Abstract
The objective of this research is to solve the current medical problems of a high incidence of fungal infections in the lungs, high misdiagnosis rate, and high mortality. In this study, firstly, the logistic regression model was used to conduct. Risk factors of pulmonary fungal infection in respiratory department were analyzed. Then a model of pulmonary fungal infection in mice was constructed, and the expression difference of Progranulin (PGRN) in serum was detected by enzyme-linked immuno sorbent assay (ELISA). The expression of PGRN in lung tissues of mice infected by pulmonary fungi was detected by Western bolt method and quantitative polymerase chain reaction (PCR). The PGRN protein and mRNA expression in the lung epithelial cells of mice were detected after the infection. Results logistic regression model was used to analyze the main risk factors affecting pulmonary infection in mice. The risk factors of pulmonary fungal infection were indent catheter, hypoproteinemia, long-term use of glucocorticoid and long-term use of antibiotics. The PGRN content in serum was obviously higher than that before pulmonary fungal infection (P < 0.01). The expression of PGRN mRNA and protein in lung tissue was obviously higher than that before infection (P < 0.01). The expression of PGRN mRNA and protein in lung tissues of the infected group was obviously higher than that of the non-infected group (P < 0.01). The expression of PGRN protein in the lung epithelial cells of mice was obviously higher at 24 h after infection than before infection (P < 0.01), and the expression of PGRN mRNA was obviously higher at 12 h after infection than before infection (P < 0.01), indicating that PGRN is highly expressed in fungal pulmonary infection and is involved in disease progression. Therefore, this study provides a new idea for the diagnosis and treatment of fungal pulmonary infection in the later stage and has a good guiding significance for the diagnosis and treatment of fungal pulmonary infection.
Keywords: Logistic regression analysis model, PGRN, Mice, Lung, Fungal infection
1. Introduction
Fungi are a class of eukaryotic organisms whose main mode of survival relies on parasitism or pythogenesis to sustain life [1], [2]. It can exist in almost any place, such as land, air, water, soil, and other external environments, and even in the human body, such as the mouth. Fungi mainly invade into the human body when the immune function of the human body declines and cause human illness because most of them are opportunistic pathogens, that is, when the human body is normal, it has a strong defense system and will not cause human illness [3], [4], [5]. Pulmonary fungal infection is a disease mainly caused by the inflammation of the lungs or bronchi caused by the invasion of fungi into the body.
PGRN is a secretory growth factor composed of 593 amino acid residues [6]. PGRN can be cut into many monomer GRN polypeptides by extracellular proteases with a molecular weight of 6 × 103 g/mol–25 × 103 g/mol. These products are called granular protein A (GRN, A), granular protein B (GRN, B), granular protein C (GRN, C), epithelial transforming growth factor (TGFE), etc. [7]. PGRN has unique physiological functions and is widely involved in various aspects of early embryo development, wound healing promotion, tumor development, host defense, and immune regulation, etc. It is one of the important functional proteins in the body [8]. Based on the above advantages, many scholars at home and abroad began to study PGRN and achieved good results in the field of inflammation treatment and immune disease treatment [9]. Previous studies have shown that the expression of PGRN was obviously up-regulated in gastric epithelial cells and cancerous tissues after infection [10], and the combination of PGRN and antibiotics in the sports system diseases of pigs infected with staphylococcus can achieve a better therapeutic effect.
According to relevant literature, it can be concluded that good results have been achieved in relevant studies based on logistic regression analysis model to analyze risk factors of pulmonary fungal infection in respiratory medicine department. As one of the important immune regulatory factors, PGRN has been widely studied in researches on immune diseases, but relatively few researches on infectious diseases. Based on the above situation, the mice with pulmonary fungal infection were taken as the research objects, and the logistic regression model was adopted to study the factors affecting pulmonary fungal infection. In addition, ELISA was used to detect the changes of PGRN content in serum of mice with lung infection. The changes of PGRN mRNA and protein expression in pulmonary epithelial cells were detected by fluorescence quantitative PCR and Western blot.
