High-risk HPVs are major viral human carcinogens responsible for approximately 5% of all human cancers. The growth of HPV-transformed cells depends on the ability of viral oncoproteins to manipulate a variety of cellular circuits, including those involved in innate immunity. Here, we show that one of these strategies relies on E7-mediated transcriptional activation of the chromatin repressor SUV39H1, which then promotes epigenetic silencing of RIG-I, cGAS, and STING genes, thereby shutting down interferon secretion in HPV-transformed cells. Pharmacological or genetic inhibition of SUV39H1 restored the innate response in HPV-transformed cells, mostly through activation of RIG-I signaling. We also show that IFN production upon transfection of poly(dA·dT) or the RIG-I agonist M8 predominantly occurs through RIG-I signaling. Altogether, the reversible nature of the modifications associated with E7-mediated SUV39H1 upregulation provides a rationale for the design of novel anticancer and antiviral therapies targeting these molecules.
KEYWORDS: HPV, RIG-I, SUV39H1, epigenetics, innate immunity
ABSTRACT
Subversion of innate immunity by oncoviruses, such as human papillomavirus (HPV), favors carcinogenesis because the mechanism(s) of viral immune evasion can also hamper cancer immunosurveillance. Previously, we demonstrated that high-risk (hr) HPVs trigger simultaneous epigenetic silencing of multiple effectors of innate immunity to promote viral persistence. Here, we expand on those observations and show that the HPV E7 oncoprotein upregulates the H3K9-specific methyltransferase, whose action shuts down the host innate immune response. Specifically, we demonstrate that SUV39H1 contributes to chromatin repression at the promoter regions of the viral nucleic acid sensors RIG-I and cGAS and the adaptor molecule STING in HPV-transformed cells. Inhibition of SUV39H1 leads to transcriptional activation of these genes, especially RIG-I, followed by increased beta interferon (IFN-β) and IFN-λ1 production after poly(dA·dT) or RIG-I agonist M8 transfection. Collectively, our findings provide new evidence that the E7 oncoprotein plays a central role in dampening host innate immunity and raise the possibility that targeting the downstream effector SUV39H1 or the RIG-I pathway is a viable strategy to treat viral and neoplastic disease.
IMPORTANCE High-risk HPVs are major viral human carcinogens responsible for approximately 5% of all human cancers. The growth of HPV-transformed cells depends on the ability of viral oncoproteins to manipulate a variety of cellular circuits, including those involved in innate immunity. Here, we show that one of these strategies relies on E7-mediated transcriptional activation of the chromatin repressor SUV39H1, which then promotes epigenetic silencing of RIG-I, cGAS, and STING genes, thereby shutting down interferon secretion in HPV-transformed cells. Pharmacological or genetic inhibition of SUV39H1 restored the innate response in HPV-transformed cells, mostly through activation of RIG-I signaling. We also show that IFN production upon transfection of poly(dA·dT) or the RIG-I agonist M8 predominantly occurs through RIG-I signaling. Altogether, the reversible nature of the modifications associated with E7-mediated SUV39H1 upregulation provides a rationale for the design of novel anticancer and antiviral therapies targeting these molecules.
INTRODUCTION
Human papillomaviruses (HPVs) are circular double-stranded DNA viruses with a small genome of approximately 8 kb. Over 200 types of HPV have been identified and classified according to whether they infect cutaneous or mucosal epithelium (http://www.ictv.global/report/papillomaviridae) (1–3). Cancer-causing HPVs are classified as high-risk (hr) types, among which the most commonly found are the α-genotypes HPV16 and HPV18, well-known to be the causative agents of cervical and anogenital cancers and heavily implicated in head and neck cancers (4, 5).
The development of HPV-associated cancers relies on the expression of two oncoproteins, E6 and E7, which are the only viral gene products consistently found in these tumors (6, 7). Although these oncoproteins do not exhibit enzymatic function, their transforming activity is mediated primarily through protein-protein interactions that ultimately favor the formation of a replication-competent environment that eventually leads to cancer (8). Specifically, hrHPV E6 targets the p53 tumor suppressor protein for degradation, thereby preventing p53 from mediating cell cycle arrest and apoptosis in response to cellular stress signals (9, 10). In contrast, hrHPV E7 promotes degradation of the retinoblastoma tumor suppressor (pRb) protein, eliciting E2F-mediated transcriptional activation of S-phase genes (2, 10). Importantly, both HPV E6 and E7 trigger epigenetic changes in chromatin by altering the expression or the enzymatic activity of a number of epigenetic modifiers, such as histone deacetylases, histone demethylases, histone acetyltransferases, and histone methyltransferases (11–19). Concomitantly, the oncogenic stimuli triggered by HPV oncoproteins cause host cells to mount an antiviral innate immune response. Nonetheless, HPVs have evolved strategies to subvert antiviral immunity in order to complete their viral life cycle and persist in the host cell (20–24).
In recent studies, we demonstrated that in NIKSmcHPV18 keratinocytes carrying episomal HPV18, as well as in HeLa cells harboring an integrated HPV18 genome, induction of both beta interferon (IFN-β) and IFN-λ1 by DNA ligands is significantly impaired compared to that of parental cells (25). Furthermore, we found that downregulation of stimulator of IFN genes (STING), cyclic GMP-AMP synthase (cGAS), and retinoic acid-inducible gene I (RIG-I) mRNA levels occurs at the transcriptional level through a novel epigenetic silencing mechanism based on the accumulation of repressive heterochromatin marks, especially H3Lys9me2 (H3K9me2), at the promoter region of these genes (25). The incorporation of histone marks in chromatin represents a dynamic balance between enzymes depositing the mark (writers) and other enzymes removing it (erasers) (26). In this regard, SUV39H1, the human homolog of the Drosophila Su(var)3-9 histone methyltransferase, is the prime histone code writer responsible for histone H3Lys9 trimethylation (H3K9me3), which marks chromatin in a closed conformation (27, 28).
In this study, we show that SUV39H1 is involved in epigenetic silencing of RIG-I, cGAS, and STING genes in hrHPV-transformed cells. Importantly, pharmacological or genetic inhibition of SUV39H1 restored the innate immune response to exogenous DNA, as reflected by the production of both IFN-β and -λ1. SUV39H1 upregulation was dependent on E7 protein expression, as demonstrated by either loss- or gain-of-function experiments. In particular, we show that loss of E7 expression in both HeLa and CaSki cells significantly enhanced IFN production upon poly(dA·dT) or RIG-I agonist M8 transfection, predominantly through RIG-I signaling.
RESULTS
SUV39H1 increases heterochromatin formation at the promoter regions of RIG-I, cGAS, and STING genes in HPV-transformed cells.
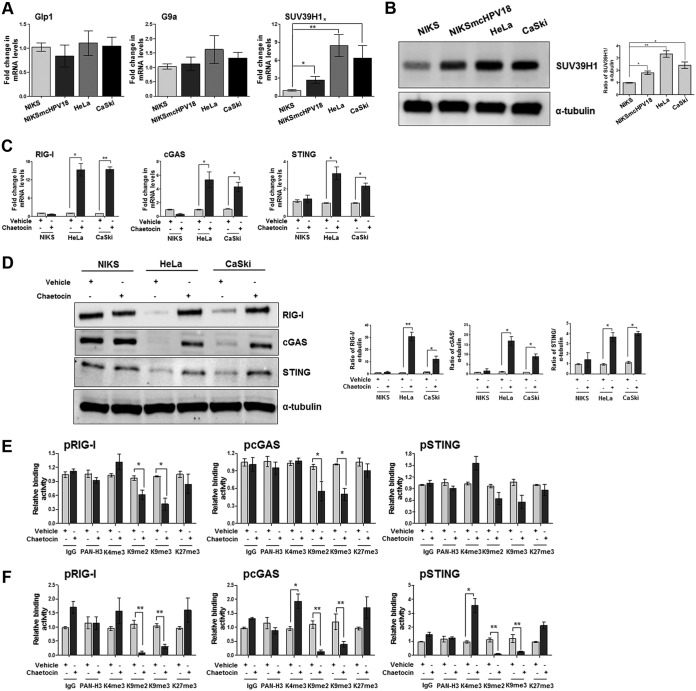
To determine which histone modifier enzyme was responsible for HPV-driven epigenetic modifications of the innate immune response, RNA extracts from NIKS, NIKSmcHPV18, or HeLa cells were analyzed for mRNA expression levels of the three major H3K9-specific methyltransferases, G9a-like protein (Glp1), G9a, and SUV39H1 (27). CaSki cells were also included in our analysis because they harbor an integrated HPV16 genome, another high-risk alpha genotype (29, 30). As shown in Fig. 1A, SUV39H1 mRNA levels were significantly upregulated in HPV-transformed versus NIKS cells, especially in HeLa and CaSki cells (8- and 6-fold, respectively), while Glp1 and G9a mRNA levels were only marginally modulated. A similar increase in SUV39H1 protein was also seen in Western blot analysis (Fig. 1B).
