Abstract
Background
Degeneration of ovarian function is an obvious feature of female aging. In addition, studies have shown that autophagy decreases with age, and DNA methylation is a hallmark epigenetic pattern during aging. However, it is not clear whether the expression and DNA methylation of autophagy genes are involved in the declines in ovarian function that occur during aging.
Results
Three groups of rats were used: 6-month-old (6 M) rats, 12-month-old (12 M) rats and 24-month-old (24 M) rats. Serum E2 levels and the mRNA and protein expression levels of Atg5, Atg12, Atg16L, Beclin1 and Lc3B were significantly decreased in aged rats. In addition, the methylation levels of the Atg5 gene were significantly increased in aged rats. The expression of the Dnmt1 and Dnmt2 genes decreased with aging; however, the expression of the Dnmt3A and Dnmt3B genes gradually increased with aging.
Conclusions
Decreased autophagic activity was involved in the declines in ovarian function in aging rats. Upregulation of the DNA methyltransferases Dnmt3A and Dnmt3B may have led to methylation of the autophagy genes Atg5 and Lc3B to ultimately cause the observed decreases in autophagic activity.
Keywords: Ovarian aging, Autophagy, DNA methylation, DNA methyltransferase
Introduction
Aging is characterized by time-dependent declines in function resulting from progressive biochemical and physiological dysregulation [1]. Compared with other tissues, the ovaries are much more severely affected by aging. The numbers of follicles and the quality of oocytes in the ovaries decrease with age, resulting in a gradual decline in fertility [2, 3]. During the transition period from perimenopause to menopause, increases in follicle-stimulating hormone (FSH) levels and decreases in estrogen levels caused by ovarian aging can lead to considerable follicular atresia and to depletion of primordial follicles [4, 5]. Previous research has revealed that the number of primordial follicles decreases at a rate of approximately 1000 follicles per month during the reproductive years of life and that only approximately 1000 follicles remain in the ovaries during the final menstrual stage (menopause) [4].
Aging is a degenerative and irreversible biological process caused by interactions among multiple factors and organs. Autophagy is a process in which cells degrade damaged organelles and biological macromolecules in lysosomes. Autophagy is a necessary process for the maintenance of intracellular homeostasis under physiological conditions, but under some conditions, such as inflammatory responses, oxidative stress, and organelle aging, autophagy may be overactivated or inhibited [6, 7]. Therefore, the regulation of intracellular autophagic activity is very important for the maintenance of normal physiological functions [8, 9]. A large body of evidence indicates that autophagic activity declines with age [10, 11]. Toth et al. has reported that loss of function of Atg1, Atg7, Atg18 and Beclin1 decreases lifespan in the nematode Caenorhabditis elegans. Komatsu et al. has reported that loss of Atg7 leads to neurodegeneration and died within 28 weeks of birth [12–14]. Tan et al. has reported that remarkable autophagy-related changes, including the accumulation of ubiquitin-positive proteins and decreases in autophagic activity, occur with age in the brains of senescence-accelerated mouse prone 8 (SAMP8) mice [15]. Abdellatif et al. has reported that cardiomyocyte-specific Atg5 ablation in aged C57BL/6 mice dramatically accelerates declines in cardiac function, resulting in shortened lifespan [16].
DNA methylation is a hallmark epigenetic pattern that occurs during aging, but aging-associated DNA methylation is not stochastic [17]. DNA methyltransferases (Dnmts) play vital roles in DNA methylation. Mammalian Dnmts are usually classified as Dnmt1, Dnmt2 or Dnmt3 (which includes four subtypes: A, B, C and L). Notably, Dnmts have different functions during the methylation process [17, 18]. Dnmt1, which is required for the maintenance of all methylation in the genome, can restore specific methylation patterns on daughter strands in accordance with the patterns on parental DNA during replication [19]. Dnmt2 methylates small transfer RNAs, and Dnmt3A and Dnmt3B are responsible for establishing DNA methylation patterns [20, 21]. Dnmt3C can protect male germ cells from retrotransposon activity, which is essential for mouse fertility [19].
Previous studies have suggested that declines in autophagy might be associated with aging. However, it is not clear whether the expression and DNA methylation of autophagy genes are involved in the declines in ovarian function that occur during aging. In this study, the expression and methylation levels of autophagy-related genes in aging rats were studied.
Results
Age-dependent declines in ovarian function
In this study, the ovaries of rats were weighed, and the ovarian index (ovarian weight/body weight) values were calculated. The results showed that ovary weight decreased with aging: the ovarian index values of 12-month-old (12 M) and 24-month-old (24 M) rats were significantly lower than those of 6-month-old (6 M) rats (Fig. 1a). In addition, the serum estradiol (E2) levels in rats were measured by radioimmunoassay (RIA). The results showed that the serum E2 levels in 12 M and 24 M rats were significantly lower than those in 6 M rats (Fig. 1b). These results confirm that the endocrine functions of the ovaries present age-dependent declines.
Fig. 1.
