The gut microbiome shows individual specificity and is affected by sex, environment, and diet; gut microbiome imbalance is related to cancer, cardiovascular diseases, and autoimmune diseases. Astronauts are faced with a challenging environment and limited diet in outer space. Recent studies indicate that the gut microbiome is altered in space simulators and space, but what happens to intestinal microorganisms when astronauts cohabitate in a self-sufficient ecosystem in which they plant and cook food is unclear. Bioregenerative life support systems (BLSSs) are ideal devices to investigate the above issues because they are closed and self-sufficient. Four healthy Chinese subjects cohabitated in a confined BLSS for 60 days, during which their physical parameters and energy/nutrient intake were recorded. We performed a metagenome-wide association study (MWAS) on 55 shotgun-sequenced fecal samples longitudinally obtained from the subjects. Alterations occurred in the gut microbial composition and function, and their relationships with energy/nutrient intake were explored.
KEYWORDS: bioregenerative life support system (BLSS), metagenome-wide association study (MWAS), individual-/sex-specific, gut microbial convergence, compositional and functional alterations
ABSTRACT
Recent studies have suggested that the gut microbiome is modified in space analogs and that human health can be affected during actual spaceflight. However, the relationship between the gut microbiome and dietary intake in simulator subjects and astronauts remains unclear. Bioregenerative life support systems (BLSSs) are confined and self-sufficient ecosystems that enable exploration of this issue. Here, we correlate changes in gut microbes to the nutrient types present in controlled diets within subjects cohabitating in a BLSS. A metagenome-wide association study (MWAS) was performed on 55 shotgun-sequenced fecal samples longitudinally obtained from healthy Chinese subjects (n = 4 in total, n = 2 per sex) subjected to a 60-day BLSS stay and a specialized diet. Each food item was categorized based on nutrient type according to the Chinese Food Ingredients List (https://wenku.baidu.com/view/3f2b628488eb172ded630b1c59eef8c75fbf9514.html?from=search). The physical parameters of each subject fluctuated within normal medical ranges. Sex- and individual-specific differences and a trend of individual convergence of the gut microbiome in the BLSS were observed. Depletion of bacterial taxa such as Faecalibacterium prausnitzii, Bifidobacterium longum, and Escherichia coli and functional modules such as short-chain fatty acid (SCFA) production, as well as an increase in an unidentified Lachnospiraceae and glutamate/tryptophan synthesis, were observed in the BLSS. Correlation analysis showed that these compositional and functional changes were associated with energy/nutrient intake during the BLSS stay. Our findings suggest that the gut microbiota is a useful indicator for monitoring health and that individual nutritive diets should be considered according to sex and individual differences in simulations or in spaceflight.
IMPORTANCE The gut microbiome shows individual specificity and is affected by sex, environment, and diet; gut microbiome imbalance is related to cancer, cardiovascular diseases, and autoimmune diseases. Astronauts are faced with a challenging environment and limited diet in outer space. Recent studies indicate that the gut microbiome is altered in space simulators and space, but what happens to intestinal microorganisms when astronauts cohabitate in a self-sufficient ecosystem in which they plant and cook food is unclear. Bioregenerative life support systems (BLSSs) are ideal devices to investigate the above issues because they are closed and self-sufficient. Four healthy Chinese subjects cohabitated in a confined BLSS for 60 days, during which their physical parameters and energy/nutrient intake were recorded. We performed a metagenome-wide association study (MWAS) on 55 shotgun-sequenced fecal samples longitudinally obtained from the subjects. Alterations occurred in the gut microbial composition and function, and their relationships with energy/nutrient intake were explored.
INTRODUCTION
Human health is dramatically affected by outer space because of its hard-vacuum conditions, microgravity, electromagnetic radiation, and the required changes in the diets of astronauts (1). During both short- and long-term spaceflights and missions, the suitable amount of food, water, and oxygen and the proper temperature required to sustain astronauts’ health must be ensured (2). Ground-based space simulators are an efficient way to explore the physical and psychological conditions of crew members during real spaceflight. The National Aeronautics and Space Administration (NASA)’s analogs, including Human Exploration Research Analog (HERA), Antarctic stations, Concordia, Aquarius/NASA Extreme Environment Mission Operations (NEEMO), Desert Research and Technology Studies (Desert RATS), NASA Space Radiation Lab (NSRL), parabolic flights, in situ resource utilization (ISRU), and Human Exploration Spacecraft Testbed for Integration and Advancement (HESTIA), are the most popular analogs used to simulate human reactions and for technological demonstrations under isolation and confinement in extreme and hostile environments to support the next generation of human exploration missions. However, these analogs are not bioregenerative and self-sufficient, and subjects must take food and other necessities with them, which is not practical for long-term and long-distance travel in space. Unlike these analogs, bioregenerative life support systems (BLSSs) are small, balanced, manmade ecosystems aimed at supporting life on space stations and spaceflights; these ecosystems include human beings, animals, plants, and microorganisms that are integrated with mechanical and physicochemical hardware to satisfy the environmental control functions of the space station and close the food cycle (3). Chinese Lunar Palace 1 (LP1) is a closed, ground-based BLSS designed to simulate the lunar environment, consisting of two plant cultivation cabins and an integrated cabin with four bedrooms, a washroom, a living room, and a waste treatment cabinet (4). LP1 integrates highly efficient plant cultivation, animal protein production, urine nitrogen recycling, and bioconversion of solid waste, representing a further step toward next-generation manned space exploration in which crew members plant and cook food themselves instead of consuming vacuum-packaged products.
The gut microbiome plays an important role in modulating human health and is susceptible to a variety of factors, such as the environment (5), diet (6), and sex (7). Changes in the composition and function of the gut microbiome are closely related to chronic diseases such as cancer, autoimmune diseases, and cardiovascular diseases (8), as well as mental disorders such as anxiety and depression (9). During space simulator experiments or real spaceflights, the crew members must stay in isolation, with limited space and altered diets. Studies have shown that long-term consumption of vacuum-packaged products may promote the growth of anaerobic bacteria, affecting the composition of the gut microbiome (10). In addition, the confined living conditions may also influence the appetites of astronauts (11) and the efficacy of energy harvest from food (12). However, few studies have investigated the influence of energy and dietary nutrient intake on the gut microbiome in either analogs or real spaceflights. BLSSs are highly closed and self-sufficient ecosystems in which there is no exchange of matter and energy, but electricity is provided via outside sources, and the crew members plant and cook food according to a special formula, making it possible to record and investigate the effect of energy/nutrient intake on the gut microbiome.