FIG 1.
Pharmacological inhibition of the H3K9-specific histone methyltransferase SUV39H1 decreases heterochromatin in hrHPV-transformed cells. (A) Transcript levels of the indicated genes were assessed by qPCR, and values were normalized to those for GAPDH, with the NIKS value set to 1. Data are presented as mean values from biological triplicates. Error bars indicate SD. *, P < 0.05; **, P < 0.01 (unpaired t test). (B) NIKS, NIKSmcHPV18, HeLa, and CaSki total cell extracts were subjected to immunoblot analysis with anti-SUV39H1 and anti-tubulin antibodies. The densitometry values of SUV39H1 were normalized to those of tubulin. Values are representative of three independent experiments. Error bars indicate SD. *, P < 0.05; **, P < 0.01 (unpaired t test). (C) NIKS, HeLa, and CaSki cells were treated with chaetocin (150 nM) or vehicle (dimethyl sulfoxide [DMSO]). After 24 h, transcript levels of the indicated genes were assessed by qPCR, and the values were normalized to those for GAPDH, with each vehicle-treated value set to 1. Data are presented as mean values from biological triplicates. Error bars indicate SD. *, P< 0.05; **, P< 0.01 (unpaired t test). (D) NIKS, HeLa, and CaSki cells were treated with chaetocin (150 nM) or vehicle (DMSO). After 24 h, total cell extracts were subjected to immunoblot analysis with anti-RIG-I, cGAS, STING, and anti-tubulin antibodies. The intensities of the bands for each antibody were quantified by densitometry, and ratios of the abundance of these proteins relative to that of tubulin were calculated. Values are representative of three independent experiments. Error bars indicate SD. *, P < 0.05; **, P < 0.01 (unpaired t test). (E and F) Extracts were prepared from HeLa (E) or CaSki cells (F) treated for 24 h with chaetocin (150 nM) or vehicle (DMSO). ChIP assay was carried out using antibodies specific for unmodified histone H3 (PAN-H3), trimethylated lysine 4 of H3 (H3K4me3), dimethylated lysine 9 of H3 (H3K9me2), trimethylated lysine 9 of H3 (H3K9me3), trimethylated lysine 27 of H3 (H3K27me3), or IgG as a control. Immunoprecipitated promoter sequences were measured by qPCR, and CT values for the samples were equated to input CT values. Values are represented as relative binding activity from three independent experiments. Error bars indicate SD. *, P < 0.05; **, P < 0.01 (unpaired t test).
To further define the mechanistic role of SUV39H1, we assessed mRNA and protein expression levels of RIG-I, cGAS, and STING genes in cells treated in the presence or absence of chaetocin, a pharmacological inhibitor of H3K9me3-specific methyltransferase (31, 32). For these experiments, HeLa and CaSki cells were chosen because they displayed higher basal levels of SUV39H1 protein than NIKSmcHPV18. As shown in Fig. 1C, RIG-I mRNA levels were significantly upregulated (15-fold) in both HeLa and CaSki cells after 24 h of chaetocin treatment, while cGAS and STING mRNA expression levels were also increased but to a lesser extent (5- and 3-fold in HeLa cells, respectively; 4- and 2.4-fold in CaSki cells, respectively). In contrast, NIKS cells, with low basal expression levels of SUV39H1 (Fig. 1A and B), did not show any significant variation in gene expression following chaetocin treatment (Fig. 1C). The same trend was also observed at the protein level for all three genes (Fig. 1D). Consistent with the aforementioned transcriptional activation, a significant decrease in H3K9me2 and H3K9me3 marks (i.e., repressive chromatin) associated with the promoter regions of RIG-I, cGAS, and STING was observed by chromatin immunoprecipitation (ChIP) assay in lysates of chaetocin-treated HeLa (Fig. 1E) and CaSki (Fig. 1F) cells. These findings indicate that the H3K9-specific methyltransferase SUV39H1, whose expression is significantly upregulated in HPV-transformed cells, is involved in the modeling of the repressive chromatin structure surrounding the RIG-I, cGAS, and STING promoters. Furthermore, pharmacological inhibition of SUV39H1 decreased the promoter-bound heterochromatin marks H3K9me2 and H3K9me3, likely switching the chromatin structure from a repressive to a permissive state.
Pharmacological and genetic inhibition of SUV39H1 activity restores IFN production in HPV-transformed cells upon poly(dA·dT) stimulation.
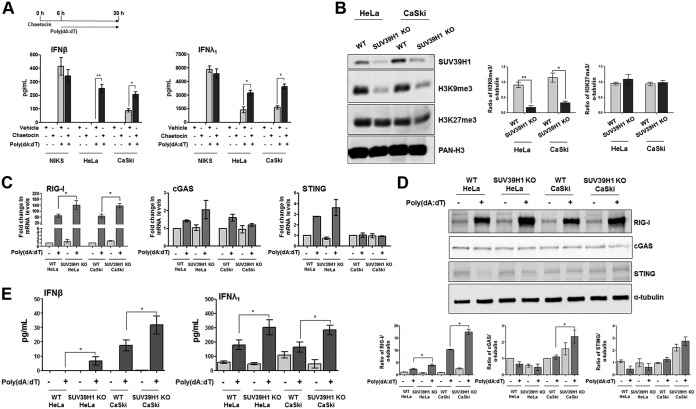
To determine if the drug-induced gain of function of RIG-I, cGAS, and STING increased IFN production upon stimulation with the DNA agonist poly(dA·dT), NIKS, HeLa, and CaSki cells were treated with chaetocin or vehicle for 6 h and transfected with poly(dA·dT) for 24 h, and supernatants were harvested to assess IFN production. Consistent with the results described above, IFN-β production was significantly higher in both chaetocin-treated HeLa and CaSki cells than in vehicle- or poly(dA·dT)-treated cells (Fig. 2A, left); a similar trend was also observed for IFN-λ1 (Fig. 2A, right). Consistent with the observed lack of SUV39H1 upregulation in NIKS cells (Fig. 1A and B), chaetocin treatment did not lead to a significant change in IFN production after poly(dA·dT) transfection (Fig. 2A).
FIG 2.
Pharmacological and genetic inhibition of SUV39H1 activity restores IFN production upon poly(dA·dT) transfection in HPV-transformed cells. (A) ELISA quantitation of IFN-β and IFN-λ1 protein in supernatants from cells treated with chaetocin (150 nM) or vehicle (DMSO) for 6 h and mock transfected or transfected with poly(dA·dT) for 24 h. Data are presented as mean values from biological triplicates. Error bars indicate SD. *, P < 0.05; **, P < 0.01 (unpaired t test). (B) Acid extracts from SUV39H1-deficient (KO) HeLa and CaSki or wild-type (WT) cells were subjected to immunoblot analysis with anti-SUV39H1, anti-H3K9me3, anti-H3K27me3, or anti-PAN-H3 antibodies. The densitometry values of protein bands were normalized to those of PAN-H3. Values are representative of three independent experiments. Error bars indicate SD. *, P < 0.05; **, P < 0.01 (unpaired t test). (C) Transcript levels of the indicated genes were assessed by qPCR in cells described in panel B, and values were normalized to those of GAPDH, with the WT mock-transfected cell value set to 1. Data are presented as mean values from biological triplicates. Error bars indicate SD. *, P < 0.05 (unpaired t test). (D) HeLa and CaSki SUV39H1 KO or control cells were subjected to immunoblot analysis with anti-RIG-I, anti-cGAS, anti-STING, or anti-tubulin antibodies. The densitometry values of protein bands were normalized to those of tubulin. Values are representative of three independent experiments. Error bars indicate SD. *, P < 0.05 (unpaired t test). (E) ELISA quantitation of IFN-β and IFN-λ1 protein in supernatants from HeLa and CaSki SUV39H1 KO or control cells mock transfected or transfected with poly(dA·dT) for 24 h. Data are presented as mean values from biological triplicates. Error bars indicate SD. *, P < 0.05 (unpaired t test).
