Fibropapillomatosis (FP) is a tumor disease associated with chelonid herpesvirus 5 (ChHV5) that is an important cause of mortality in threatened green turtles globally. FP is expanding in Florida and the Caribbean but declining in Hawaii. We show that Hawaiian turtles mount antibodies to ChHV5 mainly in response to tumors, which are the only sites of viral replication, whereas tumored and nontumored Floridian turtles are uniformly seropositive. Tumor viruses that depend on tumors for replication and spread are rare, with the only example being the retrovirus causing walleye dermal sarcoma in fish. The Hawaiian strain of ChHV5 may be the first DNA virus with such an unusual life history. Our findings, along with the fundamental differences in the life histories between Floridian turtles and Hawaiian turtles, may partly explain the differential dynamics of FP between the two regions.
KEYWORDS: fibropapilloma, green turtle, Chelonia mydas, IgY, herpesvirus, serology, fibropapillomatosis, chelonid herpesvirus 5
ABSTRACT
Fibropapillomatosis (FP) is a tumor disease associated with a herpesvirus (chelonid herpesvirus 5 [ChHV5]) that affects mainly green turtles globally. Understanding the epidemiology of FP has been hampered by a lack of robust serological assays to monitor exposure to ChHV5. This is due in part to an inability to efficiently culture the virus in vitro for neutralization assays. Here, we expressed two glycoproteins (FUS4 and FUS8) from ChHV5 using baculovirus. These proteins were immobilized on enzyme-linked immunosorbent assay plates in their native form and assayed for reactivity to two types of antibodies, full-length 7S IgY and 5.7S IgY, which has a truncated Fc region. Turtles from Florida were uniformly seropositive to ChHV5 regardless of tumor status. In contrast, in turtles from Hawaii, we detected strong antibody reactivity mainly in tumored animals, with a lower antibody response being seen in nontumored animals, including those from areas where FP is enzootic. Turtles from Hawaii actively shedding ChHV5 were more seropositive than nonshedders. In trying to account for differences in the serological responses to ChHV5 between green turtles from Hawaii and green turtles from Florida, we rejected the cross-reactivity of antibodies to other herpesviruses, differences in viral epitopes, or differences in procedure as likely explanations. Rather, behavioral or other differences between green turtles from Hawaii and green turtles from Florida might have led to the emergence of biologically different viral strains. While the strains from turtles in Florida apparently spread independently of tumors, the transmission of the Hawaiian subtype relies heavily on tumor formation.
IMPORTANCE Fibropapillomatosis (FP) is a tumor disease associated with chelonid herpesvirus 5 (ChHV5) that is an important cause of mortality in threatened green turtles globally. FP is expanding in Florida and the Caribbean but declining in Hawaii. We show that Hawaiian turtles mount antibodies to ChHV5 mainly in response to tumors, which are the only sites of viral replication, whereas tumored and nontumored Floridian turtles are uniformly seropositive. Tumor viruses that depend on tumors for replication and spread are rare, with the only example being the retrovirus causing walleye dermal sarcoma in fish. The Hawaiian strain of ChHV5 may be the first DNA virus with such an unusual life history. Our findings, along with the fundamental differences in the life histories between Floridian turtles and Hawaiian turtles, may partly explain the differential dynamics of FP between the two regions.
INTRODUCTION
Fibropapillomatosis (FP) is a tumor disease associated with a herpesvirus (chelonid herpesvirus 5 [ChHV5], genus Scutavirus, subfamily Alphaherpesvirinae) (1) that affects mainly green turtles globally (2, 3). It is one of the most important causes of green turtle stranding in Florida (4), Hawaii (5), and Brazil (6). FP causes external and internal tumors and progressive cachexia (7), leading to immunosuppression (8, 9) and opportunistic septicemias (10). FP has been most intensively studied in green turtles from Hawaii and Florida, where populations have been increasing in both regions since they became protected (11, 12); however, marked differences in the manifestation of the disease exist between the two regions. In Hawaii, FP has a case fatality rate of about 70% (13), with oral tumors being relatively common (14), but the prevalence of the disease there has been declining, for unknown reasons (15). In contrast, in Florida, FP has a much lower case fatality rate (ca. 12%) (16), oral tumors are rarely seen (17), and the prevalence of the disease is stable or expanding regionally (4, 18).
Several lines of evidence suggest that ChHV5 is a cause of FP. Epidermal intranuclear inclusions with virus-like particles with a size and a morphology consistent with those of a herpesvirus have been seen in tumors from green turtles from Florida (19) and Hawaii (20). The disease is transmissible in green turtles using cell-free tumor filtrates (21), and the disease-causing activity of these filtrates is abrogated by chloroform (22), suggesting an enveloped virus as an etiology. Molecular studies have consistently revealed herpesviral DNA in tumors from green turtles in Florida (23), Hawaii (24), and Brazil (25). More recently, herpesviral DNA has also been found in normal tissues from both tumored and nontumored green turtles in the Atlantic (26, 27). Finally, there is the precedent that herpesviruses can cause tumor diseases in other animals, such as Marek’s disease in chickens (28) and Kaposi’s sarcoma in humans (29).
The epizootiology of FP is likely tied to the life history of sea turtles. Adult female green turtles deposit eggs in the sand of the beaches where they nest, and hatchlings emerge about 3 months later and go directly to the open ocean, where they feed on invertebrates. After about 4 to 6 years, juveniles leave the open ocean to recruit to nearshore coastal habitats, where they transition from a carnivorous to a primarily herbivorous diet and there grow to adulthood, whereupon they migrate to nesting grounds to start the cycle anew (30). As the turtles age, their carapace length increases, making the straight carapace length (SCL) a good proxy for age (31). The molecular phylogeny of ChHV5 suggests that infection is acquired shortly after the juveniles migrate from the pelagic environment to nearshore foraging pastures (32). The virus is thought to be transmitted by direct contact (20), cleaner fish (33), or leeches (34).
Efforts to confirm that ChHV5 is the cause of FP in green turtles through the use of Koch’s postulates (35) have been stymied by a lack of laboratory tools. ChHV5 cannot be grown in the laboratory using standard cell monolayers (36); however, it can be grown using more laborious organotypic skin cultures, where the maturation of epidermal cells is apparently critical to allowing viral replication (37), yet these procedures are not suitable for virus isolation or the establishment of neutralization assays, which represent the gold standard for serological assays, in particular, enzyme-linked immunosorbent assay (ELISA)-based assays, for other viral infections. The inability to obtain purified virus in large quantities has also hampered efforts to generate viral antigens and develop reagents to understand the pathogenesis of the disease. Sequencing of ChHV5 (38) led to the generation of reagents that allowed the localization of ChHV5 in intranuclear inclusions in tumors from Hawaiian green turtles and the documentation of superspreaders (37). However, these studies also showed that, on average, epidermal viral shedding is intermittent, sparse, and infrequent, suggesting that this method of spread is highly inefficient. Moreover, recent studies of sea turtles in the Atlantic Ocean and eastern Pacific Ocean have shown that ChHV5 DNA is detectable by PCR in 10 to 100% of apparently healthy animals (39), thus eroding the heretofore tight link between the presence of virus and tumors (24).
A key aspect of understanding and managing viral diseases in animals is the early detection of infection, ideally, in the form of a serological test that can detect exposure to the pathogen prior to the development of clinical signs. The generation of monoclonal antibodies (MAbs) against the two major types of turtle immunoglobulins, 7S IgY and 5.7S IgY, which has a truncated Fc region (40, 41), was a significant step in allowing the development of serological assays. There have since been two attempts to develop serological assays for the detection of antibodies against ChHV5. Herbst et al. (42) used immunohistochemistry to assay the reactivity of turtle 7S IgY antibodies against intranuclear inclusions in tumor tissues in Florida green turtles from areas where FP is enzootic and areas free of FP. They found that turtles from areas where FP is enzootic were significantly more likely to have antibodies to inclusions than turtles from FP-free areas. Subsequently, Herbst et al. (43) expressed glycoprotein H from ChHV5 in a baculovirus system and used this antigen in an indirect ELISA to assay the 7S IgY reactivity of tumored and nontumored Floridian turtles to ChHV5. In contrast to the results of immunohistochemistry, the ELISA results showed that turtles that had experimentally developed FP had a 50% seroconversion rate, whereas wild turtles had an approximately 80 to 100% seroprevalence, regardless of disease status or whether or not they originated from FP-free areas or areas where FP is enzootic. Herbst et al. (43) concluded either that infection with ChHV5 is widespread in Floridian turtles or that there is antibody cross-reactivity to other herpesviruses, although cross-reactivity to a known herpesvirus in Florida responsible for lung eye trachea disease was ruled out.
