There are limited data about the proviral landscape in children exhibiting long-term suppression after early treatment, particularly in Sub-Saharan Africa where HIV-1 subtype C predominates. Investigating the sequence-intact reservoir could provide insight on the mechanisms by which intact proviruses persist and inform ongoing cure efforts. Through nearly full-length proviral amplification and sequencing (NFL-PAS), we generated 733 NFL-PAS amplicons from eight children. We showed that rare, genetically intact proviruses could be detected in children who initiated ART after 2.3 months of age. The frequency of intact proviruses was lower (P < 0.05) than that reported for HIV subtype B-infected adults treated during early HIV infection. We show that cells harboring genetically intact HIV proviruses are rare in children exhibiting long-term suppression after early treatment and may require the processing of a large number of cells to assess reservoir size. This points to the need for efficient methods to accurately quantify latent reservoirs, particularly in pediatric studies where sample availability is limited.
KEYWORDS: HIV early ART children, HIV full-length sequencing, HIV intact provirus, HIV persistence, HIV persistence children, HIV proviral landscape, HIV reservoir, HIV reservoir decay, HIV cure, human immunodeficiency virus
ABSTRACT
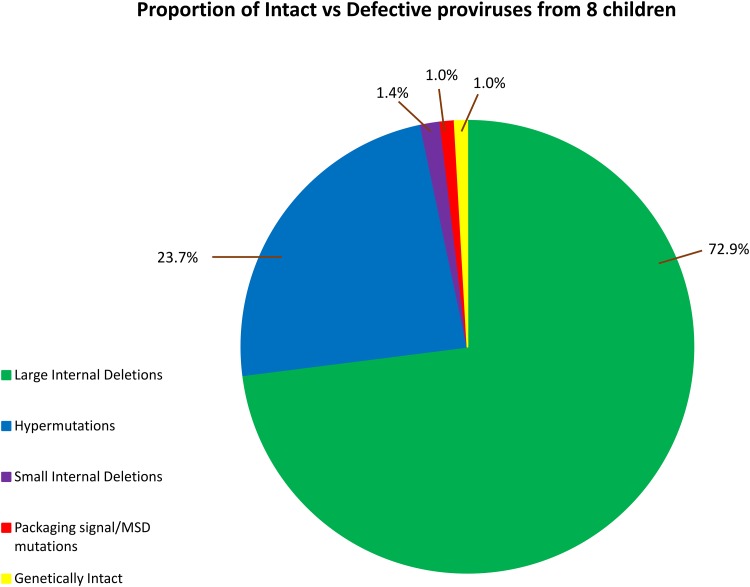
In adults starting antiretroviral therapy (ART) during acute infection, 2% of proviruses that persist on ART are genetically intact by sequence analysis. In contrast, a recent report in children treated early failed to detect sequence-intact proviruses. In another cohort of children treated early, we sought to detect and characterize proviral sequences after 6 to 9 years on suppressive ART. Peripheral blood mononuclear cells (PBMC) from perinatally infected children from the Children with HIV Early antiRetroviral (CHER) study were analyzed. Nearly full-length proviral amplification and sequencing (NFL-PAS) were performed at one time point after 6 to 9 years on ART. Amplicons with large internal deletions were excluded (<9 kb). All amplicons of ≥9 kb were sequenced and analyzed through a bioinformatic pipeline to detect indels, frameshifts, or hypermutations that would render them defective. In eight children who started ART at a median age of 5.4 months (range, 2.0 to 11.1 months), 733 single NFL-PAS amplicons were generated. Of these, 534 (72.9%) had large internal deletions, 174 (23.7%) had hypermutations, 15 (1.4%) had small internal deletions, 3 (1.0%) had deletions in the packaging signal/major splice donor site, and 7 (1.0%) were sequence intact. These 7 intact sequences were from three children who initiated ART after 2.3 months of age, one of whom had two identical intact sequences, suggestive of a cell clone harboring a replication-competent provirus. No intact proviruses were detected in four children who initiated ART before 2.3 months of age. Rare, intact proviruses can be detected in children who initiate ART after 2.3 months of age and are probably, as in adults, maintained by clonal expansion of cells infected before ART initiation.
IMPORTANCE There are limited data about the proviral landscape in children exhibiting long-term suppression after early treatment, particularly in Sub-Saharan Africa where HIV-1 subtype C predominates. Investigating the sequence-intact reservoir could provide insight on the mechanisms by which intact proviruses persist and inform ongoing cure efforts. Through nearly full-length proviral amplification and sequencing (NFL-PAS), we generated 733 NFL-PAS amplicons from eight children. We showed that rare, genetically intact proviruses could be detected in children who initiated ART after 2.3 months of age. The frequency of intact proviruses was lower (P < 0.05) than that reported for HIV subtype B-infected adults treated during early HIV infection. We show that cells harboring genetically intact HIV proviruses are rare in children exhibiting long-term suppression after early treatment and may require the processing of a large number of cells to assess reservoir size. This points to the need for efficient methods to accurately quantify latent reservoirs, particularly in pediatric studies where sample availability is limited.
INTRODUCTION
In human immunodeficiency virus (HIV)-infected individuals on long-term suppressive antiretroviral therapy (ART), HIV-1 reservoirs persist, comprised of long-lived, latent cells with replication-competent proviruses (1). These reservoirs remain hidden from the immune response but reactivate to produce new cycles of replication when therapy is interrupted (2). Reservoir establishment occurs as early as 3 days postinfection, as evident from studies in nonhuman primates showing high levels of integrated simian immunodeficiency virus (SIV) DNA in resting CD4 T cells (3). Nevertheless, initiating ART during acute infection has many benefits: it limits the establishment of reservoirs in long-lived CD4 T cell subsets, preserves anti-HIV T cell responses, and reverses chronic immune activation (4, 5). Moreover, posttreatment control is more frequent after early treatment (6).
HIV-1 DNA decays faster in individuals who start ART in acute infection than in those initiating treatment in chronic infection (7). In adults, longitudinal quantitative PCR (qPCR) measurements of proviral DNA after ART initiation reveals rapid 80 to 90% HIV-1 DNA decay in the first year on ART, likely due to the death of productively infected cells, decay of unintegrated virus, and concurrent blocking of new infections by ART (7, 8). In the second phase, years 1 to 4, there is an annual decline of 23% which represents the decay of cells with a longer half-life (7). The eventual plateau after 4 years on suppressive ART represents long-lived HIV-1-infected cells (8). In perinatally infected children starting ART within days of birth, the decay rate of HIV-1 DNA was much more rapid, with 27% of children who initiated ART within 8 days of birth reaching undetectable levels between 6 days and 4 months after the start of therapy (9). Despite this, there has been viral rebound following treatment discontinuation in almost all perinatally infected children, suggesting that replication-competent HIV proviruses are established early and persist (10).