Age-dependent declines in the ovarian index and E2 secretion in rats. (a) Ovarian index (ratio of ovarian weight to body weight). (b) Secretion of E2. E2: estradiol. The ovarian index values and the serum E2 levels of 12 M and 24 M rats were significantly lower than those of 6 M rats. 6 M: 6-month-old, 12 M:12-month-old, 24 M:24-month-old. Compared with 6 M rats, **P < 0.01,***P < 0.001,****P < 0.0001; compared with 12 M rats, ##P < 0.01
Age-dependent mRNA expression of autophagy-related genes in the ovaries of rats
Quantitative real-time PCR was used to analyze the mRNA expression of autophagy-related genes, including Atg5, Atg12, Atg16L, Beclin1 and Lc3B. As shown in Fig. 2, compared to those in 6 M and 12 M rats, the mRNA expression levels of Atg5, Atg12, Atg16L, Beclin1 and Lc3B in 24 M rats were significantly decreased. However, there were no significant differences in the mRNA expression of any autophagy-related genes between 12 M and 6 M rats. These results suggest that ovarian autophagic function is decreased in aged rats.
Fig. 2.
mRNA expression of autophagy-related genes in the ovaries of rats. (a-e) mRNA expression levels of Atg5, Atg12, Atg16L, Lc3B and Beclin1.Compared to those in 6 M and 12 M rats, the mRNA expression levels of Atg5, Atg12, Atg16L, Beclin1 and Lc3B in 24 M rats were significantly decreased. 6 M: 6-month-old, 12 M: 12-month-old, 24 M: 24-month-old. Compared with 6 M rats, *P < 0.05, **P < 0.01; compared with 12 M rats, ##P < 0.01, ###P < 0.001
Age-dependent protein expression of autophagy-related genes in the ovaries of rats
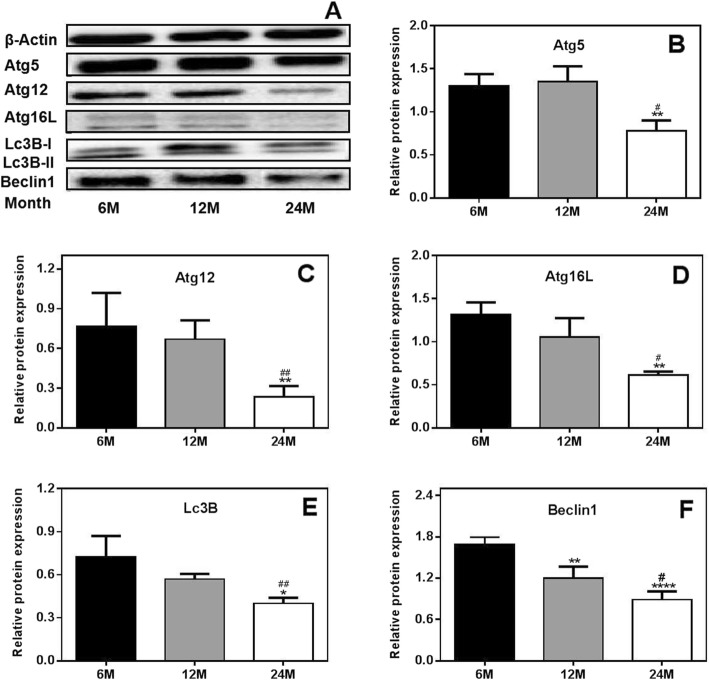
To further investigate the expression of autophagy-related genes in rat ovaries, the protein expression levels of Atg5, Atg12, Atg16L, Lc3B and Beclin1 were detected using Western blot analysis. The results showed that the protein expression levels of Atg5, Atg12, Atg16L, Beclin1 and Lc3B were significantly lower in 24 M rats than in 6 M and 12 M rats. There were no significant differences in the expression levels of autophagy-related genes between the 12 M rats and the 6 M rats (Fig. 3). These results further confirm that age-dependent reductions in autophagy occur in rat ovaries.
Fig. 3.
Protein expression of autophagy-related genes in the ovaries of rats. (a) Western blot results. (b-f) Protein expression levels of Atg5, Atg12, Atg16L, Lc3B and Beclin1. Protein expression levels of Atg5, Atg12, Atg16L, Beclin1 and Lc3B were significantly lower in 24 M rats than in 6 M and 12 M rats. 6 M: 6-month-old, 12 M: 12-month-old, 24 M: 24-month-old. Compared with 6 M rats, *P < 0.05, **P < 0.01, ****P < 0.0001; compared with 12 M rats, #P < 0.05, ##P < 0.01
Age-dependent DNA methylation is correlated with age in the ovaries of rats
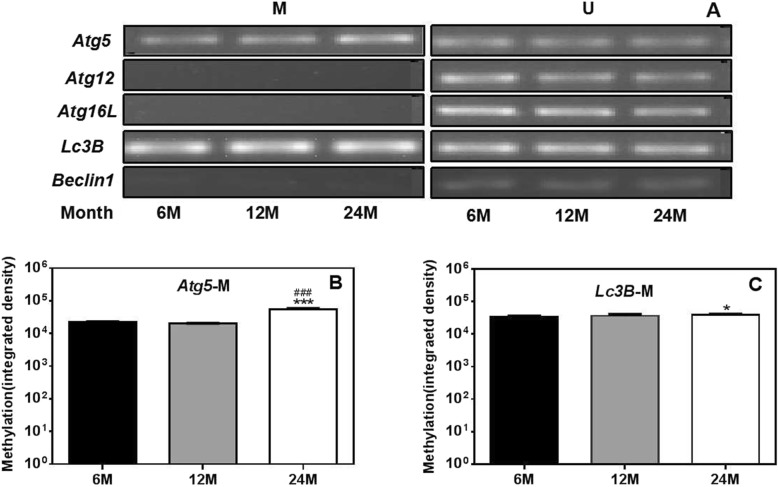
Based on the results of real-time PCR and Western blot, we found that the expression of autophagy-related genes in the ovaries of rats decreased with age. To further investigate the mechanism of the decreases in autophagic function, we detected the methylation statuses of the promoter regions of the autophagy-related genes Atg5, Atg12, Atg16L, Beclin1 and Lc3B in the ovaries of rats by methylation-specific PCR (MSP). The results are shown in Fig. 4. We found that the autophagy genes Atg5 and Lc3B were methylated in rat ovaries and that Atg5 gene methylation levels were significantly higher in the ovaries of 24 M rats than in those of 6 M and 12 M rats. In addition, the methylation levels of the Lc3B gene were significantly greater in the ovaries of 24 M rats than in those of 6 M rats. However, methylation of the Atg12, Atg16L and Beclin1 genes was not detected in the rat ovaries. These results suggest that the decreases in Atg5 and Lc3B gene expression in the ovaries of aged rats might be related to gene methylation (Fig. 4).
