Poxvirus genome replication, transcription, translation, and virion assembly occur at sites within the cytoplasm known as factories. Some poxviruses sequester infectious virions outside of the factories in inclusion bodies comprised of numerous copies of the 150-kDa ATI protein, which can provide stability and protection in the environment. We provide evidence that ATI mRNA is anchored by nascent peptides and translated at the inclusion sites rather than in virus factories. Association of ATI mRNA with inclusion bodies allows multiple rounds of local translation and prevents premature ATI protein aggregation and trapping of virions within the factory.
KEYWORDS: RNA trafficking, inclusion body, local translation, poxvirus, vaccinia virus
ABSTRACT
On-site translation of mRNAs provides an efficient means of subcellular protein localization. In eukaryotic cells, the transport of cellular mRNAs to membraneless sites usually occurs prior to translation and involves specific sequences known as zipcodes that interact with RNA binding and motor proteins. Poxviruses replicate in specialized cytoplasmic factory regions where DNA synthesis, transcription, translation, and virion assembly occur. Some poxviruses embed infectious virus particles outside of factories in membraneless protein bodies with liquid gel-like properties known as A-type inclusions (ATIs) that are comprised of numerous copies of the viral 150-kDa ATI protein. Here, we demonstrate by fluorescent in situ hybridization that these inclusions are decorated with ATI mRNA. On-site translation is supported by the localization of a translation initiation factor eIF4E and by ribosome-bound nascent chain ribopuromycylation. Nascent peptide-mediated anchoring of ribosome-mRNA translation complexes to the inclusions is suggested by release of the mRNA by puromycin, a peptide chain terminator. Following puromycin washout, relocalization of ATI mRNA at inclusions depends on RNA and protein synthesis but requires neither microtubules nor actin polymerization. Further studies show that the ATI mRNAs remain near the sites of transcription in the factory regions when stop codons are introduced near the N terminus of the ATI or large truncations are made at the N or C termini. Instead of using a zipcode, we propose that ATI mRNA localization is mediated by ribosome-bound nascent ATI polypeptides that interact with ATI protein in inclusions and thereby anchor the complex for multiple rounds of mRNA translation.
IMPORTANCE Poxvirus genome replication, transcription, translation, and virion assembly occur at sites within the cytoplasm known as factories. Some poxviruses sequester infectious virions outside of the factories in inclusion bodies comprised of numerous copies of the 150-kDa ATI protein, which can provide stability and protection in the environment. We provide evidence that ATI mRNA is anchored by nascent peptides and translated at the inclusion sites rather than in virus factories. Association of ATI mRNA with inclusion bodies allows multiple rounds of local translation and prevents premature ATI protein aggregation and trapping of virions within the factory.
INTRODUCTION
The localization of cellular proteins at specific sites is often directed by features within the protein. However, an alternate mechanism involves the targeting and on-site translation of cellular mRNAs (1–4). The asymmetric cellular distribution of mRNA was first observed in oocytes and neuronal cells. However, cytoplasmic localization of specific mRNAs occurs in many cell types. Global analysis of mRNA localization during early Drosophila embryogenesis revealed that 71% of 3,370 genes analyzed encoded subcellularly localized RNAs (5). Trafficking of mRNA occurring prior to translation is mediated by cis-acting “zipcodes” in nontranslated sequences that interact with RNA binding and motor proteins in eukaryotes (1, 6). An example is β-actin mRNA, which has zipcode elements in the 3′ untranslated region that binds in trans to zipcode binding protein 1 (IGF2BP1) and engages actin motors (7). Cotranslational mRNA trafficking mediated by the signal recognition particle occurs extensively with proteins destined for the endoplasmic reticulum. However, there are only a few examples of cotranslational targeting of mRNA to nonmembrane sites, namely, recruitment of pericentrin to the centrosome, the microtubule minus-end regulator ASPM to mitotic spindle poles of vertebrate cells (8), and the ABP140 protein to the distal pole of Saccharomyces cerevisiae (9). The presence of polyribosome-like structures around A-type inclusions (ATIs) in cells infected with cowpox virus (CPXV) might be another example of mRNA trafficking (10). However, at the time of the latter study, tools were not available to identify the mRNA or nascent protein. The present study was intended to more deeply investigate the localization and translation of the viral mRNA encoding the ATI protein.
Poxviruses are large double-stranded DNA viruses that replicate entirely within the cytoplasm of infected cells and have been used to investigate many aspects of mRNA synthesis including 5′ capping and 3′ polyadenylation (11). Studies with vaccinia virus (VACV), the prototype member of the family, established that genome replication, transcription, translation, and virion assembly occur within juxtanuclear factories (12–14). Following egress from the assembly site, some infectious mature virions (MVs) are enveloped by a double-membrane derived from endosomal and Golgi networks and transported to the cell periphery on microtubules (15–19). Additionally, MVs of certain poxviruses including CPXV, ectromelia virus, raccoonpox virus, canarypox virus, and fowlpox virus become embedded in ATIs that are distant from the virus factory (20–22). ATIs are comprised of an abundant 150-kDa protein containing multiple repeats of about 30 amino acids each (23, 24). The ATIs are dynamic, mobile bodies with liquid gel-like properties that enlarge in part by coalescence events that depend on microtubules (25). Some CPXV strains lack the A26 protein, which is necessary for embedding virions, but still make inclusions. Other orthopoxviruses, such as VACV, variola virus, monkeypox virus, and camelpox virus have truncated forms of the ATI protein that do not form large inclusions and do not embed MVs (26, 27). Embedding of MVs in ATIs may provide long-term protection in the environment and promote animal to animal transmission. Here, we use fluorescent in situ hybridization (FISH) and epitope-tagged ATI protein to demonstrate anchoring and local translation of ATI mRNA at inclusions.
RESULTS
Detection of ATI mRNA at cytoplasmic inclusion bodies.
Our plan was to express an epitope-tagged ATI protein to visualize the inclusion bodies and use FISH to localize the ATI mRNA in infected HeLa cells. Although VACV, the prototype orthopoxvirus, does not form ATIs, a previous report (28) described a recombinant VACV in which the truncated ATI open reading frame (ORF) of VACV is replaced with one encoding the full-length CPXV ATI protein. The latter virus forms typical inclusion bodies in which virus particles are embedded, providing a model for ATI formation. ATI formation is independent of virion embedding, which requires the A26 protein. To focus specifically on ATI formation in the present study, we used the recombinant VACV (vATI-HA.ΔA26) that has a C-terminal HA tag attached to the ATI protein for antibody detection by fluorescence microscopy and has the A26 gene deleted. Cellular nuclear DNA and cytoplasmic viral DNA were detected by staining with 4′,6-diamidino-2-phenylindole (DAPI). Two methods were used to prepare probes for RNA hybridization. One method used bacteriophage T7 RNA polymerase and ATI DNA templates to synthesize individual digoxigenin-labeled antisense RNA hybridization probes in vitro (29). The other method utilized chemically synthesized multiple fluorescent-labeled antisense deoxyoligonucleotide probes that bind along the length of the mRNA (30, 31).