We next used a lentiCRISPR-based approach to disrupt SUV39H1 in both HeLa and CaSki cells and confirmed protein loss by immunoblotting (Fig. 2B). Accordingly, H3K9me3 expression levels were decreased in SUV39H1 KO cells versus control cells (4- and 2.5-fold in HeLa and CaSki cells, respectively), while H3K27me3 levels remained unchanged (Fig. 2B). Consistent with the results observed in chaetocin-treated cells (Fig. 1C and D), upregulation of RIG-I mRNA expression levels upon poly(dA·dT) transfection was higher in SUV39H1 KO cells than in parental cells (2-fold in both HeLa and CaSki cells) (Fig. 2C). In contrast, poly(dA·dT) transfection failed to significantly induce both cGAS and STING mRNA levels in cells lacking SUV39H1 (Fig. 2C). The same trend was confirmed at the protein level, where RIG-I was upregulated by 1.5-fold in both poly(dA·dT)-transfected HeLa and CaSki cells compared to their similarly treated normal counterparts (Fig. 2D).
Cells next were treated with poly(dA·dT) to assess the innate immune response in terms of IFN released in the culture supernatants. As shown in Fig. 2E, IFN-β production increased by 4- and 2-fold in poly(dA·dT)-treated SUV39H1 KO HeLa and CaSki cells, respectively, compared to levels for their stimulated parental cells. A similar trend was observed for IFN-λ1 in both cell lines. Altogether, these observations demonstrate that pharmacological or genetic inhibition of SUV39H1 expression leads to modifications in the chromatin structure of the RIG-I, cGAS, and STING promoters, switching them from a repressive to a permissive status. The recovery of gene expression, especially in the case of RIG-I, then was able to restore the innate immune response to DNA ligands, as judged by the increased production of IFNs.
HPV E7 regulates SUV39H1 expression levels.
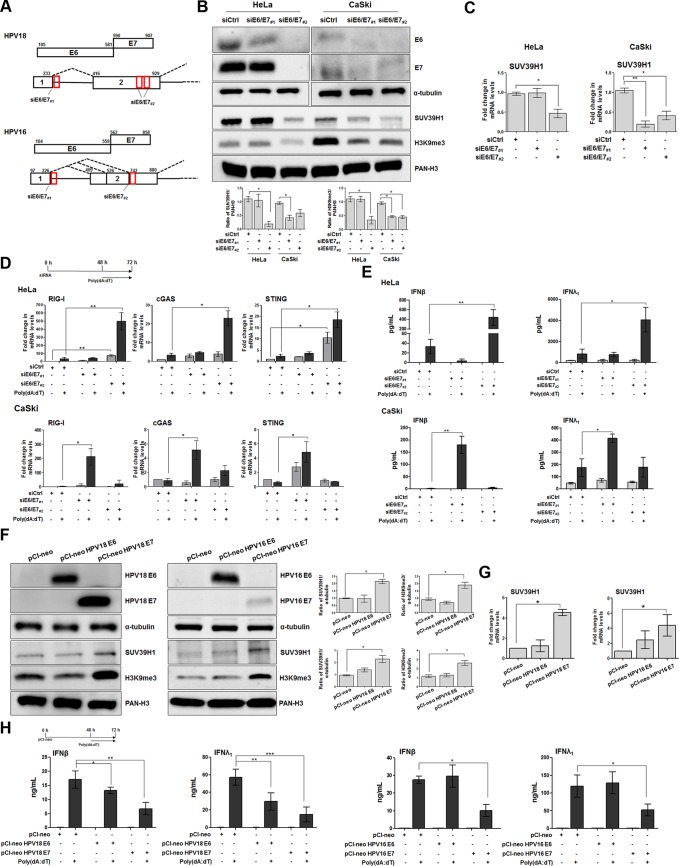
To determine which viral oncoprotein was responsible for the increase in SUV39H1 activity in hrHPV-transformed cells, E6 and E7 proteins were either silenced in HeLa and CaSki cells or overexpressed in HEK293 cells. Because E6 and E7 are transcribed as a single bicistronic pre-mRNA undergoing extensive alternative splicing, we used a short interfering RNA (siRNA) targeting the intron 1 region (siE6/E7#1), present only in unspliced RNA, which allowed us to knock down E6 expression in HeLa cells while only marginally affecting E7 expression (Fig. 3A). In addition, an exon 2-specific siRNA (siE6/E7#2) was used to simultaneously disrupt E6 and E7 expression in the same cells (33). As shown in Fig. 3B, SUV39H1 protein levels were downregulated in siE6/E7#2-transfected HeLa cells but not in cells silenced with siE6/E7#1, which were unable to inhibit E7 expression, or in siRNA control (siCtrl) cells, suggesting that E7 but not E6 regulates SUV39H1 protein expression in these cells. Consistent with SUV39H1 inhibition, total H3K9me3 protein levels were only reduced in E7-silenced cells (Fig. 3B). Assessment of the mRNA expression levels of the SUV39H1 gene confirmed that depletion of HPV oncoproteins, mainly E7 in the case of HPV18, determined a significant transcriptional inhibition of this gene (Fig. 3C).
FIG 3.
HPV E7 oncoprotein regulates SUV39H1 expression levels. (A) Diagrams of HPV16 and HPV18 E6 and E7 open read frames (ORFs; boxes with E6 and E7 labels) and bicistronic pre-mRNA transcripts with exons (boxes) and introns (lines between boxes). Numbers above the ORFs and bicistronic transcripts are nucleotide positions in each viral genome. Red boxes indicate siRNA oligonucleotide target sites. (B) Total extracts from HeLa or CaSki cells transfected with siE6/E7#1, siE6/E7#2, or siCtrl for 72 h were subjected to immunoblot analysis with anti-E6, anti-E7, or anti-tubulin, and acid extracts from the same set of samples were probed with anti-SUV39H1, anti-H3K9me3, or anti-PAN-H3. Densitometry values of protein bands were normalized to those of PAN-H3 (acid extracts) or tubulin (total extracts). Values are representative of three independent experiments. Error bars indicate SD. *, P < 0.05 (unpaired t test). (C) Transcript levels of SUV39H1 were assessed by qPCR in the cells described in panel B. Values were normalized to those of GAPDH, with siCtrl value set to 1. Data are presented as mean values from biological triplicates. Error bars indicate SD. *, P < 0.05; **, P < 0.01 (unpaired t test). (D) Transcript levels of the indicated genes were assessed by qPCR in HeLa (upper) or CaSki (lower) cells transfected with siE6/E7#1, siE6/E7#2, or siCtrl for 48 h and then mock transfected or transfected with poly(dA·dT) for 24 h. Values were normalized to those of GAPDH, with siCtrl-mock-transfected cells value set to 1. Values are representative of three independent experiments. Error bars indicate SD. *, P < 0.05; **, P < 0.01 (unpaired t test). (E) ELISA quantification of IFN-β and IFN-λ1 protein in supernatants from the cells described in panel B, mock transfected or transfected with poly(dA·dT) for 24 h. Data are presented as mean values from biological triplicates. Error bars indicate SD. *, P < 0.05; **, P < 0.01 (unpaired t test). (F) Total or acid extracts from HEK293 cells transfected with pCI-neo, pCI-neo HPV18 E6, pCI-neo HPV18 E7, pCI-neo HPV16 E6, or pCI-neo HPV16 E7 for 72 h were subjected to immunoblot analysis with anti-E6, anti-E7, or anti-tubulin antibodies (all total extracts), while anti-SUV39H1, anti-H3K9me3, or anti-PAN-H3 were used for acid extracts. Densitometry values of protein bands were normalized to those of PAN-H3 (acid extracts) or tubulin (total extracts). Values are representative of three independent experiments. Error bars indicate SD. *, P < 0.05 (unpaired t test). (G) Transcript levels of SUV39H1 were assessed by qPCR in HEK293 cells transfected with pCI-neo, pCI-neo HPV18 E6, pCI-neo HPV18 E7, pCI-neo HPV16 E6, or pCI-neo HPV16 E7 for 72 h. Values were normalized to those of GAPDH, with the pCI-neo-transfected value set to 1. Data are presented as mean values from biological triplicates. Error bars indicate SD. *, P < 0.05 (unpaired t test). (H) ELISA quantification of IFN-β and IFN-λ1 protein in supernatants from the cells described in panels F and G, mock transfected or transfected with poly(dA·dT) for 24 h. Data are presented as mean values from biological triplicates. Error bars indicate SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired t test).