The discrepancy in ELISA results and disease status for experimentally infected animals and wild animals and the high seropositivity rates in Florida might be related in part to the nonspecific reactivity of turtle antibodies to a denatured antigen. Synthesized proteins bound directly to polystyrene or other polar substrates often undergo denaturation and conformational changes that could alter epitope availability to antibodies (44). Here, we report on an ELISA using two expressed ChHV5 glycoprotein antigens immobilized in the native state and a glutathione system (45). We show that the reactivity of 7S IgY and 5.7S IgY to ChHV5 in Hawaiian turtles increased with an increase in the severity of FP, whereas Floridian turtles were uniformly seropositive, regardless of their tumor scores (TS). Moreover, turtles that were shedding virus had more antibodies than those that were not shedding. Possibilities discussed to explain the differences in the serological responses to ChHV5 between green turtles from Hawaii and green turtles from Florida include the cross-reactivity of antibodies to other herpesviruses, differences in procedures, regional differences in the viral antigen, host antibody, genetic differences in ChHV5, or the immunogenicity of ChHV5 for green turtles.
RESULTS
Antigen.
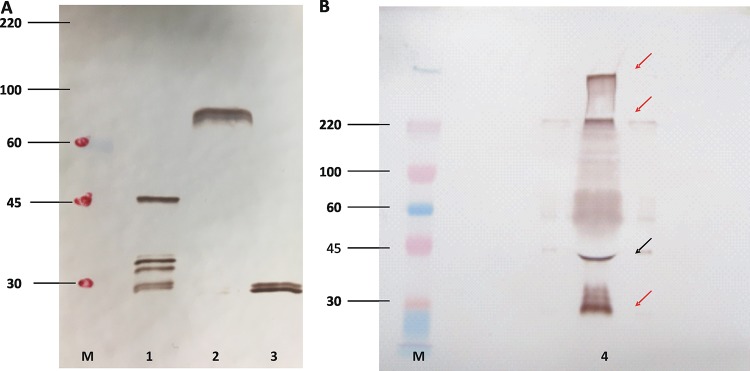
To identify and initially characterize the recombinant baculovirus-expressed antigens, infected Sf9 cell culture supernatants were assayed by polyacrylamide gel electrophoresis (PAGE), followed by Western immunoblotting. Under reducing PAGE conditions, ample amounts of proteins of the expected molecular weight were identified with the help of an MAb against the c-myc epitope, including proteins of ca. 45 kDa for the FUS4 cassette, >70 kDa for the FUS8 cassette, and <30 kDa for the c-myc-GST backbone cassette (Fig. 1). The FUS8 and the c-myc-GST cassettes gave closely migrating double bands, likely representing the glycosylated protein and its nonglycosylated precursor (46). The FUS4 cassette gave a major band of the expected size but also three smaller bands, suggesting posttranslational modifications. Under nonreducing conditions and by use of an MAb against the C-terminal glutathione S-transferase (GST) tail of the fusion protein, FUS4 resolved into a robust large (>200-kDa) band, a weaker 45-kDa band, and a 35-kDa band, the last of which is similar to that for the c-myc backbone, as seen in Fig. 1A. Thus, the FUS4 antigen appeared to comprise a combination of intact large polymers, smaller amounts of intact monomers, and the c-myc backbone with unknown amounts of residual FUS4 (Fig. 1B). Together, the blots revealed that the baculoviruses generated and secreted proteins of the expected molecular weights that remained soluble and available for binding to glutathione.
FIG 1.
Primary characterization of baculovirus-derived antigens by Western blotting. (A) Cell culture supernatants from baculovirus-inoculated Sf9 cells expressing Signal-FUS4-c-myc-GST (lane 1), Signal-FUS8-c-myc-GST (lane 2), and Signal-c-myc-GST (lane 3) were resolved on a 10% polyacrylamide gel, transferred to nitrocellulose, and probed with a monoclonal antibody against c-myc. (B) Western blot of a nonreducing gel analyzed with a monoclonal antibody against GST to further characterize the Signal-FUS4-c-myc-GST protein (lane 4). The black arrow points to the band of the expected size; red arrows point to bands of unexpected sizes. Note that the gel shown in panel B had to be overloaded to make the bands clearly visible. The positions of relevant molecular size markers are indicated (lanes M), and the numbers on the left are in kilodaltons.
Animal sizes.
The approximate age of our two plasma donor populations was addressed by measuring their straight carapace lengths (SCLs). While the SCLs for animals with a tumor score of 0 (TS0) overlapped for animals from both locations, tumored Hawaii turtles were significantly larger and older than tumored Floridian turtles (Fig. 2).
FIG 2.
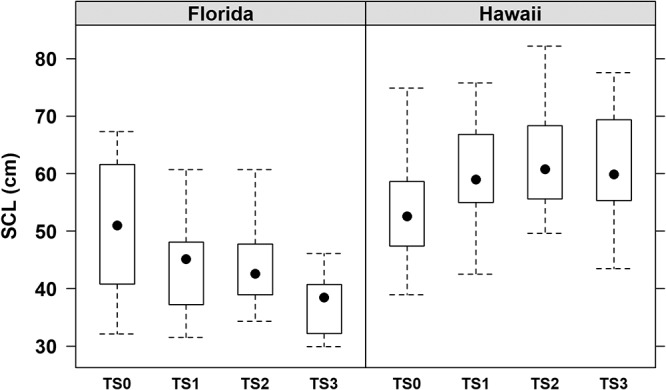
Hawaiian turtles were uniformly larger than Floridian turtles. Box plots of the straight carapace length (SCL) for green turtles from Florida and Hawaii partitioned by tumor score (TS) categories, ranging from nontumored (TS0) to severely tumored (TS3). The boxes represent the 25th and 75th percentiles, whiskers are 1.5 times the interquartile range, and the black dots are the medians.
ELISA.
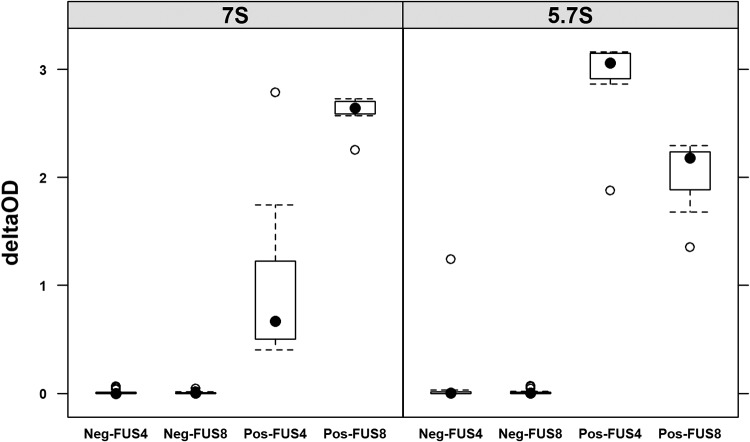
The reactivity of the positive-control plasma was compared to that of a panel of 10 putatively negative plasma samples from captive turtles originating from Sea Life Park (SLP) in Hawaii by plotting the change in the optical density (OD) at 450 nm (delta OD450). The reactivity of the positive-control plasma was the strongest for 5.7S IgY against FUS4, followed by 7S and 5.7S IgY against FUS8, with the weakest reactivity being seen for 7S IgY against FUS4; all negative-control plasma samples (from Sea Life Park) had delta OD values of close to 0, suggesting that they could be used as negative-control sera in order to define a negative cutoff value. The variability in seroreactivity was higher for the positive-control plasma than for the negative-control plasma (Fig. 3).
FIG 3.
Negative sera have uniformly low absorbances. Box plot of delta OD (OD for FUS4 or FUS8 minus the OD for the c-myc backbone) from 1 positive-control (Pos) and 10 negative-control (Neg) plasma samples tested at a 1:25 dilution for 7S or 5.7S IgY antibody reactivity against FUS4 and FUS8 on seven ELISA plates over 3 days (four plates, day 1; three plates, day 2; two plates, day 3). The boxes represent the 25th and 75th percentiles, whiskers are 1.5 times the interquartile range, black dots are medians, and open circles are outliers.
