|---|---|
| Glucose | + | + |
| DMSP as sole carbon source | + | − |
| DMSP as sole sulfur source | + | + |
| Thiosulfate | + | − |
| Taurine as sole carbon source | + | − |
| Taurine as sole sulfur source | + | − |
+, growth observed; −, no growth observed.
Pseudovibrio encodes proteins involved in the metabolism of various organic sulfur compounds (1, 3, 33–35). Accordingly, a high-affinity (ABC) transporter for taurine (KEGG module M00435) was detected in most clade A strains but was present only partially in clade B strains, as the taurine periplasmic binding protein was not encoded by these strains (Table S3 and data not shown). Consistent with this finding, taurine was utilized as source of both sulfur and carbon by P. denitrificans but not by P. hongkongensis (Table 2). Previous studies reported the ability of some Pseudovibrio strains to metabolize dimethylsulfoniopropionate (DMSP) (1, 3, 33–35). To verify whether the clade B strains retained the ability to use DMSP, we queried the strains’ proteomes via blastp and the enzymes involved in DMSP cleavage and demethylation described for strain FO-BEG1 (3). We identified a single gene per pathway in clade B (data not shown), raising the question of whether these strains can still metabolize DMSP. In our experiments, P. hongkongensis could use DMSP only as sole sulfur source, whereas P. denitrificans could utilize DMSP as both a sole carbon source and a sole sulfur source (Table 2).
Notably, although many systems (e.g., molybdate transport and spermidine and putrescine transport) were encoded in all genomes, the corresponding genes were not always identified as orthologous between clade B and clade A strains, highlighting the strong divergence between these clades.
Gene loss likely led to small genome sizes in strains of clade B.
The gene family history reconstruction suggested that the genus Pseudovibrio, as proposed today, experienced massive gene loss in the lineage leading to clade B that includes P. stylochi and P. hongkongensis (Fig. 1B). Underlining the uniqueness of this clade, the analysis predicted that the divergence in the two strains resulted in an additional >1,000 genes gained by each of them (Fig. 1B). Gene duplication played only a marginal role in genome rearrangements within the genus. In fact, the number of paralogous genes was on average around 50 per genome, with the only significant exceptions being strains PolyS9, AD26, and PHSC04, with the latter harboring 289 duplicated genes (data not shown). It is worth noting that the lineage leading to P. japonicus seems to have undergone strong genomic rearrangement, losing >2,000 and gaining >800 genes (Fig. 1B). The highest level of gene acquisition (>1,400 genes) was observed in strain PHSC04, which was among the strains with the biggest genomes and harboring the highest number of unique genes and MGEs (Fig. 1B and C; see also Table 1). MGEs seem to have played a significant role in shaping the structure of the genome of PHSC04, as more than 15% of the protein-encoding genes in this strain were located on MGEs (Fig. S3; see also Fig. S4).
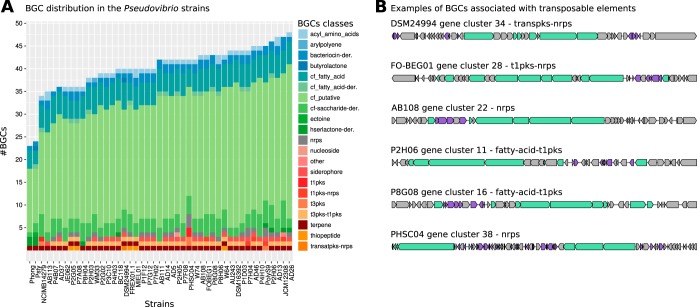
Finally, we investigated the distribution of BGCs involved in the synthesis of secondary metabolites, which have been previously reported to be abundant and diverse within the Pseudovibrio genus (1, 3, 8–10). The number of BGCs identified ranged from 23 to 48, with P. stylochi and P. hongkongensis harboring the fewest BGCs (Fig. 2A). Between 2.4% and 29.7% of the genes within BGCs were found on MGEs such as genomic islands, insertion sequences, or bacteriophages. Further, between 1.2% and 13.1% of genes within BGCs were identified as potentially horizontally acquired (Fig. S4). There was no clear difference between clade A and clade B in the proportion of BGC genes found on MGE or in horizontal gene transfer (HGT) candidates. Nevertheless, it was surprising that the strains in clade B did not harbor gene clusters encoding nonribosomal peptide synthases (NRPS) or polyketide synthases (PKS), which represent the two major classes of BGCs producing bioactive compounds. All other Pseudovibrio strains harbored these BGCs, and 26% of the NRPS or PKS genes were associated with transposable elements, suggesting that these are part of the mobilome and were potentially horizontally acquired in clade A (Fig. 2B). Unique BGCs were found in some Pseudovibrio genomes (Fig. 2A) but particularly in that of strain PHSC04. Its genome encoded three NRPS and four PKS BGCs, likely a consequence of the proliferation of the MGEs that were abundant in some of these clusters (Fig. 2B).
FIG 2.
Distribution of biosynthetic gene clusters (BGCs) in the Pseudovibrio genus. (A) Overview of the BGCs within the genus. BGCs were grouped according to the type of molecules produced by their encoded proteins. (B) Some BGCs either were detected within the mobilome of the Pseudovibrio strains or were found to be associated with transposable elements. Six examples of such clusters encoding PKS or NRPS from strains DSM24994, FO-BEG01, AB108, P2H06, P8H06, P8G08, and PHSC04 are shown, with the core biosynthetic genes highlighted in green and transposases predicted by a PFAM domain search (see Materials and Methods) highlighted in violet.
It is worth mentioning that the high level of genome fragmentation in some strains (e.g., PolyS9 [163 contigs]) can compromise the correct estimation of the number of BGCs present in a genome. In fact, if a BGC is divided into multiple fragments, it might either be missed due to absence of gene contingency or be counted multiple times in each fragment.
Overall genome relatedness indices (OGRI).
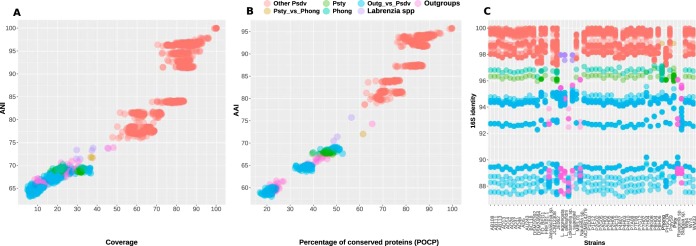
To better define the relatedness of the members of the Pseudovibrio genus and quantify the divergence of clade B from the other Pseudovibrio strains, we analyzed several OGRI (Fig. 3A and B). For comparison, we included nine additional genomes of strains from the Rhodobacteraceae family as an outgroup. All Pseudovibrio strains of clade A shared ≥76% ANI, and the two strains in clade B shared 71.8% ANI. These values dropped to <70.3% for comparisons of clade B strains to the clade A strains (Fig. 3A; see also Fig. S1). The ANI values determined in comparisons between the outgroup and the Pseudovibrio genomes ranged from 63.3% to 69.6%. Within the outgroup, the genomes of the strains belonging to the genus Labrenzia shared ANI values between 71.4% and 73.8%, whereas the ANI values determined in comparisons between different genera for the outgroup ranged from 64.4% to 73.9% (Fig. 3A).
FIG 3.