2. Materials and methods
2.1. Animals
In this experiment, C57BL/6 mice aged 6 weeks were selected as the study subjects. All the mice were purchased from Shanghai slack experimental animal co., LTD., 100 in total. 90 mice were selected to construct the model of pulmonary fungal infection, and 10 mice were selected as the control group. Preparation of experimental samples: firstly, two groups of normal mice were selected to establish the pulmonary fungal infection group and the uninfected pulmonary fungal group. Then infection was carried out. Fungal infection (candida mycoderma bacteria) was carried out on mice in the fungal infection group, and the infection multiple was 1 or 2. The mice were anesthetized and given 40 μL of infected fluid from their nostrils when they developed convulsions and gasped. Finally, the body weight and changes in the mice were recorded. Mice that confirmed successful infection were used for the experiment.
Sham operation was performed on mice, and traumatic surgical treatment including an indwelling urinary catheter, central venous catheterization, mechanical ventilation, and endotracheal intubation was performed. Patients were treated with glucocorticoids, immunosuppressants, and antibiotics. The blood protein and neutrophils of mice were measured.
2.2. Variable definition and assignment of logistic regression analysis model
The factors of pulmonary fungal infection in mice were divided into 14 related factors including basic disease (diabetes, hypertension, chronic obstructive pulmonary disease, cardiovascular and cerebrovascular diseases), creative operation (indwelling catheter, mechanical tube, central venous straight tube, trachea ventilation), long-term use of immunosuppressive agents, long-term use of glucocorticoids, long-term use of antibiotics, hypoproteinemia and neutropenia. The variable names and assignments for the factors of lung infection in mice are shown in Table 1.
Table 1.
Variable names and assignments of pulmonary infection factors in mice.
| Infectious factors | Variable names | Assignments |
|---|---|---|
| Chronic obstructive pulmonary disease | Z1 | “0” for none; “1” for the presence |
| Hypertension | Z2 | “0” for none; “1” for the presence |
| II-diabetes | Z3 | “0” for none; “1” for presence |
| Cardiovascular and cerebrovascular diseases | Z4 | “0” for none; “1” for presence |
| Indwelling catheter | Z5 | “0” for none; “1” for the presence |
| Central venous straight tube | Z6 | “0” for none; “1” for the presence |
| Mechanical ventilation | Z7 | “0” for none; “1” for presence |
| Tracheal ventilation | Z8 | “0” for none; “1” for the presence |
| Long-term use of corticosteroids | Z9 | “0” for none; “1” for the presence |
| Long-term use of immunosuppressants | Z10 | “0” for none; “1” for the presence |
| Long-term use of antibiotics | Z11 | “0” for none; “1” for the presence |
| Hypoproteinemia | Z12 | “0” for none; “1” for presence |
| Neutrophil reduction | Z13 | “0” for none; “1” for presence |
2.3. Method for detecting PGRN content in serum
Serum PGRN levels were determined by ELISA assay. Blood samples of mice in each group were collected under the condition of fasting, and serum samples of mice were obtained by centrifugation after anticoagulation treatment and stored in the refrigerator at −20 °C. The coating buffer was used to dilute the antibody to be captured to solution with working concentration and then it was added to the ELISA plate for antibody precoating. The solution was sealed with plastic wrap at 4 °C for 24 h. A phosphate buffer of 300–400 μm was added to the ELISA plate, and each hole was washed three times for about 1 min each time. The diluent was then diluted with double distilled water and incubated at room temperature for 1 h. The original liquid was discarded and the phosphate buffer of 300–400 μm was washed three times. The standard substance was prepared and diluted into 0, 62.5 pg/mL, 125 pg/mL, 250 pg/mL and 500 pg/mL. The sample lung homogenate was prepared with the original liquid. 150 μL was added to each hole and incubated for 2 h at room temperature. The phosphate buffer was washed 5 times. The antibody was incubated at room temperature for 1 h. The phosphate buffer was washed five times and 100 muons HRP secondary antibody was added to each hole. They were incubated at room temperature in dark condition for 0.5 h and washed with phosphate buffer solution for 5 times. The TMB in each hole was 100 mu L, incubated at room temperature for 15 min in dark condition, and the color change was observed. 2 mol of termination agent was added to each hole. The OD value at 490 nm was measured with a microplate reader at a reference wavelength of 570 nm. And the PGRN concentration of the sample was analyzed by the software based on the calibration data.
