How a virus survives in the arms race with its host is an intriguing question. In this study, we isolated and characterized four novel fuselloviruses, named Sulfolobus spindle-shaped viruses 19 to 22 (SSV19-22). Interestingly, SSV20-22 differ primarily in two genomic regions and are apparently convertible through homologous recombination during coinfection. Moreover, sites of single-nucleotide polymorphism (SNP) were identified throughout the genomes of SSV20-22 and, notably, enriched in certain regions, including the gene encoding the putative end filament protein VP4, which is believed to be involved in host recognition and binding by the virus.
KEYWORDS: fuselloviruses, genomic diversity, homologous recombination, single-nucleotide polymorphism, virus-host interaction
ABSTRACT
Fuselloviruses are among the most widespread and best-characterized archaeal viruses. They exhibit remarkable diversity, as the list of members of this family is rapidly growing. However, it has yet to be shown how a fuselloviral genome may undergo variation at the levels of both single nucleotides and sequence stretches. Here, we report the isolation and characterization of four novel spindle-shaped viruses, named Sulfolobus spindle-shaped viruses 19 to 22 (SSV19-22), from a hot spring in the Philippines. SSV19 is a member of the genus Alphafusellovirus, whereas SSV20-22 belong to the genus Betafusellovirus. The genomes of SSV20-SSV22 are identical except for the presence of two large variable regions, as well as numerous sites of single-nucleotide polymorphisms (SNPs) unevenly distributed throughout the genomes and enriched in certain regions, including the gene encoding the putative end filament protein VP4. We show that coinfection of the host with SSV20 and SSV22 led to the formation of an SSV21-like virus, presumably through homologous recombination. In addition, large numbers of SNPs were identified in DNA sequences retrieved by PCR amplification targeting the SSV20-22 vp4 gene from the original enrichment culture, indicating the enormous diversity of SSV20-22-like viruses in the environment. The high variability of VP4 is consistent with its potential role in host recognition and binding by the virus.
IMPORTANCE How a virus survives in the arms race with its host is an intriguing question. In this study, we isolated and characterized four novel fuselloviruses, named Sulfolobus spindle-shaped viruses 19 to 22 (SSV19-22). Interestingly, SSV20-22 differ primarily in two genomic regions and are apparently convertible through homologous recombination during coinfection. Moreover, sites of single-nucleotide polymorphism (SNP) were identified throughout the genomes of SSV20-22 and, notably, enriched in certain regions, including the gene encoding the putative end filament protein VP4, which is believed to be involved in host recognition and binding by the virus.
INTRODUCTION
Archaea are widespread on Earth and are especially well known as inhabitants in extreme environments, such as acidic hot springs, volcanic vents, and saline lakes. These organisms are infected by viruses, which, although sampled only in very few taxonomic groups so far, have demonstrated an amazing array of morphologies, including bottle shapes, droplet shapes, coil shapes, and spindle shapes, in addition to more common head-tail shapes (1–3). Spindle-shaped fuselloviruses, which infect hyperthermophilic archaea of the order Sulfolobales, are among the most widespread and best-characterized archaeal viruses. Fuselloviruses are divided into two genera, i.e., Alphafusellovirus (type species, Sulfolobus spindle-shaped virus 1, or SSV1) and Betafusellovirus (type species, Sulfolobus spindle-shaped virus 6, or SSV6) (1). As listed by International Committee on Taxonomy of Viruses (ICTV), the genus Alphafusellovirus currently comprises seven species (i.e., SSV1, SSV2, SSV4, SSV5, SSV7, SSV8, and SSV9) (4–8), whereas the genus Betafusellovirus contains two species (i.e., SSV6 and Acidianus spindle-shaped virus 1, or ASV1) (7), with SSV3 (9) and SSV10 (10) remaining to be classified. Recently, the family Fuselloviridae was enlarged drastically with the isolation of seven novel virus particles, named SSV11-15 and SSV17-18, respectively, and the identification of 27 SSVs integrated into the host genomes (11). Therefore, fuselloviruses are among the most abundant known archaeal viruses. Analysis of the available fuselloviral genomes permits the identification of core genes for this viral family (7, 11, 12). Among the core genes are those encoding the capsid proteins VP1 and VP3, an integrase, a DnaA-like protein, several putative transcriptional regulators (such as helix-turn-helix and zinc-finger proteins) and a few proteins with an unknown function. Noncore genes, most of which carry no known functions, account for over half of the total genes in each fusellovirus.
SSV1 is the first fusellovirus to be discovered, and its natural host is Sulfolobus shibatae strain B12 (13). The virus, which has been extensively studied, replicates in a UV-inducible fashion and exits the host cells via budding without cell lysis (13, 14). SSV1 carries a 15,465-bp circular double-stranded DNA (dsDNA) genome with 35 open reading frames (ORFs) (15, 16). SSV1 encodes a tyrosine family integrase, which catalyzes integration of the viral genome into and excision out of the host chromosome in a site-specific manner (17). The virus has four structural proteins, i.e., VP1 to VP4. The major capsid protein VP1, the minor capsid protein VP3, and the putative end filament protein VP4 (C792) are glycosylated (18). VP2, a highly basic protein, is capable of DNA binding (19). Alphafuselloviruses and betafuselloviruses appear to have different end filament proteins. SSV1 C792, the putative end filament protein of SSV1, differs from SSV6 B1232, the putative end filament protein of SSV6, in amino acid sequence, but the two proteins share a structural fold similar to that of the adsorption protein P2 from bacteriophage PRD1 (7). SSV1 encodes several putative transcriptional factors (20–23). Among them, SSV1 F55 functions as a molecular switch for the transcriptional regulation of the early viral genes, playing a key role in the transition from the lysogenic to the induced state of SSV1 (16, 23). SSV1 B251 is a putative DnaA-like protein whose bacterial homologue is involved in the initiation of DNA replication and the regulation of transcription of specific genes (24). SSV1 D244, a homologue of the structurally characterized SSV8 D212, is a nuclease of the PD-(D/E)XK superfamily. It is similar in structure to the archaeal Holliday junction cleavage enzyme and presumably serves a role in DNA transactions (25).
To shed light on the functions of the ORFs of SSV1, Stedman and his colleagues have introduced mutations into each of the viral ORFs (12). They demonstrated that 16 of the 35 ORFs could be disrupted without destroying virus function, and the essentiality of an ORF was generally correlated with the extent of the conservation of the ORF. Of the twelve ORFs that are unique to SSV1, eight could be mutated without a loss in infectivity (12). More recently, a similar pattern of mutation tolerance was found in SSV10 (formerly SSV-L1) (10).
The family Fuselloviridae exhibits remarkable diversity. However, it has yet to be shown how a fuselloviral genome may undergo variation at the levels of both single nucleotides and sequence stretches. Here, we report the isolation and characterization of four novel spindle-shaped viruses from a hot spring in the Philippines, which are named Sulfolobus spindle-shaped viruses 19 to 22 (SSV19-22). The genomes of SSV20-22 are identical except for the presence of two large variable regions, as well as a number of sites of single-nucleotide polymorphisms (SNPs) distributed throughout the genome but enriched in certain regions, including the gene encoding VP4, a putative end filament protein. Analysis of the total DNAs from the original enrichment culture, from which SSV20-22 were isolated, identified more SNPs in the vp4 gene. We also showed that coinfection of the host with SSV20 and SSV22 led to the formation of new viruses, presumably through homologous recombination (HR).
RESULTS
Isolation and morphology of SSV19-22.
Spindle-shaped virus-like particles (VLPs) were observed in the supernatant of an enrichment culture established in Zillig’s medium (26) with a sediment sample from a hot spring (64.1°C, pH 3.72) in Naghaso, the Philippines, by transmission electron microscopy (TEM). Two putative Sulfolobus strains (denoted E11-6 and E5), isolated by plating on solid plates and by dilution to extinction in liquid medium, respectively, were found to carry morphologically different spindle-shaped viruses. The virus infecting Sulfolobus sp. strain E11-6 was named Sulfolobus spindle-shaped virus 19 (SSV19). Despite repeated efforts, we were unable to isolate a virus-free host for SSV19. On the other hand, a virus-free host strain, named Sulfolobus sp. strain E5-1-F, was isolated from strain E5 by plating and colony picking. Strain E5-1-F was susceptible to infection by the viruses from E5, and the infected strain was named E5-1. E5-1 was found to contain a mixture of viruses, which shared similar genomic sequences but differed primarily in two regions, by viral DNA sequencing and assembly. We attempted but failed to separate and purify the viruses by screening E5-1 cells using the colony-picking approach. Thus, we amplified individual viral genomes from the infected E5-1 culture by whole-genome PCR amplification and transformed strain E5-1-F with the PCR products by electroporation. Strains containing each of the three viruses, designated SSV20, SSV21, and SSV22, respectively, were subsequently obtained. The sequences of the three viral genomes from these strains were verified by sequencing. All three viruses were found to be present in strain E5, as determined by PCR targeting the variable regions of the three viruses (see below). In addition, the three viruses were able to infect strain E5-1-F individually.
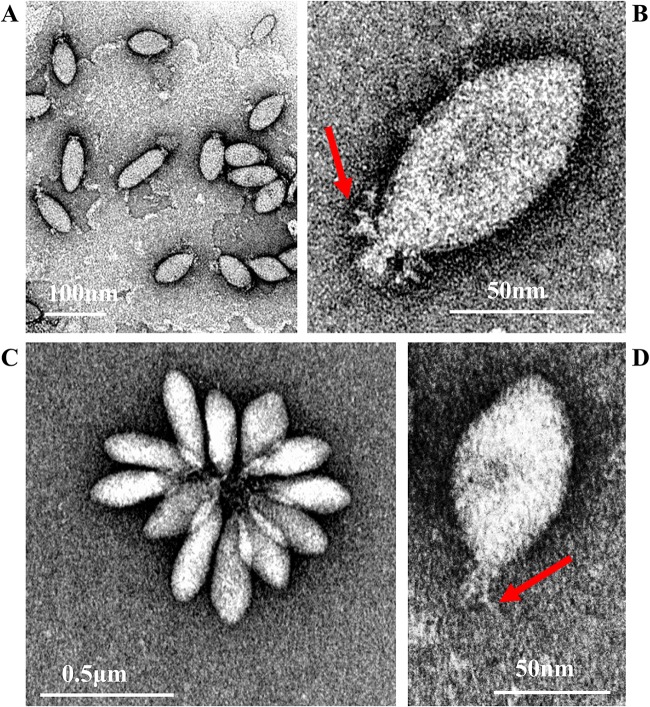
SSV20-22 are indistinguishable in shape and size, but they differ morphologically from SSV19. SSV19 is 100 to 120 nm by 30 to 40 nm in size and possesses several thick filaments on one end (Fig. 1A and B), while SSV20-22 are 80 to 100 nm by 40 to 50 nm in size with fewer and thinner tail fibers than SSV19 (Fig. 1C and D). By morphology, SSV20-22 are similar to members of the genus Alphafusellovirus, whereas SSV19 resembles SSV6 and ASV1, the known pleomorphic viruses of the genus Betafusellovirus, showing diversity in shape with both cigar-like virions and, more frequently, pear-like virions observed (Fig. 1A) (7). In addition, SSV20-22 virions occasionally join, probably through their tails, or attach to cell debris, forming a rosette-like pattern, while those of SSV19 rarely do so (Fig. 1A and C).