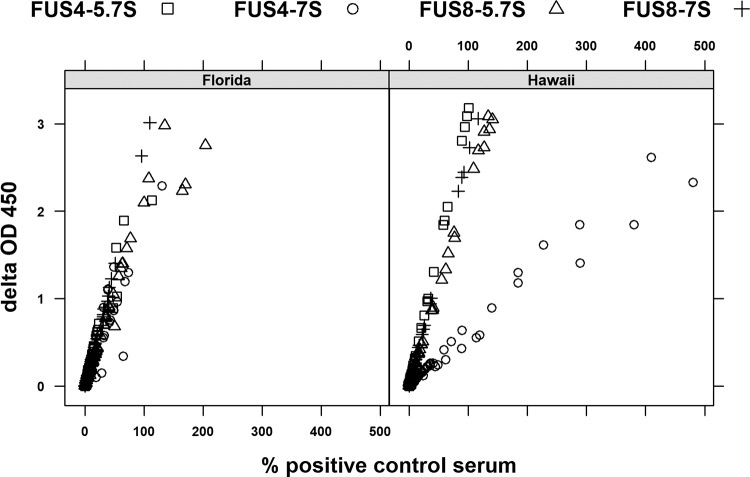
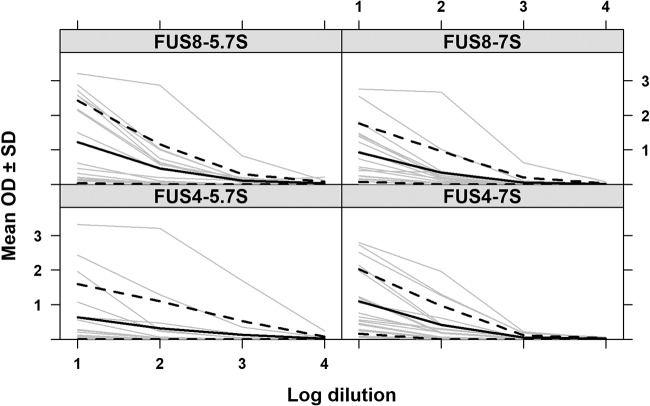
We then tested plasma from 110 Hawaiian turtles and 74 Floridian turtles (tumored and nontumored) for the reactivity of 7S and 5.7S IgY to FUS4 and FUS8 and plotted the delta OD450 values versus the percentage of the OD for the positive-control serum. For both antigens and antibody types and in both geographical locations, the two values correlated in a nearly linear fashion, and wild populations had values that represented the entire spectrum from negative to strongly positive (Fig. 4). Interestingly, most reactions were less than 100% of the value for the positive-control serum. The only exception was the reactivity of 7S IgY against FUS4 for Hawaiian turtles, where values of up to 500% of the value for the positive-control serum were observed. To address the question of whether or not the ELISA reactions corresponded to the antibody titers, a panel of plasma from 18 shedding and nonshedding Hawaiian turtles with FP was analyzed by plasma titration prior to ELISA, and a strong relationship between the serum dilution and delta OD450 values was shown (Fig. 5). Together, these data suggest that our positive-control plasma had high titers of various antibodies against ChHV5, which made it inappropriate for determining a positive cutoff value to oppose the negative cutoff value defined by the panel of plasma originating from turtles in SLP.
FIG 4.
Delta OD and the percentage of the value for the positive control correlate. Scatter plot ELISA results are expressed as delta OD450 values versus the delta OD450 values expressed as a percentage of the value for positive-control plasma for tumored and nontumored Floridian (n = 74, left) and Hawaiian (n = 110, right) green turtles assayed for the reactivity of 7S or 5.7S IgY against FUS8 or FUS4.
FIG 5.
Plasma dilutions correlate with absorbance. The plots show the delta OD450 values versus the log of serial 10-fold dilutions of plasma partitioned by antigen and antibody reactivity for 18 Hawaiian green turtles (gray), with the mean OD (solid black line) ± 1 standard deviation (dotted lines) being shown.
In the absence of an accepted gold standard and by keeping in mind that the ELISA reactivity correlated well the antibody titer, we tested two cutoff values: a delta OD value of 0.3 (representing high specificity) and a value 3 standard deviations (SD) above the mean delta OD for negative plasma (from turtles from Sea Life Park) (representing high sensitivity) (Table 1). The SLP animal population remained seronegative under both conditions, whereas a majority of animals showed reactivity with values somewhere between the two cutoff values, regardless of their tumor score (Fig. 6). Thus, using these two cutoff values, it was possible to stratify the seropositive animals into high responders and low responders.
TABLE 1.
Delta OD cutoffs used to score Hawaiian and Floridian turtles as seropositive or seronegative for antibodies to ChHV5, sensitivity or specificity of ELISA to predict tumor/nontumor status using a particular cutoff, and general cutoff type
| Assay type | Delta OD |
Cutoff type | ||
|---|---|---|---|---|
| Cutoff | Sensitivity | Specificity | ||
| All assays | 0.3 | 0.340278 | 0.926829 | High specificity |
| FUS4-5.7S (delta OD + 3 SD) | 0.030981 | 0.768293 | 0.509259 | High sensitivity |
| FUS4-7S (delta OD + 3 SD) | 0.070686 | 0.756098 | 0.592593 | |
| FUS8-5.7S (delta OD + 3 SD) | 0.06654 | 0.707317 | 0.583333 | |
| FUS8-7S (delta OD + 3 SD) | 0.025729 | 0.609756 | 0.842593 | |
FIG 6.
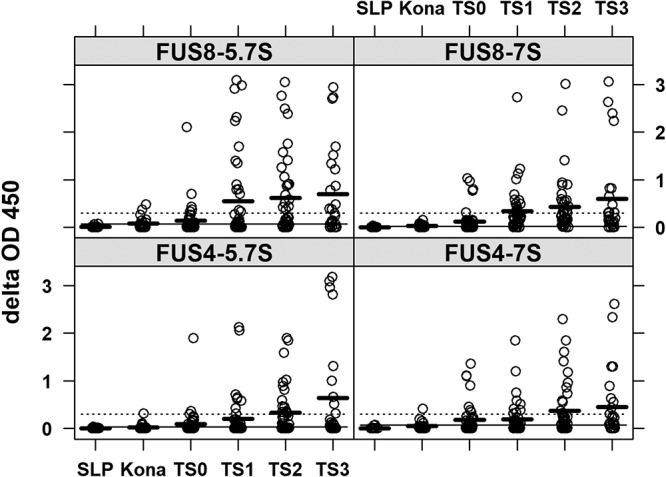
Increasing ELISA reactions with increasing tumor score. Delta OD values for different groups of animals according to tumor score categories are shown. SLP, Sea Life Park negative controls; Kona, nontumored turtles from Kona, HI (an FP-free area); TS0 to TS3, tumor scores of 0 to 3, respectively, for animals from areas in Hawaii and Florida where FP is enzootic. Thick dark lines are the mean OD values for a particular tumor score category, dotted lines are the high-specificity cutoff, and solid lines are the high-sensitivity cutoff. Sample sizes were as follows: SLP, 10 plasma samples; Kona, 20 plasma samples; TS0, 40 plasma samples; TS1, 44 plasma samples; TS2, 44 plasma samples; TS3, 26 plasma samples.
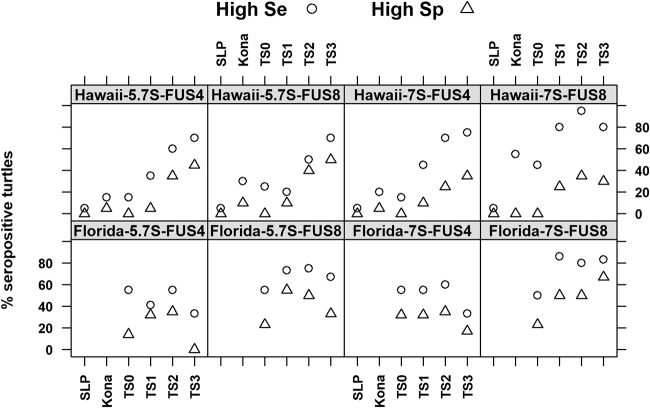
When seropositivity was assayed for each cutoff type (high sensitivity, high specificity) by tumor score and geographical region, distinct differences were seen between Floridian turtles and Hawaiian turtles. In Hawaii, the percentage of seropositive animals increased steadily with an increase in the tumor score, regardless of the cutoff, suggesting that increasing tumor scores correlate with frequent virus replication and exposure of the host to the antigen, thus leading to more pronounced antibody reactions (Fig. 7). The frequency of low responders ranged from 20% to 40% among the animals with TS0, which confirmed that the virus is able to infect animals prior to the formation of tumors. Importantly, high responders were very rare among the TS0 animals, which suggested that only a few of them had been exposed to extensive or prolonged viral replication. In contrast, Floridian turtles seemed to be uniformly seropositive, regardless of their tumor score. In particular, up to 60% of the TS0 animals from Florida were seropositive, with up to 30% high reactors being seen among them. Moreover, there was a decline in rate of seropositivity for TS3 animals for all but 7S IgY against FUS8.
FIG 7.
Contrasting seroresponses of Hawaiian and Floridian turtles. The percentages of green turtles from Hawaii and Florida testing ELISA positive (given as the percentage of seropositive turtles) for 7S or 5.7S IgY antibody reactivity to FUS4 or FUS8 according to the high-sensitivity (High Se) or high-specificity (High Sp) cutoffs for the following six groups of turtles are shown: Sea Life Park negative controls (SLP); tumor-free turtles from the Kona coast, west Hawaii (Kona); and turtles from Kaneohe Bay, Oahu, where tumors are endemic, with tumor scores (TS) ranging from 0 (TS0; no tumors) to 3 (TS3; severely tumored).
Based on the observation that increasing antibody responses correlated with increasing tumor scores for Hawaiian turtles, we hypothesized that tumor formation and viral shedding could influence the serological status for antibodies to ChHV5. Moreover, chronic exposure to viral antigens was expected to lead to increased 5.7S IgY antibody responses. To address this hypothesis, we compared the serological data for tumored turtles known to be shedding ChHV5 to those for nonshedders, as assayed by histopathology (20). For both the 7S IgY and the 5.7S IgY responses, the antibody reactions of the nonshedders remained mostly below the cutoff of a delta OD of >0.3, whereas, in comparison, more shedders had values above the cutoff. In particular, numerous high responders (those with a high 5.7S IgY response) due to chronic antigen exposure were identified among the shedders using the FUS8 antigen (Fig. 8). Interestingly, among the animals with no shedding detected at the time of necropsy, one animal (animal 21011) showed extremely high levels of 5.7S IgY antibodies against both FUS4 and FUS8. Given that only a small percentage of tumors shed virus and given the low sensitivity of histology as a tool to detect viral shedding, it is likely that the detection of shedding was missed in this animal (20).