2.4. Extraction of RNA and protein from lung tissue
After weighing the mice, they were killed with a spinal dislocation. Lung tissue samples were collected, blood was flushed, and stored in liquid nitrogen in a cryopreservation tube containing tissue preservation fluid. Lung tissue was removed, and about 50 g samples were taken with surgical scissors. The samples were placed in a mortar, and liquid nitrogen was added to grind them thoroughly. The powdered tissue was added into a centrifuge tube containing 1 mL Trizol reagent, mixed well and placed at room temperature for 5 min. 200-µm chloroform solution was added, mixed well, placed at room temperature for 2 min. and centrifuged at low temperature at 12000 rpm for 10 min. The supernatant was taken, isopropanol was added, mixed and placed at room temperature for 15 min, and centrifuged at a low temperature of 12000 rpm for 10 min. The supernatant was discarded, and 75% ethanol prepared by 0.1% sterilized DEPC was added to wash the precipitation and centrifuged at a low temperature of 12000 rpm for 10 min. Washing was repeated for 3 times. After drying, 50 µm 0.1% sterile DEPC water was added to dissolve RNA precipitation. After the concentration and purity of RNA were detected by the nucleic acid detector, it is stored in the refrigerator at −80 °C.
About 50 g of lung tissue was placed in a mortar, and 50 mL of protein lysate was added for grinding. It was moved to a centrifuge tube, added 30 µm of protein lysate, shook and mixed well, and placed in an icebox for 30 min. They were centrifuged at a low temperature of 12000 rpm for 10 min, and 100 μL supernatant was placed in a new centrifuge tube. The protein concentration was detected using the BCA kit, and the detection solution was configured on a 50:1 basis. Using 5% bovine serum protein as the standard, curves of 1 μL, 2 μL, 4 μL, 8 μL, 12 μL, 16 μL, and 20 μL were plotted. Each hole sample was injected with 2 µm, and the remaining samples were mixed with double distilled water to form 20 μL. They were incubated for 30 min at 37 °C. OD value at 562 nm was measured with the microplate reader and protein concentration was calculated. All protein samples were adjusted to uniform concentration with lysis buffer and degenerated in metal bath at 100 °C for 10 min. After cooled, it was stored in the −20 °C refrigerator.
Healthy mouse lung epithelial cells were isolated from lung tissue and cultured in a cell culture medium containing 10% fetal bovine serum and 90% high glucose DEME medium. RNA and protein were extracted from lung epithelial cells at 0 h, 1 h, 3 h, 6 h, 12 h and 24 h after fungal infection.
2.5. Detection of PGRN mRNA and protein expression in mouse lung tissue
Rt-qPCR was used to detect the difference of PGRN mRNA expression in lung tissues of normal mice and mice with pulmonary fungal infection. The extracted mouse lung RNA was used as the model, and cDNA reverse transcription was performed by using the reverse transcription kit. After cDNA was obtained from mouse lung tissues, PGRN mRNA was used as a template for quantitative detection. One μL of cDNA was placed in the eight-strand tube, and the primers of the upper and lower PGRN genes were 0.5 μL each. 10 μL TB Green reagent was added and 8 μL ddH2O was added to mix well. The octave containing the mixture was detected by fluorescence quantitative PCR.
Western blot was used to detect PGRN protein expression in the lung tissue of mice. 10% polyacrylamide separation adhesive was prepared. A sample of protein containing 6 to 14 μg was added to 50 μL per well. Electrophoresis was performed at 80 V, and the voltage was adjusted to 110 V after 30 min. The electrophoresis was stopped immediately when the proteins were completely separated. The membrane was transferred by the wet transfer method and the membrane was transferred at a constant current of 250 mA for 90 min. Polyvinylidene fluoride film was preactivated by methanol for 10 s. After the film transfer, the film was removed and put into 5% skim milk powder and shook for 1 h. The film was washed every 15 min, three times each time. The diluted PGRN primary antibody was added and placed in the labeled antibody incubation box and incubated overnight by shaking at 4 °C. TBST was washed and added with diluted secondary antibodies for incubation. TBST was used to wash the film 5 times, 5 min each time. The A and B liquids of the ECL-enhanced chemiluminescent solution were mixed and uniformly added to the PVDF membrane. The imaging equipment was scanned and stored, and the target protein was analyzed in grayscale software to evaluate the changes in protein levels.
The above methods were used to detect PGRN mRNA and protein expression in lung epithelial cells.
