Abstract
Rationale: The decades-long progression of chronic obstructive pulmonary disease (COPD) renders identifying different trajectories of disease progression challenging.
Objectives: To identify subtypes of patients with COPD with distinct longitudinal progression patterns using a novel machine-learning tool called “Subtype and Stage Inference” (SuStaIn) and to evaluate the utility of SuStaIn for patient stratification in COPD.
Methods: We applied SuStaIn to cross-sectional computed tomography imaging markers in 3,698 Global Initiative for Chronic Obstructive Lung Disease (GOLD) 1–4 patients and 3,479 controls from the COPDGene (COPD Genetic Epidemiology) study to identify subtypes of patients with COPD. We confirmed the identified subtypes and progression patterns using ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints) data. We assessed the utility of SuStaIn for patient stratification by comparing SuStaIn subtypes and stages at baseline with longitudinal follow-up data.
Measurements and Main Results: We identified two trajectories of disease progression in COPD: a “Tissue→Airway” subtype (n = 2,354, 70.4%), in which small airway dysfunction and emphysema precede large airway wall abnormalities, and an “Airway→Tissue” subtype (n = 988, 29.6%), in which large airway wall abnormalities precede emphysema and small airway dysfunction. Subtypes were reproducible in ECLIPSE. Baseline stage in both subtypes correlated with future FEV1/FVC decline (r = −0.16 [P < 0.001] in the Tissue→Airway group; r = −0.14 [P = 0.011] in the Airway→Tissue group). SuStaIn placed 30% of smokers with normal lung function at elevated stages, suggesting imaging changes consistent with early COPD. Individuals with early changes were 2.5 times more likely to meet COPD diagnostic criteria at follow-up.
Conclusions: We demonstrate two distinct patterns of disease progression in COPD using SuStaIn, likely representing different endotypes. One third of healthy smokers have detectable imaging changes, suggesting a new biomarker of “early COPD.”
Keywords: clustering, CT imaging, emphysema, bronchitis, chronic obstructive pulmonary disease
At a Glance Commentary
Scientific Knowledge on the Subject
Chronic obstructive pulmonary disease (COPD) progresses over decades, so little is known about longitudinal changes in individual patients and whether there are different patterns of disease progression in different patient subgroups.
What This Study Adds to the Field
Computational modeling of computed tomography biomarkers suggests there are two patterns of disease progression in COPD. These disease progression patterns or “subtypes” can be used to stratify individuals into two groups with distinct clinical characteristics and to stage individuals along their disease time-course. Early stages of both subtypes are identifiable in a proportion of “healthy smokers,” providing a biomarker of early COPD.
Chronic obstructive pulmonary disease (COPD) can be characterized as the consequence of a genetically susceptible individual being exposed to sufficient environmental exposures (1). The pulmonary components are heterogeneous (2) and include emphysema, small airway loss and obstruction, and larger airway inflammation. COPD progresses over decades and often remains subclinical until the later development of symptoms or exacerbations. Slow progression and heterogeneous manifestations make it challenging to construct long-term models of disease progression, as most studies collect only cross-sectional or short-term longitudinal data.
Incomplete understanding of disease progression and heterogeneity in COPD has consequences for clinical practice and drug development. First, we are currently unable to identify early stages of disease in healthy smokers, preventing interventions in early COPD where disease-modifying treatments may be most effective. Second, clinically relevant populations with severe airflow obstruction may have arrived at this point through different early mechanisms (endotypes), which may therefore have been amenable to different interventions (2).
Quantitative imaging of the lung through computed tomography offers the opportunity to better evaluate the complex relationship between structure and function in COPD. Specifically, airway wall geometry informs on chronic bronchitis, whereas emphysematous tissue destruction and gas trapping due to small airway obstruction and destruction can be quantified using density thresholds. Although this facilitates direct disease quantification, understanding the progression and heterogeneity of pathology detected by imaging measures has remained limited (3).
Previous imaging studies attempting to disentangle the heterogeneity of COPD have used clustering techniques (4, 5), probabilistic modeling (6, 7) or dimensionality reduction (8, 9). Clustering does not naturally group individuals on the same trajectory because patients at early and late stages of a cluster may look very different. Thus, these approaches confound disease subtypes with stage (see Glossary), preventing the identification of specific phenotypes independently of temporal progression. The ability to identify disease subtypes independently of disease stage has been a long-standing unmet need.
Significant progress in the understanding of neurodegenerative diseases has been made using techniques, collectively called “Disease Progression Modeling,” that reconstruct the long-term temporal progression of disease from cross-sectional data via unsupervised learning (10–15). Subtype and Stage and Inference (SuStaIn) (16) is a recent innovation arising from the study of dementia that integrates clustering and disease progression modeling, offering new ability to disentangle the heterogeneity of disease subtypes from assessment of disease stages. SuStaIn identifies subgroups of individuals (disease subtypes) with distinct progression patterns, while simultaneously reconstructing the trajectory (stage progression) of each subtype. Such data-driven progression models have not previously been applied in the field of respiratory medicine and offer a major opportunity to explain disease heterogeneity and enhance precision medicine in conditions of long natural history such as COPD.
Some results related to this study have been previously reported in the form of an abstract (17).
Methods
This is an abbreviated version of the method; please see the online supplement for further detail about each analysis step.
Definitions and Overview
Key terminology is defined in the Glossary.