In this study, four healthy Chinese volunteers (n = 2 per sex) were enrolled in the BLSS for 60 days without interruption after physical examination and psychological assessment. The energy and nutrient type of each food item were classified according to the Chinese Food Ingredients List (https://wenku.baidu.com/view/3f2b628488eb172ded630b1c59eef8c75fbf9514.html?from=search), and the physical indicators of the subjects were recorded and calculated. Fourteen fecal samples from each subject were longitudinally collected over the week before entering the BLSS, during the stay, and after leaving the BLSS. A metagenome-wide association study (MWAS) (13) was performed on 55 shotgun sequencing data sets. Our study illustrated the changes in gut microbial composition and function over time and investigated the relationships between these changes and the intake of different energy/nutrient types. Our results will not only play a crucial role in providing useful information and strategies to improve similar studies but will also provide important tips and guidance for the maintenance of human physiological and mental health in next-generation space explorations.
RESULTS
Overall changes in the gut microbiome over time.
Samples from each subject were divided into three stages: baseline (T0, BBS), during the BLSS stay (T1 to T8, DBS), and after leaving the BLSS (T9 to T13, ABS). Changes over time in the gut microbiota were examined by shotgun metagenome sequencing of DNA extracted from longitudinally collected fecal samples from the four subjects. High-quality sequencing reads (see Table S1a in the supplemental material) were aligned to the latest human gut microbial gene set (14) to obtain the gene profile. The taxonomic profiles were calculated by MetaPhlAn2 (15) and are shown in Table S1b to d. For functional changes, putative amino acid sequences translated from the gene catalog (14) were mapped to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (16), and 9,793 KEGG orthologs (KOs) were identified. Then, 104 gut metabolic modules (GMMs; evaluating gut metabolic potential and anaerobic fermentation capacity; Table S1e) and 55 gut-brain modules (GBMs; describing the relationship between the gut microbiome and mental health; Table S1f) were annotated according to Vieira-Silva et al. (17) and Valles-Colomer et al. (18).
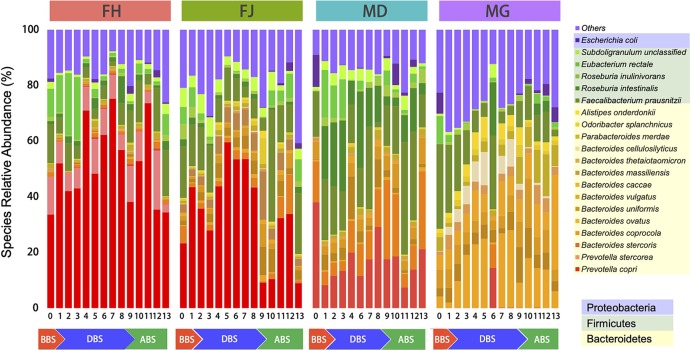
Sex- and individual-specific differences were observed in our study at the gene and genus levels by principal coordinate analysis (PCoA) (Fig. 1, gene level; Fig. S1c, genus level). Additionally, a trend of gut microbiome compositional convergence was observed during the subjects’ stay at the BLSS (Fig. 1; Fig. S2a [species level] and b [genus level]). In addition, the Shannon index displayed a slight difference among the BBS, DBS, and ABS samples (Fig. S1b to c; P < 0.05, tested with the Kruskal-Wallis rank sum test). To further illustrate the differences at the species level, GMMs and GBMs were compared among the BBS, DBS, and ABS samples (Tables S2 to S4; P < 0.05, tested with the Kruskal-Wallis rank sum test). Then, the top 20 species and genera were selected based on their mean relative abundances (Fig. 2; Fig. S3) to explore the characteristics of highly abundant intestinal microorganisms. At the genus level, female H (FH) had Prevotella as the dominant microbial genus during the experiment. For female J (FJ), Prevotella was the most abundant genus in BBS and DBS, which shifted to Bacteroides and Faecalibacterium at ABS T9 and T10. For male D (MD), the abundance of the dominant genus in BBS, Bacteroides, decreased in DBS, while the abundance of Eubacterium and Roseburia increased, and then, the dominant genus shifted to Faecalibacterium at ABS T11 and T12. The predominant genus of male G (MG) shifted from Faecalibacterium in BBS to Bacteroides as soon as he entered the facility, and this change was sustained. At the species level, Prevotella copri was the major species for female H (FH) and female J (FJ) in DBS. Male D (MD) had additional species, including Prevotella stercorea, P. copri, Roseburia intestinalis, and Faecalibacterium prausnitzii. Male G (MG) had a high relative abundance of Alistipes onderdonkii and Bacteroides spp. such as B. ovatus and B. cellulosilyticus.
FIG 1.
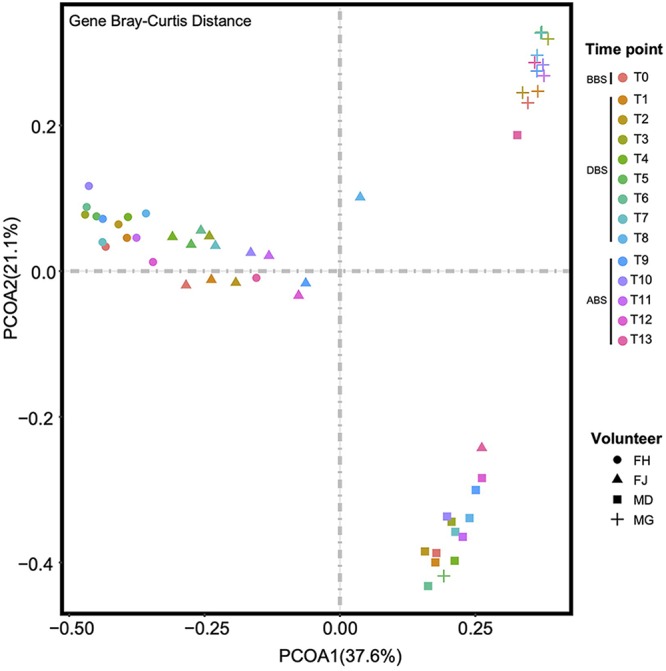
PCoA of the gut microbiome based on gene Bray-Curtis distances for all samples. Different shapes represent samples from different subjects (circle, FH; triangle, FJ; square, MD; cross, MG). Different colors indicate the time points. BBS, DBS, and ABS represent the baseline, during the BLSS stay, and after leaving the BLSS, respectively.
FIG 2.
The top 20 species of the gut microbiome based on mean relative abundance. BBS, DBS, and ABS represent the baseline, during the BLSS stay, and after leaving the BLSS, respectively. The colors of the boxes shown below the figure key indicate the phylum that each genus belongs to.