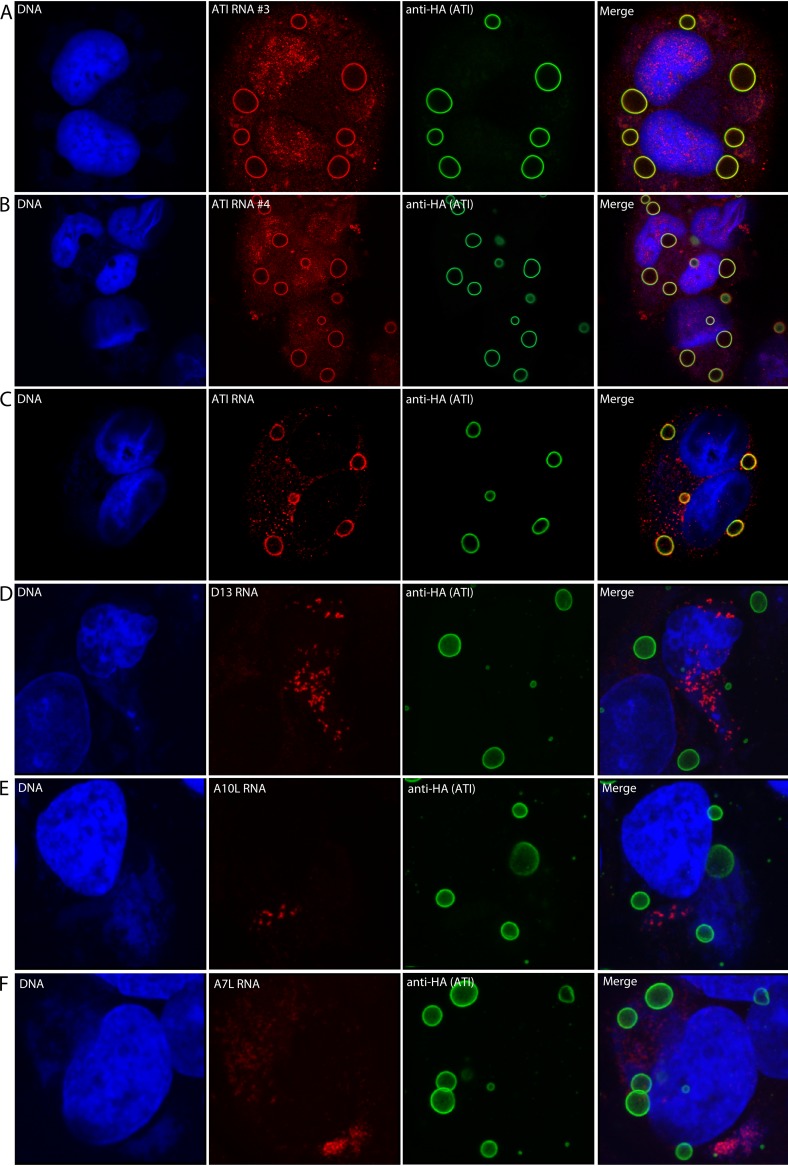
In Fig. 1A and B, individual digoxigenin antisense probes detected ATI RNA as intense rings around the ATI bodies in addition to some staining distributed over the cell. Anti-ATI antibody mainly binds to the surface of the inclusions, possibly due to prior fixation of the cells, thereby also appearing as rings (23, 28). Similar results were obtained using a pool of fluorescent-labeled deoxyoligonucleotide probes to detect ATI RNA (Fig. 1C). To evaluate the specificity of the ATI RNA localization, we used deoxyoligonucleotide probes to several other VACV mRNAs. The mRNAs encoding the D13 scaffold protein, the A10 core protein, and the A7 early transcription factor subunit were each detected in DNA factory areas with no labeling around the ATI bodies (Fig. 1D through F).
FIG 1.
Localization of ATI mRNA at inclusion bodies. HeLa cells were infected with vATI-HA.ΔA26, a recombinant VACV in which the CPXV ATI ORF with a C-terminal HA tag replaced the A25 ORF and the A26 ORF has been deleted. After 18 h, the cells were fixed and incubated with individual digoxigenin-labeled antisense ATI RNA probe 3 or 4 followed by fluorescently labeled antidigoxigenin antibody (A, B) or fluorescent antisense deoxyoligonucleotide probes to ATI (C), D13 (D), A10 (E), or A7 (F) mRNAs. The cells were also stained with anti-HA antibody, a fluorescent secondary antibody to detect the ATI protein and DAPI to label nuclear and cytoplasmic viral DNA. Panels from left to right show DNA stained with DAPI, RNA detected by FISH, ATI protein detected by antibody to the HA tag, and a merge. Panels A through C are Z sections; panels D through F are maximum intensity projections.
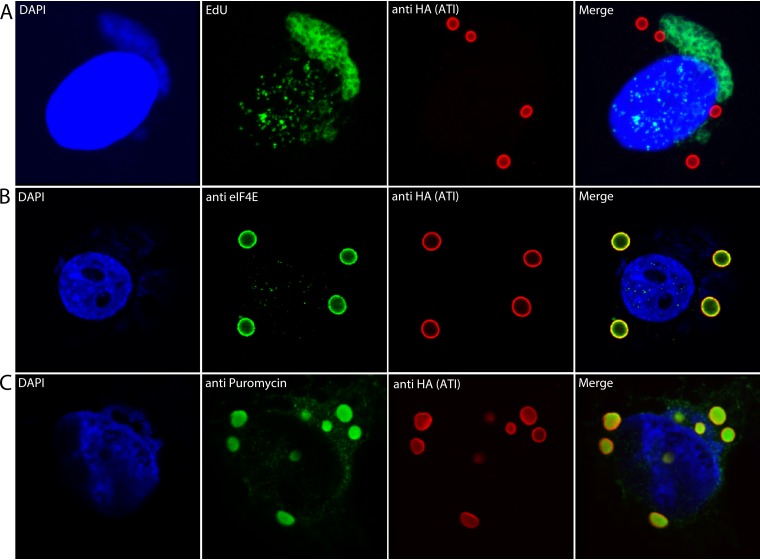
We considered the possibility that the ATI mRNA is synthesized at the inclusions, which would depend on viral DNA there. To provide more intense staining than DAPI, the cells were incubated with the thymidine analogue EdU for 2 to 8 h after infection. During this time, cellular DNA synthesis is largely shut down so EdU is incorporated mainly into viral DNA (32). Following fixation, click chemistry was used to label the DNA with Alexa Fluor 488. Confocal microscopy revealed strong staining of juxtanuclear viral factories, light staining of nuclei, but no detectable staining around inclusions (Fig. 2A). These data suggested that the ATI mRNA is synthesized in factories and subsequently localizes at sites of inclusion formation.
