African swine fever virus (AFSV), a large enveloped double-stranded DNA virus, causes a deadly infection in domestic pigs. In addition to Africa, Europe, and South America, countries in Asia, such as China, Vietnam, and Mongolia, have suffered the hazards posed by ASFV outbreaks in recent years. Until now, there has been no vaccine for protection from ASFV infection or effective treatments to cure ASF. Here, we solved the crystal structure of the ASFV dUTPase-dUMP-Mg2+ complex. The ASFV dUTPase displays a noncanonical folding pattern that differs from that of the classic homotrimeric dUTPase, in which the active site is composed of two subunits. In addition, several nonconserved residues within the 3-fold axis channel play a vital role in ASFV dUTPase homotrimer stability. Our finding on these unique structural features of the ASFV dUTPase could be explored for the design of potential specific inhibitors that target this unique enzyme.
KEYWORDS: ASFV, dUTPase, crystal structure, African swine fever virus
ABSTRACT
The African swine fever virus (ASFV) is the deadly pathogen of African swine fever (ASF) that induces high mortality, approaching 100% in domestic pigs, causes enormous losses to the global pig industry, and threatens food security. Currently, there is no effective treatment or preventive countermeasure. dUTPases (deoxyuridine 5′-triphosphate pyrophosphatases) are ubiquitous enzymes that are essential for the hydrolysis of dUTP and prevent the misincorporation of dUTP into newly synthesized DNA. Here, we present the crystal structures of the ASFV dUTPase in complex with the product dUMP and cofactor Mg2+ at a resolution of 2.2 Å. We observed that a unique “turning point” at G125 plays an unexpected critical role in the swapping region of the C-terminal segment, which is further stabilized by the interactions of the last C-terminal β strand with the β1 and β2 strands, thereby positioning the catalytic motif 5 into the active site of its own subunit instead of into a third subunit. Therefore, the ASFV dUTPase employs a novel two-subunit active site that is different than the classic trimeric dUTPase active site, which is composed of all three subunits. Meanwhile, further results confirmed that the configuration of motifs 1 to 5 has high structural homology with and a catalytic mechanism similar to that of the known trimeric dUTPases. In general, our study expands the information not only on the structural diversity of the conserved dUTPase family but also on the details needed to utilize this dUTPase as a novel target in the treatment of ASF.
IMPORTANCE African swine fever virus (AFSV), a large enveloped double-stranded DNA virus, causes a deadly infection in domestic pigs. In addition to Africa, Europe, and South America, countries in Asia, such as China, Vietnam, and Mongolia, have suffered the hazards posed by ASFV outbreaks in recent years. Until now, there has been no vaccine for protection from ASFV infection or effective treatments to cure ASF. Here, we solved the crystal structure of the ASFV dUTPase-dUMP-Mg2+ complex. The ASFV dUTPase displays a noncanonical folding pattern that differs from that of the classic homotrimeric dUTPase, in which the active site is composed of two subunits. In addition, several nonconserved residues within the 3-fold axis channel play a vital role in ASFV dUTPase homotrimer stability. Our finding on these unique structural features of the ASFV dUTPase could be explored for the design of potential specific inhibitors that target this unique enzyme.
INTRODUCTION
African swine fever virus (ASFV), a large enveloped double-stranded DNA virus, is the only member of the genus Asfivirus in the family Asfarviridae (1). The African swine fever virus causes deadly infection, inducing high mortality, approaching 100%, in domestic pigs, and causing great losses to the global pig industry and threatening food security (2–4). The extracellular enveloped ASFV virion is approximately 200 nm in diameter and contains an inner core, an inner lipid membrane, an icosahedral capsid, and an outer lipid envelope. This virus can be transmitted through direct contact with infected pigs, bites from soft ticks, and human activity (5). The early clinical signs of ASF are unspecific, are associated with fever, and can be easily confused with other diseases, making early diagnosis difficult. All these symptoms increase the difficulty of prevention. Currently, there is no effective treatment for ASF. The ASFV genome varies between approximately 170 and 193 kbp and encodes from 150 to 167 proteins (6). Genome sequence analysis has revealed that the open reading frame E165R encodes a viral dUTPase, which shares sequence similarity with other dUTPases from various species and is considered to play an essential role in maintaining the integrity of the viral DNA both by reducing the dUTP/dTTP ratio levels and by providing the substrate for thymidylate synthase.
Deoxyuridine 5′-triphosphate nucleotidohydrolase (dUTPase) catalyzes dUTP to dUMP and pyrophosphate (PPi) (7, 8). On one hand, dUMP is the substrate used by thymidylate synthase for the synthesis of dTMP; on the other hand, this biochemical pathway plays a crucial role in maintaining a low dUTP/dTTP ratio, which prevents the misincorporation of deoxyuridylate into DNA by DNA polymerases (8, 9). Based on their different oligomerization states, dUTPases can be divided into three families. In the first family, the dUTPases form homotrimers. These homotrimeric dUTPases have been found in a wide range of species, including eukaryotes, prokaryotes, some DNA viruses (such as poxvirus and staphylococcal phage) and some retroviruses. To date, a number of crystal structures of the dUTPases of this family have been observed: Homo sapiens (10), Arabidopsis thaliana (11), Plasmodium falciparum (12), Escherichia coli (13), Methanococcus jannaschii (14), Mycobacterium tuberculosis (15), vaccinia virus (16), white spot syndrome virus (WSSV) (17), equine infectious anemia virus (18), and feline immunodeficiency virus (19). The distinguishing structural feature of this family is that the trimer is assembled by three identical subunits around a 3-fold axis. Each subunit contains an eight-stranded β barrel formed by a distorted jelly-roll fold. The C-terminal β strand is typically involved in strand exchange with a neighboring monomer. The catalytic active site is formed by five conserved motifs that are distributed over the entire sequence. Divalent metal ions, preferably Mg2+, and water molecules are located in the active site, which is typically anchored by an aspartic acid (20). In most cases, the active site is formed by five motifs from all three subunits, except in WSSV and Mason-Pfizer monkey virus (17, 21).
The second family of dUTPases forms homodimers and are mostly found in protozoan pathogens, such as Trypanosoma cruzi (22) and Leishmania major (23). Because all α-helical dimeric kinetoplastid dUTPases have completely different structures compared to that of Homo sapiens, these dimeric enzymes are an attractive drug target (24, 25). The monomeric dUTPases encoded by mammalian and avian herpesviruses constitute the third and final family, which shares limited homology with the trimeric dUTPase family. It has been proposed that the monomeric dUTPase originated from the duplication of a gene encoding a trimeric enzyme (26–28).
Previous studies have shown that dUTPase regulation plays a vital role in programmed cell death in Drosophila, yeast and mammalian cells (29, 30). Furthermore, a dysfunctional dUTPase causes a novel monogenic syndrome that leads to diabetes and bone marrow failure (31). Similarly, the dUTPases of many viruses, including poxviruses, herpesviruses, and several vertebrate retroviruses, play important roles in efficient virus replication; therefore, mutations in the dUTPase sequence can significantly affect the biological and pathogenic properties of these viruses (32–36). Moreover, dUTPase has evolved anti-interferon activity in addition to its dUTPase enzymatic activity, which renders dUTPase critical for establishing an effective persistent herpesviruses infection (28, 37–39). Inhibitors of the Homo sapiens dUTPase may be further optimized as efficient anticancer chemotherapeutic compounds applied either alone or in combination with existing treatments (40, 41). In addition, knowledge of the sequence dissimilarity and the structural differences of the dUTPases of pathogens (such as herpesviruses and Plasmodium falciparum) and those of Homo sapiens lays the foundation for utilizing a dUTPase as a potential platform for antiviral design, which can be accomplished by enhancing both the affinity and selectivity for a specific viral dUTPase compared to that of the host dUTPase (10, 12, 42–44).
