ST131 is a global pathogen. This clone causes urinary tract infections and is frequently isolated from human sources. However, little is known about ST131 from environmental sources. With the widely reported increase in antibiotic concentrations found in wastewater, there is additional selection pressure for the emergence of antibiotic-resistant ST131 in this niche. The unbiased screening approach reported herein revealed that previously antibiotic-sensitive lineages of ST131 are now resistant to commonly used antibiotics present in wastewater systems and may be capable of surviving UV sterilization. This is the most comprehensive account of ST131 in the wastewater niche to date and an important step in better understanding the ecology of this global pathogen.
KEYWORDS: E. coli ST131, wastewater, clade distribution, antibiotic resistance
ABSTRACT
In the ten years since its discovery, the Escherichia coli clone sequence type 131 (ST131) has become a major international health threat, with the multidrug-resistant and extended-spectrum β-lactamase (ESBL)-producing clade C emerging as the globally dominant form. ST131 has previously been isolated from wastewater; however, most of these studies selectively screened for ESBL-producing organisms, thereby missing the majority of remaining ST131 clades. In this study, we used a high-throughput PCR-based screening strategy to comprehensively examine wastewater for the presence of ST131 over a 1-year period. Additional multiplex PCRs were used to differentiate clades and obtain an unbiased account of the total ST131 population structure within the collection. Furthermore, antimicrobial susceptibility profiles of all ST131-positive samples were tested against a range of commonly used antibiotics. From a total of over 3,762 E. coli wastewater samples, 1.86% (n = 70) tested positive for ST131, with the majority being clade A isolates. In total, 63% (n = 44) were clade A, 29% (n = 20) were clade B, 1% (n = 1) were clade C0, 6% (n = 4) were clade C1, and 1% (n = 1) were clade C2. In addition, a very high rate of resistance to commonly used antibiotics among wastewater isolates is reported, with 72.7% (n = 32) of clade A resistant to ciprofloxacin and high rates of resistance to gentamicin, sulfamethoxazole-trimethoprim, and tetracycline in clades that are typically sensitive to antibiotics.
IMPORTANCE ST131 is a global pathogen. This clone causes urinary tract infections and is frequently isolated from human sources. However, little is known about ST131 from environmental sources. With the widely reported increase in antibiotic concentrations found in wastewater, there is additional selection pressure for the emergence of antibiotic-resistant ST131 in this niche. The unbiased screening approach reported herein revealed that previously antibiotic-sensitive lineages of ST131 are now resistant to commonly used antibiotics present in wastewater systems and may be capable of surviving UV sterilization. This is the most comprehensive account of ST131 in the wastewater niche to date and an important step in better understanding the ecology of this global pathogen.
INTRODUCTION
Escherichia coli sequence type 131 (ST131) was jointly uncovered in 2008 in North America (1), Europe (2, 3), and Asia (4). This extraintestinal pathogenic E. coli (ExPEC) is responsible for millions of urinary tract infections (UTI) and bloodstream infections (BSI) annually (5). Recent bioinformatic and phylogenetic studies have categorized ST131 into five distinct lineages or clades (6). These clades are largely defined by alleles of the fimH gene, as follows: (i) clade A (fimH41), (ii) clade B (fimH22), and clade C (fimH30), further characterized into (iii) clade C0, (iv) clade C1, and (v) clade C2 (also called H30, H30R1, and H30Rx, respectively) based on the absence or presence of blaCTX-M alleles.
In less than a decade, this single clone is now recognized as one of the most successful human pathogens (5), with a global distribution on a pandemic scale (2, 7). A key factor in ST131’s global success is due to its ability to acquire resistance to antibiotics (2, 5, 6, 8). The rising prominence of CTX-M15 production and fluoroquinolone resistance among E. coli has been attributed to the clonal expansion of clade C ST131 (9). Studies have correlated the strain’s evolution of resistance to fluoroquinolones in the 1980s, when this class of antibiotics was used heavily in both human and veterinary medicine (6, 10). Additionally, ST131 has been found to acquire plasmids conferring resistance to β-lactam antibiotics and, more recently, carbapenemases (11, 12).
Despite advances in our knowledge of the genetics and evolution of ST131, progress has been comparatively slower understanding the strain’s epidemiology. Phylogenetic analysis of extensive ST131 collections have been unable to identify temporal or geographical clustering (6, 8, 13). ST131 appears to be abundantly distributed across geographical locations (2, 8), displays wide host species range (14–16), and spans potentially vast ecological environments (17), suggesting ST131 is a host generalist pathogen capable of frequent interspecies movement (13). However, large data sets are often overwhelmingly comprised of ST131 strains isolated from human clinical samples (6, 8, 13). The addition of more strains from a wider range of environments is likely to improve strain databases, such that more accurate epidemiological conclusions can be drawn from phylogenetic studies.
As an ecological niche, wastewater is a potentially rich source of ST131 for a number of reasons. (i) Although ExPEC strains mediate their pathogenicity outside the colon, the habitat of E. coli is in the gastrointestinal tract of mammals, the contents of which readily enter urban wastewater systems. (ii) Strains of E. coli are known to survive in wastewater for long periods (18–20). (iii) As an environmental niche, wastewater is an open ecosystem with consistent low levels of exposure to long half-life antibiotics, such as fluoroquinolones, that can persist at concentrations as high as the milligram per liter range in wastewater (21–23). This coexistence mediates selective pressure for the evolution of antibiotic-resistant variants of highly adaptable Enterobacteriaceae like Klebsiella spp. and ST131 in these environments. Unsurprisingly, antibiotic-resistant strains of ST131 have been isolated from wastewater in the Czech Republic (17, 24), Japan (25, 26), and France (27). However, these studies selectively screened for extended-spectrum β-lactamase (ESBL)-producing organisms, thereby omitting antibiotic-sensitive lineages of ST131 like clades A and B from analysis, giving a biased and potentially misleading representation of the true extent of ST131 in the environment.
Therefore, we hypothesized that an unbiased screen of wastewater samples would uncover a more accurate representation of the ST131 population in an urban wastewater system. Consequently, the objectives of this study were to use an unbiased high-throughput PCR-based screen to detect ST131 in wastewater, determine the relative distribution of clades among ST131-positive samples, and determine the resistance profiles to commonly used antibiotics among isolates collected over an entire year.
RESULTS
Total number of samples screened.
In total, 4,083 sewage samples containing a mixed population of microorganisms were collected at various points in the treatment process from three different wastewater treatment plants in Calgary, Canada over a 1-year period. Of these, 3,762 were found to contain E. coli as visualized on CHROMagar plates, with the remainder consisting of other Enterobacteriaceae, including Klebsiella, Enterobacter, Citrobacter, or Serratia. Of the total E. coli population, 1.86% (n = 70) tested positive for ST131.