Fig. 4.
Age-dependent DNA methylation of autophagy-related genes in the ovaries of rats. (a) Agarose gel electrophoresis of DNA methylation. (b-c) Quantitative methylation levels of Atg5 and Lc3B. .Atg5 gene and Lc3B gene methylation levels were significantly higher in the ovaries of 24 M rats than in those of 6 M rat. 6 M: 6-month-old, 12 M: 12-month-old, 24 M: 24-month-old, M: methylated bands, U: unmethylated bands. Compared with 6 M rats, *P < 0.05, ***P < 0.001; compared with 12 M rats, ###P < 0.001
Age-dependent changes in Dnmts in the ovaries of rats
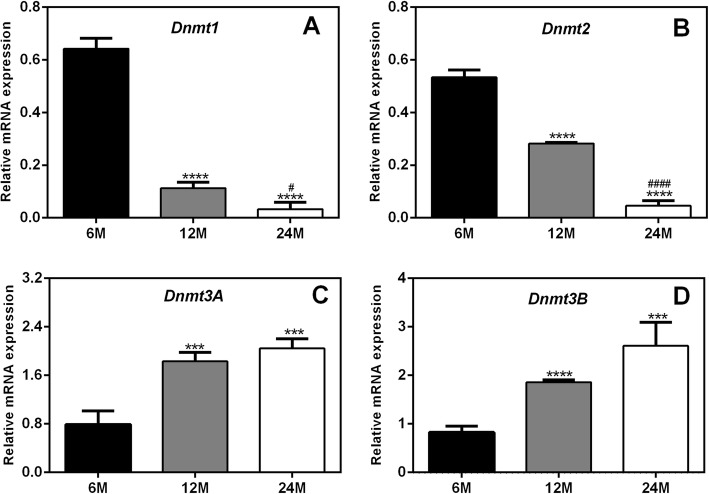
The MSP results showed that the methylation levels of the autophagy genes Atg5 and Lc3B were increased in aged rats. Because Dnmts play important roles in gene methylation, the expression levels of Dnmt1, Dnmt2, Dnmt3A and Dnmt3B in the ovaries of rats were detected by real-time PCR. As shown in Fig. 5, compared to those in 6 M rats, the expression levels of the Dnmt1 and Dnmt2 genes in 12 M and 24 M rats were significantly decreased. However, the expression levels of the Dnmt3A and Dnmt3B genes gradually increased with aging.
Fig. 5.
mRNA expression levels of Dnmt1, Dnmt2, Dnmt3A and Dnmt3B in the ovaries of rats. Compared to those in 6 M rats, the expression levels of the Dnmt1 and Dnmt2 genes in 12 M and 24 M rats were significantly decreased. However, the expression levels of the Dnmt3A and Dnmt3B genes gradually increased with aging. 6 M: 6-month-old, 12 M: 12-month-old, 24 M: 24-month-old. Compared with 6 M rats, ***P < 0.001,****P < 0.0001; compared with 12 M rats, #P < 0.05, ####P < 0.0001
The protein expression of Dnmt1, Dnmt2, Dnmt3A and Dnmt3B in the ovaries of rats was further detected by Western blot analysis. The results showed that the changes in protein expression were consistent with, albeit less significant than, the observed changes in mRNA expression. Compared to those in 6 M rats, the protein expression levels of Dnmt1 and Dnmt2 in 12 M and 24 M rats were decreased. However, the protein expression of Dnmt3A and Dnmt3B was greater in 12 M and 24 M rats than in 6 M rats. Moreover, the changes in Dnmt2 and Dnmt3A protein expression were more significant in 24 M rats than in 12 M rats (Fig. 6).
Fig. 6.