Model development used computed tomography (CT) data from COPDGene (COPD Genetic Epidemiology) phase 1 (18), comprising a cross-sectional dataset of baseline measurements from 3,479 smoking controls and 3,698 patients with COPD. We repeated the SuStaIn algorithm using baseline data from 303 smoking controls and 1,809 patients with COPD in the ECLIPSE (Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints) study (19) to verify consistency of SuStaIn output in an independent dataset. We evaluated longitudinal progression using follow-up (phase 2) COPDGene scans and data to verify the SuStaIn subtype progression patterns (reconstructed from cross-sectional data) against true longitudinal progression of individual subjects. This included 1,929 subjects with COPD and 2,158 controls who had all imaging biomarkers available from the initial scan together with measures of lung function from both time points and a second dataset of 1,675 subjects with COPD and 1,939 controls who had all imaging biomarkers available at both phases of the COPDGene study.
GLOSSARY
SUBTYPE – a group of subjects who share a particular trajectory of biomarker evolution.
STAGE – the position on a subtype trajectory of an individual subject at a specific time. In Subtype and Stage Inference (SuStaIn) this represents the degree of abnormality in imaging biomarkers, and a change in stage occurs when an imaging biomarker becomes more abnormal relative to a control population.
DISEASE PROGRESSION – change in stage with time as the natural history of the condition unfolds. We use the term in two distinct contexts:
1. GROUP (SUBTYPE) LEVEL – referring to the sequence of changes that the typical patient undergoes from start to finish.
2. INDIVIDUAL LEVEL – change in stage or severity of an individual subject as biomarkers become increasingly abnormal.
Imaging features
A set of four imaging features were derived in COPDGene: 1) emphysema, obtained using parametric response mapping (20), 2) functional small airway disease (fSAD) obtained from parametric response mapping, 3) Pi10 square root wall area (SRWA) (21), and 4) segmental airway wall thickness. CT analysis to obtain the imaging features was performed using Thirona lung quantification software (Thirona, http://www.thirona.eu) (18). There were only two imaging features available in the ECLIPSE study: emphysema and Pi10 SRWA, obtained using VIDA software (22).
Disease Progression Modeling
Given a cross-sectional dataset, SuStaIn simultaneously identifies a set of disease subtypes, each defined by a distinct trajectory of biomarker evolution with a probabilistic assignment of each subject to a subtype and stage along the corresponding trajectory. The trajectory of each subtype is described as a linear z-score model (16), consisting of a series of stages, in which each stage corresponds to a biomarker reaching a particular z-score relative to a control group. The optimal number of subtypes is determined using information criterion (a statistical technique that balances model complexity with model accuracy). This provides a population-level disease progression model, which can be used to assign individuals to subtypes and stages probabilistically. A conceptual overview is provided as Figure E1 in the online supplement (16).
Identification of COPD subtypes
We applied the SuStaIn algorithm (16) to COPD Global Initiative for Chronic Obstructive Lung Disease (GOLD)1–4 patients from the COPDGene dataset. As SuStaIn requires monotonic measurements (biomarkers that change over time in one direction only; see Discussion), we replaced fSAD, which may convert to emphysema at later stages of COPD (20), with a combined measure we term “overall tissue damage.” This was computed as the sum of fSAD and emphysema (and thus is similar to a measure of air trapping). As SuStaIn requires input features expressed as z-scores relative to a control population, we transformed each dataset into z-scores relative to the smoking controls in COPDGene. Prior to performing the z-score transformation, imaging measures were log-transformed to improve normality.
Independent evaluation of COPD subtypes
To evaluate the subtypes in an independent dataset we repeated our analysis in COPD GOLD 1–4 patients from ECLIPSE using the subset of CT metrics available from inspiratory scans and the corresponding ECLIPSE smoking controls to perform z-score transformation. As ECLIPSE only has inspiratory scans we refitted the SuStaIn algorithm to a COPDGene cross-sectional dataset consisting of baseline measurements from 4,102 smoking controls and 4,152 patients with COPD with inspiratory measurements available for emphysema and Pi10 SRWA. We refer to these data as the “Inspiratory COPDGene” dataset.
Subtyping and staging
We used the SuStaIn model (i.e., the subtype progression patterns identified using the SuStaIn algorithm) to automatically assign individuals to their most probable subtype and stage. We did this for all COPDGene patients with COPD and control subjects at each of the two visits. We repeated the same process of assigning individuals to SuStaIn subtypes and stages in the ECLIPSE and Inspiratory COPDGene datasets. We further assigned individuals from COPDGene phase 2 to subtypes and stages using the same procedure described above, identifying the subtypes and stages from the subtype progression patterns estimated using the COPDGene phase 1 dataset.
Statistical Analysis
Clinical characteristics of the subtypes
We compared the clinical characteristics of individuals assigned to each subtype using two sample t tests for continuous variables, chi-squared tests for categorical variables, and Mann-Whitney U tests for frequency data.
Relationship between SuStaIn stage and lung function
We verified that SuStaIn stage could be used as a measure of disease severity in COPD by examining whether SuStaIn stage correlated with spirometric impairment as assessed by FEV1/FVC and FEV1% predicted. We further evaluated whether a higher SuStaIn stage could be used as an indicator of future lung function decline (disease progression at an individual level) by assessing whether baseline SuStaIn stage was correlated with change in lung function between baseline and follow-up.
Longitudinal consistency of subtype and stage
Over time we would expect that subtype remains consistent but that stage will progress. We assessed whether the SuStaIn subtypes remained consistent at 5-year follow-up, quantifying consistency as the percentage of individuals in which the subtype assignment remained the same. We assessed whether individuals progressed in SuStaIn stage between baseline and follow-up by comparing the distribution of SuStaIn stages at baseline and follow-up in GOLD 1–2 and GOLD 3–4 patients using two sample t tests.