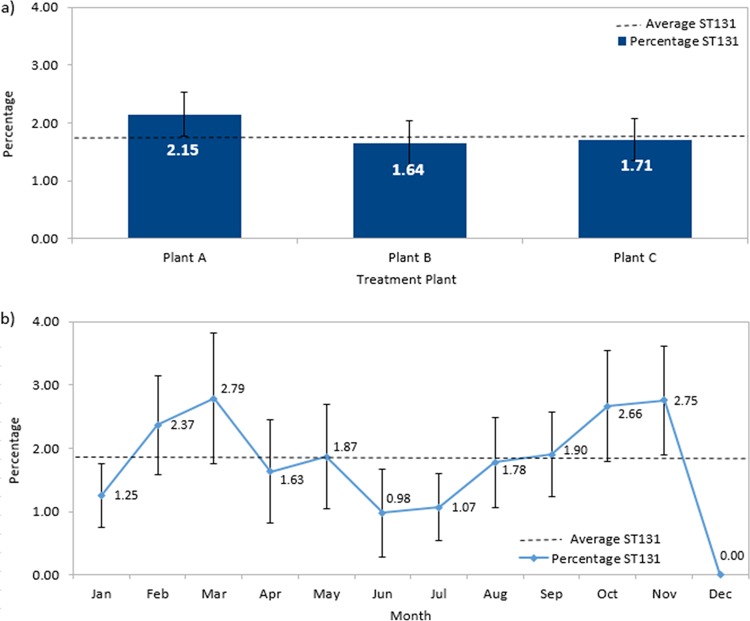
There were no significant differences between the ST131 collected from the three different wastewater treatment plants servicing the city of Calgary (Pearson χ2 test, χ = 1.084, df = 2, P = 0.582) (Fig. 1a). When ST131 strains are displayed as a percentage of total E. coli collected per month over 2015, ST131 detection remained relatively constant around the mean 1.86% mark throughout the year (Pearson χ2 test, χ = 9.608, df = 11, P = 0.566) (Fig. 1b), with the exception of December, where the absence of ST131 is likely due to less frequent sample collection during the holiday season. This is in line with other studies that considered seasonality within their data set when detecting E. coli from wastewater (28, 29) but in contrast to studies sampling directly from untreated water (30, 31), potentially highlighting the efficiency of wastewater treatment plants in maintaining a consistent reduced rate of E. coli from effluent.
FIG 1.
(a) Percentage ST131 of total E. coli samples for the three wastewater plants in Calgary. There was no difference in the number of ST131 isolates collected from the three plants. (b) Percentage ST131 of total E. coli samples identified in wastewater over time. The average ST131 detection rate is shown as a broken line. Error bars represent standard error in all cases.
Relative distribution of clades among ST131-positive samples.
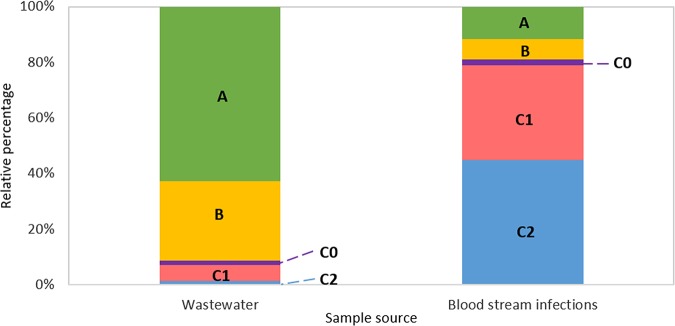
Among the 70 ST131-positive samples, 62.86% (n = 44) were clade A, 28.57% (n = 20) were clade B, 1.43% (n = 1) were clade C0, 5.71% (n = 4) were clade C1, and 1.43% (n = 1) were clade C2 (Fig. 2 and Table 1). We compared the relative distribution of ST131 clades isolated from wastewater to an independent study that collected ST131 from bloodstream isolates of infected individuals, conducted in the same geographical location over a similar time period (32). Strains from the clinical study were collected from bloodstream-infected individuals that presented to a centralized laboratory in Calgary over 2016 and were analyzed via whole-genome sequencing. Despite samples being collected from different sources, this is the most appropriate analysis due to the unbiased screening procedure performed in both studies. This study reported a relative distribution of 11.56% (n = 17) for clade A, 7.48% (n = 11) for clade B, 2.04% (n = 3) for clade C0, 34.01% (n = 50) for clade C1, and 44.9% (n = 66) for clade C2. These distributions from the two studies differ significantly (Fisher’s exact test, P = 5.13 × 10−25). Furthermore, differences between individual clades were significant for all clades except clade C0. This is of note, primarily as clade C is widely recognized as the dominant circulating form of the pathogen (2); however, the unbiased design of our study uncovered a much higher rate of clade A and B strains than clade C from wastewater.
FIG 2.
Clade distribution of ST131 isolated from different sources. The ST131 clade distribution obtained from wastewater isolates in this study was compared to the ST131 clade distribution from bloodstream isolates obtained from Peirano et al. (32).
TABLE 1.
Comparison of frequency distribution of clades between wastewater and human isolates
| Clade | Wastewater isolates (%)a (n = 70) | Human isolates (%)a (n = 147) | P value | Significance |
|---|---|---|---|---|
| A | 44 (63) | 17 (12) | 3.93 × 10−15 | <0.001 |
| B | 20 (29) | 11 (7) | 3.33 × 10−5 | <0.001 |
| C0 | 1 (1) | 3 (2) | 0.75 | NSb |
| C1 | 4 (6) | 50 (34) | 6.56 × 10−6 | <0.001 |
| C2 | 1 (1) | 66 (45) | 9.21 × 10−11 | <0.001 |
Percentage of total is given in parentheses.
NS, not significant.
Antimicrobial resistance, ESBL production, and UV survival of ST131 samples isolated from wastewater.
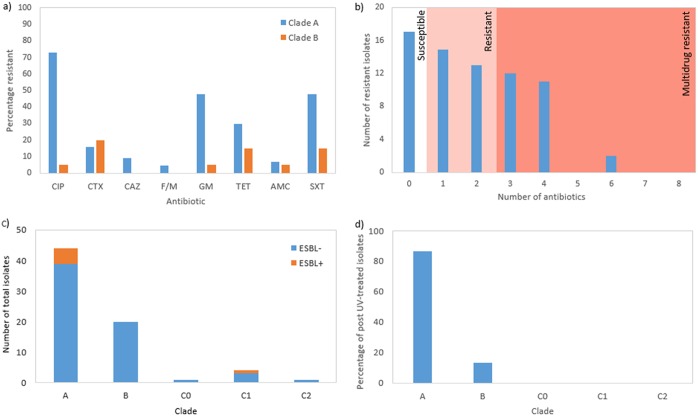
(i) Antibiotic resistance. Typically, antibiotic resistance is associated with clade C ST131 (6). As sample sizes for the three C subclades were too small for meaningful comparisons, the majority of analyses on levels of resistance apply to clade A and B strains of ST131. The rates of full resistance to each antibiotic tested in clade A and B strains are indicated in Fig. 3.