Therefore, in the face of the extremely serious epidemic outbreak in Africa, Europe, and Asia in recent years (2, 4, 45), it is imperative to identify effective druggable targets and develop therapeutic drugs. Based on the essential role of dUTPase in ASFV (36), to provide more clues on the potential of its newly discovered functions and facilitate antiviral drug designs for targeting it, we subjected the ASFV dUTPase to a structural investigation.
RESULTS
Overall structure of the ASFV dUTPase homotrimer.
The full-length ASFV dUTPase (residues 1 to 165) was expressed in E. coli with a hexahistidine tag at the N terminus. The theoretical molecular weight of the recombinant protein was approximately 20 kDa. According to the retention volume found by SEC, the observed molecular mass is approximately 67 kDa, which is close to the theoretical molecular mass of the trimer, suggesting that recombinant ASFV dUTPase exists in the trimeric form in solution.
The crystal structure of the ASFV dUTPase was determined using the molecular-replacement method and refined to a 2.2-Å resolution with a final Rwork value of 0.196 (Rfree = 0.228). The crystal structures of the ASFV dUTPase reveal a trimer with one asymmetrical unit. The final model features the full-length polypeptide, except for residues L143 to T165 in monomer A, A145 to T165 in monomer B, and T147 to T165 in monomer C, which could not be built due to the lack of interpretable electron density data, a situation indicating the high structural flexibility of the C-terminal region. All of the residues modeled in the polypeptide chain were in good agreement with the electron density map. More than 97% of all residues lay in the favorable region of the Ramachandran plot, and none of the residues lay in the disallowed region (Table 1).
TABLE 1.
Data collection and refinement statistics
| Parameter | Valuea |
|---|---|
| PDB accession no. | 6KZ6 |
| Data collection statistics | |
| X-ray source | SSRF beamline BL19U1 |
| Wavelength (Å) | 0.97915 |
| Space group | P212121 |
| Unit cell parameters (Å; °) |
a = 66.2, b = 67.3, c = 116.9 α = β = γ = 90.0 |
| Resolution range (Å) | 50.0–2.18 (2.22–2.18) |
| Unique reflections | 27,729 (1,349) |
| Completeness (%) | 100.0 (100.0) |
| Redundancy | 12.2 (9.0) |
| I/σ〈I〉 | 10.5 (1.0) |
| Rmerge (%)b | 23.9 (2.9) |
| CC1/2 | 0.99 (0.82) |
| Refinement statistics | |
| Resolution range (Å) | 33.1–2.18 (2.26–2.18) |
| Reflections used in refinement | 27,086 (2,200) |
| Reflections used for Rfree | 1,389 (106) |
| Rwork (%)c | 19.6 (26.5) |
| Rfree (%)d | 22.8 (31.2) |
| No. of nonhydrogen atoms | 3,826 |
| Protein | 3,383 |
| Ligand | 62 |
| Solvent | 381 |
| Avg B-factors | 30.3 |
| Protein | 30.2 |
| Ligand | 30.8 |
| Solvent | 31.8 |
| RMSDe | |
| Bond length (Å) | 0.008 |
| Bond angle (°) | 0.90 |
| Ramachandran (%) | |
| Favored | 97.9 |
| Allowed | 2.1 |
| Outliers | 0.0 |
Numbers in the brackets are for the highest-resolution shell.
Rmerge = ΣhΣl | Iih− 〈Ih〉 |/ΣhΣI 〈Ih〉, where 〈Ih〉 is the mean number of the observations Iih of reflection h.
Rwork = Σ(ǁFp(obs)| − |Fp(calc)ǁ)/Σ|Fp(obs)|.
Rfree is an R factor for a preselected subset (5%) of reflections that was not included in refinement.
RMSD, root mean square deviation.
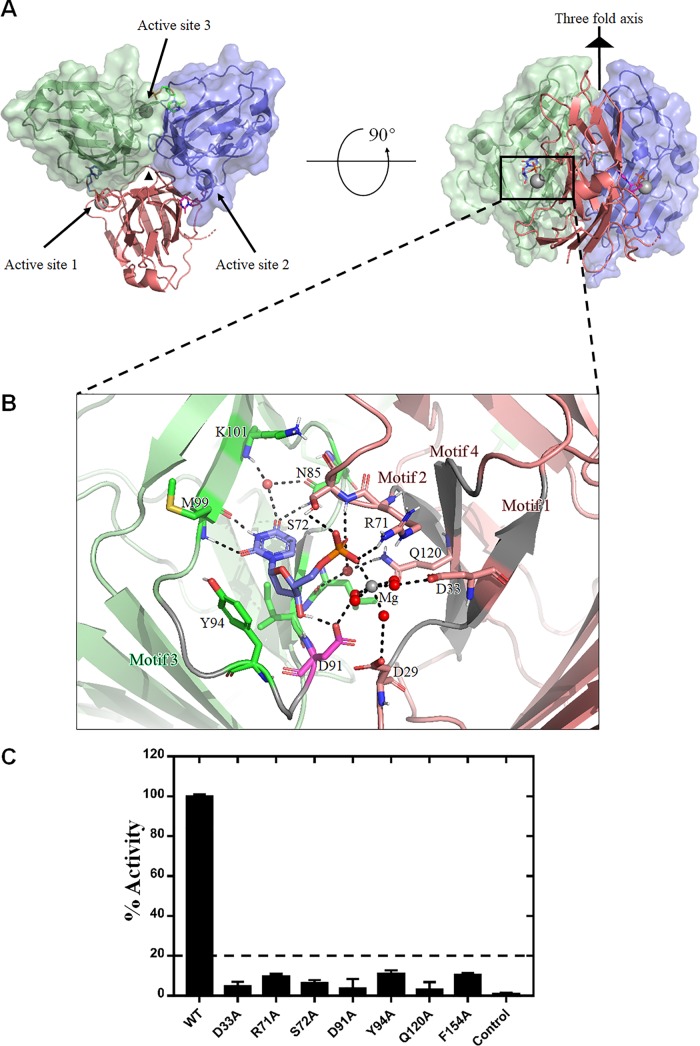
The ASFV dUTPase homotrimer has a 3-fold symmetry with a compact oligomerized assembly. The three active enzyme sites are situated at the position of the intersubunit cleft, which harbors the product, dUMP, and the enzyme cofactor, Mg2+ (Fig. 1A).
FIG 1.
Overall structure of ASFV dUTPase-dUMP-Mg2+ and investigation into the active site. (A) The top view (left) and side view (right) of the ASFV dUTPase. The monomers of the trimer are colored red, blue, and green, respectively. The 3-fold axis is indicated with a black triangle, the magnesium ion is indicated by the gray sphere, and the three individual active sites are also indicated. (B) Representation of the protein-ligand interactions at the active site of ASFV dUTPase-dUMP-Mg2+. The residues are labeled in the same colors as shown in panel A, except for D91, which participates in the nucleophilic reaction. Mg2+ is shown as a gray ball, the water molecules are shown as red balls, and the hydrogen bonds are shown as the dashed black lines. (C) An enzyme activity assay of the wild type (WT) and mutants was performed at 559 nm.
The residue composition of the ASFV dUTPase active site is conserved among the dUTPase family members.
In the active site of the ASFV dUTPase, the uracil ring is deeply buried in the active-site pocket, and the deoxyribose and α-phosphate groups are located at sites close to the periphery of the pocket. Binding of the uracil ring is achieved through several hydrogen bonds between N85, M99, and K101, the ring and one water molecule, mimicking Watson-Crick pairing (Fig. 1B). Among these binding sites, M99 and K101 are situated in the β10 strand, N85 is located in the loop domain between the β8 and β9 strands, and the conserved structural water molecule is also located as in many other trimeric dUTPase structures (10, 17, 18). The deoxyribose moiety is stabilized in position by stacking with Y94, an aromatic residue that is extremely highly conserved in almost all trimeric dUTPases. The enzyme activity of the Y94A mutant is only approximately 11.05% that of the wild-type ASFV dUTPase activity (Fig. 1C). In addition, the deoxyribose moiety also forms a hydrogen bond with the side chain hydroxyl of D91 (Fig. 1B). However, the deoxyribose of the dimeric dUTPase is anchored by hydrogen bonds with N201 and is located between the F84 and H83 residues in Trypanosoma cruzi (22).