Analysis of smoking controls
We repeated the above analyses in the COPDGene smoking control group to test whether SuStaIn subtype and stage might be useful for identification of otherwise healthy individuals at risk of developing COPD.
Results
Subject Characteristics
The baseline data of the COPDGene study participants used to develop the model are reported in Table 1. The control population (n = 3,479) was used to derive the z-scores, whereas the GOLD 1–4 patients (n = 3,698) were used to produce the subtypes.
Table 1.
Basic Demographics for the COPDGene Control and COPD Populations Used in Deriving the SuStaIn Subtype Trajectories
| Parameter | Control Subjects | Subjects with COPD |
|---|---|---|
| Subjects, n | 3,479 | 3,698 |
| Age, yr, mean (SD) | 56.90 (8.45) | 63.13 (8.61) |
| Sex, M, n (%) | 1,816 (52) | 2,087 (56) |
| Sex, F, n (%) | 1,663 (48) | 1,611 (44) |
| GOLD stage 1, n (%) | NA | 643 (17) |
| GOLD stage 2, n (%) | NA | 1,616 (44) |
| GOLD stage 3, n (%) | NA | 960 (26) |
| GOLD stage 4, n (%) | NA | 479 (13) |
| Smoking history, pack-year, mean (SD) | 37.33 (20.04) | 51.91 (26.99) |
| Exacerbations, n/yr, mean (SD) | 0.13 (0.53) | 0.64 (1.18) |
Definition of abbreviations: COPD = chronic obstructive pulmonary disease; COPDGene = COPD Genetic Epidemiology; GOLD = Global Initiative for Chronic Obstructive Lung Disease; NA = not applicable; SuStaIn = Subtype and Stage Inference.
Cross-sectional analyses in COPD
COPD subtypes
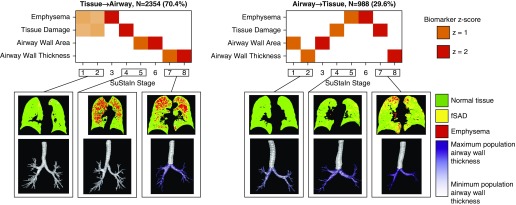
SuStaIn identified two distinct COPD progression patterns or “subtypes” (Figure 1). We have termed these “Tissue→Airway” and “Airway→Tissue.” In the Tissue→Airway group (n = 2,354, 70.4%), fSAD and emphysema are the earliest disease stages. Only subsequent to this do pathological alterations in larger airways become apparent. In the Airway→Tissue subgroup (n = 988, 29.6%), the earliest stages comprise abnormalities in larger airways, followed by fSAD and emphysema. These subtypes were reproducible in the ECLIPSE study (Figure E2 and online supplement, Results).
Figure 1.
Disease progression patterns predicted by Subtype and Stage Inference (SuStaIn). Chronic obstructive pulmonary disease is characterized by two distinct disease progression models (top row). In the Tissue→Airway subtype (70%; top left) the presence of emphysema and functional small airway disease (fSAD) initiates disease progression followed by later emergence of pathology in larger airways. (The overall tissue damage measure captures the presence of both fSAD and emphysema.) In the Airway→Tissue subtype (30%; top right), disease progression is initiated by pathology in the larger airways before the development of fSAD and emphysema. At each SuStaIn stage a new z-score event occurs when a feature transitions to a new severity level, as indexed by a z-score with respect to the control population; z-scores of z = 1 (orange) and z = 2 (red). Higher opacity represents a higher confidence in the ordering. The bottom row visualizes the parametric response mapping images and airway wall thickness values for representative patients at different SuStaIn subtypes and stages. The airway wall thickness values are visualized using a purple color scale on top of an airway tree segmentation, with the minimum value of the color scale corresponding to the first percentile of airway wall thickness values across the population and the maximum value of the color scale corresponding to the 99th percentile. In the Tissue→Airway subtype, the first individual (early stage) has early tissue damage visible at the outer edges of the lung but no airway wall changes, the second individual (middle stage) has visible tissue damage but no airway changes, and the third individual (late stage) has severe tissue damage together with airway wall thickening. In the Airway→Tissue subtype, the first individual (early stage) has early signs of airway wall thickening but no visible tissue damage, the second individual (middle stage) has clear signs of airway wall thickening but very little visible tissue damage, and the third individual (late stage) has severe airway wall thickening and tissue damage.
Clinical characteristics of the COPD subtypes
We next investigated differences in the clinical characteristics of patients between the two subtypes (Table 2). There was a smaller proportion of men in the Tissue→Airway subtype, compared with the Airway→Tissue subtype (52.3% vs. 66.5%; P < 0.001). Patients in the Tissue→Airway group had a significantly lower body mass index (BMI) than those in the Airway→Tissue group (26.65 vs. 30.54 kgm−2; P < 0.001) and a lower prevalence of chronic bronchitis (25.1% vs. 31.8%; P < 0.001). Detailed relationships between subtype, stage, breathlessness, and exacerbations are reported in Table E3. Patients in the Tissue→Airway group had marginally more severe spirometric impairment (FEV1% predicted 53.63% vs. 58.64%; P < 0.001; FEV1/FVC ratio 0.49 vs. 0.56; P < 0.001). The clinical characteristics of the SuStaIn subtypes were broadly replicable in the ECLIPSE dataset (Tables E4 and E5).