FIG 2.
Labeling of viral DNA in factories and translation of mRNA at inclusion bodies. (A) Labeling of viral DNA. HeLa cells were infected with vATI-HA.ΔA26 and incubated with EdU from 2 to 8 h. The cells were then fixed, permeabilized, and reacted with Alexa Fluor 488 azide. The cells were also stained with anti-HA antibody and a fluorescent secondary antibody to detect the ATI protein and DAPI to label nuclear and cytoplasmic viral DNA. (B) HeLa cells were infected as in Fig. 1 and incubated with mouse monoclonal anti-eIF4E and rabbit polyclonal anti-HA antibodies followed by fluorescent secondary antibodies and DAPI. (C) HeLa cells were infected as above and incubated with puromycin and cycloheximide for 30 min. After digitonin extraction and fixation, the cells were incubated with mouse monoclonal anti-puromycin and rabbit polyclonal HA antibodies followed by fluorescent secondary antibodies and DAPI. Maximum intensity projections are shown in panel A, and Z sections are shown in panels B and C. Representative images are shown.
Translation of ATI mRNA at inclusions.
The ATI promoter is exceptionally strong, and the protein is the most abundantly synthesized at late times of infection (10, 23, 33). If active translation occurs at ATIs, we would expect to find cellular translation factors there. We probed infected cells with an antibody specific for the eukaryotic translation initiation factor 4E (eIF4E), which binds to the cap structure at the 5′ ends of mRNAs, in addition to anti-HA antibody to visualize ATIs. As shown in Fig. 2B, eIF4E colocalized with the anti-HA antibody.
The method of ribosome-bound nascent chain ribopuromycylation (34) was used to directly demonstrate active translation. Puromycin is a tyrosine-tRNA mimetic that terminates translation by ribosome-catalyzed incorporation into the nascent peptide terminus (35). When puromycin is incorporated in the presence of a chain elongation inhibitor, the ribosome-bound nascent chain is immobilized and can be detected by fluorescence microscopy with a puromycin-specific monoclonal antibody. David et al. (34) previously used this technique in cells infected with VACV to show labeling of VACV factories. We detected puromycin labeling of the extended viral factory above and to the right of the nucleus and more intense labeling of the ATI bodies (Fig. 2C). Digitonin extraction, needed to remove free puromycin, may have contributed to the internal labeling of the ATI bodies with anti-HA antibody as well as puromycin.
Nascent peptides anchor mRNA at ATI sites.
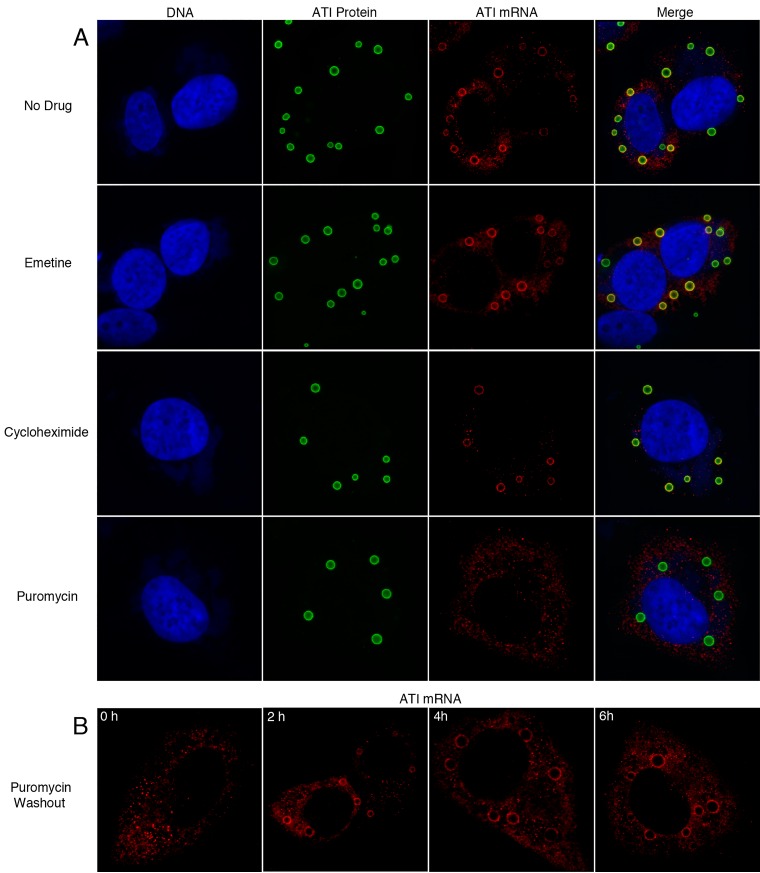
The association of ATI mRNA with inclusions could be achieved by sequences within the RNA or via the nascent protein or through other determinants. To investigate these possibilities, we compared the effects of puromycin, which releases the nascent peptide from ribosomes (36, 37), with cycloheximide (38, 39) and emetine (40), both of which inhibit translation and stabilize ribosomes on mRNA. These drugs were added to HeLa cells at 6 h after infection, and 30 min later, the cells were harvested in the presence of the antibiotics and processed for microscopic analysis of ATI RNA and proteins (Fig. 3A). ATI RNA was concentrated around the inclusions in untreated cells and cells treated with cycloheximide or emetine but not in cells treated with puromycin, where staining was distributed throughout the cytoplasm. The possible degradation of the displaced ATI RNA was not investigated. These results indicated that ribosome-associated ATI mRNA was anchored to the inclusions by the nascent peptide and released by puromycin.
FIG 3.
Displacement of ATI mRNA by puromycin. (A) HeLa cells were infected with vATI-HA.ΔA26 and at 6 h were incubated with either emetine, cycloheximide, or puromycin. After 30 min, the cells were fixed and incubated with fluorescent-labeled antisense ATI deoxyoligonucleotide probes to detect mRNA, with anti-HA antibody and secondary fluorescent antibody to detect ATI protein, and with DAPI to detect DNA. (B) HeLa cells were infected with vATI-HA.ΔA26 and at 6 h were incubated with puromycin for 30 min. The cells were then washed and incubated with drug-free medium and fixed 0, 2, 4, and 6 h and incubated with fluorescent-labeled antisense ATI deoxyoligonucleotide probes as in panel A. Z sections of typical cells are shown.
To investigate the reversibility of puromycin, the drug was washed out after a 30-min treatment. ATI RNA was detected around the preformed inclusions in some cells at 2 h and in the majority of cells after 4 and 6 h (Fig. 3B). Thus, the effects of puromycin were readily reversible.
Relocalization of mRNA around ATIs requires de novo RNA and protein synthesis.
