Abstract
Protein phosphorylation by eukaryotic protein kinases (ePKs) represents a fundamental mechanism of cell signaling in all organisms. In model vertebrates, ~10% of ePKs are classified as pseudokinases, which possess amino acid changes within the catalytic machinery of the kinase domain that distinguish them from their canonical kinase counterparts. However, pseudokinases still regulate a wide variety of signaling pathways, usually doing so in the absence of their own catalytic output. To investigate the prevalence, evolutionary relationships, and biological diversity of these pseudoenzymes, we present a comprehensive analysis of putative pseudokinase sequences in available eukaryotic, bacterial, and archaeal proteomes. We demonstrate that pseudokinases are present across all domains of life and have classified nearly 30,000 eukaryotic, 1,500 bacterial, and 20 archaeal pseudokinase sequences into 86 pseudokinase families, including ~30 families that are reported for the first time. We uncover a rich variety of pseudokinases with notable expansions not only in animals, but also in plants, fungi, and bacteria, where pseudokinases have previously received cursory attention. These expansions are accompanied by domain shuffling, which suggests roles for pseudokinases in plant innate immunity, plant-fungal interactions, and bacterial signaling. Mechanistically, the ancestral kinase fold has diverged in many distinct ways through the enrichment of unique sequence motifs to generate new families of pseudokinases in which the kinase domain is repurposed for non-canonical nucleotide binding or to stabilize unique, inactive kinase conformations. The addition of an annotated collection of predicted pseudokinase sequences to the Protein Kinase Ontology (ProKinO) represents a new minable resource for the signaling community.
Summary:
Diverse protein pseudokinases are prevalent throughout the tree of life, contributing non-catalytic functions across a variety of signaling niches.
Introduction
Protein phosphorylation catalyzed by eukaryotic protein kinases (ePKs) controls multiple aspects of prokaryotic and eukaryotic-based cell signaling (1, 2), and its dysregulation contributes to many major diseases. The conserved architecture of the eukaryotic protein kinase (ePK) domain is very well understood from both structural (3–5) and biochemical (6–8) perspectives, and the versatility of the kinase fold has been exploited many times during evolution to impart mechanistic control over diverse cell signaling processes (9, 10). A vast amount of genomic and proteomic datasets can now be mined to map the evolution of kinases and their associated signaling pathways across multiple species (11–17). In this context, some 10% of model vertebrate protein kinases contain amino acid changes at specific positions that are predicted to lead to catalytic inactivation, which led to the coining of the term ‘pseudokinase’ (5, 15, 18–21). A number of well-studied pseudokinases are thought to play central roles in signaling despite impaired catalytic function (22–26), for example through allosteric modulation of other active kinases or the transduction of cellular signals via dynamic scaffolding functions (9, 19, 21, 27–30). However, whether pseudokinases have evolved to control fundamental aspects of signaling across all organisms has never been scrutinized in depth, and much remains to be understood about the origin of pseudokinases and how they became embedded in signaling networks during prokaryotic and eukaryotic evolution.
Protein pseudokinases represent the best understood members of the growing classes of pseudoenzymes, which include pseudophosphatases (31) and pseudoproteases (32), both of which are also predicted to have lost canonical catalytic function, but nonetheless perform critical non-enzymatic roles (9, 20, 33, 34). By definition, pseudokinases lack canonical phosphotransferase activity, and they can be predicted bioinformatically by identifying sequences that lack at least one key amino acid normally required for metal and ATP binding and for catalysis (3, 7, 8, 18–20). Prominent catalytic motifs include the ‘catalytic triad’ residues, comprised of the ATP-binding β3-lysine, the catalytic aspartate within the catalytic loop HRDXXXN motif, and the metal binding aspartate of the activation loop DFG motif. Some examples of human pseudokinases with variations at these catalytic triad residues are summarized in Table 1. Importantly, loss of these canonical residues does not always abolish nucleotide-binding or phosphoryl transfer, and in some cases residual kinase activity or ATP binding may fulfill a unique functional role. However, we still define these catalytically-competent proteins as pseudokinases, in recognition of their non-canonical amino acid composition. For example, in the human EGFR-related receptor pseudokinase HER3, where the catalytic triad is conserved except for the substitution of the catalytic HRD-aspartate for asparagine, low levels of catalytic activity support HER3 irafts-autophosphorylation in vitro, although this vestigial activity is insufficient for phosphorylation of exogenous substrates in cells (22, 35, 36). In other cases, degenerated catalytic residues can be compensated by similar amino acids found elsewhere in the active site to rescue catalytic function. This is best illustrated by the With No Lysine kinases (WNKs), which lack the canonical Β3-lysine, but maintain ATP-binding and catalytic activity via a conserved compensatory lysine in the glycine rich loop (37–39). Less predictably, pseudokinases contain co-evolving amino acids that are far-removed from the active site and contribute critical non-catalytic signaling roles, as described recently for the Tribbles (TRIB) family of pseudokinases, where a co-evolved C-terminal tail docking site in the pseudokinase domain negatively regulates binding of the E3 ubiquitin-protein ligase COP1 (26, 40–42). Finally, amino acid shuffling offers new biochemical opportunities as described recently for the atypical SelO pseudokinase in which dramatic active site variations facilitate “inverted” ATP binding to support protein AMPylation instead of phosphorylation (43, 44).
Table 1.
Examples of human pseudokinases. Degraded catalytic triad residues and the amino acids that replace them in each pseudokinase are noted.
| Human pseudokinase | Degraded catalytic residue(s) |
Observed residue(s) |
|---|---|---|
| KSR1, KSR2 | K | R |
| WNK1, WNK2, WNK3, WNK4 | K | C |
| HER3 | HRD-D | N |
| JAK1 (domain2) | HRD-D | N |
| JAK2 (domain2) | HRD-D | N |
| ILK | HRD-D | A |
| TRIB3 | DFG-D | N |
| TRIB2 | DFG-D | S |
| CASK | DFG-D | G |
| GCN2 (domain2) | K, HRD-D | Y, V |
| ULK4 | K, DFG-D | L, N |
| VRK3 | HRD-D, DFG-D | N, G |
| MLKL | HRD-D, DFG-D | K, G |
| STRADB (STLK6) | HRD-D, DFG-D | S, G |
| EphB6 | K, HRD-D, DFG-D | Q S, R |
| SCYL1, SCYL2, SCYL3 | K, HRD-D, DFG-D | F, N, G |
| NRBP1, NRBP2 | K, HRD-D, DFG-D | N, N, S |
Approximately 50 protein pseudokinases are encoded by the human genome, nearly all of which are also found in rodents, suggesting conserved vertebrate-wide signaling roles (5, 15, 18). Half of vertebrate pseudokinases also have clear orthologues in well-characterized genetic model organisms, including flies and worms, supporting the assumption that pseudokinases are part of ancient genetic lineages, rather than extraneous remnants of evolutionary ‘experiments’ (13). Several pseudokinases have been analyzed in depth in human cells, including HER3 (23, 35, 45, 46), the RAF/MEK modulators KSR1/2 (24), the Janus tyrosine kinases (JAKs) (25), which contain a disease-associated pseudokinase domain positioned adjacent to an active kinase domain, and the TRIB family of pseudokinases (26, 40, 47). However, over half of the predicted human pseudokinome remains understudied at the molecular level, despite clear evidence for expression in cells. Pseudokinase-based signaling has also been described in simple model organisms (48), such as in the small genome of the intestinal parasite Giardia lamblia (16) and in commercially important plants (49, 50, 51, 52, 53). However, the origin and evolution of pseudokinases across the tree of life has not been explored in any depth.
In this resource, we present the first systematic identification of predicted pseudokinase sequences, ranging from archaea and bacteria through to simple eukaryotes, fungi, plants, and vertebrates. Based on the well understood catalytic machinery in canonical ePKs (6), we find that ePK-like pseudokinase sequences can even be detected in some archaeal and bacterial proteomes, though they are much rarer than in eukaryotic proteomes, where they appear to be ubiquitous. Corroborating previous kinome studies, we also find that the number of pseudokinases remains relatively constant among vertebrates and correlates with the relative size of the kinome. Indeed, our broad analysis permits us to establish that ~10% of ePK members should be classified as pseudokinases in swathes of vertebrate animal species. In several phyla, specific pseudokinase expansions linked to lifestyle are observed within the different kinase families, whose shared sequence signatures and domain structures permit specific functions to be deciphered. In particular, we note the expansions of interleukin-1 receptor-associated kinase (IRAK)-like pseudokinases in plants, increases of tyrosine kinase-like (TKL) pseudokinases in fungi, and a diversified family of PknB pseudokinases in bacteria. Importantly, most pseudokinases exhibit lineage-specific sequence variations that might facilitate novel modes of ATP binding, unusual catalytic outputs, and/or allosteric coupling between distal protein binding and regulatory sites. As such, pseudokinases cannot be remnants of evolution, but must instead operate as fundamental, and function-specific, signaling proteins across organisms covering some 4 billion years of evolution. Our analysis includes a minable resource and is the first comprehensive classification of pseudokinase sequences from diverse organisms, providing a conceptual starting point for future hypothesis-driven characterization of pseudokinase signaling from bacteria to humans.
Results:
Identification of pseudokinomes across the domains of life
To detect the prevalence of pseudokinases across the domains of life, we used curated multiple sequence alignment profiles of 592 protein kinase families (52, 54–58) to scan all available eukaryotic, bacterial, and archaeal proteomes in the UniProt reference proteome database (10,092 proteomes in release 2018_9) (59). Aligned sequences that lacked one or more of the canonical catalytic triad residues, namely the β3-lysineresidue, the HRD-aspartate, or the DFG-aspartate, were classified as pseudokinase sequences. Such pseudokinase sequences with non-canonical residues at the three catalytic triad positions were detected in 100% of eukaryotic proteomes analyzed, whereas only 5.8% and 2.5% of bacterial and archaeal proteomes, respectively, contained putative pseudokinase sequences (Table 2). The prevalence of kinases and pseudokinases across 93 diverse representative species throughout the domains of life is summarized in Figures 1 and 2. Consistent with previous studies, we identified 55 pseudokinases in the human kinome, including PEAK3, which was recently reported as a pseudokinase sharing similarity to the PRAG1 and PEAK1/Sgk269 pseudokinases (30, 60) (Fig. 1). We also identified a novel putative human pseudokinase (A0A1B0GUL7) that is homologous to the dual-specificity Mitogen Activated Kinase Kinase (MEK2) and possesses a pseudokinase ortholog in chimpanzee (Pan troglodytes). Other previously studied pseudokinases such as TRRAP (19, 61), SelO (43, 44), and Fam20A (29) are also considered pseudokinases, however, these atypical kinases and other small molecule kinases such as aminoglycoside kinases and lipid kinases were not considered in our analysis because they are significantly divergent from ePKs and cannot be reliably placed within the ePK evolutionary framework (see methods).
Table 2.
Detection of protein kinase and pseudokinase sequences across archaeal, bacterial, and eukaryotic proteomes.
| Archaea | Bacteria | Eukaryota | |
|---|---|---|---|
| Total # proteomes | 441 | 8818 | 833 |
| Proteomes with kinases | 149 (33.8%) | 3523 (40.0%) | 833(100%) |
| Proteomes with pseudokinases | 11 (2.49%) | 508 (5.76%) | 819 (98.3%) |
| Total # pseudokinase sequences | 20 | 1386 | 30049 |
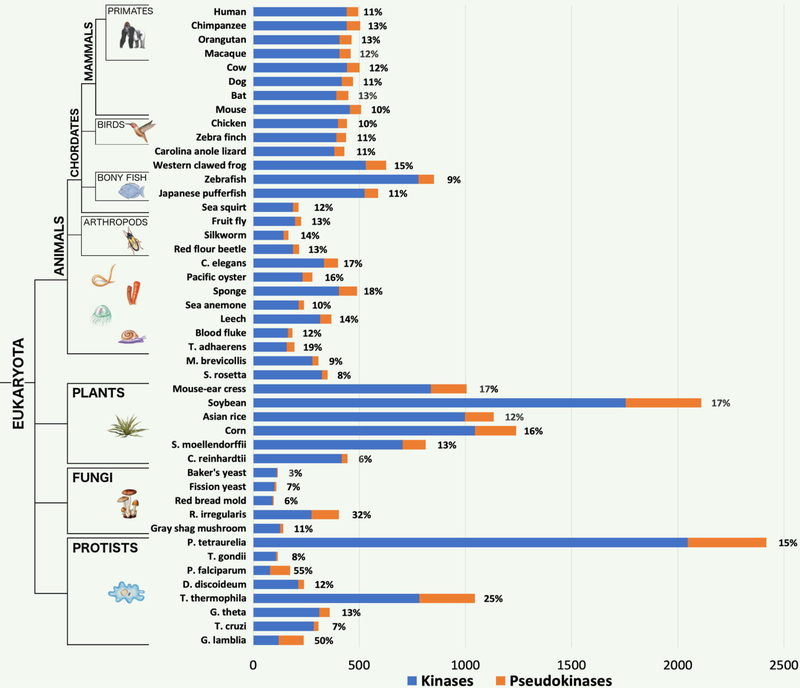
Fig. 1.