Overall genetic similarity index (A and B) and 16S rRNA gene identities (C) of the Pseudovibrio strains considered in this study. For comparison, nine additional strains belonging to the family Rhodobacteraceae were included. No gene for the 16S rRNA could be identified in the genome of Nesiotobacter exalbescens DSM 16456. Both 5S and 23S genes were detected at the edge of contigs; hence, it is likely that a complete rRNA operon was not assembled in this genome.
We calculated average amino acid identity (AAI) values using the homologous proteins as obtained from the get_homologues pipeline (36) (Fig. 3B). The two strains in clade B shared 72.1% AAI, whereas they shared only 67.5% to 68.5% AAI with strains from clade A. The comparison between all genome pairs within clade A returned AAI values ranging from 78.6% to 95.8%. Comparing the percentages of conserved proteins (POCP), clear differences could be observed. The two clade B strains had POCP values ranging from 39.5% to 48.9% compared to the clade A strains. In line with the larger AAI values, the POCP values were also higher among the Pseudovibrio strains of clade A, ranging from 62.1% to 99.7%. Comparisons of the outgroup genomes to the Pseudovibrio genomes showed AAI values ranging from 58.7% to 69.1% and POCP values ranging from 17% to 52.6%. Comparisons among the outgroup strains showed that they shared 64.4% to 74.3% AAI and 20% to 65.4% POCP (Fig. 3B). Overall, these analyses revealed that the maxima of all intergenus OGRI values calculated for the outgroups were above the maximum values detected in the comparisons between the divergent clade B and the Pseudovibrio clade A strains.
16S rRNA gene phylogeny and inferred environmental distribution.
In addition to the OGRI, we compared the 16S rRNA gene sequence identities (SI) to verify whether this marker gene reflected the genomic divergence observed within the genus (Fig. 3C). Comparisons of strains of clade B with the clade A strains displayed 16S rRNA gene SI values ranging from 96% to 97.2%, while comparisons of only the strains within clade A showed SI values of ≥97.2% (Fig. 3C). We integrated the obtained 16S rRNA gene sequences with other publicly available ones to calculate a phylogenetic tree. In total, 87 Pseudovibrio-affiliated 16S rRNA genes were used to reconstruct a tree (Fig. S5). In accordance with the results from the phylogenomic tree (Fig. 1B), P. stylochi and P. hongkongensis deeply branched off from all other Pseudovibrio sequences, forming a basal monophyletic group (Fig. S5; see also Fig. S6).
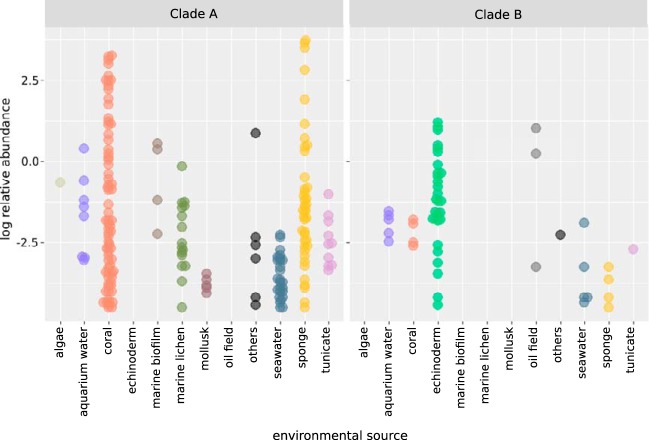
We investigated whether the reduced metabolic repertoire of clade B would be reflected by a distinct habitat range of P. hongkongensis and P. stylochi. To this end, we queried representative 16S rRNA sequences of clade A and clade B against all publicly available amplicon data sets via the IMNGS platform (37). Strain PHSC04 was detected in only 5 of the 422,880 data sets investigated, and relative abundances were around zero percent (≤0.01%); hence, it was excluded from further analyses. Environmental sources and relative abundances of P. ascidiaceicola and P. japonicus relatives were highly similar. The distribution of strain P4B07 was strikingly similar to the distributions of both of those strains, with which it shared between 98.4% and 98.6% 16S rRNA gene SI. Since these strains would be counted as individual matches even if only one of them occurred within a sample, we included only the sequence with the highest relative abundance per sample in the distribution analysis described below (Fig. 4). Clade B was almost unique to echinoderm hosts and oil field samples and was not detected in marine biofilms or in lichens, mollusks, and algae, in contrast to clade A (Fig. 4; see also Table S4). P. hongkongensis and P. stylochi were detected at maximum relative abundances of 3.4% and 1.7%, respectively, in the gut metagenome of sea urchin Lytechinus variegatus (38) (Table S4). Strikingly, no sequences related to the Pseudovibrio strains of clade A were detected in data sets obtained from sea urchins and other echinoderms (Fig. 4).
FIG 4.
Relative abundances of 16S rRNA gene sequences related to Pseudovibrio in different environmental sources. Environmental sources represented by fewer than three samples screened by the IMNGS platform are summarized as “others” for better readability. Relative abundance values of ≤0.01% are not plotted. Colors correspond to environmental source. To avoid overrepresentation of data sets with hits from two or more Pseudovibrio lineages, only the maximum relative abundance value is displayed.
Strength of purifying selection.
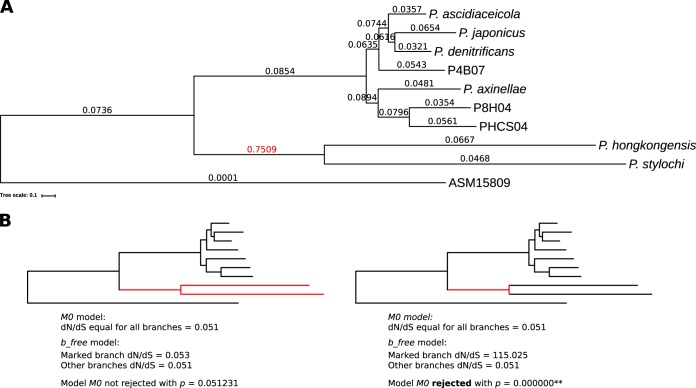
On the basis of the differences in genome size and environmental distribution, we hypothesized that divergence of the two clades was affected by different strengths of natural selection. To verify this, we determined the strength of purifying selection in a subset of Pseudovibrio lineages. We used measurements of the dN/dS ratio (the ratio of nonsynonymous to synonymous nucleotide substitutions) to assess the strength of selection, as low dN/dS values indicate stronger purifying selection. The dN/dS values for each branch on the tree were always between 0.03 and 0.09 except for the internal branch leading to lineages 8 and 9 (Fig. 5A), which had a dN/dS value of 0.75, 1 order of magnitude higher than all other values. In contrast, the dN/dS values of the external branches were below 0.07 in all lineages. A likelihood ratio test confirmed that the dN/dS value for the internal branch leading to lineages 8 and 9 was significantly different from the dN/dS values of all other branches, suggesting that this branch experienced different evolutionary pressures (Fig. 5B).
FIG 5.
dN/dS values corresponding to branches of representative Pseudovibrio genomes. (A) The tree shown represents a subset of the phylogenomic tree based on core genes (Fig. 1). A representative strain for each lineage was selected and the subtree extracted. The dN/dS values are displayed at each branch. (B) Results of a likelihood ratio test to determine whether all branches have the same dN/dS value (M0 model) or whether the marked branches (red) differ in their dN/dS values from the rest of the tree branches (b_free model). M0 was rejected when the P value was <0.05. The red marked branch(es) was tested against all other branches of the tree.