FIG 1.
Electron micrographs of SSV19 and SSV20-22. (A and B) SSV19 virions. (C and D) SSV20-22 virions. The end filaments are indicated by arrows.
Genome of SSV19.
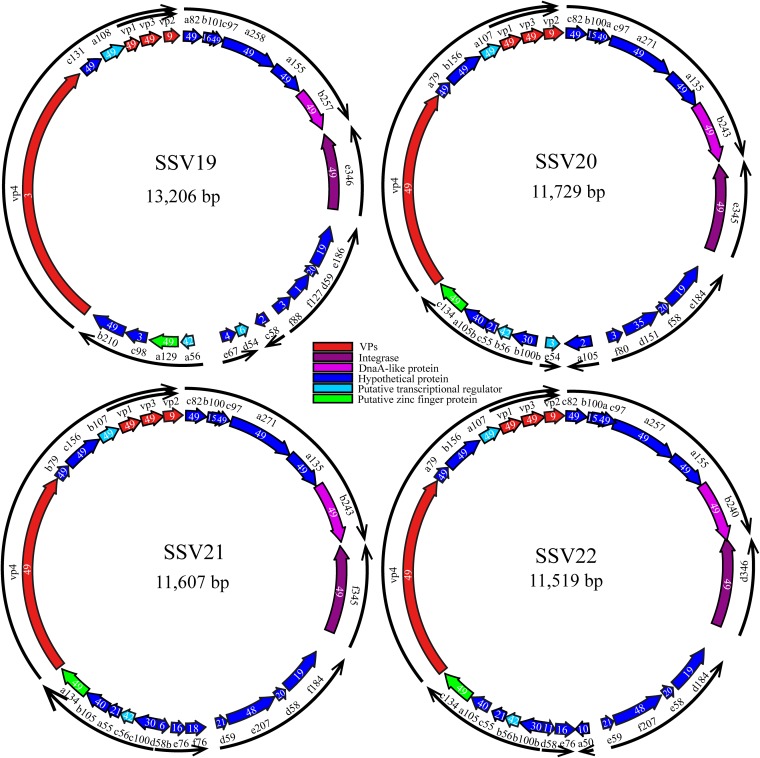
SSV19 has a circular dsDNA genome of 13,206 bp with a G+C content of 38%. A total of 24 putative ORFs were identified (Fig. 2 and Table 1). Among them, twelve (a82, c97, a258, a155, b257, e346, a129, c131, a108, vp1, vp3, and b210) are conserved among all 49 known fuselloviruses (see Table S1 in the supplemental material), and 11 (b101, vp2, vp4, e186, d59, f88, c58, d54, a56, c98, and e67) have homologues in some, but not all, of the fuselloviruses. The remaining ORF (f127), which shows no significant matches to sequences in the existing databases, is unique to SSV19.
FIG 2.
Genome maps of SSV19-22. A number in each ORF indicates the number of known fuselloviruses that possess a homologue of the ORF. The arrows of the outer ring indicate the putative transcripts, as predicted based on the previous analysis of transcription of the SSV1 genome (16, 49).
TABLE 1.
ORFs of SSV19-22a
| SSV19 | SSV20 | SSV21 | SSV22 | Annotation | Domain prediction | No. of known fuselloviruses containing a homologue | Homologue in SSV1 |
|---|---|---|---|---|---|---|---|
| a82 | c82 | c82 | c82 | Transmembrane region | 49 | SSV1 A82 | |
| c97 | c97 | c97 | c97 | Transmembrane region | 49 | SSV1 A92 | |
| a258 | a271 | a271 | a257 | Transmembrane region | 49 | SSV1 B277 | |
| a155 | a135 | a135 | a155 | Coiled coil | 49 | SSV1 A153 | |
| b257 | b243 | b243 | b240 | DnaA-like protein | AAA domain, NTP-binding motif | 49 | SSV1 B251 |
| e346 | e345 | f345 | d346 | Integrase, a tyrosine family recombinase | 49 | SSV1 D335 | |
| a129 | c134 | a134 | c134 | Putative transcription regulator | Two ZNF, C2H2 | 49 | SSV1 B129 |
| c131 | b156 | c156 | b156 | Transmembrane region | 49 | SSV1 C166 | |
| a108 | a107 | b107 | a107 | Putative transcriptional regulator | HTH domain | 49 | SSV1 B115 |
| b84 | b83 | c83 | b83 | VP1, capsid protein | 49 | VP1 | |
| b92 | c88 | a88 | c88 | VP3, capsid protein | 49 | VP3 | |
| c797 | a797 | c797 | VP4, end filament protein | Signal peptide; transmembrane region | 49 | VP4 | |
| b210 | Transmembrane region; similar to the C terminus of SSV1 C792 | 49 | VP4 | ||||
| a79 | b79 | a79 | Transmembrane region; similar to the C terminus of SSV6 C213 | 49 | SSV1 B78 | ||
| a1236 | VP4, end filament protein | 3 | |||||
| a91 | c75 | a75 | c75 | VP2, putative DNA-binding protein | 9 | VP2 | |
| b101 | b100a | b100 | b100a | 16 | SSV1 C84 | ||
| e186 | e184 | f184 | d184 | VirArc_Nuclease domain | 19 | SSV1 D244 | |
| d59 | f58 | d58 | e58 | HTH domain | 20 | SSV1 D63 | |
| d59 | e59 | 21 | |||||
| f88 | f80 | DNA binding protein | DNA binding winged-helix domain | 3 | SSV1 F112 | ||
| d151 | e207 | f207 | Coiled coil | 48 | |||
| a50 | 10 | ||||||
| f76 | 18 | ||||||
| a105 | 2 | ||||||
| e76 | e76 | 16 | |||||
| d54 | e54 | Transcriptional regulator | RHH DNA binding domain; NikR C-terminal nickel binding domain | 6 | SSV1 F55 (Tlys) | ||
| d58b | d58 | Transmembrane region | 11 | ||||
| b100b | c100 | b100b | 30 | SSV1 A100 | |||
| a56 | b56 | c56 | b56 | CopG family transcriptional regulator | RHH DNA binding domain | 42 | SSV1 C80 |
| c55 | a55 | c55 | ZnF_C2H2 | 21 | SSV1 A45 | ||
| a105b | b105 | a105 | Transmembrane region | 40 | SSV1 C102b | ||
| c98 | Transmembrane region | 3 | |||||
| c58 | Transmembrane region | 2 | |||||
| f127 | 1 | ||||||
| e67 | Transmembrane region | 4 |
A total of 49 fuselloviruses or fusellovial genomes, including SSV1-10, SSV11-15, SSV17-18, ASV1, 27 SSVs integrated into the host genomes, and SSV19-22 are compared. Genes that exist in all of these fuselloviruses/fuselloviral genomes, known as core genes, are shown in boldface.
The virus encodes the structural proteins VP1 to VP4. VP1 and VP3 share 70 to 80% and 42 to 62% sequence identity, respectively, with those of the known fuselloviruses, whereas VP2, not conserved among known fuselloviruses, is 65 to 82% similar to its homologues from SSV1, SSV6, SSV11, SSV18, SSV20, and ASV1. The putative end filament protein VP4 (A1236), which shows no sequence similarity to VP4 proteins from alphafuselloviruses (e.g., SSV1 C792), resembles VP4 proteins from betafuselloviruses (e.g., SSV6 B1232 and ASV1 A1231) in size (∼1,200 amino acid residues) and amino acid sequence (∼73% identity) (7). The protein is predicted to possess an N-terminal signal peptide and three C-terminal transmembrane regions and is likely involved in interaction with viral or host membrane. SSV19 B210 shares ∼70% identity with SSV6 C213, which resembles the C-terminal 170-amino-acid portion of the alphafuselloviral VP4 protein SSV1 C792 (7). SSV19 B257 contains an AAA domain and is annotated as a DnaA-like protein (24). SSV19 E346, an SSV1 D335 homologue, is a tyrosine family recombinase capable of site-specific integration of the viral genome into and excision from the host genome (27, 28). SSV19 D54, a homologue of SSV1 F55, is probably a transcriptional regulator involved in maintaining the lysogenic state of the virus (16, 23). Little is known about the functions of the remaining SSV19 genes (Table 1).
Genomes of SSV20-22.
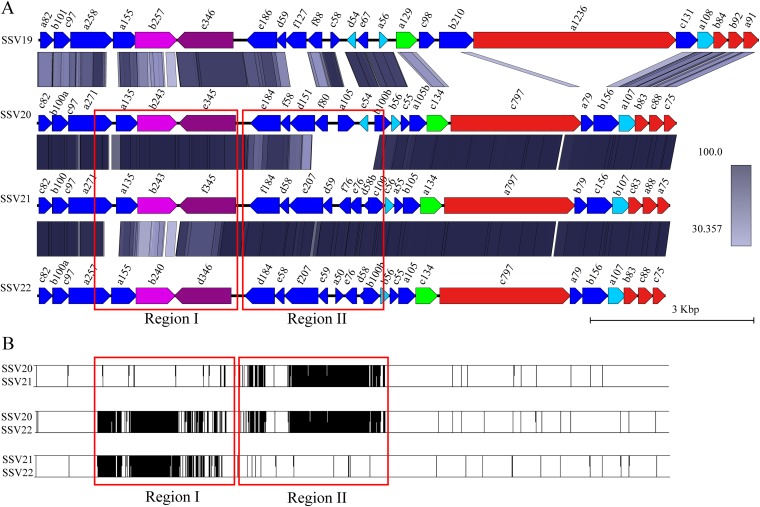
SSV20, SSV21, and SSV22 have a circular dsDNA genome of 11,729 bp (SSV20), 11,607 bp (SSV21), and 11,519 bp (SSV22) (Fig. 2). The G+C contents of the SSV20-22 genomes are 38%, 37%, and 38%, respectively. The three viruses show 53.5% (SSV20), 51.8% (SSV21), and 56.3% (SSV22) nucleotide identity with SSV19, and the nucleotide identity between two of the three viruses is 91.5% (SSV20/SSV21), 94.1% (SSV21/SSV22), or 85.9% (SSV20/SSV22). The three viruses have 25, 26, and 26 ORFs, respectively, and thirteen of them, including vp4 and a79 (as named in SSV20), are core genes. Among the remaining ORFs, the three viruses share nine proteins, i.e., B100a, A105B, C55, B56, B100b, D151, F58, E184, and C75 (VP2) (as named in SSV20) (Fig. 2, Fig. 3A, and Table 1). SSV21 and SSV22 share three additional proteins, i.e., D58b, E76, and D59 in SSV21 (Fig. 2, Fig. 3A, and Table 1). F80 in SSV20 is homologous to SSV19 F88 and SSV1 F112 (Table 1). Like SSV19 ORF D54, SSV20 E54 is a homologue of SSV1 F55 and, thus, may be involved in the maintenance of the lysogenic state.