FIG 3.
(a) Percentage of clade A and B ST131 isolates resistant to the eight antibiotics used in this study. Antibiotics included are ciprofloxacin (CIP), cefotaxime (CTX), ceftazidime (CAZ), nitrofurantoin (F/M), gentamicin (GM), tetracycline (TET), amoxicillin (AMC), and sulfamethoxazole and trimethoprim (SXT). (b) Extent of multidrug resistance in ST131 samples isolated from wastewater. The background color indicates extent of resistance, where white is susceptible to all antibiotics, pink is resistant to one or two antibiotics, and red is multidrug resistant or resistant to three or more antibiotics. (c) Number of ESBL-positive and ESBL-negative strains from all ST131 isolates. (d) Percentage of post-UV-treated ST131 samples by clade.
Among clade A ST131 strains, there was a very high prevalence of resistance to ciprofloxacin, with 72.7% (n = 32) for all A clades strains displaying full resistance. Additionally, there was also a high prevalence of resistance to three other antibiotics tested as follows: 47.7% (n = 21), 47.7% (n = 21), and 29.5% (n = 13) of all clade A strains were resistant to gentamicin, sulfamethoxazole-trimethoprim, and tetracycline, respectively. Strains were also examined to see the extent of multidrug resistance among ST131 isolated from wastewater (Fig. 3b). While 24.3% of strains (n = 17) were sensitive to all eight antibiotics tested, 40% (n = 28) were resistant to one or two antibiotics and 35.7% (n = 25) were multidrug resistant. This includes 2.9% (n = 2) of strains that were resistant to six of the eight antibiotics tested, including a clade A strain and a clade C1 strain.
(ii) ESBL production profiles. There was a low rate of ESBL-producing strains found in our collection. Overall, only 8% (n = 6) of all ST131 strains isolated from wastewater were capable of ESBL production according to CLSI guidelines (33) (Fig. 3c). ESBL production is a tightly linked feature of clade C2 ST131 (6). Interestingly, when the clades of the ESBL producers were considered, five were clade A strains and one was a C1 strain.
(iii) UV treatment survival. Our collection of samples contained isolates from water that were isolated after UV treatment. Overall, 21.4% (n = 15) of the ST131-positive samples survived UV treatment. When this is broken down by clade, 86.67% (n = 13) were clade A, 13.33% (n = 2) were clade B, and there were no clade C samples among post-UV-treated strains (Fig. 3d).
DISCUSSION
Wastewater in this study refers to all water exiting toilets or drained from domestic bathrooms or kitchens in Calgary in 2015 and does not include agricultural runoff or storm water. Like in most cities, treated wastewater output is returned back to the local river, which is the source of irrigation, drinking water, and recreational activity. Additionally, by-products of the wastewater treatment process are used for biosolid production for fertilizer and composting purposes; thus, it is crucial to know the extent to which wastewater carries human pathogens like ST131. The first objective of this study was to identify ST131 among total E. coli in wastewater in Calgary over 2015. ST131 has previously been identified from total E. coli in the river Thames and coastal bathing waters (UK) in two separate studies (34, 35), urban sewage and river water in Barcelona (Spain) (36), wastewater treatment plants and hospital wastewater in the Kansai region (Japan) (26), wastewater treatment plants in Brno (Czech Republic) (24), the wastewater treatment network in Besançon (France) (27), and coastal marine sediments in the Adriatic Sea (Italy) (37). However, our second objective was to comprehensively examine the population structure of ST131 within this environment, which has been overlooked. Given the high proportion of C clades from clinical data reported in Calgary, Canada (32), we expected to uncover a high proportion of ESBL-producing clade C ST131 with our unbiased screening approach but also an increased proportion of antibiotic-sensitive strains. Unexpectedly, we found a low rate of clade C ESBL-producing strains and a high rate of clade A strains. There are a number of reasons to explain the low frequency of clade C and relative abundance of clade A ST131 in wastewater.
Low prevalence of ST131 overall.
The low number of clade C strains uncovered in this study is likely due to the low ST131 prevalence in the Calgary wastewater system, with ST131 accounting for only 1.86% from total E. coli isolates overall. This rate is low when compared to that of other studies, which range from 73% in Brno, Czech Republic (24) to 2.4% in Besançon, France (27). Both studies sampled from wastewater treatment plants that were smaller than those in Calgary, servicing approximately 400,000 and 120,000 people, respectively, compared to 1,338,022 people in Calgary. Despite differences in the sizes of the plants, sources of samples were collected from municipal wastewater treatment plants that serviced urban and suburban areas, including hospitals and long-term care facilities, and did not include agricultural effluent in all cases. The wide range in ST131 prevalence could also be explained based on geographical location (38) or factors explained below.
Experimental design.
Typically, ST131 is isolated in studies attempting to uncover ESBL-producing species or organisms containing various CTX-M alleles present in an environment, with the vast majority of studies employing a selective screening approach to do this (24, 26, 34, 35). Understandably, a high degree of ESBL-producing ST131 is uncovered, but how this breaks down into the different clades of the organism is rarely determined. In our analysis, an unbiased screening approach allowed for the isolation of all clades of ST131, including antibiotic-sensitive strains, which would have been eliminated from analysis if ESBL or ciprofloxacin selective plates were used. This is likely to have accounted for the high rate of clade A and B strains, which are typically not associated with ESBL production.
Additionally, sampling strategy is an important consideration, as many previous studies focused on untreated water (24, 26, 27, 34–37). In our study, samples were collected before and after UV treatment to determine whether ST131 isolates were capable of surviving the entire wastewater treatment process. It is currently unknown whether the isolation of ST131 after UV treatment is due to technical limitations of the UV sterilization protocol used by treatment plants or whether UV nonsusceptibility was achieved through biological means, such as upregulated UV repair mechanisms (39) or mechanisms of UV tolerance (40).
Underestimated clade A prevalence.
Clade C may not be the most dominant form, but it is the most frequently isolated. As a consequence, there is an underestimation in the prevalence of clade A in the environment. Among nonenvironmentally focused studies, ST131 isolates are predominantly selectively screened from human or animal (either companion, farm, or wildlife) sources where only sick individuals are screened. A very limited number of studies have focused on isolation of ST131 from asymptomatic carriers potentially harboring less-pathogenic clades (41). Thus, a potentially large source of nonpathogenic and non-ESBL-producing ST131 may be underrepresented and unrealized in the current literature.
Adaptation of clade A to the environment.