In adults, the majority of proviral DNA during long-term ART is defective, with these defects accumulating within weeks of infection (11). In patients who initiated ART during chronic infection, 92% of proviral DNA was defective in contrast to 98% in those treated during acute infection (11). Nearly full-length proviral sequencing revealed large internal deletions and/or nonsense mutations due to template switching during reverse transcription, frameshifts, and guanine-to-adenine (G-to-A) hypermutations caused by cytidine deaminases APOBEC3F and APOBEC3G (11–13).
In contrast, in a cohort of 11 children from observational cohorts in the United States who exhibited long-term suppression after early treatment, no intact proviral sequences were detected from 162 nearly full-length sequences (14). This finding led us to investigate further whether sequence-intact proviruses could be detected in nine children from the Children with HIV Early antiRetroviral (CHER) trial and follow-up cohort in South Africa who exhibited long-term suppression after early treatment.
RESULTS
Obtaining intact proviral genomes.
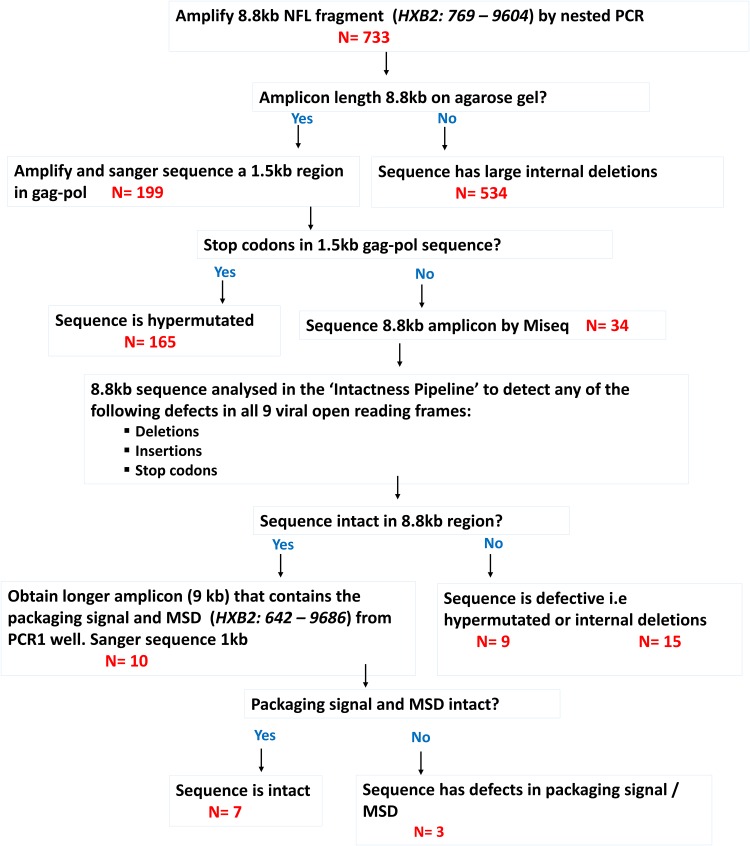
A total of 733 single-genome amplicons were obtained by NFL-PAS. Of these, 534 had large internal deletions. Of the remaining 199 amplicons, a 1.2-kb region of gag-pol was amplified and sequenced. A total of 165 amplicons were shown to be hypermutated, characterized by stop codons. Following bioinformatic analysis, 10 nearly full-length amplicons appeared to be intact, 9 were hypermutated, and 15 had small internal deletions. Although the NFL-PAS protocol had the benefit of using primers that bind in conserved regions of the HIV genome, the 8.8-kb NFL-PAS product excluded the first 793 nucleotides (nt) of the viral genome (including gag-3′ long terminal repeat [LTR]; nt 794 to 9857, HXB2 positions). A second nested PCR included the viral packaging signal (psi) and major splice donor (MSD) site (gag; nt 634 to 789, HXB2 positions). Deletions, mutations, or deleterious stop codons in these regions destroy viral infectivity (15). In the literature, deletions ranging from 8 to 98 bp have been reported in 5 to 11% of sequences (11, 15, 16). To determine whether the psi and MSD site were intact in the 10 samples, the viral packaging signal and major splice donor site were amplified and sequenced. Of these, three sequences were characterized by mutations in the psi and/or MSD region. Finally, seven sequences were shown to be genetically intact (Fig. 1, Fig. 2, and Table 1).
FIG 1.
Schematic for obtaining intact proviral genomes.
FIG 2.
Of 733 amplicons from eight children, 72.9% had large internal deletions, and 23.7% had hypermutations that rendered them defective. As the packaging signal/MSD mutations (gag leader) were assessed only in 10 cases that were provisionally intact, this proportion (1.0%) and the proportion of sequences with small internal deletions (1.4%) were adjusted using the expected rate as found in those that had a gag leader sequence. Only 1% were genetically intact.
TABLE 1.
NFL sequences per participanta
| Patient no. | Genderb | Age at ART start (mo) | HIV DNA (copies/106 PBMCs) | No. of sequences |
Total no. of single genomes | Total no. of cells assayed (×106) | |||
|---|---|---|---|---|---|---|---|---|---|
| With internal deletions | With hypermutations and stop codons | With packaging signal/MSD defects | Intact | ||||||
| ZA-009 | F | 2.0 | 186.2 | 68 | 19 | 0 | 0 | 87 | 9.2 |
| ZA-014 | M | 2.16 | 81.9 | 30 | 4 | 0 | 0 | 34 | 2.6 |
| ZA-008 | F | 2.2 | 42.3 | 89 | 101 | 0 | 0 | 190 | 6.1 |
| ZA-003 | F | 2.3 | 129.6 | 18 | 3 | 0 | 0 | 21 | 3.9 |
| ZA-015 | M | 8.5 | 247.6 | 27 | 9 | 1 | 2 | 39 | 3.9 |
| ZA-011 | F | 9.23 | 181.5 | 207 | 23 | 2 | 1 | 233 | 9.6 |
| ZA-006 | F | 9.32 | 46.7 | 56 | 15 | 0 | 4 | 75 | 6.6 |
| ZA-016 | F | 11.1 | 86.3 | 54 | 0 | 0 | 0 | 54 | 1.8 |
| Total | 5.4c | 108c | 549 | 174 | 3 | 7 | 733 | 43.7 | |
Participants had a median of 108 copies/106 PBMCs (range, 42.3 to 247.6 copies/106 PBMCs). A total of 43.7 × 106 cells were assayed. Seven intact nearly full-length sequences were detected in three of eight participants. All three participants were on continuous ART.