Dietary nutrient intake is related to gut microbiome composition.
We were interested in the effects of dietary nutrient intake on the gut microbiomes of subjects, since BLSSs are ideal environments to record and calculate the intake of various nutrients and energy sources. The physical parameters, including body mass index (BMI), blood pressure (BP), and heart rate (HR), of the subjects fluctuated within normal reference ranges (Table S5a). An average of 7 hours of total sleep time (TST) and 2 hours of deep sleep time (DST) were observed for all subjects, which is normal for adults, and the females seemed to wake up less during the night in our study (Table S5b), which might be due to an increase in P. copri abundance and tryptophan synthesis (as well as a decrease in propionate synthesis I in females in DBS), which were negatively correlated with awakenings at night (AWNs) (Fig. S4). Difference analysis (two-tailed Student’s t test, P < 0.05) revealed that all of the subjects had a higher percentage of carbohydrates and lower fat levels in DBS samples than in samples collected outside the BLSS. Additionally, all subjects consumed Tenebrio molitor as a source of protein and edible fungi at the T5, T6, and T8 time points during their stay in the BLSS (Table S5c).
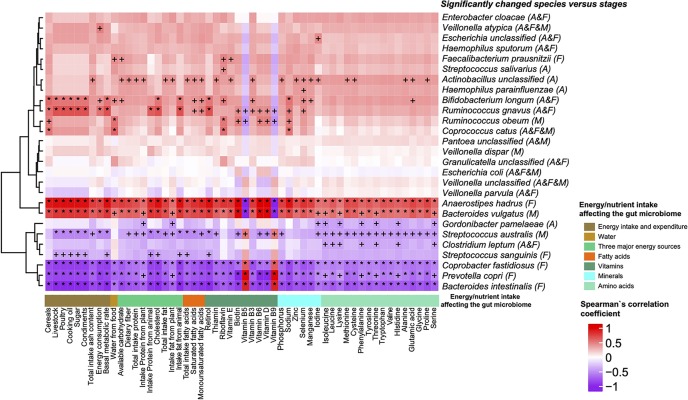
Permutational multivariate analysis of variance (PERMANOVA) was performed to analyze the influence of the intake of food energy sources (Table S5e) and various dietary nutrients (Table S5f) on the gut microbiome. The energy sources and nutrients that had a significant effect (P < 0.01, q < 0.01) on the gut microbiome are shown in Table S5d. To investigate the relationship between these significant changes in energy/nutrient intake and changes in species (P < 0.05; union set of the Kruskal-Wallis rank sum test among BBS, DBS, and ABS in all female and male samples), Spearman’s rank correlation was performed. The coefficient of association (rho) is shown by a heatmap (Fig. 3; +, P < 0.05; *, P < 0.01). Except for vitamin B5 and vitamin B9, intake of all the significantly related energy sources/nutrients was negatively correlated with Bacteroides intestinalis, P. copri, and Coprobacter fastidiosus but positively associated with Anaerostipes hadrus and Bacteroides vulgatus. The abundance of B. intestinalis, which is a human colonic microorganism and upregulates multiple endoxylanases during growth on xylan, decreased in DBS samples from females (19). The abundance of P. copri, which is involved in dietary fiber-induced improvement in glucose metabolism (20) and is enriched in some elderly rheumatoid arthritis (RA) patients (21), was increased in DBS samples from females. C. fastidiosus is a novel member of the family Porphyromonadaceae that was isolated from infant feces (22). A. hadrus is a dominant species within the human colon that produces butyrate (23) and was depleted in DBS samples from females. B. vulgatus can reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis (24) and was depleted in DBS samples from males. In addition, an unidentified Lachnospiraceae species, a fiber specialist (25), was enriched in DBS samples from all subjects. The abundance of F. prausnitzii, a well-known butyrate producer, was positively correlated with available (digestible) carbohydrate and water-from-food levels and decreased in DBS samples from females. The abundance of Clostridium leptum, which can metabolize plant polysaccharides (26), was negatively correlated with amino acid content and decreased in DBS samples from all subjects. In addition, some poorly correlated and possibly pathogenic bacteria, such as Escherichia coli (some types can cause illnesses in humans, including diarrhea, abdominal pain, fever and vomiting [27]), Enterobacter cloacae (might be opportunistic and infectious [28]), and Veillonella atypica (might cause infection [29]), were also depleted in DBS samples.
FIG 3.
Heatmap of the Spearman’s rank correlation between changed species (y axis, by the Kruskal-Wallis rank sum test among stages and Wilcoxon rank sum test between stages, P < 0.05; the letters A, F, and M in brackets after the species name indicate whether the species was significantly changed in all subjects, female subjects, and male subjects, respectively) and energy/nutrient intake, which might have an influence on the gut microbiome (x axis, assessed by PERMANOVA based on the gut microbial gene profile, q < 0.01). Red indicates positive associations, while purple indicates negative associations. +, P < 0.05; *, P < 0.01. The x axis shows energy intake and expenditure (brown), water (light brown), three major energy sources (light green), fatty acids (orange red), vitamins (dark cyan), minerals (cyan), and amino acids (turquoise) from left to right.
Functions inferred from metagenomic information.
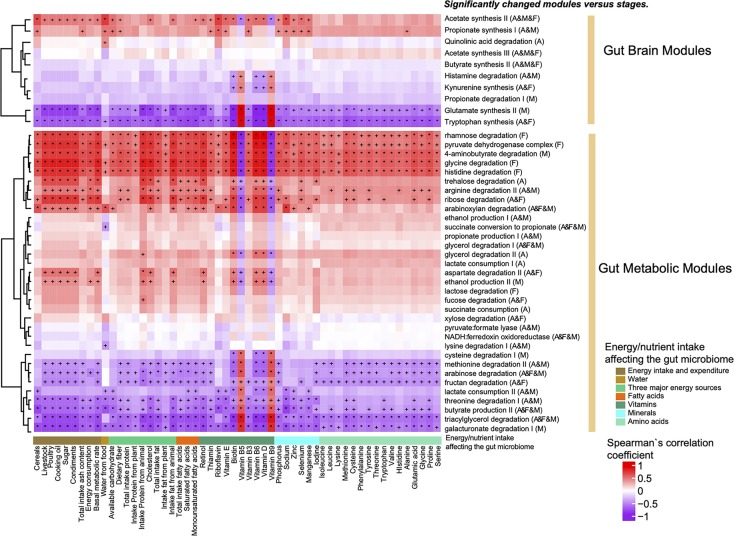
To further illustrate the associations between energy/nutrient intake and the gut microbiome, significantly changed (P < 0.05; union set of the Kruskal-Wallis rank sum test among BBS, DBS, and ABS samples at the all, female, and male levels) gut microbiome functional modules were characterized and associated with energy/nutrient intake (Fig. 4; +, P < 0.05; *, P < 0.01).