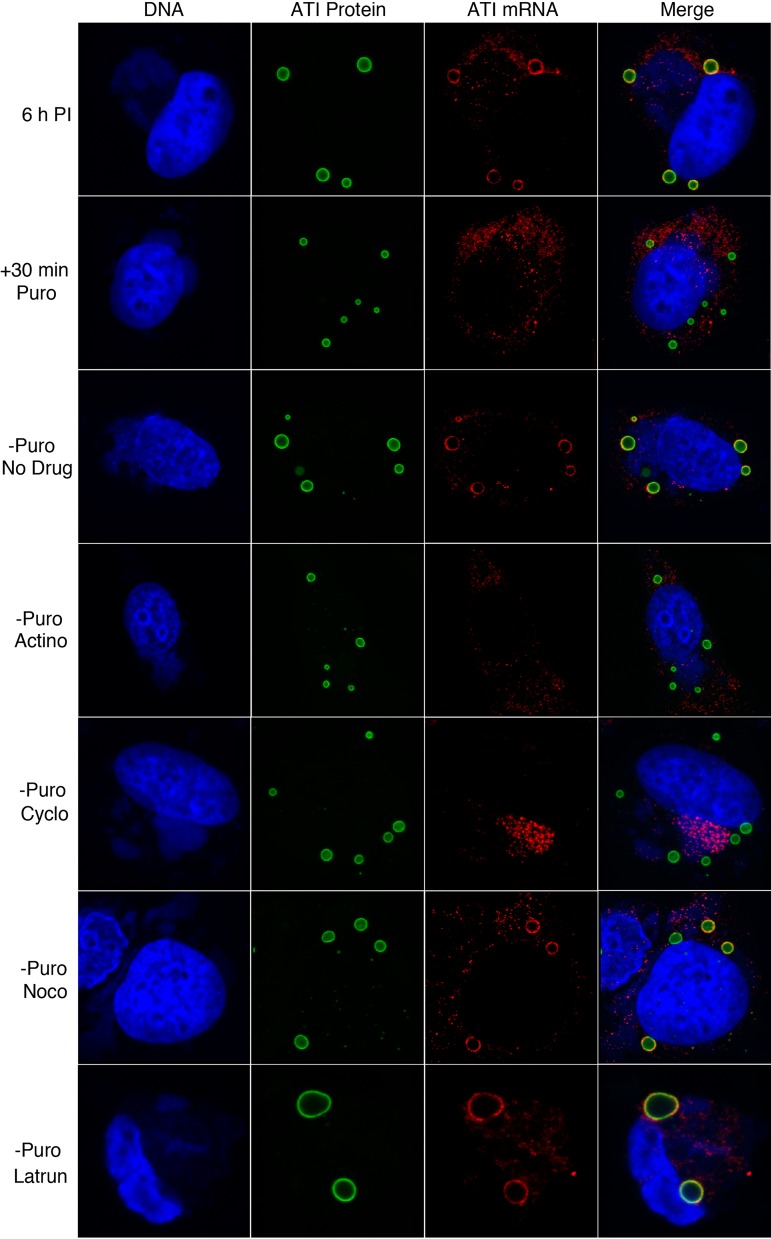
By stripping RNA from ATIs with puromycin, we could bypass the early steps of infection and specifically investigate the factors needed for RNA relocalization around existing ATIs. At 6 h after infection, the cells were incubated for 30 min in puromycin-containing medium, which was then replaced with medium without drugs as a control or with actinomycin D to inhibit new mRNA synthesis, cycloheximide to arrest protein synthesis, nocodazole to disrupt microtubules, or latrunculin to block actin polymerization (Fig. 4). After an additional 3 h, the cells were analyzed. In the absence of drugs, RNA around ATI was found in 75% of 200 cells examined. Actinomycin D prevented RNA localization around ATI, suggesting that new RNA was required. In the presence of cycloheximide, the ATI RNA remained in the virus factories indicating that protein synthesis was needed for cytoplasmic trafficking. In the presence of nocodazole, there was no reduction in the number of cells that had RNA around the ATI, indicating that microtubules were not required for localization. In the presence of latrunculin, RNA was present around ATIs in 45% of the cells examined. However, there were gross changes in the morphology of cells treated with this drug leading to loss of some cells from the coverslip, and in many cases the ATIs were increased in size apparently due to their coalescence. In summary, these data indicated that both new RNA and protein synthesis were required for localization of ATI RNA around preexisting ATIs but that neither microtubule dynamics nor actin polymerization was essential.
FIG 4.
Effects of inhibitors on relocalization of ATI mRNA around inclusions. HeLa cells were infected with vATI-HA.ΔA26 and at 6 h were incubated with puromycin for 30 min. The cells were then washed and incubated without puromycin for 3 h without drug (no drug) or with actinomycin D (Actino), cycloheximide (Cyclo), nocodazole (Noco), or latrunculin (Latrun). The cells were then fixed and probed with fluorescent-labeled antisense ATI deoxyoligonucleotide probes to detect ATI mRNA, with anti-HA antibody and secondary fluorescent antibody to detect ATI protein, and with DAPI to detect DNA. Z-sections of typical cells are shown.
Nascent ATI protein regulates localization of ATI mRNA.
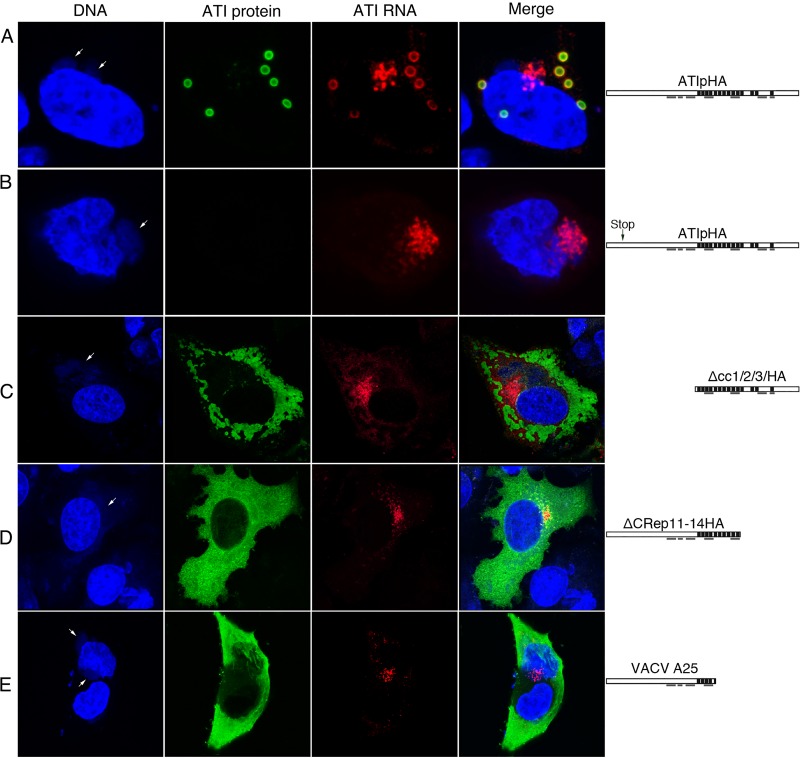
The above results suggested that protein synthesis is required for the exit of the ATI mRNA from the factory but did not distinguish between synthesis of protein generally and translation of ATI mRNA specifically. To inhibit ATI protein synthesis, we constructed two plasmids; each contained the natural ATI promoter followed by the ATI open reading frame (ORF) with a C-terminal HA tag. However, one had two stop codons near the N terminus while the other had an uninterrupted ORF. The plasmids were transfected into cells infected with a VACV that contained neither full-length nor truncated ATI ORFs. Therefore, only the transfected plasmids were capable of expressing the ATI RNA and protein. Because a VACV promoter was used for expression, the plasmids were obligatorily transcribed by the viral RNA polymerase (14). ATI bodies with associated ATI RNA formed when the plasmid with the unmodified ATI ORF was transfected, although in some cells RNA also remained in the factory (Fig. 5A). As expected, ATI were not formed when the plasmid with stop codons in the ORF was transfected; in addition, the ATI RNA remained mostly with the factory (Fig. 5B). The direct interpretation of the above results is that the nascent polypeptide is required for ATI RNA to exit the factory, although degradation of untranslated cytoplasmic RNA was not ruled out.