F, female; M, male.
Median value.
A quantitative comparison, using Fisher’s exact tests, of large internal deletions, hypermutants, psi/MSD deletions, and intact sequences from NFL data of adults with subtype B infection who started ART in acute infection revealed significantly fewer psi/MSD deletions (5% for adults versus 1% for children) and intact sequences (2% for adults versus 1% for children) (P < 0.01) (11).
Intact sequences form part of expanded suspected clones.
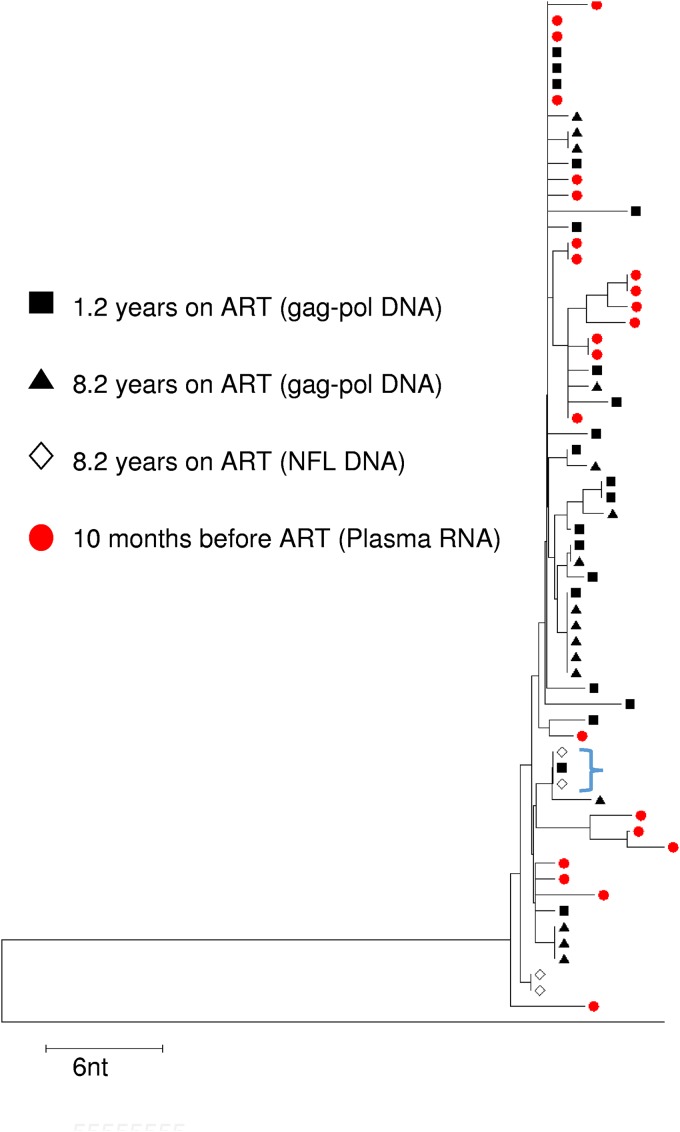
In one participant (ZA-006), two sets of intact NFL sequences were identical in the 9-kb region. When these sequences were assembled in a neighbor-joining phylogenetic tree (Fig. 3) with 1.2-kb gag-pol sequences from pretherapy plasma RNA, peripheral blood mononuclear cell (PBMC) DNA from a participant on ART for 1.2 years, and cell-associated DNA sequences from a participant on ART for 8 years, two of the intact NFL sequences formed part of a monotypic cluster (blue bracket) with a gag-pol DNA sequence from a participant on ART for 1.2 years, suggesting the persistence of a possible intact clone that was present soon after ART was initiated. Intact sequences from participant ZA-015 and ZA-011 were not identical in the 1.2-kb gag-pol region.
FIG 3.
Neighbor joining phylogenetic tree for patient ZA-006.
DISCUSSION
In this study, we detected genetically intact HIV-1 proviral sequences after 6 to 9 years of ART in children who initiated therapy with the first year of life. Our aim was to investigate proviral intactness, and we classified sequences as genetically intact when we found none of the known defects upon sequencing. Nevertheless, this does not prove replication competence, which would require identifying the integration sites, amplifying full-length HIV from the host, cloning, and transfection with production of infectious virus and which should be investigated in future studies.
In one patient, we observed two sets of identical, intact nearly full-length sequences. When aligned with gag-pol sequences obtained 1.2 years after ART initiation, two of the intact sequences formed a monotypic cluster, suggesting the persistence of an intact expanded clone. Although rare, genetically intact proviruses can be detected after several years of ART in children who were treated early, these proviruses are probably, as in adults, maintained by clonal expansion/proliferation of cells infected before ART initiation (17, 18).
A recent study found no intact HIV proviral sequences out of 164 nearly full-length sequences derived from viral outgrowth culture wells in 11 sequences obtained from children exhibiting long-term suppression who initiated ART before 6 months of age (14). The detection of intact proviruses in our study could be due to more extensive sampling. We generated 733 single-genome amplicons and found 7 (1.0%) to be intact. Whereas in the previous study proviruses were sequenced from noninduced viral outgrowth wells, we performed NFL-PAS directly on DNA from PBMC samples. On the other hand, the different findings between the two cohorts could be due to differences in the ages at the time of ART initiation. In our study we detected intact sequences from children assigned to the “deferred therapy” CHER study arm 1 who initiated ART after 8 months of age and who thus likely had a larger proportion of intact sequences; in contrast in the previous study, all participants initiated ART within 6 months of age and had no detectable intact sequences. Furthermore, the difference in viral genotypes between the two cohorts could have played a role. In our setting, HIV-1 subtype C predominated whereas the previous cohort was infected with subtype B, which supports the need for representation of pediatric cohorts across different geographical regions as differences in viral genotypes and host factors may affect HIV persistence in children.
In one child, two identical intact sequences obtained after 8 years on ART formed a monotypic cluster with a 1.2-kb gag-pol DNA sequence obtained 14.4 months after ART initiation. This suggests that clonal expansion of intact proviruses could be the mechanism that maintains the replication-competent reservoir in these children. However, one would need to perform integration site analysis in order to prove clonality as NFL-PAS sequences can be identical in gag-pol but differ elsewhere, likely due to insufficient signal in the 1.2-kb region. One such technique involving whole-genome amplification, nearly full-length sequencing, and corresponding chromosomal integration site analysis has recently been described (19). Similar identical sequences were observed in adults treated during acute infection, and it has been shown that clonal, intact proviral sequences are a source of infectious virus in viral outgrowth assays (16–18). These clonally expanded cells are a major barrier to HIV eradication and can become activated and proliferate without viral antigen expression, thus remaining hidden from the immune system or immune-based therapies (20).