FIG 4.
Heatmap of the Spearman’s rank correlation between changed functional modules (y axis, GBMs, and GMMs using the Kruskal-Wallis rank sum test among stages and Wilcoxon rank sum test between stages, P < 0.05; the letters A, F, and M in brackets after the species name indicate whether the species was significantly changed in all subjects, female subjects, and male subjects, respectively) and energy/nutrient intake, which might affect the gut microbiome (x axis, assessed with PERMANOVA based on the gut microbial gene profile; q < 0.01). Red indicates positive associations, and purple indicates negative associations. +, P < 0.05; *, P < 0.01. The x axis shows energy intake and expenditure (brown), water (light brown), three major energy sources (light green), fatty acids (orange red), vitamins (dark cyan), minerals (cyan), and amino acids (turquoise) from left to right.
Functional GMMs, including rhamnose degradation, pyruvate dehydrogenase complex, glycine degradation, and histidine degradation, which were depleted in females, and 4-aminobutyrate degradation, which was depleted in DBS samples from males, were positively related to almost all energy/nutrient intake categories except for vitamin B5 and vitamin B9. Meanwhile, the methionine degradation II pathway, arabinose degradation, fructan degradation, and triacylglycerol degradation were enriched in all individuals, the threonine degradation I pathway and the butyrate production II pathway were depleted in all individuals, and the glutamate degradation III and galacturonate degradation I pathways were depleted in DBS samples from males and showed strong negative associations with various phenotypes.
GBMs, including the glutamate degradation II pathway, the glutamate synthesis II pathway, and tryptophan synthesis, showed strong negative associations with most of the phenotypes, while the acetate synthesis II pathway and the propionate synthesis I pathway showed the opposite trend. For glutamate, which is a transmitter in the healthy brain and has excitatory effects on nerve cells (30), synthesis increased and degradation decreased in DBS samples from males. Tryptophan, an essential compound that can be synthesized by bacteria in humans, acts as a precursor to the neurotransmitter serotonin, the hormone melatonin, and vitamin B3 (31). Tryptophan synthesis was significantly elevated in DBS samples and decreased in ABS samples from all subjects. Acetate, propionate, and butyrate are important SCFAs synthesized by colonic bacteria in humans, and their synthesis decreased in DBS samples.
DISCUSSION
In short- and long-term space travel or space simulators, crew members are exposed to harsh space environments and experience changes in dietary models that can potentially affect the composition and function of their gut microbiomes, which may have a negative effect on their health. Previous studies performed on simulated or real outer-space conditions have suggested that space travel may cause both compositional (2, 32, 33) and functional (34–37) changes in the gut microbiome. However, it is not clear what happens to the human gut microbiome when humans are in a simulated outer-space environment with specialized diets and closed sources of vital necessities such as water and oxygen. This study uses an MWAS to characterize the unique features of microecological diversity, composition, and functional changes in the gut microbiota and their associations with dietary nutrient and energy intake on four healthy Chinese adults cohabitating in a confined BLSS for 60 days.
In our study, the monitored physical parameters, such as BMI, weight, sleep, BP, and HR, were shifted within normal medical ranges, which indicated that the subjects were all in a healthy physiological state. Additionally, crew members showed neither behavioral disturbance nor psychological distress during their stay in the BLSS (38). For the gut microbiome, we did not find significant changes that adversely affected human health, but a trend of gut microbial convergence was observed for each subject during their stay in the BLSS, which was in accordance with a previous long-term (6- to 12-month) space exploration study performed by Voorhies et al. (33). This might be due to either the synchronization of diets during the BLSS stay or an increase/decrease in the abundance of some bacterial taxa, such as P. copri, Eubacterium rectale, Bacteroides stercoris, and B. ovatus, as well as the interactions between diets and gut microbiomes. However, we did not observe a similar gut microbial composition across subjects, which might be a result of the short-term (60-day) cohabitation of the subjects. In addition, we found individual- and sex-specific trends in the gut microbiome in our study, which were consistent with the results obtained in a normal environment (7, 39). Not coincidentally, interindividual variability was also found by Jin et al. (40) among six Japanese men who explored Antarctica for 3 months, during which period a reduction in Bifidobacterium species abundance was observed, just as in our study. The difference was that we did not observe an increase in the abundance of Bacteroides thetaiotaomicron, which is thought to proliferate during emotional stress and might be a microbial cause of nonapparent emotional changes in subjects between being inside and outside the BLSS (38). According to our findings, cohabitation for 60 days in a confined BLSS does not compromise the compositional and functional layout of the individual- and sex-specific microbiota, suggesting the resilience of the individuality of the gut microbial ecosystem, which was consistent with the previous long-term Mars500 experiment (2).
The gut microbiome is known to be influenced by the environment and diet (41). We found certain significantly changed genera, species, and functional modules between samples collected inside and outside the BLSS, especially between DBS and ABS samples (Table S2), which suggested that the gut microbiome can be shifted by the BLSS and the specialized diet in the BLSS. This finding was consistent with the previous NASA Twins study (32), in which the gut microbial community structures of the twin subjected to long-term flight showed a significant difference between in-flight samples and the pre- and postflight combined samples, but those of the twin on the ground did not. We tried to investigate the relationship between significant changes in the gut microbiome across the BLSS and energy/nutrient intake in the BLSS based on a previous study (42). During their stay in the BLSS, all the subjects had a specialized dietary pattern, with a significantly increased intake of carbohydrates and decreased intake of fat, which has been reported to be beneficial for human health (43). Consistent with expectations, the relative abundance of bacteria that feed on fiber and carbohydrates, such as Lachnospiraceae (25) and P. copri (20), was increased in DBS samples. Interestingly, we also observed a decrease in the abundance of possible pathogenic microbes, such as E. cloacae (28) and V. atypica (29), in DBS samples, which might be due to the high-carbohydrate/low-fat diet. However, the decreased abundance of some SCFA-producing bacteria, such as B. longum (44) and F. prausnitzii (45), in DBS samples might explain the decrease in the synthesis of SCFAs, such as propionate and butyrate.