Adaptive diversification among E. coli isolates is well documented (42, 43). For ST131, a picture is beginning to emerge of each clade specializing to different niches. Many consider the evolutionary trajectory of clade C as a nascent human host specialist (2). More recently, Liu et al. investigated ST131 isolated from meat products and found that almost all samples were clade B isolates (44). If we assume that clades B and C have adapted to food animal and human hosts, respectively, the abundance of clade A in wastewater presented herein could suggest evidence of adaptation to this environment. Zhi et al. identified naturalized stress-tolerant environmental E. coli with a greater ability to survive in wastewater due to the presence of an insertion element (IS30) that upregulated uspC, producing improved motility and enhanced UV resistance in affected strains (45). PCR screens for this mutation were performed on all 70 ST131 strains isolated in our study, but only two were positive for the IS30 insertion, suggesting that there may be a variety of mutations responsible for adaptation to this particular niche. The collection of more environmental isolates in an unbiased manner and their addition to large-scale phylogenetic analyses could increase the power of epidemiological studies.
The third and final objective of this study was to determine the antibiotic susceptibilities of all of the ST131 wastewater isolates. Given the clade distribution and what is known about the resistance profiles of the different clades (6, 8), we expected antibiotic resistance to be limited to fluoroquinolone-resistant and ESBL-producing clade C isolates. Unexpectedly, there was a high rate of fluoroquinolone resistance from the clade A isolates and not from the clade C isolates.
Since the 1980s and 1990s, fluoroquinolones were the antibiotics of choice for UTI treatment in Europe and North America (46). However, up to 62% of administered fluoroquinolones are excreted nonmetabolized in urine (47), and as a result, these antibiotics are an emerging pollutant of water and soil environments (48), with high environmental concentrations promoting the evolution of resistant forms. In E. coli, resistance to fluoroquinolones arises due to the presence of point mutations in gyrA and parC (49), first observed in the 1980s when these antibiotics were prescribed heavily in human and veterinary medicine. These are defining features of the C clades of ST131 (6) and have since been linked to the rise of that clade. Our observation of fluoroquinolone resistance in clade A strains could suggest independent acquisition from resistance in clade C strains, as fluoroquinolone-resistant mutations are typically located on the ST131 chromosome rather than plasmids. However, fluoroquinolone resistance can be mediated through the acquisition of plasmids containing the aac(6’)-lb-cr gene in some cases (50). Regardless of the underlying molecular mechanisms involved, this resistant phenotype is likely to account for the high rate of clade A ST131 in wastewater.
The higher number of ESBL producers in clade A than clade C2 suggests that ESBL production in clades other than C2 is apparent, most likely as a result of horizontal transmission of plasmids among organisms in similar niches. Recent quantitative studies enumerating bacteria in water samples have found ST131 to be among the most ubiquitous ESBL-producing organisms in this niche (24, 26, 35), but this is not always the case. Dhanji et al. uncovered a high rate of fluoroquinolone-resistant but ESBL-negative ST131 isolates using a ciprofloxacin selective screening approach to uncover ST131 in the Thames river, United Kingdom (34).
There was a high occurrence of resistance to antibiotics that are commonly found at high concentrations in wastewater (51). Sulfamethoxazole-trimethoprim and gentamicin are used to treat UTIs either for uncomplicated infection (52) or intravesically for prophylaxis or recurrent infection (53), respectively. All three are found nondegraded and at high concentrations in aquatic environments (54–56), imposing selection pressure for the evolution of resistant forms. Resistance mechanisms to all three drugs are reported and understood at a molecular level (57–59). Furthermore, the antibiotics included in this study were of five different classes, meaning the evolution of resistance to one antibiotic was unlikely to mediate cross-resistance to antibiotics of other classes, and these resistance profiles are likely independent acquisition events. Intuitively, resistance profiles of isolates correlated with antibiotics that are either most commonly used to treat UTIs and/or most frequently detected in water. Overall, the presence of these antibiotics in wastewater is likely driving the high resistance levels from all clades of ST131, as indicated by our data.
Conclusion.
There were limitations to this study. First, this is a PCR-based screening approach, so we were only able to detect already known clades of ST131. No new or emerging clades could be detected in this screen. Second, genomic analysis has not yet been performed on the 70 wastewater isolates, as this is the focus of a future study. However, our aim here was to perform a high-throughput screening, and this was possible and successful with PCR. This study confirmed the presence of ST131, albeit a low one, in wastewater in Calgary, Canada. Nevertheless, a large sample size and a comprehensive unbiased screening approach uncovered the largest collection of ST131 isolated from wastewater in a single study. In-depth analysis of the population structure revealed that previously overlooked ESBL-negative clade A and B strains are the highest contributor to this contamination. These previously nonresistant clades are actively acquiring new resistances, most likely due to selection pressure from the presence of consistent low levels of commonly used antibiotics. Additionally, clade A strains of ST131 may have inherent resistances to UV sterilization. Genome sequencing and comparative genomics of these strains are logical next steps to uncover the underlying molecular mechanisms for these findings.
Wastewater treatment plants are already identified as key hot spots for emerging resistances among ST131. The addition of these samples to large-scale epidemiology studies will be invaluable in developing a more complete epidemiological understanding of this emerging global pathogen.
MATERIALS AND METHODS
Wastewater treatment plant overview.
There are three urban wastewater treatment plants (treatment plants A, B, and C) located in Calgary, AB, Canada (60), which service 966,337, 280,788, and 90,897 inhabitants, respectively, with a combined total of 1,338,022 inhabitants in 2015 overall. The three treatment plants range in size but collectively processed 465,068 m3 of wastewater per day in 2015. In all cases, treatment processes were identical. Briefly, raw wastewater (water from sinks, drains, and toilets from industrial complexes, health care facilities, and domestic households) is collected via one of three collection systems for each treatment facility. The inflow passes through headwork gates to remove large solids and then primary clarifiers to remove smaller solids. For microbial disinfection, wastewater is pumped through cloth media disks, passes through a UV disinfection step, and is released into the Bow River. The UV disinfection step uses UV lamps, ranging from 65 to 2,800 W, allowing for an average UV dose of 26 mJ/cm2 as per Alberta Environment Protection standards. The average final effluent of total suspended solids from all three wastewater plants was 6 mg/liter in 2015.
Collection of E. coli wastewater samples.
Samples were collected every week over the entire 2015 calendar year from the three different urban wastewater treatment plants in Calgary. Samples were collected before and after UV treatment by filtering water through Büchner flasks containing EZ-Pak 0.45-μm, 47-mm white gridded filters (Millipore, Burlington, MA, USA). Filter membranes were then removed and placed on m-FC agar plates, containing bile salts and rosolic acid for selective growth fecal coliforms, on which E. coli are identified as blue colonies after overnight incubation at 44.5°C. E. coli colonies are picked and grown in liquid Luria-Bertani (LB) broth in a 96-well plate format overnight at 37°C. The following day, 100 μl of 50% glycerol are added to the cultures and stored at −80°C.
High-throughput ST131 detection among E. coli wastewater samples.