FIG 3.
Comparison between members of SSV19 to SSV22. (A) Synteny analysis of proteins encoded by SSV19-22 using BLASTP (coverage, ≥50%; identity, ≥30%). (B) Pairwise alignments of genomes of SSV20-22. The vertical lines indicate sites of sequence variation. Red boxes show regions I and II.
SSV20-22 contain identical VP1 and VP2, nearly identical VP3, and slightly different VP4. VP1 and VP3 from SSV20-22 share 70 to 81% and 45 to 68% sequence identity, respectively, with their homologues from the other reported fuselloviruses (e.g., 74% and 68% identical to their counterparts from SSV19). VP2 of SSV20-22 is similar to its homologues from SSV1, SSV11, SSV18, SSV6, and ASV1 (69 to 82% identical), as well as SSV19 (80%). VP4, the putative end filament protein, is 47 to 61% identical to its homologues from other alphafuselloviruses and 27 to 28% identical to betafuselloviral SSV19 B210, ASV1 B208, and SSV6 C213 (Fig. 3A and Table 1). Compared to VP4 in SSV19, VP4 from SSV20 also contains a signal peptide at the N terminus but has more transmembrane regions (nine) distributed across the entire length of the protein.
The genomes of the three viruses differ primarily in two large variable regions, designated regions I and II, as well as at the single-nucleotide level (Fig. 3B). There are two versions for each of the two variable regions. SSV20 and SSV21 are nearly identical in region I except for several SNPs but differ in region II, SSV21 and SSV22 are very similar in region II except for a few SNPs but differ in region I, and SSV20 and SSV22 are different in both regions (Fig. 3B). Region I includes four core genes, i.e., a271, a135, b243 (DnaA-like protein), and e345 (integrase) in SSV20 or their homologues in SSV21 and SSV22, whereas region II contains seven or eight noncore genes, i.e., e184, f58, d151, and b100b or their homologues in all three viruses, d59, e76, and d58b in SSV21 and SSV22, f80, a105, and e54 in SSV20, f76 in SSV21, and a50 in SSV22 (Fig. 3A).
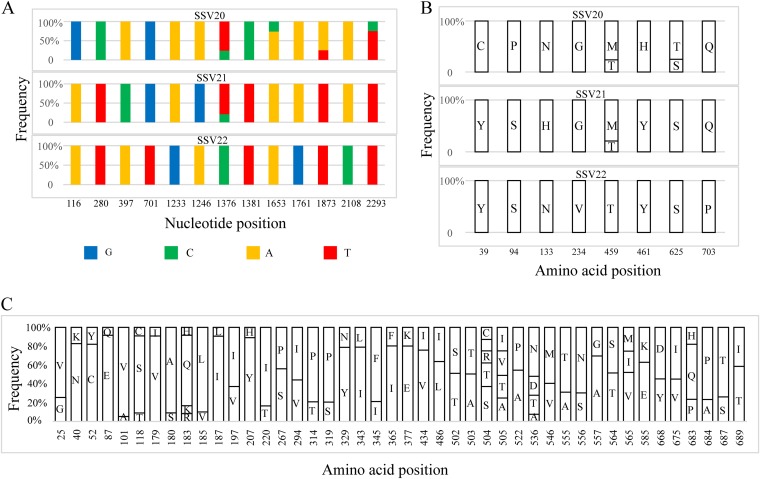
SNP sites exist over the entire viral genome sequences but are not evenly distributed, suggesting that some genes mutate more frequently than others under selective pressure. For example, while the three viruses share identical vp1 and vp2 genes for the structural proteins VP1 and VP2, respectively, and differ at a single nucleotide in vp3 (i.e., a T in SSV20/SSV21 and a C in SSV22 at the 227th nucleotide or a Val in SSV20/SSV21 and an Ala in SSV22 at the 76th amino acid residue), they contain thirteen SNP sites in vp4, the gene encoding the putative end filament protein in the three viruses (Fig. 4A). Eight of these sites in vp4 are associated with a change in amino acid residues (Fig. 4B).
FIG 4.
Distribution of SNPs and the sites of amino acid variation in VP4 of SSV20-22. (A) Thirteen SNPs in SSV20-22. The vp4 gene of SSV22 is used as the reference sequence, and the position and frequency of each SNP are indicated. (B) Amino acid variation in VP4 from SSV20-22. Amino acid residues at each site and their frequencies in the three viruses are shown. (C) Variable amino acid residues in VP4, as predicted from SNPs in the vp4 gene sequences from the enrichment culture for the isolation of SSV20-22. The bar diagram indicates the site of amino acid variation and the proportion of different amino acid residues at the site.
To determine if there were more SNPs in the DNAs of SSV20-22-like viruses in the original enrichment culture, we performed PCRs with a pair of primers encompassing the vp4 sequence (VP4-SNP-2966F/R; Table 2) as well as that targeting the vp1, vp2, and vp3 genes (VP132-SNP-1303F/R; Table 2) of SSV22 using total DNA extracted from the enrichment culture as the template. The products from two PCRs were sequenced at depths of 12,917× and 28,732×, respectively, and a mutation with a read coverage greater than 5% was regarded as an SNP. A total of 110 SNPs were identified in vp4, and 44 of them resulted in a change in amino acid residue (Fig. 4C and Tables S2 and S3). In all, mutation occurred on 4.6% of the nucleotides in vp4 or 5.5% of the amino acid residues in VP4 (Table S3). On the other hand, only a single SNP at the 7th nucleotide (7% of T and 93% of G), which corresponded to variation at the 3rd codon (GTA for Val and TTA for Leu), was identified in vp3, and none was found in vp1 or vp2 (Table S3). Sites of amino acid variation were evenly distributed throughout the polypeptide chain of VP4, including the nine transmembrane regions. Since VP4 is probably involved in the recognition of the host receptor by the virus during infection, the high rate of mutation in VP4 may allow the virus to gain advantage in overcoming host defense in nature. Our data also indicate that SSV20-22 represent only a fraction of the viral diversity in the environment where these viruses are active.
TABLE 2.
Primers used in this study
| Primer name | Primer sequence |
|---|---|
| E11-VP F | 5′-TCAGAACTGCAAGGGTTTAA-3′ |
| E11-VP R | 5′-TGACCAGCCCTAACAGTAAA-3′ |
| E5-Int F | 5’-TACCGTTTATCGTTCTCAGTAT-3’ |
| E5-Int R | 5′-TCAGTGTTGCGGAAATCTTA-3′ |
| IIA wg F | 5′-GAGGAAGGGACCAACGCAGTTACAT-3′a |
| IIA wg R | 5′-GTTGGTCCCTTCCTCTTTTGCTATCTC-3′a |
| IIB wg F | 5′-CACGGGCGTTACATCGCTCATTGCT-3′a |
| IIB wg R | 5′-GATGTAACGCCCGTGAAATCCTCGT-3′a |
| IA F | 5′-CAGTTACGCACCCAACTACAAC-3′ |
| IA R | 5′-GCTTTCATTTTCTGGATTGCTTCCT-3′ |
| IB F | 5′-GAGGTGAGACAATGCCCAACAAAAAAG-3′ |
| IB R | 5′-TCTTCATCAATGCTTCTGGCAATTCA-3′ |
| IIA F | 5′-ATGCACGAAGAAAGACTGCATAAGCGT-3′ |
| IIA R | 5′-ATGCCTAAAAAAGATAAAATCGAATGGGTAGG-3′ |
| IIB F | 5′-CAAATACTCTTTGAGGGCCTCG-3′ |
| IIB R | 5′-CATGGTTAATTGCACATAAAGAGGA-3′ |
| vp1 qPCR-F | 5′-AGCAAAGGCTGAAGGAGCAA-3′ |
| vp1 qPCR-R | 5′-GGGACTAGATTTAGCAATGTGGC-3′ |
| 1F | 5′-GAGAAGAGACAAAGTAATGATCTCGATAGCTGAGAAG-3′ |
| 1R | 5′-CTCACTACGCTTATGCAGTCTTTCTTCGTGCATTC-3′ |
| 2F | 5′-AGCATTGATGAAGATACAGGATGCAATAGACAATGAC-3′ |
| 2R | 5′-ACGAGGATTTCACGGGCGTTACATCGCTC-3′ |
| 1-784F | 5′-AGCTTGGAAGGCTGCAAAGCTTACAGC-3′ |
| 1-784R | 5′-CGCTGCTATTAGTAGCAGCAATGCTGCT-3′ |
| 2-1030F | 5′-TCAGCAAACTCCTCACTAATTAAGCCACG-3′ |
| 2-1030R | 5′-AACCTCAGGATGTAGGCCACCTTCAC-3′ |
| 9-876F | 5′-GGCTGCAAAGCTTACAGCGATTGATGCGA-3′ |
| 9-876R | 5′-CATGCCTCTCACACTGTTGAGAACATTGGA-3′ |
| 10-874F | 5′-CGTTATCGCTATACTCTTTCGCAAGCAACA-3′ |
| 10-874R | 5′-GCCTGCCAGTTAAAGAGAGGTACCTA-3′ |
| 11-848F | 5′-CTCTCACGAACACTCACCTACTATGTAGA-3′ |
| 11-848R | 5′-GTAGCAGCAATGCTGCTATTAGCGTAATAGGT-3′ |
| 12-679F | 5′-CTCCTCACTAATTATCCCGCGTGAAGCCA-3′ |
| 12-679R | 5′-GTCAAATTTGGACTCCCTGCATTACTAGCAGT-3′ |
| 13-1828R | 5′-GCGAATCCAGAGCCGAAGCAAAAGCT-3′ |
| VP4-SNP-2966F | 5′-ATGATGAGCACTCCAGATACTCCTAATAGAGAATCAG-3′ |
| VP4-SNP-2966R | 5′-TAGCCCCTATCCCCACTATTATTGCACTAATCTTC-3′ |
| VP132-SNP-1303F | 5′-GCCAAGATATTGCGAAAGAAGATACTGCAGTTGA-3′ |
| VP132-SNP-1303R | 5′-CCTTTCTTCCCTCAACCAATTGAAGAAGAAGCCT-3′ |
Overlap extension primers for the amplification of the entire viral genomes containing sequence IIA or IIB, with the overlapping sequences underlined.