FIG 8.
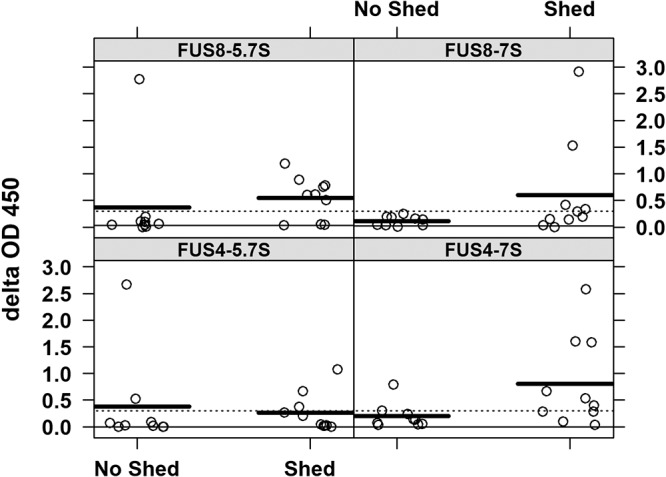
Some but not all virus shedders showed strong ELISA reactions, which inferred chronic antigen exposure. Delta OD values are for 10 shedders and 8 nonshedders of ChHV5. Thick dark lines are the mean OD values for a particular shedding category, dotted lines are the high-specificity cutoff, and solid lines are the high-sensitivity cutoff.
DISCUSSION
Using immobilized, nondenatured, baculovirus-expressed antigens, we show that green turtles in Hawaii and Florida mount detectable 7S and 5.7S IgY antibody responses to two different glycoprotein antigens of ChHV5. Using a selected panel of sera, we also established that the ELISA responses correlated well with the antibody titers. In the absence of a neutralization assay as a gold standard for the detection of antibodies against ChHV5, we tested two cutoff values for seropositivity. To favor sensitivity, we set the cutoff value at an OD of >3 SD of the mean delta OD for the negative controls, which allowed the detection of animals responding at a low level (43). To favor specificity, we set the cutoff at a delta OD of 0.3 because this OD can easily be recognized by the naked eye; this simultaneously allowed us to identify strongly responding animals (47). Regardless of the cutoff, we saw a marked difference between the antibody responses to ChHV5 of turtles from Hawaii and the antibody responses to ChHV5 of turtles from Florida. In Hawaii, the number of strong antibody responders (those reacting above a high specificity cutoff) clearly increased with the tumor score (TS), while antibody levels were lower for TS0 animals from an area where FP is enzootic (Kaneohe Bay, Oahu, HI) and lower still for animals from FP-free areas (Kona Coast, HI) and negative controls. This trend was reinforced for tumored Hawaiian turtles shedding virus, in which the levels of antibody to ChHV5 were higher than those in nonshedders. In contrast, Floridian turtles were uniformly seropositive regardless of their tumor scores, a phenomenon seen in earlier serological studies of ChHV5 in Florida green turtles using baculovirus-expressed glycoprotein H (43). In that study, differences in the serological response to ChHV5 between Hawaiian turtles and Floridian turtles might be explained in terms of the cross-reactivity of antibodies to other herpesviruses, differences in how proteins were handled during ELISA procedures, or genetic differences in the viruses that may differentially affect the immunogenicity of ChHV5 to green turtles in the two regions.
To reduce the possibilities of cross-reactivity to other herpesviruses, it seemed wise to select an alternative antigen, because glycoprotein H, the antigen used by Herbst et al. (43), is conserved across multiple families of herpesviruses and thus might include an epitope(s) for cross-reactive antibodies (48). The genomes of individual herpesviruses can encode 10 or more envelope glycoproteins, all of which have their own functional and immunogenic properties, making them more or less suitable for use for serological detection of antibodies (49, 50). FUS8, the homologue of gE in other herpesviruses, was an obvious choice, since antibodies against gE are known to be produced consistently by various hosts (51). Since the host immune response to different antigens can have different dynamics, we chose FUS4 as our second antigen. In spite of numerous necropsy surveys of nearshore (7) and pelagic (52) sea turtles, ChHV5 is the only known herpesvirus in Hawaiian green turtles, so cross-reactivity in this region was unlikely. For Florida, cross-reactivity to known herpesviruses had previously been ruled out (43). Because strong responders to ChHV5 were found among both Hawaiian and Floridian turtles, we concluded that our newly developed assays were not subject to undesired cross-reactions. In this context, viral epitope variation is also unlikely to account for the differences between Hawaii and Florida, because recent genome-wide studies of ChHV5 from Florida and ChHV5 from Hawaii have shown that the sequences of FUS4 and FUS8 in ChHV5 isolates from Florida and ChHV5 isolates from Hawaii are identical (53).
Procedural issues are unlikely to account for the differences seen between Florida and Hawaii. Previously, Herbst et al. (43) reacted plasma from Florida green turtles to baculovirus-expressed glycoprotein H coated directly on ELISA plates and assayed for 7S IgY anti-ChHV5 reactivity. Unlike our system, in which nondenatured proteins were tethered to the ELISA plate via GST fusion proteins binding to coated glutathione casein (45), coating of the proteins directly on plastic can lead to denaturation and conformational changes (44) that may bias the binding by turtle antibodies. Based on our data, we think that this possibility is unlikely, because the results obtained using our system for Floridian turtles basically replicated those of Herbst et al. (43).
Inherent differences in immune systems between Floridian turtles and Hawaiian turtles also do not explain the differences in the serological responses to ChHV5. The role of different IgYs in green turtles is not fully understood, but 7S IgY responses arise earlier than 5.7S IgY responses in Hawaiian (40) and Floridian (41) green turtles experimentally exposed to antigens. The roles of these IgYs are better known in ducks, where 5.7S IgY is important in skin sensitization and dominates in chronic immunity (54), while increased ratios of 5.7S/7S IgY dampen the phagocytic response (55). Cell-mediated immunity in Hawaiian (9) and Floridian (8) green turtles declines with an increase in the tumor score, so both populations would presumably have equivalent susceptibility to tumor formation. Finally, evidence at the protein expression level suggests that 7S IgY and 5.7S IgY expression is identical in turtles from both regions (40).
While differences in procedures, viral epitopes, or immune systems fail to explain the regional variation in the humoral immune response to ChHV5 in green turtles from Florida and Hawaii, there are several fundamental differences between the regions in how green turtles and ChHV5 interact. First, in Hawaii, the prevalence of tumors increases with straight carapace length (SCL) (56), where SCL is a proxy for turtle age (31), whereas in Florida, the incidence of FP decreases with an increase in the SCL (17). Notably, in this study, seropositive turtles in Hawaii were larger than those in Florida. Second, green turtles from Florida recruit from the pelagic environment to nearshore foraging pastures when their SCL is 20 to 30 cm (57), whereas in Hawaii, they recruit when their SCL is 35 to 40 cm (30); however, the size at maturity (SCL, >81 cm) for Hawaiian green turtles (30) does not differ much from that for Florida green turtles (SCL, 83 cm) (12). Smaller recruiting Floridian turtles would thus spend longer periods of time in nearshore foraging habitats than Hawaiian turtles with an earlier exposure to ChHV5, which is thought to be acquired after the turtles recruit from the pelagic habitat (32). This would explain the higher percentages of seropositive tumor-free animals in Florida than in Hawaii. Third, ChHV5 DNA is ubiquitous in normal and tumor tissues from healthy and diseased turtles in the western Atlantic Ocean (26, 27), which would accord with the nearly ubiquitous seropositivity regardless of disease status (43). In contrast, in central Pacific green turtles, ChHV5 DNA is limited to tumor tissues and FP-negative animals are PCR negative (24, 58), so serology would seem to track the tumor score more closely in green turtles in Hawaii. Finally, the incidence of FP is declining among turtles in Hawaii (15), whereas it is stable or FP continues to expand in Florida (4). FP has a case fatality rate of about 70% for Hawaiian green turtles (13), whereas it is about 12% for Floridian green turtles (17), suggesting that a strong humoral immune response in tumored Hawaiian green turtles does not appear to enhance survival from disease. A higher case fatality rate for FP in Hawaii that in Florida may also explain the decline of disease in that region due to the removal of tumored hosts because of mortality.
Finally, virus-tumor interactions might differ significantly between Florida and Hawaii, and data from turtles known to be shedding virus in Hawaii reveal some insights. In Hawaii, only ca. 40 to 60% of tumor-negative Hawaiian animals were seropositive, whereas 100% of turtles shedding ChHV5 (20) were seropositive. Additionally, in Hawaiian turtles, antibodies generally increased with tumor severity, suggesting that even heavily tumored animals mount a strong humoral response. Hawaiian shedders showed stronger ELISA reactions than nonshedders, shedders showed signs of chronic exposure to viral antigens (strong 5.7S IgY reactions), and ELISA reactions correlated well with higher antibody titers, all of which suggest that the antibody response to ChHV5 seems to be tightly linked to lytic virus production in Hawaiian green turtles. In contrast, Floridian turtles showed strong antibody responses even without the formation of detectable tumors. Moreover, the seroreactions of Floridian TS3 turtles even seemed to decline, suggesting that tumor formation and maintenance were not essential for their pattern of ChHV5 shedding.