For the detection of ST131 among E. coli wastewater samples, a high-throughput PCR-based screening assay was developed and applied to all 3,762 sewage samples containing E. coli collected from the three treatment plants. Cultures were established by inoculating 100 μl of LB liquid broth with frozen glycerol stocks in 96-well plates, sealed with breathable membranes (Nunc, Rochester, New York, USA), and incubated overnight at 37°C at 200 rpm. DNA was prepared by transferring 50 μl of overnight culture into another 96-well plate, adding 50 μl of Milli-Q, and sealing the plate with a 96-well PCR plate seal (Bio-Rad, Hercules, CA, USA). Heat lysis on all samples was performed in a T100 Bio-Rad thermocycler machine and consisted of heating the samples to 95°C for 10 min, followed by cooling at 4°C for 5 min. Samples were then centrifuged for 4,000 rpm for 15 min to remove cell debris, and 2 μl of supernatant was used as DNA for downstream processes.
For the PCR detection of ST131 among wastewater samples, previously published primers (Table 2) were used. PCR reagents were scaled up to screen 96 samples in each assay. All PCRs were performed with ThermoPol Taq (New England Biolabs, Ipswich, MA, USA) and consisted of an initial denaturation at 95°C for 5 min followed by 30 cycles of denaturation at 95°C for 15 s, annealing at 57°C for 20 s, and amplification at 68°C for 40 s, with a final amplification step at 68°C for 1 min. To determine the limits of ST131 DNA detection, PCRs were performed as above on a dilution series of purified genomic DNA extracted from E. coli CD306. Visible amplification was observed with DNA concentrations as low as 2.8 pg/μl or 8.5 × 10−10 pmol of genomic DNA.
TABLE 2.
Primers used in this study
| Name | Working concn (μM) | Sequence (5′ to 3′) | Identified element | Reference |
|---|---|---|---|---|
| ST131_R19-YF1 | 0.25 | AGCAACGATATTTGCCCATT | ST131 | 62 |
| ST131_R19-YR1 | 0.25 | GGCGATAACAGTACGCCATT | ||
| CladeAsp4-F | 0.25 | TGACGGGACGTGAGCAAATTA | Clade A | |
| CladeAsp4-R | 0.25 | AGTCAGACCTAGCCACCCTT | ||
| prfC-F | 0.25 | CAACGTTGAAGCAGTGTATGAG | Clade B | |
| prfC-R | 0.25 | TGACAATCGACGGCTTTAGA | ||
| C-SNP-1-F | 0.15 | CGCTGGCCAGTTATCTGAAAT | Clade C0 | |
| C-SNP-1-R | 0.15 | CCTTTCACCAACTGGGTTACT | ||
| C1-F | 0.10 | GGCCCCACAAATTGCTT | Clade C1 | |
| C1-R | 0.10 | CGCACCTCCGATACCAAA | ||
| C2-F | 0.20 | ACGGATTCAGGTAGACGATT | Clade C2 | |
| C2-R | 0.20 | CCTCACCAAAGTTGCGATTAC | ||
| flh-IS-F | 10 | CGGGGAACAAATGAGAACAC | IS30 insertion | 45 |
| fllh-IS-R | 10 | TGGAGAAACGACGCAATC |
Sample purification.
To ensure all samples that were found to contain ST131 from the wastewater treatment plants were pure, all ST131-positive samples were streaked to purity on nonselective CHROMagar orientation plates (bioMérieux, Marcy-l’Étoile, France), where E. coli colonies grow as dark pink to red colonies. After purity was confirmed, single colonies were used to inoculate 3 ml of LB liquid broth and incubated overnight at 37°C at 200 rpm, and 500-μl aliquots of pure cultures were cryogenically preserved in 50% glycerol at −80°C.
Clade identification of purified ST131-positive samples.
DNA from ST131-positive samples was used in multiplex PCRs with clade-specific primers to identify the clade to which the purified ST131-positive cultures belonged (Table 2). The clade-specific PCR program involved initial denaturation at 95°C for 5 min followed by 30 cycles of denaturation at 95°C for 40 s, annealing at 57°C for 40 s, and amplification at 68°C for 40 s, with a final amplification step at 68°C for 7 min. Samples were run on a 2.5% agarose gel.
Antimicrobial susceptibility testing of ST131-positive samples.
The antimicrobial resistance profiles of all ST131-positive isolates were determined. Samples were subjected to disk diffusion assays with eight antibiotics (Table 3). ESBL classification was based on the addition of β-lactam inhibitor clavulanic acid (10 μg) to cefotaxime and ceftazidime. Resistance profiles were assessed based on CLSI guidelines (33) and multidrug resistance classifications were based on Magiorakos et al. (61).
TABLE 3.
List of antimicrobial compounds used in this study
| Antibiotic | Classification | Concn (μg) |
|---|---|---|
| Ciprofloxacin | Fluoroquinolone | 5 |
| Cefotaxime | β-Lactam, third-generation cephalosporin | 30 |
| Ceftazidime | β-Lactam, third-generation cephalosporin | 30 |
| Gentamycin | Aminoglycoside | 10 |
| Sulfamethoxazole and trimethoprim | Sulfanilamide and dihydrofolate reductase inhibitor | 23.75/1.25 |
| Nitrofurantoin | Nitrofuran | 300 |
| Amoxycillin | β-Lactam | 20 |
| Tetracycline | Tetracycline | 30 |
Statistical analysis.
Comparisons of proportions of ST131 isolated from treatment plant or by season were analyzed by a Pearson χ2 test. Comparison of proportions of ST131 clades between clinical and wastewater samples were analyzed via Fisher’s exact test. In all cases, significance was designated for a P value of <0.05. All statistical analyses were performed in SPSS Statistics 25 (IBM Corp., Armonk, NY, USA).
ACKNOWLEDGMENTS
We thank the City of Calgary Wastewater treatment facilities, particularly Nisa Jayathilake for clarification on the treatment process and data collection. We thank the Advancing Canadian Wastewater Assets program for providing infrastructure and equipment support. We also thank Richard Moore for supervision during sample handling and storage and Diego Nobrega for help with statistical analysis.
T.D. was supported by a Canadian Institutes of Health Research (CIHR) operating grant, a Canadian Natural Sciences and Engineering Research Council (NSERC) discovery grant, the Canada Research Chair program, and a Canadian Foundation for Innovation grant. This work is supported by the Joint Programming Initiative on Antimicrobial Resistance, JPIAMR (10016015 to J.D.D.P. and R.D.).
The funders had no role in study design, data collection, interpretation, decision to publish, or manuscript preparation.
J.D.D.P. previously received funding from Merck and Astra Zeneca. The remaining authors have no conflicts of interest.