We found that the majority (99%) of HIV-1 proviruses were defective either due to large internal deletions or hypermutation that led to stop codons, similar to what has been reported in both children and adults (11, 14, 16). The large internal deletions are a result of errors during reverse transcription while G-to-A hypermutations are induced by the host cell’s cytidine deaminases APOBEC3F and APOBEC3G (12, 13). The proportion of intact proviruses in our cohort (1%) was closer to the 2% reported in subtype B-infected adults who initiated ART during acute infection but distinct from the 12% reported in adults who initiated ART during chronic infection (11, 15). However, it is not known if the children in our study were infected intrauterine or perinatally.
Our cohort consisted of three children who were fully suppressed on ART and five children who either had periods of ART interruption (arm 2 and arm 3) or were poorly suppressed for a brief period. All 7 intact sequences were from three children who were part of the deferred-ART arm 1 of the CHER trial and therefore began ART when CD4 levels were <25%, an indication of disease progression. These children, however, remained fully suppressed on ART following commencement although two individuals had delayed initial HIV viral load suppression. In contrast, children without detectable intact proviruses started ART before 2.3 months but had subsequent periods of poor suppression or therapy interruption. A recent study showed that the proportion of intact versus defective proviruses remained the same before and after brief ART interruptions of about 3 months (21). In our study, some patients’ therapy was interrupted for up to 1 year, yet these participants did not have detectable intact proviruses. These findings corroborate previous studies showing that the largest proportion of the long-surviving reservoir is established before therapy initiation, with limited replenishment during periods of therapy failure or interruption (7, 21, 22). Early treatment initiation may therefore be the most important factor for reducing the reservoir size, whereas short, monitored periods of interruption, necessary to assess the effect of curative interventions, may not have long-term effects on reservoirs.
In children in whom no intact proviruses have been detected by NFL-PAS, intact proviruses may nevertheless persist. During the CHER trial, all but one child whose therapy was interrupted (arm 2 and arm 3) eventually had rebound viremia regardless of how early ART was initiated, providing evidence that infectious virus was present (23). The one participant with posttherapy control or remission initiated ART at 2 months, interrupted therapy after 40 weeks, and has maintained viral suppression for 8.5 years (23). This child was recruited at a different site and therefore not included in our study. It is not known if intact proviruses are present in this child or if control is due to unique immune responses. Another French child from the ANRS EPF-CO10 pediatric cohort received a 6-week course of zidovudine at birth and initiated ART at 3 months of age; ART was discontinued after 5 years, and the child has maintained virologic control for over 11 years (24). The mechanisms of long-term viral remission after early initiation of ART in infants are unknown and warrant further investigation to understand which host immune factors play a role. In contrast, infectious virus could be recovered from viral outgrowth assays on samples from children up to 2 years of age who started ART at a median age of 8.1 weeks (25). In children who started ART between 0.1 and 2.6 months of age, infectious virus was not recoverable, and DNA was not detectable, suggesting a smaller reservoir (26). However, this does not constitute a cure. Mathematical models and studies in adult cohorts suggest that a very small reservoir may be associated with delayed viral rebound (27–29). In the case of the Mississippi baby who initiated therapy within 30 h of birth, virus rebounded 27 months after therapy cessation (10). In contrast, in another infant who received immediate antiretroviral therapy and continued ART for 4 years, virus rebounded within 7 days of therapy interruption (30). This shows that the reservoir is seeded very early during infection and persists over long periods on ART, despite being undetectable by culture and molecular methods.
Due to limited sample availability, we could not perform viral outgrowth assays to prove that intact proviruses were infectious. Although the quantitative viral outgrowth assay (qVOA) is considered the gold standard for detecting replication-competent viruses, not all intact proviruses are reactivated in cell culture, and it has recently been reported that qVOA underestimates reservoir size by 25-fold in patients treated during acute infection and by 27-fold in chronic-phase-treated patients (11). Therefore, molecular assays, such as those used in this study, provide a sensitive estimate of the reservoir and are even more appropriate for pediatric studies for which sample availability is often limited. There is, however, a need for more efficient methods that can generate nearly full-length amplicons and differentiate between intact and defective proviruses. A recent study highlights one such approach in which a quantitative PCR assay utilizes multiple probes (based on the known distribution of defects in patients receiving long-term treatment) to simultaneously differentiate between intact and defective proviruses (31).
We studied eight children who had sufficient samples and a sufficiently high proviral load. However, as is the case with most pediatric studies, there was limited sample availability, and relatively small amounts of PBMCs (<10 million cells) were tested in each individual. Therefore, the absence of detected sequence-intact nearly full-length proviruses in five participants does not exclude their presence. Furthermore, our findings may not be fully representative of reservoirs in subtype C-infected children exhibiting long-term suppression. In addition, the NFL-PAS assay is cumbersome and not feasible in children with ultralow proviral loads, such as in infants treated very early (9).
This study adds to the limited data on children treated early, especially those with subtype C infection. The larger number of amplicons generated than in prior studies may have increased the chance of finding intact sequences. Although rare, intact proviruses were detectable after 6 to 9 years in subtype C-infected children treated early. As seen in adults, these proviruses are probably maintained by clonal expansion/proliferation of cells infected before ART initiation. Future work could include linking intact proviral sequences to CD4 T cell clones and to plasma virus that rebounds after therapy cessation or due to latency-reversing agents. There is also a need for more efficient sequencing of full-length proviruses, possibly involving third-generation sequencing platforms (32).
MATERIALS AND METHODS
Study participants.
During the CHER trial, HIV-infected infants who were asymptomatic and had baseline CD4 percentages of ≥25% were randomized to deferred ART (arm 1), according to concurrent guidelines, or to immediate time-limited ART for either 40 (arm 2) or 96 (arm 3) weeks. ART was initiated in all three arms for CD4 percentages of <25% or Centers for Disease Control stage C or severe protocol-defined stage B disease (33). The participants have been enrolled since December 2014 in an ongoing follow-up study at the Family Centre for Research with Ubuntu (FAM-CRU) in Tygerberg Academic Hospital, Cape Town, South Africa. Peripheral blood was collected from eight perinatally HIV-infected children. The eight participants were selected on the basis of having sufficient sample material and HIV cell-associated DNA loads higher than 40 copies/106 PBMCs at the 6- to 7-year sampling to increase the likelihood of obtaining nearly full-length amplicons. The median age of ART initiation was 5.4 months (range, 2.0 to 11.1 months) (Table 2). Two children had periods when ART was interrupted as per the CHER trial. Periods of interruption ranged from 4 months to 1 year. One child was poorly suppressed for the first 2.5 years on ART, probably due to poor adherence. The remaining four participants were fully suppressed on ART. At the time of testing, participants had been on ART for a median of 8.1 years (range, 6.9 to 9.1 years), and the median cell-associated HIV DNA load was 108 copies/106 PBMCs (range, 42.3 to 247.6 copies/106 PBMCs). All eight participants maintained undetectable viral loads within the most recent 36 months of samples studied.