FIG 5.
Export of ATI mRNA from factory region is translation dependent. (A to D) HeLa cells were transfected with a plasmid expressing the uninterrupted or mutated ATI ORF with a C-terminal HA tag and after 4 h were infected with VACV vΔA25ΔΑ26, which lacks either the CPXV ATI ORF or the truncated VACV ATI ORF, for an additional 6 h. ATI RNA was detected by FISH using fluorescently labeled deoxyoligonucleotide antisense ATI RNA probes, ATI protein with anti-HA antibody, followed by fluorescent secondary antibody and DNA with DAPI. Diagrams of the expressed ATI proteins are shown on the right of each row. Repeat sequences are indicated by black bars, and coiled-coil domains are underlined. Arrows point to cytoplasmic factories. (E) Localization of ATI protein and mRNA expressed by VACV with the naturally truncated A25 ATI ORF. After 16 h, the cells were fixed and analyzed with fluorescent-labeled deoxyoligonucleotide antisense ATI RNA probes and antibody to the ATI protein followed by a fluorescent secondary antibody and DAPI. Arrows point to cytoplasmic factories. Maximum projections of typical cells are shown.
mRNA encoding N- or C-terminally truncated ATI protein localizes in the factory.
Next, we investigated the effects of truncations on the localization of the ATI protein and mRNA. Previous studies showed that N-terminal or C-terminal truncations of the ATI ORF prevented the formation of typical inclusion bodies, resulting in the dispersal of the ATI protein in the cytoplasm (28). To determine the effect of these truncations on the localization of the ATI mRNA, plasmids encoding truncated ATI proteins were transfected into cells that were infected with a VACV without either the CPXV ATI ORF or the truncated VACV A25 ORF. The ATI protein lacking the N-terminal 609 amino acids but retaining all 14 repeats of the 1,284-amino acid ATI protein was distributed in amorphous aggregates throughout the cytoplasm, whereas much of the RNA was detected near the DNA factories (Fig. 5C). A similar result was obtained when the ATI protein lacked the C terminus including 4 repeats (Fig. 5D). We also took advantage of the natural C-terminal truncation of the ATI in VACV strain WR, which is caused by stop codons, the first of which occurred within the fifth repeat of the A25 ORF (26). This 94-kDa ATI protein detected with antibody was distributed throughout the cytoplasm, whereas the mRNA detected by FISH was concentrated within or near the factory (Fig. 5E).
DISCUSSION
The present investigation was stimulated by an earlier study that showed electron microscopic evidence for the presence of polyribosomes around ATI that form during CPXV infection (10). This was an intriguing observation since poxvirus mRNA is synthesized and translated in cytoplasmic factories (14, 34, 41), which are usually distant from ATI inclusions. In the present study, the ATI mRNA was identified by FISH and resolved around the inclusions, whereas several other viral mRNAs examined remained near DNA factories. Furthermore, translation at inclusion sites was suggested by colocalization of the translation initiation factor eIF4E and substantiated by detection of puromycylated nascent peptide chains associated with the ATI. The nascent peptide was responsible for anchoring the RNA to the ATI since the RNA was displaced by puromycin, which dissociates peptides from ribosomes, but not by the translation inhibitors cycloheximide and emetine that stabilize the ribosomes on mRNA. These results contrast with previous findings for pretranslational actin mRNA localization in which neither the anchoring nor the trafficking of mRNA was affected by puromycin or cycloheximide (42) and are similar to examples of cotranslational eukaryotic mRNA localization (8, 9).
The possibility that the ATI RNA is synthesized at inclusion sites was considered; however, viral DNA could not be detected there by DAPI staining or labeling with the thymidine analog EdU. The ATI mRNA is likely synthesized in factories, the site of viral DNA synthesis and accumulation. This raises the question of how ATI mRNA localizes to the inclusions. Unlike cellular mRNAs that commonly have 5′ and 3′ untranslated sequences capable of accommodating a zipcode, poxvirus genes including the ATI are closely packed and have minimal or no untranslated sequences. Moreover, ATI RNA expressed from a plasmid that contained only the promoter and the precisely defined ORF localized around inclusions. Therefore, either an RNA zipcode resides within the ORF itself, or more likely another mechanism enables mRNA translocation. By stripping ATI RNA from the inclusions with puromycin, we determined the requirements for RNA relocalization after removal of the drug. In the absence of an inhibitor, ATI mRNA reappeared around the inclusions within 2 h. Actinomycin D, a transcription inhibitor, prevented relocalization indicating that de novo synthesized RNA was required. When protein synthesis was inhibited by cycloheximide, the newly synthesized ATI mRNA was detected at DNA factories where transcription occurs. Furthermore, the ATI mRNA localized near the transcription sites when stop codons were introduced near the N terminus of the ORF. The possibility that untranslated ATI RNA outside of the factory has a short half-life was not excluded. However, taken together, these data indicated that the nascent peptide is a key determinant for the release of ATI mRNA from the factory as well as anchoring mRNA at inclusion sites.
The structure of the ATI protein has not yet been determined. However, the 1,284-amino acid protein has a large number of charged amino acids and fourteen 30-amino acid repeats that are believed to be involved in their self-association to form inclusion bodies (23, 24). An ATI protein with an N-terminal truncation that resulted in deletion of the first 609 amino acids with retention of all 14 repeats or C-terminal truncations that deleted 4 to 9 repeats were abundantly synthesized and dispersed in amorphous aggregates throughout the cytoplasm rather than forming discrete nearly spherical inclusions (25). Despite the cytoplasmic location of the truncated ATI proteins, the ATI mRNA was detected largely at or near the viral factories, indicating that it was not translocated or was unstable elsewhere.