Together, these data suggest that behavioral differences between turtles from Florida and turtles from Hawaii have led to the evolution of distinct viral subtypes which differ in their transmission and spreading properties. The Floridian ChHV5 subtype seems to be transmitted independently of tumor formation, a fact confirmed by the consistent detection of ChHV5 DNA in normal tissues. In contrast, tumor formation seems to play a major role in the dissemination of the Hawaiian subtype, with productively infected cells exclusively being found in tumor tissue and with healthy tissue remaining free of ChHV5 (37, 58). Tumorigenic viruses requiring tumor formation for replication and transmission are rare. The only example is walleye dermal sarcoma virus, a retrovirus responsible for skin tumors in fish that is dependent on tumor formation for transmission (59). The Hawaiian subtype of ChHV5 may be the first DNA virus whose transmission depends on prior tumor formation.
MATERIALS AND METHODS
Turtle plasma.
Plasma for Hawaiian green turtles originated from 10 captive turtles raised at Sea Life Park (SLP), which has no history of FP; 20 wild turtles from west Hawaii (Kona/Kohala, HI), where FP has been historically absent; and 80 turtles from Kaneohe Bay, Oahu, HI, where FP is enzootic (9). Turtles from Kaneohe Bay were scored for tumor severity (20 turtles for each tumor score category), ranging from 0 (TS0; no tumors) to 3 (TS3; heavily tumored), as described previously (60). During capture or at necropsy, the straight carapace length of the animals was measured to the nearest 0.1 cm with calipers. Seventy-four Florida turtle plasma samples originated from turtles in the Indian River Lagoon, and the severity of FP in these turtles was given a score of from 0 to 3 (16), with 20 animals having TS0, 24 each having TS1 and TS2, and 6 having TS3. Plasma was collected from the Hawaiian turtles between 1998 and 1999 and from the Floridian turtles between 2001 and 2006. The samples were frozen at −70°C. Plasma from turtles with known virus shedding status came from turtles that underwent necropsy between 2003 and 2016, where tumors were exhaustively examined by histology for the presence of epidermal intranuclear inclusions (20). This sample set comprised 9 plasma samples from nonshedders and 10 from shedders. All animal sampling procedures were done under National Oceanic and Atmospheric Administration, National Marine Fisheries, permits and local state permits (Hawaii and Florida) with appropriate IACUC approvals.
Baculovirus antigen.
We chose two ChHV5 proteins as our antigens, FUS4 and FUS8. The choice of FUS4 was based on its partial homology to gD, the receptor-binding protein of many alphaherpesviruses (38) that may be important for the ability of herpesvirus to infect cells (61). We chose FUS8 because its gE homologues in other herpesviruses elicit strong immune responses (62, 63).
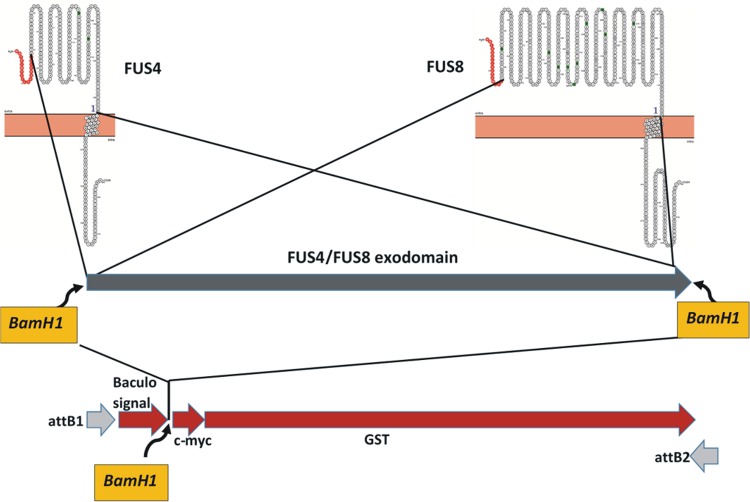
To express the antigens of interest in the baculovirus system, three synthetic constructs were made. For the first two constructs, the amino acid sequences of FUS4 and FUS8 were selected and retrotranslated in silico into nucleotide sequences that best fit the codon usage of insect cells. Then, BamHI restriction enzyme sites were added to either end of the constructs. A third construct was flanked by attB1 sites for site-directed recombination and comprised a codon usage-optimized open reading frame for a synthetic baculovirus signal peptide sequence, SP1-2 (64), followed by a BamHI site, a c-myc tag, and a C-terminal glutathione S-transferase (Fig. 9). All three constructs were produced synthetically (GenScript, NJ, USA) and provided as pUC57 clones. Construct three had flanking attB sites and was transferred to pDONR221 (Life Technologies) by a BP (recombination between attB and attP sites) reaction according to standard gateway recombination protocols, before it was opened by BamHI restriction enzyme digestion. The fragments of interest were excised from the first two constructs by BamHI restriction enzyme digestion and ligated separately into the opened pDONR221 vectors. The religated vector without an insert served as a control. In summary, three donor vectors, designated A, B, and C, were generated in pDONR221 (Table 2). Each construct was verified by nucleotide sequencing.
FIG 9.
Cloning strategy. The amino acid sequences of extracellular domains (exodomains) of the FUS4 and FUS8 glycoproteins (top), depicted here with their signal peptides (red), transmembrane region, and intracellular regions, were selected for gene synthesis. The synthetic constructs (blue arrow, middle) were bracketed by BamHI restriction enzyme sites. A third synthetic construct (red arrows, bottom), comprising a baculovirus signal sequence, an internal BamHI restriction enzyme site, a c-myc tag, and a GST tail, flanked by attB recombination sites, was cloned into pDONR221 before two variants were constructed. One variant had the FUS4 exodomain inserted into the BamHI site, and the other had the FUS8 exodomain inserted into the BamHI site.
TABLE 2.
pDONR221 constructs used for generating recombinant baculoviruses
| Designation | Fusion protein | Predicted mol wt (kDa) |
|---|---|---|
| pDONR221A | Signal-FUS4-c-myc-GST | 48 |
| pDONR221B | Signal-FUS8-c-myc-GST | 73.4 |
| pDONR221C | Signal-c-myc-GST | 27.3 |
Using standard Gateway protocols (Life Technologies), each construct was transferred by an LR reaction (catalyzing the in vitro recombination between an entry clone flanked by attL sites and a destination vector containing attR sites) to the pDEST8 vector, which conferred upstream of the cassette the baculovirus (Autographa californica multiple nucleopolyhedrovirus) polyhedrin promoter and downstream of the cassette the simian virus 40 late polyadenylation signal, as well as flanking sequences for transposition (Tn7) into the bacmid-cloned baculovirus genome. The resulting baculovirus bacmids were again verified by nucleotide sequencing before Sf9 cells were transfected to reconstitute replicating baculoviruses.
Baculoviruses were reconstituted by transfecting the bacmid DNAs into Sf9 cells. One microgram of DNA and 4 μl of the FuGene transfection reagent (Promega) were added to 25 μl of Grace’s insect medium with no fetal calf serum (FCS) (Life Technologies). The whole mix was transferred to a tube containing 500 μl of TNM-FH medium (Grace’s insect cell medium supplemented with 10% FCS), and the mixture was added to adherent Sf9 cells in 24-well plates (200,000 cells/well). The cells were incubated at 27°C, and the medium was collected after 96 h and centrifuged at 1,000 × g at 4°C for 10 min. The supernatant was harvested as the passage 0 (P0) viral stock solution. A 200-μl aliquot was stored at −80°C, and the rest was kept at 4°C for further expansion of the viral stocks. For passage 1 (P1) baculovirus amplification, 4 × 106 Sf9 cells cultured in 5 ml TNM-FH medium in T25 flasks were inoculated with 100 μl of the P0 viral stock solution and incubated at 27°C. The P1 viral stock was harvested 7 days after infection. Passage 2 (P2) baculoviral amplification was done in the same way, but using the P1 viral stock as the inoculum.
To determine the viral titer of the P2 viral stock, Sf9 cells were seeded on a 96-well plate (40,000 cells per well), inoculated with a 10-fold dilution series of the P2 viral stock in a volume of 100 μl, and incubated at 27°C. After 96 h, the cells were fixed in 4% paraformaldehyde for 15 min and then simultaneously blocked and permeabilized with phosphate-buffered saline (PBS) supplemented with 0.1% Triton X-100, 3% horse plasma, and 1% bovine serum albumin. Cells were incubated for 45 min at 37°C with an anti-c-myc MAb (Thermo Fisher) diluted 1:1,000 in blocking solution and then with a secondary horseradish peroxidase (HRP)-linked goat anti-mouse IgG (H+L) conjugate (Thermo Fisher) diluted 1:1,000 for 45 min at 37°C. Red color development was done with a 3-amino-9-ethylcarbazole substrate kit (Vector Laboratories) and was used to identify cells expressing the desired c-myc-tagged recombinant protein. The Reed and Muench method (65) was used to calculate the 50% endpoint dilution of the viral stocks.