Table 2.
Demographics of Patients in the Tissue→Airway and Airway→Tissue Subtypes
| Feature | Tissue→Airway | Airway→Tissue | P Value |
|---|---|---|---|
| Number of patients, n (%) | 2,354 (70.4%) | 988 (29.6%) | |
| Sex, M, n (%) | 1,230 (52.3) | 657 (66.5) | P < 0.001 |
| Sex, F, n (%) | 1,124 (47.7) | 331 (33.5) | |
| Age, yr, mean (SD) | 63.18 (8.14) | 63.17 (9.49) | P = 0.92 |
| BMI, kg/m2, mean (SD) | 26.65 (5.43) | 30.54 (6.28) | P < 0.001 |
| FEV1, % predicted, mean (SD) | 53.63 (23.05) | 58.64 (17.74) | P < 0.001 |
| FEV1/FVC ratio, mean (SD) | 0.49 (0.14) | 0.56 (0.11) | P < 0.001 |
| GOLD stage 1, n (%) | 340 (14.4) | 103 (10.4) | P < 0.001 |
| GOLD stage 2, n (%) | 908 (38.6) | 559 (56.6) | |
| GOLD stage 3, n (%) | 680 (28.9) | 273 (27.6) | |
| GOLD stage 4, n (%) | 426 (18.1) | 53 (5.4) | |
| Smoking history, pack-year, mean (SD) | 53.10 (26.42) | 50.35 (26.12) | P = 0.006 |
| Exacerbations, n/yr, mean (SD) | 0.71 (1.23) | 0.62 (1.16) | P = 0.018 |
| Chronic bronchitis, n (%) | 591 (25.1) | 314 (31.8) | P < 0.001 |
| Percent emphysema, mean (SD) | 15.17 (13.67) | 4.08 (6.46) | P < 0.001 |
| Percent fSAD, mean (SD) | 28.89 (11.86) | 20.18 (12.83) | P < 0.001 |
| Percent tissue damage, mean (SD) | 44.06 (20.79) | 24.26 (17.57) | P < 0.001 |
| Airway wall area percentage, mean (SD) | 51.94 (6.81) | 61.77 (6.21) | P < 0.001 |
| Pi10 SRWA, mm, mean (SD) | 2.52 (0.48) | 3.13 (0.56) | P < 0.001 |
| Airway wall thickness, mm, mean (SD) | 1.06 (0.19) | 1.34 (0.21) | P < 0.001 |
Definition of abbreviations: BMI = body mass index; fSAD = functional small airway disease; GOLD = Global Initiative for Chronic Obstructive Lung Disease; SRWA = square root wall area.
We report two sample t tests for continuous variables, chi-squared test for categorical variables, and Mann-Whitney U test results for frequency data. Only patients at Subtype and Stage Inference stages ≥1 were included.
Relationship of COPD subtype stage with baseline lung function
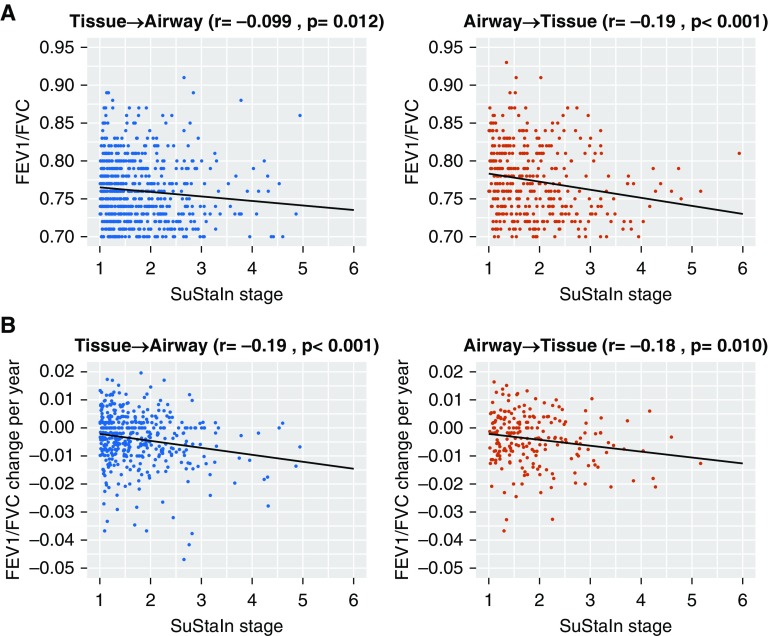
We investigated whether SuStaIn stage could be used as a measure of disease severity in COPD by examining correlations between SuStaIn stage and baseline spirometry. We found a significant correlation between SuStaIn stage and FEV1/FVC (Figure 2A) and FEV1% predicted (Figure E3A). The relationship was stronger in the Tissue→Airway group: SuStaIn stage correlation with FEV1/FVC and FEV1% predicted r = −0.63 (P < 0.001) and r = −0.66 (P < 0.001), respectively. In the Airway→Tissue group, the correlation coefficients were −0.58 (P < 0.001) for FEV1/FVC and −0.51 (P < 0.001) for FEV1% predicted. The relationship between SuStaIn stage and baseline lung function was nonlinear in the Tissue→Airway subtype and linear in the Airway→Tissue subtype (online supplement, Results). The correlations between baseline lung function and SuStaIn stage were replicable in the ECLIPSE dataset (Figures E4 and E5).
Figure 2.