2.6. Statistical method
SPSS20.0 statistical software was used for statistical analysis. Descriptive statistical methods were used for counting data, which were expressed in percentages and constituent ratios. The chi-square test was performed on the count data conforming to the normal distribution, Significance t-test was used for the measurement data in line with normal distribution, which was expressed as mean ± standard deviation (x ± s). The variables with statistically significant differences in the univariate analysis were applied to logistic multivariate regression analysis, and the factors of pulmonary fungal infection were obtained by statistical method, and P < 0.05 indicated statistically significant.
3. Results and discussion
3.1. Mouse fungal infections and distribution of survey samples
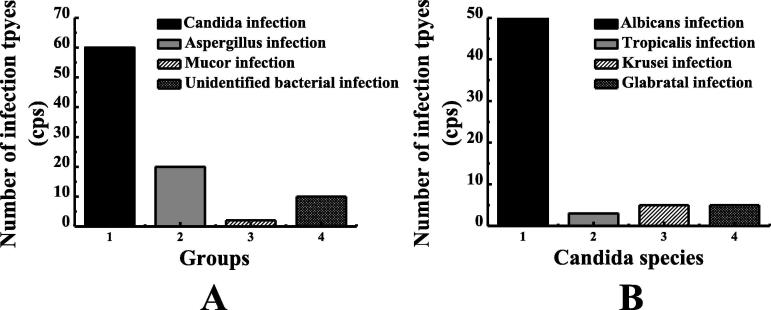
In the fungal infection group, there were 68 mice with primary foundational diseases, 7 with immune system diseases, 3 with blood system diseases, 3 with diabetes, and 9 with no foundational diseases, a total of 90 mice were collected. In the non-fungal infection group, there were 70 cases of primary basic diseases, 3 cases of immunological diseases, 4 cases of consumption diseases, 2 cases of diabetes, 1 case of cerebral infarction, 1 case of nephropathy, and 9 cases with no foundational diseases, a total of 90 mice. According to the statistics of 90 mice with pulmonary infection, as shown in Fig. 1, there were 60 cases of Candida infection, 20 cases of aspergillus infection, 2 cases of mucor infection, and 8 cases of unconfirmed bacterial infection. Among them, as shown in Fig. 1B, there were 50 cases of candida Albicans infection, 2 cases of candida tropicalis infection, 4 cases of candida krusei infection, and 4 cases of Candida glabrata infection. The distribution of infection types of the samples is shown in Fig. 1A and B.
Fig. 1.
Type of fungal infection in mice and type of candida infection. Note: figure A showed the type of fungal infection. Figure B showed the genus type of candida.
3.2. Univariate logistic regression analysis model results
Table 2 shows the results of univariate analysis of mice with pulmonary fungal infection. From the statistical results, there was no statistically significant difference in the incidence of chronic obstructive pulmonary disease (P = 0.316), hypertension (P = 0.890), diabetes (P = 0.243), cerebrovascular disease (P = 0.637), neutropenia (P = 0.198), mechanical ventilation (P = 0.136), tracheal ventilation (P = 0.191) and long-term use of immunosuppressants (P = 0.161) between the fungal infection group and the non-fungal infection group. There were statistically significant differences in an indwelling catheter (P = 0.001), long-term use of glucocorticoids (P = 0.001), long-term use of antibiotics (P < 0.001), and hypoproteinemia (P = 0.013), which were risk factors for pulmonary fungal infection in mice. This is consistent with the results of Mu et al.
Table 2.
Univariate analysis of mice with pulmonary fungal infection.
| Infectious factor | Odds ratio | 95% confidence interval for the odds ratio | P-value |
|---|---|---|---|
| Chronic obstructive pulmonary disease | 2.145 | 1.138–4.039 | 0.316 |
| Hypertension | 1.050 | 0.539–2.034 | 0.890 |
| II-diabetes | 1.700 | 0.701–3.951 | 0.243 |
| Cardiovascular disease | 1.187 | 0.590–2.371 | 0.637 |
| Indwelling catheter | 3.610 | 1.621–8.050 | 0.001 |
| Central venous straight tube | 3.167 | 1.182–8.519 | 0.025 |
| Mechanical ventilation | 2.519 | 1.073–5.928 | 0.136 |
| Tracheal ventilation | 2.030 | 0.725–5.760 | 0.191 |
| Long-term use of corticosteroids | 3.261 | 1.687–8.822 | 0.001 |
| Long-term use of immunosuppressants | 3.855 | 0.637–16.629 | 0.161 |
| Long-term use of antibiotics | 3.788 | 1.904–7.529 | <0.001 |
| Hypoproteinemia | 2.155 | 1.158 = 4.014 | 0.013 |
| Neutropenia | 3.861 | 0.781–19.101 | 0.198 |
3.3. Results of multivariate logistic regression analysis
Indwelling catheter, hypoproteinemia, long-term use of glucocorticoids and long-term use of antibiotics were risk factors for pulmonary fungal infection in mice, as shown in Table 3.