TABLE 2.
Participant characteristicsa
| Participant no. | Gender | CHER study arm | Age at ART start (mo) | ART treatment history | Longitudinal viral load history | Duration of suppression up to 6- to 7-yr sample (yr) | Duration of ART at sample (yr) | HIV cell-associated DNA (copies/106 PBMCs) |
|---|---|---|---|---|---|---|---|---|
| ZA-009 | F | Arm 2 | 2.0 | Continuous | Viral blips for first 6 mo on ART; has subsequently maintained undetectable viral load | 5.4 | 9.1 | 186.2 |
| ZA-014 | M | Arm 2 | 2.2 | Interrupted after 9.9 mo; reinitiated after 13 mo | Viremic for 1 yr following interruption; viral load undetectable 3 mo after ART reinitiation; has subsequently maintained undetectable viral load | 4.8 | 6.9 | 81.9 |
| ZA-008 | F | Arm 3 | 2.2 | Continuous | Viral load suppressed for 2 yr on ART; viremic after 2.2 yr on ART; viral load undetectable 5 mo later; has subsequently maintained undetectable viral load | 4.2 | 7.0 | 42.3 |
| ZA-003 | F | Arm 3 | 2.3 | Interrupted after 2 yr; reinitiated after 4 mo | Viremic 4 mo after ART interruption; viral load undetectable 5 mo after ART reinitiation; has subsequently maintained undetectable viral load | 8.1 | 8.6 | 129.6 |
| ZA-015 | M | Arm 1 | 8.5 | Continuous | Has maintained undetectable viral load | 6.5 | 7.9 | 247.6 |
| ZA-011 | F | Arm 1 | 9.2 | Continuous | Has maintained undetectable viral load | 5.1 | 7.3 | 181.5 |
| ZA-006 | F | Arm 1 | 9.3 | Continuous | Has maintained undetectable viral load | 7.0 | 8.2 | 46.7 |
| ZA-016 | F | Arm 1 | 11.1 | Continuous | Poorly suppressed for first 2.5 yr on ART; has subsequently maintained undetectable viral load | 4.9 | 8.8 | 86.3 |
| Median value | 5.4 | 5.3 | 8.1 | 108.0 |
With the exception of participant ZA-009, ART for all participants consisted of zidovudine/lamivudine/lopinavir-ritonavir. After 7.7 years, the ART for participant ZA-009 consisted of zidovudine/lamivudine/efavirenz.
Ethical considerations.
Ethical approval was obtained from the Human Research Ethics Committee of Stellenbosch University (ethics approvals N13/04/046 and M14/07/029). Written informed consent was obtained from the parents or legal guardians on behalf of the study participants.
Plasma viral load quantification.
Plasma viral loads were quantified using a Roche Amplicor HIV Monitor assay, version 1.0 (Roche Diagnostics), or an Abbott Diagnostics Real Time HIV-1 assay (Abbott Laboratories).
Total cell-associated DNA quantification.
Peripheral blood mononuclear cells (PBMCs) were isolated from whole blood by Ficoll density gradient centrifugation. Total HIV-1 cell-associated DNA was quantified using a real-time PCR assay targeting a conserved region of the pol gene as previously described (34). The number of cells assayed in each sample was determined by quantifying the ccr5 gene.
NFL-PAS.
Genomic DNA was extracted from 1.25 × 106 PBMCs as previously described, but the method was modified by omitting template fragmentation by sonification in order to preserve the integrity of longer fragments (34). Genomic DNA was diluted to a proviral endpoint, and nearly full-length single-genome amplification and sequencing (NFL-PAS) were performed with Ranger mix (Bioline) and the following previously described primers, with minor modifications for HIV-1 subtype C: prenested PCR Li_OuterF (5′-AAATCTCTAGCAGTGGCGCCCGAACAG-3′); Li_OuterR (5′-TGAGGGATCTCTAGTTACCAGAGTC-3′); nested PCR Li_InnerF (5′-GCGGAGGCTAGAAGGAGAGAGATGG-3′); Li_InnerR (5′-GCACTCAAGGCAAGCTTTATTGAGGCTTA-3′) (35). For prenested PCR, 2 μl of extracted DNA was added to 10 μM forward and reverse primers, 1× Ranger mix, and nuclease-free water up to a final volume of 10 μl. Cycling conditions were 95°C for 2 min and 30 cycles at 98°C for 10 s, 61°C for 10 min, and 72°C for 10 min, followed by a hold at 4°C. Prenested PCR products were diluted by adding 80 μl of 5 mM Tris-HCl. For the nested PCR, 2 μl of diluted DNA was added to 10 μM forward and reverse primers, 1× Ranger mix, and nuclease-free water up to a final volume of 10 μl. Cycling conditions were 95°C for 2 min and 30 cycles at 98°C for 10 s, 65.5°C for 10 min, and 72°C for 10 min, followed by a hold at 4°C.
First, the assay was validated to determine the effect of background cellular DNA on detecting low-copy-number targets using the HIV-1 LAV-infected 8E5 T cell line that contains one integrated copy of HIV per cell. DNA from 8E5 cells was spiked into various amounts of HIV-negative DNA to mimic patient samples where there is a low HIV-1 cell-associated DNA load in a high background of cellular genomic DNA. Next, for each patient, single-genome amplicons were obtained at a dilution, and <30% of PCR replicates were positive.
PCRs on 96-well plates were stained with EZ vison blue light DNA dye (Amresco, PA) and viewed under UV light. All fluorescing wells were run on a 1% sodium borate (SB; boric acid, sodium hydroxide, and distilled water) agarose gel to assess amplicon size. Samples that were 8.8 kb were selected for further analysis while shorter fragments were considered to have large internal deletions.
Selection of 8.8-kb fragments for MiSeq.
To detect sequences with a G-to-A hypermutation and exclude products in which more than one template was present in the reaction mixture, a 1.5-kb region spanning P6, protease, and the first 900 nt of reverse transcriptase (p6-PR-RT; nt 1893 to 3408, HXB2 positions) was sequenced from each 8.8-kb amplicon as previously described (36). Sequences were aligned using ClustalW and assessed for evidence of hypermutation in Hypermut (https://www.hiv.lanl.gov/content/sequence/HYPERMUT/background.html). Sequences without evidence of G-to-A hypermutation were selected for next-generation sequencing by Illumina MiSeq (Illumina, CA). These samples were gel extracted from a 1% SB agarose gel using a NucleoTrap gel extraction kit (Macherey-Nagel, Germany) according to the manufacturer’s instructions.