Protein expression of Dnmt1, Dnmt2, Dnmt3A and Dnmt3B in the ovaries of rats.(a) Western blot results. (b-e) Protein expression levels of Dnmt1, Dnmt2, Dnmt3A and Dnmt3B. Compared to those in 6 M rats, the protein expression levels of Dnmt1 and Dnmt2 in 12 M and 24 M rats were decreased. However, the protein expression of Dnmt3A and Dnmt3B was greater in 12 M and 24 M rats than in 6 M rats. 6 M: 6-month-old, 12 M: 12-month-old, 24 M: 24-month-old. Compared with 6 M rats, *P < 0.05,***P < 0.001; compared with 12 M rats, #P < 0.05
Based on the above results, we hypothesize that the increases in Dnmt3A and Dnmt3B, but not Dnmt1 and Dnmt2, might be related to increases in the methylation of the autophagy genes Atg5 and Lc3B in aged rats.
Discussion
The aging process of ovaries is characterized by morphological atrophy and functional declines in reproductive hormones (especially estrogen). The rat is the most widely used model animal in studies on ovarian function. Rats exhibit periodic follicular development and sex hormone changes in a process called the estrous cycle, which is similar to the human menstrual cycle [5, 22]. The secretion of ovarian hormones decreases in an age-dependent manner in rats; Acuna et al. found that the estrogen levels of 8- to 14-month-old rats are significantly lower than those of 6 M rats [23]. In this study, we measured ovarian weight and estrogen levels in 6 M, 12 M and 24 M rats. The results showed that with increasing age, ovary weight decreased gradually, and serum E2 levels decreased significantly. Consistent with our results, Nie et al. reported that the ovaries of aged (40-week-old) female C57BL/6 mice exhibit structural and functional deterioration and that secretion of E2 significantly decreases with consecutive superovulations in these naturally age-deteriorated ovaries [24].
Autophagy is widely involved in biological processes such as growth, development, apoptosis and aging [25, 26]. The autophagy-related genes Atg, Beclin1 and Lc3B are the core molecules that maintain and induce autophagy [27]. Different Atg genes participate in different steps of autophagy [28]. For example, activation of Atg1 promotes the initiation of autophagosome formation [29]; the Atg12 conjugation system (Atg12-Atg5-Atg16L complex) is necessary for autophagosome formation, phagophore elongation and cargo recognition [30, 31]; and Atg7 is an essential catalyst for autophagosome assembly. The conversion of Lc3B-I to Lc3B-II is also necessary for autophagosome formation [32]. It has been established that autophagy in various tissues decreases gradually with age and that autophagy progression is closely related to the expression of various autophagy-related genes [14].
Therefore, in this study, we detected the expression of autophagy-related molecules at the gene and protein levels. We found that the mRNA and protein expression levels of Atg5, Atg12, Atg16L, Beclin1 and Lc3B were significantly decreased in aged rats, suggesting that ovarian aging in these rats was accompanied by decreases in autophagic function. Khlil et al. observed that the mRNA and protein expression levels of Atg5 and Lc3B were significantly reduced in macrophages from aged mice [33]. In addition, Glatigny et al. noted decreased accumulation of the Lc3B-II, Beclin1 and Atg5 proteins in the hippocampi of 16-month-old mice [34]. In another study, decreased expression levels of Lc3B and Beclin1 were observed in the kidneys of aged rats [35]. Based on the above results, we suspect that ovarian aging in rats was accompanied by reductions in autophagic activity.
Gene expression levels are negatively correlated with DNA methylation levels [36, 37]. Stubbs et al. found considerable CpG methylation in the cerebral cortices, lungs, hearts and other tissues of aged mice [37]. In this study, we found DNA methylation changes associated with the Lc3B and Atg5 genes among all the downregulated autophagy-related genes in the ovaries of aged rats. Thus, we speculate that the decreases in autophagy gene expression in the ovaries of aged rats may be at least partly due to DNA methylation of some of these genes.
DNA methylation, or the addition of a methyl group onto DNA, is catalyzed by Dnmts, including Dnmt1, Dnmt2, and Dnmt3 (Dnmt3A and Dnmt3B). Dysregulation of DNA methylation has been implicated in diseases including neurological diseases, inflammatory diseases and cancers [38, 39]. In this study, we found that both the mRNA and protein expression levels of Dnmt3A and Dnmt3B were significantly increased in aged rats and that the increased expression of these genes might be related to increased methylation of the autophagy genes Atg5 and Lc3B. We also found that the expression of Dnmt1 and Dnmt2 in rat ovaries gradually decreased with aging; however, these decreases were not found to be related to the methylation of autophagy-related genes, indicating that Dnmt1 and Dnmt2 may regulate ovarian aging through other mechanisms. Previous studies have shown that Dnmt1 can regulate the functions of various types of stem cells, including embryonic stem cells and osteoblasts [40, 41]. We speculate that the role of Dnmt1 in aging may be related to the proliferation and dysfunction of stem cells [42]. The findings of several published studies are consistent with our data. For example, Fasolino et al. observed that Dnmt3A expression significantly increases in the striatum throughout aging in a mouse model of aging-related changes in DNA methylation [43]. In addition, Casillas et al. found that while Dnmt1 expression declines during aging, Dnmt3B expression increases during aging and in neoplastically transformed human fibroblasts [44]. Furthermore, Zhou et al. confirmed that the mRNA and protein expression levels of Dnmt3A and Dnmt3B are significantly elevated and that those of Dnmt1 are significantly downregulated in oligodendrocyte precursor cells from 16-month-old rats [45].