The same siRNA sets were also used in CaSki cells, in which selected ablation was not achieved, however, given that both siE6/E7#1 and siE6/E7#2 were able to knock down both oncoproteins, albeit to different extents (Fig. 3B). As expected, transfection of either siE6/E7#1 or siE6/E7#2, both capable of shutting down E7 protein expression, but not siCtrl resulted in downregulation of SUV39H1 protein levels. Fittingly, H3K9me3 protein expression was significantly inhibited in both E7-silenced cells but not in siCtrl-transfected cells.
Altogether, these findings indicate that E7 plays a major role in SUV39H1 transcriptional activation and in the ensuing epigenetic silencing of the innate response. Accordingly, mRNA expression levels of RIG-I, cGAS, and STING genes were significantly increased after poly(dA·dT) transfection in siE6/E7#2-silenced HeLa cells (Fig. 3D, upper). Of note, transcriptional activation of the RIG-I gene was induced 70-fold in unstimulated and 15-fold in siE6/E7#2-transfected HeLa cells (Fig. 3D, upper). A similar trend was also observed in E6/E7-silenced CaSki cells: 9-fold induction in unstimulated and 110-fold induction in stimulated siE6/E7#1-transfected CaSki cells (Fig. 3D, lower). Importantly, siE6/E7#1 generally led to a more robust transcriptional activation of all three genes compared to levels in cells transfected with siE6/E7#2, in good agreement with the stronger SUV39H1 inhibition shown in Fig. 3A and B.
Consistent with the restoration of PRR expression, IFN-β and IFN-λ1 production was significantly higher in siE6/E7#2- versus siCtrl-transfected HeLa cells following poly(dA·dT) stimulation (18-fold for IFN-β and 10-fold for IFN-λ1, respectively) (Fig. 3E, upper). IFN production was also significantly enhanced in siE6/E7#1-transfected CaSki cells stimulated with poly(dA·dT) compared to similarly treated siCtrl cells (140-fold for IFN-β and 2.5-fold for IFN-λ1) (Fig. 3E, lower).
In parallel, HEK293 cells expressing either E6 or E7 from HPV18 or HPV16 were evaluated for SUV39H1 expression and global H3K9 trimethylation. Interestingly, SUV39H1 expression was increased in all E7-expressing cells compared to that of control cells (2- and 2.4-fold induction in HEK293 cells expressing HPV18 and HPV16, respectively) (Fig. 3F), while SUV39H1 expression was unchanged in E6-expressing cells. Consistently, E7, but not E6, reproducibly increased total H3K9me3 marks (Fig. 3F). Furthermore, SUV39H1 was upregulated at the mRNA level in E7-expressing cells, indicating that the induction occurred at the transcriptional level (Fig. 3G). Lastly, poly(dA·dT)-mediated IFN-β and IFN-λ1 production was significantly inhibited in both HPV18 E6- and E7-expressing cells, although to a much higher extent in E7- than E6-expressing cells (Fig. 3H, 1st and 2nd panels). In contrast, a clear-cut picture emerged in the case of HPV16, where only the E7 protein significantly downregulated the release of both IFN-β and IFN-λ1 upon poly(dA·dT) transfection compared to control cells (Fig. 3H, 3rd and 4th panels).
RIG-I is essential to regain the innate immune response in hrHPV-transformed cells.
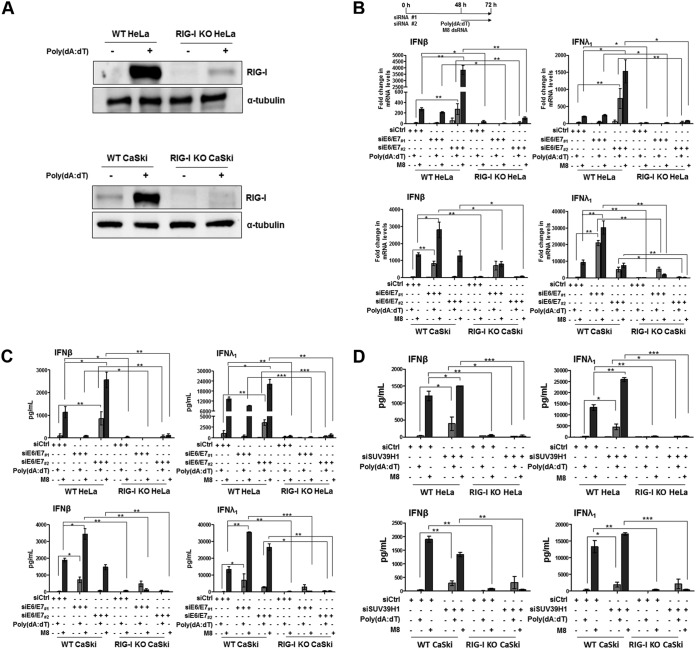
To further test the hypothesis that RIG-I activation in response to E7-mediated downregulation of SUV39H1 is responsible for restoration of IFN inducibility, we asked whether knockdown of RIG-I expression by lentiCRISPR would prevent IFN gene upregulation and protein secretion in HeLa and CaSki cells. RIG-I disruption (RIG-I knockout [KO]) was confirmed by immunoblotting under basal conditions or upon poly(dA·dT) transfection (Fig. 4A). In addition, cells were also stimulated with the sequence-optimized 5′pppRNA RIG-I specific agonist M8 (34–36). IFN transcriptional activation and secretion then was measured under basal conditions or in E6/E7-silenced cells in the presence or absence of the aforementioned stimuli (Fig. 4B and C). Notably, M8 turned out to be a much stronger IFN inducer than poly(dA·dT) in either cell line, especially in the case of IFN-β. Similar to what was observed for poly(dA·dT), M8 treatment of E6/E7-depleted cells induced higher levels of both IFN-β and IFN-λ1 at either the mRNA or protein level than wild-type (WT) cells. Consistent with the results reported in Fig. 3D, E7 silencing by siE6/E7#2 in HeLa or siE6/E7#1 in CaSki cells reestablished agonist-mediated IFN-β and IFN-λ1 transcription and secretion (Fig. 4B and C, respectively). In contrast, IFN inducibility in response to either agonist in RIG-I KO HeLa cells was not restored, indicating that RIG-I is required for IFN induction in HPV-transformed cells. Intriguingly, siE6/E7#1-transfected RIG-I KO CaSki cells displayed a significant reduction in IFN production by M8 compared with that of siCtrl-transfected or siE6/E7#1-transfected WT CaSki cells, especially with regard to IFN-β (Fig. 4C, lower). On the other hand, following poly(dA·dT) transfection, siE6/E7#1-treated RIG-I KO CaSki cells showed levels of IFN-β/IFN-λ1 secretion comparable to those observed in treated WT CaSki cells (Fig. 4C, lower), implying that the absence of RIG-I signaling is compensated for by the cGAS-STING pathway.
