For at least 30 years, it has been known that HIV-1 Vpr, a protein carried in the virion, is important for efficient infection of primary macrophages. Vpr is also a determinant of the pathogenic effects of HIV-1 in vivo. A number of cellular proteins that interact with Vpr have been identified. So far, it has not been possible to associate these proteins with altered viral replication in macrophages or to explain why Vpr is carried in the virus particle. Here, we show that Vpr mitigates the antiviral effects of REAF, a protein highly expressed in primary macrophages and one that inhibits virus replication during reverse transcription. REAF is degraded by Vpr within 30 min of virus entry in a manner dependent on the nuclear localization of Vpr and its interaction with the cell’s protein degradation machinery.
KEYWORDS: HIV, innate immunity, REAF, RPRD2, restriction factors, Vpr
ABSTRACT
The human immunodeficiency virus type 1 (HIV-1) accessory protein Vpr enhances viral replication in both macrophages and, to a lesser extent, cycling T cells. Virion-packaged Vpr is released in target cells shortly after entry, suggesting it is required in the early phase of infection. Previously, we described REAF (RNA-associated early-stage antiviral factor; RPRD2), a constitutively expressed protein that potently restricts HIV replication at or during reverse transcription. Here, we show that a virus without an intact vpr gene is more highly restricted by REAF and, using delivery by virus-like particles (VLPs), that Vpr alone is sufficient for REAF degradation in primary macrophages. REAF is more highly expressed in macrophages than in cycling T cells, and we detected, by coimmunoprecipitation assay, an interaction between Vpr protein and endogenous REAF. Vpr acts quickly during the early phase of replication and induces the degradation of REAF within 30 min of viral entry. Using Vpr F34I and Q65R viral mutants, we show that nuclear localization and interaction with cullin 4A-DBB1 (DCAF1) E3 ubiquitin ligase are required for REAF degradation by Vpr. In response to infection, cells upregulate REAF levels. This response is curtailed in the presence of Vpr. These findings support the hypothesis that Vpr induces the degradation of a factor, REAF, that impedes HIV infection in macrophages.
IMPORTANCE For at least 30 years, it has been known that HIV-1 Vpr, a protein carried in the virion, is important for efficient infection of primary macrophages. Vpr is also a determinant of the pathogenic effects of HIV-1 in vivo. A number of cellular proteins that interact with Vpr have been identified. So far, it has not been possible to associate these proteins with altered viral replication in macrophages or to explain why Vpr is carried in the virus particle. Here, we show that Vpr mitigates the antiviral effects of REAF, a protein highly expressed in primary macrophages and one that inhibits virus replication during reverse transcription. REAF is degraded by Vpr within 30 min of virus entry in a manner dependent on the nuclear localization of Vpr and its interaction with the cell’s protein degradation machinery.
INTRODUCTION
Human immunodeficiency virus type 1 (HIV-1) infects CD4+ T cells and macrophages in vivo. HIV-1 has four nonstructural accessory genes: nef, vif, vpu, and vpr. nef, vif, and vpu diminish host innate immunity. A function for Vpr has been elusive, but it is required for efficient replication in macrophages and for pathogenesis in vivo (1, 2). A widely acknowledged but poorly understood Vpr-mediated phenotype is the induction of cell cycle arrest at the G2/M phase using the cullin 4A-DBB1 (DCAF1) E3 ubiquitin ligase and the recruitment of an unknown substrate for proteasomal degradation. A large number of Vpr substrates have been reported (3–11). Yan et al. (12) showed that helicase-like transcription factor (HLTF) weakly restricts replication of HIV-1 in T cells (12). HLTF was shown previously to be downmodulated by Vpr (8, 12). Furthermore, Greenwood et al. reported that Vpr promotes large-scale remodeling of approximately 2,000 cellular proteins, including those that bind nucleic acids and others involved with the cell cycle (13).
Substantial quantities of Vpr are incorporated into viral particles and released from the major capsid protein (CA) after entry into the cell (14, 15). The timing of Vpr release coincides with the initiation of reverse transcription, a process that transcribes the RNA genome into DNA for subsequent integration into the host cell DNA (16). The early release of Vpr from the CA implies it has an early function prior to integration events. When considering the role of Vpr in cell tropism and pathogenesis, the investigation of proteins that have a direct effect on viral replication is a priority.
Here, we investigate RNA-associated early-stage antiviral factor (REAF) (also known as regulation of nuclear pre-mRNA domain-containing protein 2 [RPRD2]), originally described as an unknown restriction to HIV replication called lentiviral restriction 2 (Lv2) (17, 18). Lv2 was first shown to restrict the replication of HIV-2, and subsequently it was shown to inhibit the replication of HIV-1 and simian immunodeficiency virus (SIV) during reverse transcription (19). Lv2/REAF restriction is cell type dependent (18, 20–23) and active in certain cell types, including HeLa-CD4 cells and primary macrophages (17, 18). The susceptibility of the virus to Lv2 is determined by both the viral envelope (Env) and capsid (CA) (22, 23). REAF was identified in a whole-genome small interfering RNA (siRNA) screen for HIV-1 restriction factors. REAF limits the completion of proviral DNA synthesis and integration of the viral genome (17). Subsequently, REAF was demonstrated to form a major component of Lv2 (18).
Here, we show that within 30 min of cellular entry, only HIV-1 that contains Vpr can induce the degradation of REAF and rescue efficient viral replication in primary macrophages. Using Vpr mutant viruses, we demonstrate that the nuclear localization of Vpr and its ability to interact with DCAF1 E3 ubiquitin ligase are requirements for REAF degradation. Downmodulation of REAF by Vpr in the early phase of infection is transient, and reexpression to basal levels is achieved by approximately 1 h. After infection with HIV-1 or treatment with polyriboinosinic-polyribocytidylic acid [poly(I·C)] or lipopolysaccharide (LPS), cells respond by increasing REAF levels. In the case of viral infection, this response is curtailed in the presence of Vpr. Therefore, our results support the hypothesis that Vpr induces the degradation of a cellular protein, REAF, that impedes HIV-1 infection in macrophages during reverse transcription.
(This article was submitted to an online preprint archive [24].)
RESULTS
HIV-1 Vpr interacts with REAF and overcomes restriction.
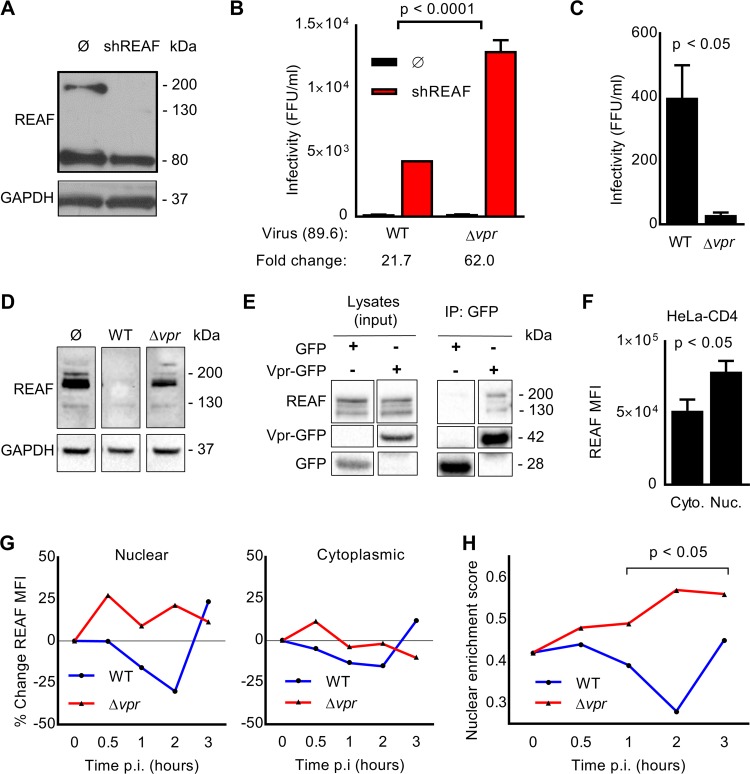
REAF restricts HIV-1 replication in HeLa-CD4 cells (17, 19). We sought to determine if a viral accessory gene could overcome REAF, and so we tested the infectivity of HIV-1 89.6WT (wild type) and mutants with vpr (89.6Δvpr), vif (89.6Δvif), or vpu (89.6Δvpu) deleted in these cells. Preventing REAF expression using short hairpin RNA (shRNA) (HeLa-CD4 shRNA-REAF cells) (Fig. 1A) revealed its potent antiviral effect. Despite a standard input for each virus (50 focus-forming units [FFU]/ml, as measured on HeLa-CD4 cells), there was significantly greater rescue of HIV-1 89.6Δvpr (>3 fold; P < 0.0001) than of HIV-1 89.6WT (Fig. 1B). The prevention of REAF expression using shRNA alleviates the need for Vpr. Conversely, there was no significantly greater rescue for either HIV-1 89.6Δvif or HIV-1 89.6Δvpu than for HIV-1 89.6WT (data not shown). Thus, vpr potently overcomes the restriction imposed by REAF.