Conclusion
In this study, we found that decreased autophagic activity was involved in the declines in ovarian function in aging rats. Upregulation of the Dnmt3A and Dnmt3B may have led to methylation of the autophagy genes Atg5 and Lc3B to ultimately cause the observed decreases in autophagic activity.
Methods
Animals and sample collection
Female Sprague-Dawley (SD) rats were obtained from the experimental animal center of Harbin Medical University. The rats were divided into three groups (n = 8/group): the 6-month-old (6 M, adult) rat group, the 12-month-old (12 M, menopausal) rat group, and the 24-month-old (24 M, aged) rat group. The rats were anesthetized with 10% chloral hydrate, serum samples were obtained by centrifuging blood samples at 3000 rpm for 15 min at 4 °C, and the ovaries were either embedded in paraffin for histological analysis or frozen in liquid nitrogen and stored at − 80 °C for molecular analysis. The protocols for all animal experiments were approved by the Institutional Animal Care and Use Committee (IACUC) of Harbin Medical University.
Experimental plan
Female SD rats were divided into three groups (n = 8/group): the 6 M, 12 M and 24 M groups. Serum was collected and used for measurement of the hormone estradiol (E2) by radioimmunoassay (RIA). The mRNA and protein expression of autophagy genes in ovarian tissue was examined by real-time PCR and Western blot analysis. The DNA methyltransferases and methylation levels of autophagy genes were examined by real-time PCR and methylation-specific PCR (MSP).
Hormone detection
The levels of E2 in serum were detected using commercial RIA kits (Sino-UK Institute of Biological Technology, Beijing, China).
Western blot analysis
The protein expression levels of autophagy-related genes were determined by Western blot analysis. Proteins were extracted from ovaries using RIPA lysis buffer, and the protein concentrations were determined by BCA protein assay. Total protein (20 μg) was electrophoresed on 10% or 12% SDS-PAGE gels and transferred onto PVDF membranes (Immobilon-P, Millipore, USA). The membranes were blocked for 2 h in 5% milk/TBST and then incubated overnight at 4 °C with primary antibodies. β-Actin (1:6000), Atg5 (1:1000), Lc3B (1:1000), Atg16L (1:1000), Atg12 (1:1000) and Beclin1 (1:1000) primary antibodies were purchased from Cell Signaling Technology; Dnmt1 (1:250), Dnmt3A(1:250) and Dnmt3B (1:500) primary antibodies were purchased from Affinity Biosciences; and a Dnmt2 (1:500) primary antibody was purchased from Santa Cruz.. The following day, the membranes were incubated with horseradish peroxidase (HRP)-conjugated goat anti-rabbit or goat anti-mouse IgG (Santa Cruz, USA) for 1 h at room temperature. The blot bands were visualized using Pierce ECL Western Blot Substrate (Engreen Biosystem, China). β-Actin was used as the endogenous control and the relative band density was quantified by densitometry using ImageJ software.
Real-time PCR
The mRNA expression levels of autophagy-related genes were determined by real-time PCR (Applied Biosystems 7500). Total RNA was extracted from ovaries using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). First-strand cDNA was synthesized from the total RNA using a PrimeScript 1st Strand cDNA Synthesis Kit (Takara Bio Inc., China). The PCR products were amplified using a SYBR Premix Ex TaqII Kit (Takara Bio Inc., China) in 20 μL mixtures containing 1 μL of cDNA template and 0.5 μM forward and reverse primers (Table 1). β-Actin was used as the endogenous control, and the data were evaluated by the 2-ΔΔCt method.
Table 1.
Sequences of the Primers Used in Real-Time PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Atg5 | CACTGGGACTTCTGCTCCTG | TTCTTCAACCAAGCCAAACC |
| Atg12 | AAACGAAGAAATGGGCTGTG | GAAGGGGCAAAGGACTGATT |
| Atg16L | CTGTGCTTTTCCCGTCTTTC | GCCCTGATTTGGTTTCCAC |
| Lc3B | GGTGTTTTTCTCCTGGTTTGG | GCACTTGGACTTCAGCCTTC |
| Beclin1 | GCCTCTGAAACTGGACACG | CCTCTTCCTCCTGGCTCTCT |
| Dnmt1 | CGACGACGCTAAGGACGATGATG | GCCTTGTTGCTCGCCTCTGTC |
| Dnmt2 | GCGGTTGCGAGAGGATGGAAC | ACGTCAATAGCAGCCACCACATG |
| Dnmt3B | AGACCAGAGGCCGCAGATCAAG | TCCGCTTCACCATCTCCATCTCC |
| Dnmt3A | TGCCAGTCATCCGCCACCTC | CTCCGTCCTCTCGTTCTTGGTG |
MSP
The methylation levels of the promoter regions of autophagy-related genes were assessed using MSP. Genomic DNA was extracted from ovaries using a MiniBEST Universal Genomic DNA Extraction Kit (Takara Bio Inc., China) and then treated with an EZ DNA Methylation-Gold Kit (Zymo Research, USA) for bisulfite conversion. The converted DNA was used as a template for MSP amplification with specific primers (Table 2) designed by Methylprimer online (http://www.urogene.org/methprimer/indexl.html). The MSP products were analyzed on 3% agarose gels, and quantitative analyses were performed using ImageJ software.