FIG 4.
RIG-I is crucial for the innate immune response in hrHPV-transformed cells. (A) Total extracts from HeLa and CaSki RIG-I-deficient (RIG-I KO) or wild-type (WT) cells, mock transfected or transfected with poly(dA·dT) for 24 h, were subjected to immunoblot analysis with anti-RIG-I or anti-tubulin antibodies. One representative Western blot from three independent triplicates is shown. (B) Transcript levels of IFN-β and IFN-λ1 genes were assessed by qPCR in RIG-I KO HeLa (upper) and CaSki (lower) or WT cells transfected with siE6/E7#1, siE6/E7#2, or siCtrl for 48 h and then mock transfected or transfected with poly(dA·dT) or M8 for 24 h. Values were normalized to those of GAPDH, with the value for siCtrl-mock-transfected cells set to 1. Values are representative of three independent experiments. Error bars indicate SD. *, P < 0.05; **, P < 0.01 (unpaired t test). (C) ELISA quantitation of IFN-β and IFN-λ1 protein in supernatants from the cells described in panel B. Data are presented as mean values from biological triplicates. Error bars indicate SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired t test). (D) ELISA quantification of IFN-β and IFN-λ1 protein in supernatants from RIG-I KO HeLa (upper) and CaSki (lower) or WT cells, transfected with siCtrl or siSUV39H1 for 48 h and mock transfected or transfected with poly(dA·dT) or M8 for 24 h. Data are presented as mean values from biological triplicates. Error bars indicate SD. *, P < 0.05; **, P < 0.01; ***, P < 0.001, (unpaired t test).
Similar results were obtained by directly silencing the SUV39H1 gene in both parental and RIG-I KO cells (Fig. 4D). Once again, M8-mediated induction of IFN-β and IFN-λ1 secretion was significantly enhanced in SUV39H1-silenced WT HeLa cells compared to that of parental cells and more abundant than that observed in poly(dA·dT)-transfected cells (Fig. 4D, upper). A similar trend was observed in siSUV39H1-transfected WT CaSki cells for IFN-λ1 in response to poly(dA·dT) or M8 transfection (Fig. 4D, lower right). In the case of IFN-β (Fig. 4D, lower left), the enhancement in siSUV39H1-transfected WT CaSki cells was more evident upon poly(dA·dT) transfection than M8 transfection. Consistent with the results shown in Fig. 4C (upper), in RIG-I KO HeLa cells, IFN production was almost abolished in response to either poly(dA·dT) or M8 transfection; in siSUV39H1 RIG-I KO CaSki cells the activity of M8 was dramatically reduced, whereas that of poly(dA·dT)-treated cells remained similar to that observed in siSUV39H1-transfected WT CaSki cells stimulated with poly(dA·dT) (Fig. 4D, lower).
Altogether, these findings clearly indicate that the RIG-I pathway plays a functional role in IFN production in hrHPV-transformed cells. Furthermore, RIG-I signaling can be substantially upregulated by inhibiting either E7 or SUV39H1 expression.
DISCUSSION
We recently reported that downregulation of RIG-I, cGAS, and STING mRNA levels in hrHPV-harboring cells occurs at the transcriptional level through a novel epigenetic silencing mechanism, as shown by the presence of repressive heterochromatin marks at the promoter region of these genes (25). In the present study, we expand on those findings and show that in hrHPV-transformed cells the increase in the repressive H3K9me2 and H3K9me3 marks is achieved through transcriptional induction of the H3K9-specific methyltransferase SUV39H1 (37). Specifically, we demonstrate that both pharmacological inhibition and gene silencing of SUV39H1 negatively affect the binding of H3K9me2 and H3K9me3 to the promoter region of RIG-I, cGAS, and STING genes. The reduction of these two repressive marks at the promoter regions of the aforementioned genes was closely followed by gene transcriptional activation, even in the absence of any exogenous stimulus. When SUV39H1 KO cells were treated with poly(dA·dT), the release of both IFN-β and IFN-λ1 was significantly increased compared to that of stimulated parental cells. Of note, the impact of SUV39H1 activity on chromatin structure in HeLa cells harboring an integrated HPV18 was similar to that observed in CaSki cells containing an integrated HPV16, indicating that these two high-risk genotypes, accounting for the majority of HPV-related cancers (2), have developed evolutionarily conserved strategies in order to epigenetically overturn key players of the innate immune response.
We also demonstrate that SUV39H1 upregulation is predominantly dependent on E7 protein expression, as demonstrated by either loss- or gain-of-function experiments. In particular, we show that loss of E7 expression in both HeLa and CaSki cells boosts the innate immune response through inhibition of SUV39H1 activity and transcriptional activation of genes upstream of the IFN cascade, especially RIG-I, which is followed by a substantial increase in IFN production upon poly(dA·dT) transfection.
The importance of the RIG-I pathway in hrHPV-transformed cells also emerged when we used a strong RIG-I agonist, M8 (34–36). Under basal conditions, M8 treatment of HeLa and CaSki cells was sufficient to achieve robust IFN production compared to poly(dA·dT)-stimulated cells, indicating that the host immune response was strictly dependent on the intrinsic performance of the agonist. Consistent with the results obtained in poly(dA·dT)-transfected cells, this induction was further enhanced in SUV39H1- or E7-depleted cells. Knockdown of the RIG-I gene in hrHPV-transformed cells ablates IFN induction after M8 but not poly(dA·dT) transfection, further confirming that inhibition of SUV39H1 activity preferentially rescues the RIG-I pathway rather than the cGAS-STING pathway.
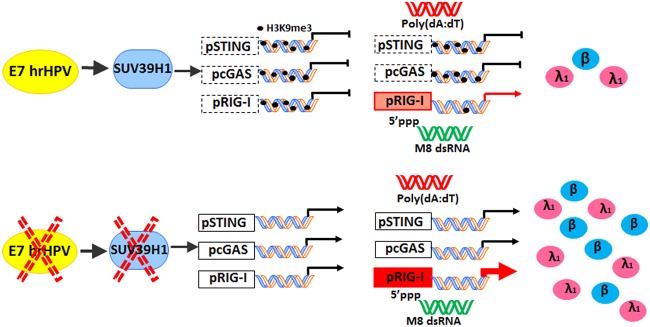
Collectively, these findings demonstrate that drug-targeted activation of the RIG-I signaling pathway may be a feasible option to trigger the innate immune response in HPV-transformed cells, which could improve the effectiveness of existing anticancer therapies (38–44). In summary, the present study describes a novel mechanism whereby impairment of the innate immune response in hrHPV-transformed cells occurs through E7-mediated transcriptional downregulation of RIG-I, cGAS, and STING and is dependent on SUV39H1 activity. As summarized in Fig. 5, our findings show an unprecedented role of SUV39H1 methyltransferase in switching the chromatin status from permissive to repressive, which in turn dampens the innate immune response in hrHPV-transformed cells.
FIG 5.
Schematic model representing the impact of SUV39H1 activity on the promoter region of RIG-I, cGAS, and STING genes under basal conditions or after transfection with poly(dA·dT) or the RIG-I agonist M8. In the lower panel, the same cellular circuits are illustrated under conditions where E7 or SUV39H1 is inhibited.
MATERIALS AND METHODS
Cell culture, plasmids, transfection, and treatments.
The spontaneously immortalized human keratinocyte cell line NIKS (Stratatech Corporation) were cultured in the presence of J2 3T3 fibroblast feeders as previously described (45). HeLa and HEK293 cells were grown in Dulbecco’s modified Eagle’s medium (Sigma-Aldrich) and CaSki cells in RPMI (Thermo Fisher Scientific), both supplemented with 10% fetal bovine serum (Sigma-Aldrich). NIKSmcHPV18 cells, stably harboring a high viral load of HPV18 episomal genomes, were obtained and cultured as previously described (25).