Relationship between Subtype and Stage Inference (SuStaIn) stage and lung function. (A) Scatterplot of cross-sectional spirometry versus SuStaIn stage for the Tissue→Airway and Airway→Tissue subtypes. A linear and a quadratic model are fitted to the data via a least-squares estimation to gauge the relationship between SuStaIn stage and markers of lung function. In the Tissue→Airway subtype, there is a visible nonlinear relationship between lung function and SuStaIn stage, with a more rapid decrease in lung function at earlier SuStaIn stages. The decline in lung function in the Airway→Tissue subgroup is linear and less rapid at earlier SuStaIn stages. (B) Scatterplot of measured decline in spirometry versus baseline SuStaIn stage for the Tissue→Airway and Airway→Tissue subtypes in Global Initiative for Chronic Obstructive Lung Disease (GOLD) 1–2 subjects. In both the Tissue→Airway and Airway→Tissue subtypes, SuStaIn stage at baseline correlated with future decline in lung function measured using FEV1/FVC.
Longitudinal analyses in COPD
Relationship of SuStaIn stage with longitudinal decline in lung function
We tested whether baseline SuStaIn stage correlated with future decline in lung function in the subset of individuals with spirometry available at both time points (patient characteristics reported in Table E6). Earlier SuStaIn stages were associated with more rapid future, measured individual level progression of FEV1/FVC ratio and FEV1% predicted. Considering the annualized change in spirometry after 5-year follow-up in GOLD 1–2 patients (Figure 2B for FEV1/FVC ratio and Figure E3B for FEV1% predicted), we found that baseline SuStaIn stage correlated with rate of decline in FEV1/FVC and FEV1% predicted in both subtypes: r = −0.16 (P < 0.001) and r = −0.14 (P = 0.011) for baseline SuStaIn stage and change in FEV1/FVC in the Tissue→Airway and Airway→Tissue groups, respectively; and r = −0.20 (P < 0.001) and r = −0.14 (P = 0.011) between baseline SuStaIn stage and change in FEV1% predicted. In GOLD 3–4 patients assigned to the Tissue→Airway subtype there was no significant correlation between baseline SuStaIn stage and change in FEV1/FVC (r = −0.001; P = 0.98) or FEV1% predicted (r = −0.019; P = 0.69). In GOLD 3–4 patients assigned to the Airway→Tissue subtype there was a significant correlation between baseline SuStaIn stage and change in FEV1/FVC (r = −0.23; P = 0.005), but this was not reflected in the FEV1% predicted measure (r = −0.15; P = 0.069).
Stability of COPD subtype and progression of COPD stage over time
SuStaIn assumes that individuals belong to a single disease subtype, progressing only in stage with time. We verified that the SuStaIn subtypes remained the same at 5-year follow-up using a longitudinal validation dataset consisting of COPDGene individuals who had all imaging biomarkers available at both phases (Tables E7–E9). The assignment to Tissue→Airway and Airway→Tissue subtypes remained consistent in 1,283/1,472 (87%) individuals. SuStaIn stages showed a strong correlation between baseline and follow-up, but individuals tended to progress in stage within each subtype (Figure E6), giving confidence that the model is a good representation of disease. Individual stage progression was more rapid in GOLD 1–2 patients than GOLD 3–4 patients (online supplement, Results), supporting the clinically important hypothesis that disease activity is greatest earlier in disease, whereas spirometrically more severe disease may be considered less active.
Analyses in control smokers without COPD
Early detection of individuals at risk for COPD in the control population
We hypothesized that a subset of the smoking control population would exhibit features of early COPD SuStaIn stages despite spirometry within the normal range. The majority of control patients were staged at SuStaIn stage 0 (n = 2,457, 71%). By considering control subjects assigned a stage >0, we were able to identify a group of control subjects (29%) with imaging abnormalities. There were 641 control subjects (18% of the control population) in the Tissue→Airway subtype and 381 subjects (11% of the control population) in the Airway→Tissue subtype. Moreover, within each respective subtype, there were 37 (6%) and 40 (10%) individuals at SuStaIn stages ≥3.
Relationship of SuStaIn stage with lung function in the control population
We tested whether nonzero SuStaIn stage could be used as a marker of early disease in the control population by testing for associations with lung function. SuStaIn stage was associated with baseline lung function and longitudinal decline in lung function in the control population (see Figure 3 for FEV1/FVC ratio, Figure E7 for FEV1% predicted, and online supplement, Results).
Figure 3.
Relationship between lung function and Subtype and Stage Inference (SuStaIn) stage in smoking controls. Baseline SuStaIn stage is associated with cross-sectional and longitudinal changes in airflow obstruction in smoking controls. (A) Scatterplot of baseline values FEV1/FVC versus SuStaIn stage in the control population. (B) Scatterplot of longitudinal change in FEV1/FVC per year versus SuStaIn stage in the control population.
Longitudinal SuStaIn subtype and stage in the control population
We tested the consistency of the SuStaIn subtype assignments in the smoking controls at 5-year follow-up (Table E10). At 5-year follow-up the assignment to Tissue→Airway and Airway→Tissue subtypes remained consistent in 86% of individuals. We verified that the SuStaIn stages were broadly similar at follow-up in the control population. The SuStaIn stages at baseline and follow-up showed a strong correlation (Figure E8): r = 0.48 (P < 0.001) in the Tissue→Airway subgroup, and r = 0.61 (P < 0.001) in the Airway→Tissue subgroup.