REFERENCES
- 1.Coque TM, Novais Â, Carattoli A, Poirel L, Pitout J, Peixe L, Baquero F, Cantón R, Nordmann P. 2008. Dissemination of clonally related Escherichia coli strains expressing extended-spectrum β-lactamase CTX-M-15. Emerg Infect Dis 14:195–200. doi: 10.3201/eid1402.070350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nicolas-Chanoine M-H, Bertrand X, Madec J-Y. 2014. Escherichia coli ST131, an intriguing clonal group. Clin Microbiol Rev 27:543–574. doi: 10.1128/CMR.00125-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nicolas-Chanoine MH, Blanco J, Leflon-Guibout V, Demarty R, Alonso MP, Caniça MM, Park YJ, Lavigne JP, Pitout J, Johnson JR. 2008. Intercontinental emergence of Escherichia coli clone O25:H4-ST131 producing CTX-M-15. J Antimicrob Chemother 61:273–281. doi: 10.1093/jac/dkm464. [DOI] [PubMed] [Google Scholar]
- 4.Yumuk Z, Afacan G, Nicolas-Chanoine M-H, Sotto A, Lavigne J-P. 2008. Turkey: a further country concerned by community-acquired Escherichia coli clone O25-ST131 producing CTX-M-15. J Antimicrob Chemother 62:284–288. doi: 10.1093/jac/dkn181. [DOI] [PubMed] [Google Scholar]
- 5.Pitout JDD, DeVinney R. 2017. Escherichia coli ST131: a multidrug-resistant clone primed for global domination. F1000Res 6:195. doi: 10.12688/f1000research.10609.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ben Zakour NL, Alsheikh-Hussain AS, Ashcroft MM, Nhu NTK, Roberts LW, Stanton-Cook M, Schembri MA, Beatson SA. 2016. Sequential acquisition of virulence and fluoroquinolone resistance has shaped the evolution of Escherichia coli ST131. mBio 7:e00347-16. doi: 10.1128/mBio.00347-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers BA, Sidjabat HE, Paterson DL. 2011. Escherichia coli O25b-ST131: a pandemic, multiresistant, community-associated strain. J Antimicrob Chemother 66:1–14. doi: 10.1093/jac/dkq415. [DOI] [PubMed] [Google Scholar]
- 8.Petty NK, Ben Zakour NL, Stanton-Cook M, Skippington E, Totsika M, Forde BM, Phan M-D, Gomes Moriel D, Peters KM, Davies M, Rogers BA, Dougan G, Rodriguez-Baño J, Pascual A, Pitout JDD, Upton M, Paterson DL, Walsh TR, Schembri MA, Beatson SA. 2014. Global dissemination of a multidrug resistant Escherichia coli clone. Proc Natl Acad Sci U S A 111:5694–5699. doi: 10.1073/pnas.1322678111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banerjee R, Johnson JR. 2014. A new clone sweeps clean: the enigmatic emergence of Escherichia coli sequence type 131. Antimicrob Agents Chemother 58:4997–5004. doi: 10.1128/AAC.02824-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, Turner P, Anson LW, Kasarskis A, Batty EM, Kos V, Wilson DJ, Phetsouvanh R, Wyllie D, Sokurenko E, Manges AR, Johnson TJ, Price LB, Peto TEA, Johnson JR, Didelot X, Walker AS, Crook DW. 2016. Evolutionary history of the global emergence of the Escherichia coli epidemic clone ST131. mBio 7:e02162-15. doi: 10.1128/mBio.02162-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peirano G, Pitout J. 2010. Molecular epidemiology of Escherichia coli producing CTX-M β-lactamases: the worldwide emergence of clone ST131 O25: H4. Int J Antimicrob Agents 35:316–321. doi: 10.1016/j.ijantimicag.2009.11.003. [DOI] [PubMed] [Google Scholar]
- 12.Nordmann P, Naas T, Poirel L. 2011. Global spread of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 17:1791–1798. doi: 10.3201/eid1710.110655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNally A, Oren Y, Kelly D, Pascoe B, Dunn S, Sreecharan T, Vehkala M, Välimäki N, Prentice MB, Ashour A, Avram O, Pupko T, Dobrindt U, Literak I, Guenther S, Schaufler K, Wieler LH, Zhiyong Z, Sheppard SK, McInerney JO, Corander J. 2016. Combined analysis of variation in core, accessory and regulatory genome regions provides a super-resolution view into the evolution of bacterial populations. PLoS Genet 12:e1006280. doi: 10.1371/journal.pgen.1006280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathers AJ, Peirano G, Pitout J. 2015. The role of epidemic resistance plasmids and international high-risk clones in the spread of multidrug-resistant Enterobacteriaceae. Clin Microbiol Rev 28:565–591. doi: 10.1128/CMR.00116-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manageiro V, Clemente L, Jones-Dias D, Albuquerque T, Ferreira E, Caniça M. 2015. CTX-M-15-producing Escherichia coli in Dolphin, Portugal. Emerg Infect Dis 21:2249–2251. doi: 10.3201/eid2112.141963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yair Y, Gophna U. 2018. Pandemic bacteremic Escherichia coli strains: evolution and emergence of drug-resistant pathogens, p 163–180. In Escherichia coli, a versatile pathogen. Springer, New York, NY. [DOI] [PubMed] [Google Scholar]
- 17.Jamborova I, Johnston BD, Papousek I, Kachlikova K, Micenkova L, Clabots C, Skalova A, Chudejova K, Dolejska M, Literak I, Johnson JR. 2018. Extensive genetic commonality among wildlife, wastewater, community, and nosocomial isolates of Escherichia coli sequence type 131 (H30R1 and H30Rx subclones) that carry blaCTX-M-27 or blaCTX-M-15. Antimicrob Agents Chemother 62:e00519-18. doi: 10.1128/AAC.00519-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kussell E, Kishony R, Balaban NQ, Leibler S. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169:1807–1814. doi: 10.1534/genetics.104.035352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Van Elsas JD, Semenov AV, Costa R, Trevors JT. 2011. Survival of Escherichia coli in the environment: fundamental and public health aspects. ISME J 5:173–183. doi: 10.1038/ismej.2010.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Berthe T, Ratajczak M, Clermont O, Denamur E, Petit F. 2013. Evidence for coexistence of distinct Escherichia coli populations in various aquatic environments and their survival in estuary water. Appl Environ Microbiol 79:4684–4693. doi: 10.1128/AEM.00698-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Renew JE, Huang C-H. 2004. Simultaneous determination of fluoroquinolone, sulfonamide, and trimethoprim antibiotics in wastewater using tandem solid phase extraction and liquid chromatography–electrospray mass spectrometry. J Chromatogr A 1042:113–121. doi: 10.1016/j.chroma.2004.05.056. [DOI] [PubMed] [Google Scholar]