Kinome and pseudokinome sizes evaluated in 46 eukaryotic species. Blue bars represent the number of kinases detected in the proteome of each species, and orange bars represent the number of pseudokinases. Percentages indicate the fraction of kinases from each proteome that were determined to be pseudokinases. The tree on the left indicates major evolutionary kingdoms and phyla.
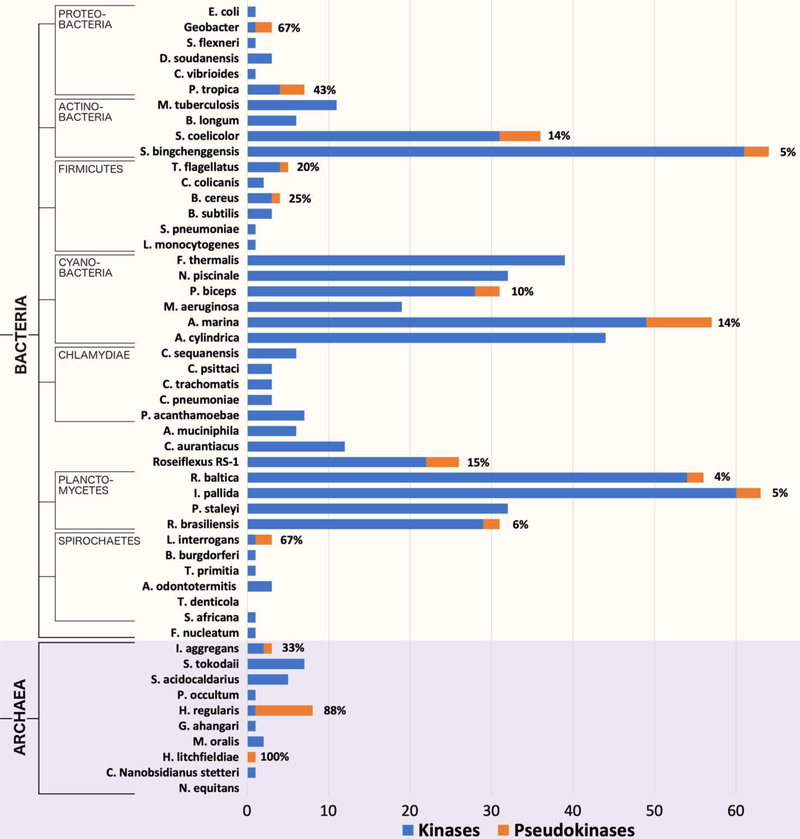
Fig. 2.
Kinome and pseudokinome sizes evaluated in 51 bacterial and archaeal species. Blue bars represent the number of kinases detected in the proteome of each species, and orange bars represent the number of pseudokinases. Percentages indicate the fraction of kinases from each proteome that were determined to be pseudokinases. The tree on the left indicates major evolutionary kingdoms and phyla.
Pseudokinase complements of kinomes (hereafter referred to as pseudokinomes) are nearly always proportional to the size of the kinome in vertebrate species (Fig. 1), which is consistent with previous estimates that pseudokinases generally account for ~10% of kinome content (18). However, both kinome and pseudokinome sizes are much more diverse across other (non-vertebrate) eukaryotic clades. For example, pseudokinase sequences account for between 8–17% of plant kinomes, which are often drastically expanded in size when compared to metazoan kinomes. Moreover, a mycorrhizal fungal species, Rhizophagus irregularis, and two protist species, Paramecium tetraurelia and Tetrahymena thermophila, have significantly expanded kinomes respective to other fungi and protists, and the pseudokinomes of R. irregularis and T. thermophila are also markedly expanded, comprising 32% and 25% of each kinome respectively. We also note a remarkable expansion of pseudokinases in eukaryotic pathogens, including Plasmodium falciparum and Giardia lamblia, which possess relatively small kinomes (16, 62), but contain highly expanded pseudokinomes that account for more than a half of their kinomes (Fig. 1).
Also unprecedented was the varied detection of pseudokinases across diverse bacterial phyla with high sequence similarity to pknB kinases (Fig. 2). Bacterial kinomes and pseudokinomes exhibit a large amount of diversity in size, particularly when compared to those in eukaryotes. For example, we note the large expansion of pseudokinases in Streptomyces coelicolor, which has 31 protein kinases and 5 pseudokinases, whereas the proteomes of Shigella flexneri and Escherichia coli like most bacterial proteomes lack pseudokinases, containing only 1 protein kinase sequence each. In contrast, the proteomes of some bacterial species, including Treponema denticola, do not contain any detectable protein kinase sequences. Nonetheless, pseudokinases were detected in at least one species for every bacterial phylum that we examined, except in Chlamydiae, suggesting that pseudokinases are not confined to any specific bacterial classifications such as gram-negative or gram-positive bacteria, but rather are found across diverse bacterial phyla.
Remarkably, a small number of ePK sequences were also detected in archaea (Fig. 2, Table S1), where 11 archaeal proteomes contained putative pseudokinase sequences. In particular, while most of these archaeal proteomes contained only one or two pseudokinase sequences, 7 out of the 8 ePK sequences detected in Halorientalis regularis were identified as pseudokinases. Sequences and alignments for the kinomes and pseudokinomes of every organism represented in Figures 1 and 2 are provided in the Supplementary Material (Data file S1). Additionally, the number of ePK and pseudokinase sequences detected in the 10,092 proteomes currently available in the UniProt reference proteome database is provided in Table S1.
Classification of pseudokinase sequences
We next classified pseudokinase sequences into evolutionarily related clusters using an optimal multiple-category Bayesian Partitioning with Pattern Selection (omcBPPS) algorithm, which classifies sequences based on patterns of amino acid conservation and variation in a large multiple sequence alignment (see methods) (63, 64). Due to the large number of pseudokinase sequences analyzed, we first classified a diverse representative set of 26,273 pseudokinase sequences (see methods for details). Some of the well-known metazoan pseudokinase families such as the JAK and TRIB families were found to fall into distinct pseudokinase sequence clusters (26). The identified pseudokinase clusters were incorporated within an existing evolutionary hierarchy of kinase groups and families (18) (see methods), and a resulting hierarchy of 592 sequence profiles containing both canonical kinase families and pseudokinase families were used to detect, classify, and align all pseudokinase sequences from the UniProt reference proteome database.
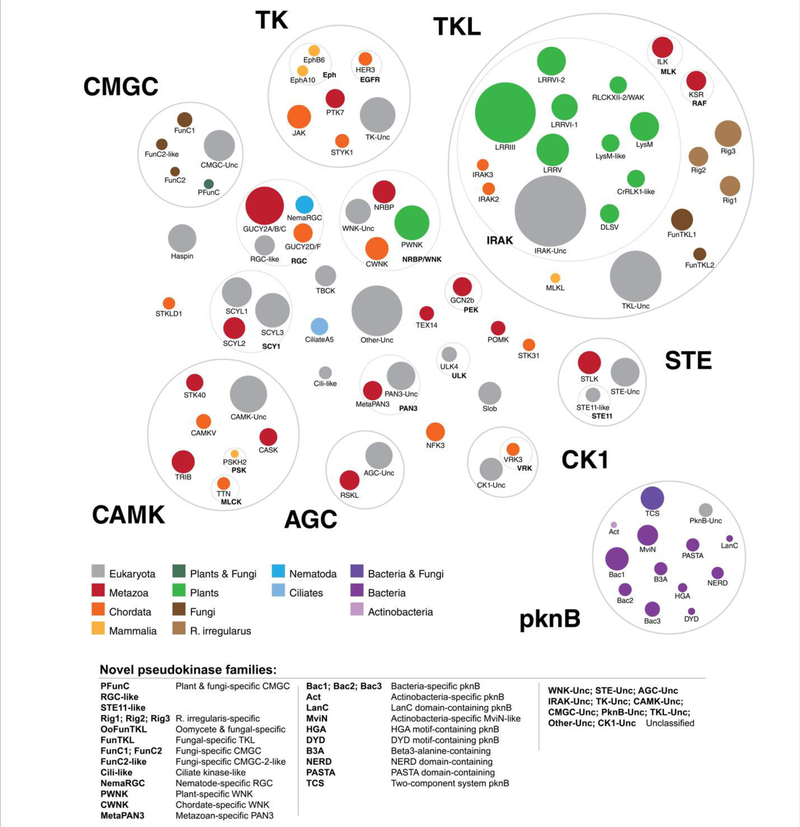
Overall, we detected pseudokinase families across all major kinase groups (Fig. 3). Whereas some kinase groups defined by Manning and colleagues (18), such as the STE and AGC groups, possess relatively few pseudokinases, the Tyrosine Kinase-Like (TKL) group is highly enriched with pseudokinases. This increase is caused by the diversification of TKL pseudokinase sequences in various plant and fungal species, where the general expansion of canonical TKL kinases has previously been noted (52, 65). We also identify species-specific divergence of pseudokinase sequences, such as the divergence of metazoan-specific poly(A)-nuclease deadenylation complex subunit 3 (PAN3) from other eukaryotic PAN3, and the diversification of plant WNK kinases from the classical chordate WNK kinases. In contrast, some pseudokinase clusters comprise orthologs found across diverse taxonomic groups, such as Haspin, which is found across diverse eukaryotes, and the TRIB family of pseudokinases, which is found across diverse metazoa, where they function as regulators of protein ubiquitylation (26). Detailed taxonomic annotations for each pseudokinase family are provided in Table S2.
Fig. 3.
A new classification of pseudokinase families. Each colored circle represents a distinct pseudokinase family, which is colored according to the taxonomic group(s) in which it is found. Kinase groups and families from the human kinome classification (18) are depicted by gray circles and labeled in bold font.
Notably, using our integrated evolutionary hierarchy of pseudokinase clusters with previously established kinase groups and families, we can now identify some canonical kinase sequences conserving the catalytic triad residues that classify into pseudokinase families. For example, four sequences containing the canonical HRD motif classified with the HER3 pseudokinases, which typically contain a conserved HRN motif that severely blunts catalysis. These sequences were identified to be HER3 orthologs in four separate rodent species (Mus musculus, Rattus norvegicus, Cricetulus griseus, and Mesocricetus auratus) (66). Similarly, while orthologs of the Kinase Suppressor of Ras (KSR) family can be detected across diverse metazoan species, only chordate KSR’s are classified as predicted pseudokinases due to the replacement of the β3-lysineby an arginine within the canonical ‘VAIK’ motif. In addition, we often identify large expansions of pseudokinase families in plant species that are accompanied by the presence of few canonical sequence members. For example, in black cottonwood (Populus trichocarpa), we identify one canonical PWNK member containing the β3-lysine(‘VAWK’ motif), along with 15 pseudokinase members containing a ‘VAWN’ motif characteristic of PWNK pseudokinases. The clustering of canonical sequences within pseudokinase families is summarized in Table S3, and alignments of these canonical sequences are provided in Data file S3.
Additionally, our in-depth analysis led to the identification of many ‘novel’ pseudokinase families, which are listed in Table 3. These include the plant lysin motif (LysM)-like pseudokinase family and the bacterial HGA motif-containing pknB pseudokinase family, as well as unclassified pseudokinase families that do not readily fall within the identified pseudokinase clusters (which we term the tyrosine kinase (TK) unclassified, TKL unclassified, STE unclassified, CK1 unclassified, AGC unclassified, CAMK unclassified, CMGC unclassified, PknB unclassified, and Other unclassified pseudokinase families) (Fig. 3). As noted previously, pseudokinases in the TKL group appear to have expanded significantly, particularly in non-metazoan species. Consistently, five distinct fungal-specific pseudokinase families have emerged in the TKL group, of which three (termed Rig 1–3) are currently found predominantly in the species R. irregularis, which has a significantly expanded kinome and pseudokinome compared to other fungi (Fig. 1 and Fig. S1). In addition, IRAK pseudokinases (part of the TKL group) are massively expanded in plants, which classify into 9 unique families. This mirrors the well-known plant-specific expansions of the canonical IRAK (also known as RLK/Pelle) kinase family, which have previously been classified into 65 subfamilies (52). A surprising finding from our analysis was the detection of over 1,200 putative bacterial PknB pseudokinases, which we have classified into 12 distinct clusters. Some bacterial pseudokinase clusters are specific to selected bacterial phyla, such as the Actinobacteria-specific pknB pseudokinase family (which we term Act), whereas other families such as the NERD domain-containing pknB pseudokinase family are found more broadly across diverse bacterial phyla. Sequences and alignments for each pseudokinase family identified here are available through the Protein Kinase Ontology (ProKinO, http://vulcan.cs.uga.edu/prokino/about/browser) and in the Supplementary Material (Data file S2). In the following sections, we build on the first comprehensive pseudokinase catalogue by examining the putative functions of plant IRAK pseudokinases and pseudokinases in R. irregularis and in sequenced bacteria.