Progression to COPD in the control population
Finally, we tested whether those controls assigned to Tissue→Airway and Airway→Tissue subtypes had a greater individual risk of disease progression compared with those who were normal (SuStaIn stage 0), as measured by a classification of GOLD stage 1 or greater at follow-up. Of the SuStaIn 0 controls, 8.7% progressed to GOLD stage 1 at follow-up, compared with 23.0% of the Tissue→Airway subtype and 20.9% of the Airway→Tissue subtype. This represents a significantly higher rate of progression to COPD among those assigned to SuStaIn subtypes compared with those with normal imaging metrics (P < 0.001, chi-squared test) and therefore that SuStaIn provides a biomarker of early COPD.
Discussion
We report the first application of SuStaIn in COPD, replicating the subtypes identified by SuStaIn in a separate cohort at baseline, and over time in the original cohort. SuStaIn identifies two distinct patterns (subtypes) of COPD disease progression, and early stages of both subtypes were detectable in 29% of healthy smokers. Seventy percent of subjects with COPD comprise a Tissue→Airway subtype that follows a progression model in which abnormalities in the small airways (fSAD) and emphysema develop before measurable changes in larger airways. A minority of subjects (30%) comprise an Airway→Tissue subtype in which disease starts in the larger airways before the later development of emphysema and small airway dysfunction. SuStaIn disease stages correlate with cross-sectional and longitudinal markers of spirometric impairment, with greater loss of lung function at earlier SuStaIn stages of disease. The assignment to Tissue→Airway and Airway→Tissue subtypes remained consistent at 5-year follow-up, whereas individuals tended to progress in stage. Progression through stages was more rapid in earlier disease. We therefore identify two distinct patterns of subtype progression in COPD, of potential utility in the clinic and clinical trials, and provide a biomarker of early COPD in smoking controls.
The long natural history of COPD, over decades, has prevented any single study reporting on longitudinal disease progression in individual patients. Disease progression modeling provides a potential solution. Our findings are important for a number of reasons. First, we show that different subjects are on different disease trajectories and may therefore represent distinct endotypes requiring different interventions. Second, we provide early identification of people at risk of developing COPD, while spirometry is still normal. Reducing the future burden of COPD requires both early identification of smokers likely to develop the condition and targeted therapy. Finally, our modeling suggests that later stages of COPD progress more slowly and therefore that disease activity may be greatest in early disease, where treatment and prevention should be targeted.
The Tissue→Airway and Airway→Tissue subtypes we have defined mirror, to some extent, recognized descriptions of COPD, while providing a novel imaging biomarker for early disease stratification. Historically, typical phenotypes of COPD have been referred to as “pink puffers” and “blue bloaters” (23). The relative presence of chronic bronchitis or emphysema in addition to significant differences in BMI characterized these classic phenotypes. Such features are also seen in our results, with patients in the Tissue→Airway subtype having a significantly lower BMI and lower incidence of chronic bronchitis compared with those in the Airway→Tissue subtype.
Various studies have shown that inflammatory changes in the small airways are fundamental processes driving the progression and severity of COPD (24). Our results also suggest that the small airways, emphysema, and bronchitis are the principal drivers of COPD progression but that these occur in different proportions and at different times in the two different groups. Just as Hogg and colleagues (24) showed that a cascade of inflammatory processes lead to small airway disease and lung function impairment, it is possible that the distinct subtypes we have identified are a function of distinct inflammatory mechanisms (25) with consequent differences in progression patterns. The ability of SuStaIn to separate patients into distinct subtypes at early stages could enable the characterization of different COPD endotypes.
SuStaIn posits that cross-sectional patient measurements arise from different stages along a disease time course and that there are distinct groups of individuals (disease subtypes) that undergo different patterns of disease progression. The assumption is that variation in both subtype and stage produces heterogeneity in observed disease biomarkers. Previous findings align with this assumption. The study by Vestbo and colleagues (2) demonstrates highly variable decline in FEV1 in 3-year longitudinal data. As lung function impairment arises from the bulk effect of complex pathological abnormalities in lung structure, different proportions and types of structural damage could explain this variability across patients. The fact that we observed different rates of FEV1 decline within different subtypes supports this explanation. We therefore demonstrate that changes measured solely by imaging may be used to disentangle subtypes of patients who experience different trajectories of lung function impairment, imperceptible with bulk physiological measurements. Early-life factors might also affect the trajectory of lung function decline and risk of developing COPD, but information on these is unfortunately not available in the COPDGene and ECLIPSE cohorts.
Previous research has provided a strong case for early detection of COPD, yet this remains challenging in practice. Fletcher and Peto (26) described the rate of lung function decline in COPD, suggesting slow decline at onset followed by a more rapid phase in advanced disease. Recent studies have suggested that faster decline in lung function impairment occurs earlier in disease (27), particularly in mild-to-moderate COPD (27, 28). These results are mirrored in studies showing that smokers may develop emphysema on CT before abnormal lung function (29, 30). Undetected structural alterations may be critical in the early, accelerated decline of lung function and the subsequent course of COPD. Our results support this, as we show that early, undetected pathological changes are present in a proportion of healthy smokers, whereas lung function decline is accelerated at earlier stages of disease in the Tissue→Airway subtype. Moreover, our work adds a new dimension to existing models of disease progression in COPD (26, 27) by disentangling how lung function changes with disease across the COPD population, helping to explain heterogeneity in lung function decline (2).