Table 3.
Results of pulmonary fungal infections by logistic multivariate regression analysis.
| Infection factors | Regression coefficients | Standard deviation | Χ2 value | P-value | Odds ratio | OR 95% confidence interval |
|---|---|---|---|---|---|---|
| Hypoglycemic protein | 1.147 | 0.570 | 4.050 | 0.045 | 3.140 | 1.031–9.561 |
| Long-term use of corticosteroids | 1.214 | 0.479 | 6.245 | 0.011 | 3.372 | 1.298–8.741 |
| Long-term use of antibiotics | 1.145 | 0.411 | 7.790 | 0.003 | 3.139 | 1.407–7.011 |
| Indwelling catheter | 0.875 | 0.372 | 5.635 | 0.020 | 2.391 | 1.163–4.905 |
It can be concluded from Table 3 that the P-value of hypoglycemic protein is 0.045, and the odds ratio is 3.140. The long-term use of glucocorticoids has a P value of 0.011, an odds ratio of 3.372, long-term use of antibiotics has a P value of 0.003 and an odds ratio of 3.139, and the indwelling catheter has a P value of 0.020 and an odds ratio of 2.391. They are risk factors for fungal infections of the lungs.
In this study, the long-term use of hormones was found to have 4.121 times the risk of pulmonary fungal infection in mice that did not use hormones. However, currently, glucocorticoids are used in the treatment of bronchial asthma and idiopathic pulmonary interstitial fibrosis. Although hormones can improve lung function, they can also inhibit the ability of white blood cells to devour, which will greatly increase the occurrence of fungal infection. As can be concluded from Table 2, Table 3, the long-term use of antibiotics has an odds ratio of 3.788 and a Χ2 value of 7.790. This result indicates that long-term use of antibiotics also increases the probability of fungal infections. Long-term use of antibiotics will change the types of bacteria in the body, leading to the imbalance of the microbial ecology of the body. Once the microbial ecology of the body is out of balance, it will provide an opportunity for the cultivation and reproduction of fungi, which will lead to the increase of the incidence of fungal infection. Low blood protein refers to a decrease in the rate of protein synthesis in the body caused by various external factors, which leads to a decrease in serum protein caused by decreased synthesis. Low serum protein levels can lead to reduced immunity, which can lead to fungal infections and complications that can cause multiple organ failure. Long-term use of indurated catheter is also an important risk factor for pulmonary fungal infection. Indwalling catheter is a creative procedure that can lead to urinary tract infection, which in turn destroys the normal physiological environment of the urinary system, further reducing the ability to resist pathogens entering the body, causing urinary tract fungal infections and creating new infections based on this infection. When fungi are not under immediate control in the body, they proliferate and travel through lymphatic vessels and blood to the organs of the body, causing fungal infections.