FIG 1.
HIV-1 Vpr interacts with REAF and overcomes restriction. (A) REAF protein in HeLa-CD4 cells (Ø) and HeLa-CD4 shRNA-REAF cells (shREAF). GAPDH was a loading control. (B) Infectivity of HIV-1 89.6WT and HIV-1 89.6Δvpr in HeLa-CD4 cells (Ø) and HeLa-CD4 shRNA-REAF cells (shREAF). Viral inputs were equivalent at approximately 50 FFU/ml, measured on the HeLa-CD4 cells. The error bars indicate the standard deviations of the means derived from a range of duplicate titrations. Fold changes in FFU are indicated. (C) Foci of infection resulting from equal p24 inputs (1 ng) of HIV-1 89.6WT or HIV-1 89.6Δvpr in HeLa-CD4 cells. The error bars indicate the standard deviations of the means derived from a range of duplicate titrations. (D) REAF protein in HeLa-CD4 cells 24 h postchallenge with HIV-1 89.6WT or HIV-1 89.6Δvpr. GAPDH was a loading control. (E) HEK-293T cells were transfected with Vpr-GFP expression plasmid or GFP control vector, expression was analyzed by Western blotting (left), and protein was immunoprecipitated (IP) with anti-GFP beads. Coimmunoprecipitated REAF was detected in the Vpr-GFP precipitation (right). (F) Nuclear (Nuc.) and cytoplasmic (Cyto.) REAF MFI in HeLa-CD4 cells measured by imaging flow cytometry. The error bars represent standard deviations of the means of replicates. (G) Percent change in REAF MFI from time 0 in the nucleus (left) and cytoplasm (right) of HeLa-CD4 cells over time after challenge with HIV-1 89.6WT or HIV-1 89.6Δvpr. The results are representative of three independent experiments. p.i., postinfection. (H) Nuclear enrichment scores of HeLa-CD4 cells over time postchallenge with HIV-1 89.6WT or HIV-1 89.6Δvpr. A lower nuclear enrichment score indicates that a lower proportion of overall REAF is located in the nucleus, as calculated with IDEAS software. The results are representative of three independent experiments. Where quantitative comparisons are made, the data were derived from the same blot or blots processed together.
We are not aware of previous reports that Vpr overcomes known or unknown HIV-1 restrictions in HeLa-CD4 cells. Therefore, we confirmed that the mutant HIV-1 89.6Δvpr is restricted in HeLa-CD4 cells compared to HIV-1 89.6WT. Figure 1C shows that, despite equal viral inputs (measured by enzyme-linked immunosorbent assay [ELISA] of the viral protein p24), significantly fewer foci of infection (FFU) resulted from challenge with HIV-1 89.6Δvpr than from challenge with HIV-1 89.6WT. Further support of a role for Vpr in overcoming REAF is evidenced in Fig. 1D. When HeLa-CD4 cells were challenged with HIV-1 89.6WT (which has an intact vpr gene), REAF protein was downmodulated. The observed downmodulation was dependent on the presence of Vpr, as HIV-1 89.6Δvpr is incapable of degrading REAF. Moreover, Fig. 1E shows that Vpr and REAF interact with each other, either directly or indirectly as part of a complex, as they were coimmunoprecipitated.
Vpr is released from the capsid and enters the nucleus shortly after infection (11). Imaging flow cytometry combines traditional flow cytometry with microscopy, facilitating the evaluation of both the overall level and subcellular localization of proteins in large populations of cells (25, 26). Using imaging flow cytometry, we determined the relative subcellular localization of REAF in HeLa-CD4 cells. The analysis revealed that REAF is more highly expressed in the nuclear region than in the cytoplasmic region of cycling HeLa-CD4 cells (Fig. 1F).
Previously, we reported that REAF affects the production of reverse transcripts early in infection and that at this critical time point REAF is transiently downmodulated in HeLa-CD4 cells (19). Also using imaging flow cytometry, we looked at the subcellular distribution of REAF at the early time points following HIV-1 infection. REAF protein was quantified by imaging flow cytometry in the cytoplasm and nuclei of HeLa-CD4 cells over the first 3 h of infection with either HIV-1 89.6WT or HIV-1 89.6Δvpr. Following challenge with HIV-1 89.6Δvpr, REAF levels increased within 0.5 h in both the nucleus (∼25%) (Fig. 1G, left) and cytoplasm (∼10%) (Fig. 1G, right). Nuclear levels remained high for 3 h. Conversely, in the presence of Vpr (HIV-1 89.6WT), this increase in REAF was curtailed; instead, there was a steady decline from 0.5 to 2 h. The decline was most apparent in the nucleus, with ∼20% reduction by 1 h and ∼30% at 2 h. By 3 h, levels of REAF protein had recovered. The virus carries limited quantities of Vpr (16), which potentially explains why there is a pause in REAF downmodulation. Lower levels of REAF were also observed in the cytoplasm over time after infection with HIV-1 89.6WT, but to a much lesser extent (Fig. 1G, right). The nuclear enrichment score (NES) is a comparison of the intensity of REAF fluorescence inside the nucleus (defined using DAPI [4′,6-diamidino-2-phenylindole]) to the total fluorescence intensity of REAF in the entire cell (defined using bright-field images). The lower the score, the less REAF is in the nucleus relative to that in the cell overall. Imaging flow cytometry software determined the NES over time after infection with HIV-1 89.6WT or HIV-1 89.6Δvpr (Fig. 1H). By 1 to 2 h, significant segregation emerged; in the presence of Vpr, relative nuclear levels of REAF were suppressed. The greatest segregation occurred 2 h postinfection.
Fluctuations in subcellular REAF expression after HIV-1 infection are Vpr dependent.
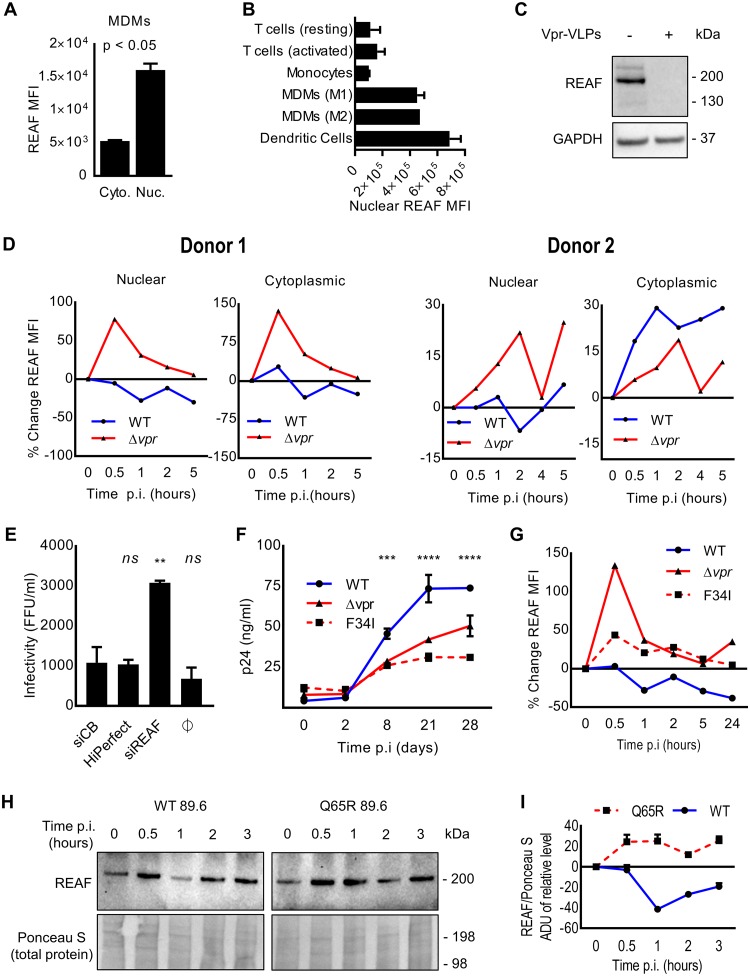
Macrophages are targets for HIV infection in vivo (27, 55, 56). Vpr has been shown, to varying degrees, to be more beneficial for replication in these cells than in cycling T cells (27–31). For these reasons, we investigated REAF effects in monocyte-derived macrophages (MDMs). Using imaging flow cytometry, we determined that, similar to HeLa-CD4 cells, MDMs have significantly greater quantities of REAF in the nucleus than in the cytoplasm (Fig. 2A). Nuclear levels of REAF were also compared in a number of primary cell types using imaging flow cytometry (Fig. 2B). Compared with either monocytes or resting/activated T cells, both MDMs and dendritic cells (DCs) highly express nuclear REAF. As shown in Fig. 2C, MDMs were treated with virus-like particles (VLPs) containing Vpr, and Western blotting confirmed that Vpr downmodulates REAF in MDMs and that Vpr alone is sufficient to induce this downmodulation.
FIG 2.