- 22.Larsson E, Al-Hamimi S, Jönsson JÅ. 2014. Behaviour of nonsteroidal anti-inflammatory drugs and eight of their metabolites during wastewater treatment studied by hollow fibre liquid phase microextraction and liquid chromatography mass spectrometry. Sci Total Environ 485:300–308. doi: 10.1016/j.scitotenv.2014.03.055. [DOI] [PubMed] [Google Scholar]
- 23.Berglund B, Fick J, Lindgren P. 2015. Urban wastewater effluent increases antibiotic resistance gene concentrations in a receiving northern European river. Environ Toxicol Chem 34:192–196. doi: 10.1002/etc.2784. [DOI] [PubMed] [Google Scholar]
- 24.Dolejska M, Frolkova P, Florek M, Jamborova I, Purgertova M, Kutilova I, Cizek A, Guenther S, Literak I. 2011. CTX-M-15-producing Escherichia coli clone B2-O25b-ST131 and Klebsiella spp. isolates in municipal wastewater treatment plant effluents. J Antimicrob Chemother 66:2784–2790. doi: 10.1093/jac/dkr363. [DOI] [PubMed] [Google Scholar]
- 25.Gomi R, Matsuda T, Yamamoto M, Chou P-H, Tanaka M, Ichiyama S, Yoneda M, Matsumura Y, Gomi R, Matsuda T, Yamamoto M, Chou P-H, Tanaka M, Ichiyama S, Yoneda M, Matsumura Y. 2018. Characteristics of carbapenemase-producing Enterobacteriaceae in wastewater revealed by genomic analysis. Antimicrob Agents Chemother 62:e02501-17. doi: 10.1128/AAC.02501-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gomi R, Matsuda T, Matsumura Y, Yamamoto M, Tanaka M, Ichiyama S, Yoneda M. 2017. Occurrence of clinically important lineages, including the sequence type 131 C1-M27 subclone, among extended-spectrum-β-lactamase-producing Escherichia coli in wastewater. Antimicrob Agents Chemother 61:e00564-17. doi: 10.1128/AAC.00564-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bréchet C, Plantin J, Sauget M, Thouverez M, Talon D, Cholley P, Guyeux C, Hocquet D, Bertrand X. 2014. Wastewater treatment plants release large amounts of extended-spectrum β-lactamase-producing Escherichia coli into the environment. Clin Infect Dis 58:1658–1665. doi: 10.1093/cid/ciu190. [DOI] [PubMed] [Google Scholar]
- 28.Hoelle J, Johnson JR, Johnston BD, Kinkle B, Boczek L, Ryu H, Hayes S. 2019. Survey of US wastewater for carbapenem-resistant Enterobacteriaceae. J Water Health 17:219–226. doi: 10.2166/wh.2019.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liang S, Hall G, Champagne P. 2018. Monitoring seasonal variations in treatment performance of a wastewater stabilization pond with algal blooms and pH fluctuations. WDSA/CCWI Joint Conference Proceedings, Kingston, ON, Canada. [Google Scholar]
- 30.Diwan V, Hanna N, Purohit M, Chandran S, Riggi E, Parashar V, Tamhankar A, Stålsby Lundborg C. 2018. Seasonal variations in water-quality, antibiotic residues, resistant bacteria and antibiotic resistance genes of Escherichia coli isolates from water and sediments of the Kshipra River in Central India. Int J Environ Res Public Health 15:1281. doi: 10.3390/ijerph15061281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ornelas Van Horne Y, Parks J, Tran T, Abrell L, Reynolds KA, Beamer PI. 2019. Seasonal variation of water quality in unregulated domestic wells. Int J Environ Res Public Health 16:1569. doi: 10.3390/ijerph16091569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peirano G, Pitout JDD, Church DL, Holland S. 2018. Population structure of extra-intestinal Escherichia coli (ExPEc) causing bloodstream infections in a centralized Canadian region. European Congress of Clinical Microbiology and Infectious Disease, Madrid, Spain. [DOI] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. 2018. Performance standards for antimicrobial susceptibility testing; 29th informational supplement. CLSI M100-S29. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 34.Dhanji H, Murphy NM, Akhigbe C, Doumith M, Hope R, Livermore DM, Woodford N. 2011. Isolation of fluoroquinolone-resistant O25b:H4-ST131 Escherichia coli with CTX-M-14 extended-spectrum β-lactamase from UK river water. J Antimicrob Chemother 66:512–516. doi: 10.1093/jac/dkq472. [DOI] [PubMed] [Google Scholar]
- 35.Leonard AFC, Zhang L, Balfour AJ, Garside R, Hawkey PM, Murray AK, Ukoumunne OC, Gaze WH. 2018. Exposure to and colonisation by antibiotic-resistant E. coli in UK coastal water users: environmental surveillance, exposure assessment, and epidemiological study (Beach Bum Survey). Environ Int 114:326–333. doi: 10.1016/j.envint.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 36.Colomer-Lluch M, Mora A, López C, Mamani R, Dahbi G, Marzoa J, Herrera A, Viso S, Blanco JE, Blanco M, Alonso MP, Jofre J, Muniesa M, Blanco J. 2013. Detection of quinolone-resistant Escherichia coli isolates belonging to clonal groups O25b:H4-B2-ST131 and O25b:H4-D-ST69 in raw sewage and river water in Barcelona, Spain. J Antimicrob Chemother 68:758–765. doi: 10.1093/jac/dks477. [DOI] [PubMed] [Google Scholar]
- 37.Vignaroli C, Luna GM, Pasquaroli S, Di Cesare A, Petruzzella R, Paroncini P, Biavasco F. 2013. Epidemic Escherichia coli ST131 and Enterococcus faecium ST17 in coastal marine sediments from an Italian beach. Environ Sci Technol 47:13772–13780. doi: 10.1021/es4019139. [DOI] [PubMed] [Google Scholar]
- 38.Hocquet D, Muller A, Bertrand X. 2016. What happens in hospitals does not stay in hospitals: antibiotic-resistant bacteria in hospital wastewater systems. J Hosp Infect 93:395–402. doi: 10.1016/j.jhin.2016.01.010. [DOI] [PubMed] [Google Scholar]
- 39.Witkin EM. 1976. Ultraviolet mutagenesis and inducible DNA repair in Escherichia coli. Bacteriol Rev 40:869–907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sale JE, Lehmann AR, Woodgate R. 2012. Y-family DNA polymerases and their role in tolerance of cellular DNA damage. Nat Rev Mol Cell Biol 13:141–152. doi: 10.1038/nrm3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nicolas-Chanoine M-H, Gruson C, Bialek-Davenet S, Bertrand X, Thomas-Jean F, Bert F, Moyat M, Meiller E, Marcon E, Danchin N, Noussair L, Moreau R, Leflon-Guibout V. 2013. 10-Fold increase (2006–11) in the rate of healthy subjects with extended-spectrum β-lactamase-producing Escherichia coli faecal carriage in a Parisian check-up centre. J Antimicrob Chemother 68:562–568. doi: 10.1093/jac/dks429. [DOI] [PubMed] [Google Scholar]