Virion composition.
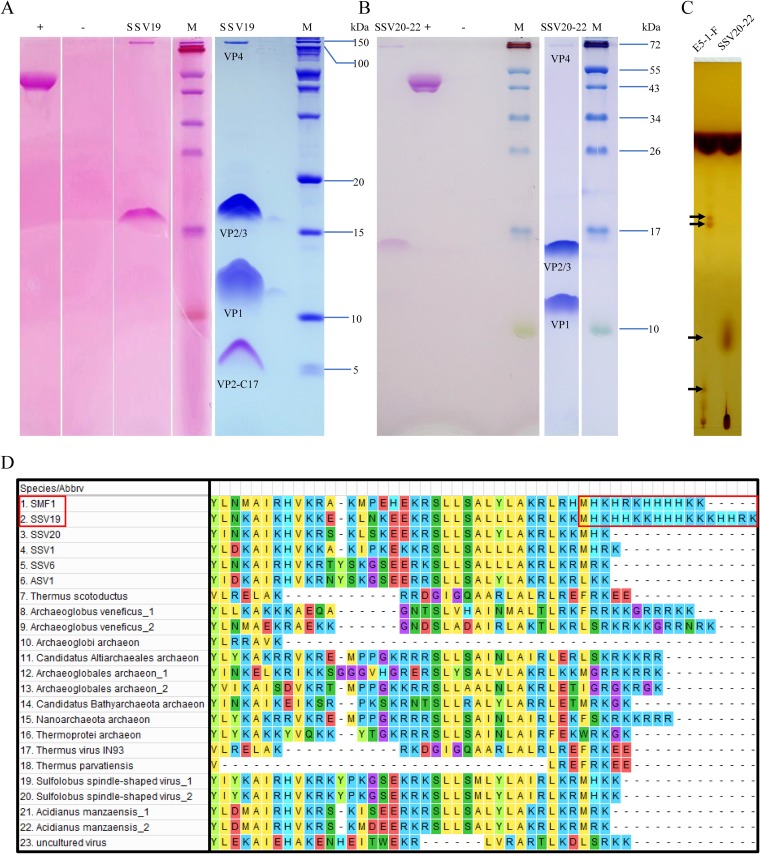
The structural proteins of SSV19-22 were separated by SDS-PAGE and analyzed by mass spectrometry (MS) and N-terminal sequencing. Each virus contains four structural proteins, as expected from the genome annotation. As shown in Fig. 5A and B, VP1 is the major capsid protein, since it is the most abundant of the four proteins in both viruses. The homologue of VP1 in SSV1 was shown to be essential for SSV1 infectivity (12). Both SSV20-22–VP4 (C797, homologue of SSV1 C792) and SSV19-VP4 (A1236, homologue of SSV6 B1232) were detected in minute amounts in the virions. VP2 and VP3, which were of similar molecular masses, comigrated on the SDS-PAGE gel, as confirmed by N-terminal amino acid sequencing. VP2, a highly basic protein, is believed to bind viral DNA within the capsid, although it is not well conserved among fuselloviruses and deletion of the gene for VP2 in SSV1 did not result in the loss of infectivity of the progeny virions (12). Deletion of the highly conserved minor capsid VP3 in SSV1 resulted in morphological change but did not affect the infectivity of the virus (12). It was noticed that these structural proteins, especially VP2 and VP3, did not migrate as far on the gel as expected based on their molecular masses. It is possible that posttranslational modifications were responsible for the aberrant migration pattern of the proteins on the gel. Indeed, we found that VP2 and/or VP3 as well as VP4, but not VP1, from SSV19 and SSV20-22 were glycosylated (Fig. 5A and B). By comparison, VP1, VP3, and VP4 were glycosylated in SSV1 (18). Interestingly, there was a band at the position of a 5-kDa protein in the SSV19 sample on the gel (Fig. 5A). The protein contained the last 17 amino acid residues of VP2 (denoted VP2-C17) and, therefore, was likely the cleavage product of VP2. VP2-C17 has a very unusual HK-rich sequence (i.e., MHKHHKKHHHKKKHHRK) (Fig. 5D), which has not been found in other known VP2-containing fuselloviruses (e.g., SSV1, SSV6, ASV1, and SSV20-22). It is worth noting that a VP2-like protein from Sulfolobales Mexican fusellovirus 1 (SMF1), whose genome was assembled from the metagenome of a sample taken from a Mexican hot spring (29), has a similar C-terminal sequence (i.e., MHKHRKHHHHKK) (Fig. 5D). It is unclear where this peptide is located in the virion. Since it is rich in basic amino acid residues, VP2-C17 may be involved in DNA binding.
FIG 5.
Protein and lipid analyses of SSV19 and SSV20-22 virions. (A and B) Tricine–SDS-PAGE and glycoprotein staining of the virions of SSV19 (A) and SSV20-22 (B). Purified SSV20-22 virions were subjected to electrophoresis in a 14% tricine–SDS-polyacrylamide gel. The gel was stained with Coomassie brilliant blue (right) or with a glycoprotein staining kit (left). Glycosylated proteins were stained pink. +, positive control (horseradish peroxidase provided by the manufacturer); −, negative control (soybean trypsin inhibitor provided by the manufacturer); M, molecular mass markers. (C) Thin-layer chromatography of lipids extracted from SSV20-22 virions and the host cells. Differences between the lipids from SSV20-22 and those from the host cells are indicated by arrows. (D) Sequence alignment of the C-terminal regions of VP2-like proteins. Homologues of SSV19-VP2 with an e value of ≤10−3 were selected and aligned with Muscle. The C-terminal regions of SSV19 and SMF1 VP2 with similar HK repeats are shown in a red box.
As revealed by thin-layer chromatography, SSV20-22 contains both lipids identical to and those different from its host, suggesting that the virus serves roles in the synthesis or modification of the lipids (Fig. 5C). However, there are no clues as to how these processes take place.
Virus hosts.
SSV19 and SSV20-22 appeared to have a very narrow host range. Strain E5-1-F, the virus-free host for SSV20-22, was resistant to infection by SSV19. We also tested the ability of SSV19 and SSV20-22 to infect Sulfolobus solfataricus P2 (now renamed Saccharolobus solfataricus P2), Sulfolobus tokodai 7 (renamed Sulfurisphaera tokodaii 7), Sulfolobus islandicus REY15A, Sulfolobus tengchongenesis RT8-4, and Sulfolobus sp. strain A20. None of these strains were infected by any of the viruses.
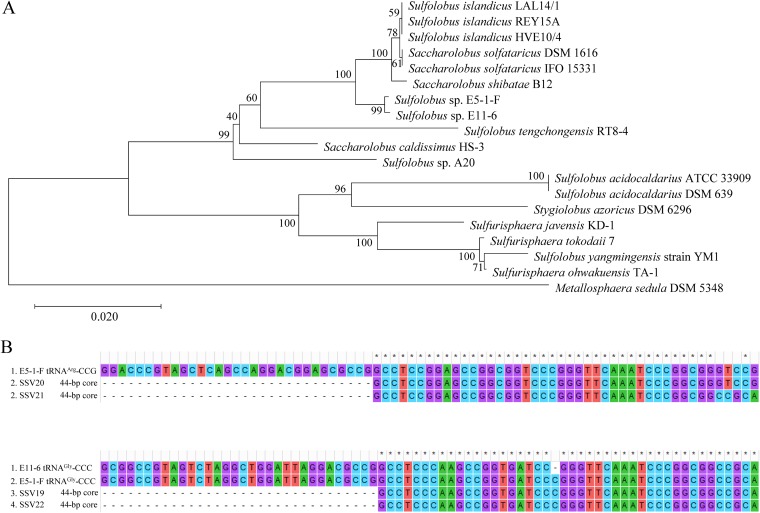
Sequence analysis reveals that Sulfolobus sp. strain E11-6 is ∼99.8% identical to Sulfolobus sp. strain E5-1-F at the 16S rRNA gene sequence level, and both strains are closely related to Sulfolobus solfataricus strain IFO 15331 (renamed Saccharolobus solfataricus strain IFO 15331), Sulfolobus shibatae B12 (renamed Saccharolobus shibatae B12), and Sulfolobus solfataricus strain DSM 1616 (renamed Saccharolobus solfataricus strain DSM 1616) (98.5 to 98.7% identity). A phylogenetic tree constructed by the neighbor-joining method shows that E5-1-F and E11-6 strains form a branch with S. solfataricus, S. shibatae, and S. islandicus (Fig. 6A).
FIG 6.
Phylogenetic analysis of viral hosts and sequence alignment of the sites of integration. (A) A 16S rRNA gene-based phylogenetic tree of Sulfolobus sp. strain E5-1-F, Sulfolobus sp. strain E11-6, and 17 additional Sulfolobus strains constructed by using the neighbor-joining method (45). The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) are shown next to the branches (50). The evolutionary distances are in units of the number of base substitutions per site. (B) Sequence alignment of the sites of integration of SSV19-SSV22 with the 3′ terminal part of the tRNA genes, which contain the potential sites of viral integration, from their hosts.