Fluctuations in subcellular REAF expression after HIV-1 infection are Vpr dependent. (A) Nuclear and cytoplasmic REAF MFI in MDMs measured by imaging flow cytometry. The error bars represent standard deviations of the means of replicates. (B) Nuclear REAF MFI in the indicated primary cell types measured by imaging flow cytometry. The error bars represent standard deviations of the means of the results for two blood donors. (C) REAF protein in MDMs treated with empty or Vpr-containing VLPs. GAPDH was a loading control. The VLP inputs were equivalent at 100 ng of p24. (D) Percent changes in subcellular REAF MFIs in MDMs from time 0 measured by imaging flow cytometry after challenge with HIV-1 89.6WT or HIV-1 89.6Δvpr. Data from two donors are presented. p.i., postinfection. (E) Infectivity of HIV-1 89.6WT in MDMs transfected with siRNA-REAF. HiPerfect (transfection reagent) and siCB were negative controls. **, P < 0.01; ns, not significant; one-way ANOVA and post hoc Dunnett’s test. (F) Infectivity of HIV-1 89.6WT compared with HIV-1 89.6Δvpr and HIV-1 89.6F34I in MDMs. p24 antigen concentrations over 28 days postinfection are indicated. Viral inputs were equivalent at 50 ng of p24. The error bars represent standard deviations of the means of duplicates (***, P < 0.001; ****, P < 0.0001; two-way ANOVA; the same results were obtained for HIV-1 89.6WT versus HIV-1 89.6Δvpr and HIV-1 89.6F34I). The data are representative of at least two independent experiments. (G) Percent change in total cellular REAF MFI from time 0 in MDMs after challenge with HIV-1 89.6WT, HIV-1 89.6Δvpr, or HIV-1 89.6F34I. The results are representative of three independent experiments. (H) REAF protein, measured by Western blotting, in MDMs challenged with HIV-1 89.6WT or HIV-1 89.6Q65R over time. Ponceau S staining of nitrocellulose membrane was a loading control. (I) Associated densitometry, where the error bars represent standard deviations of means of analyses performed in triplicate.
We investigated the ability of Vpr to degrade REAF in MDMs early in infection. The subcellular fluctuations in REAF mean fluorescence intensity (MFI) were measured by imaging flow cytometry in large populations of target cells (>5,000). In the presence of Vpr (HIV-1 89.6WT), nuclear REAF decreased within 2 h of viral infection of macrophages from two donors (Fig. 2D), similar to what was observed in HeLa-CD4 cells (Fig. 1G). In contrast, also in both donors, nuclear REAF rapidly increased from as early as 0.5 h, when the virus does not contain Vpr (HIV-1 89.6Δvpr) (Fig. 2D). For the cytoplasmic compartment, a similar picture emerged for REAF fluctuation. In both donors, when Vpr was absent, REAF levels increased rapidly within 0.5 h of infection (Fig. 2D). This cytoplasmic increase was curtailed in donor 1 when Vpr was present. In donor 2, the loss of nuclear REAF after HIV-1 89.6WT infection was paralleled by an increase in cytoplasmic REAF. Similar kinetics of total REAF protein fluctuation were measured by Western blotting in the presence or absence of Vpr in MDMs from two further donors (data not shown).
We sought to determine if knockdown of REAF in primary macrophages results in an increase in susceptibility to HIV-1 infection. As shown in Fig. 2E, primary MDMs were treated with siRNA targeting REAF (siREAF) or a control protein (siCB). Cells lacking REAF were found to be significantly (P < 0.0001) more susceptible to infection with HIV-1 89.6. We confirmed previous reports that HIV-1 replication in MDMs is more efficient in the presence of Vpr (27, 29). Figure 2F shows that HIV-1 89.6Δvpr has restricted replication in MDMs compared with the wild-type virus expressing Vpr (HIV-1 89.6WT).
To investigate further the relationship between nuclear REAF and Vpr, we generated a virus with a substitution within Vpr (F34I). HIV-1 89.6F34I is incapable of localizing to the nuclear membrane or of interacting with the nuclear transport protein importin-α and nucleoporins (29). Like HIV-1 89.6Δvpr, the mutant virus (HIV-1 89.6F34I) replicates less efficiently in MDMs (Fig. 2F). Using imaging flow cytometry, the respective abilities of the three viruses (HIV-1 89.6WT, HIV-1 89.6Δvpr, and HIV-1 89.6F34I) to downmodulate total REAF protein were investigated in MDMs (Fig. 2G). As expected, there was a loss of total REAF from 0.5 h after HIV-1 89.6WT infection (Fig. 2G) with a transient recovery at around 2 h. The opposite occurred in the absence of Vpr (HIV-1 89.6Δvpr); REAF levels increased after infection. The increase in REAF levels was most potent after 0.5 h of infection with HIV-1 89.6Δvpr. HIV-1 89.6F34I, similar to HIV-1 89.6 Δvpr, could no longer deplete REAF in MDMs (Fig. 2G).
Other targets of Vpr have been reported. It recruits SLX4-SLX1/MUS81-EME1 endonucleases to DCAF1, activating MUS81 degradation and triggering arrest in G2/M (9). It also degrades HLTF, a protein recently shown to enhance infection of HIV-1 in T-cell lines (8, 12). To determine if the depletion of REAF requires the association of Vpr with DCAF1, we generated another mutant virus with a different substitution within vpr, Q65R. Previously, the Q65R mutation was shown to ablate the association between DCAF1 and Vpr and the ability of Vpr to induce arrest at G2/M (32–34). Fig. 2H and I show that this mutant, compared to HIV-1 89.6WT, is unable to downmodulate REAF. We cannot rule out inhibition of synthesis or increased nuclear export, in addition to degradation, as possibilities.
Expression of REAF during the cell cycle varies.
A phenotype of HIV-1 Vpr is the ability to induce cell cycle arrest at G2/M in cycling T cells (27, 35–37). The failure of both HIV-1 89.6F34I and HIV-1 89.6Q65R to efficiently induce G2/M arrest (29, 32, 38–40) and our observation that they cannot downmodulate REAF (Fig. 2G, H, and I) prompted us to investigate REAF and the cell cycle.
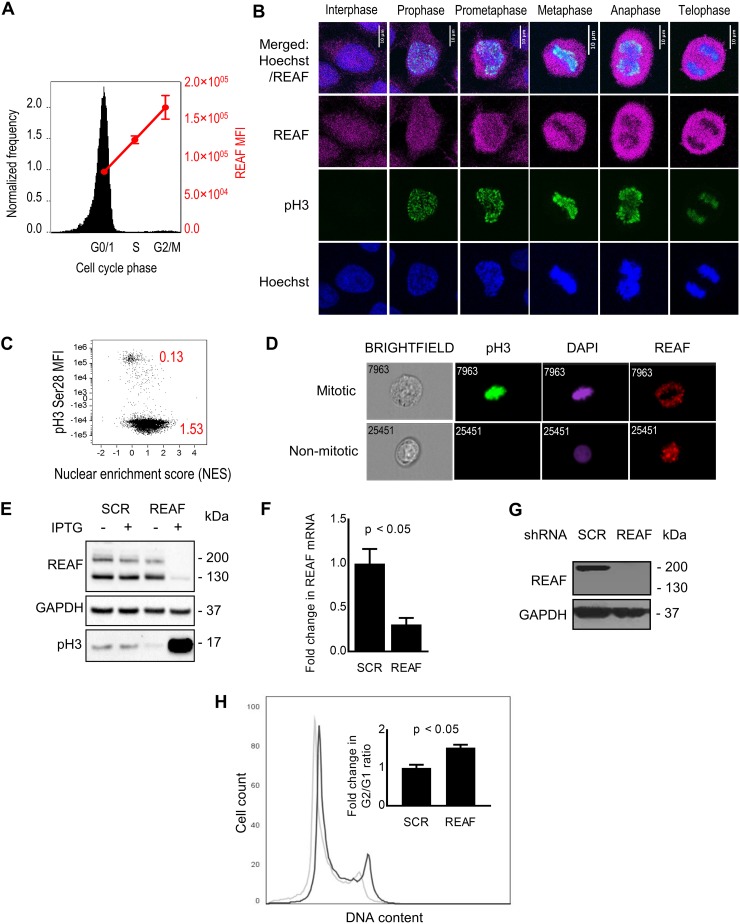
First, we determined the expression levels of REAF at various phases of the cell cycle using imaging flow cytometry (Fig. 3A). REAF protein levels were lowest in G0/G1, increased through S phase, and peaked in G2/M. Confocal microscopy of cycling cells agreed with the quantitative analysis shown in Fig. 3A; overall REAF levels appeared greater in mitotic cells (Fig. 3B). There was also apparent exclusion of REAF from the nuclear region of the cell during mitosis (particularly during metaphase, anaphase, and telophase). Quantitative analysis by imaging flow cytometry of cycling cells confirmed that the mitotic population had a lower nuclear enrichment score (0.13) than the nonmitotic cells (1.53), indicating a lower intensity of REAF in the nucleus than in the cell as a whole (Fig. 3C). Representative images of subcellular REAF in mitotic and nonmitotic cells from imaging flow cytometry are presented in Fig. 3D.
FIG 3.