DISCUSSION
In recent decades, increasing efforts have been directed to characterize the microbiome of marine invertebrates, especially sponges, as they play a crucial role in maintaining ocean health and harbor a treasury of bioactive metabolites, often produced by their associated microbes (1, 39–46). Strains belonging to the genus Pseudovibrio have been repeatedly isolated during these campaigns and have been studied due to their ability to inhibit the growth of many bacterial pathogens (1, 2, 8, 9). Small-scale comparative genomic studies have pointed out genomic differences within the genus and suggested potential taxonomic inconsistencies (7, 10). In recent years, phylogenomics has helped elucidate phylogenetic relationships and facilitated accurate (re)classification of microbes, going beyond 16S rRNA gene-based approaches (18–21). Following this trend, we performed a robust phylogenomic reconstruction to verify the genomic architecture and the degree of relatedness of the bacteria within the genus Pseudovibrio, considering the evolutionary trajectories that have shaped their diversity. Clear differences emerged within the genus, suggesting the presence of a basal monophyletic clade containing P. stylochi and P. hongkongensis (lineages 8 and 9 forming clade B) (Fig. 1B).
The genus Pseudovibrio is characterized by high genomic diversity as shown by the large open pan-genome and high number of accessory and unique genes (Fig. 1A and C). We observed a striking difference in genome sizes (Table 1) between clade B and clade A, raising the question of whether this difference has evolved through gene loss or acquisition. The genomic architectures of clade A and clade B have been similarly shaped by HGT (∼2% of coding DNA sequences [CDS]; see Fig. S4 in the supplemental material). This, together with the gene family reconstruction that we performed (Fig. 1), supports the view that clade B diverged from clade A through gene loss from an ancestor with a large genome. Instead, the larger genomes in clade A might be maintained by balancing gene gain and loss. This is in line with current theories indicating that bacterial accessory genes are likely to be lost via drift if they are neutral or nearly neutral in nature, resulting in a reduction in genome size (47). In contrast, accessory genes remain in a genome or population if they can provide a benefit and are thus maintained by natural selection (26, 47). Therefore, in clade A, natural selection might maintain larger bacterial genomes and counteract genome erosion and loss of gene functions via genetic drift (47, 48).
The latter has likely affected clade B. Although neither lineage 8 nor lineage 9 of clade B showed increased dN/dS values, the high value obtained for the internal branch leading to clade B indicates relaxed purifying selection, which supports the view that the extensive gene loss that resulted from genetic drift occurred before the split into the two species. It is worth noting that the high dN/dS value could also derive from the high divergence of the two clades, leading to inaccurate dN/dS estimation. Taken together, our data indicate that the larger genomes in clade A have been maintained by natural selection and that, in contrast, the process of natural selection was relaxed in the branch leading to clade B. This raises the question of which factors would drive such distinct evolutionary trajectories. Acquisition and conservation of a broad repertoire of accessory genes can be beneficial under environmental conditions that are variable or heterogeneous. Considering the wider niche breadth of strains within clade A than within clade B (Fig. 4), the implied selective pressure to maintain a large and versatile genome in clade A would allow these bacteria to exploit a diversity of environmental conditions. For strains of clade B, with a more restricted habitat range, a broad repertoire of genes might not provide any benefit, resulting in the loss of genes through genetic drift.
The results of the comparative genomics and growth experiments indicate that strains in clade A and clade B differ in their ability to utilize inorganic and organic sulfur compounds. SOX pathway-encoding Pseudovibrio species, such as P. denitrificans, but not SOX-lacking P. stylochi and P. hongkongensis may use thiosulfate as supplemental energy source, a common feature among marine heterotrophs (49). Likewise, taurine was not used by P. hongkongensis but was used by P. denitrificans. Taurine is an abundant antioxidant in marine invertebrates, such as sponges (50–52), from which the majority of Pseudovibrio strains have been isolated (1, 39, 42, 43) (for examples, see references 1, 13, and 40). This compound can act as an osmolyte (51) or as a carbon and sulfur storage compound and can be used to detoxify hydrogen sulfide (53). However, to the best of our knowledge, taurine has not been detected in marine flatworms; hence, it might not be relevant as a substrate for P. hongkongensis and other clade B strains. Taken together, these data suggest that the lack of both inorganic sulfur metabolism and a high-affinity taurine transporter in clade B strains might be owed to the adaptation to the flatworm host.
While P. hongkongensis did not use taurine, it used DMSP as a sole sulfur source but not as a sole carbon source. DMSP has been previously reported to occur in marine flatworms, likely produced by their symbiotic algae (54, 55). Moreover, Stylochus flatworms may accumulate DMSP by preying on phytoplankton-feeding oysters and barnacles (56, 57). Interestingly, our analysis revealed that P. hongkongensis and P. stylochi occur in the sea urchin Lytechinus variegatus (Fig. 4; see also Table S4 in the supplemental material), which might also take up significant amounts of DMSP via grazing on seagrass and algae (58), both of which are able to produce DMSP (59–62). As such, the strains of clade B might be exposed to significant amounts of DMSP because of the lifestyle and feeding strategies of their hosts. The restricted host range in clade B (Fig. 4) may provide rather stable conditions (e.g., constant availability of DMSP), decreasing the need for genes to access a variety of different sulfur sources. The consequently lower selective pressure (Fig. 5) for keeping a broad repertoire of genes in clade B may have led to the loss of genes and pathways in sulfur metabolism compared to clade A.
The strong differences in genome sizes (Table 1), the differences in encoded functional categories and metabolic features (Fig. 1 and 2; see also Table S3), and the strong phylogenetic divergence (Fig. 1) prompted us to verify whether the strains of P. stylochi and P. hongkongensis would need reclassification. We used this extensive genomic data set to compare OGRIs among the Pseudovibrio strains, also considering their relationship with other members of the Rhodobacteraceae family. Consistent with the phylogenomic analyses, the comparison of OGRIs that we performed showed divergence between the clade B strains and the other isolates (Fig. 3). Although the clade B strains shared >96% identity with other isolates at the 16S rRNA level, placing them into the same genus, there is evidence that the 16S rRNA gene has low phylogenetic resolution for species and genus discrimination in some bacterial groups (15–17). In fact, although Yarza and colleagues set the 16S rRNA gene identity threshold for genus to 94.5%, they also clearly stated that these thresholds are minimal and indicative values (63). Similarly, although the majority of OGRI differences between clade B strains and the other Pseudovibrio species were around the cutoff values for genus definition, AAI/ANI thresholds do not have to be considered to be strict boundaries but rather approximate values and might need adjustment for the bacterial group of interest (64). In fact, AAI and ANI between clade B and clade A were within the range of intergenus values calculated for the other strains of the Rhodobacteraceae family (Fig. 3). Finally, clade B shared POCP values of <50% with strains of clade A, which were even lower than the those that some members of the Rhodobacteraceae family shared with Pseudovibrio clade A strains (Fig. 3B). POCP values of <50% have been reported to represent a robust parameter for genus definition (65), suggesting that clade B strains P. hongkongensis and P. stylochi should be regarded as a new genus. A high-quality taxon description should be based on thorough analyses of phylogenetic affiliation, genomic coherence, and phenotypic characterization (22). Following these criteria, the phylogenomic, comparative genomics, and phenotyping analyses we performed provide compelling evidence for the reclassification of the Pseudovibrio stylochi and Pseudovibrio hongkongensis species into a new genus, Polycladidibacter gen. nov.