Infection/inflammation and cardiovascular diseases are the main threats faced by astronauts. Increased abundance of RA-related bacteria, such as C. leptum, and decreased abundance of Enterobacter spp. and B. vulgatus in DBS samples might also be risk factors for infection/inflammation and acute arterial events (24, 46). Additionally, the decreased capacity for SCFA synthesis during the BLSS stay may also indicate a decreased capacity for anti-inflammatory activity. Sleep disorders represent another problem faced by astronauts in space, not only affecting cognitive abilities and overall health but also disrupting the body’s circadian rhythm, leading to fatigue and mood changes, as well as metabolic disorders, heart disease, and gastrointestinal problems and, in turn, to accidents on the job. The increased relative abundance of Haemophilus parainfluenzae and Haemophilus sputorum (Fig. S4a), which were negatively correlated with TST but positively correlated with AWNs, in DBS samples from all the subjects might have affected the sleep quality of the subjects. In addition, alterations in some GBMs, such as the synthesis of propionate, butyrate, and glutamate, in our study also indicated that the gut microbiome may play a role in mental disorders such as anxiety (47) and depression (48) in those confined in the BLSS, although there were no obvious emotional changes in the subjects (38). In addition, specific links between significantly changed gut microbes/functional modules and energy/nutrient intake passed from BBS to DBS to ABS samples, as described in Results, also show that the gut microbiota should be monitored in those confined in BLSSs.
Notably, the general and sex-specific trends described in this study do not necessarily reflect what happens in other subjects because of the individual-specific resilience of the gut microbiome. We will consider adding more space-related risk factors, such as weightlessness and electromagnetic radiation, to the BLSS to simulate a more realistic space environment. In addition, the crops and fruits in this experiment need proper temperature and humidity conditions, and we will introduce some drought-tolerant and hypoxia- and low-temperature-resistant plants such as millet and highland barley into the BLSS in the future. Moreover, the protein supply in the experiment mainly came from T. molitor cultivated in the BLSS and vacuum-compressed chicken and pork brought in from outside; we will try to cultivate some other animals that can provide nutrition to humans to achieve complete closure of the BLSS. In addition, we believe that it would be beneficial to add more samples to the BBS group in the future.
In summary, our results revealed the compositional and functional changes in the gut microbiota and their relationships with dietary nutrient and energy intake in a BLSS. Sex- and individual-specific differences in the gut microbiome indicated that sex and individuality should be considered when designing similar simulated experiments or real space missions. Moreover, changes in some specific bacteria, including B. longum, F. prausnitzii, E. coli, and E. cloacae, and functional modules, such as SCFA production, also indicated that it might be helpful to balance the gut microbiome and maintain subject health by providing prebiotics/probiotics and individual dietary guidance depending on individual gut microbiome characteristics.
MATERIALS AND METHODS
Statement of institutional review board approval.
This study was approved by the Bioethics Committee of Beihang University and BGI-Shenzhen. All subjects voluntarily participated in this project and signed an informed consent form.
Volunteer enrollment, sample collection, and phenotype determination.
Four healthy volunteers (female H [FH], female J [FJ], male D [MD], and male G [MG]) with ages from 23 to 27 years and BMIs from 18.5 to 22.9 were enrolled after taking a general physical examination and profile of mood states (POMS) psychological assessment. Fecal samples were collected weekly during the BLSS stay. One and five samples were obtained before and after the BLSS stay, respectively. In total, 14 samples were obtained for each subject, immediately stored at –80°C, and transferred to the laboratory on dry ice when the experiment was complete.
Various phenotypic metadata (Table S5a and b) were recorded during the BLSS stay, including body indexes (body weight [BW], fat-free body weight [FFBW], BMI, systolic BP [SBP], diastolic BP [DBP], HR), sleep indicators (TST, DST, AWN), total carbohydrate, fat, and protein intake ratios during the whole experiment (Table S5c), total energy intake and consumption, and basal metabolic rate (BMR). Consumption of multitudinous dietary nutrients, including water, vitamins, minerals, amino acids, fatty acids, and fat/protein from animals/plants was recorded via a food frequency questionnaire (FFQ) (Table S5e to h) and calculated according to the Chinese Food Ingredients List (version 2015).
DNA preparation and metagenomic sequencing.
DNA extraction was performed using the phenol/trichloromethane method after thawing samples on ice. Extracts were treated with DNase-free RNase, and DNA quality was measured using agarose gel electrophoresis and a Qubit 3.0 fluorometer (Thermo Fisher, Waltham, MA, USA). A paired-end metagenomic sequencing strategy was performed on a BGI-SEQ500 platform (49) (insert size, 350 bp; read length, 100 bp).
Quality control and host genome filtering.
The raw reads that had 50% low-quality bases (quality ≤20) or more than five ambiguous bases were excluded. The remaining reads were mapped to the human genome (hg19) using SOAP v2.22 (-m, 100; -x, 600; -v, 7; -p, 6; -l, 30; -r, 1; -M, 4; -c, 0.95), and the matching reads were removed (49). An average of 6.5 gigabytes of data was generated for each sample (Table S2).
Taxonomic profiling of metagenomic samples.
The high-quality nonhuman reads were defined as clean reads and aligned against the latest human gut microbial 11.4-M (14) gene catalog through SOAP v2.22 (-m, 100; -x, 600; -v, 7; -p, 6; -l, 30; -r, 1; -M, 4; -c, 0.9) to generate the gene profile. MetaPhlAn2 (15) (–input_type fastq –ignore_viruses –nproc 6) was used to generate phylum, genus, and species profiles from the clean reads.
Acquisition of gut metabolic/brain module profiles.
Putative amino acid sequences were translated from the gene catalogs and aligned against the proteins or domains in the KEGG databases (release 79.0, with animal and plant genes removed) using BLASTP v2.26 (default parameters except -m, 8; -e, 1e-5; -F, -a, 6; and -b, 50). Each protein was assigned to a KEGG ortholog group on the basis of the highest scoring annotated hit(s) containing at least one segment pair scoring over 60 bits. The relative abundance profile of KOs was determined by summing the relative abundance of genes from each KO using the mapped reads per sample (40). The abundance of each GMM (-a, 2; -d, GMM.v1.07.txt; -s, average) and GBM (default parameters) was calculated as shown previously (17, 18).
PERMANOVA of the effects of various phenotypic metadata on the gut microbiome.
PERMANOVA (50) was performed on the gene abundance profile of the samples to assess the effect of the intake of each energy and nutrient type listed in Table S5e to h, and the results are shown in Table S5d. We used Bray-Curtis distance and 9,999 permutations in R (v3.10, vegan package, R Project for Statistical Computing) (51). The P values were adjusted with the Benjamini-Hochberg correction.
Richness and diversity analysis.