Table 2.
Sequences of the Primers Used in Methylation-Specific PCR
| Gene | Forward primer | Reverse primer |
|---|---|---|
| Atg5-M | GTTGGTTTAAGAAGGAAATTAGCG | CCCGAACTACTCCTAAACGTA |
| Atg5-U | GGTTTAAGAAGGAAATTAGTGGAT | ACAAAACCCAAACTACTCCTAAACATA |
| Atg12-M | ATAGATTAGTTAGGGCGAGTAGCG | CCCGACTATTTCCTATCGAA |
| Atg12-U | AGATTAGTTAGGGTGAGTAGTGAGT | TCAACCCAACTATTTCCTATCAA |
| Atg16L-M | GTAAGGAAATAGATATTAGCGGAT | GACCGAAAACGTCGTAAAAC |
| Atg16L-U | AAGTAGTAAGGAAATAGATATTAGTGGAT | AAACTCAACCAAAAACATCATAAAAC |
| Lc3B-M | GGAGATATATAAGGGAAGTGATCGTC | GACGCTATTTAAAAATCTTCTCG |
| Lc3B-U | AGGAGATATATAAGGGAAGTGATTGTT | CCAACACTATTTAAAAATCTTCTCA |
| Beclin1-M | TTTATAAGAGAGTATGGACGGTTTC | CCGATCGACTAACTAAAAACTTC |
| Beclin1-U | AATTTATAAGAGAGTATGGATGGTTTT | CCAATCAACTAACTAAAAACTTCC |
Statistics
All data in this study are expressed as the mean ± standard deviation. Statistical calculations were performed by one-way ANOVA using GraphPad Prism version 6.0 (GraphPad Software Inc.). P < 0.05 was considered to indicate a statistically significant difference.
Acknowledgements
Not applicable.
Abbreviations
- 12 M
12-month-old
- 24 M
24-month-old
- 6 M
6-month-old
- Dnmt1
DNA methyltransferases1
- Dnmt2
DNA methyltransferases 2
- Dnmt3A
DNA methyltransferases 3A
- Dnmt3B
DNA methyltransferases 3B
- Dnmts
DNA methyltransferases
- E2
Estradiol
- FSH
Follicle-stimulating hormone
- MSP
Methylation-specific PCR
Authors’ contributions
LQY designed study, acqusited data, drafted manuscript. CMH and WJ acqusited and analysed data. GQ, GXC, JXT and XSS acqusited data. ZH designed study, drafted and revised manuscript. All authors have read and approved the final manuscript.”
Funding
This work was supported by grants from the Chinese National Natural Science Foundation (HZ, 81571957), Harbin Medical University graduate innovative research projects (QYL, YJSCX2016-2HYD), and the Fundamental Research Funds for the Provincial Universities (JW, 2017-KYYWF-0297; MHC, 2019).
Availability of data and materials
Not applicable.
Ethics approval and consent to participate
This study makes use of rats, and the experimental protocol for the use of animal was approved by the Institutional Animal Care and Use Committee (IACUC) of Harbin Medical University.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Azzu V, Valencak TG. Energy metabolism and ageing in the mouse: a mini-review. Gerontol. 2017;63(4):327–336. doi: 10.1159/000454924. [DOI] [PubMed] [Google Scholar]
- 2.Cruz G, Fernandois D, Paredes AH. Ovarian function and reproductive senescence in the rat: role of ovarian sympathetic innervation. Reprod. 2017;153(2):R59–R68. doi: 10.1530/REP-16-0117. [DOI] [PubMed] [Google Scholar]
- 3.Wilkosz P, Greggains GD, Tanbo TG, Fedorcsak P. Female reproductive decline is determined by remaining ovarian reserve and age. PLoS One. 2014;9(10):e108343. doi: 10.1371/journal.pone.0108343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Q, Geng X, Zheng W, Tang J, Xu B, Shi Q. Current understanding of ovarian aging. Sci China Life Sci. 2012;55(8):659–669. doi: 10.1007/s11427-012-4352-5. [DOI] [PubMed] [Google Scholar]
- 5.Finch CE. The menopause and aging, a comparative perspective. J Steroid Biochem Mol Biol. 2014;142:132–141. doi: 10.1016/j.jsbmb.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen M, Jiang Y, Guan Z, Cao Y, Li L, Liu H, Sun SC. Protective mechanism of FSH against oxidative damage in mouse ovarian granulosa cells by repressing autophagy. Autophagy. 2017;13(8):1364–1385. doi: 10.1080/15548627.2017.1327941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Murphy KR, Baggett B, Cooper LL, Lu Y. J OU, Sedivy JM, Terentyev D, Koren G: enhancing autophagy diminishes aberrant Ca(2+) homeostasis and Arrhythmogenesis in aging rabbit hearts. Front Physiol. 2019;10:1277. doi: 10.3389/fphys.2019.01277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lapaquette P, Guzzo J, Bretillon L, Bringer MA. Cellular and molecular connections between autophagy and inflammation. Mediat Inflamm. 2015;2015:398483. doi: 10.1155/2015/398483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nagar R. Autophagy: a brief overview in perspective of dermatology. Indian J Dermatol, Venereol leprology. 2017;83(3):290–297. doi: 10.4103/0378-6323.196320. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh AK, Mau T, O'Brien M, Garg S, Yung R. Impaired autophagy activity is linked to elevated ER-stress and inflammation in aging adipose tissue. Aging. 2016;8(10):2525–2537. doi: 10.18632/aging.101083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhou J, Chong SY, Lim A, Singh BK, Sinha RA, Salmon AB, Yen PM. Changes in macroautophagy, chaperone-mediated autophagy, and mitochondrial metabolism in murine skeletal and cardiac muscle during aging. Aging. 2017;9(2):583–599. doi: 10.18632/aging.101181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Toth ML, Sigmond T, Borsos E, Barna J, Erdelyi P, Takacs-Vellai K, Orosz L, Kovacs AL, Csikos G, Sass M, et al. Longevity pathways converge on autophagy genes to regulate life span in Caenorhabditis elegans. Autophagy. 2008;4(3):330–338. doi: 10.4161/auto.5618. [DOI] [PubMed] [Google Scholar]
- 13.Komatsu M, Waguri S, Chiba T, Murata S, Iwata J, Tanida I, Ueno T, Koike M, Uchiyama Y, Kominami E, et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nat. 2006;441(7095):880–884. doi: 10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 14.Martinez-Lopez N, Athonvarangkul D, Singh R. Autophagy and aging. Adv Exp Med Biol. 2015;847:73–87. doi: 10.1007/978-1-4939-2404-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan CC, Yu JT, Tan MS, Jiang T, Zhu XC, Tan L. Autophagy in aging and neurodegenerative diseases: implications for pathogenesis and therapy. Neurobiol Aging. 2014;35(5):941–957. doi: 10.1016/j.neurobiolaging.2013.11.019. [DOI] [PubMed] [Google Scholar]
- 16.Abdellatif M, Sedej S, Carmona-Gutierrez D, Madeo F, Kroemer G. Autophagy in cardiovascular aging. Circ Res. 2018;123(7):803–824. doi: 10.1161/CIRCRESAHA.118.312208. [DOI] [PubMed] [Google Scholar]
- 17.Pal S, Tyler JK. Epigenetics and aging. Sci Adv. 2016;2(7):e1600584. doi: 10.1126/sciadv.1600584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Masser DR, Hadad N, Porter H, Stout MB, Unnikrishnan A, Stanford DR, Freeman WM. Analysis of DNA modifications in aging research. GeroScience. 2018;40(1):11–29. doi: 10.1007/s11357-018-0005-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barau J, Teissandier A, Zamudio N, Roy S, Nalesso V, Herault Y, Guillou F, Bourc'his D. The DNA methyltransferase DNMT3C protects male germ cells from transposon activity. Sci. 2016;354(6314):909–912. doi: 10.1126/science.aah5143. [DOI] [PubMed] [Google Scholar]
- 20.Uysal F, Ozturk S, Akkoyunlu G. Superovulation alters DNA methyltransferase protein expression in mouse oocytes and early embryos. J Assist Reprod Genet. 2018;35(3):503–513. doi: 10.1007/s10815-017-1087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang W, Xu J. DNA methyltransferases and their roles in tumorigenesis. Biomarker research. 2017;5:1. doi: 10.1186/s40364-017-0081-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burger HG. Physiology and endocrinology of the menopause. Med. 2006;34(1):27–30. doi: 10.1383/medc.2006.34.1.27. [DOI] [Google Scholar]
- 23.Acuna E, Fornes R, Fernandois D, Garrido MP, Greiner M, Lara HE, Paredes AH. Increases in norepinephrine release and ovarian cyst formation during ageing in the rat. Reprod Biol Endocrinol : RB&E. 2009;7:64. doi: 10.1186/1477-7827-7-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nie X, Dai Y, Zheng Y, Bao D, Chen Q, Yin Y, Fu H, Hou D. Establishment of a mouse model of premature ovarian failure using consecutive superovulation. Cell Physiol Biochem : Int J Exp Cell Physiol, Biochem Pharmacol. 2018;51(5):2341–2358. doi: 10.1159/000495895. [DOI] [PubMed] [Google Scholar]
- 25.Ravanan P, Srikumar IF, Talwar P. Autophagy: the spotlight for cellular stress responses. Life Sci. 2017;188:53–67. doi: 10.1016/j.lfs.2017.08.029. [DOI] [PubMed] [Google Scholar]
- 26.Mizushima N, Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147(4):728–741. doi: 10.1016/j.cell.2011.10.026. [DOI] [PubMed] [Google Scholar]
- 27.Dong Y, Chen H, Gao J, Liu Y, Li J, Wang J. Molecular machinery and interplay of apoptosis and autophagy in coronary heart disease. J Mol Cell Cardiol. 2019;136:27–41. doi: 10.1016/j.yjmcc.2019.09.001. [DOI] [PubMed] [Google Scholar]