Taxonomy.
Polycladidibacter (Po.ly.cla.di.di.bac’ter; N.L. neut. pl. n. Polycladida order of marine flatworms; N.L. masc. n. bacter a rod, N.L. masc. n. Polycladidibacter a rod isolated from a marine flatworm of the order Polycladida).
The genus is described following the description of Pseudovibrio stylochi given by Zhang and colleagues in 2016 (25) with the following additions. The members of this genus form a basal monophyletic group in a phylogenetic tree built from a subset of 552 marker genes for conserved proteins. The closest phylogenetic neighbor to the genus is the genus Pseudovibrio. On the basis of comparative genomic analyses and growth experiments using P. hongkongensis strain UST20140214-015BT = KCTC 42383 = MCCC 1K00451, reduced sulfur compounds are not oxidized via the SOX pathway and taurine is not utilized as a carbon or sulfur source. DMSP is utilized as a sole sulfur source but not as a carbon source. The type species is Polycladidibacter hongkongensis comb. nov.
Description of Polycladidibacter hongkongensis comb. nov. (basonym: Pseudovibrio hongkongensis).
The description is as given previously by Xu et al. (24) with the following additions. The phylogenetic position based on 552 marker genes for conserved proteins is in accordance with the genus description. It is unable to oxidize reduced sulfur compounds using the SOX pathway. Taurine is not utilized as sulfur or carbon source. DMSP is utilized as a sole source of sulfur. The type strain is UST20140214-015BT = KCTC 42383 = MCCC 1K00451.
Description of Polycladidibacter stylochi comb. nov. (basonym: Pseudovibrio stylochi).
The description is as given previously by Zhang et al. (25) with the following additions. The phylogenetic position based on 552 marker genes for conserved proteins is in accordance with the genus description. The type strain is UST20140214-052T = KCTC 42384 = MCCC 1K00452.
MATERIALS AND METHODS
Comparative genomic analyses.
All available genome assemblies classified as belonging to the Pseudovibrio genus were downloaded from NCBI (66) or the ENA archive (67). To avoid bias due to differences in gene calling and annotation, all assemblies were newly annotated using Prokka v1.12 (68). Completeness and contamination of each Pseudovibrio proteome were checked using CheckM v1.0.13 (69) and a set of 528 marker proteins characteristic of the family Rhodobacteraceae, as by default in the CheckM reference data set. For strains having multiple assemblies, the one showing the highest degree of completeness, the lowest level of contamination, and the lowest number of contigs was chosen.
Homologous gene families were calculated via the use of get_homologues pipeline v03012018 (36), using Diamond v0.8.25 (70) and OrthoMCL v1.4 (71) and the following parameters: -S 50 -C 50 -t 0. The genome of the bacterium Labrenzia alexandrii DFL-11 (GenBank assembly accession no. GCA_000158095.2) was included in this analysis and used as the outgroup. The nucleotide sequences of the genes belonging to the core genome were analyzed using the get_phylomarker pipelines v2.2.8, identifying high-quality marker genes for robust phylogenomic analyses (72). This pipeline applies a set of sequential filters to exclude recombinant gene alignments and those producing anomalous or poorly resolved trees. To compute a phylogenetic tree, 552 marker genes were selected and used. Within the get_phylomarker pipelines, the final concatenated alignment based on codons was used to search for the best model of evolution via ModelFinder (73) and to construct a phylogenetic tree with IQ-Tree (74) and the best model of evolution GTR+F+ASC+R4. The best tree was determined by performing bootstrapping with 100 replicates using the UFBoot2 ultrafast bootstrapping algorithm (75), and approximate Bayes branch support values were estimated. Compositional analyses for the estimation of the pan-genome dimensions were performed using the get_homologues pipeline and omitting the outgroup. Finally, AAI and POCP were calculated using the -A flag and -P flag via get_homologues v0301209, respectively. The POCP calculation was modified from that previously described by Qin et al. (65) as follows: [(C1 + C2)/(T1 + T2)]*100% (where C1 and C2 represent the conserved number of proteins and T1 and T2 represent the total number of proteins in the two compared genomes, respectively).
A representative protein sequence for each homologous gene family forming the pan-genome was used to retrieve COG functional annotations (28), using the pipeline BPGA (76). The COG annotation was then extended to all proteins of each homologous family. Functional annotation based on KEGG (30) was obtained, submitting all protein sequences of the Pseudovibrio genomes to eggNOG v4.5.1 (77). The functional annotations were further used for enrichment analyses to verify whether particular functional categories were significantly enriched in any genomes. This was also done for specific subsets of genes that showed distinct distribution patterns among the strains, including (i) genes not present in the divergent species P. stylochi and P. hongkongensis but found in ≥95% of Pseudovibrio genomes; (ii) genes uniquely found in the genomes of both divergent strains; and (iii) genes uniquely found in the genome of strain PHSC04. All enrichment analyses, performed using the functional annotations retrieved from COG and KEGG, were done using the enricher function in the R (78) package clusterProfileR (79) with the genes of all Pseudovibrio genomes serving as the background. The absence or presence of specific metabolic features in P. stylochi and P. hongkongensis was verified by querying their proteomes using blastp and the proteins of the ABC transporter for taurine (TauABC) from Agrobacterium sp. (GenBank identifiers [IDs] PVE65474, CVI23808, and EHH08539) and the proteins of the SOX and DMSP pathways described previously in strain Pseudovibrio sp. FO-BEG1 (3). Finally, the gene sets following the distribution patterns described above were also mapped onto KEGG pathways using the BlastKOALA platform (80) to verify the presence of specific cellular KEGG modules. Secondary metabolite biosynthetic gene clusters were detected using the standalone version of antismash v4.1.0 and the ClusterFinder algorithm (81, 82).
The reconstruction of gene family history and ancestral gene family sizes was performed using COUNT software (83). We used maximum likelihood (ML) birth-and-death models (84). ML reconstruction does not consider sequence information but relies solely on the copy numbers of the homologous gene families. The model accounts for lineage- and family-specific gene loss and duplication rates. Here, rates were optimized using a gain-loss-duplication model and a Poisson distribution at the root of the phylogenetic tree, allowing different gain-loss-duplication rates for different lineages. Rates were optimized iteratively as suggested in the software manual using a maximum of 100 optimization rounds. COUNT does not require an outgroup species, but the rooted topology of the phylogenomic tree was used.
ANI was calculated via blastn within the pipeline pyani v0.2.7 (85). 16S rRNA gene identities were calculated after recovering all 16S rRNA genes from the annotated genomes, aligning them via the use of T-coffee v111.00.d625267, and using the function -output sim in the T-coffee suite (86). For comparison, we included the following additional Rhodobacteraceae family genomes in the calculation of the overall genome relatedness indices (OGRI; AAI, ANI, and POCP) and of 16S rRNA identity: Stappia stellulata DSM 5886 (GenBank assembly ID: GCA_000423705), Ruegeria sp. TM1040 (GenBank assembly ID: GCA_000014065), Nesiotobacter exalbescens DSM 16456 (GenBank assembly ID: GCA_000422785), Nautella sp. ECSMB14104 (GenBank assembly ID: GCA_000972285), Labrenzia suaedae strain DSM 22153 (GenBank assembly ID: GCA_900142725), Loktanella sp. DSM 29012 (GenBank assembly ID: GCA_900110705); Jannaschia sp. CCS1 (GenBank assembly ID: GCF_000013565), Labrenzia aggregata IAM 12614 (GenBank assembly ID: GCF_000168975), and Labrenzia alexandrii DFL-11 (GenBank assembly ID: GCA_000158095). All genomes were processed as described for the Pseudovibrio strains. Data was then analyzed using R and plotted using the ggplot2 package (87).