Alpha diversity (within samples) at the gene and genus levels was quantified with the Shannon index using relative gene abundance profiles. Beta diversity (between samples) at the gene and genus levels was calculated based on Bray-Curtis distance (R v3.2.5, vegan package 2.4-4).
Correlation analysis.
Spearman’s rank correlation was used to evaluate the relationships between the changed phenotypes (evaluated with PERMANOVA) and the changed gut microbiota (at the species level) as well as similarities between samples. Coefficient of association (rho) results are shown by heatmaps (Fig. 3 and 4), and the significance is indicated by the P value (+, P < 0.05; *, P < 0.01).
Statistical analysis.
The changes in the gut microbiome composition and functional modules for subjects among different stages (BBS, DBS, and ABS) were evaluated using the Kruskal-Wallis rank sum test. The ratios of carbohydrate, fat, and protein intake inside and outside the BLSS were calculated using Student’s t test.
Data availability.
The high-quality nonhuman reads that support the findings of this study have been deposited in the CNSA (https://db.cngb.org/cnsa/) of CNGBdb under the accession number CNP0000408.
Supplementary Material
ACKNOWLEDGMENTS
J. Chen and Q. Wang were both responsible for sequencing data processing, data interpretation, figure presentation, and article architecture design. In addition, J. Chen was responsible for experimental design, DNA extraction and determination of samples, shotgun sequencing, phenotypic classification, and manuscript writing; Q. Wang focused on data production and presentation. Z. Hao was responsible for experimental design, sample collection, phenotype determination, and calculations.
This study was funded by the Shenzhen Municipal Government of China (grants JCYJ20160229172757249 and JCYJ20170818111103886) and the National Natural Science Foundation of China (grant 81670606).
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Schwendner P, Mahnert A, Koskinen K, Moissl-Eichinger C, Barczyk S, Wirth R, Berg G, Rettberg P. 2017. Preparing for the crewed Mars journey: microbiota dynamics in the confined Mars500 habitat during simulated Mars flight and landing. Microbiome 5:129. doi: 10.1186/s40168-017-0345-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Turroni S, Rampelli S, Biagi E, Consolandi C, Severgnini M, Peano C, Quercia S, Soverini M, Carbonero FG, Bianconi G, Rettberg P, Canganella F, Brigidi P, Candela M. 2017. Temporal dynamics of the gut microbiota in people sharing a confined environment, a 520-day ground-based space simulation, MARS500. Microbiome 5:39. doi: 10.1186/s40168-017-0256-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Skoog AI. 1984. BLSS: a contribution to future life support. Adv Space Res 4:251–262. doi: 10.1016/0273-1177(84)90569-6. [DOI] [PubMed] [Google Scholar]
- 4.Fu Y, Li L, Xie B, Dong C, Wang M, Jia B, Shao L, Dong Y, Deng S, Liu H, Liu G, Liu B, Hu D, Liu H. 2016. How to establish a bioregenerative life support system for long-term crewed missions to the moon or Mars. Astrobiology 16:925–936. doi: 10.1089/ast.2016.1477. [DOI] [PubMed] [Google Scholar]
- 5.Rothschild D, Weissbrod O, Barkan E, Kurilshikov A, Korem T, Zeevi D, Costea PI, Godneva A, Kalka IN, Bar N, Shilo S, Lador D, Vila AV, Zmora N, Pevsner-Fischer M, Israeli D, Kosower N, Malka G, Wolf BC, Avnit-Sagi T, Lotan-Pompan M, Weinberger A, Halpern Z, Carmi S, Fu J, Wijmenga C, Zhernakova A, Elinav E, Segal E. 2018. Environment dominates over host genetics in shaping human gut microbiota. Nature 555:210–215. doi: 10.1038/nature25973. [DOI] [PubMed] [Google Scholar]
- 6.Conlon MA, Bird AR. 2014. The impact of diet and lifestyle on gut microbiota and human health. Nutrients 7:17–44. doi: 10.3390/nu7010017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haro C, Rangel-Zúñiga OA, Alcalá-Díaz JF, Gómez-Delgado F, Pérez-Martínez P, Delgado-Lista J, Quintana-Navarro GM, Landa BB, Navas-Cortés JA, Tena-Sempere M, Clemente JC, López-Miranda J, Pérez-Jiménez F, Camargo A. 2016. Intestinal microbiota is influenced by gender and body mass index. PLoS One 11:e0154090. doi: 10.1371/journal.pone.0154090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marietta E, Horwath I, Balakrishnan B, Taneja V. 2019. Role of the intestinal microbiome in autoimmune diseases and its use in treatments. Cell Immunol 339:50–58. doi: 10.1016/j.cellimm.2018.10.005. [DOI] [PubMed] [Google Scholar]
- 9.Mayer EA, Tillisch K, Gupta A. 2015. Gut/brain axis and the microbiota. J Clin Invest 125:926–938. doi: 10.1172/JCI76304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mills J, Donnison A, Brightwell G. 2014. Factors affecting microbial spoilage and shelf-life of chilled vacuum-packed lamb transported to distant markets: a review. Meat Sci 98:71–80. doi: 10.1016/j.meatsci.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz MB, Just DR, Chriqui JF, Ammerman AS. 2017. Appetite self-regulation: environmental and policy influences on eating behaviors. Obesity (Silver Spring) 25 Suppl 1:S26–S38. doi: 10.1002/oby.21770. [DOI] [PubMed] [Google Scholar]