MiSeq library preparation.
MiSeq library preparation and sequencing were performed at the Institute for Microbial Biotechnology and Metagenomics (IMBM) at the University of Western Cape. An Illumina Nextera XT DNA library prep kit (Illumina, CA) was used. In a single reaction, DNA was enzymatically fragmented, and adapters were added to the template. The DNA was then purified and amplified by a PCR that indexed the samples by adding different primer pairs to individual samples. A 300-cycle MiSeq reagent kit, version 2 (Illumina, CA), was used to sequence the library. A 10% PhiX spike was included for a low-diversity library as recommended by the manufacturer.
Bioinformatic analysis of MiSeq data.
All reads with the same index were assembled to form a consensus sequence. Next, sequences of < 8.8 kb or samples with mixed templates were excluded from further analysis. The remaining sequences were translated and analyzed in the nine viral open reading frames (ORFs). A sequence was determined to be intact if within these ORFs, there were no stop codons, frameshift mutations, or hypermutations that could preclude viral infectivity.
Amplification and sequencing of the HIV packaging signal and major splice donor site.
A forward primer was designed to bind upstream of the NFL product (gag Pr55; nt 642, HXB2 position). For each limiting dilution that yielded an 8.8-kb intact sequence, prenested PCR product was added to a nested PCR, using the newly designed forward primer NFL_alt_FW (5′-CCGAACAGGGACBHGAAAGCGAA-3′) and the prenested reverse primer Li_Outer_R (5′-TGAGGGATCTCTAGTTACCAGAGTC-3′) to generate an amplicon that included the gag leader region. For each reaction mixture, 5 μl of DNA was added to 10 μM forward and reverse primers, 1× Ranger mix, and nuclease-free water up to a final volume of 25 μl. Cycling conditions were 95°C for 2 min and 30 cycles at 98°C for 10 s, 61.5°C for 10 min, and 72°C for 10 min, followed by a hold at 4°C. The product was 9,044 bp in length (gag Pr55-3′ LTR; nt 642 to 9686, HXB2 position). The 9,044-bp fragment was gel extracted from 0.8% Tris-acetate EDTA (TAE) buffer using a NucleoTrap gel extraction kit (Macherey-Nagel, Germany). A 1-kb region containing the packaging signal (psi) and major splice donor (MSD) site was sequenced by automated Sanger sequencing (ABI 3730xl; Applied Biosystems). The 1-kb length ensured a 750-bp overlap of the region that was previously sequenced by MiSeq and confirmed that the same variant was sequenced. All sequences that were determined to be intact contained an intact packaging signal (psi) and Rev-responsive element (RRE).
Data availability.
Accession numbers for sequences obtained in this study have been deposited in GenBank under accession numbers MN611462 to MN611468.
ACKNOWLEDGMENTS
We thank the participants and caregivers for their willingness to take part in this study as well as the clinic staff for their assistance.
This study was supported by grants from the National Cancer Institute (1U01CA200441-01), the National Institute of Mental Health (1R01MH105134-01), the Polio Research Foundation, Leidos Biomedical Research Corporation (HHSN261200800001E), the National Institute of Allergy and Infectious Diseases (NIAID; 5UM1AI126603, Martin Delaney Collaboratory), and the South African Medical Research Council Collaborating Centre for HIV Laboratory Research. Support for the CHER study was provided by the U.S. NIAID through the CIPRA network (grant U19AI53217), Departments of Health of the Western Cape and Gauteng, South Africa, and GlaxoSmithKline/Viiv Healthcare.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
G.U.V.Z., M.F.K., M.F.C., J.W.M., and M.G.K. participated in the conception and design of the study. E.K.H. and K.J. developed the NFL-PAS protocol. M.G.K. generated near-full-length sequences and quantified DNA levels. I.W., B.K.-M., M.J.B., W.-S.H., S.E., M.G.K., and W.S. analyzed data. All authors interpreted data. M.G.K. drafted the manuscript, including the figures and tables. All authors revised the manuscript critically for intellectual content and have provided approval for the final submission.
J.W.M. is a consultant for Gilead Sciences, Inc., a shareholder of Cocrystal Pharma, Inc., and has received research support from Gilead Sciences, Inc., and Janssen Pharmaceuticals, Inc. All other authors declare that they have no competing interests.
REFERENCES
- 1.Chun T, Stuyver L, Mizell SB, Ehler LA, Mican JAM, Baseler M, Lloyd AL, Nowak MA, Fauci AS. 1997. Presence of an inducible HIV-1 latent reservoir during highly active antiretroviral therapy. Proc Natl Acad Sci U S A 94:13193–13197. doi: 10.1073/pnas.94.24.13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Huang S, Ren Y, Thomas AS, Chan D, Mueller S, Ward AR, Patel S, Bollard CM, Cruz CR, Karandish S, Truong R, Macedo AB, Bosque A, Kovacs C, Benko E, Piechocka-Trocha A, Wong H, Jeng E, Nixon DF, Ho YC, Siliciano RF, Walker BD, Jones RB. 2018. Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J Clin Invest 128:876–889. doi: 10.1172/JCI97555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Whitney JB, Hill AL, Sanisetty S, Penaloza-MacMaster P, Liu J, Shetty M, Parenteau L, Cabral C, Shields J, Blackmore S, Smith JY, Brinkman AL, Peter LE, Mathew SI, Smith KM, Borducchi EN, Rosenbloom DIS, Lewis MG, Hattersley J, Li B, Hesselgesser J, Geleziunas R, Robb ML, Kim JH, Michael NL, Barouch DH. 2014. Rapid seeding of the viral reservoir prior to SIV viremia in Rhesus monkeys. Nature 512:74–77. doi: 10.1038/nature13594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jain V, Hartogensis W, Bacchetti P, Hunt PW, Hatano H, Sinclair E, Epling L, Lee TH, Busch MP, McCune JM, Pilcher CD, Hecht FM, Deeks SG. 2013. Antiretroviral therapy initiated within 6 months of HIV infection is associated with lower T-cell activation and smaller HIV reservoir size. J Infect Dis 208:1202–1211. doi: 10.1093/infdis/jit311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ananworanich J, Puthanakit T, Suntarattiwong P, Chokephaibulkit K, Kerr SJ, Fromentin R, Bakeman W, Intasan J, Mahanontharit A, Sirivichayakul S, Chomont N, HIV-NAT 194 Study Group. 2013. Reduced markers of HIV persistence and restricted HIV-specific immune responses after early antiretroviral therapy in children. AIDS 28:1–6. doi: 10.1097/QAD.0000000000000178. [DOI] [PubMed] [Google Scholar]