The single phosphate group of dUMP extensively interacts with several residues of the dUTPase, the water molecules and Mg2+ (Fig. 1B). Residues R71 and S72 directly interact with the phosphate group through the side chain hydroxyl and main-chain nitrogen. Residues Q120, D33, D29, and D91 interact with the phosphate group via water molecules and Mg2+, forming an irregular octahedral configuration by five water molecules and a phosphate group that coordinates with one Mg2+. In general, similar to other classic trimeric dUTPases (10, 13, 19), the structural information implies that key residue interactions with phosphate groups play a critical role in the enzyme activity initiation process. This observation was further confirmed by a mutant enzyme activity assay. The results showed that the R71A and S72A mutants, which form direct hydrogen bonds with the α-phosphate group, exhibited approximately 6.5 to 9.6% the activity of the wild-type ASFV dUTPase activity (Fig. 1C). Moreover, the enzyme activities of the D33A, D91A, and Q120A mutants were almost totally abolished (Fig. 1C). Previous studies showed that, in the EIAV dUTPase, D72 is involved in the nucleophilic attack response during dUTP hydrolysis (18). By analyzing the structure of the ASFV dUTPase, we found that D91 is located at the position corresponding to D72 of the EIAV dUTPase (Fig. 1B). Taken together, the structural and mutagenesis results suggest that ASFV dUTPase shares a similar catalytic mechanism with classic trimeric dUTPases.
The ASFV dUTPase active site is composed of five highly conserved motifs.
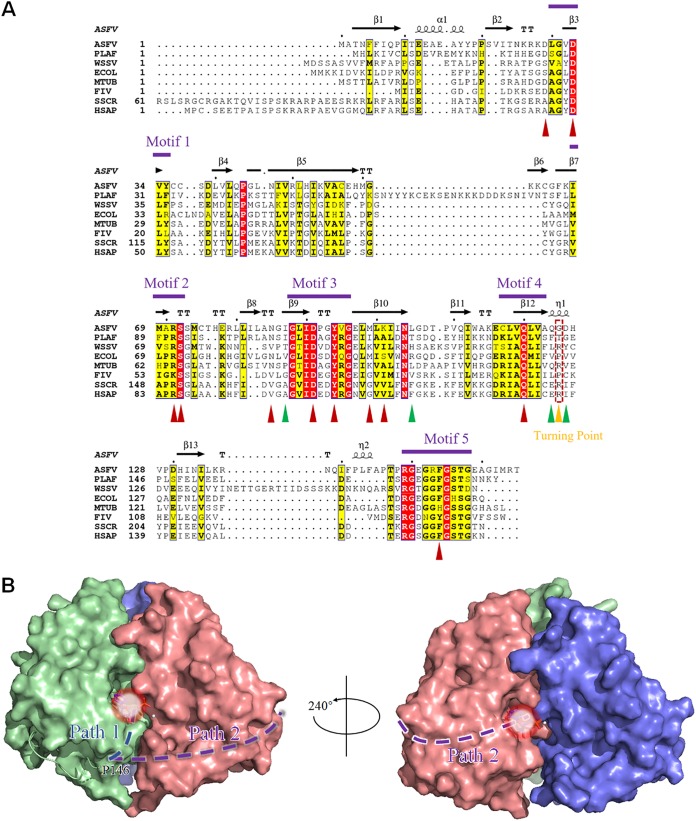
As previously described, the typical class I trimer dUTPase generally contains five conserved motifs (20). Therefore, we aligned the protein sequence of the ASFV dUTPase with seven other dUTPase protein sequences. The results showed that the ASFV dUTPase also possesses five strictly conserved motifs. Among them, motif 1 contains residues D29 to Y35, corresponding to the β3 strand; motif 2 contains residues I68 to S72, corresponding to the β7 strand; motif 3 contains residues G88-G96, corresponding to the β9 strand and the following loop; and motif 4 contains residues C107-A113, corresponding to the β12 strand. Motif 5 comprises residues R149-G158 located at the C terminus of the polypeptide chain (Fig. 2A).
FIG 2.
(A) Sequence alignment of the trimeric dUTPases and the motifs of the ASFV dUTPase. PLAF, Plasmodium falciparum; WSSV, white spot syndrome virus; ECOL, E. coli; MTUB, M. tuberculosis; FIV, feline immunodeficiency virus; SSCR, Sus scrofa; HSAP, Homo sapiens. Strictly and relatively conserved residues are highlighted in red and yellow boxes, respectively. The secondary structure elements of the ASFV dUTPase are shown above the alignment. The positions of motifs 1 to 5 are indicated. The residues coordinating to dUMP are marked with upright red triangles, and the residue interactions across the 3-fold axis are marked with upright green triangles, whereas the position of the turning point is indicated by a gold triangle and a dotted box. (B) Proposed novel C-terminal orientation of the ASFV dUTPase. The C terminus is hypothesized to reach its own subunit (path 1), instead of reaching out to the third subunit in the classic manner (path 2). The light white circular area indicates the position of the active site.
In the trimeric dUTPase family, these five motifs constitute the typical enzymatic active center of dUTPase. Among them, motifs 1, 2, and 4 are in one subunit, and motif 3 is in the neighboring subunit. Motif 5 is located in the C terminus of the polypeptide chain, which bypass the second subunit to reach the enzyme active pocket formed by the second subunit and the third subunit, where it participates in the dUTP hydrolysis reaction. Generally, motif 5 undergoes a transition from disordered to ordered upon substrate binding or through a substrate analog combination. Furthermore, motif 5 contains a P-loop-like (a phosphate-binding loop-like) motif, usually with a conserved GXXXXGK amino acid sequence, where X refers to any amino acid (46). The presence of this motif enables the correct binding of dUTP, thus ensuring efficient dUTPase activity. Notably, the fourth X position is highly conserved for the F residue and plays a critical role as a “Phe-lid,” which caps the uracil recognition pocket by stacking over the uracil ring. It has been indicated that this “Phe-lid” may hold the uracil in place, while water is excluded from the active site (10). Adjacent to the P-loop-like motif, there is a completely conserved R residue, named “the arginine finger,” which enables the structural organization of the active site to be efficiently catalyzed by the coordination of its nucleotide (47).
According to the ASFV dUTPase crystal structure, motifs 1, 2, and 4 with motif 3 constitute the enzyme pocket; motifs 1, 2, and 4 are from the same subunit, and motif 3 is from another subunit (Fig. 1B). The compositional characteristics of motifs 1 to 4 are similar to those of the known classic homotrimeric dUTPases. In addition, residues that play an important role in enzyme activity are mainly located in these motifs. D29 and D33 are located in motif 1, R71 and S72 are located on motif 2, D91 and Y94 are located in motif 3, and Q120 is located in motif 4 (Fig. 1B, Fig. 2A). However, the C terminus of the ASFV dUTPase containing motif 5 in all the monomers cannot be built due to a break in the continuous electron density, which indicates the high flexibility of this region. Nevertheless, the primary sequence alignment of several dUTPases indicates that ASFV dUTPase motif 5 retains a highly conserved P-loop-like motif structure, which plays an important role in dUTP hydrolysis. However, in our ASFV dUTPase-dUMP-Mg2+ complex structure, the C-terminal residues after P146 were invisible due to the lack of defined electron density. To further clarify the function of this region, we generated a mutant for the “Phe-lid,” F154A. The results from the enzyme assay showed that the F154A mutant retained only approximately 10.5% of the activity of the wild-type enzyme (Fig. 1C). According to our current structure, the ASFV C terminus may take one of two proposed routes to reach the active site: in one route it directly reaches the active site formed by a subunit from which it originates, and in the other it winds around the neighboring subunit to reach the next active site (Fig. 2B). However, the latter route requires at least 10 residues, while there are two residues between P146 and R149 (motif 5 of the ASFV dUTPase contains ten residues, from R149 to G158). Therefore, the next active site may be out of reach for motif 5, as indicated by the latter assumption; hence, it is very likely that the C-terminal segment of the ASFV dUTPase adopts the first route to reach the active site of its own subunit.