For large-scale antigen production, 4 × 107 mimic Sf9 cells (Life Technologies) were seeded per T150 flask, inoculated with the P2 viral stock at a multiplicity of infection (MOI) of 5, overlaid with a final volume of 40 ml of serum-free medium (SFM-900 III; Life Technologies), and incubated at 27°C for 7 days. Protease inhibitor (cOmplete Mini protease inhibitor; Roche) was added to the medium after harvesting of the cell culture supernatant.
Western immunoblotting.
At various stages of the project, the desired recombinant proteins were assayed by polyacrylamide gel electrophoresis (PAGE) (66) and Western immunoblotting (67). For this purpose, we used equal volumes of cell culture supernatant and 2× Laemmli buffer (Bio-Rad Laboratories) containing sodium dodecyl sulfate (SDS; Bio-Rad Laboratories). 2-Mercaptoethanol (BME; Sigma) was either added (for reducing conditions) or omitted (for nonreducing conditions) before denaturing and solubilizing the samples by incubating at 95°C for 5 min. The samples were then resolved on 10% polyacrylamide gels, transferred to nitrocellulose membranes, and probed with an anti-c-myc MAb at 1:5,000, followed by incubation with goat anti-mouse immunoglobulin-HRP conjugate (1:1,000; Southern Biotech). Color development was done with 3,3′-diaminobenzidine tetrahydrochloride (DAB; Sigma).
ELISA.
The concept of the ELISA was to bind the FUS4 and FUS8 via glutathione S-transferase (GST) to glutathione casein-coated plates, ensuring that the proteins remained in their native state (45). Glutathione casein was made as described previously (45). To standardize the coating of the antigen, a series of checkerboard assays was done by coating round-bottom ELISA plates (Falcon) with glutathione casein at concentrations ranging from 1:2,000 to 1:32,000, diluted 2-fold serially at 50 μl/well, and the plates were incubated overnight at 4°C. After washing three times with PBS–0.3% Tween 20 (PBST), the plates were blocked in PBS–0.3% Tween 20 with 2% milk (PBST-M), washed, and then coated with FUS4 or FUS8 antigen culture supernatants at dilutions ranging from 1:12.5 to 1:256,000, diluted 2-fold serially, and incubated for 1 h at 37°C. Following washes in PBST, antigen coating was assayed by reacting a mouse anti-c-myc MAb (Invitrogen) diluted 1:5,000 for 1 h at 37°C, and the plates were washed and then reacted with HRP-conjugated goat anti-mouse immunoglobulin (Southern Biotech) at 1:1,000 for 1 h at 37°C. After washing, color development was done with 3,3′,5,5′-tetramethylbenzidine peroxidase (TMB) substrate (KPL), which was stopped after 20 min with 1 M phosphoric acid, whereupon the absorbance at 450 nm was read after 10 min. From this procedure, we found the optimal coating concentration to be 1:3,000 for glutathione casein and 1:50 for FUS4, FUS8, and the c-myc backbone.
For the ELISA, the antigens (FUS4, FUS8 and the c-myc backbone) were coated on glutathione casein-coated plates at the optimal calculated concentrations described above. Turtle plasma was diluted 1:25 in high-salt (0.5 M NaCl) borate buffer–0.3% Tween 20 with 2% milk. The plates were then incubated with green turtle anti-7S IgY or anti-5.7S IgY MAbs (40) at 0.2 μg/ml diluted in PBST-M, followed by washing and incubation with polyclonal goat anti-mouse immunoglobulin diluted 1:1,000 in PBST-M. Color development and reading of the absorbance were performed as described above, except that the reaction was stopped after 30 min. OD values for FUS4 or FUS8 were corrected for the absorbance of the c-myc backbone by subtracting the OD of the c-myc backbone from the OD of FUS4 or FUS8 (delta OD).
A panel of plasma samples from FP-positive green turtles was assayed by ELISA for 7S and 5.7S IgY reactivity against FUS4 and FUS8, and from this, we selected one plasma sample, referred to here as the “positive control,” that had overall strong reactivity to both antigens for both antibodies to serve as a standard positive control to control for plate-to-plate variation. ODs were expressed as a percentage of the positive-control OD according to the following formula: [(delta OD of FUS4 or FUS8)/(delta OD of the positive control)] × 100. To confirm that the ELISA ODs related to the plasma titers, we assayed a panel of plasma samples at serial 10-fold dilutions ranging from 1:10 to 1:10,000. These samples originated from turtles confirmed to be shedding (n = 10) or not shedding (n = 9) ChHV5 in tumors, as confirmed by histopathology (20).
In the absence of a gold standard for the detection of antibodies against ChHV5, we used two types of cutoff points to score turtles as seropositive: (i) the mean delta OD for negative control plasma plus 3 standard deviations (43), calculated from 20 plasma samples from negative-control turtles from Sea Life Park assayed for each antigen/antibody type combination (FUS4 + 7S IgY, FUS8 + 7S IgY, FUS4 + 5.7S IgY, and FUS8 + 5.7S IgY), and (ii) a delta OD of >0.3, regardless of assay type, because this difference can easily be recognized by eye and because such an approach has previously been used in the context of papillomavirus serology (47). Calculations of sensitivity and specificity made by the use of these cutoffs showed that the use of 3 SD presented a relatively high sensitivity and a low specificity, whereas the use of a delta OD of >0.3 presented a low sensitivity and a high specificity (Table 2). These cutoff values, i.e., a delta OD of >0.3 and 3 SD above the mean delta OD value for the negative controls, are referred to here as high-specificity and high-sensitivity cutoffs, respectively. The straight carapace lengths of the Florida and Hawaii turtles were compared for each tumor score category using the t test with a Bonferroni adjustment (68) to an alpha level of significance to account for the number of comparisons (0.05/4). Analyses were done with R software (69).
Data availability.
All data from this study are available from the U.S. Geological Survey in Excel format at https://doi.org/10.5066/P969TAIJ.
ACKNOWLEDGMENTS
M.A.’s contribution to this research was supported by the Wyss Charitable Endowment. A.W. was supported by a private donation from the late Robert Wyler to M.A. (grant F-52601-10-01), as well as by general funds allocated to M.A. by the University of Zurich.
Brian Stacy and two anonymous reviewers provided constructive comments.
Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. government.
IAP World Services is under contract to the U.S. Geological Survey.