Chaetocin (150 nM) was obtained from Sigma-Aldrich. Poly(dA·dT) (1.25 μg/ml) (InvivoGen) was transfected into cells using Lipofectamine 3000 according to the manufacturer’s instructions (Thermo Fisher Scientific). M8 5′pppRNAs (100 ng/ml) was generated as previously described (35) and transfected using Lipofectamine RNAiMax transfection reagent as recommended by the manufacturer (Thermo Fisher Scientific).
HeLa and CaSki cells were transfected with siRNA using Lipofectamine RNAiMax transfection reagent (Invitrogen). The following siRNAs were used: SUV39H1 (M-009604-02-0005; siGENOME SMARTpool siRNA) and control siRNA (D-001206-13-05; siGENOME nontargeting siRNA pool) were purchased from Dharmacon; siRNAs against RIG-I and E6/E7#1 HPV18 were synthesized by Dharmacon, whereas siRNAs against E6/E7#2 HPV18, E6/E7#1 HPV16, and E6/E7#2 HPV16 were synthesized by Sigma-Aldrich. E6/E7#1 HPV18 siRNA sequences were kindly provided by Lawrence Banks and are available through him (46). The siRNA sequences were the following: HPV18 E6E7#1 forward, 5′-CUCUGUGUAUGGAGACACAU-3′; HPV18 E6E7#1 reverse, 5′-AUGUGUCUCCAUACACAGAG-3′; HPV18 E6E7#2 a forward, 5′-CGAUGAAAUAGAUGGAGUU-3′; HPV18 E6E7#2 a reverse, 5′-AACUCCAUCUAUUUCAUCG-3′; HPV18 E6E7#2 b forward, 5′-GCUAGUAGUAGAAAGCUCA-3′; HPV18 E6E7#2 b reverse, 5′-UGAGCUUUCUACUACUAGC-3′; HPV16 E6E7#1 forward, 5′-GACAGAGCCCAUUACAAUA-3′; HPV16 E6E7#1 reverse, 5′-UAUUGUAAUGGGCUCUGUC-3′; HPV16 E6E7#2 forward, 5′-GCACACACGUAGACAUUCG-3′; and HPV16 E6E7#2 reverse, 5′-CGAAUGUCUACGUGUGUGC-3′.
HPV16 and HPV18 E6 or E7 genes were subcloned into pCI-neo mammalian expression vector (Promega) within compatible SalI/EcoRI or XbaI/EcoRI restriction enzyme sites, respectively. The primer sequences used for these amplifications were the following: HPV18 E6 EcoRI_forward, 5′-cgGAATTCATGGCGCGCTTTGAGGAT-3′; HPV18 E6 SalI_reverse, 5′-acgcGTCGACTTATACTTGTGTTTCTCTGCG-3′; HPV18 E7 EcoRI_forward, 5′-cgGAATTCATGCATGGACCTAAGGCA-3′; HPV18 E7 XbaI_reverse, 5′-gcTCTAGATTACTGCTGGGATGCACAC-3′; HPV16 E6 EcoRI_forward, 5′-cgGAATTCATGCACCAAAAGAGAACT-3′; HPV16 E6 SalI_reverse, 5′-acgcGTCGACTTACAGCTGGGTTTCTCTACG-3′; HPV16 E7 EcoRI_forward, 5′-cgGAATTCATGCATGGAGATACACCT-3′; HPV16 E7 XbaI_reverse, 5′-gcTCTAGATTATGGTTTCTGAGAACAGAT-3′.
All constructs were sequenced (Eurofins), and overexpression was confirmed by Western blotting. HEK293 cells were transiently transfected with pCI-neo vector expressing HPV16 or HPV18 E6, E7, or empty vector control (1 μg) using Lipofectamine 3000 according to the manufacturer’s instructions (Thermo Fisher Scientific).
Quantitative nucleic acid analysis.
Real-time quantitative reverse transcription (qRT)-PCR analysis was performed on a CFX96 real-time system (Bio-Rad Laboratories Srl). Total RNA was extracted using TRI Reagent (Sigma-Aldrich), and 1 μg was retrotranscribed using an iScript cDNA synthesis kit (Bio-Rad Laboratories Srl). Reverse-transcribed cDNAs were amplified in duplicate using SensiFast SYBR (Bioline) for cellular genes. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) housekeeping gene was used to normalize for variations in cDNA levels. The reaction conditions consisted of an enzyme activation cycle of 30 s at 95°C, 40 cycles of 10 s of denaturation at 95°C, and 10 s of annealing at 60°C. The primers sequences used to amplify the indicated genes were the following: Glp1 forward, 5′-CTGACACAGAGGACAGGAAGC-3′; Glp1 reverse, 5′-TCTCGAACTTCTCTGGGATCTT-3′; G9a forward, 5′-TCCGACAGCAAGTCTGAAGTT-3′; G9a reverse, 5′-TGACTGATTCCCTGACTCCTC-3′; SUV39H1 forward, 5′-CATGGAGTACGTGGGAGAGAT-3′; SUV39H1 reverse, 5′-CCTGACGGTCGTAGATCTGG-3′; GAPDH forward, 5′-AGTGGGTGTCGCTGTTGAAGT-3′; GAPDH reverse, 5′-AACGTGTCAGTGGTGGACCTG-3′; RIG-I forward, 5′-CTGGACCCTACCTACATCCTG-3′; RIG-I reverse, 5′-GGCATCCAAAAAGCCACGG-3′; cGAS forward, 5′-ATGCAAAGGAAGGAAATGGT-3′; cGAS reverse, 5′-TTTAAACAATCTTTCCTGCAACA-3′; STING forward, 5′-GAGCAGGCCAAACTCTTCTG-3′; STING reverse, 5′-TGCCCACAGTAACCTCTTCC-3′; IFN-β forward, 5′-GTCTCCTCCAAATTGCTCTC-3′; IFN-β reverse, 5′-ACAGGAGCTTCTGACACTGA-3′; IFN-λ1 forward, 5′-CGCCTTGGAAGAGTCACTCA-3′; IFN-λ1 reverse, 5′-GAAGCCTCAGGTCCCAATTC-3′.
Immunoblotting.
Whole-cell protein extracts were prepared and subjected to immunoblot analysis as previously described (47). The nuclear proteins were obtained by acidic extraction by resuspending cell pellets in 200 μl of ice-cold lysis buffer (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol, and 200 mM HCl) supplemented with protease (Sigma-Aldrich) and phosphatase inhibitor cocktail (Active Motif). Cells were kept on ice for 30 min, and then the histone-enriched supernatants were collected by centrifugation at 4°C. Samples were subsequently precipitated with eight volumes of acetone overnight, centrifuged, and air dried, and pellets were resuspended in deionized water.
The following antibodies were used: rabbit monoclonal antibody anti-SUV39H1 (diluted 1:1,000; 702443; Thermo Fisher Scientific), rabbit polyclonal antibodies anti-cGAS (diluted 1:500; HPA031700; Sigma-Aldrich), RIG-I (diluted 1:10,000; 06-1040; Merck Millipore), anti-H3K9me3 (diluted 1:500; 07-442; Merck Millipore), anti-H3K27me3 (diluted 1: 20,000; 07-449; Merck Millipore), anti-HPV18 E6 (diluted 1:250; GTX132687; GeneTex), anti-HPV18 E7 (diluted 1:500; GTX133412; GeneTex), anti-HPV16 E6 (diluted 1:500; GTX32686; GeneTex), and anti-HPV16 E7 (diluted 1:500; GTX133411; GeneTex) and mouse monoclonal antibody (MAb) anti-STING (1:1,500; MAB7169; R&D Systems). MAb against α-tubulin (diluted 1:4,000; 39527; Active Motif) and rabbit antibody against unmodified histone H3 (diluted 1:15,000; 06-755; Merck Millipore) were used as a control for protein loading. Immunocomplexes were detected using sheep anti-mouse or donkey anti-rabbit immunoglobulin antibodies conjugated to horseradish peroxidase (HRP) (GE Healthcare Europe GmbH) and visualized by enhanced chemiluminescence (Super Signal West Pico; Thermo Fisher Scientific). Images were acquired, and densitometry of the bands was performed using Quantity One software (version 4.6.9; Bio-Rad Laboratories Srl). Densitometry values were normalized using the corresponding loading controls.