ASFV adopts a novel folding pattern compared to those in the trimeric dUTPase family.
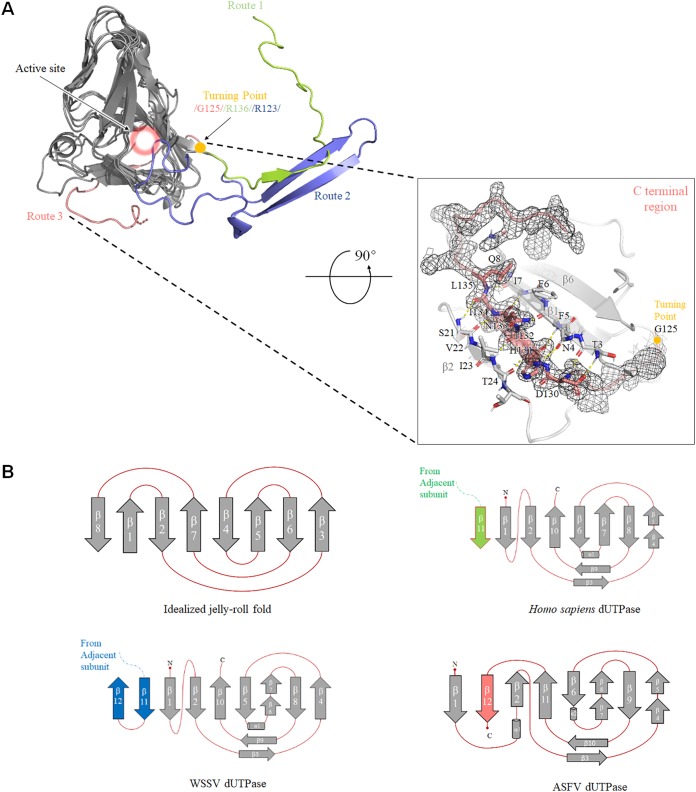
Previous studies have shown that the C terminus can take two different routes to reach the active site. In canonical homotrimeric dUTPases, including that of Homo sapiens, FIV, and EIAV, the C terminus containing motif 5 of each subunit is swapped at the domain-swapping region to reach the active site (10, 18, 19). However, in the recently discovered dUTPase structure of the WSSV, the pre-V insert forms an unusual β-hairpin structure in the domain-swapping region and thereby facilitates another orientation for the C-terminal segment, positioning the catalytic motif 5 into the active site of its own subunit instead of into a third subunit (17). Interestingly, we found that the ASFV dUTPase adopts a novel C-terminal region folding-swapping path, thus forming a completely new route to reach its catalytic pocket. We superimposed the crystal dUTPase structure from Homo sapiens, WSSV, and ASFV (Fig. 3A). The core domain of the ASFV dUTPase (from β1 to β11) shares a highly structural homology; however, the C terminus swapping region displays an extreme discrepancy in the orientation of the main chain. The Homo sapiens dUTPase C terminus extends outside the core domain and interchanges among the three subunits; however, the ASFV dUTPase C terminus executes a sharp turn and stretches backward from the “turning point” and wraps around its own subunit and into the catalytic pocket. Here, we define these three different configurations as route 1, route 2, and route 3 (Fig. 3A).
FIG 3.
(A) Three-dimensional (3D) structure and topology diagrams comparing Homo sapiens, WSSV, and ASFV dUTPases. A superposition of Homo sapiens (PDB 2HQU), WSSV (PDB 5Y5P), and ASFV dUTPase 3D structures is shown. Similar portions of the structure are shown as dark gray, and the unique C-terminal orientations are shown in salmon (ASFV), light green (Homo sapiens), and light blue (WSSV). The critical turning point is shown as a gold ball, and the active site is indicated by a pink halo (upper left). The β-turn is shown as a stick model. The C-terminal region of the ASFV dUTPase is displayed in the lower right frame, and the last β-sheet is stabilized by hydrogen-bonds with β1 and β2 strands. The electron density map (2Fo-Fc contoured at 1.0 σ) around residues after G125 is shown as a black mesh. (B) Homo sapiens, WSSV, and ASFV dUTPase have a very similar topology as the idealized jelly-roll fold.
According to the results from the primary sequence alignment, the “turning point” in the ASFV dUTPase is located at G125, while R136 is located at this position in the Homo sapiens dUTPase, and R123 is located at this position in the WSSV dUTPase. To our knowledge, many trimeric dUTPases that adopt a route 1 or route 2 C-terminal orientation also harbor an amino acid with bulky side chains at this site (Fig. 2A). However, G125 makes a typical β-turn, in which the carbonyl oxygen atom of A123 forms a strong hydrogen bond (1.9 Å) with the amide hydrogen atom of D126. Furthermore, this sharp turn structure causes a 180° flip in the direction of its own C terminus. Moreover, the last β strand is packed alongside the β1 strand and runs antiparallel to the β2 strand, which greatly enhances the stability of this fold. Residues D130 to L135 in the last β strand form seven hydrogen bonds with the T3-I7 residues of the β1 strand and four hydrogen bonds with the S21-I23 residues of the β2 strand. These hydrogen-bonding interactions maintain the stable conformation formed by the last β strand with the β1 and β2 strands (Fig. 3A).
From the perspective of structure topology, the overall fold of the trimeric dUTPase subunit resembles a jelly-roll fold. Compared to the standard jelly-roll β barrel, Homo sapiens, and WSSV dUTPase, ASFV dUTPase utilizes different manners to build the distorted β-barrel jelly-roll fold. In general, the fold of the Homo sapiens dUTPase consists primarily of an eight-stranded antiparallel β jelly roll, which is formed, in order, by β2, β10, β5, β6/7, β8, and β4, plus the β1 N-terminal strand and the adjacent parallel β11 C-terminal strand. In contrast, in the WSSV dUTPase, in addition to the distorted jelly-roll fold formed by β11, β1, β2, β10, β5, β6/7, β8, and β4, the position of β12 is on the outside because of its interaction with β11. Similar to those in the Homo sapiens dUTPase, β11 and β12 are from the adjacent subunit. However, in the ASFV dUTPase, β1, β12, β2, β11, β6, β7/8, β9, and β4/5 form a modified jelly-roll fold, and β12 is located next to the β barrel, where it is maintained through hydrogen bond interactions with the β1 and β2 strands (Fig. 3B).
The ASFV dUTPase trimer is stabilized by several nonconserved residues.
The driving force for the stabilization of the dUTPase homotrimer could be indicated by three major areas: the subunit-subunit interface, contacts along the perimeter of the trimer, and interactions along the 3-fold axis and contacts made through the C-terminal “arm” exchange (20). Compared to the C terminus in the canonical dUTPase, including that of Homo sapiens (10), E. coli (13), EIAV (18), and FIV (19), and the noncanonical dUTPases, such as WSSV (17), the ASFV dUTPase C terminus completely wraps around the subunit in which it belongs, not around an adjacent subunit. As discussed above, the ASFV dUTPase loses the third type of interaction force because the C terminus does not interact with the other subunits. The results from a Protein Interfaces, Structures and Assemblies (PISA) analysis showed that the accessible surface area buried in the interface was 6,550 Å2, which corresponds to approximately one-third of the accessible surface area of all the subunits (18,650 Å2), indicating a stable trimer in solution.