REFERENCES
- 1.Davison AJ, Eberle R, Ehlers B, Hayward GS, McGeoch DJ, Minson AC, Pellett PE, Roizman B, Studdert MJ, Thiry E. 2009. The order Herpesvirales. Arch Virol 154:171–177. doi: 10.1007/s00705-008-0278-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Herbst LH. 1994. Fibropapillomatosis of marine turtles. Annu Rev Fish Dis 4:389–425. doi: 10.1016/0959-8030(94)90037-X. [DOI] [Google Scholar]
- 3.Jones K, Ariel E, Burgess G, Read M. 2016. A review of fibropapillomatosis in green turtles (Chelonia mydas). Vet J 212:48–57. doi: 10.1016/j.tvjl.2015.10.041. [DOI] [PubMed] [Google Scholar]
- 4.Foley AM, Schroeder BA, Redlow AE, Fick-Child KJ, Teas WG. 2005. Fibropapillomatosis in stranded green turtles (Chelonia mydas) from the eastern United States (1980–98): trends and associations with environmental factors. J Wildl Dis 41:29–41. doi: 10.7589/0090-3558-41.1.29. [DOI] [PubMed] [Google Scholar]
- 5.Chaloupka M, Work TM, Balazs GH, Murakawa SKK, Morris RM. 2008. Cause-specific temporal and spatial trends in green sea turtle strandings in the Hawaiian Archipelago (1982–2003). Mar Biol 154:887–898. doi: 10.1007/s00227-008-0981-4. [DOI] [Google Scholar]
- 6.Rodenbusch CR, Baptistotte C, Werneck MR, Pires TT, Melo MTD, de Ataíde MW, dos Reis K, Testa P, Alieve MM, Canal CW. 2014. Fibropapillomatosis in green turtles Chelonia mydas in Brazil: characteristics of tumors and virus. Dis Aquat Organ 111:207–217. doi: 10.3354/dao02782. [DOI] [PubMed] [Google Scholar]
- 7.Work TM, Balazs GH, Rameyer RA, Morris R. 2004. Retrospective pathology survey of green turtles (Chelonia mydas) with fibropapillomatosis from the Hawaiian Islands, 1993–2003. Dis Aquat Organ 62:163–176. doi: 10.3354/dao062163. [DOI] [PubMed] [Google Scholar]
- 8.Cray C, Varella R, Bossart GD, Lutz P. 2001. Altered in vitro immune responses in green turtles (Chelonia mydas) with fibropapillomatosis. J Zoo Wildl Med 32:436–440.2.0.CO;2] [DOI] [PubMed] [Google Scholar]
- 9.Work TM, Rameyer RA, Balazs GH, Cray C, Chang SP. 2001. Immune status of free-ranging green turtles with fibropapillomatosis from Hawaii. J Wildl Dis 37:574–581. doi: 10.7589/0090-3558-37.3.574. [DOI] [PubMed] [Google Scholar]
- 10.Work TM, Balazs GH, Wolcott M, Morris RM. 2003. Bacteraemia in Hawaiian green turtles, Chelonia mydas, with fibropapillomatosis. Dis Aquat Organ 53:41–46. doi: 10.3354/dao053041. [DOI] [PubMed] [Google Scholar]
- 11.Balazs GH, Chaloupka M. 2004. Thirty-year recovery trend in the once depleted Hawaiian green turtle stock. Biol Conservation 117:491–498. doi: 10.1016/j.biocon.2003.08.008. [DOI] [Google Scholar]
- 12.Ehrhart LM, Redfoot WE, Bagley DA. 2007. Marine turtles of the central region of the Indian River Lagoon system, Florida. Florida Sci 70:415–434. [Google Scholar]
- 13.Bennett P, Keuper-Bennett U, Balazs GH. 1999. Photographic evidence for the regression of fibropapillomas afflicting green turtles at Honokawai, Maui, in the Hawaiian islands, p 37–39. Proc 19th Ann Symp Sea Turtle Biol Conservation , South Padre Island, TX. [Google Scholar]
- 14.Aguirre AA, Balazs G, Spraker T, Murakawa S, Zimmerman B. 2002. Pathology of oropharyngeal fibropapillomatosis in green turtles, Chelonia mydas. J Aquat Anim Health 14:298–304. doi:. [DOI] [PubMed] [Google Scholar]
- 15.Chaloupka M, Balazs GH, Work TM. 2009. Rise and fall over 26 years of a marine epizootic in Hawaiian green sea turtles. J Wildl Dis 45:1138–1142. doi: 10.7589/0090-3558-45.4.1138. [DOI] [PubMed] [Google Scholar]
- 16.Hirama S, Ehrhart L. 1999. Prevalence and severity of green turtle fibropapillomatosis in the Indian River Lagoon. Florida Sci 62:35. [Google Scholar]
- 17.Hirama S, Ehrhart LM. 2007. Description, prevalence and severity of green turtle fibropapillomatosis in three developmental habitats on the east coast of Florida. Florida Sci 70:435–448. [Google Scholar]
- 18.Hargrove S, Work T, Brunson S, Foley AM, Balazs G. Proceedings of the 2015 International Summit on Fibropapillomatosis: global status, trends, and population impacts, p 1–85. U.S. Department of Commerce, Washington, DC. [Google Scholar]
- 19.Jacobson ER, Buergelt C, Williams B, Harris RK. 1992. Herpesvirus in cutaneous fibropapillomas of the green turtle Chelonia mydas. Dis Aquat Org 12:1–6. doi: 10.3354/dao012001. [DOI] [Google Scholar]
- 20.Work TM, Dagenais J, Balazs GH, Schettle N, Ackermann M. 2015. Dynamics of virus shedding and in-situ confirmation of chelonid herpesvirus 5 in Hawaiian green turtles with fibropapillomatosis. Vet Pathol 52:1195–1220. doi: 10.1177/0300985814560236. [DOI] [PubMed] [Google Scholar]
- 21.Herbst LH, Jacobson ER, Moretti R, Brown T, Sundberg JP, Klein PA. 1995. Experimental transmission of green turtle fibropapillomatosis using cell-free tumor extracts. Dis Aquat Organisms 22:1–12. doi: 10.3354/dao022001. [DOI] [Google Scholar]
- 22.Herbst LH, Moretti R, Brown T, Klein PA. 1996. Sensitivity of the transmissible green turtle fibropapillomatosis agent to chloroform and ultracentrifugation conditions. Dis Aquat Organisms 25:225–228. doi: 10.3354/dao025225. [DOI] [Google Scholar]
- 23.Lackovich JK, Brown DR, Homer BL, Garber RL, Mader DR, Moretti RH, Patterson AD, Herbst LH, Oros J, Jacobson ER, Curry SS, Klein PA. 1999. Association of herpesvirus with fibropapillomatosis of the green turtle Chelonia mydas and the loggerhead turtle Caretta caretta in Florida. Dis Aquat Organisms 37:89–97. doi: 10.3354/dao037089. [DOI] [PubMed] [Google Scholar]
- 24.Quackenbush SL, Work TM, Balazs GH, Casey RN, Rovnak J, Chaves A, DuToit L, Baines JD, Parrish CR, Bowser PR, Casey JW. 1998. Three closely related herpesviruses are associated with fibropapillomatosis in marine turtles. Virology 246:392–399. doi: 10.1006/viro.1998.9207. [DOI] [PubMed] [Google Scholar]
- 25.Rodenbusch CR, Almeida LL, Marks FS, Ataíde MW, Alievi MM, Tavares M, Pereira RA, Canal CW. 2012. Detection and characterization of fibropapilloma associated herpesvirus of marine turtles in Rio Grande do Sul, Brazil. Pesq Vet Bras 32:1179–1183. doi: 10.1590/S0100-736X2012001100018. [DOI] [Google Scholar]
- 26.Alfaro-Nunez A, Bojesen AM, Bertelsen MF, Wales N, Balazs GH, Gilbert MT. 2016. Further evidence of chelonid herpesvirus 5 (ChHV5) latency: high levels of ChHV5 DNA detected in clinically healthy marine turtles. PeerJ 4:e2274. doi: 10.7717/peerj.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Page-Karjian A, Torres F, Zhang J, Rivera S, Diez C, Moore PA, Moore D, Brown C. 2012. Presence of chelonid fibropapilloma-associated herpesvirus in tumored and non-tumored green turtles, as detected by polymerase chain reaction, in endemic and non-endemic aggregations, Puerto Rico. Springerplus 1:35. doi: 10.1186/2193-1801-1-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schat K, Nair V. 2013. Marek’s disease In Swayne DE, Glisson JR, McDougald LR, Nolan LK, Suarez DL, Nair V (ed), Diseases of poultry. John Wiley & Sons, Inc, New York, NY. [Google Scholar]
- 29.Mesri EA, Cesarman E, Boshoff C. 2010. Kaposi’s sarcoma and its associated herpesvirus. Nat Rev Cancer 10:707–719. doi: 10.1038/nrc2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balazs GH. 1980. Synopsis of biological data of the green turtle in the Hawaiian islands. Report NOAA-TM-NMFS National Oceanic and Atmospheric Administration, Washington, DC. [Google Scholar]
- 31.Frazer NB, Ehrhart LM. 1985. Preliminary growth models for green, Chelonia mydas, and loggerhead, Caretta caretta, turtles in the wild. Copeia 1985:73–79. doi: 10.2307/1444792. [DOI] [Google Scholar]
- 32.Ene A, Su M, Lemaire S, Rose C, Schaff S, Moretti R, Lenz J, Herbst L. 2005. Distribution of chelonid fibropapillomatosis-associated herpesvirus variants in Florida: molecular genetic evidence for infection of turtles following recruitment to neritic developmental habitats. J Wildl Dis 41:489–497. doi: 10.7589/0090-3558-41.3.489. [DOI] [PubMed] [Google Scholar]
- 33.Lu Y, Yu Q, Zamzow J, Wang Y, Losey GS, Balazs GH, Nerurkar VR, Yanagihara R. 2000. Detection of green turtle herpesviral sequence in saddleback wrasse Thalassoma duperrey: a possible mode of transmission of green turtle fibropapilloma. J Aquat Anim Health 12:58–63. doi:. [DOI] [PubMed] [Google Scholar]
- 34.Greenblatt RJ, Work TM, Balazs GH, Sutton CA, Casey RN, Casey JW. 2004. The Ozobranchus leech is a candidate mechanical vector for the fibropapilloma-associated turtle herpesvirus found latently infecting skin tumors on Hawaiian green turtles (Chelonia mydas). Virology 321:101–110. doi: 10.1016/j.virol.2003.12.026. [DOI] [PubMed] [Google Scholar]
- 35.Koch R. 1882. Die Aetiologie der Tuberculose. Berlin Klin Wochenschr 19:221–230. [Google Scholar]