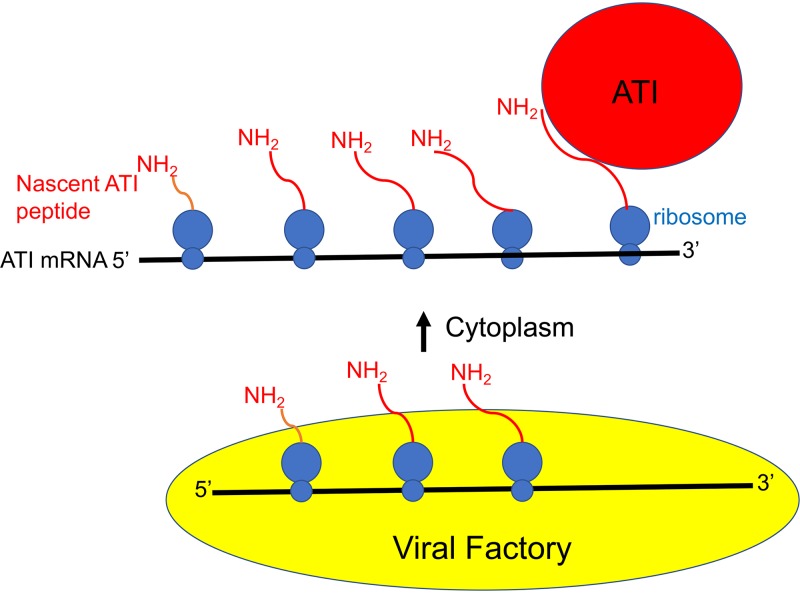
In summary, we have demonstrated local translation of ATI RNA at inclusions. However, our data do not fit the eukaryotic zipcode mRNA trafficking model for the following reasons: (i) zipcodes are mostly present in the 3′ untranslated region and occasionally in the 5′ untranslated, but the ATI coding region is sufficient for localization; (ii) zipcode localization does not require translation whereas the ATI RNA localization does; and (iii) zipcode localization involves actin polymerization whereas ATI RNA does not. According to the model depicted in Fig. 6, viral DNA is transcribed within or adjacent to the factory where translation initiation begins. The complex consisting of ribosomes, ATI mRNA, and nascent polypeptide is released into the cytoplasm. Interaction between the nascent ATI polypeptide and ATI protein in the inclusion anchors the mRNA-ribosome complex, which may reach the inclusion sites by random or other unknown mechanism, as inhibitors of microtubule dynamics and actin polymerization did not prevent this. We suggest that local translation of ATI mRNA at inclusions prevents premature aggregation of the ATI protein near transcription sites and allows the sequestering of virions outside of viral factories.
FIG 6.
Model for ATI mRNA anchoring and local translation at inclusion bodies. Transcription and translation of VACV DNA occurs within or adjacent to the viral factory. A translation complex comprised of ATI mRNA, associated ribosomes, and nascent peptides is shown emerging from the factory. Interactions between nascent ATI peptides and ATI proteins anchors the complex at inclusions where multiple cycles of translation occur.
MATERIALS AND METHODS
Cells and viruses.
HeLa cells were grown in Dulbecco’s minimum essential medium (DMEM) supplemented with 2 mM l-glutamine, 100 units penicillin, and 100 μg streptomycin per ml (Quality Biological, Inc.) and containing 10% fetal bovine serum (FBS) (Sigma-Aldrich). Recombinant viruses vATI+A26−, and vATI−A26− were described previously (25, 28). Standard methods were used for propagation and titration of VACV (43).
Plasmids.
The plasmids ΔNcc1/2/3HA and ΔCRep11-14HA have been described previously (28). The plasmid ΑΤΙ(ΗΑ) with 2 stop codons was constructed with the Q5 site-directed mutagenesis kit (New England BioLabs).
FISH hybridization probes.
To make digoxigenin-labeled probes, DNA primers of 50 nucleotides containing the T7 promoter followed by sequences complementary to segments of the CPXV ATI ORF were synthesized and used as templates for the T7 RNA polymerase using the Dig RNA labeling kit and digoxigenin-UTP (Sigma-Aldrich). Stellaris DNA probes conjugated to Quasar 570 of approximately 22 nucleotides complementary to the CPXV ATI and other VACV ORFs were obtained from Biosearch Technologies.
Inhibitors.
Translation inhibitors were used at the following concentrations and were obtained from Sigma-Aldrich: puromycin (50 μg/ml), cycloheximide (300 μg/ml), and emetine (5 μM). A 50× concentration of actinomycin (Calbiochem) was prepared by dissolving 250 mg in 800 μl of dimethyl sulfoxide (DMSO). Nocodazole (33.2 μM) and latrunculin (1 μM) were from Sigma-Aldrich and Cayman Chemicals, respectively.
Antibodies.
Sheep antidigoxigenin (catalog number 11333089001; Sigma-Aldrich), mouse anti-HA monoclonal antibody (MAb) (catalog number 901502; BioLegend) and rabbit anti-HA polyclonal (catalog number 902301; BioLegend), mouse MAb to anti-eIF4E (catalog number 9976; Santa Cruz Biotechnology), and Alexa Fluor 488 (Invitrogen) and Alexa Fluor 594 (Invitrogen) conjugated to anti-IgG of appropriate species were obtained from indicated commercial sources. Anti-puromycin mouse MAb 12D10 (44) and rabbit anti-ATI were provided by Jon Yewdell and David Pickup, respectively.
In situ hybridization and fluorescence microscopy.
The procedure was modified from Singer lab protocols (http://www.singerlab.org/protocols). HeLa cells were plated on 12-mm glass circular coverslips in a 24-well dish and the next day infected with 3 PFU/cell of VACV for 18 h. Cells were washed 3× with Dulbecco’s phosphate-buffered saline (DPBS), followed by fixation with 4% formaldehyde (Sigma-Aldrich). After 4 washes in DPBS, cells were permeabilized in 70% ethanol overnight at 4°C. Cells were rehydrated for 10 min in 2× saline sodium citrate (SSC) buffer (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) with 30% formamide. Cells were then hybridized overnight at 37°C in 300 μl of a mixture containing 50 ng of probe, 10% dextran sulfate (Sigma-Aldrich), 2 mM vanadyl-ribonucleoside complex (Sigma-Aldrich), 0.02% RNase-free bovine serum albumin (BSA) (Amresco), 40 μg Escherichia coli tRNA (Sigma-Aldrich), 2× SSC, and 30% formamide. Cells were washed 3× for 30 min at 37°C in 0.1× SSC with 30% formamide and then incubated with sheep antidigoxigenin for 4 h at 37°C in 2× SSC, 8% formamide, 2 mM vanadyl-ribonucleoside complex, and 0.2% RNase-free BSA (Sigma-Aldrich) and washed 3× for 20 min in 2× SSC, 8% formamide at room temperature followed by Alexa Fluor 488 (Invitrogen) and washed again. To detect ATI protein, cells were incubated for 4 h at 37°C with anti-HA antibody and diluted 1:100 in 2× SSC, 8% formamide, 2 mM vanadyl-ribonucleoside complex, and 0.2% BSA followed by Alexa Fluor 594 in the same buffer. To detect DNA, the cells were then incubated with 5 ng/ml DAPI for 30 min followed by washing and mounting onto slides using ProLong gold. Analysis was done using a Leica SP5 confocal microscope, and brightness and contrast were adjusted using Adobe Photoshop.