16S rRNA gene phylogenetic trees and environmental distribution of Pseudovibrio strains.
16S rRNA gene sequences were retrieved from the published genomes used in the comparative genomic analyses of this study (see above). Complementing this data set, published 16S rRNA gene sequences affiliated with the genus Pseudovibrio and relatives were downloaded from the SILVA database (release 132 SSU Ref NR 99) (88). Sequences of isolated strains were subsampled, keeping only the single sequence where sequence quality, alignment quality, and pintail quality were closest to 100% per strain. When SILVA quality markers were equal for sequences of the same strain, the longer sequence was kept for further analyses. Sequences of uncultured organisms were also included. 16S rRNA gene sequences of Labrenzia spp. and Stappia spp. served as an outgroup. 16S rRNA gene sequences were aligned using the SINA (v1.2.11) Aligner (89). The best model of evolution (K2P+I+G4) was determined with ModelFinder (73) and applied to calculate a maximum likelihood phylogenetic tree using IQ-Tree (74) and ultrafast bootstrap (UFBoot2) (75) with 1,000 replicates.
The environmental distribution of Pseudovibrio lineages and the divergent lineages formed by P. hongkongensis and P. stylochi were then investigated using the IMNGS platform, which systematically screens and processes the SRA for prokaryotic 16S rRNA gene amplicon data sets and builds sample-specific sequence databases (37). The 16S rRNA sequences of the available Pseudovibrio type strains of P. ascidiaceicola, P. axinellae, P. denitrificans, and P. japonicus were used to investigate the environmental distribution of the genus Pseudovibrio. PHSC04 was added due to its distinct genomic features, and P4B07 was chosen because it formed an independent lineage in the phylogenomic tree (Fig. 1B). 16S rRNA sequences of both P. hongkongensis and P. stylochi were used to verify the environmental distribution of the divergent lineages. 16S rRNA gene sequences were queried against all data sets accessible by the IMNGS platform (accessed June 2019) using a similarity threshold of 99% and a minimum length of 300 bp. Data sets with only a single sequence match were excluded.
Detection of mobile genetic elements.
We annotated the PFAM domains in each protein encoded in the analyzed genomes using the pfamscan.pl pipeline (90) with default parameters. To identify restriction-modification systems, we modified previously published lists (10, 91) of PFAM domains by screening the PFAM database (90) using the search terms “restriction” and “restriction-modification” with subsequent curation of the obtained PFAM annotation for these domains to exclude unreliable hits (see Table S5 in the supplemental material). Similarly, we created a PFAM domain list for transposases, using the search term “transposase,” and used it to select all associated proteins within the analyzed genomes (Table S6).
Phage contigs or prophages were predicted with VirSorter v 1.0.3 (92), and only phage signals of higher confidence levels 1 and 2 were considered. Insertion sequences were detected using ISEScan v 1.6 with default settings (93). Genomic islands were predicted with IslandViewer 4 (94) by aligning the Pseudovibrio genomes against the complete genome of Labrenzia aggregata strain RMAR6-6.
HGT candidates were determined by their dinucleotide frequencies as implemented in the CompareM v0.0.23 pipeline (95). We regarded those genes that were identified as “deviant” in the compareM analysis, according to the “Hotelling T-squared statistic” value of >37.89, as potentially horizontally acquired.
Calculation of dN/dS.
To determine differences in the strengths of purifying selection for the Pseudovibrio lineages, we selected a representative genome from each of the nine lineages. We extracted the phylogenomic tree for only these strains (including the ASM15809 outgroup from the phylogenomic tree shown in Fig. 1B) using Dendroscope 3 (96) (Fig. 5). We used codeml from the PAML package (97) as implemented in the ete3 toolkit (98) to calculate dN/dS for each branch of the tree based on the alignment of the 552 phylogenetic marker genes. We performed a likelihood ratio test with the null hypothesis of the M0 model that assumes equal dN/dS values for all branches in a tree against an alternative model, b_free, that allows the marked branches to have a dN/dS value different from those seen with the other (background) branches. We tested two scenarios to determine (i) whether lineages 8 and 9 and their internal branch had the same dN/dS value as all other branches in the tree and (ii) whether only the internal branch leading to lineages 8 and 9 was different from all other branches in the tree (Fig. 5).
In addition, we calculated the dN/dS value for each lineage, allowing each branch to result in a different dN/dS value, by choosing the fb model with default settings (ω = 0.7; Fig. 5). Running the same model while adjusting the ω value to 0.2 and 1.2 resulted in the same dN/dS values for all branches except for minor changes in two branches by less than 0.001.
Growth experiments.
Cultures of Pseudovibrio denitrificans DN34 (32) and Pseudovibrio hongkongensis UST20140214-015B (24) were grown in artificial seawater containing the following (in grams per liter): NaCl, 26.4; MgCl2·6H2O, 5.7; KCl, 0.66; CaCl2·2H2O, 1.47; KBr, 0.1; H3BO3, 0.02; SrCl2·6H2O, 0.033; NaF, 0.003; NaHCO3, 0.19. Also, we added 6.8 g of MgSO4·7H2O as a sulfur source except when testing for utilization of other sulfur sources. KH2PO4 and NH4Cl were added to reach final concentrations of 2.7 mM and 2.8 mM, respectively. The medium was amended (1:1,000 [vol/vol]) with trace metal, vitamins (including vitamin B12), and selenium-tungstate solutions as described previously by Widdel (99). To test different carbon sources for growth, cultures of P. denitrificans and P. hongkongensis were amended with glucose (2 mM) or DMSP (2.4 mM) or taurine (6 mM), corresponding to 12 mM carbon each. Taurine was tested as a sole source of sulfur at a final concentration of 0.9 mM with 2 mM glucose in sulfur-free medium. Thiosulfate utilization as an additional electron donor was tested by adding 23.9 mM thiosulfate to medium containing 2 mM glucose. We reasoned that if the strains oxidized thiosulfate, sulfate would be produced. Hence, in the absence of any other sulfur source, the produced sulfate should sustain bacterial growth. An absence of growth under this condition would indicate that the strain did not oxidize thiosulfate. Utilization of DMSP as the sole source of sulfur was tested by adding 2.1 mM DMSP to sulfur-free medium containing 0.2 mM glucose as a carbon source. All cultures were incubated under oxic conditions in the dark at 28°C. Growth of cultures was evaluated by visual inspection of changes in turbidity.
Data availability.
GenBank accession numbers representing data determined in this article are listed in Table 1.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. Oriente and A. Oren for their help with Latin nomenclature.