3.4. Changes of PGRN expression in serum of mice after fungal infection
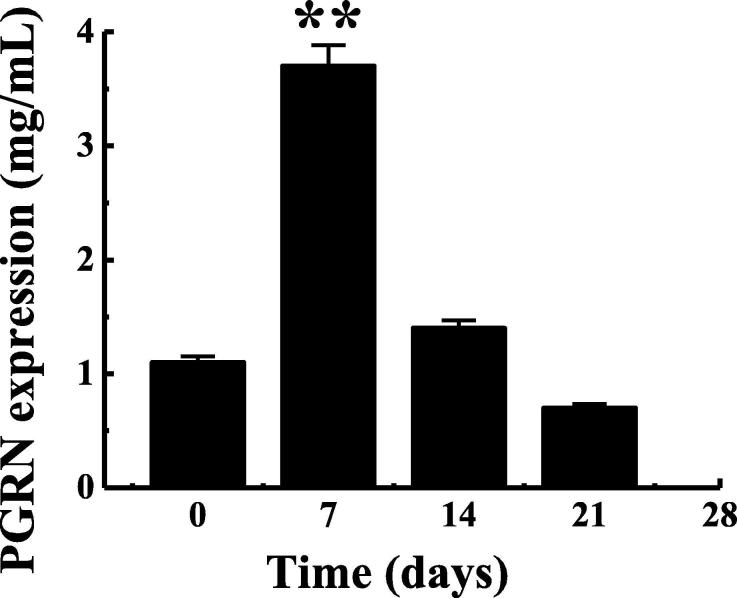
The ELISA method was used to detect the PGRN content in the serum of the fungal-infected mice, as shown in Fig. 2. As can be concluded from Fig. 2, After 7 days of fungal infection, PGRN content in the serum of mice was obviously higher than that before infection (P < 0.01), while at 14 days of infection, PGRN content had recovered to the original level. However, PGRN content showed a trend of decrease at 21d after infection, but there was no significant difference from that before infection (P > 0.05). This was basically consistent with the result that Gong et al. detected that serum PGRN level in patients with chronic HBV infection increased with the passage of infection time by Elisa.
Fig. 2.
Changes of PGRN content in the serum of mice with pulmonary fungal infection. Note: ** indicated that the expression was obviously different from that on the 0d, P < 0.01.
3.5. Changes of PGRN expression in pulmonary tissues of mice with fungal infection
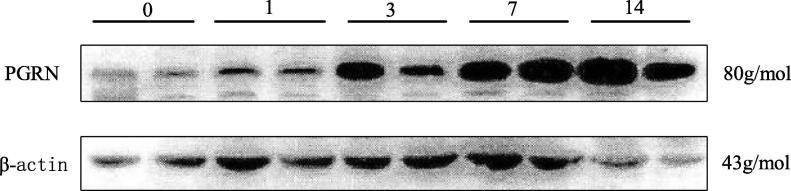
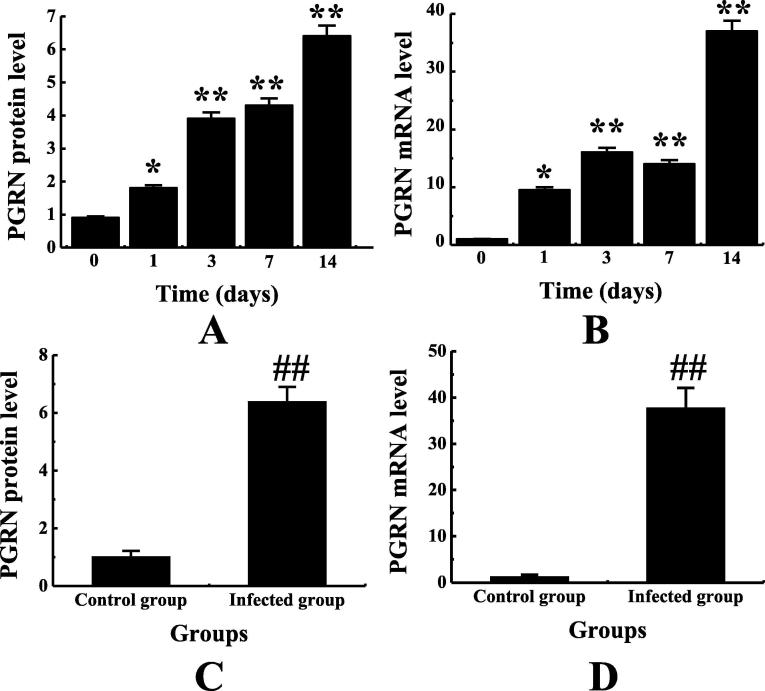
The total protein and mRNA were extracted from the lung tissues of mice infected with the fungus, PGRN protein and mRNA in the lung tissues of fungus-infected mice were detected. According to Fig. 3, with the passage of time, the expression of PGRN protein in the lung tissue of mice showed a trend of gradual up-regulation. According to Fig. 4A and 4B, PGRN protein and mRNA in lung tissues of mice were increased to different degrees on 1d, 3d, 7d and 14d after pulmonary fungal infection compared with that on 0d, and obviously higher on the first day than that on 0d(P < 0.05). On the third, seventh and 14th day, the expression was obviously higher than that on the 0th day (P < 0.01), and the expression was the highest on the 14th day. The PGRN protein and mRNA expressions in the lung tissues of the non-infected group and the infected group were compared. According to Fig. 4C and 4D, the PGRN protein and mRNA expressions in the lung tissues of the infected group were obviously higher than those in the non-infected group (P < 0.01). This is basically consistent with the results of Li et al. 's study that PGRN expression was obviously up-regulated in myocardial tissue in the mouse model of myocarditis after CVB3 infection.