Depletion of REAF and G2/M accumulation. (A) Imaging flow cytometry of cell cycle phase and REAF protein in DAPI-stained primary monocytes. (B) Confocal microscopy of subcellular REAF in HeLa-CD4 cells. Phospho-histone H3 (Ser28) (pH3) staining and chromatin morphology (Hoechst 33342) were used for cell cycle phase identification. (C) Imaging flow cytometry of subcellular REAF in cycling HeLa-CD4 cells. A lower nuclear enrichment score (red) indicates a lower proportion of overall REAF in the nucleus. Phospho-histone H3 (Ser28) staining confirmed that mitotic cells had a lower score of 0.13. (D) Representative images of subcellular REAF in mitotic and nonmitotic cells. (E) REAF protein in THP-1 cells with IPTG-inducible shRNA targeting REAF (shREAF) or a scrambled control sequence (shSCR). Phospho-histone H3 (Ser10/Thr11) was a mitotic marker, and GAPDH was a loading control. (F) Fold change in the mRNA transcript level in PM1 shREAF cells normalized to PM1 shSCR cells as measured by qPCR. (G) REAF protein in PM1 cells expressing shRNA targeting REAF (shREAF) and PM1 cells expressing a scrambled control sequence (shSCR). GAPDH was a loading control. (H) Flow cytometry of cell cycle phase in PI-stained PM1 shREAF (black line) and PM1 shSCR (gray line) cells. The plot is representative of three biological replicates. The inset shows the fold change in the G2/G1 ratio in PM1 shREAF cells normalized to PM1 shSCR cells. The error bars represent standard deviations of the means of three biological replicates.
To determine if the G2/M arrest phenotype induced by Vpr could be related to its ability to downmodulate REAF, we generated inducible THP-1 and PM1 cell lines that upon induction produced shRNA targeting either REAF or a scrambled control sequence (SCR). After knockdown of REAF in THP-1 cells, there was a clear increase in the expression of the mitotic marker phosphorylated histone H3 (Ser10/Thr11) (Fig. 3E). However, when measured more quantitatively by DNA content analysis in PM1 cells, the potency of the G2/M arrest appeared weak compared to the levels previously described (29, 37). REAF downmodulation in PM1 cells was confirmed by a reduction in mRNA and protein (Fig. 3F and G). Cell cycle phase profiles were determined by flow cytometry (Fig. 3H). The increase in the G2/G1 ratio of cells with REAF knocked down, although small (Fig. 3H, inset), was comparable to those in other studies where individual reported targets of Vpr were knocked down (13). In agreement with Greenwood et al., we contend that downmodulation of more than one protein may be required to produce the strong Vpr-induced G2/M arrest reported (13, 29, 37).
REAF is not IFN stimulated or under positive selection.
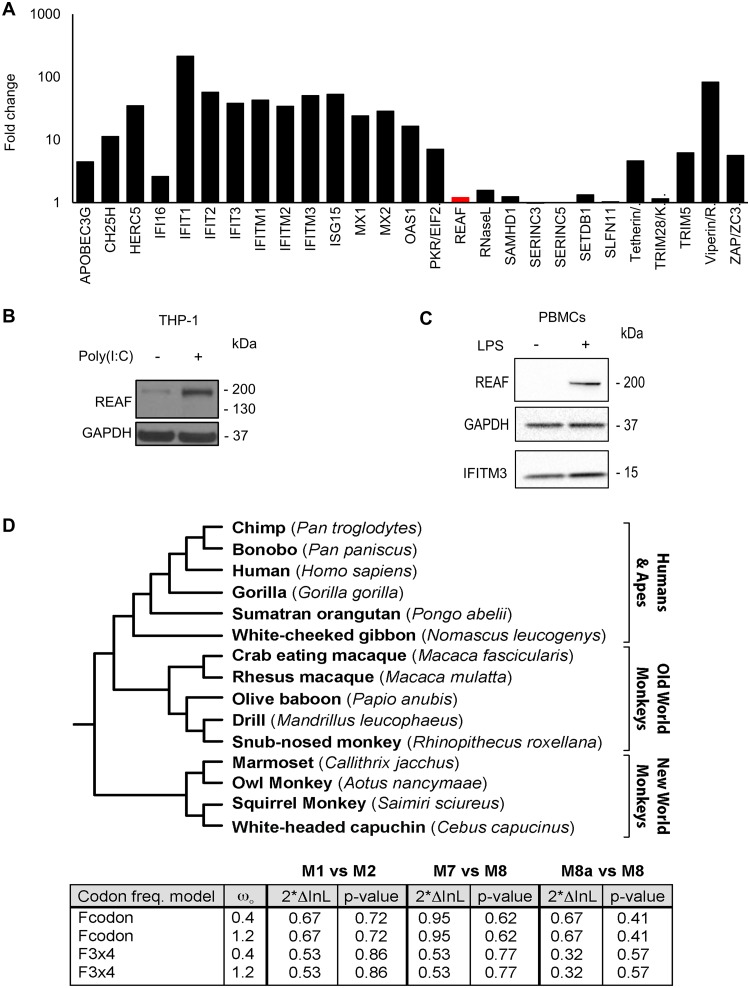
Interferon alpha (IFN-α) is central to innate immune responses and is known to induce many HIV-1 restriction factors (41, 42). We used RNA microarray analysis to determine if IFN-α upregulated REAF mRNA in MDMs. Figure 4A shows that IFN-α induced mRNA upregulation of many known antiviral genes, including those encoding the HIV restriction factors APOBEC3G, MX2, tetherin, and viperin (41, 43, 44), but with little or no upregulation of REAF mRNA. Nevertheless, antiviral factors are also often upregulated in response to pathogen-associated molecular patterns (PAMPs). Poly(I·C) is a double-stranded RNA used to stimulate molecular pattern recognition pathways associated with viral infection. Figure 4B shows that poly(I·C) induces REAF in THP-1 cells, a macrophage line. LPS, another PAMP that is Toll-like receptor 4 (TLR4) specific (45), also induces the upregulation of REAF expression in peripheral blood mononuclear cells (PBMCs) (Fig. 4C).
FIG 4.
REAF is not IFN stimulated or under positive selection. (A) RNA microarray analysis determined changes in REAF mRNA compared to other antiviral factors in MDMs treated with IFN-α (500 IU/ml). (B) REAF protein in PMA-differentiated THP-1 cells after poly(I·C) treatment for 48 h. GAPDH was a loading control. (C) REAF protein in PBMCs after LPS treatment. GAPDH was a loading control, and IFITM3 was a positive control for LPS-induced upregulation. (D) REAF DNA sequences from 15 extant primate species (tree length, 0.2 substitutions per site along all branches of the phylogeny) (top) were analyzed using the PAML package for signatures of positive natural selection (bottom). Initial seed values for ω (ωO) and different codon frequency models were used in the maximum-likelihood simulation. Twice the differences in the natural logarithms of the likelihoods (2*InL) of the two models were calculated and evaluated using the chi-squared critical value. The P value indicates the confidence with which the null model (M1, M7, or M8a) can be rejected in favor of the model of positive selection (M2 or M8).
Restriction factors are often under evolutionary positive selection at sites that interact with virus. We compared REAF DNA sequences from 15 extant primate species using the PAML package (see below) for signatures of positive natural selection. We found no evidence of positive selection of REAF in the primate lineage (Fig. 4D).
DISCUSSION
The deletion of vpr in HIV-1 leads to impairment of its replication in both HeLa-CD4 cells and primary macrophages. A number of experiments presented here point to a role for Vpr in the counterrestriction of the antiviral protein REAF. First, HIV-1 replication is significantly enhanced by knockdown of REAF in either HeLa-CD4 cells or primary macrophages, and this phenotype is more pronounced for viruses lacking vpr. Second, REAF is downmodulated early after infection in a manner dependent on both the presence of Vpr and, as demonstrated by vpr single point mutations, the localization of Vpr to the nuclear envelope and its interaction with a nuclear-localized E3 ubiquitin ligase, DCAF1. Third, using VLPs, we showed that Vpr alone is sufficient to downmodulate REAF in MDMs. Finally, by coimmunoprecipitation, we demonstrated that REAF and Vpr physically interact, either directly or indirectly as part of a complex. Taken together, our results highlight the importance of the relationship between REAF and the HIV-1 accessory protein Vpr.
Others have shown a specific requirement for Vpr in the efficient infection of nondividing cells and less so in cycling T cells (12, 27, 29). The requirement for Vpr in macrophage infection is substantiated here; reduced viral replication was observed after infection of MDMs with either HIV-1 89.6Δvpr or HIV-1 89.6F34I compared to HIV-1 89.6WT infection. This is the first demonstration of vpr-alleviated impairment of HIV-1 replication in primary macrophages. Recently Yan et al. showed that HLTF, a reported target of Vpr, restricts replication of HIV-1 in T cells, while Lahouassa et al. also reported a Vpr-dependent loss of HLTF at 6 h postinfection (8, 12). HLTF downmodulation occurs concomitantly with that of REAF as early as 0.5 h postinfection (data not shown). Interestingly, HLTF and REAF were identified in the same screen for proteins that interact with single-stranded DNA (46).