Our findings are clinically and statistically significant despite the limited precision of some CT metrics. The attenuation value of a voxel is dependent on several factors, such as radiation dose, scanner modality, the reconstruction kernel, and inspiration level (31). CT scans in COPDGene were not spirometrically gated. Variations in inspiration across patients may cause errors in the measurement of emphysema. Moreover, measurements relating to the airway tree are averages of six bronchial paths in the upper and intermediate zones of the lung (18). Nonetheless, we demonstrate that the SuStaIn subtype trajectories derived from these imaging metrics are reproducible in both a separate cohort and in the same cohort over time and have strong stratification capabilities in separating individuals with distinct clinical characteristics and patterns of lung function decline. SuStaIn does assume that progression is one directional and that disease cannot regress—it is not known if this may occur in early stages of disease, and the explanation for CT abnormalities in a proportion of people with normal spirometry requires further study.
In conclusion, we report the first use of SuStaIn to study disease progression in COPD, as an exemplar chronic respiratory disease. Using this technique, we report the following novel findings. First, there are two distinct subtypes of COPD—the majority of patients develop small airway disease and emphysema before large airway wall changes, but a significant minority (30%) develop large airway wall changes first. Second, the relationship with lung function in these subtypes is different, with a more rapid initial decline in lung function (greater disease activity) observed in the Tissue→Airway group. This may explain the heterogeneity observed in FEV1 decline across COPD populations. Finally, the technique suggests that a group of healthy subjects with early COPD at risk of disease progression can be identified using CT biomarkers. In heterogeneous long-term conditions such as COPD there is a real need to better stratify patients for targeted therapy. SuStaIn provides a novel technique to achieve this and a mechanism for detection of early disease.
Supplementary Material
Acknowledgments
Acknowledgment
COPDGene Investigators – Core Units
Administrative Center: James D. Crapo (PI), Edwin K. Silverman (PI), Barry J. Make, and Elizabeth A. Regan.
Genetic Analysis Center: Terri Beaty, Ferdouse Begum, Peter J. Castaldi, Michael Cho, Dawn L. DeMeo, Adel R. Boueiz, Marilyn G. Foreman, Eitan Halper-Stromberg, Lystra P. Hayden, Craig P. Hersh, Jacqueline Hetmanski, Brian D. Hobbs, John E. Hokanson, Nan Laird, Christoph Lange, Sharon M. Lutz, Merry-Lynn McDonald, Margaret M. Parker, Dandi Qiao, Elizabeth A. Regan, Edwin K. Silverman, Emily S. Wan, Sungho Won, Phuwanat Sakornsakolpat, and Dmitry Prokopenko.
Imaging Center: Mustafa Al Qaisi, Harvey O. Coxson, Teresa Gray, MeiLan K. Han, Eric A. Hoffman, Stephen Humphries, Francine L. Jacobson, Philip F. Judy, Ella A. Kazerooni, Alex Kluiber, David A. Lynch, John D. Newell, Jr., Elizabeth A. Regan, James C. Ross, Raul San Jose Estepar, Joyce Schroeder, Jered Sieren, Douglas Stinson, Berend C. Stoel, Juerg Tschirren, Edwin Van Beek, Bram van Ginneken, Eva van Rikxoort, George Washko, and Carla G. Wilson.
PFT QA Center, Salt Lake City, UT: Robert Jensen.
Data Coordinating Center and Biostatistics, National Jewish Health, Denver, CO: Douglas Everett, Jim Crooks, Camille Moore, Matt Strand, and Carla G. Wilson.
Epidemiology Core, University of Colorado Anschutz Medical Campus, Aurora, CO: John E. Hokanson, John Hughes, Gregory Kinney, Sharon M. Lutz, Katherine Pratte, and Kendra A. Young.
Mortality Adjudication Core: Surya Bhatt, Jessica Bon, MeiLan K. Han, Barry J. Make, Carlos Martinez, Susan Murray, Elizabeth A. Regan, Xavier Soler, and Carla G. Wilson.
Biomarker Core: Russell P. Bowler, Katerina Kechris, and Farnoush Banaei-Kashani.
COPDGene Investigators – Clinical Centers
Ann Arbor VA, Ann Arbor, MI: Jeffrey L. Curtis, Carlos H. Martinez, and Perry G. Pernicano.
Baylor College of Medicine, Houston, TX: Nicola Hanania, Philip Alapat, Mustafa Atik, Venkata Bandi, Aladin Boriek, Kalpatha Guntupalli, Elizabeth Guy, Arun Nachiappan, and Amit Parulekar.
Brigham and Women’s Hospital, Boston, MA: Dawn L. DeMeo, Craig P. Hersh, Francine L. Jacobson, M.D., and George Washko.
Columbia University, New York, NY: R. Graham Barr, John Austin, Belinda D’Souza, Gregory D. N. Pearson, Anna Rozenshtein, and Byron Thomashow.
Duke University Medical Center, Durham, NC: Neil MacIntyre, Jr., H. Page McAdams, and Lacey Washington.
HealthPartners Research Institute, Minneapolis, MN: Charlene McEvoy and Joseph Tashjian.
Johns Hopkins University, Baltimore, MD: Robert Wise, Robert Brown, Nadia N. Hansel, Karen Horton, Allison Lambert, and Nirupama Putcha.
Los Angeles Biomedical Research Institute at Harbor UCLA Medical Center, Torrance, CA: Richard Casaburi, Alessandra Adami, Matthew Budoff, Hans Fischer, Janos Porszasz, Harry Rossiter, and William Stringer.
Michael E. DeBakey VA Medical Center, Houston, TX: Amir Sharafkhaneh and Charlie Lan.