- 36.Work TM, Dagenais J, Balazs GH, Schumacher J, Lewis TD, Leong JC, Casey RN, Casey JW. 2009. In vitro biology of fibropapilloma-associated turtle herpesvirus and host cells in Hawaiian green turtles (Chelonia mydas). J Gen Virol 90:1943–1950. doi: 10.1099/vir.0.011650-0. [DOI] [PubMed] [Google Scholar]
- 37.Work TM, Dagenais J, Weatherby TM, Balazs GH, Ackermann M. 2017. In vitro replication of Chelonid herpesvirus 5 in organotypic skin cultures from Hawaiian green turtles (Chelonia mydas). J Virol 91:e00404-17. doi: 10.1128/JVI.00404-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ackermann M, Koriabine M, Hartmann-Fritsch F, de Jong PJ, Lewis TD, Schetle N, Work TM, Dagenais J, Balazs GH, Leong JC. 2012. The genome of chelonid herpesvirus 5 harbors atypical genes. PLoS One 7:e46623. doi: 10.1371/journal.pone.0046623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alfaro-Núñez A, Bertelsen MF, Bojesen AM, Rasmussen I, Zepeda-Mendoza L, Olsen MT, Gilbert M. 2014. Global distribution of chelonid fibropapilloma-associated herpesvirus among clinically healthy sea turtles. BMC Evol Biol 14:206. doi: 10.1186/PREACCEPT-1568873981260399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Work TM, Dagenais J, Breeden R, Schneemann A, Sung J, Hew B, Balazs GH, Berestecky JM. 2015. Green turtles (Chelonia mydas) have novel asymmetrical antibodies. J Immunol 195:5452–5456. doi: 10.4049/jimmunol.1501332. [DOI] [PubMed] [Google Scholar]
- 41.Herbst LH, Klein PA. 1995. Monoclonal antibodies for the measurement of class-specific antibody responses in the green turtle, Chelonia mydas. Vet Immunol Immunopathol 46:317–335. doi: 10.1016/0165-2427(94)05360-5. [DOI] [PubMed] [Google Scholar]
- 42.Herbst LH, Greiner EC, Ehrhart LM, Bagley DA, Klein PA. 1998. Serological association between spirorchidiasis, herpesvirus infection, and fibropapillomatosis in green turtles from Florida. J Wildl Dis 34:496–507. doi: 10.7589/0090-3558-34.3.496. [DOI] [PubMed] [Google Scholar]
- 43.Herbst LH, Lemaire S, Ene AR, Heslin DJ, Ehrhart LM, Bagley DA, Klein PA, Lenz J. 2008. Use of baculovirus-expressed glycoprotein H in an enzyme-linked immunosorbent assay developed to assess exposure to chelonid fibropapillomatosis-associated herpesvirus and its relationship to the prevalence of fibropapillomatosis in sea turtles. Clin Vaccine Immunol 15:843–851. doi: 10.1128/CVI.00438-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darst SA, Robertson CR, Berzofsky JA. 1988. Adsorption of the protein antigen myoglobin affects the binding of conformation-specific monoclonal antibodies. Biophys J 53:533–539. doi: 10.1016/S0006-3495(88)83133-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sehr P, Zumbach K, Pawlita M. 2001. A generic capture ELISA for recombinant proteins fused to glutathione S-transferase: validation for HPV serology. J Immunol Methods 253:153–162. doi: 10.1016/s0022-1759(01)00376-3. [DOI] [PubMed] [Google Scholar]
- 46.Ackermann M, Longnecker R, Roizman B, Pereira L. 1986. Identification, properties, and gene location of a novel glycoprotein specified by herpes simplex virus 1. Virology 150:207–220. doi: 10.1016/0042-6822(86)90280-1. [DOI] [PubMed] [Google Scholar]
- 47.Lange CE, Tobler K, Favrot C, Muller M, Nothling JO, Ackermann M. 2009. Detection of antibodies against epidermodysplasia verruciformis-associated canine papillomavirus 3 in sera of dogs from Europe and Africa by enzyme-linked immunosorbent assay. Clin Vaccine Immunol 16:66–72. doi: 10.1128/CVI.00346-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Szpara ML, Gatherer D, Ochoa A, Greenbaum B, Dolan A, Bowden RJ, Enquist LW, Legendre M, Davison AJ. 2014. Evolution and diversity in human herpes simplex virus genomes. J Virol 88:1209–1227. doi: 10.1128/JVI.01987-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Banks TA, Hariharan MJ, Rouse BT. 1998. Assessing cell-mediated immune responses to HSV in murine systems. Methods Mol Med 10:327–343. doi: 10.1385/0-89603-347-3:327. [DOI] [PubMed] [Google Scholar]
- 50.Belknap EB, Walters LM, Kelling C, Ayers VK, Norris J, McMillen J, Hayhow C, Cochran M, Reddy DN, Wright J, Collins JK. 1999. Immunogenicity and protective efficacy of a gE, gG and US2 gene-deleted bovine herpesvirus-1 (BHV-1) vaccine. Vaccine 17:2297–2305. doi: 10.1016/s0264-410x(98)00466-6. [DOI] [PubMed] [Google Scholar]
- 51.Kaashoek MJ, Rijsewijk FA, Oirschot JT. 1996. Persistence of antibodies against bovine herpesvirus 1 and virus reactivation two to three years after infection. Vet Microbiol 53:103–110. doi: 10.1016/s0378-1135(96)01238-2. [DOI] [PubMed] [Google Scholar]
- 52.Work TM, Balazs GH. 2010. Pathology and distribution of sea turtles landed as bycatch in the Hawaii-based North Pacific pelagic longline fishery. J Wildl Dis 46:422–432. doi: 10.7589/0090-3558-46.2.422. [DOI] [PubMed] [Google Scholar]
- 53.Morrison CL, Iwanowicz L, Work TM, Fahsbender E, Breitbart M, Adams C, Iwanowicz D, Sanders L, Ackermann M, Cornman RS. 2018. Genomic evolution, recombination, and inter-strain diversity of chelonid alphaherpesvirus 5 from Florida and Hawaii green sea turtles with fibropapillomatosis. PeerJ 6:e4386. doi: 10.7717/peerj.4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Warr GW, Magor KE, Higgins DA. 1995. IgY: clues to the origin of modern antibodies. Immunol Today 16:392–398. doi: 10.1016/0167-5699(95)80008-5. [DOI] [PubMed] [Google Scholar]
- 55.Humphrey BD, Calvert CC, Klasing KC. 2004. The ratio of full length IgY to truncated IgY in immune complexes affects macrophage phagocytosis and the acute phase response of mallard ducks (Anas platyrhynchos). Dev Comp Immunol 28:665–672. doi: 10.1016/j.dci.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 56.Chaloupka M, Balazs G. 2005. Modelling the effect of fibropapilloma disease on the somatic growth dynamics of Hawaiian green sea turtles. Mar Biol 147:1251–1260. doi: 10.1007/s00227-005-0026-1. [DOI] [Google Scholar]
- 57.Mendonça MT, Ehrhart LM, Mendonca MT. 1982. Activity, population size and structure of immature Chelonia mydas and Caretta caretta in Mosquito Lagoon, Florida. Copiea 1982:161–167. doi: 10.2307/1444280. [DOI] [Google Scholar]
- 58.Quackenbush SL, Casey RN, Murcek RJ, Paul TA, Work TM, Limpus CJ, Chaves A, duToit L, Perez JV, Aguirre AA, Spraker TR, Horrocks JA, Vermeer LA, Balazs GH, Casey JW. 2001. Quantitative analysis of herpesvirus sequences from normal and fibropapillomas of marine turtles with real time PCR. Virology 287:105–111. doi: 10.1006/viro.2001.1023. [DOI] [PubMed] [Google Scholar]
- 59.Rovnak J, Quackenbush SL. 2010. Walleye dermal sarcoma virus: molecular biology and oncogenesis. Viruses 2:1984–1999. doi: 10.3390/v2091984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Work TM, Balazs GH. 1999. Relating tumor score to hematology in green turtles with fibropapillomatosis in Hawaii. J Wildl Dis 35:804–807. doi: 10.7589/0090-3558-35.4.804. [DOI] [PubMed] [Google Scholar]
- 61.Tran LC, Kissner JM, Westerman LE, Sears AE. 2000. A herpes simplex virus 1 recombinant lacking the glycoprotein G coding sequences is defective in entry through apical surfaces of polarized epithelial cells in culture and in vivo. Proc Natl Acad Sci U S A 97:1818–1822. doi: 10.1073/pnas.020510297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Weiss M, Brum MCS, Anziliero D, Weiblen R, Flores EF. 2015. A glycoprotein E gene-deleted bovine herpesvirus 1 as a candidate vaccine strain. Braz J Med Biol Res 48:843–851. doi: 10.1590/1414-431X20154243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jacobs L. 1994. Glycoprotein E of pseudorabies virus and homologous proteins in other alphaherpesvirinae. Arch Virol 137:209–228. doi: 10.1007/bf01309470. [DOI] [PubMed] [Google Scholar]
- 64.Futatsumori-Sugai M, Tsumoto K. 2010. Signal peptide design for improving recombinant protein secretion in the baculovirus expression vector system. Biochem Biophys Res Commun 391:931–935. doi: 10.1016/j.bbrc.2009.11.167. [DOI] [PubMed] [Google Scholar]
- 65.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Hyg 27:493–497. [Google Scholar]
- 66.Laemmli U. 1970. Cleavage of structural proteins during assembly of the head bacteriophage T4. Nature 227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 67.Towbin H, Staehelin T, Gordon J. 1979. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A 76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Rice WR. 1989. Analyzing tables of statistical tests. Evolution 43:223–225. doi: 10.1111/j.1558-5646.1989.tb04220.x. [DOI] [PubMed] [Google Scholar]
- 69.R Core Team. 2017. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: https://www.R-project.org/. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All data from this study are available from the U.S. Geological Survey in Excel format at https://doi.org/10.5066/P969TAIJ.