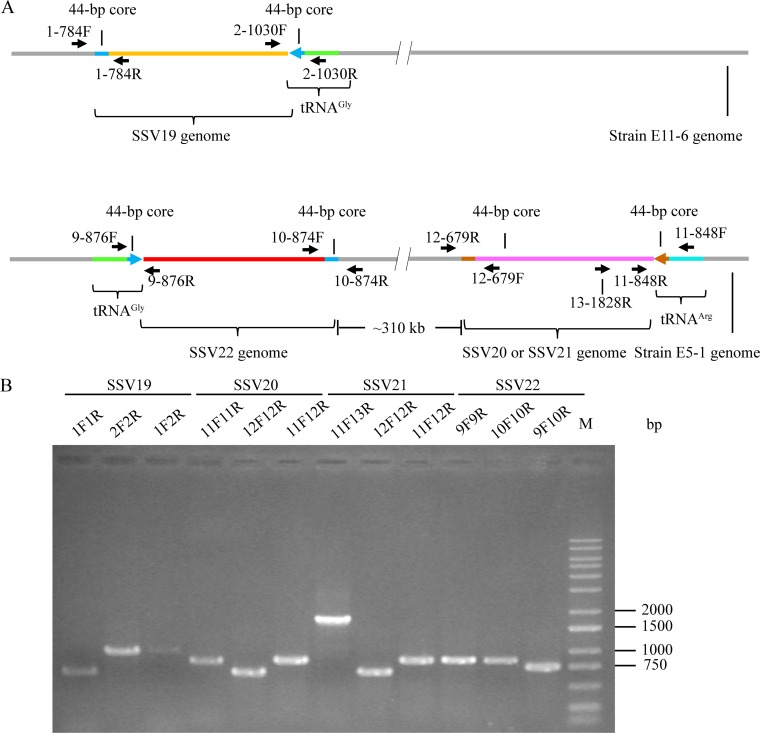
We subsequently sequenced and compared the genomes of E11-6 and E5-1-F strains, focusing on their integration sites and CRISPR spacers. In the E5-1-F genome, the 3′-end sequences in two tRNA genes, i.e., tRNAArg (CCG) and tRNAGly (CCC), were found to match perfectly and nearly perfectly with the 44-bp core sequence of the viral attP site of SSV20/SSV21 and of SSV22, respectively (Fig. 6B). In the E11-6 genome, the 3′-end sequence of the tRNAGly (CCC) gene matches nearly perfectly with the 44-bp core sequence of the SSV19 attP site (Fig. 6B). Therefore, we conclude that these tRNA genes contain integration sites of the corresponding viral genomes. We then examined the occupancy of the integration sites by PCR with primers targeting sequences flanking the sites of integration. We were able to identify both occupied and unoccupied integration sites in E11-6 and the infected E5-1-F strain (Fig. 7). It seems that the site of integration was more frequently occupied in E11-6 than in the infected E5-1-F (Fig. 7B). SSV20-22 integrated at two different sites (SSV20/SSV21 in the tRNAArg and SSV22 in the tRNAGly genes). In a similar observation, S. islandicus M.06.0.8 was found to contain two sites for the integration by SSVs (11). The two integration sites are in opposite orientation and separated by ∼310 kb in the E5-1-F genome (Fig. 7A). If both sites were occupied simultaneously by an integrated viral genome, HR between the two viral sequences, which were highly similar, could conceivably occur, leading to inversion of the host DNA sequence between the two sites. However, no inversion of the intervening region between the two sites was detected by PCR. As shown in the PCR assays, neither of the integration sites was fully occupied (Fig. 7B). Thus, it remains to be determined if the two sites could be simultaneously occupied or what would be the chances of double occupancy.
FIG 7.
Detection of the integration of SSV19-22 DNA into their host genomes. (A) A diagram showing the sites of integration for SSV19-22 as well as the positions of primers used in the analysis of viral integration. The tRNA genes, in which integration occurred, and primer pairs flanking the attP sites are indicated. (B) Occupancy of the integration sites on the host chromosomes. DNA extracted from virus-infected host cells was amplified using specific primer pairs for integrated and unoccupied sites. PCR products were loaded onto a 1.0% agarose gel. After electrophoresis, the gel was photographed under UV light. The expected sizes of the PCR products were 784 bp (1F/1R), 1,030 bp (2F/2R), 1,033 bp (1F/2R), 876 bp (9F/9R), 874 bp (10F/10R), 758 bp (9F/10R), 848 bp (11F/11R), 679 bp (12F/12R), 860 bp (11F12R), and 1,828 bp (11F/13R), respectively. M, molecular weight standards.
Putative CRISPR spacers were predicted with CRISPRCasFinder (https://crisprcas.i2bc.paris-saclay.fr) (30, 31). A huge difference was noticed between the two strains in the number of spacers identified. There are 14 CRISPRs with 463 spacers in E5-1-F but only two CRISPRs with 11 spacers in E11-6. While E5-1-F possesses an array of CRISPR-CAS systems, including type IA, IIIA, IIIU, ID, and IIIB, E11-6 has only one incomplete, and probably nonfunctional, CRISPR-CAS system. Notably, a single spacer in the E5-1-F genome (i.e., 5′-CAAATTGAAAAATAATGTCTTCAATTGTGATTTGGCCTTT-3′) matches perfectly with a sequence in SSV19 ORF f88, providing a possible explanation for the failure of SSV19 to infect E5-1-F.
Infection of the host by SSV20-22.
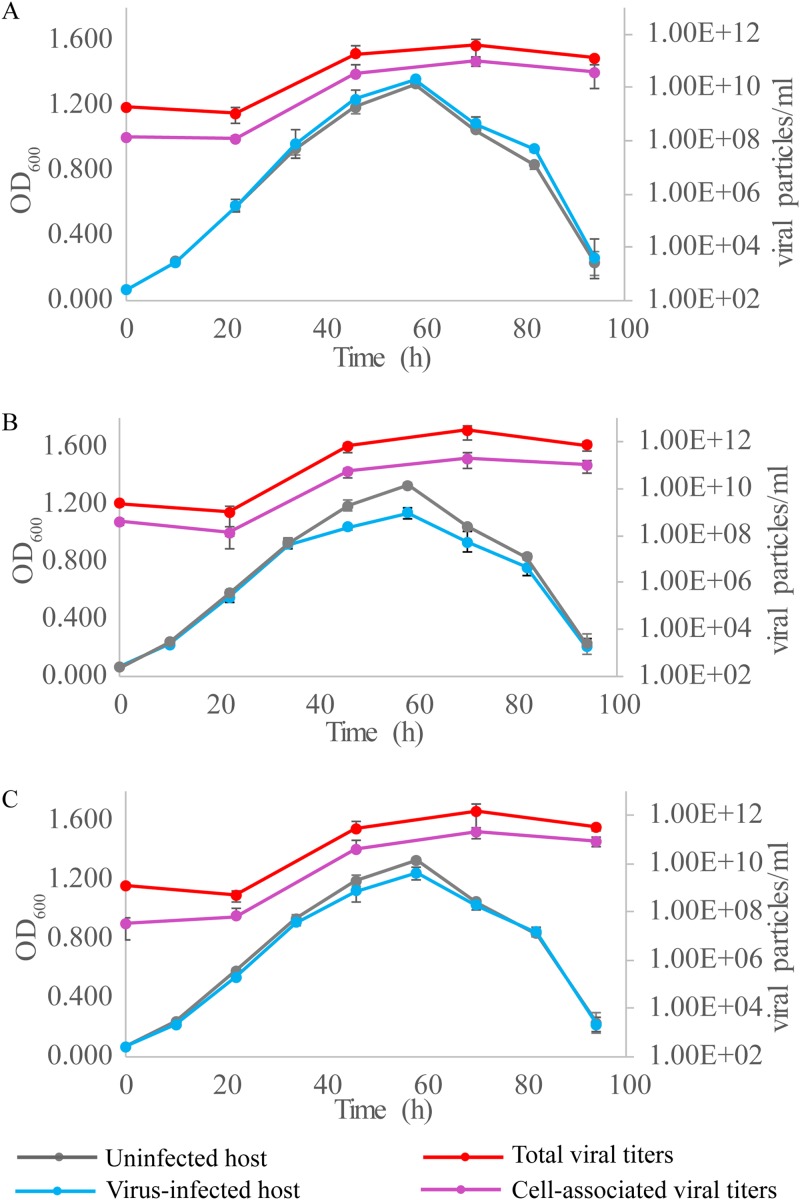
We then tested the effect of SSV20-22 infection on the growth of the host cells. Sulfolobus sp. strain E5-1-F was infected with the three viruses separately at a multiplicity of infection (MOI) of ∼5. The copy numbers of the total and cell-associated viruses in the culture were determined by quantitative PCR (qPCR) (Table S4). All three viruses showed very little effect on the infected host during the logarithmic phase of cell growth. Both the growth rate and the maximum cell density of the host cells were not significantly affected by infection with any of the viruses (Fig. 8). A slight decrease in maximum cell density was observed only in the culture infected with SSV21 (Fig. 8B). Despite the lack of a significant lag phase for the infected host cells, the number of virus particles in the culture did not increase until after 20 h of incubation, suggesting a substantial eclipse period for viral replication. The titers of the virus particles in the infected cultures peaked at 1011 to ∼1012 copies/ml, as quantified by qPCR, or about 2 to ∼3 orders of magnitude greater than the concentrations of the host cells (∼109 cells/ml). The fractions of the virus particles found associated with the cells or sedimentable cellular materials were 25% (SSV20), 7% (SSV21), and 14% (SSV22). Since a significant fraction of the virus particles were attached to the cell surface, as observed under TEM, the vast majority of the total virus particles detected were released from the host cells. As suggested by the identical curves for the stationary phases of the infected and uninfected cultures, the release of the virus particles was not accompanied by the lysis of the host cells.
FIG 8.
Infection of Sulfolobus sp. strain E5-1-F by SSV20-22. Strain E5-1 was grown to the exponential phase (OD600 of ∼0.2). The cells were infected with SSV20 (A), SSV21 (B), or SSV22 (C) at an MOI of ∼5. The OD600 of the culture was monitored. DNAs were extracted from the culture and the harvested cells. The viral copy number was determined by qPCR. Each data point represents an average from three independent measurements with the means and standard deviations indicated.
Recombination between the genomes of SSV20 and SSV22.