This research was supported by the Austrian Science Fund (FWF), project number 31010.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Romano S. 2018. Ecology and biotechnological potential of bacteria belonging to the genus Pseudovibrio. Appl Environ Microbiol 84:1–16. doi: 10.1128/AEM.02516-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crowley SP, O'Gara F, O'Sullivan O, Cotter PD, Dobson ADW. 2014. Marine Pseudovibrio sp. as a novel source of antimicrobials. Mar Drugs 12:5916–5929. doi: 10.3390/md12125916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bondarev V, Richter M, Romano S, Piel J, Schwedt A, Schulz-Vogt HN. 2013. The genus Pseudovibrio contains metabolically versatile bacteria adapted for symbiosis. Environ Microbiol 15:2095–2113. doi: 10.1111/1462-2920.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Romano S, Schulz-Vogt HN, González JM, Bondarev V. 2015. Phosphate limitation induces drastic physiological changes, virulence-related gene expression, and secondary metabolite production in Pseudovibrio sp. strain FO-BEG1. Appl Environ Microbiol 81:3518–3528. doi: 10.1128/AEM.04167-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schwedt A, Seidel M, Dittmar T, Simon M, Bondarev V, Romano S, Lavik G, Schulz-Vogt HN. 2015. Substrate use of Pseudovibrio sp. growing in ultra-oligotrophic seawater. PLoS One 10:e0121675. doi: 10.1371/journal.pone.0121675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Romano S, Bondarev V, Kölling M, Dittmar T, Schulz-Vogt HN. 14 March 2017, posting date Phosphate limitation triggers the dissolution of precipitated iron by the marine bacterium Pseudovibrio sp. FO-BEG1. Front Microbiol doi: 10.3389/fmicb.2017.00364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Romano S, Fernàndez-Guerra A, Reen FJ, Glöckner FO, Crowley SP, O’Sullivan O, Cotter PD, Adams C, Dobson ADW, O’Gara F. 30 March 2016, posting date Comparative genomic analysis reveals a diverse repertoire of genes involved in prokaryote-eukaryote interactions within the Pseudovibrio genus. Front Microbiol doi: 10.3389/fmicb.2016.00387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Naughton LM, Romano S, O’Gara F, Dobson ADW. 18 August 2017, posting date Identification of secondary metabolite gene clusters in the Pseudovibrio genus reveals encouraging biosynthetic potential toward the production of novel bioactive compounds. Front Microbiol doi: 10.3389/fmicb.2017.01494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Versluis D, McPherson K, van Passel MWJ, Smidt H, Sipkema D. 2017. Recovery of previously uncultured bacterial genera from three Mediterranean sponges. Mar Biotechnol (NY) 19:454–468. doi: 10.1007/s10126-017-9766-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alex A, Antunes A. 2018. Genus-wide comparison of Pseudovibrio bacterial genomes reveal diverse adaptations to different marine invertebrate hosts. PLoS One 13:e0194368. doi: 10.1371/journal.pone.0194368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fróes AM, Freitas TC, Vidal L, Appolinario LR, Leomil L, Venas T, Campeão ME, Silva CJF, Moreira APB, Berlinck RGS, Thompson FL, Thompson CC. 2018. Genomic attributes of novel symbiont Pseudovibrio brasiliensis sp. nov. isolated from the sponge Arenosclera brasiliensis. Front Mar Sci 5:81. doi: 10.3389/fmars.2018.00081. [DOI] [Google Scholar]
- 12.Klenk HP, Göker M. 2010. En route to a genome-based classification of Archaea and Bacteria? Syst Appl Microbiol 33:175–182. doi: 10.1016/j.syapm.2010.03.003. [DOI] [PubMed] [Google Scholar]
- 13.Thompson CC, Amaral GR, Campeão M, Edwards RA, Polz MF, Dutilh BE, Ussery DW, Sawabe T, Swings J, Thompson FL. 2015. Microbial taxonomy in the post-genomic era: rebuilding from scratch? Arch Microbiol 197:359–370. doi: 10.1007/s00203-014-1071-2. [DOI] [PubMed] [Google Scholar]
- 14.Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P. 2018. A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nat Biotechnol 36:996–1004. doi: 10.1038/nbt.4229. [DOI] [PubMed] [Google Scholar]
- 15.Janda JM, Abbott SL. 2007. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45:2761–2764. doi: 10.1128/JCM.01228-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breider S, Scheuner C, Schumann P, Fiebig A, Petersen J, Pradella S, Klenk H-P, Brinkhoff T, Göker M. 11 August 2014, posting date Genome-scale data suggest reclassifications in the Leisingera-Phaeobacter cluster including proposals for Sedimentitalea gen. nov. and Pseudophaeobacter gen. nov. Front Microbiol doi: 10.3389/fmicb.2014.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nouioui I, Carro L, García-López M, Meier-Kolthoff JP, Woyke T, Kyrpides NC, Pukall R, Klenk HP, Goodfellow M, Göker M. 22 August 2018, posting date Genome-based taxonomic classification of the phylum Actinobacteria. Front Microbiol doi: 10.3389/fmicb.2018.02007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mateo-Estrada V, Graña-Miraglia L, López-Leal G, Castillo-Ramírez S. 2019. Phylogenomics reveals clear cases of misclassification and genus-wide phylogenetic markers for Acinetobacter. Genome Biol Evol 11:2531–2541. doi: 10.1093/gbe/evz178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gupta RS, Son J, Oren A. 2019. A phylogenomic and molecular markers based taxonomic framework for members of the order Entomoplasmatales: proposal for an emended order Mycoplasmatales containing the family Spiroplasmataceae and emended family Mycoplasmataceae comprised of six genera. Antonie Van Leeuwenhoek 112:561–588. doi: 10.1007/s10482-018-1188-4. [DOI] [PubMed] [Google Scholar]
- 20.Graña-Miraglia L, Arreguín-Pérez C, López-Leal G, Muñoz A, Pérez-Oseguera A, Miranda-Miranda E, Cossío-Bayúgar R, Castillo-Ramírez S. 24 October 2018, posting date Phylogenomics picks out the par excellence markers for species phylogeny in the genus Staphylococcus. PeerJ doi: 10.7717/peerj.5839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Prabha R, Singh DP. 15 February 2019, posting date Cyanobacterial phylogenetic analysis based on phylogenomics approaches render evolutionary diversification and adaptation: an overview of representative orders. 3 Biotech doi: 10.1007/s13205-019-1635-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosselló-Móra R, Amann R. 2015. Past and future species definitions for Bacteria and Archaea. Syst Appl Microbiol 38:209–216. doi: 10.1016/j.syapm.2015.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. 30 November 2018, posting date High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun doi: 10.1038/s41467-018-07641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xu Y, Li Q, Tian R, Lai Q, Zhang Y. 2015. Pseudovibrio hongkongensis sp. nov., isolated from a marine flatworm. Antonie Van Leeuwenhoek 108:127–132. doi: 10.1007/s10482-015-0470-y. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Y, Li Q, Tian R, Lai Q, Xu Y. 2016. Pseudovibrio stylochi sp. nov., isolated from a marine flatworm. Int J Syst Evol Microbiol 66:2025–2029. doi: 10.1099/ijsem.0.000984. [DOI] [PubMed] [Google Scholar]