Fig. 3.
Electrophoretogram of PGRN protein in lung tissue of mice infected with pulmonary fungi.
Fig. 4.
Changes of PGRN protein and mRNA expression in lung tissues of mice infected with pulmonary fungi and their comparison with non-infected mice. Note: figure A showed the change of PGRN protein expression. Figure B showed the change of PGRN mRNA expression. Figure C showed the comparison of PGRN protein between the non-infected group and the infected group. Figure D showed the comparison of PGRN mRNA between the non-infected group and the infected group. * indicated that there was a significant difference between the expression on the 0d and that on the 0d, P < 0.05. ** indicated that the expression on the 0d was obviously different from that on the 0d, P < 0.01. ## indicated that there was an extremely significant difference between the infected group and the infected group, P < 0.01.
3.6. Changes of PGRN expression in epithelial cells of mice with fungal infection
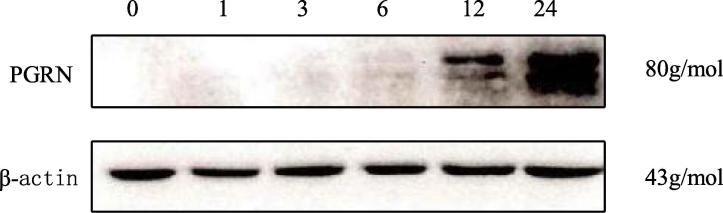
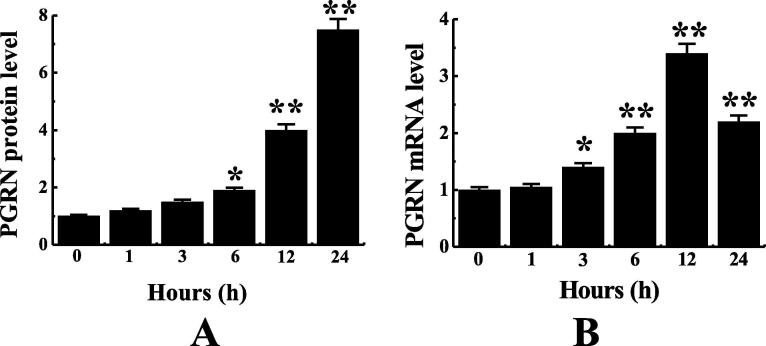
Cell level experiments were conducted on mice in the fungal infection group, as shown in Fig. 5. PGRN in the lung epithelial cells of mice in the pulmonary fungal infection group increased obviously with the passage of time, and the PGRN content in the epithelial cells was 6.5 times higher 24 h after infection than that at 0 h after infection and twice that of PGRN at 12 h after infection. Changes of PGRN mRNA and protein expression in lung epithelial cells at different times were detected. According to Fig. 6A, the expression of PGRN protein in cells after 12 and 24 h of fungal infection was obviously higher than that after 0 h of infection (P < 0.01). The expression of PGRN protein in the cells after 6 h infection was obviously higher than that after 0 h infection (P < 0.05).However, the expression of PGRN protein was the highest 24 h after infection. According to Fig. 6B, the expression of PGRN mRNA in the cells at 3 h after infection was obviously higher than that at 0 h after infection (P < 0.05). The expression of PGRN mRNA was obviously higher at 6 h, 12 h, and 24 h after infection than at 0 h after infection (P < 0.01).However, the expression of PGRN mRNA was the highest after 12 h infection.
Fig. 5.
Electrophoretogram of PGRN protein in pulmonary epithelial cells of fungal infected mice.
Fig. 6.
Changes of PGRN protein and mRNA in epithelial cells of mice infected by pulmonary fungi for different periods. Note: * meant there was a significant difference from 0 h, P < 0.05; ** indicated that there was an extremely significant difference compared with 0 h, P < 0.01.