For FISH with Stellaris probes, the infected and washed cells were fixed for 10 min in 3.7% formaldehyde in DPBS, washed 4× in DPBS, and placed in 70% ethanol overnight. Ethanol was removed, and cells were washed 4× at 5-min intervals with wash buffer containing 10% formamide in 2× SSC buffer. Fluorescently labeled antisense deoxyoligonucleotides (Bioresearch Technologies) were resuspended in Tris-EDTA (TE) buffer (10 mM Tris-HCl, 1 mM sodium EDTA, pH 8.0) to give a final solution of 25 μM. The probe was diluted 1:100 with hybridization buffer (100 mg/ml dextran sulfate, 10% formamide in 2× SSC), added to cells, and incubated in a humidified incubator at 37°C overnight. The remaining steps to detect ATI protein and DNA were carried out as described above.
EdU labeling.
EdU at a final concentration of 10 μM was added to cells at 2 h after infection. At 8 h, the cells were washed three times with PBS containing Mg2+ and Ca2+ and fixed with 4% paraformaldehyde in DPBS for 15 min at room temperature. After fixation, the cells were washed three times in DPBS with 1% BSA. Cells were permeabilized with 0.1% Triton X-100 in DPBS with 1% BSA for 20 min at room temperature and then washed three times in DPBS with 1% BSA and subjected to click reaction with Alexa Fluor 488 azide according to the Click-iT EdU imaging kit protocol (Thermo Fisher).
Ribopuromycylation.
The procedure followed that of David et al. (34) with minor modifications. HeLa cells at ∼80% confluence were infected with 3 PFU/cell of virus for 7 h. The cells were incubated with 91 μM puromycin and 208 μM emetine in DMEM with 7.5% FBS for 5 min at 37°C. The following extraction procedures were performed on ice with prechilled reagents. Cells were incubated for 2 min with 500 μl/well of permeabilization buffer (50 mM Tris-HCl, pH 7.5, 5 mM MgCl2, 25 mM KCl, 355 μM cycloheximide, EDTA-free protease inhibitors [Sigma-Aldrich], and 10 U/ml RNaseOut [Thermo Fisher] containing 0.015% digitonin [Sigma-Aldrich]). After this extraction step, cells were washed once with 500 μl of permeabilization buffer without digitonin and fixed with 500 μl of 3% paraformaldehyde (Electron Microscopy Sciences) for 15 min at room temperature. The fixed cells were washed 4× with DPBS and maintained at 4°C before immunofluorescence staining.
ACKNOWLEDGMENTS
We thank Amanda Howard for initiating this study, Andrea Weisberg for help in preparing the figures, and other members of our laboratory for support. Jon Yewdell and David Pickup kindly provided anti-puromycin and anti-ATI antibodies, respectively.
This research was funded by the Division of Intramural research, NIAID, NIH.
B.M. planned experiments, analyzed data, and wrote manuscript. G.C.K. carried out experiments, analyzed data, and made figures.
REFERENCES
- 1.Jung H, Gkogkas CG, Sonenberg N, Holt CE. 2014. Remote control of gene function by local translation. Cell 157:26–40. doi: 10.1016/j.cell.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parton RM, Davidson A, Davis I, Weil TT. 2014. Subcellular mRNA localisation at a glance. J Cell Sci 127:2127–2133. doi: 10.1242/jcs.114272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ryder PV, Lerit DA. 2018. RNA localization regulates diverse and dynamic cellular processes. Traffic 19:496–502. doi: 10.1111/tra.12571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.St Johnston D. 1995. The intracellular localization of messenger RNAs. Cell 81:161–170. doi: 10.1016/0092-8674(95)90324-0. [DOI] [PubMed] [Google Scholar]
- 5.Lecuyer E, Yoshida H, Parthasarathy N, Alm C, Babak T, Cerovina T, Hughes TR, Tomancak P, Krause HM. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 131:174–187. doi: 10.1016/j.cell.2007.08.003. [DOI] [PubMed] [Google Scholar]
- 6.Andreassi C, Riccio A. 2009. To localize or not to localize: mRNA fate is in 3’UTR ends. Trends Cell Biol 19:465–474. doi: 10.1016/j.tcb.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 7.Kislauskis EH, Zhu X, Singer RH. 1994. Sequences responsible for intracellular localization of beta-actin messenger RNA also affect cell phenotype. J Cell Biol 127:441–451. doi: 10.1083/jcb.127.2.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sepulveda G, Antkowiak M, Brust-Mascher I, Mahe K, Ou T, Castro NM, Christensen LN, Cheung L, Jiang X, Yoon D, Huang B, Jao LE. 2018. Co-translational protein targeting facilitates centrosomal recruitment of PCNT during centrosome maturation in vertebrates. Elife 7:e34959. doi: 10.7554/eLife.34959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kilchert C, Spang A. 2011. Cotranslational transport of ABP140 mRNA to the distal pole of S. cerevisiae. EMBO J 30:3567–3580. doi: 10.1038/emboj.2011.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ichihashi Y, Dales S. 1973. Biogenesis of poxviruses: relationship between a translation complex and formation of A-type inclusions. Virology 51:297–319. doi: 10.1016/0042-6822(73)90430-3. [DOI] [PubMed] [Google Scholar]
- 11.Moss B. 2013. Poxviridae, p 2129–2159. In Knipe DM, Howley PM (ed), Fields virology, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 12.Cairns J. 1960. The initiation of vaccinia infection. Virology 11:603–623. doi: 10.1016/0042-6822(60)90103-3. [DOI] [PubMed] [Google Scholar]
- 13.Dales S, Mosbach EH. 1968. Vaccinia as a model for membrane biogenesis. Virology 35:564–583. doi: 10.1016/0042-6822(68)90286-9. [DOI] [PubMed] [Google Scholar]
- 14.Katsafanas GC, Moss B. 2007. Colocalization of transcription and translation within cytoplasmic poxvirus factories coordinates viral expression and subjugates host functions. Cell Host Microbe 2:221–228. doi: 10.1016/j.chom.2007.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hollinshead M, Rodger G, Van Eijl H, Law M, Hollinshead R, Vaux DJ, Smith GL. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J Cell Biol 154:389–402. doi: 10.1083/jcb.200104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward BM, Moss B. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J Virol 75:11651–11663. doi: 10.1128/JVI.75.23.11651-11663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smith GL, Law M. 2004. The exit of vaccinia virus from infected cells. Virus Res 106:189–197. doi: 10.1016/j.virusres.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 18.Ward BM. 2005. Visualization and characterization of the intracellular movement of vaccinia virus intracellular mature virions. J Virol 79:4755–4763. doi: 10.1128/JVI.79.8.4755-4763.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sivan G, Weisberg AS, Americo JL, Moss B. 2016. Retrograde transport from early endosomes to the trans-Golgi network enables membrane wrapping and egress of vaccinia virions. J Virol 90:8891–8905. doi: 10.1128/JVI.01114-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Marchal J. 1930. Infectious ectromelia. A hitherto undescribed virus disease of mice. J Pathol 33:713–728. doi: 10.1002/path.1700330317. [DOI] [Google Scholar]