Table 3. List of novel pseudokinase families.
Representative members of novel pseudokinase families are listed.
| PsK family | Kinase family |
Kinase group |
Example member(s) |
|---|---|---|---|
| TK-Unclassified | TK | Camponotus floridanus putative tyrosine-protein kinase Wsck (E2ACX7_CAMFO) | |
| LysM | IRAK | TKL | Arabidopsis thaliana LYK5/LysM-containing receptor-like kinase 5/At2g33580 (LYK5_ARATH) |
| LysM-Iike | IRAK | TKL | Arabidopsis thaliana Protein kinase superfamily protein/At3g57120 (Q8VYG5_ARATH) |
| DLSV | IRAK | TKL | Arabidopsis thaliana Cysteine-rich receptor-like protein kinase 45/Cysteine-rich RLK45/At4g11890 (CRK45_ARATH) |
| RLCKXII-2/WAK | IRAK | TKL | Arabidopsis thaliana Non-functional pseudokinase ZED1/hopZ-ETI-deficient 1/At3g57750 (ZED1_ARATH) |
| CrRLK1-Iike | IRAK | TKL | Cajanus cajan Receptor-like protein kinase ANXUR2/KK1_043523 (A0A151QYL0_CAJCA) |
| LRRIII | IRAK | TKL | Arabidopsis thaliana PRK1/Pollen receptor-like kinase 1/At5g35390 (PRK1_ARATH) |
| LRRV | IRAK | TKL | Arabidopsis thaliana SUB/STRUBBELIG/At1g11130 (SUB_ARATH) |
| LRRVI-1 | IRAK | TKL | Arabidopsis thaliana Leucine-rich repeat protein kinase family protein/MUA22.21/At5g14210 (A0A178US29_ARATH) |
| LRRVI-2 | IRAK | TKL | Arabidopsis thaliana MDIS1/Male discoverer 1/At5g45840 (MDIS1_ARATH) |
| Rig1 | TKL | Rhizophagus irregularis Rad53p/RirG_173180 (A0A015K1H5_9GLOM) | |
| Rig2 | TKL | Rhizophagus irregularis Skt5p/RirG_180750 (A0A015 ISZ5_9GLOM ) | |
| Rig3 | TKL | Rhizophagus irregularis Ssk22p/RirG_232660 (A0A015 IBX1_9GLOM ) | |
| FunTKL1 | TKL | Rhizoctonia solani Uncharacterized protein/RSAG8_12895 (A0A066UWY2_9HOMO) | |
| FunTKL2 | TKL | Penicillium subrubescens Uncharacterized protein/PENSUB_13048 (A0A1Q5SUF1_9EURO) | |
| IRAK-Unclassified | TKL | Arabidopsis thaliana BIR2/BAK1-interacting receptor-like kinase 2/At3g28450 (BIR2_ARATH) | |
| STE11-like | STE | Arabidopsis thaliana Protein kinase superfamily protein/At2g40580 (O22877_ARATH) | |
| STE-Unclassified | AGC | Homo sapiens Uncharacterized protein (A0A1B0GUL7_HUMAN); Pan troglodytes Uncharacterized protein (A0A2I3RK43_PANTR) | |
| AGC-Unclassified | AGC | Paramecium tetraurelia hypothetical protein/GSPATT00021006001 (A0DWE7_PARTE) | |
| CAMK-Unclassified | CAMK | Paramecium tetraurelia Uncharacterized protein/GSPATT00003535001 (A0E5V5_PARTE) | |
| PFunC | CMGC | Sorghum bicolor Uncharacterized protein/SORBI_3005G165300 (A0A1Z5RK15_SORBI) | |
| FunC1 | CMGC | Penicillium patulum Uncharacterized protein/PGRI_024730 (A0A135LI09_PENPA) | |
| FunC2 | CMGC | Penicillium subrubescens SRSF protein kinase 3/PENSUB_10992 (A0A1Q5T6W4_9EURO) | |
| FunC2-like | CMGC | Coprinopsis cinerea CMGC/SRPK protein kinase/CC1G_09673 (A8P9H1_COPC7) | |
| NemaRGC | RGC | Other | Caenorhabditis elegans Receptor-type guanylate cyclase gcy-1/AH6.1 (GCY1_CAEEL) |
| RGC-like | RGC | Other | Amphimedon queenslandica Guanylate cyclase (A0A1X7VH93_AMPQE) |
| CWNK | WNK | Other | Homo sapiens WNK1/Protein kinase with no lysine 1 (WNK1_HUMAN) |
| PWNK | WNK | Other | Arabidopsis thaliana AtWNK1/Protein kinase with no lysine 1/At3g04910 (WNK1_ARATH) |
| WNK-Unclassified | WNK | Other | Caenorhabditis elegans Serine/threonine-protein kinase WNK/Protein kinase with no lysine 1/C46C2.1 (WNK_CAEEL) |
| Cili-like | Other | Neurospora crassa Mitotic spindle checkpoint component mad3/NCU10043 (V5IRN2_NEUCR) | |
| MetaPAN3 | PknB | Homo sapiens PAN3/Poly(A)-nuclease deadenylation complex subunit 3 (PAN3_HUMAN) | |
| Bac1 | PknB | Streptomyces aurantiacus JA 4570 Uncharacterized protein/STRAU_1734 (S3ZNQ3_9ACTN) | |
| Bac2 | PknB | Frankia sp. EI5c Serine/threonine protein kinase /UG55_100447 (A0A166T1F0_9ACTN) | |
| Bac3 | PknB | Trichodesmium erythraeum (strain IMS101) Serine/threonine protein kinase with TPR repeats/Tery_4781 (Q10VJ1_TRIEI) | |
| Act | PknB | Streptomyces sp. NTK 937 Aminoglycoside phosphotransferase/DT87_12690 (A0A069K2D6_9ACTN) | |
| MviN | PknB | Mycobacterium shimoidei Transmembrane serine/threonine-protein kinase D PknD/STPK D/BHQ16_10175 (A0A1E3TG29_MYCSH) | |
| LanC | PknB | Streptomyces vietnamensis Uncharacterized protein /SVTN_34785 (A0A0B5I836_9ACTN) | |
| DYD | PknB | Frankia sp. (strain EANlpec) Nucleic acid binding OB-fold tRNA/helicase-type/Franean1_4264 (A8L9E5_FRASN) | |
| HGA | PknB | Actinosynnema mirum (strain ATCC 29888) Uncharacterized protein/Amir_3792 (C6WD53_ACTMD) | |
| B3A | PknB | Lentisphaera araneosa HTCC2155 Serine/threonine-protein kinase/LNTAR_17493 (A6DFI7_9BACT) | |
| PASTA | PknB | Firmicutes bacterium CAG:94 PASTA domain protein/BN815_00934 (R6ZDX7_9FIRM) | |
| NERD | PknB | Singulisphaera acidiphila (strain ATCC BAA-1392) Nuclease-like protein/protein kinase family protein/Sinac_1959 (L0DAB9_SINAD) | |
| TCS | PknB | Rhodopirellula islandica Two-component signal transduction/RISK_006087 (A0A0J1B5P6_RHOIS) |
A massive, plant-specific IRAK pseudokinase expansion
The plant IRAK kinases have been previously classified into 65 subfamilies (52). We now identify 9 unique IRAK pseudokinase families (Fig. 3), which are conserved across multiple plant species (Table S2), including two pseudokinase families that resemble the lysin-motif receptor-like (LysM RLK) kinases, termed LysM and LysM-like, and the leucine-rich repeat receptor-like (LRR RLK) pseudokinase families, termed LRRIII, LRRV, LRRV1–1, and LRRVI-2. In addition, we define three “mixed” pseudokinase families that contain domains homologous to multiple previously defined IRAK subfamilies, such as RLCKXII-2/WAK and the CrRLK1-like family, which are summarized in Table S2. The widespread distribution of these new plant pseudokinase families across diverse plant species is summarized in Table S4.
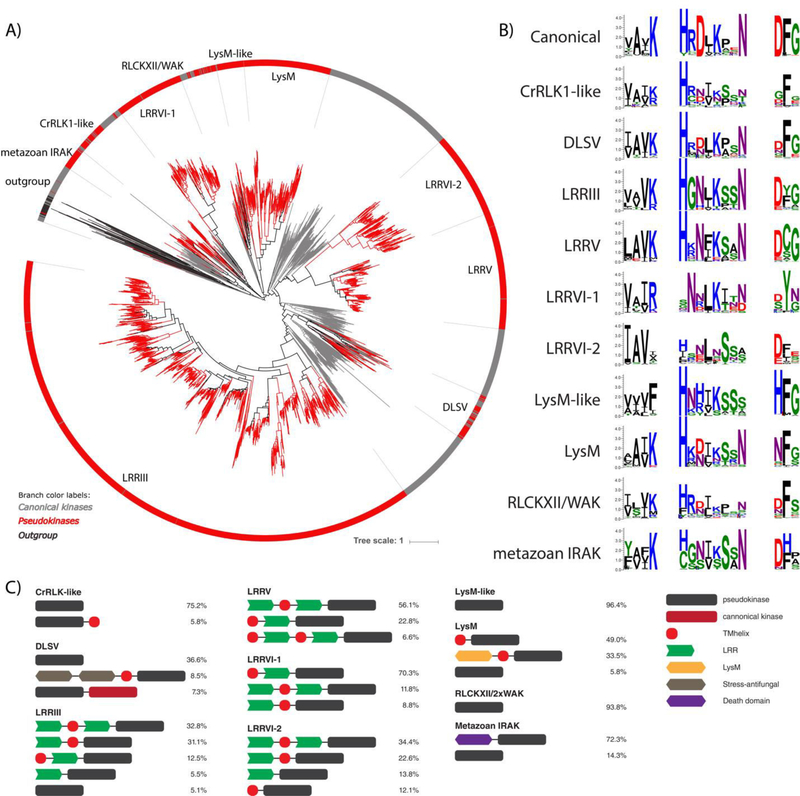
To specifically examine the evolutionary expansion of IRAK pseudokinases in plants, we constructed a rooted phylogenetic tree of both canonical and pseudokinase IRAK sequences using diverse non-IRAK TKL sequences and non-TKL sequences as an outgroup. The plant IRAK pseudokinase families form distinct clades in the phylogenetic tree that clearly distinguish them from metazoan IRAK pseudokinase sequences (Fig. 4A). In addition, metazoan IRAK members are cytoplasmic and typically contain a death domain N-terminal to the kinase domain (Fig. 4C), which is not observed in any plant IRAK kinases. We note that the three mixed pseudokinase families we identified (RLCKXII-2/WAK, CrRLKl-like, DLSV) form distinct monophyletic clades in the phylogenetic tree, indicating close homology and common descent, despite their homology to multiple IRAK subfamilies. In addition, the IRAK pseudokinases are generally divergent from canonical plant IRAK members, as shown by the long branch lengths separating the pseudokinase family clades from the canonical IRAK sequences (red lines) (Fig. 4A). In some pseudokinase families, several canonical sequences cluster within pseudokinase family clades (i.e. RLCKXII-2/WAK, CrRLKl-like, DLSV, and LRRIII, and LRRV families), indicating that these pseudokinase families also have very close, likely catalytically active, homologs.
Fig. 4.
Plant-specific IRAK pseudokinase families. (A) Phylogenetic tree of catalytically active and pseudokinase members of the IRAK family. The 9 plant IRAK pseudokinase families are labeled, and IRAK pseudokinase sequences are shown in red. Canonical IRAK sequences are shown in gray. Outgroup sequences are shown in black. (B) Sequence logos of catalytic motifs for IRAK pseudokinase families. (C) Unique domain structures observed in plant IRAK pseudokinase families. The most common domain structures observed in each family are shown (occurring >5%), with frequencies of each domain structure indicated.