The above experiments prove that PGRN is involved in the expression change in the process of pulmonary fungal infection in mice, and PGRN is involved in the development of fungal infectious diseases through mouse models and cell level experiments.
4. Conclusion
In this experiment, Multivariate logistic regression analysis showed that the risk of long-term use of hormones for pulmonary fungal infection was 4.112 times higher than that of mice without hormones. Long-term antibiotic mice had 3.962 times the risk of infection compared with non-antibiotic mice, suggesting that drug therapy also increased the risk of fungal infection. Hypoalbumin and indurated urinary tube all increased the risk of pulmonary fungal infection in mice. PGRN was highly expressed after fungal infection in the lungs of mice. The content of PGRN in serum, total protein and mRNA increased obviously with the passage of time, which verified that PGRN was involved in the expression changes during the pulmonary fungal infection of mice. The involvement of PGRN in the development of fungal infectious diseases was demonstrated by mice model and cell level experiments, which suggested that PGRN was involved in the development of pulmonary infection in mice.
In this research, multiple logistic regression analysis models were used to analyze the factors of pulmonary fungal infection of PGRN in mice. The factors affecting pulmonary infection in mice were analyzed and predicted by statistical method, which provided guidance for the diagnosis and treatment of pulmonary fungal infection in the future, and provided theoretical basis for the diagnosis, treatment, and rehabilitation of pulmonary fungal infection patients.
Footnotes
Peer review under responsibility of King Saud University.
References
- 1.Vanherp L., Poelmans J., Hillen A. Bronchoscopic fibered confocal fluorescence microscopy for longitudinal in vivo assessment of pulmonary fungal infections in free-breathing mice[J] Sci. Rep. 2018;8(1):3009–3010. doi: 10.1038/s41598-018-20545-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tweedle J.L., Deepe J.G. TNF-α antagonism reveals a gut/lung axis that amplifies regulatory T cells in a pulmonary fungal infection.[J] Infect. Immun. 2018;86(6):9–18. doi: 10.1128/IAI.00109-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhou X., Sun L., Bracko O. Impaired prosaposin lysosomal trafficking in frontotemporal lobar degeneration due to progranulin mutations[J] Nat. Commun. 2017;8:77–78. doi: 10.1038/ncomms15277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo Q., Xu L., Li H. Progranulin causes adipose insulin resistance via increased autophagy resulting from activated oxidative stress and endoplasmic reticulum stress[J] Lipids Health Dis. 2017;16(1):25–26. doi: 10.1186/s12944-017-0425-6. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 5.Tripathi D., Welch E., Cheekatla S.S. Alcohol enhances type 1 interferon-α production and mortality in young mice infected withMycobacterium tuberculosis[J] PLoS Pathog. 2018;14(8):174–175. doi: 10.1371/journal.ppat.1007174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mu F.G., He H.L., Li J. Risk factors for invasive pulmonary fungal infection in children[J] Chinese J. Contemporary Pediatr. 2014;16(8):779. [PubMed] [Google Scholar]
- 7.Johnson D.C. Chronic Candidal bronchitis: a consecutive series[J] Open Respiratory Med. J. 2012;6(1):145–149. doi: 10.2174/1874306401206010145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nakamura N., Tanaka C., Takeuchi-Kaneko Y. Transmission of antibiotic-resistance markers by hyphal fusion suggests partial presence of parasexuality in the root endophytic fungus Glutinomyces brunneus[J] Mycol. Progr. 2019;18(3):453–462. [Google Scholar]
- 9.Jiang S., Zhang J. Recent advances in the pathogenesis and treatment of hypoproteinemia after surgical stress] [J. Zhonghua Wei Zhong Bing Ji Jiu Yi Xue. 2017;29(3):284. doi: 10.3760/cma.j.issn.2095-4352.2017.03.020. [DOI] [PubMed] [Google Scholar]
- 10.Li L., Li L., Xiao L. Progranulin ameliorates coxsackievirus-B3-induced viral myocarditis by downregulating Th1 and Th17 cells[J] Exp. Cell Res. 2018;367(2):241–250. doi: 10.1016/j.yexcr.2018.04.001. [DOI] [PubMed] [Google Scholar]