To further characterize the stability of the ASFV dUTPase, we performed a detailed analysis of this homotrimer. The interface between the adjacent subunits in the trimer comprises residues from a number of different parts in the sequence. The DIMPLOT program of LigPlot+ (48) revealed that 23 hydrogen bonds are formed between chains A and B, 33 hydrogen bonds are formed between chains A and C, and 24 hydrogen bonds are formed between chains B and C. A number of hydrogen bonds are formed via water molecules, and these hydrogen bonds, which are located at the subunit-subunit interface, play an important role in trimer stability.
As determined from the known structure, there are considerable differences in the mode by which the trimer is associated along the 3-fold axis, despite the conserved overall quaternary structure that forms the basis of the trimer. In general, the formation of aromatic clusters presumably serves to strengthen the association between the subunits in the EIAV, E. coli and Homo sapiens dUTPases (13, 32, 40). However, there are no aromatic residues that interact across the 3-fold axis in the ASFV dUTPase. At one end of the channel, the L46, L80, I82, I103, and L105 residues from each subunit form a hydrophobic region. In addition to these interactions, the side chains of the I87 residue in each of the three subunits point to the 3-fold axis in another hydrophobic interaction. At the other end of the channel, the Q124, G125, and D126 residues from the three subunits form a marvelous and stable hydrogen-bonded network (Fig. 4A). Moreover, the sequence alignment reveals that these residues are unique in ASFV (Fig. 2A). A Thermofluor assay results show that the Tm value of the ASFV dUTPase is approximately 84°C (Fig. 4B), 7.8°C higher than that of the WSSV dUTPase and 17.2°C higher than that of the E. coli dUTPase (17), indicating a much more stable trimer. We also monitored the thermal stability of the three mutants. The Tm values for the I87D, L105N, and D126G mutants are approximately 55, 46.5, and 46.5°C, respectively, which are significantly lower than those of the wild type (Fig. 4B). The reduced thermal stability assay confirms the unique structural feature for ASFV dUTPase homotrimer.
FIG 4.
Intermolecular interactions of the monomers of the ASFV dUTPase. (A) Interactions along the 3-fold axis. The hydrophobic interactions of L105 and I87 residues and a hydrogen-bond interactions network are displayed. The residues are shown as a stick model, and the hydrogen bonds are shown as dashed yellow lines. The black triangle represents the 3-fold axis. (B) Results from the thermal stability test of the ASFV dUTPase. The Tm is identified by plotting the first derivative of the fluorescence emission as a function of temperature (−dF/dT), and the Tm is represented at the lowest part of the curve.
DISCUSSION
We present here the ASFV dUTPase-dUMP-Mg2+ complex crystal structure. We also present that the recombinant ASFV dUTPase forms a homotrimer proven to be in a functional state for enzyme activity. The ASFV dUTPase shares a trimeric structure similar to that of the classic dUTPases that are categorized into the class I dUTPase family and include those of Homo sapiens (10), Escherichia coli (13), FIV (19), EIAV (18), Mycobacterium tuberculosis (15), and WSSV (17); in the main body of all of these dUTPases, a distorted jelly-roll fold is formed by eight β strands. The homotrimer has a 3-fold axis, and along this axis is a channel. The interactions within this channel play an important role in homotrimer stability. According to the crystal structure of the ASFV dUTPase, the homotrimer interaction is stabilized by electrostatic interactions and hydrophobic interactions.
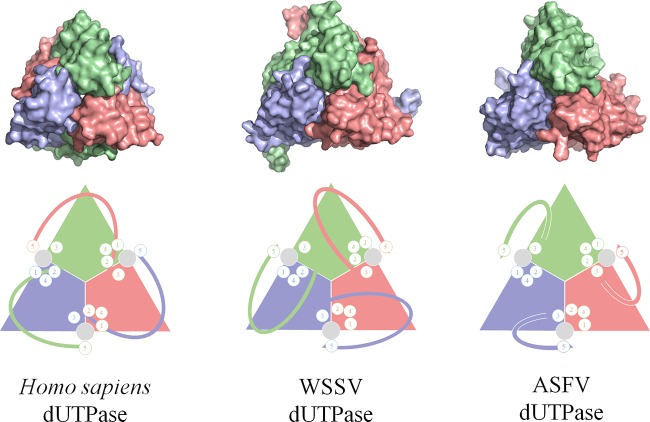
The primary sequence alignment of the ASFV dUTPase shows high similarity in the formation of the active site to that of other homotrimeric dUTPases, which contain five strictly conserved motifs (Fig. 5). In a particularly active site, motifs 1, 2, and 4, which interact with the phosphate moiety of the substrate, potentially in coordination with a metal ion, are all in one subunit. Motif 3, which binds to the nucleoside portion of the substrate, comes from another subunit (20). However, in contrast to the high structural homology of motifs 1 to 4, the configuration of motif 5 varies among different species. Motif 5 is provided by a third subunit in one form, mainly in the classic dUTPases, such as those in Homo sapiens (10), E. coli (13), and FIV (19); however, in another variant, motif 5 is in the subunit where motif 3 is located, including in the dUTPases of ASFV and WSSV (17). The WSSV dUTPase C terminus takes a go-and-return mode, reaching the subunit to which it belongs due to a “pre-5 insertion,” a unique sequence between motifs 4 and 5 (17); however, the ASFV dUTPase C terminus adopts a novel folding pattern that correctly places motif 5 into the active site.
FIG 5.
Comparison of active-site assembly among Homo sapiens, WSSV, and ASFV dUTPases. Homo sapiens dUTPase assembles each active site with all three subunits, whereas the WSSV and ASFV dUTPases assemble each active site with two adjacent subunits. The upper and lower panels display the crystal structures and schematic cartoons, respectively. In the lower panel, the circled numbers 1 to 5 represent the motif positions around the active site.
The C terminus in most trimeric dUTPases extends from motif 4 outside the distorted jelly-roll fold body. In contrast, the ASFV dUTPase C terminus folds back at the vicinity of motif 4 instead of extending outward. This β-turn forms at the turning point, G125, and the hydrogen bonds formed by the β13 strand with the β1 and β2 strands have important roles in the backward folding of the ASFV dUTPase C terminus. We first observed that the β-turn formed at the turning point G125 in the ASFV dUTPase completely flips the orientation of the C terminus. Most importantly, this novel manner of folding ensures the efficient enzymatic activity of the ASFV dUTPase (36).
In the canonical dUTPase, such as that of Homo sapiens, the individual active site is formed by all three subunits. Motifs 1, 2, and 4 are contributed by one subunit and motif 3 by another, while motif 5 is provided by the third subunit. Distinct from the canonical dUTPases, the C-terminal region of the WSSV is reversed by a β-hairpin in the swapping region and thus orients toward its own subunit. In a remarkable finding, the active site of the WSSV is actually formed by only two subunits: the catalytic motifs 1, 2, and 4 from one subunit and motifs 3 and 5 from the other. Similarly, the active site pocket of the ASFV dUTPase is also composed of these five conserved motifs contributed by two subunits (Fig. 5). It is notable that, although the noncanonical two-subunit active site is formed in a similar manner, the C terminus of ASFV adopts an orientation different from that of WSSV. Therefore, our findings on the ASFV dUTPase expands our knowledge of dUTPases in which the active site is composed of two subunits.
Lopata et al. (49) first studied the catalytic mechanism of the Mycobacterium tuberculosis dUTPase using a combination of computational, biochemical, and biophysical methods. The results showed that small loop movements in the C terminus of the dUTPase enable the nucleotides to be shuttled between the binding pocket and the solvent. Conserved motif 5 undergoes a disorder-to-order transition upon ligand binding to organize the active site into a catalytically competent conformation (17, 50, 51). The conserved residues, such as a “Phe-lid” and the arginine finger of motif 5, play vital roles in the stability of the substrate conformation. In our structure, motif 5 was not built due to the lack of continuous interpretable electron density data; however, this break in electron density may imply the high flexibility of motif 5 in the ASFV dUTPase. Nevertheless, we propose a reasonable homotrimer model for the ASFV dUTPase according to the dUTPase-dUMP-Mg2+ complex structure and the inferences described above.