- 12.Endesfelder D, Engel M, Davis-Richardson AG, Ardissone AN, Achenbach P, Hummel S, Winkler C, Atkinson M, Schatz D, Triplett E, Ziegler AG, zu Castell W. 2016. Towards a functional hypothesis relating anti-islet cell autoimmunity to the dietary impact on microbial communities and butyrate production. Microbiome 4:17. doi: 10.1186/s40168-016-0163-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang J, Jia H. 2016. Metagenome-wide association studies: fine-mining the microbiome. Nat Rev Microbiol 14:508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- 14.Xie H, Guo R, Zhong H, Feng Q, Lan Z, Qin B, Ward KJ, Jackson MA, Xia Y, Chen X, Chen B, Xia H, Xu C, Li F, Xu X, Al-Aama JY, Yang H, Wang J, Kristiansen K, Wang J, Steves CJ, Bell JT, Li J, Spector TD, Jia H. 2016. Shotgun metagenomics of 250 adult twins reveals genetic and environmental impacts on the gut microbiome. Cell Syst 3:572–584e3. doi: 10.1016/j.cels.2016.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Truong DT, Franzosa EA, Tickle TL, Scholz M, Weingart G, Pasolli E, Tett A, Huttenhower C, Segata N. 2015. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nat Methods 12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 16.Du J, Yuan Z, Ma Z, Song J, Xie X, Chen Y. 2014. KEGG-PATH: Kyoto Encyclopedia of Genes and Genomes-based pathway analysis using a path analysis model. Mol Biosyst 10:2441–2447. doi: 10.1039/c4mb00287c. [DOI] [PubMed] [Google Scholar]
- 17.Vieira-Silva S, Falony G, Darzi Y, Lima-Mendez G, Garcia Yunta R, Okuda S, Vandeputte D, Valles-Colomer M, Hildebrand F, Chaffron S, Raes J. 2016. Species-function relationships shape ecological properties of the human gut microbiome. Nat Microbiol 1:16088. doi: 10.1038/nmicrobiol.2016.88. [DOI] [PubMed] [Google Scholar]
- 18.Valles-Colomer M, Falony G, Darzi Y, Tigchelaar EF, Wang J, Tito RY, Schiweck C, Kurilshikov A, Joossens M, Wijmenga C, Claes S, Van Oudenhove L, Zhernakova A, Vieira-Silva S, Raes J. 2019. The neuroactive potential of the human gut microbiota in quality of life and depression. Nat Microbiol 4:623–632. doi: 10.1038/s41564-018-0337-x. [DOI] [PubMed] [Google Scholar]
- 19.Wang K, Pereira GV, Cavalcante JJ, Zhang M, Mackie R, Cann I. 2016. Bacteroides intestinalis DSM 17393, a member of the human colonic microbiome, upregulates multiple endoxylanases during growth on xylan. Sci Rep 6:34360. doi: 10.1038/srep34360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kovatcheva-Datchary P, Nilsson A, Akrami R, Lee YS, De Vadder F, Arora T, Hallen A, Martens E, Björck I, Bäckhed F. 2015. Dietary fiber-induced improvement in glucose metabolism is associated with increased abundance of Prevotella. Cell Metab 22:971–982. doi: 10.1016/j.cmet.2015.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Scher JU, Sczesnak A, Longman RS, Segata N, Ubeda C, Bielski C, Rostron T, Cerundolo V, Pamer EG, Abramson SB, Huttenhower C, Littman DR. 2013. Expansion of intestinal Prevotella copri correlates with enhanced susceptibility to arthritis. Elife 2:e01202. doi: 10.7554/eLife.01202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shkoporov AN, Khokhlova EV, Chaplin AV, Kafarskaia LI, Nikolin AA, Polyakov VY, Shcherbakova VA, Chernaia ZA, Efimov BA. 2013. Coprobacter fastidiosus gen. nov., sp. nov., a novel member of the family Porphyromonadaceae isolated from infant faeces. Int J Syst Evol Microbiol 63:4181–4188. doi: 10.1099/ijs.0.052126-0. [DOI] [PubMed] [Google Scholar]
- 23.Kant R, Rasinkangas P, Satokari R, Pietila TE, Palva A. 2015. Genome sequence of the butyrate-producing anaerobic bacterium Anaerostipes hadrus PEL 85. Genome Announc 3:e00224-15. doi: 10.1128/genomeA.00224-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshida N, Emoto T, Yamashita T, Watanabe H, Hayashi T, Tabata T, Hoshi N, Hatano N, Ozawa G, Sasaki N, Mizoguchi T, Amin HZ, Hirota Y, Ogawa W, Yamada T, Hirata KI. 2018. Bacteroides vulgatus and Bacteroides dorei reduce gut microbial lipopolysaccharide production and inhibit atherosclerosis. Circulation 138:2486–2498. doi: 10.1161/CIRCULATIONAHA.118.033714. [DOI] [PubMed] [Google Scholar]
- 25.Chung WS, Walker AW, Louis P, Parkhill J, Vermeiren J, Bosscher D, Duncan SH, Flint HJ. 2016. Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biol 14:3. doi: 10.1186/s12915-015-0224-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabeerdoss J, Sankaran V, Pugazhendhi S, Ramakrishna BS. 2013. Clostridium leptum group bacteria abundance and diversity in the fecal microbiota of patients with inflammatory bowel disease: a case-control study in India. BMC Gastroenterol 13:20. doi: 10.1186/1471-230X-13-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang SC, Lin CH, Aljuffali IA, Fang JY. 2017. Current pathogenic Escherichia coli foodborne outbreak cases and therapy development. Arch Microbiol 199:811–825. doi: 10.1007/s00203-017-1393-y. [DOI] [PubMed] [Google Scholar]
- 28.Davin-Regli A, Pagès J-M. 2015. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front Microbiol 6:392. doi: 10.3389/fmicb.2015.00392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Egland PG, Palmer RJ Jr, Kolenbrander PE. 2004. Interspecies communication in Streptococcus gordonii-Veillonella atypica biofilms: signaling in flow conditions requires juxtaposition. Proc Natl Acad Sci U S A 101:16917–16922. doi: 10.1073/pnas.0407457101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou Y, Danbolt NC. 2014. Glutamate as a neurotransmitter in the healthy brain. J Neural Transm (Vienna) 121:799–817. doi: 10.1007/s00702-014-1180-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Slominski A, Semak I, Pisarchik A, Sweatman T, Szczesniewski A, Wortsman J. 2002. Conversion of L-tryptophan to serotonin and melatonin in human melanoma cells. FEBS Lett 511:102–106. doi: 10.1016/s0014-5793(01)03319-1. [DOI] [PubMed] [Google Scholar]
- 32.Garrett-Bakelman FE, Darshi M, Green SJ, Gur RC, Lin L, Macias BR, McKenna MJ, Meydan C, Mishra T, Nasrini J, Piening BD, Rizzardi LF, Sharma K, Siamwala JH, Taylor L, Vitaterna MH, Afkarian M, Afshinnekoo E, Ahadi S, Ambati A, Arya M, Bezdan D, Callahan CM, Chen S, Choi AMK, Chlipala GE, Contrepois K, Covington M, Crucian BE, De Vivo I, Dinges DF, Ebert DJ, Feinberg JI, Gandara JA, George KA, Goutsias J, Grills GS, Hargens AR, Heer M, Hillary RP, Hoofnagle AN, Hook VYH, Jenkinson G, Jiang P, Keshavarzian A, Laurie SS, Lee-McMullen B, Lumpkins SB, MacKay M, Maienschein-Cline MG, et al. 2019. The NASA Twins Study: a multidimensional analysis of a year-long human spaceflight. Science 364:eaau8650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voorhies AA, Mark Ott C, Mehta S, Pierson DL, Crucian BE, Feiveson A, Oubre CM, Torralba M, Moncera K, Zhang Y, Zurek E, Lorenzi HA. 2019. Study of the impact of long-duration space missions at the International Space Station on the astronaut microbiome. Sci Rep 9:9911. doi: 10.1038/s41598-019-46303-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shao D, Yao L, Riaz MS, Zhu J, Shi J, Jin M, Huang Q, Yang H. 2017. Simulated microgravity affects some biological characteristics of Lactobacillus acidophilus. Appl Microbiol Biotechnol 101:3439–3449. doi: 10.1007/s00253-016-8059-6. [DOI] [PubMed] [Google Scholar]