We next examined the relative level of degradation of the canonical catalytic motifs and the domain organizations of the major IRAK pseudokinase families, allowing us to expand on the potential functions of some IRAK pseudokinases. The LysM pseudokinase family shares sequence homology in the kinase domain with known catalytically active LysM RLKs and conserves a similar domain architecture, which is characterized by an intracellular kinase domain, and one or more extracellular LysM domains (Fig. 4C). In plants, LysM RLKs play diverse sensing functions, recognizing chitin oligosaccharides in plant defense responses towards fungi (67), as well as binding peptidoglycans on bacterial cell walls to aid recognition of symbiotic bacteria (68). Recent studies of two LysM pseudokinases, MtNFP and LjNFR5 from Medicago, demonstrate the importance MtNFP and LjNFR5 during the Rhizobium pre-infection response and to the specificity of Rhizobium-legume symbiosis, respectively. Despite their lack of catalytic activity, these LysM pseudokinases are believed to contribute to their appropriate signaling pathways via interactions with other active LysM RLKs (69). We also detect another distinct family, which we term the LysM-like pseudokinase family, which shares sequence homology in the kinase domain with active LysM RLKs, but lacks LysM and transmembrane domains, suggesting a uniquely cytoplasmic function for this family (Fig. 4C). Moreover, the LysM and LysM-like families share different patterns of residue conservation, as identified in our pattern-based classification, and they form distinct clades in the phylogenetic tree (Fig. 4A), suggesting that they likely have divergent functions.
The LRRV pseudokinase family comprises the Strubbelig receptor family, which consists solely of pseudokinases, with 9 members in Arabidopsis thaliana alone (70). The best characterized member of this family, Strubbelig (SUB), plays roles in organ development and cellular morphogenesis, however, the mechanism by which it contributes to cell signaling despite a lack of catalytic activity, and the functions of other LRRV members are not yet known (70, 71). While the domain organizations for most IRAK pseudokinase families such as LRRV are rather well conserved, pseudokinases in the DLSV family possess diverse domain architectures, indicating that a common pseudokinase domain has co-evolved with a variety of different protein domains in order to diversify biological function. For example, DLSV pseudokinases homologous to the DUF26 IRAK subfamily contain extracellular domains associated with salt-stress and antifungal responses (72, 73) (Fig. 4C). Although DUF26 kinases have been associated with plant-specific functions in ROS/redox signaling and stress adaptation (74), to our knowledge, pseudokinase complements of DUF26 kinases have not been described previously. Other DLSV pseudokinases exhibit tandem pseudokinase and canonical kinase domains (Fig. 4C). Previously described IRAK pseudokinases in plants include MDIS2 (MRH1) (75), ZED1 (51), RSK1 (76), BSK8 (77), BIR2 (49), SOBIR1 (78), and CRN (79), and their placement in the expanded IRAK pseudokinase classification is described in Table S5.
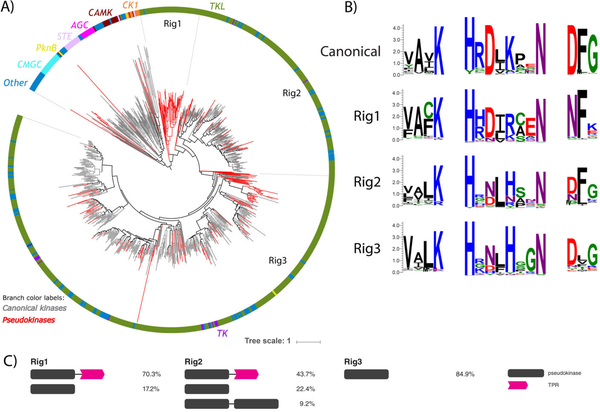
Expansion of TKL pseudokinases in Rhizophagus irregularis
We identified and analyzed 3 novel fungal pseudokinase families (termed Rig1, Rig2, and Rig3), which are currently comprised predominantly of sequences from a single species, the commercially important soil inoculant Rhizophagus irregularis (Fig. 3). This organism has a highly expanded proteome when compared to other fungal species, including an expanded kinome (65, 80). Notably, the R. irregularis fungal kinome comprises ~2.6% of the entire proteome, a larger proportion than is observed for any other fungal kinome analyzed (Fig. S1). Even more remarkably, 32% of these kinases possess pseudokinase domains (Fig. 1) that most closely resemble the TKL kinases. Interestingly, several lines of evidence suggest that the genes coding these pseudokinases are expressed at the protein level (65, 81), suggesting that protein pseudokinases must contribute to an important biological function in R. irregularis.
Sequence comparisons to known TKL kinase families demonstrated that R. irregularis-specific TKL kinases are divergent from canonical TKL families and are most homologous to the leucine rich repeat kinase (LRRK) family of TKL’s. LRRK’s are intracellular kinases distinct from IRAK LRR RLK’s and are conserved in metazoans and expanded in the slime-mold Dictyostelium discoidium, but are otherwise absent in fungi. In order to understand the evolutionary events that led to the expansion of these novel pseudokinase families, we conducted a phylogenetic analysis of the entire R. irregularis kinome, which, like the human kinome (18), consists of 7 major ePK groups. R. irregularis kinase sequences from the AGC, CMGC, STE, CAMK, and CK1 groups form mostly separate monophyletic clades in a phylogenetic tree of the R. irregularis kinome, whereas TK sequences are clustered within various groupings of TKL’s (Fig. 5A). Interestingly, the R. irregularis kinome additionally includes pknB-like kinase sequences, which are typically associated with bacteria. However, as reflected by the phylogenetic tree (Fig. 5A), TKL sequences comprise the majority of the R. irregularis kinome. Thus, the expanded kinome of R. irregularis can be attributed to a substantial expansion amongst TKL kinases. Furthermore, we found that 166 of the 183 pseudokinases (90%) identified in R. irregularis are TKL-like, indicating that R. irregularis pseudokinases emerged primarily from within the TKL group. Of the three distinct pseudokinases families, Rig1 forms a monophyletic group comprised entirely of pseudokinases, whereas Rig2 and Rig3 cluster into clades including both pseudokinase and active kinase members. Rig2 and Rig3 are the most diverse families in R. irregularis, and both cluster with canonical sequences from the TK and “Other” groups, based on the human kinome classification (18).
Fig. 5.
Rhizophagus irregularis-specific TKL pseudokinase families. (A) Phylogenetic tree of the R. irregularis kinome. Canonical kinase branches are colored in gray and pseudokinases in red. Major kinase groups are labelled using different colors in the outer circle. The 3 major R. irregularis specific pseudokinase families are labelled as Rig1, Rig2 and Rig3. (B) Sequence logos of catalytic motifs for Rig1, Rig2, and Rig3 pseudokinase families. (C) The most common domain structures observed in Rig pseudokinase families are shown (occurring >5%), with frequencies of each domain structure indicated.
Further analysis of the domain organization in these pseudokinase sequences revealed that Rig 1 and Rig2 pseudokinases often have an additional putative tetratricopeptide (TPR) domain C-terminal to the pseudokinase domain, whereas Rig3 pseudokinases are single domain pseudokinases (Fig. 5C). TPR repeats are short structural motifs that are classically involved in mediating protein-protein interactions crucial for cell signaling. Apart from the TPR repeat regions, no additional domains were found in the pseudokinase members of the 3 clades.
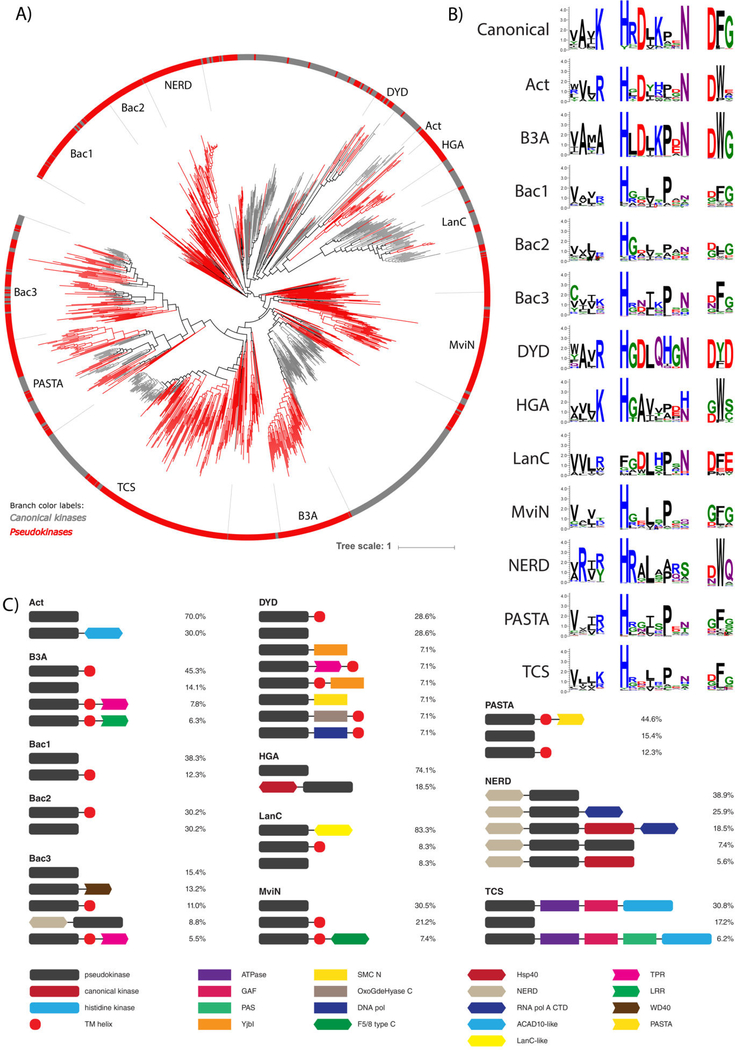
PknB-like pseudokinases conserved in bacteria
We identified 12 unique families of bacterial pseudokinases that are most closely related to the pknB group of canonical prokaryotic protein kinases (Fig. 3) and have been classified and named based on their conserved domains, taxonomic specificity, and/or their similarity to previously identified kinases. Three bacterial pseudokinase families (DYD, HGA, B3A) were named based on the unique conservation of noncanonical catalytic motifs. In order to examine the evolutionary relationships between these families, we built a phylogenetic tree of the 1,145 pknB pseudokinase sequences identified in this study combined with a representative set of canonical pknB kinases (Fig. 6A). Each bacterial pseudokinase family falls into a distinct clade and mostly segregates away from canonical kinase sequences.
Fig. 6.
Bacterial pknB pseudokinase families. (A) Phylogenetic tree of pknB canonical kinases and pseudokinases. The 12 pknB-related pseudokinase families are labeled and shown in red branches. Representative canonical pknB kinases are shown in gray. (B) Sequence logos of catalytic motifs in pknB pseudokinase families. (C) Unique domain structures observed in bacterial pseudokinase families. The most common domain structures observed in each family are shown (occurring >5%), with percentages indicating the frequency of each domain structure.
Analysis of the catalytic motifs for each pknB pseudokinase family shows that different pknB pseudokinase families diverge in the canonical catalytic motifs in a variety of ways (Fig. 7B). For example, the MviN and PASTA families exhibit the most extreme degeneration of catalytic residues and lack all three canonical residues associated with catalysis (β3-lysine, HRD-aspartate, DFG-aspartate), in addition to the conserved magnesium-binding asparagine located at the end of the catalytic loop. Conversely, many bacterial pseudokinase families appear to conserve the catalytic aspartates of the HRD and DFG motifs, as well as the magnesium-binding asparagine residue; this is the case for the Act, DYD, B3A, and LanC pseudokinase families. Of these, the Act, DYD, and LanC families all possess a chemically comparable arginine residue in place of the ATP-binding β3-lysine, and thus might still retain ATP-binding and catalytic functions, as demonstrated for canonical kinases such as Aurora A (82).
Fig. 7.