Last, but not least, our structure facilitates the design of a structure-based antivirus against ASFV. Previous research has shown that ASFV dUTPase is a highly specific enzyme that is required for efficient replication and serves as a potential antiviral target (36). Given the low primary sequence similarity (approximately 23%) between Sus scrofa and ASFV, the unique structural features of the ASFV dUTPase could be explored as a possible guideline for the design of ASFV-specific inhibitors.
In conclusion, our crystal structure of the ASFV dUTPase reveals a novel folding pattern in the trimeric dUTPase family and expands our knowledge of the structural diversity within the dUTPase family. Moreover, the structural and functional information we present paves the way for further development for ASFV-specific inhibitors.
MATERIALS AND METHODS
Protein production.
The codon-optimized wild-type cDNA of ASFV dUTPase (GenBank accession no. AXZ95902.1) was synthesized by GENEWIZ. The full-length dUTPase was cloned into pET-28a (Novagen) vector with NdeI and HindIII restriction sites using cloning primers. The sequences of the primers were as follows: forward, 5′-GGAATTCCATATGATGGCGACGAACTTCTTCAT-3′; and reverse, 5′-CCCTCGAGTTAGGTGCGCATGATGCCCGCT-3′. The accuracy of the inserts was verified by sequencing.
The recombinant plasmid of ASFV dUTPase was transformed into E. coli strain BL21(DE3) (TransGen Biotech, Beijing, China). The cells were cultured at 37°C in 800 ml of Luria-Bertani medium containing 100 μg/ml ampicillin. Once the optical density at 600 nm reached 0.5 to 0.6, the culture was transferred to 16°C. Protein expression was induced by the addition of 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for an additional 16 to 18 h. The cells were harvested, resuspended in lysis buffer (20 mM HEPES [pH 7.5], 500 mM NaCl, 10 mM MgCl2), and homogenized with a low-temperature ultrahigh pressure cell disrupter (Union-Biotech). The lysate was centrifuged at 25,000 × g for 30 min at 4°C to remove cell debris. The supernatant was loaded twice onto a Ni-NTA column preequilibrated with lysis buffer. Resin was washed seven to eight times with 100 ml of wash buffer (20 mM HEPES, 200 mM NaCl, 10 mM MgCl2, 20 mM imidazole [pH 7.5]), ASFV dUTPase protein was eluted with elution buffer containing 20 mM HEPES, 200 mM NaCl, 500 mM imidazole, and 10 mM MgCl2 (pH 7.5). The protein was further purified on a Superdex 200 (GE Healthcare) column equilibrated with 20 mM HEPES-NaOH (pH 7.5), 200 mM NaCl, 10 mM MgCl2, and 1 mM dithiothreitol. SDS-PAGE analysis revealed >95% purity of the final purified recombinant protein. Fractions from the single major peak were pooled and concentrated to 10 mg/ml for crystallization.
Crystallization.
Initial crystallization conditions were screened by the sitting-drop vapor-diffusion method using commercial crystal screening kits at 16°C, including the Index, Crystal Screen, PEG/Ion, and Salt/RX kits from Hampton Research and Wizard I-IV kits from Emerald BioSystems. The complex was prepared by mixing dUTPase (N-terminal His tag; 4/8 mg/ml) with dUMP (Meilunbio, China) in a 1:5 molar ratio with 5 mM MgCl2 at 16°C. This protein complex and reservoir solutions were mixed in a ratio of 1:1, and all conditions were equilibrated against 100 μl of reservoir solution in a 48-well format plate. Small crystals of dUTPase appeared after 1 day in 0.2 M sodium chloride, 0.1 M HEPES (pH 7.5), and 25% (wt/vol) polyethylene glycol 3350. The final pyramid-like crystals were grown in 0.16 M sodium chloride, 0.08 M HEPES (pH 7.5), 0.02 M sodium citrate tribasic dihydrate, and 17% (wt/vol) polyethylene glycol 3350. Crystals were harvested and cryoprotected by plunging them into liquid nitrogen for X-ray data collection.
X-ray data collection and structure determination.
The data were collected at beamline BL19U (SSRF, China) under cryogenic conditions at 100 K. The data sets were processed using the HKL3000 package. The structure was solved by the Molecular Replacement program in the PHENIX software suite with Plasmodium falciparum dUTPase (PDB 1VYQ) as the initial search model. The models were built into the modified experimental electron density using COOT and refined in PHENIX. The dUMP molecule was modeled at the final stage of refinement, and the model quality was verified using the program MolProbity in PHNIX. Structural models were drawn using the program PyMOL (https://pymol.org/2/).
Mutant protein production.
Active sites D33, R71, S72, D91, Y94, Q120, and F154 were mutated individually to alanine to measure the dUTPase mutant protein protease enzyme activity. The residue I87 was mutated to aspartic acid, L105 was mutated to asparagine, and D126 was mutated to leucine. The site-directed mutations of ASFV dUTPase were generated by using a Fast Mutagenesis System (TransGen Biotech, China). The sequences of all the mutation constructs were verified by DNA sequencing (Tsingke, China). The sequences of the primers are not shown here. All the mutant proteins were expressed and purified following the procedure used for the wild-type protein.
Enzyme activity measurement.
The enzyme activities of dUTPases and their mutants were measured by a phenol red pH indicator at 559-nm assay as previously described (52), in which protons released in the dUTPase reaction were detected. Briefly, the measurements were performed in 1 mM HEPES (pH 7.5) buffer containing 150 mM KCl, 40 μM Phenol Red, 5 mM MgCl2, 20 nM dUTPase, and 20 μM dUTP on a UV/Vis spectrometer (Perkin-Elmer lambda35) with 10-mm-path-length cuvettes at 25°C. The data processing and image generation were completed using Prism 7.0 (GraphPad).
Thermal stability assay.
A Thermofluor assay (53) was performed on a quantitative PCR instrument (CFX96; Bio-Rad). The fluorescence (excitation wavelength, 470 nm; emission wavelength, 570 nm) of the protein solution (4 μM protein in 50 mM HEPES [pH 7.0] plus 5× SYPRO Orange dye [Sigma] in a final volume of 20 μl) was measured as a function of temperature at a climbing rate of 0.5°C/min from 25 to 95°C. The data were analyzed using Prism 7.0 (GraphPad).
Data availability.
Atomic coordinates and structure factors for ASFV dUTPase have been deposited in the Protein Data Bank under accession number 6KZ6.
ACKNOWLEDGMENTS
This study was supported by grants from the National Natural Science Foundation of China (grants 31670731 and 31870733), the National Key R&D Program of China (grants 2018YFE0200402 and 2017YFC840300), and Projects of International Cooperation and Exchanges NSFC (grant 81520108019); by the Young Elite Scientist Sponsorship Program by CAST; and by the Science and Technology Innovation Achievements and Team Building Foundation of Nankai University (grant ZB19500403).