- 26.Sela I, Wolf YI, Koonin EV. 2016. Theory of prokaryotic genome evolution. Proc Natl Acad Sci U S A 113:11399–11407. doi: 10.1073/pnas.1614083113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Galperin MY, Makarova KS, Wolf YI, Koonin EV. 2015. Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res 43:D261–D269. doi: 10.1093/nar/gku1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jäckle O. 2018. Evolution and physiology of the Paracatenula symbiosis. University of Bremen, Bremen, Germany. [Google Scholar]
- 30.Kanehisa M. 1996. Toward pathway engineering: a new database of genetic and molecular pathways. Sci Technol Japan 59:34–38. [Google Scholar]
- 31.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shieh WY, Lin YT, Jean WD. 2004. Pseudovibrio denitrificans gen. nov., sp. nov., a marine, facultatively anaerobic, fermentative bacterium capable of denitrification. Int J Syst Evol Microbiol 54:2307–2312. doi: 10.1099/ijs.0.63107-0. [DOI] [PubMed] [Google Scholar]
- 33.Garren M, Son K, Raina JB, Rusconi R, Menolascina F, Shapiro OH, Tout J, Bourne DG, Seymour JR, Stocker R. 2014. A bacterial pathogen uses dimethylsulfoniopropionate as a cue to target heat-stressed corals. ISME J 8:999–1007. doi: 10.1038/ismej.2013.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Raina J-B, Tapiolas D, Motti CA, Foret S, Seemann T, Tebben J, Willis BL, Bourne DG. 2016. Isolation of an antimicrobial compound produced by bacteria associated with reef-building corals. PeerJ 4:e2275. doi: 10.7717/peerj.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Raina JB, Clode PL, Cheong S, Bougoure J, Kilburn MR, Reeder A, Forêt S, Stat M, Beltran V, Thomas-Hall P, Tapiolas D, Motti CM, Gong B, Pernice M, Marjo CE, Seymour JR, Willis BL, Bourne DG. 4 April 2017, posting date Subcellular tracking reveals the location of dimethylsulfoniopropionate in microalgae and visualises its uptake by marine bacteria. Elife doi: 10.7554/eLife.23008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Contreras-Moreira B, Vinuesa P. 2013. GET_HOMOLOGUES, a versatile software package for scalable and robust microbial pangenome analysis. Appl Environ Microbiol 79:7696–7701. doi: 10.1128/AEM.02411-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lagkouvardos I, Joseph D, Kapfhammer M, Giritli S, Horn M, Haller D, Clavel T. 2016. IMNGS: a comprehensive open resource of processed 16S rRNA microbial profiles for ecology and diversity studies. Sci Rep 6:33721–33729. doi: 10.1038/srep33721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de Lamarck JBM. 1816. Histoire naturelle des animaux sans vertèbres. Tome troisième. Deterville/Verdière, Paris, France. [Google Scholar]
- 39.Muscholl-Silberhorn A, Thiel V, Imhoff JF. 2008. Abundance and bioactivity of cultured sponge-associated bacteria from the Mediterranean Sea. Microb Ecol 55:94–106. doi: 10.1007/s00248-007-9255-9. [DOI] [PubMed] [Google Scholar]
- 40.Sertan-De Guzman AA, Predicala RZ, Bernardo EB, Neilan BA, Elardo SP, Mangalindan GC, Tasdemir D, Ireland CM, Barraquio WL, Concepcion GP. 2007. Pseudovibrio denitrificans strain Z143-1, a heptylprodigiosin-producing bacterium isolated from a Philippine tunicate. FEMS Microbiol Lett 277:188–196. doi: 10.1111/j.1574-6968.2007.00950.x. [DOI] [PubMed] [Google Scholar]
- 41.Rypien KL, Ward JR, Azam F. 2010. Antagonistic interactions among coral-associated bacteria. Environ Microbiol 12:28–39. doi: 10.1111/j.1462-2920.2009.02027.x. [DOI] [PubMed] [Google Scholar]
- 42.Santos OCS, Pontes P, Santos JFM, Muricy G, Giambiagi-deMarval M, Laport MS. 2010. Isolation, characterization and phylogeny of sponge-associated bacteria with antimicrobial activities from Brazil. Res Microbiol 161:604–612. doi: 10.1016/j.resmic.2010.05.013. [DOI] [PubMed] [Google Scholar]
- 43.Penesyan A, Tebben J, Lee M, Thomas T, Kjelleberg S, Harder T, Egan S. 2011. Identification of the antibacterial compound produced by the marine epiphytic bacterium Pseudovibrio sp. D323 and related sponge-associated bacteria. Mar Drugs 9:1391–1402. doi: 10.3390/md9081391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson JR, Rivera HE, Closek CJ, Medina M. 7 January 2015, posting date Microbes in the coral holobiont: partners through evolution, development, and ecological interactions. Front Cell Infect Microbiol doi: 10.3389/fcimb.2014.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Webster NS, Thomas T. 2016. Defining the sponge hologenome. mBio 7:e00135-16. doi: 10.1128/mBio.00135-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Esteves AIS, Cullen A, Thomas T. 1 March 2017, posting date Competitive interactions between sponge-associated bacteria. FEMS Microbiol Ecol doi: 10.1093/femsec/fix008. [DOI] [PubMed] [Google Scholar]
- 47.Bobay L-M, Ochman H. 2018. Biological species in the viral world. Proc Natl Acad Sci U S A 115:6040–6045. doi: 10.1073/pnas.1717593115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Koonin EV, Wolf YI. 2008. Genomics of bacteria and archaea: the emerging dynamic view of the prokaryotic world. Nucleic Acids Res 36:6688–6719. doi: 10.1093/nar/gkn668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Spring S. 2014. Function and evolution of the sox multienzyme complex in the marine gammaproteobacterium Congregibacter litoralis. ISRN Microbiol 2014:597418–597411. doi: 10.1155/2014/597418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Carbone M, Núñez-Pons L, Ciavatta ML, Castelluccio F, Avila C, Gavagnin M. 2014. Occurrence of a taurine derivative in an antarctic glass sponge. Nat Prod Commun 9:469–470. [PubMed] [Google Scholar]
- 51.Allen JA, Garrett MR. 1971. Taurine in marine invertebrates. Adv Mar Biol 9:205–253. doi: 10.1016/S0065-2881(08)60343-0. [DOI] [Google Scholar]
- 52.Huang R, Peng Y, Zhou X, Yang X, Liu Y. 2013. A new taurine derivative from South China Sea marine sponge Axinella sp. Nat Prod Res 27:1537–1541. doi: 10.1080/14786419.2012.733389. [DOI] [PubMed] [Google Scholar]
- 53.Joyner JL, Peyer SM, Lee RW. 2003. Possible roles of sulfur-containing amino acids in a chemoautotrophic bacterium-mollusc symbiosis. Biol Bull 205:331–338. doi: 10.2307/1543296. [DOI] [PubMed] [Google Scholar]
- 54.Van Bergeijk SA, Stal LJ. 2001. Dimethylsulfoniopropionate and dimethylsulfide in the marine flatworm Convoluta roscoffensis and its algal symbiont. Mar Biol 138:209–216. doi: 10.1007/s002270000444. [DOI] [Google Scholar]
- 55.Van Alstyne KL, Puglisi MP. 2007. DMSP in marine macroalgae and macroinvertebrates: distribution, function, and ecological impacts. Aquat Sci 69:394–402. doi: 10.1007/s00027-007-0888-z. [DOI] [Google Scholar]
- 56.Hopkins SH. 1949. Preliminary survey of the literature on Stylochus and other flatworms associated with oysters. Tex A M Res Found Proj 9:1–16. [Google Scholar]
- 57.Landers WS, Rhodes EW. 1970. Some factors influencing predation by the flatworm, Stylochus ellipticus (Girard), on oysters. Chesapeake Sci 11:55–60. doi: 10.2307/1351343. [DOI] [Google Scholar]
- 58.Souza CF, de Oliveira AS, Pereira RC. 2008. Feeding preference of the sea urchin Lytechinus variegatus (Lamarck, 1816) on seaweeds. Braz J Oceanogr 56:239–247. doi: 10.1590/S1679-87592008000300008. [DOI] [Google Scholar]
- 59.Dacey JWH, Wakeham SG. 1986. Oceanic DMS: production during zooplankton grazing on phytoplankton. Science 233:1314–1316. doi: 10.1126/science.233.4770.1314. [DOI] [PubMed] [Google Scholar]