ChIP assay.
ChIP assays were performed as previously described (45). Immunoprecipitation was performed with 3 μg of unmodified histone H3 (06-755), dimethyl-histone H3 (Lys4; 07-030), dimethyl-histone H3 (Lys9; 07-441), trimethyl-histone H3 (Lys9; 07-442), and trimethyl-histone H3 (Lys27; 07-449) antibodies, all purchased from Merck Millipore (Merck Millipore SpA). Threshold cycle (CT) values for the samples were equated to input CT values to provide percentages of input for comparison, and these were normalized to the enrichment level of unmodified histone H3 for each cell line. The primers used to amplify RIG-I, cGAS, and STING promoters were the following: pRIG-I forward, 5′-TCTCCCCAAGTTTATCTGTAGGT-3′; pRIG-I reverse, 5′-TGAATCCCTGTGCATTTCCA-3′; pSTING forward, 5′-TAGCTCTTGTTACCGGAA-3′; pSTING reverse, 5′-ATGATTCCCCGTCTCATA-3′; pcGAS forward, 5′-CTCTTGCGGCCTTTTCCTG-3′; and pcGAS reverse, 5′-CTCTCTGCATGGCCTTTCC-3′.
ELISA.
The cytokines secreted in the culture supernatants were analyzed using single analyte human enzyme-linked immunosorbent assay (ELISA) kits for IFN-β (DY814-05; DuoSet ELISA human IFN-β; R&D Systems) and IFN-λ1 (DY7246; DuoSet ELISA human IL-29/IFN-λ1; R&D Systems) according to the manufacturer’s instructions. All absorbance readings were measured at 450 nm using a Victor X4 multilabel plate reader (Perkin Elmer).
Generation of SUV39H1 and RIG-I knockout HeLa and CaSki cells.
SUV39H1 or RIG-I (DDX58, for DExD-Hbox helicase 58) knockout cells were generated with CRISPR/Cas9 technology using single guide RNA (sgRNA) obtained from Applied Biological Materials Inc. (no. K2317005_SUV39H1, K0575405_DDX58, and K010_ scrambled sgRNA; All-in-One Lentivectors). To produce viral particles, HEK293T cells were transfected with an All-in-One Lentivector set encoding Cas9 and SUV39H1, DDX58, or scrambled sgRNAs alongside 2nd Generation Packaging System Mix (Applied Biological Materials, Inc.) using Lipofectamine 2000 (Invitrogen). Viral supernatants were harvested at 72 h postinfection and used to transduce cells by infection in the presence of 8 mg/ml Polybrene. Transduced HeLa or CaSki cells were selected with puromycin (4 μg/ml) 48 h postinfection over the course of 14 days postransduction. After selection, successful knockout was confirmed by immunoblotting.
Statistical analysis.
All statistical tests were performed using GraphPad Prism version 5.00 for Windows (GraphPad Software). The data are presented as means ± standard deviations (SD). For comparisons consisting of two groups, means were compared using two-tailed Student’s t tests. Differences were considered statistically significant at a P value of <0.05.
ACKNOWLEDGMENTS
We thank Marcello Arsura for critically reviewing the manuscript.
This work was supported by the Italian Ministry for University and Research-MIUR (PRIN 2017 to C.B. and M.G.) and the AGING Project–Department of Excellence–DIMET, University of Piemonte Orientale.
REFERENCES
- 1.Van Doorslaer K, Ruoppolo V, Schmidt A, Lescroël A, Jongsomjit D, Elrod M, Kraberger S, Stainton D, Dugger KM, Ballard G, Ainley DG, Varsani A. 2017. Unique genome organization of non-mammalian papillomaviruses provides insights into the evolution of viral early proteins. Virus Evol 3:vex027. doi: 10.1093/ve/vex027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Doorbar J, Quint W, Banks L, Bravo IG, Stoler M, Broker TR, Stanley MA. 2012. The biology and life-cycle of human papillomaviruses. Vaccine 30(Suppl 5):F55–F70. doi: 10.1016/j.vaccine.2012.06.083. [DOI] [PubMed] [Google Scholar]
- 3.Egawa N, Egawa K, Griffin H, Doorbar J. 2015. Human papillomaviruses; epithelial tropisms, and the development of neoplasia. Viruses 7:3863–3890. doi: 10.3390/v7072802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Galloway DA, Laimins LA. 2015. Human papillomaviruses: shared and distinct pathways for pathogenesis. Curr Opin Virol 14:87–92. doi: 10.1016/j.coviro.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Groves IJ, Coleman N. 2015. Pathogenesis of human papillomavirus-associated mucosal disease. J Pathol 235:527–538. doi: 10.1002/path.4496. [DOI] [PubMed] [Google Scholar]
- 6.Hoppe-Seyler K, Bossler F, Braun JA, Herrmann AL, Hoppe-Seyler F. 2018. The HPV E6/E7 oncogenes: key factors for viral carcinogenesis and therapeutic targets. Trends Microbiol 26:158–168. doi: 10.1016/j.tim.2017.07.007. [DOI] [PubMed] [Google Scholar]
- 7.Moody CA, Laimins LA. 2010. Human papillomavirus oncoproteins: pathways to transformation. Nat Rev Cancer 10:550–560. doi: 10.1038/nrc2886. [DOI] [PubMed] [Google Scholar]
- 8.McLaughlin-Drubin ME, Münger K. 2009. Oncogenic activities of human papillomaviruses. Virus Res 143:195–208. doi: 10.1016/j.virusres.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Talis AL, Huibregtse JM, Howley PM. 1998. The role of E6AP in the regulation of p53 protein levels in human papillomavirus (HPV)-positive and HPV-negative cells. J Biol Chem 273:6439–6445. doi: 10.1074/jbc.273.11.6439. [DOI] [PubMed] [Google Scholar]
- 10.Mittal S, Banks L. 2017. Molecular mechanisms underlying human papillomavirus E6 and E7 oncoprotein-induced cell transformation. Mutat Res Rev Mutat Res 772:23–35. doi: 10.1016/j.mrrev.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 11.Rincon-Orozco B, Halec G, Rosenberger S, Muschik D, Nindl I, Bachmann A, Ritter TM, Dondog B, Ly R, Bosch FX, Zawatzky R, Rösl F. 2009. Epigenetic silencing of interferon-kappa in human papillomavirus type 16-positive cells. Cancer Res 69:8718–8725. doi: 10.1158/0008-5472.CAN-09-0550. [DOI] [PubMed] [Google Scholar]
- 12.McLaughlin-Drubin ME, Crum CP, Münger K. 2011. Human papillomavirus E7 oncoprotein induces KDM6A and KDM6B histone demethylase expression and causes epigenetic reprogramming. Proc Natl Acad Sci U S A 108:2130–2135. doi: 10.1073/pnas.1009933108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Durzynska J, Lesniewicz K, Poreba E. 2017. Human papillomaviruses in epigenetic regulations. Mutat Res Rev Mutat Res 772:36–50. doi: 10.1016/j.mrrev.2016.09.006. [DOI] [PubMed] [Google Scholar]
- 14.Soto D, Song C, McLaughlin-Drubin ME. 2017. Epigenetic alterations in human papillomavirus-associated cancers. Viruses 9:248. doi: 10.3390/v9090248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Soto DR, Barton C, Munger K, McLaughlin-Drubin ME. 2017. KDM6A addiction of cervical carcinoma cell lines is triggered by E7 and mediated by p21CIP1 suppression of replication stress. PLoS Pathog 13:e1006661. doi: 10.1371/journal.ppat.1006661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gautam D, Johnson BA, Mac M, Moody CA. 2018. SETD2-dependent H3K36me3 plays a critical role in epigenetic regulation of the HPV31 life cycle. PLoS Pathog 14:e1007367. doi: 10.1371/journal.ppat.1007367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Brehm A, Nielsen SJ, Miska EA, McCance DJ, Reid JL, Bannister AJ, Kouzarides T. 1999. The E7 oncoprotein associates with Mi2 and histone deacetylase activity to promote cell growth. EMBO J 18:2449–2458. doi: 10.1093/emboj/18.9.2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langsfeld ES, Bodily JM, Laimins LA. 2015. The deacetylase sirtuin 1 regulates human papillomavirus replication by modulating histone acetylation and recruitment of DNA damage factors NBS1 and Rad51 to viral genomes. PLoS Pathog 11:e1005181. doi: 10.1371/journal.ppat.1005181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Munger K, Jones DL. 2015. Human papillomavirus carcinogenesis: an identity crisis in the retinoblastoma tumor suppressor pathway. J Virol 89:4708–4711. doi: 10.1128/JVI.03486-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hong S, Laimins LA. 2017. Manipulation of the innate immune response by human papillomaviruses. Virus Res 231:34–40. doi: 10.1016/j.virusres.2016.