- 42.Shaik S, Ranjan A, Tiwari SK, Hussain A, Nandanwar N, Kumar N, Jadhav S, Semmler T, Baddam R, Islam MA, Alam M, Wieler LH, Watanabe H, Ahmed N, Shaik S, Ranjan A, Tiwari SK, Hussain A, Nandanwar N, Kumar N, Jadhav S, Semmler T, Baddam R, Islam MA, Alam M, Wieler LH, Watanabe H, Ahmed N. 2017. Comparative genomic analysis of globally dominant ST131 clone with other epidemiologically successful extraintestinal pathogenic Escherichia coli (ExPEc) lineages. mBio 8:e01596-17. doi: 10.1128/mBio.01596-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Finn TJ, Shewaramani S, Leahy SC, Janssen PH, Moon CD. 2017. Dynamics and genetic diversification of Escherichia coli during experimental adaptation to an anaerobic environment. PeerJ 5:e3244. doi: 10.7717/peerj.3244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu CM, Stegger M, Aziz M, Johnson TJ, Waits K, Nordstrom L, Gauld L, Weaver B, Rolland D, Statham S, Horwinski J, Sariya S, Davis GS, Sokurenko E, Keim P, Johnson JR, Price LB, Liu CM, Stegger M, Aziz M, Johnson TJ, Waits K, Nordstrom L, Gauld L, Weaver B, Rolland D, Statham S, Horwinski J, Sariya S, Davis GS, Sokurenko E, Keim P, Johnson JR, Price LB. 2018. Escherichia coli ST131-H22 as a foodborne uropathogen. mBio 9:e00470-18. doi: 10.1128/mBio.00470-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhi S, Banting G, Li Q, Edge TA, Topp E, Sokurenko M, Scott C, Braithwaite S, Ruecker NJ, Yasui Y, McAllister T, Chui L, Neumann NF. 2016. Evidence of naturalized stress-tolerant strains of Escherichia coli in municipal wastewater treatment plants. Appl Environ Microbiol 82:5505–5518. doi: 10.1128/AEM.00143-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Naber KG. 2000. Survey on antibiotic usage in the treatment of urinary tract infections. J Antimicrob Chemother 46:49–52. doi: 10.1093/jac/46.suppl_1.49. [DOI] [PubMed] [Google Scholar]
- 47.Golet EM, Xifra I, Siegrist H, Alder AC, Giger W. 2003. Environmental exposure assessment of fluoroquinolone antibacterial agents from sewage to soil. Environ Sci Technol 37:3243–3249. doi: 10.1021/es0264448. [DOI] [PubMed] [Google Scholar]
- 48.Teglia CM, Perez FA, Michlig N, Repetti MR, Goicoechea HC, Culzoni MJ. 2019. Occurrence, distribution and ecological risk of fluoroquinolones in rivers and wastewaters. Environ Toxicol Chem 38:2305–2313. doi: 10.1002/etc.4532. [DOI] [PubMed] [Google Scholar]
- 49.Lindgren PK, Karlsson Å, Hughes D. 2003. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother 47:3222–3232. doi: 10.1128/aac.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Peirano G, Van Der Bij AK, Freeman JL, Poirel L, Nordmann P, Costello M, Tchesnokova VL, Pitout J. 2014. Characteristics of Escherichia coli sequence type 131 isolates that produce extended-spectrum β-lactamases: global distribution of the H30-Rx sublineage. Antimicrob Agents Chemother 58:3762–3767. doi: 10.1128/AAC.02428-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kusari S, Prabhakaran D, Lamshöft M, Spiteller M. 2009. In vitro residual anti-bacterial activity of difloxacin, sarafloxacin and their photoproducts after photolysis in water. Environ Pollut 157:2722–2730. doi: 10.1016/j.envpol.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 52.Raz R, Chazan B, Kennes Y, Colodner R, Rottensterich E, Dan M, Lavi I, Stamm W, Israeli Urinary Tract Infection Group . 2002. Empiric use of trimethoprim-sulfamethoxazole (TMP-SMX) in the treatment of women with uncomplicated urinary tract infections, in a geographical area with a high prevalence of TMP-SMX–resistant uropathogens. Clin Infect Dis 34:1165–1169. doi: 10.1086/339812. [DOI] [PubMed] [Google Scholar]
- 53.Saux NL, Robinson J. 2019. Aminoglycosides—alive and well in treatment of pediatric infections: a case of benefit versus risk. Off J Assoc Med Microbiol Infect Dis Canada 4:1–5. doi: 10.3138/jammi.2018.09.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Löffler D, Ternes TA. 2003. Analytical method for the determination of the aminoglycoside gentamicin in hospital wastewater via liquid chromatography–electrospray-tandem mass spectrometry. J Chromatogr A 1000:583–588. doi: 10.1016/s0021-9673(03)00059-1. [DOI] [PubMed] [Google Scholar]
- 55.Ryan CC, Tan DT, Arnold WA. 2011. Direct and indirect photolysis of sulfamethoxazole and trimethoprim in wastewater treatment plant effluent. Water Res 45:1280–1286. doi: 10.1016/j.watres.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 56.Kim S, Eichhorn P, Jensen JN, Weber AS, Aga DS. 2005. Removal of antibiotics in wastewater: effect of hydraulic and solid retention times on the fate of tetracycline in the activated sludge process. Environ Sci Technol 39:5816–5823. doi: 10.1021/es050006u. [DOI] [PubMed] [Google Scholar]
- 57.Heuer H, Krögerrecklenfort E, Wellington EMH, Egan S, van Elsas JD, van Overbeek L, Collard J-M, Guillaume G, Karagouni AD, Nikolakopoulou TL, Smalla K. 2002. Gentamicin resistance genes in environmental bacteria: prevalence and transfer. FEMS Microbiol Ecol 42:289–302. doi: 10.1111/j.1574-6941.2002.tb01019.x. [DOI] [PubMed] [Google Scholar]
- 58.Eliopoulos GM, Huovinen P. 2001. Resistance to trimethoprim-sulfamethoxazole. Clin Infect Dis 32:1608–1614. doi: 10.1086/320532. [DOI] [PubMed] [Google Scholar]
- 59.Roberts MC. 2005. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett 245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- 60.The City of Calgary. 2018. Wastewater treatment online tour. The City of Calgary, Calgary, AB, Canada: https://www.calgary.ca/UEP/Water/Pages/Water-and-wastewater-systems/Wastewater-system/Wastewater-treatment-tour.aspx. [Google Scholar]
- 61.Magiorakos A-P, Srinivasan A, Carey RB, Carmeli Y, Falagas ME, Giske CG, Harbarth S, Hindler JF, Kahlmeter G, Olsson-Liljequist B, Paterson DL, Rice LB, Stelling J, Struelens MJ, Vatopoulos A, Weber JT, Monnet DL. 2012. Multidrug‐resistant, extensively drug‐resistant and pandrug‐resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect 18:268–281. doi: 10.1111/j.1469-0691.2011.03570.x. [DOI] [PubMed] [Google Scholar]
- 62.Matsumura Y, Pitout JDD, Peirano G, Devinney R, Noguchi T, Yamamoto M, Gomi R, Matsuda T, Nakano S, Nagao M, Tanaka M, Ichiyama S. 2017. Rapid identification of different Escherichia coli sequence type 131 clades. Antimicrob Agents Chemother 61:e00179-17. doi: 10.1128/AAC.00179-17. [DOI] [PMC free article] [PubMed] [Google Scholar]