- 28.Huang F, Wang BR, Wang YG. Role of autophagy in tumorigenesis, metastasis, targeted therapy and drug resistance of hepatocellular carcinoma. World J Gastroenterol. 2018;24(41):4643–4651. doi: 10.3748/wjg.v24.i41.4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hurley JH, Young LN. Mechanisms of autophagy initiation. Annu Rev Biochem. 2017;86:225–244. doi: 10.1146/annurev-biochem-061516-044820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. EMBO J. 2012;31(22):4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy. 2013;9(3):424–425. doi: 10.4161/auto.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schaaf MB, Keulers TG, Vooijs MA, Rouschop KM. LC3/GABARAP family proteins: autophagy-(un)related functions. FASEB J : Official Publication of the Fed Am Soc Exp Biol. 2016;30(12):3961–3978. doi: 10.1096/fj.201600698R. [DOI] [PubMed] [Google Scholar]
- 33.Khalil H, Tazi M, Caution K, Ahmed A, Kanneganti A, Assani K, Kopp B, Marsh C, Dakhlallah D, Amer AO. Aging is associated with hypermethylation of autophagy genes in macrophages. Epigenetics. 2016;11(5):381–388. doi: 10.1080/15592294.2016.1144007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Glatigny M, Moriceau S, Rivagorda M, Ramos-Brossier M, Nascimbeni AC, Lante F, Shanley MR, Boudarene N, Rousseaud A, Friedman AK, et al. Autophagy is required for memory formation and reverses age-related memory decline. Curr Biol : CB. 2019;29(3):435–448. doi: 10.1016/j.cub.2018.12.021. [DOI] [PubMed] [Google Scholar]
- 35.Diao C, Wang L, Liu H, Du Y, Liu X. Aged kidneys are refractory to autophagy activation in a rat model of renal ischemia-reperfusion injury. Clin Interv Aging. 2019;14:525–534. doi: 10.2147/CIA.S197444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bell JT, Pai AA, Pickrell JK, Gaffney DJ, Pique-Regi R, Degner JF, Gilad Y, Pritchard JK. DNA methylation patterns associate with genetic and gene expression variation in HapMap cell lines. Genome Biol. 2011;12(1):R10. doi: 10.1186/gb-2011-12-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stubbs TM, Bonder MJ, Stark AK, Krueger F, Team BIAC. von Meyenn F, Stegle O, Reik W. Multi-tissue DNA methylation age predictor in mouse. Genome Biol. 2017;18(1):68. doi: 10.1186/s13059-017-1203-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roubroeks JAY, Smith RG, van den Hove DLA, Lunnon K. Epigenetics and DNA methylomic profiling in Alzheimer's disease and other neurodegenerative diseases. J Neurochem. 2017;143(2):158–170. doi: 10.1111/jnc.14148. [DOI] [PubMed] [Google Scholar]
- 39.Bhat S, Kabekkodu SP, Varghese VK, Chakrabarty S, Mallya SP, Rotti H, Pandey D, Kushtagi P, Satyamoorthy K. Aberrant gene-specific DNA methylation signature analysis in cervical cancer. Tumour Biol : J Int Soc Oncodev Biol Med. 2017;39(3):1010428317694573. doi: 10.1177/1010428317694573. [DOI] [PubMed] [Google Scholar]
- 40.Hainer SJ, McCannell KN, Yu J, Ee LS, Zhu LJ, Rando OJ, Fazzio TG. DNA methylation directs genomic localization of Mbd2 and Mbd3 in embryonic stem cells. eLife. 2016;5. [DOI] [PMC free article] [PubMed]
- 41.Yu S, Pu Y, Jin H, Dian J, Ge T, Zhihe Z: [role of DNA methylation in regulation of osteogenic differentiation of stem cells]. Hua xi kou qiang yi xue za zhi = Huaxi kouqiang yixue zazhi = West Chin J Stomatol 2016, 34(5):526–530. [DOI] [PMC free article] [PubMed]
- 42.Ren X, Xiao X, Wang B, Zhang Y, Yi M, Li J, Shi W. Role of DNA methyltransferase 1 in mouse skin aging. Zhong nan da xue xue bao Yi xue ban = J Cent S Univ Med Sci. 2018;43(4):415–420. doi: 10.11817/j.issn.1672-7347.2018.04.013. [DOI] [PubMed] [Google Scholar]
- 43.Fasolino M, Liu S, Wang Y, Zhou Z. Distinct cellular and molecular environments support aging-related DNA methylation changes in the substantia nigra. Epigenomics. 2017;9(1):21–31. doi: 10.2217/epi-2016-0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Casillas MA, Jr, Lopatina N, Andrews LG, Tollefsbol TO Transcriptional control of the DNA methyltransferases is altered in aging and neoplastically-transformed human fibroblasts. Mol Cell Biochem. 2003;252(1–2):33–43. doi: 10.1023/A:1025548623524. [DOI] [PubMed] [Google Scholar]
- 45.Zhou J, Wu YC, Xiao BJ, Guo XD, Zheng QX, Wu B. Age-related changes in the global DNA methylation profile of Oligodendrocyte progenitor cells derived from rat spinal cords. Curr Med Sci. 2019;39(1):67–74. doi: 10.1007/s11596-019-2001-y. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.