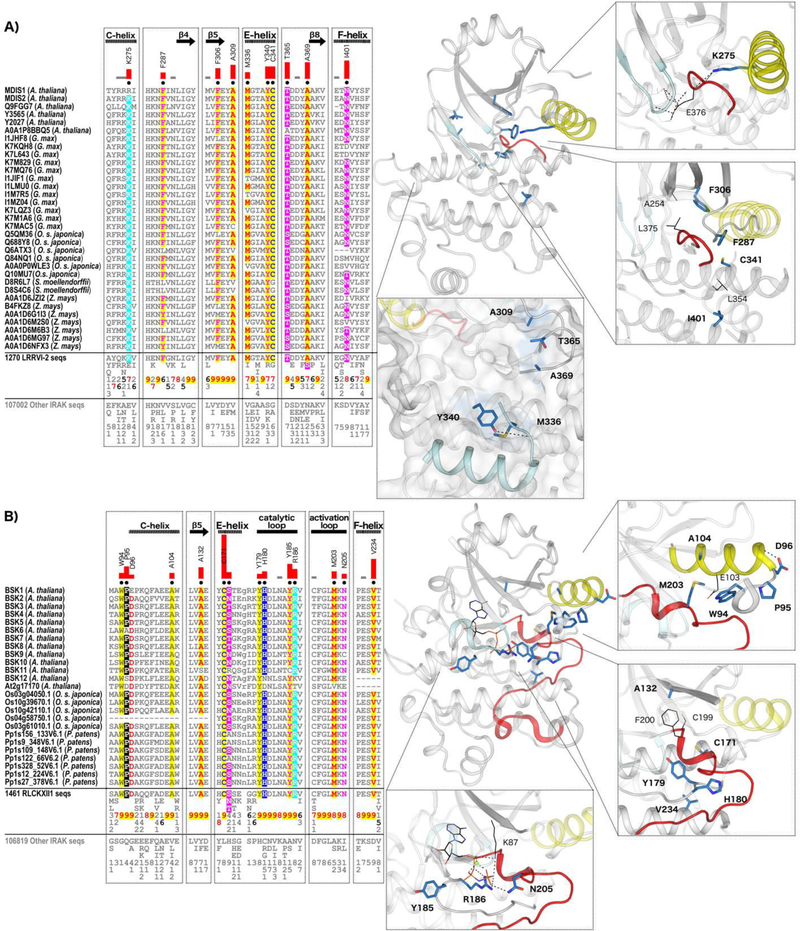
IRAK pseudokinase-specific features contribute to unique conformations in key catalytic regions. (A) LRRVI-2 pseudokinase family-specific sequence motifs. In the alignment, columns are highlighted where amino acids are highly conserved in LRRVI-2 pseudokinase family sequences and non-conserved and/or biochemically dissimilar in other IRAK sequences. Red bar lengths quantify the degree of divergence between LRRVI-2 and other IRAK sequences. Column-wise amino acid and insertion/deletion frequencies are indicated in integer tenths where a “5” indicates an occurrence of 50–60% in the given (weighted) sequence set. Columns used by the Bayesian partitioning procedure to sort LRRVI-2 sequences from other IRAK sequences are marked with black dots. Kinase secondary structures are annotated above the alignment. In the structure, the glycine-rich loop is colored in light cyan, the C-helix in yellow, and the activation loop in red. Family-specific residues are shown in blue sticks. Residues occurring in canonical catalytic motifs are shown in black lines. Hydrogen bonds are shown in dashed black lines. (B) RLCKXII-1 pseudokinase family-specific sequence motifs.
Analysis of the common domain organizations of bacterial pknB pseudokinase families reveals that, like eukaryotic pseudokinases, bacterial pseudokinases also co-occur with protein signaling domains involved in a wide range of biological functions (Fig. 6C). Two families in particular, termed NERD and TCS, have highly conserved domain architectures of particular interest. For example, the domain architecture of NERD is characterized by an N-terminal NERD domain, which is predicted to function as a nuclease (83), followed by a pknB pseudokinase domain and often a C-terminal canonical (catalytically active) pknB kinase domain. This multi-domain architecture is also observed in PglW, a constituent of the Streptomyces coelicolor A(3)2 phage growth limitation system (84). The active kinase domain of the PglW is catalytically competent (85), whereas the function of the pseudokinase domain remains unknown. Additionally, we identified putative protein domains that resemble the C-terminal domain of the bacterial RNA polymerase α-subunit in ~45% of the NERD family members; this domain classically functions in DNA binding and protein-protein interactions in bacterial RNA polymerases (86). The co-occurence of these nucleic acid-associated domains with pseudokinase domains suggests that the NERD pseudokinase family may be involved in signaling pathways relevant to transcriptional regulation. We also note the unique domain structure within the TCS family, which has clear orthologs in both bacteria and fungi (Fig. 3, Table S2) and contains a pseudokinase domain that often co-occurs with other protein domains typically found in two-component signaling systems involving histidine and aspartate phosphorylation. Nearly 40% of TCS family pseudokinases contain an N-terminal pseudokinase domain followed by an ATPase domain, a GAF-domain, and a C-terminal histidine kinase domain (Fig. 6C). This domain architecture is reminiscent of two-component system proteins that have been previously identified in bacterial species from the Cyanobacteria, Proteobacteria, and Spirochaete phyla, as well as fungal species such as Candida albicans and Schizosaccharomyces pombe (87–92). These proteins have been associated with a wide range of functions, including nitrogen metabolism and glycolipid synthesis in Anabaena sp. PCC7120, hyphal development and pathogenicity in C. albicans, and cell cycle regulation and oxidative stress response in S.pombe (87, 91, 93–101). To our knowledge, our analysis is the first to reveal pseudokinase domains that are likely to be associated with two-component signaling in bacteria.
Sequence and structural basis for pseudokinase evolutionary divergence: a case study on two plant IRAK pseudokinase families
Conformational flexibility in pseudokinases has permitted the structurally conserved ancestral protein kinase fold to be ‘re-purposed’ for multiple cellular signaling roles, including the evolution of new ways through which to bind and modulate cellular targets. Using the LRRVI-2 and RLCKXII-l pseudokinase families as examples, we evaluated how evolution has constrained sequences in different pseudokinase families to disrupt canonical ATP-binding and constrain kinase domain conformations in a multitude of ways that serve to abolish catalytic activity. This leads us to propose novel molecular functions that may have evolved in LRRVI-2 pseudokinases through the selection of unique motifs on the surface of the protein.
The LRRVI-2 pseudokinase family includes the previously described pseudokinase, MDIS2, which interacts with the plant potassium channel AKT2 (75) associated with root hair formation (102). However, little is known about the molecular functions of other pseudokinases in this family. Using the crystal structure of a LRRVI-2 pseudokinase from Zea mays (GRMZM2G135359), we examined the structural role of LRRVI-2 specific motifs. ATP binding in the GRMZM2G135359 structure appears to be completely inhibited due to the complete obstruction of the ATP-binding site by the activation loop. This activation loop conformation is stabilized by a LRRVI-2-specific lysine (Lys 275) that replaces the ATP-binding C-helix glutamate (Fig. 7A) and hydrogen bonds to the backbone of the activation loop. Notably, a glutamate in the activation loop “DLE” motif, which replaces the canonical DFG motif, hydrogen bonds to the glycine rich loop to occlude ATP binding. The inhibitory activation loop conformation is additionally stabilized by hydrophobic interactions between LRRVI-2-specific residues including a phenylalanine in the gatekeeper position on the β5 strand (Phe306), a phenylalanine in the αC-β4 loop (Phe287), and a cysteine in the E-helix (Cys341). These residues form hydrophobic interactions with Ala254, which replaces the ATP-binding β3-lysine, and with Leu375, which replaces the DFG-phenylalanine. Together these hydrophobic interactions appear to stabilize the C-helix in an inactive, outward conformation and the activation loop in an autoinhibitory conformation that occludes ATP-binding. In addition, a LRRVI-2 family-specific asparagine replaces the canonical F-helix aspartate, which typically participates in a switch-like mechanism and stabilizes the active conformation of the kinase domain by forming key hydrogen bonds with the catalytic loop backbone (103).
The F-helix aspartate is typically conserved as an asparagine in LRRVI-2 pseudokinases, although the GRMZM2G135359 structure has a hydrophobic isoleucine at this position, and other LRRVI-2 members are noted to have a valine, methionine or aspartate (Fig. 7A). Mutation of the F-helix-aspartate residue to an asparagine or a leucine in canonical kinases such as Aurora A abrogates activity (103), thus, substitution of the F-helix-aspartate to an asparagine or hydrophobic residue is predicted to inactivate LRRVI-2 pseudokinases. Likewise, variations in the catalytic loop (replacement of the HRD motif by LRN motif) may contribute to the observed inactive structural conformation in LRRVI-2. In addition, we found that several LRRVI-2 family-specific motifs occur well outside of the catalytically important regions, including the surface of the protein near the N- and C-terminal tails. We note one such example in the GRMZM2G135359 structure, where LRRVI-2-specific residues appear to dock the I-helix and C-terminal tail onto the backside of the kinase domain. Specifically, a methionine (Met336) and a tyrosine (Tyr340) on the E-helix tether the C-tail through hydrophobic and hydrogen bonding interactions, respectively (Fig. 7A). Another cluster of LRRVI-2-specific, surface-exposed residues (A309, T365, A369) may also participate in tethering the C-tail and extend this interaction to the hinge region of the kinase domain.
To further investigate the evolutionary basis for IRAK pseudokinase functional specialization, we next quantified the evolutionary constraints imposed on the RLCKXII-1 family of pseudokinases, which comprise the brassinosteroid signaling kinases (BSKs). BSKs are involved in regulating plant growth and physiology in response to brassinosteroid hormone signals. The crystal structure of BSK8 has been determined (77) and shown to bind the non-hydrolzyable ATP analog AMP-PNP, despite the atypical DFG motif (CFG in BSK8) conformation. In addition to an unusual glycine-rich loop structure and the presence of a conserved small amino acid at the gatekeeper position (Ala132) (77), our studies also reveal family-specific variations in both the active site (Tyr185, Arg186, Asn205) and in allosteric regions such as the F-helix-aspartate (Val234), which provide clues to the unusual mode of AMP-PNP binding in RLCKXII-1 (Fig. 7B). Family-specific replacement of the canonical HRD-motif histidine and arginine in the catalytic loop (Tyr179and His180) facilitates unique hydrogen bonding and hydrophobic interactions that stabilize the activation loop in a unique inactive conformation (Fig. 7B). Other RLCKXII-1 features contribute to unique inactive conformations of the C-helix via hydrophobic packing interactions with the ATP-binding C-helix glutamate (Glu103, Ala104, Met203, and Trp94) and by promoting a unique secondary structure of the β3-αC loop and C-helix (Pro95 and Asp96). These findings add to the seemingly limitless ways in which ePK superfamily members can evolve new sequence features to affect kinase conformations and to ultimately modulate signaling outputs from the kinase domain.
Discussion
Using a large-scale bioinformatics analysis, we have significantly expanded the classification of pseudokinases, which are found to exist in diverse living organisms. We identified a total of 86 putative pseudokinase families and demonstrated that pseudokinases are present across the tree of life. Our analysis strongly suggests that pseudokinases are polyphyletic, emerging through numerous events during the course of protein kinase evolution, presumably for different biological niches and signaling roles through non-catalytic functions, the broad extent of which is revealed by our analysis.
Pseudokinases have evolved in all the major ePK groups, however, the TKL group is particularly enriched with pseudokinases, largely due to expansions of TKL kinases in plants and fungi. These TKL group expansions occur in the IRAK family in plants, whereas TKL expansions in fungi, including R. irregularis, comprise distinct pseudokinase families unrelated to any known TKL families in other organisms, corroborating previous descriptions of ‘unclassifiable’ kinases in fungi (104, 105). Why have TKL’s in particular been selected during evolution for such marked kinome and pseudokinome expansions in both plants and fungi? One possible explanation is the lack of TKs in these organisms, which, in metazoa, evolved and duplicated to play crucial phosphotyrosine-dependent roles in multicellular signal transduction (106). Whereas tyrosine kinases comprise most of the receptor protein kinase repertoire in metazoans, the IRAK family of TKL’s comprise a receptor kinase-like repertoire in plants, and thus the expansion of IRAK kinases and pseudokinases in plants may be analogous to the expansion of receptor tyrosine kinases in metazoa. In line with this view, LysM pseudokinases in Medicago are believed to contribute to Rhizobium interactions by interacting with active LysM RLK members, which is reminiscent of metazoan tyrosine kinases and tyrosine pseudokinases, such as the HER3 pseudokinase which allosterically modulates closely related, canonical EGFR family members (36, 69). Thus, the expansion of IRAK pseudokinases in plants mirrors the expansion of canonical IRAK kinases, perhaps due to regulatory interactions between co-evolved kinases and pseudokinases. Mechanistically, plant IRAK kinases are known to play vital roles in plant-fungi interactions, both during symbiotic interactions as well as during pathogen defense, suggesting that the expansion of fungal TKL kinases and pseudokinases may have arisen due to a close symbiotic co-evolution. R. irregularis participates in arbuscular mycorrhizal symbiotic relationships with more than two-thirds of all known plant species (65), and recent studies have evaluated the roles of the expanded R. irregularis proteome (107) as well as its kinome (81, 108) in this symbiotic relationship. Nevertheless, additional investigation of the contribution of R. irregularis pseudokinases in plant-fungal symbiosis is warranted. Furthermore, an understanding of how the symbiotic or infectious nature of host-pathogen interactions operate in the context of bacterial and eukaryotic pseudokinomes is likely to yield important information explaining how such relationships emerged and were propagated during the cellular ‘wiring’ of both physiological and pathological signaling pathways. As such, the prevalence of pseudokinases in some pathogenic protists such as Plasmodium falciparum and Giardia lamblia and in some bacteria suggest possible roles in pathogenicity. An understanding of the extent and biological niches for symbiotic and pathogenic pseudokinases will also create further opportunities for pseudokinase-based targeting with small molecules in the future.