- 6.Sáez-Cirión A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, Potard V, Versmisse P, Melard A, Prazuck T, Descours B, Guergnon J, Viard JP, Boufassa F, Lambotte O, Goujard C, Meyer L, Costagliola D, Venet A, Pancino G, Autran B, Rouzioux C, ANRS VISCONTI Study Group. 2013. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog 9:e1003211. doi: 10.1371/journal.ppat.1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Buzon M, Martin-Gayo E, Pereyra F, Ouyang Z, Sun H, Li JZ, Piovoso M, Shaw A, Dalmau J, Zangger N, Martinez-Picado J, Zurakowski R, Yu XG, Telenti A, Walker BD, Rosenberg ES, Lichterfeld M. 2014. Long-term antiretroviral treatment initiated at primary HIV-1 infection affects the size, composition, and decay kinetics of the reservoir of HIV-1-infected CD4 T cells. J Virol 88:10056–10065. doi: 10.1128/JVI.01046-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Besson GJ, Lalama CM, Bosch RJ, Gandhi RT, Bedison MA, Aga E, Riddler SA, McMahon DK, Hong F, Mellors JW. 2014. HIV-1 DNA decay dynamics in blood during more than a decade of suppressive antiretroviral therapy. Clin Infect Dis 59:1312–1321. doi: 10.1093/cid/ciu585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Veldsman KA, Maritz J, Isaacs S, Katusiime MG, Janse Van Rensburg A, Laughton B, Mellors JW, Cotton MF, van Zyl GU. 2018. Rapid decline of HIV-1 DNA and RNA in infants starting very early antiretroviral therapy may pose a diagnostic challenge. AIDS 32:629–634. doi: 10.1097/QAD.0000000000001739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Luzuriaga K, Gay H, Ziemniak C, Sanborn K, Somasundaran M, Rainwater-Lovett K, Mellors JW, Rosenbloom DIS, Persaud D. 2015. Viremic relapse following prolonged antiretroviral-free HIV-1 remission in a perinatally infected child. N Engl J Med 372:786–788. doi: 10.1056/NEJMc1413931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bruner KM, Murray AJ, Pollack RA, Soliman MG, Laskey SB, Capoferri AA, Lai J, Strain MC, Lada SM, Hoh R, Ho YC, Richman DD, Deeks SG, Siliciano JD, Siliciano RF. 2016. Defective proviruses rapidly accumulate during acute HIV-1 infection. Nat Med 22:1043–1049. doi: 10.1038/nm.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kieffer TL, Kwon P, Nettles RE, Han Y, Ray SC, Siliciano RF. 2005. G-A hypermutation in protease and reverse transcriptase regions of human immunodeficiency virus type 1 residing in resting CD4+ T cells in vivo. J Virol 79:1975–1980. doi: 10.1128/JVI.79.3.1975-1980.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Abram ME, Ferris AL, Shao W, Alvord WG, Hughes SH. 2010. Nature, position, and frequency of mutations made in a single cycle of HIV-1 replication. J Virol 84:9864–9878. doi: 10.1128/JVI.00915-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rainwater-Lovett K, Ziemniak C, Watson D, Luzuriaga K, Siberry G, Petru A, Chen Y, Uprety P, McManus M, Ho YC, Lamers SL, Persaud D. 2017. Paucity of intact non-induced provirus with early, long-term antiretroviral therapy of perinatal HIV infection. PLoS One 12:e0170548. doi: 10.1371/journal.pone.0170548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ho Y-C, Shan L, Hosmane NN, Wang J, Laskey SB, Rosenbloom DIS, Lai J, Blankson JN, Siliciano JD, Siliciano RF. 2013. Replication-competent noninduced proviruses in the latent reservoir increase barrier to HIV-1 cure. Cell 155:540–551. doi: 10.1016/j.cell.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hiener B, Horsburgh BA, Eden J-S, Barton K, Schlub TE, Lee E, von Stockenstrom S, Odevall L, Milush JM, Liegler T, Sinclair E, Hoh R, Boritz EA, Douek D, Fromentin R, Chomont N, Deeks SG, Hecht FM, Palmer S. 2017. Identification of genetically intact HIV-1 proviruses in specific CD4+ T cells from effectively treated participants. Cell Rep 21:813–822. doi: 10.1016/j.celrep.2017.09.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bui JK, Sobolewski MD, Keele BF, Spindler J, Musick A, Wiegand A, Luke BT, Shao W, Hughes SH, Coffin JM, Kearney MF, Mellors JW. 2017. Proviruses with identical sequences comprise a large fraction of the replication-competent HIV reservoir. PLoS Pathog 13:e1006283. doi: 10.1371/journal.ppat.1006283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simonetti FR, Sobolewski MD, Fyne E, Shao W, Spindler J, Hattori J, Anderson EM, Watters SA, Hill S, Wu X, Wells D, Su L, Luke BT, Halvas EK, Besson G, Penrose KJ, Yang Z, Kwan RW, Van Waes C, Uldrick T, Citrin DE, Kovacs J, Polis MA, Rehm CA, Gorelick R, Piatak M, Keele BF, Kearney MF, Coffin JM, Hughes SH, Mellors JW, Maldarelli F. 2016. Clonally expanded CD4+ T cells can produce infectious HIV-1 in vivo. Proc Natl Acad Sci U S A 113:1883–1888. doi: 10.1073/pnas.1522675113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Einkauf KB, Lee GQ, Gao C, Sharaf R, Sun X, Hua S, Chen SM, Jiang C, Lian X, Chowdhury FZ, Rosenberg ES, Chun TW, Li JZ, Yu XG, Lichterfeld M. 2019. Intact HIV-1 proviruses accumulate at distinct chromosomal positions during prolonged antiretroviral therapy. J Clin Invest 129:988–998. doi: 10.1172/JCI124291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang Z, Gurule EE, Brennan TP, Gerold JM, Kwon KJ, Hosmane NN, Kumar MR, Beg SA, Capoferri AA, Ray SC, Ho YC, Hill AL, Siliciano JD, Siliciano RF. 2018. Expanded cellular clones carrying replication-competent HIV-1 persist, wax, and wane. Proc Natl Acad Sci 115:E2575–E2584. doi: 10.1073/pnas.1720665115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarridge KE, Blazkova J, Einkauf K, Petrone M, Refsland EW, Justement JS, Shi V, Huiting ED, Seamon CA, Lee GQ, Yu XG, Moir S, Sneller MC, Lichterfeld M, Chun TW. 2018. Effect of analytical treatment interruption and reinitiation of antiretroviral therapy on HIV reservoirs and immunologic parameters in infected individuals. PLoS Pathog 14:e1006792. doi: 10.1371/journal.ppat.1006792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miller V, Sabin C, Hertogs K, Bloor S, Martinez-Picado J, D'Aquila R, Larder B, Lutz T, Gute P, Weidmann E, Rabenau H, Phillips A, Staszewski S. 2000. Virological and immunological effects of treatment interruptions in HIV-1 infected patients with treatment failure. AIDS 14:2857–2867. doi: 10.1097/00002030-200012220-00007. [DOI] [PubMed] [Google Scholar]