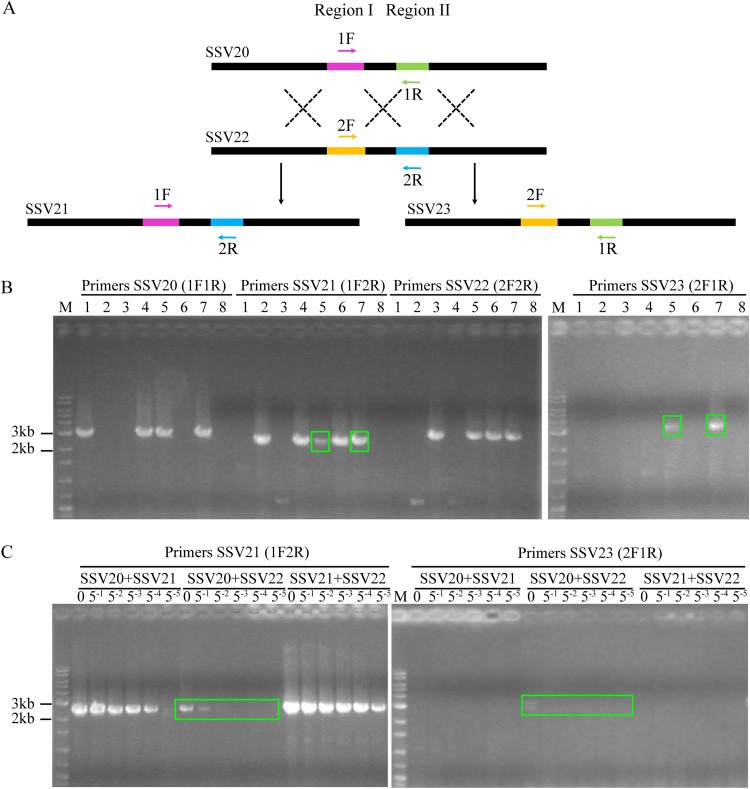
The genome of SSV21 differs primarily in region II from that of SSV20 and region I from that of SSV22. In other words, the SSV21 genome appears to result from recombination between the SSV20 and the SSV22 genomes with respect to the two regions. To determine if and how readily this process occurred in vivo, we coinfected strain E5-1-F with a mixture of SSV20/21, SSV21/22, SSV20/22, or SSV20/21/22 at a virus ratio of 1:1 or 1:1:1 and an MOI of 5. After incubation for 48 h, total DNA was extracted from the infected cultures, and PCR was performed with primer pairs designed to identify the two regions (Fig. 9A and Table 2). As expected, the specific PCR fragment for SSV21 was detected in DNA from the culture coinfected with SSV20/SSV22 or SSV20/SSV21/SSV22 and not from the other cultures (Fig. 9B). Furthermore, a specific fragment of another possible virus, denoted SSV23, which was detected by PCR in the enrichment culture that led to the identification of SSV20-22 but evaded isolation, was also detected in the cultures coinfected with SSV20/SSV22 or SSV20/SSV21/SSV22 (Fig. 9B). Apparently, recombination between the viral genomes of SSV20 and SSV22 resulted in the reassortment of regions I and II, generating the recombinant viral genomes. Further PCR tests with a serial dilution of the template DNA showed that the recombinant viral genomes were at least 2 orders of magnitude less abundant than the parental viral genomes in quantity (Fig. 9C).
FIG 9.
Coinfection of Sulfolobus sp. strain E5-1-F by SSV20-22. (A) A diagram showing proposed crossovers between the three viruses. Primer pairs designed to detect the HR events are indicated. (B) Agarose gel electrophoresis of the products of recombination between SSV20 to SSV22. Strain E5-1 was grown to the exponential phase (OD600 of ∼0.2). The cells were infected or coinfected with different viruses at an MOI of ∼5. Lanes 1 to 8: SSV20, SSV21, SSV22, SSV20 plus SSV21, SSV20 plus SSV22, SSV21 plus SSV22, SSV20 plus SSV21 plus SSV22, and a no virus control. After incubation for 48 h, total DNA was extracted from the infected culture. PCRs were performed with indicated primer pairs using the total DNA as the template. PCR products were subjected to electrophoresis in 1.0% agarose gel. Recombination products are shown in green boxes. (C) Estimation of the frequency of recombination between SSV20 and SSV22. The total DNA extracted from the coinfected culture was serially diluted by 51- to 55-fold. PCRs targeting SSV21 and SSV23 were performed using the serially diluted total DNA as the template. Undiluted DNA is indicated by a zero; M, molecular weight standards.
DISCUSSION
The family Fuselloviridae has expanded rapidly over the years. In this study, we have added four new members, i.e., SSV19-22, to the list to bring the number of known fuselloviruses to 49 (see Table S1 in the supplemental material). SSV19 harbors a genome of ∼13.2 kb, while SSV20-22 have genomes of ∼11.5 to 11.7 kb and, thus, are among the smallest of all known fuselloviruses. In comparison, the genome sizes of SSV1 and SSV2, two extensively studied fuselloviruses, are 15,465 and 14,759 bp, respectively. Accordingly, the numbers of ORFs in SSV19 (24 ORFs) and SSV20-22 (25, 26, and 26 ORFs, respectively) are ∼30% fewer than those in SSV1 (35 ORFs) and SSV2 (35 ORFs). Based on our study and those of others, the number of core genes shared by all known fuselloviruses now stands at twelve or thirteen (12). Obviously, the availability of SSV20-22 with a drastically reduced number of noncore genes (13, 14) will facilitate the investigation of fusellovirus biology, especially the interactions between a fusellovirus and its host at the molecular level and, in the longer term, the modification and exploitation of fuselloviruses for synthetic biological applications.
In the present study, we show for the first time interconversion among different fusellovirus isolates, or variants, which are highly related at the genomic level, by homologous recombination. Variants were previously reported in the viral isolates of S. islandicus rod-shaped virus 1 (SIRV1) (32). The genomes of the SIRV1 variants were closely related and differed mainly in gene order, gene size, and gene content at localized genomic sites. In a population of variants, the predominant one changed when different host strains were infected. The change in the proportion of different variants in a population was believed to be caused by a novel and putative intron-driven mechanism (33). In addition, a putative archaeal virus, HAV1, assembled from the metagenomic sequences of an environmental sample, was also shown to have variants with sequence differences resulting from insertions, deletions, and other alterations, some of which were caused by recombination through direct repeats (34). Moreover, some fuselloviruses appeared to form chimeras through recombination, presumably serving as a mechanism for the generation of viral diversity (7, 11).
SSV20-22 differ primarily in two genomic regions (regions I and II) but otherwise are highly similar in the remaining genome sequences, except for SNPs. Each region has two versions. Switching between the two versions may alter the interaction of the virus with the host. For example, the two versions of region I encode two different integrases. A change in region I might lead to the integration of a virus into a different site in the host genome. The three viruses also differ at the nucleotide level with a number of SNPs detected. By analyzing DNA sequences extracted from the original enrichment culture, we found that the extent of single-nucleotide polymorphism in SSV20-22 was probably far greater than that demonstrated by the comparison of the genome sequence of the three viruses. Moreover, SNPs were not evenly distributed but enriched in genes such as vp4, which encodes the tail filament protein. No specific patterns of detected point mutations were apparent. Since VP4 is a putative end filament protein, it may serve an important role in host receptor recognition. The high rate of mutation in VP4 may help the virus to overcome host defense.
Extensive variation in the SSV20-22 genome at the levels of both long DNA stretches and single nucleotides appears to depend primarily on efficient HR in the host cell. We found that coinfection of the host strains with SSV20 and SSV22 led to the production of SSV21-like virus with respect to the two variable regions. It was reported that S. islandicus was able to incorporate efficiently short, multiply mismatched donor DNA in a continuous or discontinuous manner by diverse HR events, and the minimal size of homology allowing a specific marker replacement was as short as 10 bp (35). Therefore, the DNA recombination system in Sulfolobus may allow exchange of SNPs between coinfecting SSV20 and SSV22. Taken together, our data suggest that, once a host cell is coinfected with different viruses, efficient HR between their genomic sequences of various sizes would occur, producing progeny virions with a diverse array of genotypes.
Viruses and their hosts are known to be engaged in an everlasting arms race during the course of evolution. The presence of variants could conceivably enable a virus to adapt to infecting its evolving host (36, 37). The complex virus-host interactions would ultimately drive the coevolution of viruses and their hosts.
MATERIALS AND METHODS
Isolation of viruses and host strains.
A sample of sediment collected from an acidic hot spring (GPS coordinates 13°07′48″N, 123°54′36″E; 64.1°C, pH 3.72) in Naghaso, Philippines, was inoculated into Zillig’s medium (26). After incubation with shaking for 7 days at 75°C, the culture was centrifuged at 5,000 × g for 30 min at 4°C. The supernatant was filtered through a 0.45-μm filter (Merck Millipore), and the filtrate was centrifuged at 120,000 × g for 1 h at 4°C. The pellet was resuspended in Zillig’s basal salts (26), and the suspension was examined under TEM for the presence of virus-like particles.
To isolate virus-containing and virus-free strains, the enrichment culture was subjected to strain purification by double-layer plating on solid plates and by dilution to extinction in liquid medium. To purify strains by plating, the enrichment culture was diluted and plated on PSCVY plates (38) with 0.2% Gelrite in the top layer and 0.8% Gelrite in the bottom layer. Single colonies were picked and inoculated into liquid PSCVY medium. After incubation, the culture supernatant was examined by TEM. Cultures containing virus particles were purified again by plating. A virus-carrying strain, denoted Sulfolobus sp. strain E11-6, was isolated and shown to contain SSV19. To isolate a virus-free strain, strain E11-6 was subcultured numerous times and plated. A large number of single colonies were screened in liquid culture by TEM and PCR, using a primer pair of E11-VP F/R (Table 2), designed on the basis of the SSV19 viral capsid genes. However, no virus-free host strain for SSV19 was obtained.
To purify strains by dilution to extinction, the enrichment culture was subjected to 10-fold serial dilution in Zillig’s medium, and the dilutions were incubated with shaking for ∼4 days at 75°C. The highest dilutions, which turned turbid after incubation, were tested for the presence of virus particles by TEM. A virus-carrying culture, named E5, was identified and plated on a PSCVY plate. After screening colonies by TEM and PCR, using a primer pair E5-Int F/R (Table 2) which targeted the integrase gene from viral DNA of E5, a virus-free strain, denoted Sulfolobus sp. strain E5-1-F, was obtained. The strain was infected with the virus from strain E5. The infected strain, denoted Sulfolobus sp. strain E5-1, was purified by two rounds of colony picking. Strain E5-1 contained at least four different viruses, as revealed by viral genome sequencing and assembly. The four putative viral genomes differed primarily in two regions (regions I and II), and each of the two regions had two alternative sequences (i.e., IA and IB for region I and IIA and IIB for region II). In other words, among the four viral genomes, two shared region I and differed in region II, while the other two shared region II and differed in region I. We were unsuccessful in purifying individual viruses by picking single colonies. Taking advantage of the small sizes of the viral genomes, the whole-genome PCR amplification approach was employed to isolate individual viruses. Viral genome sequences from strain E5-1 were amplified by PCR using LA taq (TaKaRa) with the overlap extension primer pair IIA-wg F/R or IIB-wg F/R (Table 2), which were designed to amplify the entire viral genomes containing sequence IIA or sequence IIB, respectively. PCR products, which might result from the amplification of viral genomes containing different region I sequences, were introduced into strain E5-1-F by electroporation, and the transformants were plated on PSCVY plates. Colonies were screened for those containing virus particles by TEM. Strains containing a single virus were obtained by PCR with primers permitting the distinction of viral genomes with different combinations of regions I and II (i.e., IA F/R, IB F/R, IIA F/R, and IIB F/R; Table 2). Strains carrying a single virus, designated SSV20, SSV21, and SSV22, respectively, were purified by repeated plating and colony picking. However, we were unable to obtain a strain containing SSV23, identified in strain E5-1.
Purification of virions.