REFERENCES
- 1.Alonso C, Borca M, Dixon L, Revilla Y, Rodriguez F, Escribano JM, ICTV Report Committee. 2018. ICTV virus taxonomy profile: Asfarviridae. J Gen Virol 99:613–614. doi: 10.1099/jgv.0.001049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Normile D. 2019. African swine fever marches across much of Asia. Science 364:617–618. doi: 10.1126/science.364.6441.617. [DOI] [PubMed] [Google Scholar]
- 3.Sanchez-Cordon PJ, Montoya M, Reis AL, Dixon LK. 2018. African swine fever: a re-emerging viral disease threatening the global pig industry. Vet J 233:41–48. doi: 10.1016/j.tvjl.2017.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cwynar P, Stojkov J, Wlazlak K. 2019. African swine fever status in Europe. Viruses 11:310. doi: 10.3390/v11040310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dixon LK, Sun H, Roberts H. 2019. African swine fever. Antiviral Res 165:34–41. doi: 10.1016/j.antiviral.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 6.Salas ML, Andres G. 2013. African swine fever virus morphogenesis. Virus Res 173:29–41. doi: 10.1016/j.virusres.2012.09.016. [DOI] [PubMed] [Google Scholar]
- 7.Bertani LE, Haggmark A, Reichard P. 1961. Synthesis of pyrimidine deoxyribonucleoside diphosphates with enzymes from Escherichia coli. J Biol Chem 236:PC67–PC68. [PubMed] [Google Scholar]
- 8.Greenberg GR, Somerville RL. 1962. Deoxyuridylate kinase activity and deoxyuridine triphosphatase in Escherichia coli. Proc Natl Acad Sci U S A 48:247–257. doi: 10.1073/pnas.48.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Freeman L, Buisson M, Tarbouriech N, Van der Heyden A, Labbé P, Burmeister WP. 2009. The flexible motif V of Epstein-Barr virus deoxyuridine 5′-triphosphate pyrophosphatase is essential for catalysis. J Biol Chem 284:25280–25289. doi: 10.1074/jbc.M109.019315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mol CD, Harris JM, Mcintosh EM, Tainer JA. 1996. Human dUTP pyrophosphatase: uracil recognition by a β-hairpin and active sites formed by three separate subunits. Structure 4:1077. doi: 10.1016/s0969-2126(96)00114-1. [DOI] [PubMed] [Google Scholar]
- 11.Inoguchi N, Chaiseeda K, Yamanishi M, Kim MK, Jang Y, Bajaj M, Chia CP, Becker DF, Moriyama H. 2015. Structural insights into the mechanism defining substrate affinity in Arabidopsis thaliana dUTPase: the role of tryptophan 93 in ligand orientation. BMC Res Notes 8:784. doi: 10.1186/s13104-015-1760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whittingham JL, Leal I, Nguyen C, Kasinathan G, Bell E, Jones AF, Berry C, Benito A, Turkenburg JP, Dodson EJ, Perez LMR, Wilkinson AJ, Johansson NG, Brun R, Gilbert IH, Pacanowska DG, Wilson KS. 2005. dUTPase as a platform for antimalarial drug design: structural basis for the selectivity of a class of nucleoside inhibitors. Structure 13:329–338. doi: 10.1016/j.str.2004.11.015. [DOI] [PubMed] [Google Scholar]
- 13.Cedergren-Zeppezauer ES, Larsson G, Nyman PO, Dauter Z, Wilson KS. 1992. Crystal structure of a dUTPase. Nature 355:740–743. doi: 10.1038/355740a0. [DOI] [PubMed] [Google Scholar]
- 14.Huffman JL, Li H, White RH, Tainer JA. 2003. Structural basis for recognition and catalysis by the bifunctional dCTP deaminase and dUTPase from Methanococcus jannaschii. J Mol Biol 331:885–896. doi: 10.1016/s0022-2836(03)00789-7. [DOI] [PubMed] [Google Scholar]
- 15.Chan S, Segelke B, Lekin T, Krupka H, Cho US, Kim MY, So M, Kim CY, Naranjo CM, Rogers YC, Park MS, Waldo GS, Pashkov I, Cascio D, Perry JL, Sawaya MR. 2004. Crystal structure of the Mycobacterium tuberculosis dUTPase: insights into the catalytic mechanism. J Mol Biol 341:503–517. doi: 10.1016/j.jmb.2004.06.028. [DOI] [PubMed] [Google Scholar]
- 16.Samal A, Schormann N, Cook WJ, DeLucas LJ, Chattopadhyay D. 2007. Structures of vaccinia virus dUTPase and its nucleotide complexes. Acta Crystallogr D Biol Crystallogr 63:571–580. doi: 10.1107/S0907444907007871. [DOI] [PubMed] [Google Scholar]
- 17.Zang K, Li F, Ma Q. 2018. The dUTPase of white spot syndrome virus assembles its active sites in a noncanonical manner. J Biol Chem 293:1088–1099. doi: 10.1074/jbc.M117.815266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dauter Z, Persson R, Rosengren AM, Nyman PO, Wilson KS, Cedergren-Zeppezauer ES. 1999. Crystal structure of dUTPase from equine infectious anaemia virus; active site metal binding in a substrate analogue complex. J Mol Biol 285:655–673. doi: 10.1006/jmbi.1998.2332. [DOI] [PubMed] [Google Scholar]
- 19.Prasad GS, Stura EA, McRee DE, Laco GS, Hasselkus-Light C, Elder JH, Stout CD. 1996. Crystal structure of dUTP pyrophosphatase from feline immunodeficiency virus. Protein Sci 5:2429–2437. doi: 10.1002/pro.5560051205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Persson R, Cedergren-Zeppezauer ES, Wilson KS. 2001. Homotrimeric dUTPases; structural solutions for specific recognition and hydrolysis of dUTP. Curr Protein Pept Sci 2:287–300. doi: 10.2174/1389203013381035. [DOI] [PubMed] [Google Scholar]
- 21.Veronika NP, Orsolya B, Mónika F, István S, Iva P, Michalea R, Helena Z, Dmitri S, Maxim P, Veronika H. 2006. Flexible segments modulate co-folding of dUTPase and nucleocapsid proteins. Nucleic Acids Res 35:495–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Harkiolaki M, Dodson EJ, Bernier-Villamor V, Turkenburg JP, González-Pacanowska D, Wilson KS. 2004. The crystal structure of Trypanosoma cruzi dUTPase reveals a novel dUTP/dUDP binding fold. Structure 12:41–53. doi: 10.1016/j.str.2003.11.016. [DOI] [PubMed] [Google Scholar]
- 23.Hemsworth GR, Moroz OV, Fogg MJ, Scott B, Bosch-Navarrete C, González-Pacanowska D, Wilson KS. 2011. The crystal structure of the Leishmania major deoxyuridine triphosphate nucleotidohydrolase in complex with nucleotide analogues, dUMP, and deoxyuridine. J Biol Chem 286:16470–16481. doi: 10.1074/jbc.M111.224873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Corinne N, Ganasan K, Isabel LC, Alexander MB, Marcel K, Reto B, Ruiz-Pérez LM, Johansson NG, Dolores GP, Gilbert IH. 2005. Deoxyuridine triphosphate nucleotidohydrolase as a potential antiparasitic drug target. J Med Chem 48:5942. doi: 10.1021/jm050111e. [DOI] [PubMed] [Google Scholar]
- 25.Stout CD. 2004. Induced fit, drug design, and dUTPase. Structure 12:2–3. doi: 10.1016/j.str.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 26.Baldo AM, McClure MA. 1999. Evolution and horizontal transfer of dUTPase-encoding genes in viruses and their hosts. J Virol 73:7710–7721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tarbouriech N, Buisson M, Seigneurin JM, Cusack S, Burmeister WP. 2005. The monomeric dUTPase from Epstein-Barr virus mimics trimeric dUTPases. Structure 13:1299–1310. doi: 10.1016/j.str.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Ariza ME, Glaser R, Williams MV. 2014. Human herpesviruses-encoded dUTPases: a family of proteins that modulate dendritic cell function and innate immunity. Front Microbiol 5:504. doi: 10.3389/fmicb.2014.00504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nation MD, Guzder SN, Giroir LE, Deutsch WA. 1989. Control of Drosophila deoxyuridine triphosphatase: existence of a developmentally expressed protein inhibitor. Biochem J 259:593–596. doi: 10.1042/bj2590593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Williams D, Norman G, Khoury C, Metcalfe N, Briard J, Laporte A, Sheibani S, Portt L, Mandato CA, Greenwood MT. 2011. Evidence for a second messenger function of dUTP during Bax mediated apoptosis of yeast and mammalian cells. Biochim Biophys Acta 1813:315–321. doi: 10.1016/j.bbamcr.2010.11.021. [DOI] [PubMed] [Google Scholar]