- 35.Klaus DM, Howard HN. 2006. Antibiotic efficacy and microbial virulence during space flight. Trends Biotechnol 24:131–136. doi: 10.1016/j.tibtech.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 36.Jiang P, Green SJ, Chlipala GE, Turek FW, Vitaterna MH. 2019. Reproducible changes in the gut microbiome suggest a shift in microbial and host metabolism during spaceflight. Microbiome 7:113. doi: 10.1186/s40168-019-0724-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Singh NK, Wood JM, Karouia F, Venkateswaran K. 2018. Succession and persistence of microbial communities and antimicrobial resistance genes associated with International Space Station environmental surfaces. Microbiome 6:204. doi: 10.1186/s40168-018-0585-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao Z, Zhu Y, Feng S, Meng C, Hu D, Liu H, Liu H. 2019. Effects of long term isolation on the emotion change of “Lunar Palace 365” crewmembers. Science Bull 64:881–884. doi: 10.1016/j.scib.2019.05.019. [DOI] [PubMed] [Google Scholar]
- 39.Gilbert JA. 2015. Our unique microbial identity. Genome Biol 16:97. doi: 10.1186/s13059-015-0664-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jin JS, Touyama M, Yamada S, Yamazaki T, Benno Y. 2014. Alteration of a human intestinal microbiota under extreme life environment in the Antarctica. Biol Pharm Bull 37:1899–1906. doi: 10.1248/bpb.b14-00397. [DOI] [PubMed] [Google Scholar]
- 41.Tasnim N, Abulizi N, Pither J, Hart MM, Gibson DL. 2017. Linking the gut microbial ecosystem with the environment: does gut health depend on where we live? Front Microbiol 8:1935. doi: 10.3389/fmicb.2017.01935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hao Z, Li L, Fu Y, Liu H. 2018. The influence of bioregenerative life-support system dietary structure and lifestyle on the gut microbiota: a 105-day ground-based space simulation in Lunar Palace 1. Environ Microbiol 20:3643–3656. doi: 10.1111/1462-2920.14358. [DOI] [PubMed] [Google Scholar]
- 43.Singh RK, Chang HW, Yan D, Lee KM, Ucmak D, Wong K, Abrouk M, Farahnik B, Nakamura M, Zhu TH, Bhutani T, Liao W. 2017. Influence of diet on the gut microbiome and implications for human health. J Transl Med 15:73. doi: 10.1186/s12967-017-1175-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ríos-Covián D, Ruas-Madiedo P, Margolles A, Gueimonde M, de Los Reyes-Gavilán CG, Salazar N. 2016. Intestinal short chain fatty acids and their link with diet and human health. Front Microbiol 7:185. doi: 10.3389/fmicb.2016.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Benus RF, van der Werf TS, Welling GW, Judd PA, Taylor MA, Harmsen HJ, Whelan K. 2010. Association between Faecalibacterium prausnitzii and dietary fibre in colonic fermentation in healthy human subjects. Br J Nutr 104:693–700. doi: 10.1017/S0007114510001030. [DOI] [PubMed] [Google Scholar]
- 46.Jie Z, Xia H, Zhong SL, Feng Q, Li S, Liang S, Zhong H, Liu Z, Gao Y, Zhao H, Zhang D, Su Z, Fang Z, Lan Z, Li J, Xiao L, Li J, Li R, Li X, Li F, Ren H, Huang Y, Peng Y, Li G, Wen B, Dong B, Chen JY, Geng QS, Zhang ZW, Yang H, Wang J, Wang J, Zhang X, Madsen L, Brix S, Ning G, Xu X, Liu X, Hou Y, Jia H, He K, Kristiansen K. 2017. The gut microbiome in atherosclerotic cardiovascular disease. Nat Commun 8:845. doi: 10.1038/s41467-017-00900-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnorr SL, Bachner HA. 2016. Integrative therapies in anxiety treatment with special emphasis on the gut microbiome. Yale J Biol Med 89:397–422. [PMC free article] [PubMed] [Google Scholar]
- 48.Chen JJ, Bai SJ, Li WW, Zhou CJ, Zheng P, Fang L, Wang HY, Liu YY, Xie P. 2018. Urinary biomarker panel for diagnosing patients with depression and anxiety disorders. Transl Psychiatry 8:192. doi: 10.1038/s41398-018-0245-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fang C, Zhong H, Lin Y, Chen B, Han M, Ren H, Lu H, Luber JM, Xia M, Li W, Stein S, Xu X, Zhang W, Drmanac R, Wang J, Yang H, Hammarstrom L, Kostic AD, Kristiansen K, Li J. 2018. Assessment of the cPAS-based BGISEQ-500 platform for metagenomic sequencing. Gigascience 7:1–8. doi: 10.1093/gigascience/gix133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang X, Zhang D, Jia H, Feng Q, Wang D, Liang D, Wu X, Li J, Tang L, Li Y, Lan Z, Chen B, Li Y, Zhong H, Xie H, Jie Z, Chen W, Tang S, Xu X, Wang X, Cai X, Liu S, Xia Y, Li J, Qiao X, Al-Aama JY, Chen H, Wang L, Wu QJ, Zhang F, Zheng W, Li Y, Zhang M, Luo G, Xue W, Xiao L, Li J, Chen W, Xu X, Yin Y, Yang H, Wang J, Kristiansen K, Liu L, Li T, Huang Q, Li Y, Wang J. 2015. The oral and gut microbiomes are perturbed in rheumatoid arthritis and partly normalized after treatment. Nat Med 21:895–905. doi: 10.1038/nm.3914. [DOI] [PubMed] [Google Scholar]
- 51.Zapala MA, Schork NJ. 2006. Multivariate regression analysis of distance matrices for testing associations between gene expression patterns and related variables. Proc Natl Acad Sci U S A 103:19430–19435. doi: 10.1073/pnas.0609333103. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The high-quality nonhuman reads that support the findings of this study have been deposited in the CNSA (https://db.cngb.org/cnsa/) of CNGBdb under the accession number CNP0000408.