The transient nature and timing of REAF depletion shown here are consistent with its ability to impede the production of reverse transcripts early in infection (19). After an initial downmodulation of REAF following infection, REAF depletion was paused, perhaps due to the limited quantities of Vpr carried in the virus particle (16). Recently, Greenwood et al. carried out a whole-cell proteomics screen for factors up- or downmodulated by Vpr in T cells. They identified almost 2,000 proteins affected, underlining the promiscuous activity of Vpr (13). In light of these findings, it is important that attention be directed to those reported Vpr targets that affect replication of HIV-1 in primary cells.
Our model is that Vpr is carried into the cell by HIV-1 in limited quantities, but sufficient to downmodulate REAF in the time frame required for reverse transcription to proceed unhindered. Interestingly, nuclear localization of Vpr is also required for the downmodulation of REAF, perhaps similar to the Vpx-mediated depletion of the reverse transcription inhibitor SAMHD1 (whose degradation is initiated in the nucleus) (47). Localization of Vpr to the nuclear region is a requirement for interaction with REAF and DCAF1, and this results in the degradation of REAF. We propose that REAF is linked to the innate immune response, as treatment of cells with poly(I·C) or LPS induces its expression. Furthermore, HIV-1 replication without an intact vpr gene induces the expression of REAF to high levels in primary macrophages as early as 30 min postinfection.
A poorly understood event is the induction by Vpr of cell cycle arrest in the G2/M phase after infection. We report here that the loss of REAF from cycling cells contributes to an accumulation of the population in G2/M. However, the levels of G2/M induction are weak compared to those in early reports (29, 37). Thus, we contend that Vpr-induced downmodulation of more than one protein by Vpr may be required for the complete induction of G2/M arrest seen previously. In support of this, G2/M arrest was only weakly induced when Greenwood et al. independently knocked down several Vpr-targeted proteins, such as MCM10, SMN1, CDCA2, and ZNF267 (13).
REAF is unlike the evolving HIV restriction factors, such as APOBEC3G, SAMHD1, TRIM5, and BST2/tetherin, and is more similar to SERINC3 and -5, which are not under positive selection (48, 49). REAF has many properties of restriction factors (43, 50). It interacts with HIV-1 reverse transcripts, impeding reverse transcription (19). It is germ line encoded, constitutively expressed in cells, regulated by the proteasome system, suppressed by an accessory protein (Vpr), and upregulated by poly(I·C) and LPS. Our results support the current model for Vpr activity, which is that it induces the degradation of proteins involved in an unknown restriction of HIV-1. We propose that REAF may be a crucial component of a Vpr-targeted restriction system that is active against HIV-1.
MATERIALS AND METHODS
Ethics statement.
Leukocyte cones, from which PBMCs were isolated, were obtained from the NHS blood transfusion service at St. George’s Hospital, London, United Kingdom. The donors were anonymous, and thus, patient consent was not required. The local ethical approval reference number is 06/Q0603/59.
Cell lines.
HEK-293T cells (ATCC); PM1, THP-1, C8166, and HeLa-CD4 cells (all National Institute for Biological Standards and Control [NIBSC] AIDS Reagents); and HeLa-CD4 shRNA-REAF cells (previously described [17]) were maintained at 37°C in 5% CO2. The cells were cultured in Dulbecco’s modified Eagle medium (DMEM) (ThermoFisher) supplemented with 5 to 10% fetal bovine serum (FBS) and appropriate antibiotics (all from Thermo Fisher). HeLa-CD4 shRNA-REAF cells were selected for resistance to puromycin in medium supplemented with 10 μg/ml puromycin (Invitrogen).
The isopropyl-β-d-1-thiogalactopyranoside (IPTG)-inducible vector pLKO-IPTG-3xLacO (Sigma) was used to express shRNAs targeted against REAF (Mission TRCN0000141116; Sigma). Additionally, a nontarget (scrambled) control was prepared. Viral particles for cell line transductions were prepared by cotransfecting HEK-293T cells with pLKO-IPTG-3xLacO, the Gag/Pol packaging vector pLP1, a Rev expression vector (pLP2), and the vesicular stomatitis virus G protein (VSV-G) expression vector pVPack-VSV-G (Stratagene). After 72 h, the virus was clarified by low-speed centrifugation and passed through a 0.45-μm-pore-size filter. THP-1 and PM1 cells were transduced by treating with viral particles in the presence of 8 g/ml Polybrene for 72 h, after which resistant colonies were selected and maintained with 2 μg/ml puromycin. Culturing cells in the presence of 1 mM IPTG for 72 h induced expression of shRNAs.
Transfections and virus/VLP production.
The infectious molecular clone for HIV-1 89.6 was obtained from the Centre for AIDS Research (NIBSC, United Kingdom). Infectious full-length and chimeric HIV clones were prepared by linear polyethylenimine 25K (Polysciences), Lipofectamine 2000 (Invitrogen), or Lipofectamine 3000 (Invitrogen) transfection of HEK-293T cells. VLPs were produced by linear polyethylenimine 25K (Polysciences) transfection of HEK-293T cells. The VLP packaging vector was a gift from N. Landau, and its production is described in reference 57.
The plasmid construct HIV-1 89.6Δvpr was generated from the HIV-1 89.6 molecular clone, using overlap extension PCR (42). Clones were confirmed by plasmid sequencing (Source BioScience). Primer sequences are available upon request. HIV-1 p89.6 F34I and Q65R vpr mutants were made by site-directed mutagenesis (Agilent) of the p89.6 plasmid. HEK-293T cells were plated at 2 × 104/cm2 in 10-cm dishes (for virus and VLP production) 48 h prior to transfection. For virus/VLP production, the supernatant was harvested 72 h posttransfection and cleared of cell debris by centrifugation at 500 × g for 5 min. All viruses were amplified with C8166 cells for 48 h prior to harvest.
Titration of replication-competent virus.
HeLa-CD4 cells were seeded at 1.5 × 104 cells/well in 48-well plates to form adherent monolayers of cells. The cell monolayers were challenged with serial 1:5 dilutions of virus, and the titer was assessed after 48 h by in situ intracellular staining of HIV-1 p24 to identify individual foci of viral replication (FFU), as described previously (51). For infection time course experiments, 400 to 500 μl of 1 × 105 FFU/ml (HeLa-CD4 cells) or 3 × 103 FFU/ml (MDMs) virus was added per well to cells cultured in 6-well trays for 24 h (HeLa-CD4 cells) or 7 days (MDMs). For Fig. 2F and H, cells were challenged with 50 ng p24 in 6-well plates with 2 × 106 MDMs per well. For Fig. 2F, supernatants were harvested on days 0, 2, 8, 21, and 28 postchallenge, and p24 concentrations were analyzed by ELISA.
p24 ELISA.
ELISA plates were precoated with 5 μg/ml sheep anti-HIV-1 p24 antibody (Aalto Bio Reagents) at 4°C overnight. Viral supernatants were treated with 1% Empigen BB (Merck) for 30 min at 56°C and then plated at 1:10 dilution in Tris-buffered saline (TBS) on precoated plates and incubated for 3 h at room temperature. Alkaline phosphatase-conjugated mouse anti-HIV-1 p24 monoclonal antibody (Aalto Bio Reagents) in TBS, 20% sheep serum, 0.05% (vol/vol) Tween 20 was then added and incubated for 1 h at room temperature. After 4 washes with phosphate-buffered saline (PBS)-0.01% (vol/vol) Tween 20 and 2 washes with ELISA Light washing buffer (ThermoFisher), CSPD substrate with Sapphire II enhancer (ThermoFisher) was added and incubated for 30 min at room temperature before chemiluminescence detection using a plate reader.
cDNA synthesis and qPCR.
Total RNA was extracted from PM1 cells using a ReliaPrep RNA kit (Promega). One-step reverse transcription-quantitative PCR (qPCR) (Quantbio) using TaqMan probes detected amplified transcripts. Data acquired by an Agilent Mx3000 was analyzed with MxPro software.
Gene expression RNA microarray.
Prior to microarray analysis, RNA from MDMs was prepared using an Illumina TotalPrep RNA amplification kit (Ambion) according to the manufacturer’s instructions. The probes were hybridized on an Illumina HT12v3 bead array following the manufacturer’s standard hybridization and scanning protocols. Raw measurements were processed with GenomeStudio software (Illumina) and quantile normalized.
IFN, poly(I·C), and LPS treatments.
MDMs were treated with IFN (500 IU/ml) for 24 h before harvest for RNA extraction. Recombinant IFN-α was purchased from Sigma (IFN-αA/D human) and is a combination of human subtypes 1 and 2. THP-1 cells were treated with poly(I·C) (25 μg/ml; HMW/LyoVec; Invitrogen) for 48 h before analysis by Western blotting. Prior to poly(I·C) treatment, THP-1 cells were treated with phorbol 12-myristate 13-acetate (PMA) (62 ng/ml) for 3 days and then with PMA-free DMEM for 2 days to allow differentiation and recovery. PBMCs isolated from healthy blood donors were treated with LPS (10 ng/ml) for 24 h before analysis by Western blotting.