A strain containing a virus was grown in liquid PSCVY medium for 5 days with shaking at 75°C (optical density at 600 nm [OD600] of ∼1.0). The culture was centrifuged at 5,000 × g for 30 min at 4°C, and the supernatant was filtered through a 0.45-μm filter and centrifuged at 120,000 × g for 1 h at 4°C. The pellet was resuspended in Zillig’s basal salts and subjected to two runs of cesium chloride density gradient centrifugation, with a final CsCl concentration of 0.45 g/ml at 200,000 × g for 24 h at 4°C. The purified virions were washed by ultrafiltration through a 30-kDa centrifugal filter unit (Merck Millipore).
Electron microscopy.
Virions were stained with 2% (wt/vol) uranyl acetate and observed under a JEM-1400 transmission electron microscope (JEOL).
DNA isolation, sequencing, and sequence analysis.
Purified virus particles were digested with proteinase K (0.6 mg/ml) at 50°C for 3 h in the presence of 2% SDS. Cetyltrimethylammonium bromide (CTAB) and NaCl were added to 1% and 0.8 M, respectively. After incubation at 65°C for 10 min, the sample was extracted with phenol-chloroform-isoamyl alcohol (25:24:1), and the DNA was precipitated with 70% ethanol at –20°C for 2 h, centrifuged, dried, and dissolved in Tris-EDTA buffer. Viral DNA was sequenced by using the Illumina HiSeq-PE150 (SSV19) and -PE250 (SSV20-22) system at Microbial Genome Research Center, Institute of Microbiology, Chinese Academy of Sciences, China. The genome sequences were assembled using the SPAdes 3.11 package (39). The genome was annotated by RAST (40–42) as well as ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/) using the genetic code for Bacterial, Archaeal, and Plant Plastid. Alignments were performed by using the Basic Local Alignment Search Tool (BLASTP) at NCBI. Gene domain architectures were predicted with SMART (http://smart.embl-heidelberg.de). Pairwise alignment of ORFs of SSV19-22 with the other forty-five fuselloviruses (SSV1-SSV10, ASV1, SSV11-15, SSV17-18, and 27 integrated fuselloviral genomes) were performed via BLASTP (NCBI) with an e value of ≤10−3.
Sulfolobus sp. strain E11-6 and Sulfolobus sp. strain E5-1-F were sequenced by PacBio RSII technology and assembled by hierarchical genome assembly processing at Beijing Institute of Genomics, Chinese Academy of Sciences.
Phylogenetic analysis.
Phylogenetic analysis of 16S rRNA genes from the two viral hosts and 17 additional Sulfolobus strains was conducted in MEGA7 (43). Sequences were aligned using ClustalW (44), and the phylogenetic tree was constructed using the neighbor-joining method (45) with a bootstrap of 1,000 replicates. The evolutionary distances were computed using the maximum composite likelihood method (46). All positions containing gaps and missing data were eliminated.
Protein analysis.
SSV19 virions (∼200 μg) and SSV20-22 virions (∼300 μg), purified by CsCl density gradient centrifugation, were loaded on a 14% tricine SDS-PAGE gel (47). The gel was stained with Coomassie brilliant blue G250. The proteins were digested in-gel with trypsin (12.5 ng/μl), and the resulting peptides were identified by an AB Sciex 5800 TOF/TOF (AB Sciex, Foster City, CA). For N-terminal amino acid sequencing, proteins in the gel were electrophoretically transferred to a polyvinylidene difluoride membrane (Merck Millipore). The protein bands then were sliced, and the proteins were subjected to N-terminal amino acid sequencing at the Protein Sequencing Center, School of Life Sciences, Peking University, China.
Lipid analysis.
Thin-layer chromatography of the lipids from virions and host cells was performed as described previously, with modifications (48). Strain E5-1-F was cultured to logarithmic phase (OD600 of 0.3 to 0.4), and cells (from 200 ml culture) were harvested, washed, and resuspended in Zillig’s basal salts. Purified virions (∼12 mg/ml) were also prepared. Samples of the cells and the virions were lyophilized in a Speed-Vac concentrator, resuspended in chloroform-methanol (1:1), sonicated, and incubated at 65°C for 16 h. After centrifugation, the supernatant was dried to powders. The dried extracts were redissolved in chloroform-methanol-H2O (75:24:1, 100 μl), and the samples were subjected to thin-layer chromatography on a Merck silica 60 plate in chloroform-methanol-H2O (75:24:1) for 3 h at 10°C. After the run, the plate was dipped with iodine overnight in a sealed environment.
Infection experiments and quantification of viral DNA.
Strain E5-1-F, grown exponentially (OD600 of ∼0.2 to 0.4) in PSCVY medium, was infected at an MOI of ∼5 with purified SSV20, SSV21, or SSV22. The cultures were incubated with shaking at 75°C. Total DNAs were extracted from the cultures (for total virus counts) and from the harvested cells (for cell-associated virus counts), and the viral DNA was quantified by qPCR with a virus-specific primer pair, vp1 qPCR-F/R (Table 2), on a LightCycler 480 fluorescence quantitative PCR instrument (Roche Life Science) (Table S4). To prepare a standard curve for virus quantification, the vp1 PCR fragment (165 bp) was cloned into the pEASY-T1 cloning vector (TransGen Biotech). The plasmid then was serially diluted and amplified as the known control to generate a standard curve relating the copy number of viral DNA with CT (threshold cycle) values. A qPCR mixture (20 μl) contained 10 μl of 2× TB Green Premix Ex Taq II (Tli RNaseH Plus; TaKaRa Bio), 0.8 μl each of the vp1 primers (10 μM), 2 μl of DNA sample, and 6.4 μl of ultrapure water. A two-step qPCR program was used with the following parameters: 1 cycle of 95°C, 30 s; 35 cycles of 95°C, 5 s, and 60°C, 30 s; 1 cycle of 95°C, 5 s, 60°C, 1 min, and 95°C, 0 min; 1 cycle of cooling to 50°C, 30 s. The efficiency of qPCR was ∼96%, with an r2 value of >0.99 (Table S4). Data were analyzed with LightCycler 480 software, version 1.5.1.62, using the Fit Points method to calculate the CT values according to the software instructions. Specificity of the qPCRs was tested by gel electrophoresis through 2.0% agarose and by melting curve analysis. For coinfection experiments, the infected cultures were grown with shaking for 3 days at 75°C. The viral genomes were identified by PCR using the indicated primer pairs, 1F/1R, 1F/2R, 2F/1R, and 2F/2R (Table 2).
Data availability.
The genome sequences of SSV19 and SSV20-22 are available in the GenBank database under accession numbers MN496305 (SSV19), MN496306 (SSV20), MN496307 (SSV21), and MN496308 (SSV22). The accession numbers of two host strains available in the GenBank database are CP045687 (Sulfolobus sp. strain E5-1-F) and CP045706 (Sulfolobus sp. strain E11-6).
Supplementary Material
ACKNOWLEDGMENTS
We thank Kenneth M. Stedman, from Portland State University, OR, for providing information on the annotation of SSV1-SSV3 and SSV10, Carlo A. Arcilla, from the National Institute of Geological Sciences, and Ramonito Solis from the University of the Philippines and the Resource Management Department, Energy Development Corporation Ortigas Center, the Philippines, for assistance with sample collection, and Yaxin Zhu, from the State Key Laboratory of Microbial Resources, Institute of Microbiology, Chinese Academy of Sciences, for assistance in the extraction of genomic DNA from Sulfolobus strains.
This work was supported by the National Natural Science Foundation of China (NSFC) grant 31970170.
Footnotes
Supplemental material is available online only.
REFERENCES
- 1.Wang H, Peng N, Shah SA, Huang L, She Q. 2015. Archaeal extrachromosomal genetic elements. Microbiol Mol Biol Rev 79:117–152. doi: 10.1128/MMBR.00042-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Krupovic M, Cvirkaite-Krupovic V, Iranzo J, Prangishvili D, Koonin EV. 2018. Viruses of archaea: structural, functional, environmental and evolutionary genomics. Virus Res 244:181–193. doi: 10.1016/j.virusres.2017.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prangishvili D, Bamford DH, Forterre P, Iranzo J, Koonin EV, Krupovic M. 2017. The enigmatic archaeal virosphere. Nat Rev Microbiol 15:724–739. doi: 10.1038/nrmicro.2017.125. [DOI] [PubMed] [Google Scholar]
- 4.Schleper C, Kubo K, Zillig W. 1992. The particle SSV1 from the extremely thermophilic archaeon Sulfolobus is a virus: demonstration of infectivity and of transfection with viral DNA. Proc Natl Acad Sci U S A 89:7645–7649. doi: 10.1073/pnas.89.16.7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stedman KM, She Q, Phan H, Arnold HP, Holz I, Garrett RA, Zillig W. 2003. Relationships between fuselloviruses infecting the extremely thermophilic archaeon Sulfolobus: SSV1 and SSV2. Res Microbiol 154:295–302. doi: 10.1016/S0923-2508(03)00074-3. [DOI] [PubMed] [Google Scholar]
- 6.Peng X. 2008. Evidence for the horizontal transfer of an integrase gene from a fusellovirus to a pRN-like plasmid within a single strain of Sulfolobus and the implications for plasmid survival. Microbiology 154:383–391. doi: 10.1099/mic.0.2007/012963-0. [DOI] [PubMed] [Google Scholar]
- 7.Redder P, Peng X, Brugger K, Shah SA, Roesch F, Greve B, She Q, Schleper C, Forterre P, Garrett RA, Prangishvili D. 2009. Four newly isolated fuselloviruses from extreme geothermal environments reveal unusual morphologies and a possible interviral recombination mechanism. Environ Microbiol 11:2849–2862. doi: 10.1111/j.1462-2920.2009.02009.x. [DOI] [PubMed] [Google Scholar]
- 8.Wiedenheft B, Stedman K, Roberto F, Willits D, Gleske AK, Zoeller L, Snyder J, Douglas T, Young M. 2004. Comparative genomic analysis of hyperthermophilic archaeal Fuselloviridae viruses. J Virol 78:1954–1961. doi: 10.1128/jvi.78.4.1954-1961.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Stedman KM, Clore A, Combet-Blanc Y. 2006. Biogeographical diversity of archaeal viruses, p 131–143. In Logan NA, Lappin-Scott HM, Oyston PCF (ed), Prokaryotic diversity: mechanisms and significance. Cambridge University Press, Cambridge, United Kingdom. [Google Scholar]