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Westrich JA, Warren CJ, Pyeon D. 2017. Evasion of host immune defenses by human papillomavirus. Virus Res 231:21–33. doi: 10.1016/j.virusres.2016.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Krump NA, You J. 2018. Molecular mechanisms of viral oncogenesis in humans. Nat Rev Microbiol 16:684–698. doi: 10.1038/s41579-018-0064-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang C, Pauli EK, Biryukov J, Feister KF, Meng M, White EA, Münger K, Howley PM, Meyers C, Gack MU. 2018. The human papillomavirus E6 oncoprotein targets USP15 and TRIM25 to suppress RIG-I-mediated innate immune signaling. J Virol 92:e01737-17. doi: 10.1128/JVI.01737-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau L, Gray EE, Brunette RL, Stetson DB. 2015. DNA tumor virus oncogenes antagonize the cGAS-STING DNA-sensing pathway. Science 350:568–571. doi: 10.1126/science.aab3291. [DOI] [PubMed] [Google Scholar]
- 25.Albertini S, Lo Cigno I, Calati F, De Andrea M, Borgogna C, Dell'Oste V, Landolfo S, Gariglio M. 2018. HPV18 persistence impairs basal and DNA ligand-mediated IFN-β and IFN-λ(1) production through transcriptional repression of multiple downstream effectors of pattern recognition receptor signaling. J Immunol 200:2076–2089. doi: 10.4049/jimmunol.1701536. [DOI] [PubMed] [Google Scholar]
- 26.Zhang T, Cooper S, Brockdorff N. 2015. The interplay of histone modifications–writers that read. EMBO Rep 16:1467–1481. doi: 10.15252/embr.201540945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fritsch L, Robin P, Mathieu JR, Souidi M, Hinaux H, Rougeulle C, Harel-Bellan A, Ameyar-Zazoua M, Ait-Si-Ali S. 2010. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell 37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 28.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, Opravil S, Doyle M, Sibilia M, Jenuwein T. 2001. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell 107:323–337. doi: 10.1016/S0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 29.Meissner JD. 1999. Nucleotide sequences and further characterization of human papillomavirus DNA present in the CaSki, SiHa and HeLa cervical carcinoma cell lines. J Gen Virol 80:1725–1733. doi: 10.1099/0022-1317-80-7-1725. [DOI] [PubMed] [Google Scholar]
- 30.Xu F, Cao M, Shi Q, Chen H, Wang Y, Li X. 2015. Integration of the full-length HPV16 genome in cervical cancer and Caski and Siha cell lines and the possible ways of HPV integration. Virus Genes 50:210–220. doi: 10.1007/s11262-014-1164-7. [DOI] [PubMed] [Google Scholar]
- 31.Kaniskan HÜ, Konze KD, Jin J. 2015. Selective inhibitors of protein methyltransferases. J Med Chem 58:1596–1629. doi: 10.1021/jm501234a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. 2005. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat Chem Biol 1:143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 33.Tang S, Tao M, McCoy JP Jr, Zheng ZM. 2006. The E7 oncoprotein is translated from spliced E6*I transcripts in high-risk human papillomavirus type 16- or type 18-positive cervical cancer cell lines via translation reinitiation. J Virol 80:4249–4263. doi: 10.1128/JVI.80.9.4249-4263.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goulet ML, Olagnier D, Xu Z, Paz S, Belgnaoui SM, Lafferty EI, Janelle V, Arguello M, Paquet M, Ghneim K, Richards S, Smith A, Wilkinson P, Cameron M, Kalinke U, Qureshi S, Lamarre A, Haddad EK, Sekaly RP, Peri S, Balachandran S, Lin R, Hiscott J. 2013. Systems analysis of a RIG-I agonist inducing broad spectrum inhibition of virus infectivity. PLoS Pathog 9:e1003298. doi: 10.1371/journal.ppat.1003298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiang C, Beljanski V, Yin K, Olagnier D, Ben Yebdri F, Steel C, Goulet ML, DeFilippis VR, Streblow DN, Haddad EK, Trautmann L, Ross T, Lin R, Hiscott J. 2015. Sequence-specific modifications enhance the broad-spectrum antiviral response activated by RIG-I agonists. J Virol 89:8011–8025. doi: 10.1128/JVI.00845-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Beljanski V, Chiang C, Kirchenbaum GA, Olagnier D, Bloom CE, Wong T, Haddad EK, Trautmann L, Ross TM, Hiscott J. 2015. Enhanced influenza virus-like particle vaccination with a structurally optimized RIG-I agonist as adjuvant. J Virol 89:10612–10624. doi: 10.1128/JVI.01526-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rea S, Eisenhaber F, O'Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtler K, Ponting CP, Allis CD, Jenuwein T. 2000. Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406:593–599. doi: 10.1038/35020506. [DOI] [PubMed] [Google Scholar]
- 38.Patel SA, Minn AJ. 2018. Combination cancer therapy with immune checkpoint blockade: mechanisms and strategies. Immunity 48:417–433. doi: 10.1016/j.immuni.2018.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Grivennikov SI, Greten FR, Karin M. 2010. Immunity, inflammation, and cancer. Cell 140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hopcraft SE, Damania B. 2017. Tumour viruses and innate immunity. Philos Trans R Soc B 372:20160267. doi: 10.1098/rstb.2016.0267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shekarian T, Valsesia-Wittmann S, Brody J, Michallet MC, Depil S, Caux C, Marabelle A. 2017. Pattern recognition receptors: immune targets to enhance cancer immunotherapy. Ann Oncol 28:1756–1766. doi: 10.1093/annonc/mdx179. [DOI] [PubMed] [Google Scholar]
- 42.Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. 2013. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer 13:759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 43.Langsfeld E, Laimins LA. 2016. Human papillomaviruses: research priorities for the next decade. Trends Cancer 2:234–240. doi: 10.1016/j.trecan.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Smola S. 2017. Immunopathogenesis of HPV-associated cancers and prospects for immunotherapy. Viruses 9:254. doi: 10.3390/v9090254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lo Cigno I, De Andrea M, Borgogna C, Albertini S, Landini MM, Peretti A, Johnson KE, Chandran B, Landolfo S, Gariglio M. 2015. The nuclear DNA sensor IFI16 acts as a restriction factor for human papillomavirus replication through epigenetic modifications of the viral promoters. J Virol 89:7506–7520. doi: 10.1128/JVI.00013-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ganti K, Massimi P, Manzo-Merino J, Tomaić V, Pim D, Playford MP, Lizano M, Roberts S, Kranjec C, Doorbar J, Banks L. 2016. Interaction of the human papillomavirus E6 oncoprotein with sorting nexin 27 modulates endocytic cargo transport pathways. PLoS Pathog 12:e1005854. doi: 10.1371/journal.ppat.1005854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gugliesi F, Mondini M, Ravera R, Robotti A, de Andrea M, Gribaudo G, Gariglio M, Landolfo S. 2005. Up-regulation of the interferon-inducible IFI16 gene by oxidative stress triggers p53 transcriptional activity in endothelial cells. J Leukoc Biol 77:820–829. doi: 10.1189/jlb.0904507. [DOI] [PubMed] [Google Scholar]