Western blotting.
Cells were harvested and lysed in 30 to 50 μl of radioimmunoprecipitation assay (RIPA) buffer supplemented with NaF (5 μM), Na2VO3 (5 μM), β-glycerophosphate (5 μM), and 1× protease inhibitor cocktail (Cytoskeleton). The protein concentration of each sample was determined using a bicinchoninic acid (BCA) protein assay kit (Pierce); 12.5 to 70 μg of total protein was separated by SDS-PAGE (4 to 12% Bis-Tris gels; Invitrogen) at 120 V for 1 h 45 min in MOPS (morpholinepropanesulfonic acid) SDS running buffer (Invitrogen). The separated proteins were transferred onto nitrocellulose membranes (0.45-μm pore size; GE Healthcare) at 45 V for 2 h in ice-cold 20% (vol/vol) methanol NuPAGE transfer buffer (ThermoFisher). Vpr affects GAPDH levels in macrophages, so Ponceau S was used as a loading control (58). After transfer, the membranes were stained for total protein using Ponceau S staining solution (0.1% [wt/vol] Ponceau in 5% [vol/vol] acetic acid), washed 3 times for 5 min each time on an orbital shaker in distilled water (dH2O), and imaged using the ChemiDoc gel imaging system. The membranes were blocked for 1 h at room temperature in 5% (wt/vol) nonfat milk powder in TBS-T buffer. Specific proteins were detected with primary antibodies by incubation with membranes overnight at 4°C and with secondary antibodies for 1 h at room temperature. All the antibodies were diluted in blocking buffer. Proteins were visualized using ECL Prime Western blotting detection reagent (GE Healthcare), imaged either using the ChemiDoc gel imaging system (Bio-Rad) or by exposure to CL-XPosure films (ThermoScientific), and developed. In all places where quantitative comparisons were made, such as for Fig. 1D and E, blots were derived from the same blot or blots processed together.
Antibodies.
Primary rabbit polyclonal antibody to REAF (RbpAb-RPRD2) has been previously described (19). For imaging flow cytometry and confocal microscopy, RbpAb-RPRD2 was detected using goat anti-rabbit IgG conjugated with Alexa Fluor 647 (Invitrogen). Fluorescein isothiocyanate (FITC)-labeled anti-phospho-histone H3 (Ser28) Alexa 488 (BD Bioscience) was used for imaging flow cytometry and confocal microscopy. MsmAb-GFP (green fluorescent protein) (Abcam) was detected by anti-mouse IgG antibody conjugated to horseradish peroxidase (HRP) (GE Healthcare) for Western blotting. Also for Western blotting, RbpAb-RPRD2, RbmAb-IFITM3 (EPR5242; Insight Biotechnology), RbpAb-GAPDH (glyceraldehyde-3-phosphate dehydrogenase), and RbmAb-phospho-histone H3 (Ser10/Thr11) were detected with donkey anti-rabbit IgG conjugated to HRP (GE Healthcare).
Immunoprecipitation.
HEK-293T cells, transfected with either Vpr-GFP expression plasmid or GFP control expression vector, were lysed 72 h posttransfection in RIPA buffer supplemented with NaF (5 μM), Na2VO3 (5 μM), β-glycerophosphate (5 μM), and 1× protease inhibitor cocktail (Cytoskeleton). The total protein concentration was determined using a BCA protein assay kit (Pierce). GFP-TRAP magnetic agarose beads were equilibrated in ice-cold dilution buffer (10 mM Tris-Cl, pH 7.5, 150 mM NaCl, 0.5 mM EDTA; Chromotek) according to the manufacturer’s instructions. Cell lysates containing 100 μg of total protein were incubated with 10 μl of equilibrated beads for 2 h at 4°C with gentle agitation. The beads were washed three times with PBS with Tween 20 (PBST) buffer before analysis of immunoprecipitated protein by Western blotting.
Magnetic separation of primary human lymphocytes.
PBMCs were isolated from leukocyte cones (NHS Blood Transfusion Service, St. George’s Hospital, London, United Kingdom) by density gradient centrifugation with Lymphoprep density gradient medium (Stemcell Technologies). Peripheral monocytes were isolated from PBMCs using human CD14+ magnetic beads (Miltenyi Biotech) according to the manufacturer’s instructions. CD4+ T cells were isolated from the flowthrough using a human CD4+ T cell isolation kit (Miltenyi Biotech). CD14+ monocytes and CD4+ T cells were either differentiated or fixed directly after isolation for intracellular staining. To obtain M1 and M2 macrophages (M1/M2 MDMs), monocytes were treated with either granulocyte-macrophage colony-stimulating factor (GM-CSF) (100 ng/ml; Peprotech) or macrophage colony-stimulating factor (M-CSF) (100 ng/ml) for 7 days, with medium replenished on day 4. To obtain DCs, monocytes were treated with GM-CSF (50 ng/ml) and interleukin 4 (IL-4) (50 ng/ml) for 7 days, with medium replenished on day 4. Activated CD4+ T cells were obtained by stimulating freshly isolated CD4+ T cells at 1 × 106/ml with T cell activator CD3/CD28 Dynabeads (ThermoFisher), at a bead/cell ratio of 1, for 7 days. The magnetic beads were removed prior to intracellular staining and imaging flow cytometry.
Immunofluorescence.
HeLa-CD4 cells were plated at 2 × 104/cm2 in 8-well chamber slides for confocal microscopy. The cells were washed with PBS and fixed in 2% paraformaldehyde-PBS for 10 min at room temperature. The fixed cells were permeabilized in 0.2% Triton X-100–PBS for 20 min at room temperature and incubated with primary antibodies in PBS–0.1% Triton X-100–2% BSA overnight at 4°C. After 3 washes in PBS, the cells were labeled with secondary antibodies in the same buffer for 1 h at room temperature and washed 3 times with PBS. Nuclei were counterstained with Hoechst 33342 (2 μM; ThermoFisher) for 5 min at room temperature. The labeled cells were mounted with ProLong Diamond antifade mountant (ThermoFisher) and analyzed on a laser scanning confocal microscope (LSM 710; Carl Zeiss). Images were acquired with ZEN software and analyzed with ImageJ.
Imaging flow cytometry.
Cells were fixed in Fix and Perm solution A (Nordic MUbio) for 30 min and permeabilized with 0.2% Triton X-100–PBS. MDMs were blocked with human serum (1%). The staining buffer used was 0.1% Triton X-100, 0.5% FBS. Nuclei were counterstained with DAPI (1 μg/ml) for 2 h. Imaging flow cytometry was performed using an Amnis ImageStream Mark II flow cytometer (Merck) and INSPIRE software (Amnis). A minimum of 5,000 events were collected for each sample. IDEAS software (Amnis) was used for analysis and to determine the NES. The NES is a comparison of the intensity of REAF fluorescence inside the nucleus (defined using the exclusively nuclear stain DAPI) to the total fluorescence intensity of REAF in the entire cell (defined using bright-field images). A lower nuclear enrichment score indicates that a smaller proportion of overall REAF is located within the nucleus.
Statistics.
Statistical significance in all experiments was calculated by Student's t test (two tailed) or analysis of variance (ANOVA) (indicated). Data are represented as means and standard deviations (error bars). GraphPad Prism and Excel were used for calculation and illustration of graphs.
Cell cycle analysis.
Cell cycle phase distribution was determined by analysis of the DNA content via either flow cytometry (BD FACS Canto II) or imaging flow cytometry. Cells were fixed in Fix and Perm solution A (Nordic MUbio) and stained with DAPI (1 μg/ml) before analysis by imaging flow cytometry. Cell lysates were assessed by Western blotting using the anti-phospho-histone H3 (Ser10/Thr11) antibody as an additional mitotic marker. Chromatin morphology and anti-phospho-histone H3 (Ser28) were used to determine the cells in indicated phases of the cell cycle and mitosis in confocal microscopy experiments. The cell cycle status of PM1 cells was determined via propidium iodide (PI) staining using FxCycle PI/RNase solution (ThermoFisher). Stained cells were analyzed on an NxT flow cytometer (ThermoFisher).
Evolutionary analysis.
To ascertain the evolutionary trajectory of REAF, we analyzed DNA sequence alignments of REAF proteins from 15 species of extant primates using codeml (as implemented by PAML 4.2) (52). The evolution of REAF was compared to several NSsites models of selection, M1, M7, and M8a (neutral models with site classes of dN/dS ratios [ratios of nonsynonymous to synonymous evolutionary changes] of <1 or ≤1) and M2 and M8 (positive-selection models allowing an additional site class with dN/dS ratios of >1). Two models of codon frequencies (F61 and F3x4) and two different seed values for dN/dS ratios (ω) were used in the maximum-likelihood simulations. Likelihood ratio tests were performed to evaluate which model of evolution the data fit significantly better. The P value indicates the confidence with which the null model (M1, M7, or M8a) can be rejected in favor of the model of positive selection (M2 and M8). The alignment of REAF was analyzed by GARD to confirm the lack of recombination during REAF evolution (53). Neither positively selected sites nor signatures of episodic diversifying selection were detected within REAF by additional evolutionary analysis by REL and FEL or MEME (54).