- 21.Downie AW. 1939. A study of the lesions produced experimentally by cowpox virus. J Pathol 48:361–379. doi: 10.1002/path.1700480212. [DOI] [Google Scholar]
- 22.Kato S, Takahashi M, Kameyama S, Kamahora J. 1959. A study on the morphological and cyto-immunological relationship between the inclusions of variola, cowpox, rabbitpox, vaccinia (variola origin) and vaccinia IHD, and a consideration of the term “Guarnieri body”. Biken’s J 2:353–363. [Google Scholar]
- 23.Patel DD, Pickup DJ, Joklik WK. 1986. Isolation of cowpox virus A-type inclusions and characterization of their major protein component. Virology 149:174–189. doi: 10.1016/0042-6822(86)90119-4. [DOI] [PubMed] [Google Scholar]
- 24.Funahashi S, Sato T, Shida H. 1988. Cloning and characterization of the gene encoding the major protein of the A-type inclusion body of cowpox virus. J Gen Virol 69:35–47. doi: 10.1099/0022-1317-69-1-35. [DOI] [PubMed] [Google Scholar]
- 25.Howard AR, Moss B. 2012. Formation of orthopoxvirus cytoplasmic A-type inclusion bodies and the embedding of virions are dynamic processes requiring microtubules. J Virol 86:5905–5914. doi: 10.1128/JVI.06997-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Amegadzie BY, Sisler JR, Moss B. 1992. Frame-shift mutations within the vaccinia virus A-type inclusion protein gene. Virology 186:777–782. doi: 10.1016/0042-6822(92)90046-r. [DOI] [PubMed] [Google Scholar]
- 27.Meyer H, Rziha H-J. 1993. Characterization of the gene encoding the A-type inclusion protein of camelpox virus and sequence comparison with other orthopoxviruses. J Gen Virol 74:1679–1684. doi: 10.1099/0022-1317-74-8-1679. [DOI] [PubMed] [Google Scholar]
- 28.Howard AR, Weisberg AS, Moss B. 2010. Congregation of orthopoxvirus virions in cytoplasmic A-type inclusions is mediated by interactions of a bridging protein (A26p) with a matrix protein (ATIp) and a virion membrane-associated protein (A27p). J Virol 84:7592–7602. doi: 10.1128/JVI.00704-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sive HL, Grainger RM, Harland RM. 2007. Synthesis and purification of digoxigenin-labeled RNA probes for in situ hybridization. CSH Protoc 2007:4778. doi: 10.1101/pdb.prot4778. [DOI] [PubMed] [Google Scholar]
- 30.Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. 2008. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods 5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orjalo AV Jr, Johansson HE. 2016. Stellaris(R) RNA fluorescence in situ hybridization for the simultaneous detection of immature and mature long noncoding RNAs in adherent cells. Methods Mol Biol 1402:119–134. doi: 10.1007/978-1-4939-3378-5_10. [DOI] [PubMed] [Google Scholar]
- 32.Senkevich TG, Katsafanas G, Weisberg A, Olano LR, Moss B. 2017. Identification of vaccinia virus replisome and transcriptome proteins by isolation of proteins on nascent DNA coupled with mass spectrometry. J Virol 91:e01015-17. doi: 10.1128/JVI.01015-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patel DD, Pickup DJ. 1987. Messenger RNAs of a strongly-expressed late gene of cowpox virus contains a 5’-terminal poly(A) leader. EMBO J 6:3787–3794. doi: 10.1002/j.1460-2075.1987.tb02714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.David A, Dolan BP, Hickman HD, Knowlton JJ, Clavarino G, Pierre P, Bennink JR, Yewdell JW. 2012. Nuclear translation visualized by ribosome-bound nascent chain puromycylation. J Cell Biol 197:45–57. doi: 10.1083/jcb.201112145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pestka S. 1971. Inhibitors of ribosome functions. Annu Rev Microbiol 25:487–562. doi: 10.1146/annurev.mi.25.100171.002415. [DOI] [PubMed] [Google Scholar]
- 36.Yarmolinsky MB, Haba GL. 1959. Inhibition by puromycin of amino acid incorporation into protein. Proc Natl Acad Sci U S A 45:1721–1729. doi: 10.1073/pnas.45.12.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Joklik WK, Becker Y. 1965. Studies on the genesis of polyribosomes. I. Origin and significance of the subribosomal particles. J Mol Biol 13:496–510. doi: 10.1016/s0022-2836(65)80112-7. [DOI] [PubMed] [Google Scholar]
- 38.Godchaux W III, Adamson SD, Herbert E. 1967. Effects of cycloheximide on polyribosome function in reticulocytes. J Mol Biol 27:57–72. doi: 10.1016/0022-2836(67)90351-8. [DOI] [PubMed] [Google Scholar]
- 39.Lodish HF. 1971. Alpha and beta globin messenger ribonucleic acid. Different amounts and rates of initiation of translation. J Biol Chem 246:7131–7718. [PubMed] [Google Scholar]
- 40.Grollman AP. 1968. Inhibitors of protein biosynthesis. V. Effects of emetine on protein and nucleic acid biosynthesis in HeLa cells. J Biol Chem 243:4089–4094. [PubMed] [Google Scholar]
- 41.Walsh D, Arias C, Perez C, Halladin D, Escandon M, Ueda T, Watanabe-Fukunaga R, Fukunaga R, Mohr I. 2008. Eukaryotic translation initiation factor 4F architectural alterations accompany translation initiation factor redistribution in poxvirus-infected cells. Mol Cell Biol 28:2648–2658. doi: 10.1128/MCB.01631-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sundell CL, Singer RH. 1990. Actin mRNA localizes in the absence of protein synthesis. J Cell Biol 111:2397–2403. doi: 10.1083/jcb.111.6.2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cotter CA, Earl PL, Wyatt LS, Moss B. 2017. Preparation of cell cultures and vaccinia virus stocks. Curr Protoc Mol Biol 117:1–18. [DOI] [PubMed] [Google Scholar]
- 44.Schmidt EK, Clavarino G, Ceppi M, Pierre P. 2009. SUnSET, a nonradioactive method to monitor protein synthesis. Nat Methods 6:275–277. doi: 10.1038/nmeth.1314. [DOI] [PubMed] [Google Scholar]