- 10.Goodman DA, Stedman KM. 2018. Comparative genetic and genomic analysis of the novel fusellovirus Sulfolobus spindle-shaped virus 10. Virus Evol 4:vey022. doi: 10.1093/ve/vey022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pauly MD, Bautista MA, Black JA, Whitaker RJ. 2019. Diversified local CRISPR-Cas immunity to viruses of Sulfolobus islandicus. Philos Trans R Soc Lond B Biol Sci 374:20180093. doi: 10.1098/rstb.2018.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Iverson EA, Goodman DA, Gorchels ME, Stedman KM. 2017. Extreme mutation tolerance: nearly half of the archaeal fusellovirus sulfolobus spindle-shaped virus 1 genes are not required for virus function, including the minor capsid protein gene vp3. J Virol 91:e02406-16. doi: 10.1128/JVI.02406-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Martin A, Yeats S, Janekovic D, Reiter WD, Aicher W, Zillig W. 1984. SAV 1, a temperate u.v.-inducible DNA virus-like particle from the archaebacterium Sulfolobus acidocaldarius isolate B12. EMBO J 3:2165–2168. doi: 10.1002/j.1460-2075.1984.tb02107.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quemin ER, Chlanda P, Sachse M, Forterre P, Prangishvili D, Krupovic M. 2016. Eukaryotic-like virus budding in Archaea. mBio 7:e01439-16. doi: 10.1128/mBio.01439-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Palm P, Schleper C, Grampp B, Yeats S, McWilliam P, Reiter WD, Zillig W. 1991. Complete nucleotide sequence of the virus SSV1 of the archaebacterium Sulfolobus shibatae. Virology 185:242–250. doi: 10.1016/0042-6822(91)90771-3. [DOI] [PubMed] [Google Scholar]
- 16.Fusco S, She Q, Bartolucci S, Contursi P. 2013. T(lys), a newly identified Sulfolobus spindle-shaped virus 1 transcript expressed in the lysogenic state, encodes a DNA-binding protein interacting at the promoters of the early genes. J Virol 87:5926–5936. doi: 10.1128/JVI.00458-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Reiter WD, Palm P, Yeats S. 1989. Transfer RNA genes frequently serve as integration sites for prokaryotic genetic elements. Nucleic Acids Res 17:1907–1914. doi: 10.1093/nar/17.5.1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quemin ER, Pietila MK, Oksanen HM, Forterre P, Rijpstra WI, Schouten S, Bamford DH, Prangishvili D, Krupovic M. 2015. Sulfolobus spindle-shaped virus 1 contains glycosylated capsid proteins, a cellular chromatin protein, and host-derived lipids. J Virol 89:11681–11691. doi: 10.1128/JVI.02270-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Reiter W-D, Palm P, Henschen A, Lottspeich F, Zillig W, Grampp B. 1987. Identification and characterization of the genes encoding three structural proteins of the Sulfolobus virus-like particle SSV1. Mol Gen Genet 206:144–153. doi: 10.1007/BF00326550. [DOI] [Google Scholar]
- 20.Lawrence CM, Menon S, Eilers BJ, Bothner B, Khayat R, Douglas T, Young MJ. 2009. Structural and functional studies of archaeal viruses. J Biol Chem 284:12599–12603. doi: 10.1074/jbc.R800078200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Menon SK, Maaty WS, Corn GJ, Kwok SC, Eilers BJ, Kraft P, Gillitzer E, Young MJ, Bothner B, Lawrence CM. 2008. Cysteine usage in Sulfolobus spindle-shaped virus 1 and extension to hyperthermophilic viruses in general. Virology 376:270–278. doi: 10.1016/j.virol.2008.03.026. [DOI] [PubMed] [Google Scholar]
- 22.Kraft P, Oeckinghaus A, Kummel D, Gauss GH, Gilmore J, Wiedenheft B, Young M, Lawrence CM. 2004. Crystal structure of F-93 from Sulfolobus spindle-shaped virus 1, a winged-helix DNA binding protein. J Virol 78:11544–11550. doi: 10.1128/JVI.78.21.11544-11550.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fusco S, She Q, Fiorentino G, Bartolucci S, Contursi P. 2015. Unravelling the role of the F55 regulator in the transition from lysogeny to UV induction of Sulfolobus spindle-shaped virus 1. J Virol 89:6453–6461. doi: 10.1128/JVI.00363-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Koonin EV. 1992. Archaebacterial virus SSV1 encodes a putative DnaA-like protein. Nucleic Acids Res 20:1143. doi: 10.1093/nar/20.5.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Menon SK, Eilers BJ, Young MJ, Lawrence CM. 2010. The crystal structure of D212 from Sulfolobus spindle-shaped virus ragged hills reveals a new member of the PD-(D/E)XK nuclease superfamily. J Virol 84:5890–5897. doi: 10.1128/JVI.01663-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zillig W, Kletzin A, Schleper C, Holz I, Janekovic D, Hain J, Lanzendorfer M, Kristjansson JK. 1993. Screening for Sulfolobales, their plasmids and their viruses in Icelandic solfataras. Syst Appl Microbiol 16:609–628. doi: 10.1016/S0723-2020(11)80333-4. [DOI] [Google Scholar]
- 27.Muskhelishvili G, Palm P, Zillig W. 1993. SSV1-encoded site-specific recombination system in Sulfolobus shibatae. Mol Gen Genet 237:334–342. doi: 10.1007/bf00279436. [DOI] [PubMed] [Google Scholar]
- 28.Muskhelishvili G. 1993. The archaeal SSV integrase promotes intermolecular excisive recombination in vitro. Syst Appl Microbiol 16:605–608. doi: 10.1016/S0723-2020(11)80332-2. [DOI] [Google Scholar]
- 29.Servín-Garcidueñas LE, Peng X, Garrett RA, Martínez-Romero E. 2013. Genome sequence of a novel archaeal fusellovirus assembled from the metagenome of a Mexican hot spring. Genome Announc 1:e0016413. doi: 10.1128/genomeA.00164-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yoshida H, Fukushima Y, Goto M, Tsuyuki Y, Takahashi T. 2019. Analysis of the type II-A CRISPR-Cas System in Streptococcus canis isolated from diseased companion animals and one human patient in Japan. Jpn J Infect Dis 72:261–265. doi: 10.7883/yoken.JJID.2018.492. [DOI] [PubMed] [Google Scholar]
- 31.Couvin D, Bernheim A, Toffano-Nioche C, Touchon M, Michalik J, Neron B, Rocha EPC, Vergnaud G, Gautheret D, Pourcel C. 2018. CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res 46:W246–W251. doi: 10.1093/nar/gky425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang C, Whitaker RJ. 2018. Microhomology-mediated high-throughput gene inactivation strategy for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Appl Environ Microbiol 84:e02167-17. doi: 10.1128/AEM.02167-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Prangishvili D, Arnold HP, Gotz D, Ziese U, Holz I, Kristjansson JK, Zillig W. 1999. A novel virus family, the Rudiviridae: structure, virus-host interactions and genome variability of the sulfolobus viruses SIRV1 and SIRV2. Genetics 152:1387–1396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng X, Kessler A, Phan H, Garrett RA, Prangishvili D. 2004. Multiple variants of the archaeal DNA rudivirus SIRV1 in a single host and a novel mechanism of genomic variation. Mol Microbiol 54:366–375. doi: 10.1111/j.1365-2958.2004.04287.x. [DOI] [PubMed] [Google Scholar]
- 35.Garrett RA, Prangishvili D, Shah SA, Reuter M, Stetter KO, Peng X. 2010. Metagenomic analyses of novel viruses and plasmids from a cultured environmental sample of hyperthermophilic neutrophiles. Environ Microbiol 12:2918–2930. doi: 10.1111/j.1462-2920.2010.02266.x. [DOI] [PubMed] [Google Scholar]
- 36.Paez-Espino D, Sharon I, Morovic W, Stahl B, Thomas BC, Barrangou R, Banfield JF. 2015. CRISPR immunity drives rapid phage genome evolution in Streptococcus thermophilus. mBio 6:e00262-15. doi: 10.1128/mBio.00262-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han P, Deem MW. 2017. Non-classical phase diagram for virus bacterial coevolution mediated by clustered regularly interspaced short palindromic repeats. J R Soc Interface 14:20160905. doi: 10.1098/rsif.2016.0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng L, Zhu H, Chen Z, Liang YX, She Q. 2009. Unmarked gene deletion and host-vector system for the hyperthermophilic crenarchaeon Sulfolobus islandicus. Extremophiles 13:735–746. doi: 10.1007/s00792-009-0254-2. [DOI] [PubMed] [Google Scholar]
- 39.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA, Formsma K, Gerdes S, Glass EM, Kubal M, Meyer F, Olsen GJ, Olson R, Osterman AL, Overbeek RA, McNeil LK, Paarmann D, Paczian T, Parrello B, Pusch GD, Reich C, Stevens R, Vassieva O, Vonstein V, Wilke A, Zagnitko O. 2008. The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T, Edwards RA, Gerdes S, Parrello B, Shukla M, Vonstein V, Wattam AR, Xia F, Stevens R. 2014. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Res 42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brettin T, Davis JJ, Disz T, Edwards RA, Gerdes S, Olsen GJ, Olson R, Overbeek R, Parrello B, Pusch GD, Shukla M, Thomason JA, Stevens R, Vonstein V, Wattam AR, Xia F. 2015. RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci Rep 5:8365. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thompson JD, Gibson TJ, Higgins DG. 2002. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2:Unit 2.3. [DOI] [PubMed] [Google Scholar]
- 45.Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 46.Tamura K, Nei M, Kumar S. 2004. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci U S A 101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schagger H. 2006. Tricine-SDS-PAGE. Nat Protoc 1:16–22. doi: 10.1038/nprot.2006.4. [DOI] [PubMed] [Google Scholar]
- 48.Arnold HP, Zillig W, Ziese U, Holz I, Crosby M, Utterback T, Weidmann JF, Kristjanson JK, Klenk HP, Nelson KE, Fraser CM. 2000. A novel lipothrixvirus, SIFV, of the extremely thermophilic crenarchaeon Sulfolobus. Virology 267:252–266. doi: 10.1006/viro.1999.0105. [DOI] [PubMed] [Google Scholar]
- 49.Frols S, Gordon PM, Panlilio MA, Schleper C, Sensen CW. 2007. Elucidating the transcription cycle of the UV-inducible hyperthermophilic archaeal virus SSV1 by DNA microarrays. Virology 365:48–59. doi: 10.1016/j.virol.2007.03.033. [DOI] [PubMed] [Google Scholar]
- 50.Felsenstein J. 1985. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The genome sequences of SSV19 and SSV20-22 are available in the GenBank database under accession numbers MN496305 (SSV19), MN496306 (SSV20), MN496307 (SSV21), and MN496308 (SSV22). The accession numbers of two host strains available in the GenBank database are CP045687 (Sulfolobus sp. strain E5-1-F) and CP045706 (Sulfolobus sp. strain E11-6).