- 23.Violari A, Cotton MF, Kuhn L, Schramm DB, Paximadis M, Loubser S, Shalekoff S, Da Costa Dias B, Otwombe K, Liberty A, McIntyre J, Babiker A, Gibb D, Tiemessen CT. 2019. A child with perinatal HIV infection and long-term sustained virological control following antiretroviral treatment cessation. Nat Commun 10:1–11. doi: 10.1038/s41467-019-08311-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frange P, Faye A, Avettand-Fenoël V, Bellaton E, Descamps D, Angin M, David A, Caillat-Zucman S, Peytavin G, Dollfus C, Le Chenadec J, Warszawski J, Rouzioux C, Sáez-Cirión A, ANRS EPF-CO10 Pediatric Cohort and the ANRS EP47 VISCONTI study group. 2016. HIV-1 virological remission lasting more than 12 years after interruption of early antiretroviral therapy in a perinatally infected teenager enrolled in the French ANRS EPF-CO10 paediatric cohort: a case report. Lancet HIV 3:e49–54. doi: 10.1016/S2352-3018(15)00232-5. [DOI] [PubMed] [Google Scholar]
- 25.Persaud D, Palumbo PE, Ziemniak C, Hughes MD, Alvero CG, Luzuriaga K, Yogev R, Capparelli EV, Chadwick EG. 2012. Dynamics of the resting CD4+ T-cell latent HIV reservoir in infants initiating HAART less than 6 months of age. AIDS 26:1483–1490. doi: 10.1097/QAD.0b013e3283553638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bitnun A, Samson L, Chun T-W, Kakkar F, Brophy J, Murray D, Justement S, Soudeyns H, Ostrowski M, Mujib S, Harrigan PR, Kim J, Sandstrom P, Read SE. 2014. Early initiation of combination antiretroviral therapy in HIV-1 Infected newborns can achieve sustained virologic suppression with low frequency of CD4+ T cells carrying HIV in peripheral blood. Clin Infect Dis 59:1012–1019. doi: 10.1093/cid/ciu432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Steingrover R, Pogány K, Fernandez Garcia E, Jurriaans S, Brinkman K, Schuitemaker H, Miedema F, Lange JM, Prins JM. 2008. HIV-1 viral rebound dynamics after a single treatment interruption depends on time of initiation of highly active antiretroviral therapy. AIDS 22:1583–1588. doi: 10.1097/QAD.0b013e328305bd77. [DOI] [PubMed] [Google Scholar]
- 28.Williams JP, Hurst J, Stöhr W, Robinson N, Brown H, Fisher M, Kinloch S, Cooper D, Schechter M, Tambussi G, Fidler S, Carrington M, Babiker A, Weber J, Koelsch KK, Kelleher AD, Phillips RE, Frater J, SPARTACTrial Investigators. 2014. HIV-1 DNA predicts disease progression and post-treatment virological control. Elife 3:e03821. doi: 10.7554/eLife.03821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hill AL, Rosenbloom DIS, Fu F, Nowak MA, Siliciano RF. 2014. Predicting the outcomes of treatment to eradicate the latent reservoir for HIV-1. Proc Natl Acad Sci U S A 111:13475–13480. doi: 10.1073/pnas.1406663111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Butler KM, Gavin P, Coughlan S, Rochford A, Mc Donagh S, Cunningham O, Poulsom H, Watters SA, Klein N. 2015. Rapid viral rebound after 4 years of suppressive therapy in a seronegative HIV-1 infected infant treated from birth. Pediatr Infect Dis J 34:e48–e51. doi: 10.1097/INF.0000000000000570. [DOI] [PubMed] [Google Scholar]
- 31.Bruner KM, Wang Z, Simonetti FR, Bender A, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, Bertagnolli LN, Capoferri AA, Kufera JT, Timmons A, Nobles C, Gregg J, Wada N, Ho YC, Zhang H, Margolick JB, Blankson JN, Deeks SG, Bushman FD, Siliciano JD, Laird GM, Siliciano RF. 2019. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566:120–125. doi: 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Parikh UM, McCormick K, Van Zyl G, Mellors JW. 2017. Future technologies for monitoring HIV drug resistance and cure. Curr Opin HIV AIDS 12:182–189. doi: 10.1097/COH.0000000000000344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cotton MF, Violari A, Otwombe K, Panchia R, Dobbels E, Rabie H, Josipovic D, Liberty A, Lazarus E, Innes S, van Rensburg AJ, Pelser W, Truter H, Madhi SA, Handelsman E, Jean-Philippe P, McIntyre JA, Gibb DM, Babiker AG, CHER Study Team. 2013. Early time-limited antiretroviral therapy versus deferred therapy in South African infants infected with HIV: results from the children with HIV early antiretroviral (CHER) randomised trial. Lancet 382:1555–1563. doi: 10.1016/S0140-6736(13)61409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hong F, Aga E, Cillo A, Yates AL, Besson G, Fyne E, Koontz DL, Jennings C, Zheng L, Mellors JW. 2016. Novel assays to measure total cell-associated HIV-1 DNA and RNA. J Clin Microbiol 54:902–911. doi: 10.1128/JCM.02904-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salminen MO, Koch C, Sanders-Buell E, Ehrenberg PK, Michael NL, Carr JK, Burke DS, McCutchan FE. 1995. Recovery of virtually full-length HIV-1 provirus of diverse subtypes from primary virus cultures using the polymerase chain reaction. Virology 213:80–86. doi: 10.1006/viro.1995.1548. [DOI] [PubMed] [Google Scholar]
- 36.Palmer S, Kearney M, Maldarelli F, Elias K, Bixby CJ, Bazmi H, Rock D, Falloon J, Davey RT Jr, Dewar RL, Metcalf JA, Hammer S, Mellors JW, Coffin JM. 2005. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J Clin Microbiol 43:403–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Accession numbers for sequences obtained in this study have been deposited in GenBank under accession numbers MN611462 to MN611468.