Examining the clustering of canonical kinase sequences within pseudokinase families suggests that some pseudokinase families include very closely-related, and potentially catalytically-active, amino acid sequences (Table S3). While it is currently believed that pseudokinases most likely evolved from gene-duplicated canonical kinases (33), the clustering of canonical sequences within pseudokinase families does not rule out the possibility that some catalytically-active kinases might have evolved from ‘pseudokinases’, or other poorly defined ancestral non-enzymatic proteins, as recently reported for other pseudoenzymes (109–111). Indeed, some pseudokinase families have quite subtle chemical substitutions at the catalytic triad positions and might therefore be poised to ‘revert’ to a catalytically-active enzyme in response to a random mutagenic event or appropriate evolutionary pressure. An artificially-guided evolutionary pathway for reversion has been demonstrated for the relatively well-understood pseudokinase CASK, with five steps required to regenerate catalytic activity comparable to a canonical CAMK homolog (112). Likewise, pseudokinase families such as Rig3, HER3, STKLD1, and PSKH2 have conserved all canonical catalytic motifs except for the HRD-aspartate, which is normally substituted to asparagine; canonical members are likely to have evolved in these families by a simple substitution ‘back’ to the canonical HRD-aspartate. Alternatively, kinases can retain low (or very low) levels of catalytic activity even with aspartate-to-asparagine substitutions in the HRD motif (36, 113, 114), suggesting that pseudokinase families with few relatively benign catalytic motif substitutions may sometimes represent low activity kinases with the capacity for signaling, and may explain why canonical sequences sometimes cluster within pseudokinase families. A case for latent catalytic activity can also be made for pseudokinase families such as Act, DYD, and LanC, which conserve the catalytic triad residues except for a benign lysine-to-arginine substitution in the B3-strand. Nevertheless, the detection of pseudokinase families that have been evolutionarily retained across diverse species suggest that these predicted pseudokinases cannot be mere ‘remnants of evolution’, but rather that they play important biological roles, either through non-canonical, enzyme-based signaling or, in the majority of cases, via non-catalytic functions that await discovery.
Pseudokinases are likely to have evolved due to a relaxed constraint on usually invariant catalytic residues. However, in this study, our examination of the conserved sequence motifs associated with pseudokinase evolutionary divergence reveals variations not only in catalytic motifs, but also in conserved ‘non-catalytic’ regions distal from the pseudo-active site (Fig. 5). For example, the observation of pseudokinase family-specific variations at the highly conserved F-helix aspartate suggests that this allosteric region is indeed important for kinase domain activation, as proposed in previous work (103, 115). This region of the pseudokinase domain, which lies far away from the catalytic machinery, has been refashioned to perturb catalytic activity, in a similar fashion to the classical catalytic triad residues of ePKs that originally-defined pseudokinases. In addition, by comparing two plant IRAK pseudokinase families, we found that inactive kinase domain conformations are stabilized in divergent ways between different pseudokinase families through distinct sets of sequence motifs that have been selectively constrained during evolution. The conservation of pseudokinase family-specific motifs on the surface of the kinase domain further suggest that pseudokinase families have evolved novel interactions with other proteins domains or with flexible linker regions in their ‘substrates’. To evidence this, we identified a patch of LRRVI-2 family-specific residues on the surface of the pseudokinase domain that helps tether the C-terminal tail. In terms of regulation, one of the tethering residues is a tyrosine, whose phosphorylation could impart a switch-like function to alter tethering of the C-tail, similar to that observed in the SRC family kinases (116), or for recruiting tyrosine phosphorylated proteins via the binding of SH2 domains (1, 117). Regulatory tyrosine phosphorylation by dual-specificity LRR RLK’s has recently been recognized in plants, where it likely represents a fundamental signaling role despite the lack of conventional tyrosine kinases encoded in plant kinomes (118–120). The tethering and untethering of flexible flanking linkers is also emerging as a common theme in canonical kinase regulation (55, 121–124) and for modulation of kinase-protein interactions (26, 55, 125, 126). Consistently, these flexible segments are often evolutionary hot-spots for neofunctionalization among signaling proteins (116, 127).
This comprehensive curated resource represents a comparative analysis of >30,000 pseudokinase sequences, which includes numerous representative species covering archaea, bacteria, protists, fungi, plants, and animals. It provides a new conceptual framework for characterizing pseudokinase evolution, and for the experimental dissection of pseudokinase-dependent lifestyles ranging across a very wide variety of model organisms. Our sub-classification of pseudokinases, which includes more than 30 previously unrecognized families, points to fundamental roles for pseudokinases in nearly all biological systems, where re-use of the versatile protein kinase fold has permitted a vast array of noncatalytic signaling mechanisms, many of which might be targeted therapeutically. Our data also represent a useful starting-point for the evaluation of other types of pseudoenzymes in diverse biological systems and sets the stage for future evolutionary analyses of other pseudoenzyme families across the kingdoms of life.
Materials and Methods:
Detection of pseudokinase sequences
Protein kinase sequences were extracted from the NCBI non-redundant (nr) (downloaded 18/04/04) and UniProt reference proteome databases (Release 2018_09) (59). Protein kinase sequences were identified and aligned using previously curated profiles of diverse eukaryotic and eukaryotic-like protein kinases (2, 52, 54, 57, 58) and the rapid and accurate alignment procedure MAPGAPS (56). Some sequences contained many low complexity regions (for example in P. falciparum and T. thermophila), and therefore a filter was used to mask these low complexity regions during eukaryotic kinase domain detection. Sequences that did not span from at least the β3-lysineto the G-helix (like many atypical kinases and small molecule kinases) were deemed fragmentary and removed. Two additional positions were examined to filter out atypical and small molecule kinases: the APE motif glutamate at the end of the activation loop and an arginine at the beginning of the I-helix. These residues are ePK-specific (not found in atypical and small molecule kinases) and form a conserved salt bridge in ePKs (2), thus ePK sequences lacking both residues were deemed to be non-ePKs and were removed. Pseudokinases were then identified as those lacking at least one of the three key catalytic residues: the β3-lysine, HRD-aspartate, and DFG-aspartate.
Bayesian pattern-based classification of pseudokinase sequences
We used the pseudokinomes extracted from the UniProt proteomes as well as additional diverse pseudokinase sequences extracted from the NCBI nr protein database as input into the optimal multiple-category Bayesian Partitioning with Pattern Selection algorithm (omcBPPS) (63, 64). To remove closely related sequences, we purged the nr pseudokinase sequences using 75% sequence identity cutoff (22,152 total nr pseudokinase sequences), and restrained the number of UniProt proteomes to 83 diverse representative archaeal, bacterial, and eukaryotic species (4,121 total UniProt sequences). A combined total of 26,273 distinct sequences of aligned pseudokinase domains was then used as an input for Bayesian pattern-based clustering procedure (omcBPPS) using a cluster size cut off of 50 sequences to identify the major sequence families(63, 64).
Based on this clustering, we initially identified 68 unique pseudokinase clusters. Some human pseudokinases were not classified by the algorithm due to the limited number of sequences (<50 sequences). In these cases, we ran separate clustering (omcBPPS analyses) within these groups using a minimum size of 15 sequences per cluster, which allowed us to further sub-classify these clusters. From these sub-classifications, we took only the clusters containing human pseudokinases in order to cover the entire human pseudokinome in our classification, yielding a total of 77 unique pseudokinase clusters in total.
The 77 pseudokinase clusters were incorporated within an existing hierarchical profile of ePK sequences, yielding 592 total ePK sequence profiles using MAPGAPS (2, 52, 57, 58). Using the resulting hierarchical sequence profile, we then classified all sequences from each of the 10,092 proteomes available in the UniProt proteomes database to all kinase/pseudokinase families in the profile. Pseudokinase sequences that did not classify into the 77 pseudokinase clusters were placed within one of the 9 unclassified pseudokinase families based on sequence similarity to the major kinase groups (i.e. TK-, TKL-, STE-, CK1-, AGC-, CAMK-, CMGC-, Other-, and PknB-unclassified groups), resulting in a total of 86 pseudokinase families. For 38 pseudokinase families, canonical kinase sequences clustered into the pseudokinase family. For these cases, canonical sequences were removed from the pseudokinase sequence set and are noted in Table S3. Canonical sequences that clustered within the 38 pseudokinase families are provided in Data file S3. Pseudokinase family alignments were manually evaluated for possible misalignments, which are noted in Table S2.
Phylogenetic tree building
To understand the relationships between the identified pseudokinase families, Hidden Markov Models (HMMs) were built using alignments for each family, and HMM-to-HMM distances were computed. From these distances, a distance matrix was created to build a neighbor joining tree, which was used to approximate the distances of families in Fig. 3. This method was implemented using pHMM-Tree (128).
The IRAK, R.irregularis, and pknB phylogenetic trees were built from aligned kinase domains using FastTree version 2.1.10 using default settings, which implements the JTT ML model and calculates local support values for internal nodes via the Shimodaira-Hasegawa test (129,130). The rooted IRAK phylogenetic tree was created using a total of 7,647 diverse plant and metazoan IRAK sequences. These sequences included all plant IRAK pseudokinase sequences (5,405 sequences), canonical plant kinase sequences purged at 80% sequence identity (1,894 sequences), metazoan IRAK pseudokinases purged at 90% sequence identity (113 sequences), and canonical metazoan kinases purged at 80% sequence identity (145 sequences). Also included were diverse non-IRAK TKL sequences (41 sequences) and non-TKL kinase sequences (49 sequences), which were used as an outgroup to root the tree. The unrooted R. irregularis phylogenetic tree was created using all 787 members of its kinome. The unrooted pknB phylogenetic tree was created using 769 representative pseudokinase sequences assigned to a pknB-related family (removed 376 divergent sequences with conservation log odds score <−10) in addition to a set of representative canonical pknB kinase sequences (511 sequences). iTOL was used to generate the final trees (131).
Weblogo creation
Weblogos were created using version 3.6 of the WebLogo 3 online server (132). Amino acids were colored according to their biochemical properties: basic in blue, acidic in red, amide groups in purple, nonpolar residues in black, and polar or uncharged residues in green.
Determination of domain organizations
Additional domains for each of the IRAK, R. irregularis, and pknB-related pseudokinase families were initially identified using NCBI’s Batch Web CD-Search Tool against the CDD database with an expected value threshold of 0.01 and a maximum of 500 hits per CD-search (133, 134). Transmembrane (TM) helices were identified using TMHMM 2.0 (135) and the Phobius web server (136). TM-helices were annotated as such only if both TMHMM and Phobius predicted the same TM-helix region within a 10-residue margin. We verified kinase domain hits using our manually curated protein kinase profiles and removed any overlapping domain predictions.
Pattern analysis of plant IRAK families
To detect sequence motifs associated with the evolution of IRAK pseudokinase families, we used sequence sets from the omcBPPS classification of IRAK pseudokinase families as seed alignments and used mcBPPS (137) to optimally partition 136,068 diverse IRAK sequences extracted from the NCBI nr database (including active sequences) into either pseudokinase family sets or a “background” set that includes unclassified IRAK sequences. Some pseudokinase families have very close active homologs, thus separate seed alignments for canonical IRAK families were also included to ensure that pseudokinase partitions did not include active members. mcBPPS identifies the amino acid patterns that most distinguishes each pseudokinase partition from other sequences, and the 30 most statistically significant patterns for each family were analyzed using available crystal structures.
Analysis of fungal proteomes and kinomes
To compare the kinome and proteome sizes across different fungal species, we analyzed a total of 448 fungal proteomes obtained from the EnsemblFungi database (138). Kinases and pseudokinases were identified using previously curated multiple protein alignment profiles and examination of the β3-lysine, HRD-aspartate, and DFG-aspartate positions, as detailed above.
Supplementary Material
Data file S1. Alignments of model organism kinomes and pseudokinomes
Data file S2. Sequences and alignments of pseudokinase families
Data file S3. Alignments of canonical sequences classified into pseudokinase families
Fig. S1. Expansions of fungal proteomes and kinomes. The proteome and kinome sizes for 449 fungal species/strains are shown, with each species colored by the fungal phylum to which it belongs. Dot sizes represent the proportion of the proteome that is comprised by kinases. The point for R. irregularis is labeled.
Table S1. (Microsoft Excel format). Kinase and pseudokinase sequence counts detected in 10,092 archaeal, bacterial, and eukaryotic proteomes
Table S2. (Microsoft Excel format). Catalog and annotation of pseudokinase families
Table S3. (Microsoft Excel format). Catalog and annotation of canonical sequences classified into pseudokinase families
Table S4. (Microsoft Excel format). Distribution of plant IRAK pseudokinase families across diverse plant species
Table S5. (Microsoft Excel format). Known plant IRAK pseudokinases and their classifications
Acknowledgements:
Members of N. K.’s laboratory are acknowledged for helpful discussions.