Data availability.
All RNA microarray data are available in the Gene Expression Omnibus (GEO) database with accession number GSE54455.
ACKNOWLEDGMENTS
This work was partly supported by an MRC Senior Non-Clinical Fellowship awarded to A.M. (G117/547) and Ph.D. studentships awarded by QMUL Life Sciences Institute (LSI) (C.E.J.) and The Rosetrees Trust (J.M.G. and C.E.J.; M665 and M275). R.D.S. was supported by the Wellcome Trust-University of Edinburgh Institutional Strategic Support Fund.
The monoclonal antibodies to HIV-1 p24 (EVA365 and EVA366) were provided by the EU Program EVA Centre for AIDS Reagents, NIBSC, United Kingdom (AVIP contract number LSHP-CT-2004-503487). The Wellcome Trust (101604/Z/13/Z) funded the purchase of an Amnis ImageStream imaging flow cytometer. We thank N. Landau for the kind gift of VPL constructs.
REFERENCES
- 1.Eckstein DA, Sherman MP, Penn ML, Chin PS, De Noronha CM, Greene WC, Goldsmith MA. 2001. HIV-1 Vpr enhances viral burden by facilitating infection of tissue macrophages but not nondividing CD4+ T cells. J Exp Med 194:1407–1419. doi: 10.1084/jem.194.10.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Malim MH, Emerman M. 2008. HIV-1 accessory proteins—ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398. doi: 10.1016/j.chom.2008.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, DeLucia M, Ahn J. 2016. SLX4-SLX1 protein-independent down-regulation of MUS81-EME1 protein by HIV-1 viral protein R (Vpr). J Biol Chem 291:16936–16947. doi: 10.1074/jbc.M116.721183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schrofelbauer B, Yu Q, Zeitlin SG, Landau NR. 2005. Human immunodeficiency virus type 1 Vpr induces the degradation of the UNG and SMUG uracil-DNA glycosylases. J Virol 79:10978–10987. doi: 10.1128/JVI.79.17.10978-10987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Romani B, Shaykh Baygloo N, Aghasadeghi MR, Allahbakhshi E. 2015. HIV-1 Vpr protein enhances proteasomal degradation of MCM10 DNA replication factor through the Cul4-DDB1[VprBP] E3 ubiquitin ligase to induce G2/M cell cycle arrest. J Biol Chem 290:17380–17389. doi: 10.1074/jbc.M115.641522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Maudet C, Sourisce A, Dragin L, Lahouassa H, Rain J-C, Bouaziz S, Ramirez BC, Margottin-Goguet F. 2013. HIV-1 Vpr induces the degradation of ZIP and sZIP, adaptors of the NuRD chromatin remodeling complex, by hijacking DCAF1/VprBP. PLoS One 8:e77320. doi: 10.1371/journal.pone.0077320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lv L, Wang Q, Xu Y, Tsao LC, Nakagawa T, Guo H, Su L, Xiong Y. 2018. Vpr targets TET2 for degradation by CRL4(VprBP) E3 ligase to sustain IL-6 expression and enhance HIV-1 replication. Mol Cell 70:961–970 e965. doi: 10.1016/j.molcel.2018.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lahouassa H, Blondot M-L, Chauveau L, Chougui G, Morel M, Leduc M, Guillonneau F, Ramirez BC, Schwartz O, Margottin-Goguet F. 2016. HIV-1 Vpr degrades the HLTF DNA translocase in T cells and macrophages. Proc Natl Acad Sci U S A 113:5311–5316. doi: 10.1073/pnas.1600485113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laguette N, Brégnard C, Hue P, Basbous J, Yatim A, Larroque M, Kirchhoff F, Constantinou A, Sobhian B, Benkirane M. 2014. Premature activation of the SLX4 complex by Vpr promotes G2/M arrest and escape from innate immune sensing. Cell 156:134–145. doi: 10.1016/j.cell.2013.12.011. [DOI] [PubMed] [Google Scholar]
- 10.Hrecka K, Hao C, Shun MC, Kaur S, Swanson SK, Florens L, Washburn MP, Skowronski J. 2016. HIV-1 and HIV-2 exhibit divergent interactions with HLTF and UNG2 DNA repair proteins. Proc Natl Acad Sci U S A 113:E3921–E3930. doi: 10.1073/pnas.1605023113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hofmann S, Dehn S, Businger R, Bolduan S, Schneider M, Debyser Z, Brack-Werner R, Schindler M. 2017. Dual role of the chromatin-binding factor PHF13 in the pre- and post-integration phases of HIV-1 replication. Open Biol 7:170115. doi: 10.1098/rsob.170115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yan J, Shun MC, Zhang Y, Hao C, Skowronski J. 2019. HIV-1 Vpr counteracts HLTF-mediated restriction of HIV-1 infection in T cells. Proc Natl Acad Sci U S A 116:9568–9577. doi: 10.1073/pnas.1818401116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Greenwood EJD, Williamson JC, Sienkiewicz A, Naamati A, Matheson NJ, Lehner PJ. 2019. Promiscuous targeting of cellular proteins by Vpr drives systems-level proteomic remodeling in HIV-1 infection. Cell Rep 27:1579–1596 e1577. doi: 10.1016/j.celrep.2019.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen EA, Terwilliger EF, Jalinoos Y, Proulx J, Sodroski JG, Haseltine WA. 1990. Identification of HIV-1 vpr product and function. J Acquir Immune Defic Syndr 3:11–18. [PubMed] [Google Scholar]
- 15.Bachand F, Yao XJ, Hrimech M, Rougeau N, Cohen EA. 1999. Incorporation of Vpr into human immunodeficiency virus type 1 requires a direct interaction with the p6 domain of the p55 gag precursor. J Biol Chem 274:9083–9091. doi: 10.1074/jbc.274.13.9083. [DOI] [PubMed] [Google Scholar]
- 16.Desai TM, Marin M, Sood C, Shi J, Nawaz F, Aiken C, Melikyan GB. 2015. Fluorescent protein-tagged Vpr dissociates from HIV-1 core after viral fusion and rapidly enters the cell nucleus. Retrovirology 12:88. doi: 10.1186/s12977-015-0215-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marno KM, O’Sullivan E, Jones CE, Diaz-Delfin J, Pardieu C, Sloan RD, McKnight A. 2017. RNA-associated early-stage antiviral factor is a major component of Lv2 restriction. J Virol 91:e01228-16. doi: 10.1128/JVI.01228-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKnight A, Griffiths DJ, Dittmar M, Clapham P, Thomas E. 2001. Characterization of a late entry event in the replication cycle of human immunodeficiency virus type 2. J Virol 75:6914–6922. doi: 10.1128/JVI.75.15.6914-6922.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marno KM, Ogunkolade BW, Pade C, Oliveira NMM, O'Sullivan E, McKnight Á. 2014. Novel restriction factor RNA-associated early-stage anti-viral factor (REAF) inhibits human and simian immunodeficiency viruses. Retrovirology 11:3. doi: 10.1186/1742-4690-11-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Harrison IP, McKnight A. 2011. Cellular entry via an actin and clathrin-dependent route is required for Lv2 restriction of HIV-2. Virology 415:47–55. doi: 10.1016/j.virol.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Marchant D, Neil SJ, Aubin K, Schmitz C, McKnight A. 2005. An envelope-determined, pH-independent endocytic route of viral entry determines the susceptibility of human immunodeficiency virus type 1 (HIV-1) and HIV-2 to Lv2 restriction. J Virol 79:9410–9418. doi: 10.1128/JVI.79.15.9410-9418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reuter S, Kaumanns P, Buschhorn SB, Dittmar MT. 2005. Role of HIV-2 envelope in Lv2-mediated restriction. Virology 332:347–358. doi: 10.1016/j.virol.2004.11.025. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz C, Marchant D, Neil SJ, Aubin K, Reuter S, Dittmar MT, McKnight A. 2004. Lv2, a novel postentry restriction, is mediated by both capsid and envelope. J Virol 78:2006–2016. doi: 10.1128/jvi.78.4.2006-2016.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibbons JM, Marno KM, Pike R, Lee W-YJ, Jones CE, Ogunkolade BW, Pardieu C, Bryan A, Fu RM, Warnes G, Rowley PA, Sloan RD, McKnight Á. 2019. HIV-1 accessory protein Vpr interacts with REAF/RPRD2 to mitigate its antiviral activity. bioRxiv doi: 10.1101/408161. [DOI]
- 25.Zuba-Surma EK, Kucia M, Abdel-Latif A, Lillard JW Jr, Ratajczak MZ. 2007. The ImageStream system: a key step to a new era in imaging. Folia Histochem Cytobiol 45:279–290. [PubMed] [Google Scholar]