- 60.Dacey JWH, King GM, Lobel PS. 1994. Herbivory by reef fishes and the production of dimethylsulfide and acrylic acid. Mar Ecol Prog Ser 112:67–74. doi: 10.3354/meps112067. [DOI] [Google Scholar]
- 61.Borges AV, Champenois W. 2015. Seasonal and spatial variability of dimethylsulfoniopropionate (DMSP) in the Mediterranean seagrass Posidonia oceanica. Aquat Bot 125:72–79. doi: 10.1016/j.aquabot.2015.05.008. [DOI] [Google Scholar]
- 62.Yoch DC. 2002. Dimethylsulfoniopropionate: its sources, role in the marine food web, and biological degradation to dimethylsulfide. Appl Environ Microbiol 68:5804–5815. doi: 10.1128/aem.68.12.5804-5815.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yarza P, Yilmaz P, Pruesse E, Glöckner FO, Ludwig W, Schleifer KH, Whitman WB, Euzéby J, Amann R, Rosselló-Móra R. 2014. Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences. Nat Rev Microbiol 12:635–645. doi: 10.1038/nrmicro3330. [DOI] [PubMed] [Google Scholar]
- 64.Konstantinidis KT, Rosselló-Móra R, Amann R. 2017. Uncultivated microbes in need of their own taxonomy. ISME J 11:2399–2406. doi: 10.1038/ismej.2017.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Qin QL, Xie B, Zhang XY, Chen XL, Zhou BC, Zhou J, Oren A, Zhang YZ. 2014. A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215. doi: 10.1128/JB.01688-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.National Center for Biotechnology Information. 1988. National Center for Biotechnology Information (NCBI). National Library of Medicine, Bethesda, MD. [Google Scholar]
- 67.Harrison PW, Alako B, Amid C, Cerdeño-Tárraga A, Cleland I, Holt S, Hussein A, Jayathilaka S, Kay S, Keane T, Leinonen R, Liu X, Martínez-Villacorta J, Milano A, Pakseresht N, Rajan J, Reddy K, Richards E, Rosello M, Silvester N, Smirnov D, Toribio AL, Vijayaraja S, Cochrane G. 2019. The European Nucleotide Archive in 2018. Nucleic Acids Res 47:D84–D88. doi: 10.1093/nar/gky1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 69.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. 2015. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 71.Li L, Christian R, Stoeckert J, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vinuesa P, Ochoa-Sánchez LE, Contreras-Moreira B. 1 Ma Y 2018, posting date GET_PHYLOMARKERS, a software package to select optimal orthologous clusters for phylogenomics and inferring pan-genome phylogenies, used for a critical geno-taxonomic revision of the genus Stenotrophomonas. Front Microbiol doi: 10.3389/fmicb.2018.00771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kalyaanamoorthy S, Minh BQ, Wong TKF, Von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Nguyen LT, Schmidt HA, Von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Le SV. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. doi: 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chaudhari NM, Gupta VK, Dutta C. 2016. BPGA-an ultra-fast pan-genome analysis pipeline. Sci Rep 6:24373–24310. doi: 10.1038/srep24373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, Von Mering C, Bork P. 2016. EGGNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 79.Yu G. 2018. clusterProfiler: universal enrichment tool for functional and comparative study. bioRxiv 10.1101/256784. [DOI]
- 80.Kanehisa M, Sato Y, Morishima K. 2016. BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J Mol Biol 428:726–731. doi: 10.1016/j.jmb.2015.11.006. [DOI] [PubMed] [Google Scholar]
- 81.Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K, Pati A, Godfrey PA, Koehrsen M, Clardy J, Birren BW, Takano E, Sali A, Linington RG, Fischbach MA. 2014. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158:412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Blin K, Wolf T, Chevrette MG, Lu X, Schwalen CJ, Kautsar SA, Suarez Duran HG, De Los Santos ELC, Kim HU, Nave M, Dickschat JS, Mitchell DA, Shelest E, Breitling R, Takano E, Lee SY, Weber T, Medema MH. 2017. AntiSMASH 4.0—improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res 45:W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Csurös M. 2010. Count: evolutionary analysis of phylogenetic profiles with parsimony and likelihood. Bioinformatics 26:1910–1912. doi: 10.1093/bioinformatics/btq315. [DOI] [PubMed] [Google Scholar]
- 84.Nei M, Rooney AP. 2005. Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. 2016. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods 8:12–24. doi: 10.1039/C5AY02550H. [DOI] [Google Scholar]
- 86.Notredame C, Higgins DG, Heringa J. 2000. T-coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 87.Wickham H. 2016. ggplot2: elegant graphics for data analysis, 2nd ed Springer-Verlag, New York, NY. [Google Scholar]
- 88.Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig W, Peplies J, Glöckner FO. 2007. SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Pruesse E, Peplies J, Glöckner FO. 2012. SINA: accurate high-throughput multiple sequence alignment of ribosomal RNA genes. Bioinformatics 28:1823–1829. doi: 10.1093/bioinformatics/bts252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, Sonnhammer ELL, Hirsh L, Paladin L, Piovesan D, Tosatto SCE, Finn RD. 2019. The Pfam protein families database in 2019. Nucleic Acids Res 47:D427–D432. doi: 10.1093/nar/gky995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Croucher NJ, Coupland PG, Stevenson AE, Callendrello A, Bentley SD, Hanage WP. 19 November 2014, posting date Diversification of bacterial genome content through distinct mechanisms over different timescales. Nat Commun doi: 10.1038/ncomms6471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Roux S, Enault F, Hurwitz BL, Sullivan MB. 2015. VirSorter: mining viral signal from microbial genomic data. PeerJ 3:e985. doi: 10.7717/peerj.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xie Z, Tang H. 2017. ISEScan: automated identification of insertion sequence elements in prokaryotic genomes. Bioinformatics 33:3340–3347. doi: 10.1093/bioinformatics/btx433. [DOI] [PubMed] [Google Scholar]
- 94.Bertelli C, Laird MR, Williams KP, Lau BY, Hoad G, Winsor GL, Brinkman F. 2017. IslandViewer 4: expanded prediction of genomic islands for larger-scale datasets. Nucleic Acids Res 45:W30–W35. doi: 10.1093/nar/gkx343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Parks D. 2018. CompareM: a toolbox for comparative genomics. https://github.com/dparks1134/CompareM.
- 96.Huson DH, Scornavacca C. 2012. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol 61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 97.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 98.Huerta-Cepas J, Serra F, Bork P. 2016. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol 33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Widdel F. 1983. Methods for enrichment and pure culture isolation of filamentous gliding sulfate-reducing bacteria. Arch Microbiol 134:282–285. doi: 10.1007/BF00407803. [DOI] [Google Scholar]
- 100.Conway JR, Lex A, Gehlenborg N. 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33:2938–2940. doi: 10.1093/bioinformatics/btx364. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
GenBank accession numbers representing data determined in this article are listed in Table 1.