Funding: This work was supported by the National Institutes of Health (R01GM1149 to N.K.) and the National Science Foundation (MCB-1149106 to N.K.). UGA-Liverpool travel fellowship for A.K. is acknowledged. This work was also funded by a BBSRC Tools and Resources Development Fund award (BB/N021703/1), a Royal Society Research Grant and North West Cancer Research grants (CR1088 and CR1097) to P. A. E.
Footnotes
Competing interests: The authors declare that they have no conflicts of interest with the contents of this article.
Data and materials availability: Sequence data has been made available through ProKinO (http://vulcan.cs.uga.edu/prokino/about/browser).
References:
- 1.Cohen P, The origins of protein phosphorylation. Nat Cell Biol 4, E127–130 (2002). [DOI] [PubMed] [Google Scholar]
- 2.Kannan N, Taylor SS, Zhai Y, Venter JC, Manning G, Structural and functional diversity of the microbial kinome. PLoS Biol 5, e17 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Knighton DR et al. , Crystal structure of the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 407–414 (1991). [DOI] [PubMed] [Google Scholar]
- 4.Knighton DR et al. , Structure of a peptide inhibitor bound to the catalytic subunit of cyclic adenosine monophosphate-dependent protein kinase. Science 253, 414–420 (1991). [DOI] [PubMed] [Google Scholar]
- 5.Scheeff ED, Eswaran J, Bunkoczi G, Knapp S, Manning G, Structure of the pseudokinase VRK3 reveals a degraded catalytic site, a highly conserved kinase fold, and a putative regulatory binding site. Structure 17, 128–138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Adams JA, Kinetic and catalytic mechanisms of protein kinases. Chem Rev 101, 2271–2290 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Hanks SK, Quinn AM, Hunter T, The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241, 42–52 (1988). [DOI] [PubMed] [Google Scholar]
- 8.Hanks SKH, T.;, The eukaryotic protein kinase superfamily-kinase catalytic domain structure and classification FASEB journal 9, 576–596 (1995). [PubMed] [Google Scholar]
- 9.Murphy JM, Mace PD, Eyers PA, Live and let die: insights into pseudoenzyme mechanisms from structure. Curr Opin Struct Biol 47, 95–104 (2017). [DOI] [PubMed] [Google Scholar]
- 10.Scheeff ED, Bourne PE, Structural evolution of the protein kinase-like superfamily. PLoS Comput Biol 1, e49 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kostich M. et al. , Human members of the eukaryotic protein kinase family. Genome Biol 3, RESEARCH0043 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wilson LJ et al. , New Perspectives, Opportunities, and Challenges in Exploring the Human Protein Kinome. Cancer Res 78, 15–29 (2018). [DOI] [PubMed] [Google Scholar]
- 13.Manning G, Plowman GD, Hunter T, Sudarsanam S, Evolution of protein kinase signaling from yeast to man. Trends Biochem Sci 27, 514–520 (2002). [DOI] [PubMed] [Google Scholar]
- 14.Manning G, Young SL, Miller WT, Zhai Y, The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan. Proc Natl Acad Sci U S A 105, 9674–9679 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caenepeel S, Charydczak G, Sudarsanam S, Hunter T, Manning G, The mouse kinome: discovery and comparative genomics of all mouse protein kinases. Proc Natl Acad Sci U S A 101, 11707–11712 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manning G. et al. , The minimal kinome of Giardia lamblia illuminates early kinase evolution and unique parasite biology. Genome Biol 12, R66 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zulawski M, The Arabidopsis Kinome: phylogeny and evolutionary insights into functional diversification. BMC genomics 15, (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manning G, Whyte DB, Martinez R, Hunter T, Sudarsanam S, The protein kinase complement of the human genome. Science 298, 1912–1934 (2002). [DOI] [PubMed] [Google Scholar]
- 19.Boudeau J, Miranda-Saavedra D, Barton GJ, Alessi DR, Emerging roles of pseudokinases. Trends Cell Biol 16, 443–452 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Eyers Patrick A., J. M. Murphy, Dawn of the dead: protein pseudokinases signal new adventures in cell biology. Biochemical Society Transactions 41, 969–974 (2013). [DOI] [PubMed] [Google Scholar]
- 21.Reiterer V, Eyers PA, Farhan H, Day of the dead: pseudokinases and pseudophosphatases in physiology and disease. Trends Cell Biol 24, 489–505 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Murphy JM et al. , A robust methodology to subclassify pseudokinases based on their nucleotide-binding properties. Biochem J 457, 323–334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mendrola JM, Shi F, Park JH, Lemmon MA, Receptor tyrosine kinases with intracellular pseudokinase domains. Biochem Soc Trans 41, 1029–1036 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dhawan NS, Scopton AP, Dar AC, Small molecule stabilization of the KSR inactive state antagonizes oncogenic Ras signalling. Nature 537, 112–116 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Babon JJ, Lucet IS, Murphy JM, Nicola NA, Varghese LN, The molecular regulation of Janus kinase (JAK) activation. Biochem J 462, 1–13 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eyers PA, Keeshan K, Kannan N, Tribbles in the 21st Century: The Evolving Roles of Tribbles Pseudokinases in Biology and Disease. Trends Cell Biol 27, 284–298 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zeqiraj E, van Aalten DM, Pseudokinases-remnants of evolution or key allosteric regulators? Curr Opin Struct Biol 20, 772–781 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sierla M. et al. , The Receptor-like Pseudokinase GHR1 Is Required for Stomatal Closure. Plant Cell 30, 2813–2837 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang H. et al. , Structure and evolution of the Fam20 kinases. Nat Commun 9, 1218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lecointre C. et al. , Dimerization of the Pragmin Pseudo-Kinase Regulates Protein Tyrosine Phosphorylation. Structure 26, 545–554 e544 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Chen MJ, Dixon JE, Manning G, Genomics and evolution of protein phosphatases. Sci Signal 10, (2017). [DOI] [PubMed] [Google Scholar]
- 32.Zettl M, Adrain C, Strisovsky K, Lastun V, Freeman M, Rhomboid family pseudoproteases use the ER quality control machinery to regulate intercellular signaling. Cell 145, 79–91 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eyers PA, Murphy JM, The evolving world of pseudoenzymes: proteins, prejudice and zombies. BMC Biol 14, 98 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murphy JM, Farhan H, Eyers PA, Bio-Zombie: the rise of pseudoenzymes in biology. Biochemical Society Transactions 45, 537–544 (2017). [DOI] [PubMed] [Google Scholar]
- 35.Novotny CJ et al. , Overcoming resistance to HER2 inhibitors through state-specific kinase binding. Nat Chem Biol 12, 923–930 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shi F, Telesco SE, Liu Y, Radhakrishnan R, Lemmon MA, ErbB3/HER3 intracellular domain is competent to bind ATP and catalyze autophosphorylation. Proc Natl Acad Sci U S A 107, 7692–7697 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu B. et al. , WNK1, a novel mammalian serine/threonine protein kinase lacking the catalytic lysine in subdomain II. J Biol Chem 275, 16795–16801 (2000). [DOI] [PubMed] [Google Scholar]
- 38.Lee AY et al. , Protein kinase WNK3 regulates the neuronal splicing factor Fox-1. Proc Natl Acad Sci U S A 109, 16841–16846 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ahlstrom R, Yu AS, Characterization of the kinase activity of a WNK4 protein complex. Am J Physiol Renal Physiol 297, F685–692 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Eyers PA, TRIBBLES: A Twist in the Pseudokinase Tail. Structure 23, 1974–1976 (2015). [DOI] [PubMed] [Google Scholar]
- 41.Uljon S. et al. , Structural Basis for Substrate Selectivity of the E3 Ligase COP1. Structure 24, 687–696 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jamieson SA et al. , Substrate binding allosterically relieves autoinhibition of the pseudokinase TRIB1. Sci Signal 11, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sheetz JB, Lemmon MA, Flipping ATP to AMPlify Kinase Functions. Cell 175, 641–642 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sreelatha A. et al. , Protein AMPylation by an Evolutionarily Conserved Pseudokinase. Cell 175, 809–821 e819 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sergina NV et al. , Escape from HER-family tyrosine kinase inhibitor therapy by the kinase-inactive HER3. Nature 445, 437–441 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Claus J. et al. , Inhibitor-induced HER2-HER3 heterodimerisation promotes proliferation through a novel dimer interface. Elife 7, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bailey FP et al. , The Tribbles 2 (TRB2) pseudokinase binds to ATP and autophosphorylates in a metal-independent manner. Biochem J 467, 47–62 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Labesse G. et al. , ROP2 from Toxoplasma gondii: a virulence factor with a protein-kinase fold and no enzymatic activity. Structure 17, 139–146 (2009). [DOI] [PubMed] [Google Scholar]
- 49.Blaum BS et al. , Structure of the pseudokinase domain of BIR2, a regulator of BAK1-mediated immune signaling in Arabidopsis. J Struct Biol 186, 112–121 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Gish LA, Clark SE, The RLK/Pelle family of kinases. Plant J 66, 117–127 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lewis JD et al. , The Arabidopsis ZED1 pseudokinase is required for ZAR1-mediated immunity induced by the Pseudomonas syringae type III effector HopZ1a. Proc Natl Acad Sci U S A 110, 18722–18727 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lehti-Shiu MD, Shiu SH, Diversity, classification and function of the plant protein kinase superfamily. Philos Trans R Soc Lond B Biol Sci 367, 2619–2639 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu PL, Du L, Huang Y, Gao SM, Yu M, Origin and diversification of leucine-rich repeat receptor-like protein kinase (LRR-RLK) genes in plants. BMC Evol Biol 17, 47 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bateman A. et al. , The Pfam protein families database. Nucleic Acids Res 32, D138–141 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kannan N, Haste N, Taylor SS, Neuwald AF, The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci U S A 104, 1272–1277 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Neuwald AF, Rapid detection, classification and accurate alignment of up to a million or more related protein sequences. Bioinformatics 25, 1869–1875 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.McSkimming DI et al. , KinView: a visual comparative sequence analysis tool for integrated kinome research. Mol Biosyst 12, 3651–3665 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Talevich E, Mirza A, Kannan N, Structural and evolutionary divergence of eukaryotic protein kinases in Apicomplexa. BMC Evol Biol 11, 321 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.UniProt C, UniProt: a hub for protein information. Nucleic Acids Res 43, D204–212 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Patel O. et al. , Structure of SgK223 pseudokinase reveals novel mechanisms of homotypic and heterotypic association. Nat Commun 8, 1157 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McMahon SB, Van Buskirk HA, Dugan KA, Copeland TD, Cole MD, The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94, 363–374 (1998). [DOI] [PubMed] [Google Scholar]
- 62.Ward P, Equinet L, Packer J, Doerig C, Protein kinases of the human malaria parasite Plasmodium falciparum: the kinome of a divergent eukaryote. BMC Genomics 5, 79 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Neuwald AF, A Bayesian sampler for optimization of protein domain hierarchies. J Comput Biol 21, 269–286 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Neuwald AF, Evaluating, comparing, and interpreting protein domain hierarchies. J Comput Biol 21, 287–302 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tisserant E. et al. , Genome of an arbuscular mycorrhizal fungus provides insight into the oldest plant symbiosis. Proceedings of the National Academy of Sciences of the United States of America 110, 20117–20122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sierke SL, Cheng K, Kim HH, Koland JG, Biochemical characterization of the protein tyrosine kinase homology domain of the ErbB3 (HER3) receptor protein. Biochem J 322 ( Pt 3), 757–763 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Miya A. et al. , CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci U S A 104, 19613–19618 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Radutoiu S. et al. , Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592 (2003). [DOI] [PubMed] [Google Scholar]
- 69.Pietraszewska-Bogiel A. et al. , Interaction of Medicago truncatula lysin motif receptor-like kinases, NFP and LYK3, produced in Nicotiana benthamiana induces defence-like responses. PLoS One 8, e65055 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Eyuboglu B. et al. , Molecular characterisation of the STRUBBELIG-RECEPTOR FAMILY of genes encoding putative leucine-rich repeat receptor-like kinases in Arabidopsis thaliana. BMC Plant Biol 7, 16 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Chevalier D. et al. , STRUBBELIG defines a receptor kinase-mediated signaling pathway regulating organ development in Arabidopsis. Proceedings of the National Academy of Sciences 102, 9074–9079 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Miyakawa T, Miyazono K, Sawano Y, Hatano K, Tanokura M, Crystal structure of ginkbilobin-2 with homology to the extracellular domain of plant cysteine-rich receptor-like kinases. Proteins 77, 247–251 (2009). [DOI] [PubMed] [Google Scholar]