- 31.Dos Santos RS, Daures M, Philippi A, Romero S, Marselli L, Marchetti P, Senee V, Bacq D, Besse C, Baz B, Marroqui L, Ivanoff S, Masliah-Planchon J, Nicolino M, Soulier J, Socie G, Eizirik DL, Gautier JF, Julier C. 2017. dUTPase (DUT) is mutated in a novel monogenic syndrome with diabetes and bone marrow failure. Diabetes 66:1086–1096. doi: 10.2337/db16-0839. [DOI] [PubMed] [Google Scholar]
- 32.Lichtenstein DL, Rushlow KE, Cook RF, Raabe ML, Swardson CJ, Kociba GJ, Issel CJ, Montelaro RC. 1995. Replication in vitro and in vivo of an equine infectious anemia virus mutant deficient in dUTPase activity. J Virol 69:2881–2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pyles RB, Sawtell NM, Thompson RL. 1992. Herpes simplex virus type 1 dUTPase mutants are attenuated for neurovirulence, neuroinvasiveness, and reactivation from latency. J Virol 66:6706–6713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steagall WK, Robek MD, Perry ST, Fuller FJ, Payne SL. 1995. Incorporation of uracil into viral DNA correlates with reduced replication of EIAV in macrophages. Virology 210:302–313. doi: 10.1006/viro.1995.1347. [DOI] [PubMed] [Google Scholar]
- 35.Turelli P, Petursson G, Guiguen F, Mornex JF, Vigne R, Querat G. 1996. Replication properties of dUTPase-deficient mutants of caprine and ovine lentiviruses. J Virol 70:1213–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Oliveros M, Garcia-Escudero R, Alejo A, Vinuela E, Salas ML, Salas J. 1999. African swine fever virus dUTPase is a highly specific enzyme required for efficient replication in swine macrophages. J Virol 73:8934–8943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ariza ME, Glaser R, Kaumaya PT, Jones C, Williams MV. 2009. The EBV-encoded dUTPase activates NF-κB through the TLR2 and MyD88-dependent signaling pathway. J Immunol 182:851–859. doi: 10.4049/jimmunol.182.2.851. [DOI] [PubMed] [Google Scholar]
- 38.Madrid AS, Ganem D. 2012. Kaposi’s sarcoma-associated herpesvirus ORF54/dUTPase downregulates a ligand for the NK activating receptor NKp44. J Virol 86:8693–8704. doi: 10.1128/JVI.00252-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams MV, Cox B, Ariza ME. 2016. Herpesviruses dUTPases: a new family of pathogen-associated molecular pattern (PAMP) proteins with implications for human disease. Pathogens 6:2. doi: 10.3390/pathogens6010002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.McIntosh EM, Ager DD, Gadsden MH, Haynes RH. 1992. Human dUTP pyrophosphatase: cDNA sequence and potential biological importance of the enzyme. Proc Natl Acad Sci U S A 89:8020–8024. doi: 10.1073/pnas.89.17.8020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yano W, Yokogawa T, Wakasa T, Yamamura K, Fujioka A, Yoshisue K, Matsushima E, Miyahara S, Miyakoshi H, Taguchi J, Chong KT, Takao Y, Fukuoka M, Matsuo K. 2018. TAS-114, a first-in-class dual dUTPase/DPD inhibitor, demonstrates potential to improve therapeutic efficacy of fluoropyrimidine-based chemotherapy. Mol Cancer Ther 17:1683–1693. doi: 10.1158/1535-7163.MCT-17-0911. [DOI] [PubMed] [Google Scholar]
- 42.Recio E, Musso-Buendía A, Vidal AE, Ruda GF, Kasinathan G, Nguyen C, Ruiz-Pérez LM, Gilbert IH, González-Pacanowska D. 2011. Site-directed mutagenesis provides insights into the selective binding of trityl derivatives to Plasmodium falciparum dUTPase. Eur J Med Chem 46:3309–3314. doi: 10.1016/j.ejmech.2011.04.052. [DOI] [PubMed] [Google Scholar]
- 43.Hampton SE, Baragaña B, Schipani A, Bosch-Navarrete C, Musso-Buendía JA, Recio E, Kaiser M, Whittingham JL, Roberts SM, Shevtsov M, Brannigan JA, Kahnberg P, Brun R, Wilson KS, González-Pacanowska D, Johansson NG, Gilbert IH. 2011. Design, synthesis, and evaluation of 5′-diphenyl nucleoside analogues as inhibitors of the Plasmodium falciparum dUTPase. ChemMedChem 6:1816–1831. doi: 10.1002/cmdc.201100255. [DOI] [PubMed] [Google Scholar]
- 44.Harris JM, McIntosh EM, Muscat GE. 1999. Structure/function analysis of a dUTPase: catalytic mechanism of a potential chemotherapeutic target. J Mol Biol 288:275–287. doi: 10.1006/jmbi.1999.2680. [DOI] [PubMed] [Google Scholar]
- 45.Simulundu E, Sinkala Y, Chambaro HM, Chinyemba A, Banda F, Mooya LE, Ndebe J, Chitanga S, Makungu C, Munthali G, Fandamu P, Takada A, Mweene AS. 2018. Genetic characterization of African swine fever virus from 2017 outbreaks in Zambia: identification of p72 genotype II variants in domestic pigs. Onderstepoort J Vet Res 85:e1–e5. doi: 10.4102/ojvr.v85i1.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saraste M, Sibbald PR, Wittinghofer A. 1990. The P-loop: a common motif in ATP- and GTP-binding proteins. Trends Biochem Sci 15:430–434. doi: 10.1016/0968-0004(90)90281-f. [DOI] [PubMed] [Google Scholar]
- 47.Nagy GN, Suardiaz R, Lopata A, Ozohanics O, Vekey K, Brooks BR, Leveles I, Toth J, Vertessy BG, Rosta E. 2016. Structural characterization of arginine fingers: identification of an arginine finger for the pyrophosphatase dUTPases. J Am Chem Soc 138:15035–15045. doi: 10.1021/jacs.6b09012. [DOI] [PubMed] [Google Scholar]
- 48.Laskowski RA, Swindells MB. 2011. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J Chem Inf Model 51:2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 49.Lopata A, Leveles I, Bendes AA, Viskolcz B, Vertessy BG, Jojart B, Toth J. 2016. A hidden active site in the potential drug target Mycobacterium tuberculosis dUTPase is accessible through small amplitude protein conformational changes. J Biol Chem 291:26320–26331. doi: 10.1074/jbc.M116.734012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Varga B, Barabás O, Kovári J, Tóth J, Hunyadi-Gulyás E, Klement E, Medzihradszky KF, Tölgyesi F, Fidy J, Vértessy BG. 2007. Active site closure facilitates juxtaposition of reactant atoms for initiation of catalysis by human dUTPase. FEBS Lett 581:4783–4788. doi: 10.1016/j.febslet.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 51.Garcia-Nafria J, Timm J, Harrison C, Turkenburg JP, Wilson KS. 2013. Tying down the arm in bacillus dUTPase: structure and mechanism. Acta Crystallogr D Biol Crystallogr 69:1367–1380. doi: 10.1107/S090744491300735X. [DOI] [PubMed] [Google Scholar]
- 52.Balázs V, Orsolya B, Eniko T, Nikolett N, Péter N, Vértessy BG. 2008. Active site of mycobacterial dUTPase: structural characteristics and a built-in sensor. Biochem Biophys Res Commun 373:8–13. doi: 10.1016/j.bbrc.2008.05.130. [DOI] [PubMed] [Google Scholar]
- 53.Niesen FH, Berglund H, Vedadi M. 2007. The use of differential scanning fluorimetry to detect ligand interactions that promote protein stability. Nat Protoc 2:2212–2221. doi: 10.1038/nprot.2007.321. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Atomic coordinates and structure factors for ASFV dUTPase have been deposited in the Protein Data Bank under accession number 6KZ6.