- 26.Han Y, Gu Y, Zhang AC, Lo YH. 2016. Review: imaging technologies for flow cytometry. Lab Chip 16:4639–4647. doi: 10.1039/c6lc01063f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connor RI, Chen BK, Choe S, Landau NR. 1995. Vpr is required for efficient replication of human immunodeficiency virus type-1 in mononuclear phagocytes. Virology 206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 28.Heinzinger NK, Bukrinsky MI, Haggerty SA, Ragland AM, Kewalramani V, Lee MA, Gendelman HE, Ratner L, Stevenson M, Emerman M. 1994. The Vpr protein of human immunodeficiency virus type 1 influences nuclear localization of viral nucleic acids in nondividing host cells. Proc Natl Acad Sci U S A 91:7311–7315. doi: 10.1073/pnas.91.15.7311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vodicka MA, Koepp DM, Silver PA, Emerman M. 1998. HIV-1 Vpr interacts with the nuclear transport pathway to promote macrophage infection. Genes Dev 12:175–185. doi: 10.1101/gad.12.2.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Balliet JW, Kolson DL, Eiger G, Kim FM, McGann KA, Srinivasan A, Collman R. 1994. Distinct effects in primary macrophages and lymphocytes of the human immunodeficiency virus type 1 accessory genes vpr, vpu, and nef: mutational analysis of a primary HIV-1 isolate. Virology 200:623. doi: 10.1006/viro.1994.1225. [DOI] [PubMed] [Google Scholar]
- 31.Chen R, Le Rouzic E, Kearney JA, Mansky LM, Benichou S. 2004. Vpr-mediated incorporation of UNG2 into HIV-1 particles is required to modulate the virus mutation rate and for replication in macrophages. J Biol Chem 279:28419–28425. doi: 10.1074/jbc.M403875200. [DOI] [PubMed] [Google Scholar]
- 32.Jacquot G, Le Rouzic E, Maidou-Peindara P, Maizy M, Lefrere JJ, Daneluzzi V, Monteiro-Filho CM, Hong D, Planelles V, Morand-Joubert L, Benichou S. 2009. Characterization of the molecular determinants of primary HIV-1 Vpr proteins: impact of the Q65R and R77Q substitutions on Vpr functions. PLoS One 4:e7514. doi: 10.1371/journal.pone.0007514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le Rouzic E, Belaidouni N, Estrabaud E, Morel M, Rain JC, Transy C, Margottin-Goguet F. 2007. HIV1 Vpr arrests the cell cycle by recruiting DCAF1/VprBP, a receptor of the Cul4-DDB1 ubiquitin ligase. Cell Cycle 6:182–188. doi: 10.4161/cc.6.2.3732. [DOI] [PubMed] [Google Scholar]
- 34.Le Rouzic E, Morel M, Ayinde D, Belaidouni N, Letienne J, Transy C, Margottin-Goguet F. 2008. Assembly with the Cul4A-DDB1DCAF1 ubiquitin ligase protects HIV-1 Vpr from proteasomal degradation. J Biol Chem 283:21686–21692. doi: 10.1074/jbc.M710298200. [DOI] [PubMed] [Google Scholar]
- 35.He J, Choe S, Walker R, Di Marzio P, Morgan DO, Landau NR. 1995. Human immunodeficiency virus type 1 viral protein R (Vpr) arrests cells in the G2 phase of the cell cycle by inhibiting p34cdc2 activity. J Virol 69:6705–6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogel ME, Wu LI, Emerman M. 1995. The human immunodeficiency virus type 1 vpr gene prevents cell proliferation during chronic infection. J Virol 69:882–888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jowett JB, Planelles V, Poon B, Shah NP, Chen ML, Chen IS. 1995. The human immunodeficiency virus type 1 vpr gene arrests infected T cells in the G2 + M phase of the cell cycle. J Virol 69:6304–6313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacquot G, Le Rouzic E, David A, Mazzolini J, Bouchet J, Bouaziz S, Niedergang F, Pancino G, Benichou S. 2007. Localization of HIV-1 Vpr to the nuclear envelope: impact on Vpr functions and virus replication in macrophages. Retrovirology 4:84. doi: 10.1186/1742-4690-4-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.DeHart JL, Zimmerman ES, Ardon O, Monteiro-Filho CM, Argañaraz ER, Planelles V. 2007. HIV-1 Vpr activates the G2 checkpoint through manipulation of the ubiquitin proteasome system. Virol J 4:57. doi: 10.1186/1743-422X-4-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maudet C, Bertrand M, Le Rouzic E, Lahouassa H, Ayinde D, Nisole S, Goujon C, Cimarelli A, Margottin-Goguet F, Transy C. 2011. Molecular insight into how HIV-1 Vpr protein impairs cell growth through two genetically distinct pathways. J Biol Chem 286:23742–23752. doi: 10.1074/jbc.M111.220780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Goujon C, Malim MH. 2010. Characterization of the alpha interferon-induced postentry block to HIV-1 infection in primary human macrophages and T cells. J Virol 84:9254–9266. doi: 10.1128/JVI.00854-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cheney KM, McKnight A. 2010. Interferon-alpha mediates restriction of human immunodeficiency virus type-1 replication in primary human macrophages at an early stage of replication. PLoS One 5:e13521. doi: 10.1371/journal.pone.0013521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Doyle T, Goujon C, Malim MH. 2015. HIV-1 and interferons: who’s interfering with whom? Nat Rev Microbiol 13:403–413. doi: 10.1038/nrmicro3449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Malim MH, Bieniasz PD. 2012. HIV restriction factors and mechanisms of evasion. Cold Spring Harb Perspect Med 2:a006940. doi: 10.1101/cshperspect.a006940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hoshino K, Takeuchi O, Kawai T, Sanjo H, Ogawa T, Takeda Y, Takeda K, Akira S. 1999. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J Immunol 162:3749–3752. [PubMed] [Google Scholar]
- 46.Marechal A, Li JM, Ji XY, Wu CS, Yazinski SA, Nguyen HD, Liu S, Jimenez AE, Jin J, Zou L. 2014. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol Cell 53:235–246. doi: 10.1016/j.molcel.2013.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brandariz-Nunez A, Valle-Casuso JC, White TE, Laguette N, Benkirane M, Brojatsch J, Diaz-Griffero F. 2012. Role of SAMHD1 nuclear localization in restriction of HIV-1 and SIVmac. Retrovirology 9:49. doi: 10.1186/1742-4690-9-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.McLaren PJ, Gawanbacht A, Pyndiah N, Krapp C, Hotter D, Kluge SF, Gotz N, Heilmann J, Mack K, Sauter D, Thompson D, Perreaud J, Rausell A, Munoz M, Ciuffi A, Kirchhoff F, Telenti A. 2015. Identification of potential HIV restriction factors by combining evolutionary genomic signatures with functional analyses. Retrovirology 12:41. doi: 10.1186/s12977-015-0165-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Murrell B, Vollbrecht T, Guatelli J, Wertheim JO. 2016. The evolutionary histories of antiretroviral proteins SERINC3 and SERINC5 do not support an evolutionary arms race in primates. J Virol 90:8085–8089. doi: 10.1128/JVI.00972-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kluge SF, Sauter D, Kirchhoff F. 2015. SnapShot: antiviral restriction factors. Cell 163:774–774 e771. doi: 10.1016/j.cell.2015.10.019. [DOI] [PubMed] [Google Scholar]
- 51.McKnight A, Clapham PR, Weiss RA. 1994. HIV-2 and SIV infection of nonprimate cell lines expressing human CD4: restrictions to replication at distinct stages. Virology 201:8–18. doi: 10.1006/viro.1994.1260. [DOI] [PubMed] [Google Scholar]
- 52.Yang Z. 2007. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol 24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 53.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SD. 2006. GARD: a genetic algorithm for recombination detection. Bioinformatics 22:3096–3098. doi: 10.1093/bioinformatics/btl474. [DOI] [PubMed] [Google Scholar]
- 54.Pond SL, Frost SD. 2005. DataMonkey: rapid detection of selective pressure on individual sites of codon alignments. Bioinformatics 21:2531–2533. doi: 10.1093/bioinformatics/bti320. [DOI] [PubMed] [Google Scholar]
- 55.Koenig S, Gendelman HE, Orenstein JM, Dal Canto MC, Pezeshkpour GH, Yungbluth M, Janotta F, Aksamit A, Martin MA, Fauci AS. 1986. Detection of AIDS virus in macrophages in brain tissue from AIDS patients with encephalopathy. Science 233:1089–1093. doi: 10.1126/science.301903. [DOI] [PubMed] [Google Scholar]
- 56.Joseph SB, Arrildt KT, Sturdevant CB, Swanstrom R. 2015. HIV-1 target cells in the CNS. J Neurovirol 21:276–289. doi: 10.1007/s13365-014-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bobadilla S, Sunseri N, Landau NR. 2013. Efficient transduction of myeloid cells by an HIV-1-derived lentiviral vector that packages the Vpx accessory protein. Gene Ther 20:514–520. doi: 10.1038/gt.2012.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Barrero CA, Datta PK, Sen S, Deshmane S, Amini S, Khalili K, Merali S. 2013. HIV-1 Vpr modulates macrophage metabolic pathways: a SILAC-based quantitative analysis. PLoS One 8:e68376. doi: 10.1371/journal.pone.0068376. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All RNA microarray data are available in the Gene Expression Omnibus (GEO) database with accession number GSE54455.