- 73.Zhang L. et al. , Identification of an apoplastic protein involved in the initial phase of salt stress response in rice root by two-dimensional electrophoresis. Plant Physiol 149, 916–928 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Bourdais G. et al. , Large-Scale Phenomics Identifies Primary and Fine-Tuning Roles for CRKs in Responses Related to Oxidative Stress. PLoS Genet 11, e1005373 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sklodowski K. et al. , The receptor-like pseudokinase MRH1 interacts with the voltage-gated potassium channel AKT2. Sci Rep 7, 44611 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Huard-Chauveau C. et al. , An atypical kinase under balancing selection confers broad-spectrum disease resistance in Arabidopsis. PLoS Genet 9, e1003766 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grutter C, Sreeramulu S, Sessa G, Rauh D, Structural characterization of the RLCK family member BSK8: a pseudokinase with an unprecedented architecture. J Mol Biol 425, 4455–4467 (2013). [DOI] [PubMed] [Google Scholar]
- 78.Gao M. et al. , Regulation of cell death and innate immunity by two receptor-like kinases in Arabidopsis. Cell Host Microbe 6, 34–44 (2009). [DOI] [PubMed] [Google Scholar]
- 79.Nimchuk ZL, Tarr PT, Meyerowitz EM, An evolutionarily conserved pseudokinase mediates stem cell production in plants. Plant Cell 23, 851–854 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kuo A, Kohler A, Martin FM, Grigoriev IV, Expanding genomics of mycorrhizal symbiosis. Front Microbiol 5, 582 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Handa Y. et al. , RNA-seq Transcriptional Profiling of an Arbuscular Mycorrhiza Provides Insights into Regulated and Coordinated Gene Expression in Lotus japonicus and Rhizophagus irregularis. Plant Cell Physiol 56, 1490–1511 (2015). [DOI] [PubMed] [Google Scholar]
- 82.Haydon CE et al. , Identification of novel phosphorylation sites on Xenopus laevis Aurora A and analysis of phosphopeptide enrichment by immobilized metal-affinity chromatography. Mol Cell Proteomics 2, 1055–1067 (2003). [DOI] [PubMed] [Google Scholar]
- 83.Grynberg M, Godzik A, NERD: a DNA processing-related domain present in the anthrax virulence plasmid, pXO1. Trends Biochem Sci 29, 106–110 (2004). [DOI] [PubMed] [Google Scholar]
- 84.Sumby PS, M.C., Genetics of the phage growth limitation (Pgl) system ofStreptomyces coelicolor A3(2) Molecular microbiology 44, 489–500 (2002). [DOI] [PubMed] [Google Scholar]
- 85.Hoskisson PAS, P.; Smith M, The phage growth limitation system in Streptomyces coelicolor A(3)2 is a toxin/antitoxin system, comprising enzymes with DNA methyltransferase, protein kinase and ATPase activity. Virology, (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ebright RH, Busby S, The Escherichia coli RNA polymerase alpha subunit: structure and function. Curr Opin Genet Dev 5, 197–203 (1995). [DOI] [PubMed] [Google Scholar]
- 87.Aoyama K, Aiba H, Mizuno T, Genetic analysis of the His-to-Asp phosphorelay implicated in mitotic cell cycle control: involvement of histidine-kinase genes of Schizosaccharomyces pombe. Biosci Biotechnol Biochem 65, 2347–2352 (2001). [DOI] [PubMed] [Google Scholar]
- 88.Ashby MK, Houmard J, Cyanobacterial two-component proteins: structure, diversity, distribution, and evolution. Microbiol Mol Biol Rev 70, 472–509 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Calera JA, Choi GH, Calderone RA, Identification of a putative histidine kinase two-component phosphorelay gene (CaHK1) in Candida albicans. Yeast 14, 665–674 (1998). [DOI] [PubMed] [Google Scholar]
- 90.Krupa AS, N., Diversity in domain architectures of Ser/Thr kinases and their homologues in prokaryotes. BMC genomics 6, 129 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Phalip VL, J.; Zhang C, HistK a cyanobacterial protein with both a serine/threonine kinase domain and a histidine kinase domain: implication for the mechanism of signal transduction. The biochemical journal 360, 639–644 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhang X. et al. , Genome-wide survey of putative serine/threonine protein kinases in cyanobacteria. BMC Genomics 8, 395 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Buck V. et al. , Peroxide sensors for the fission yeast stress-activated mitogen-activated protein kinase pathway. Mol Biol Cell 12, 407–419 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Calera JA, Zhao XJ, De Bernardis F, Sheridan M, Calderone R, Avirulence of Candida albicans CaHK1 mutants in a murine model of hematogenously disseminated candidiasis. Infect Immun 67, 4280–4284 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Calera JAC, R., Flocculation of hyphae is associated with a deletion in the putative CaHKl two-component histidine kinase gene from Candida albicans Microbiology 145, 1431–1442 (1999). [DOI] [PubMed] [Google Scholar]
- 96.Cheng Y. et al. , A pair of iron-responsive genes encoding protein kinases with a Ser/Thr kinase domain and a His kinase domain are regulated by NtcA in the Cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 188, 4822–4829 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kruppa M. et al. , The role of the Candida albicans histidine kinase [CHK1) gene in the regulation of cell wall mannan and glucan biosynthesis. FEMS Yeast Res 3, 289–299 (2003). [DOI] [PubMed] [Google Scholar]
- 98.Quinn J. et al. , Two-component mediated peroxide sensing and signal transduction in fission yeast. Antioxid Redox Signal 15, 153–165 (2011). [DOI] [PubMed] [Google Scholar]
- 99.Shi L. et al. , Two genes encoding protein kinases of the HstK family are involved in synthesis of the minor heterocyst-specific glycolipid in the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 189, 5075–5081 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Torosantucci A, Deletion of the Two-Component Histidine Kinase Gene (CHK1) of Candida albicans Contributes to Enhanced Growth Inhibition and Killing by Human Neutrophils In Vitro. Infection and Immunity 70, 985–987 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yamada-Okabe T. et al. , Roles of three histidine kinase genes in hyphal development and virulence of the pathogenic fungus Candida albicans. J Bacteriol 181, 7243–7247 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jones MA, Raymond MJ, Smirnoff N, Analysis of the root-hair morphogenesis transcriptome reveals the molecular identity of six genes with roles in root-hair development in Arabidopsis. Plant J 45, 83–100 (2006). [DOI] [PubMed] [Google Scholar]
- 103.Oruganty K, Talathi NS, Wood ZA, Kannan N, Identification of a hidden strain switch provides clues to an ancient structural mechanism in protein kinases. Proc Natl Acad Sci U S A 110, 924–929 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hunter T, Plowman GD, The protein kinases of budding yeast: six score and more. Trends Biochem Sci 22, 18–22 (1997). [DOI] [PubMed] [Google Scholar]
- 105.Stajich JE et al. , Insights into evolution of multicellular fungi from the assembled chromosomes of the mushroom Coprinopsis cinerea (Coprinus cinereus). Proc Natl Acad Sci U S A 107, 11889–11894 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Miller WT, Tyrosine kinase signaling and the emergence of multicellularity. Biochim Biophys Acta 1823, 1053–1057 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Werner GDA, Zhou Y, Pieterse CMJ, Kiers ET, Tracking plant preference for higher-quality mycorrhizal symbionts under varying CO2 conditions over multiple generations. Ecol Evol 8, 78–87 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nanjareddy K, Arthikala MK, Gomez BM, Blanco L, Lara M, Differentially expressed genes in mycorrhized and nodulated roots of common bean are associated with defense, cell wall architecture, N metabolism, and P metabolism. PLoS One 12, e0182328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Todd AEO, C.A.; Thornton JM, Sequence and structural differences between enzyme and nonenzyme homologs. Structure 10, 1435–1451 (2002). [DOI] [PubMed] [Google Scholar]
- 110.Kaltenbach M. et al. , Evolution of chalcone isomerase from a noncatalytic ancestor. Nat Chem Biol 14, 548–555 (2018). [DOI] [PubMed] [Google Scholar]
- 111.Clifton BE et al. , Evolution of cyclohexadienyl dehydratase from an ancestral solute-binding protein. Nature Chemical Biology 14, 542–547 (2018). [DOI] [PubMed] [Google Scholar]
- 112.Mukherjee K, Sharma M, Jahn R, Wahl MC, Sudhof TC, Evolution of CASK into a Mg2+-sensitive kinase. Sci Signal 3, ra33 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Williams DM, Cole PA, Proton demand inversion in a mutant protein tyrosine kinase reaction. J Am Chem Soc 124, 5956–5957 (2002). [DOI] [PubMed] [Google Scholar]
- 114.Skamnaki VT et al. , Catalytic mechanism of phosphorylase kinase probed by mutational studies. Biochemistry 38, 14718–14730 (1999). [DOI] [PubMed] [Google Scholar]
- 115.Mohanty S. et al. , Hydrophobic Core Variations Provide a Structural Framework for Tyrosine Kinase Evolution and Functional Specialization. PLoS Genet 12, e1005885 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Harrison SC, Variation on an Src-like theme. Cell 112, 737–740 (2003). [DOI] [PubMed] [Google Scholar]
- 117.Williams JG, Zvelebil M, SH2 domains in plants imply new signalling scenarios. Trends Plant Sci 9, 161–163 (2004). [DOI] [PubMed] [Google Scholar]
- 118.Oh MH et al. , Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc Natl Acad Sci U S A 106, 658–663 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Luan S, Tyrosine phosphorylation in plant cell signaling. Proc Natl Acad Sci U S A 99, 11567–11569 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Perraki A. et al. , Phosphocode-dependent functional dichotomy of a common co-receptor in plant signalling. Nature, (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hubbard SR , Juxtamembrane autoinhibition in receptor tyrosine kinases. Nat Rev Mol Cell Biol 5, 464–471 (2004). [DOI] [PubMed] [Google Scholar]
- 122.Kwon A, John M, Ruan Z, Kannan N, Coupled regulation by the juxtamembrane and sterile alpha motif (SAM) linker is a hallmark of ephrin tyrosine kinase evolution. J Biol Chem 293, 5102–5116 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Mirza A, Mustafa M, Talevich E, Kannan N, Co-conserved features associated with cis regulation of ErbB tyrosine kinases. PLoS One 5, e14310 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Plaza-Menacho I. et al. , RET Functions as a Dual-Specificity Kinase that Requires Allosteric Inputs from Juxtamembrane Elements. Cell Rep 17, 3319–3332 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Gajiwala KS, EGFR: tale of the C-terminal tail. Protein Sci 22, 995–999 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Ma B, Tsai CJ, Haliloglu T, Nussinov R, Dynamic allostery: linkers are not merely flexible. Structure 19, 907–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brown CJ, Johnson AK, Dunker AK, Daughdrill GW, Evolution and disorder. Curr Opin Struct Biol 21, 441–446 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Huo L. et al. , pHMM-tree: phylogeny of profile hidden Markov models. Bioinformatics 33, 1093–1095 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Price MN, Dehal PS, Arkin AP, FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26, 1641–1650 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Price MN, Dehal PS, Arkin AP, FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Letunic I, Bork P, Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44, W242–245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Crooks GE, Hon G, Chandonia JM, Brenner SE, WebLogo: a sequence logo generator. Genome Res 14, 1188–1190 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Marchler-Bauer A. et al. , CDD/SPARCLE: functional classification of proteins via subfamily domain architectures. Nucleic Acids Res 45, D200–D203 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Marchler-Bauer A. et al. , CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39, D225–229 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Krogh A, Larsson B, von Heijne G, Sonnhammer EL, Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305, 567–580 (2001). [DOI] [PubMed] [Google Scholar]
- 136.Kall L, Krogh A, Sonnhammer EL, Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res 35, W429–432 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Neuwald AF, Surveying the manifold divergence of an entire protein class for statistical clues to underlying biochemical mechanisms. Stat Appl Genet Mol Biol 10, Article 36 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kersey PJ et al. , Ensembl Genomes 2018: an integrated omics infrastructure for non-vertebrate species. Nucleic Acids Res 46, D802–D808 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data file S1. Alignments of model organism kinomes and pseudokinomes
Data file S2. Sequences and alignments of pseudokinase families
Data file S3. Alignments of canonical sequences classified into pseudokinase families
Fig. S1. Expansions of fungal proteomes and kinomes. The proteome and kinome sizes for 449 fungal species/strains are shown, with each species colored by the fungal phylum to which it belongs. Dot sizes represent the proportion of the proteome that is comprised by kinases. The point for R. irregularis is labeled.
Table S1. (Microsoft Excel format). Kinase and pseudokinase sequence counts detected in 10,092 archaeal, bacterial, and eukaryotic proteomes
Table S2. (Microsoft Excel format). Catalog and annotation of pseudokinase families
Table S3. (Microsoft Excel format). Catalog and annotation of canonical sequences classified into pseudokinase families
Table S4. (Microsoft Excel format). Distribution of plant IRAK pseudokinase families across diverse plant species
Table S5. (Microsoft Excel format). Known plant IRAK pseudokinases and their classifications