Minneapolis VA Health Care System, Minneapolis, MN: Christine Wendt and Brian Bell.
Morehouse School of Medicine, Atlanta, GA: Marilyn G. Foreman, Eugene Berkowitz, and Gloria Westney.
National Jewish Health, Denver, CO: Russel P. Bowler and David A. Lynch.
Reliant Medical Group, Worcester, MA: Richard Rosiello and David Pace.
Temple University, Philadelphia, PA: Gerard Criner, David Ciccolella, Francis Cordova, Chandra Dass, Gilbert D’Alonzo, Parag Desai, Michael Jacobs, Steven Kelsen, Victor Kim, A. James Mamary, Nathaniel Marchetti, Aditi Satti, Kartik Shenoy, Robert M. Steiner, Alex Swift, Irene Swift, and Maria Elena Vega-Sanchez.
University of Alabama, Birmingham, AL: Mark Dransfield, William Bailey, Surya Bhatt, Anand Iyer, Hrudaya Nath, and J. Michael Wells.
University of California, San Diego, CA: Joe Ramsdell, Paul Friedman, Xavier Soler, and Andrew Yen.
University of Iowa, Iowa City, IA: Alejandro P. Comellas, Karin F. Hoth, John D. Newell, Jr., and Brad Thompson.
University of Michigan, Ann Arbor, MI: MeiLan K. Han, Ella A. Kazerooni, and Carlos H. Martinez.
University of Minnesota, Minneapolis, MN: Joanne Billings, Abbie Begnaud, and Tadashi Allen.
University of Pittsburgh, Pittsburgh, PA: Frank Sciurba, Jessica Bon, Divay Chandra, Carl Fuhrman, and Joel Weissfeld.
University of Texas Health Science Center, San Antonio, San Antonio, TX: Antonio Anzueto, Sandra Adams, Diego Maselli-Caceres, and Mario E. Ruiz.
Footnotes
Supported by Engineering and Physical Sciences Research Council (EPSRC) grants EP/H046410/1 and EP/K502959/1 (F.J.S.B.); University College London Hospitals (UCLH) NIHR Research Capability Funding Senior Investigator Award under grant RCF107/DH/2014 (F.J.S.B. and D.J.H.); EPSRC Doctoral Prize Fellowship and MRC Skills Development Fellowship (A.L.Y.); EPSRC Centre For Doctoral Training in Medical Imaging grant EP/L016478/1 and Industrial Fellowship from the Royal Commission for the Exhibition of 1851 (B.R.); an industrial CASE studentship with funding from GlaxoSmithKline Research and Development, Agreement BIDS3000032413 (B.R.); the European Union’s Horizon 2020 research and innovation program under grant agreement 666992 (D.C.A.); EPSRC grants M020533, M006093, and J020990 (D.C.A.). This work was supported by the UCLH NIHR Biomedical Research Centre. The COPDGene Study was supported by Award U01 HL089897 and Award U01 HL089856 from the NHLBI. The content is solely the responsibility of the authors and does not necessarily represent the official views of the NHLBI or the NIH. The COPDGene project is also supported by the Chronic Obstructive Pulmonary Disease Foundation through contributions made to an Industry Advisory Board comprised of AstraZeneca, Boehringer Ingelheim, GlaxoSmithKline, Novartis, Pfizer, Siemens, and Sunovion. The ECLIPSE study was sponsored by GlaxoSmithKline. The study sponsor did not place any restrictions regarding statements made in this manuscript. A Steering Committee and a Scientific Committee comprising academic and sponsor representatives developed the original ECLIPSE study design, had full access to the study data, and were responsible for decisions regarding publications.
A complete list of COPDGene (COPD Genetic Epidemiology) Investigators members may be found before the beginning of the References.
Author Contributions: All authors meet criteria for authorship as recommended by the International Committee of Medical Journal Editors. A.L.Y., F.J.S.B., D.J.H., D.C.A., and J.R.H. designed the study. A.L.Y. and F.J.S.B. performed the modeling and statistical analysis and wrote the initial manuscript. COPDGene Investigators including M.K.H., C.J.G., and D.A.L. assisted with collection and analysis of COPDGene data. B.R. designed and generated visualizations for Figure 1. All authors contributed to the production of the final manuscript with revision for important intellectual content.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201908-1600OC on October 28, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
Contributor Information
Collaborators: for the COPDGene Investigators, James D. Crapo, Edwin K. Silverman, Barry J. Make, Elizabeth A. Regan, Terri Beaty, Ferdouse Begum, Peter J. Castaldi, Michael Cho, Dawn L. DeMeo, Adel R. Boueiz, Marilyn G. Foreman, Eitan Halper-Stromberg, Lystra P. Hayden, Craig P. Hersh, Jacqueline Hetmanski, Brian D. Hobbs, John E. Hokanson, Nan Laird, Christoph Lange, Sharon M. Lutz, Merry-Lynn McDonald, Margaret M. Parker, Dandi Qiao, Emily S. Wan, Sungho Won, Phuwanat Sakornsakolpat, Dmitry Prokopenko, Mustafa Al Qaisi, Harvey O. Coxson, Teresa Gray, MeiLan K. Han, Eric A. Hoffman, Stephen Humphries, Francine L. Jacobson, Philip F. Judy, Ella A. Kazerooni, Alex Kluiber, David A. Lynch, John D. Newell, Jr., James C. Ross, Raul San Jose Estepar, Joyce Schroeder, Jered Sieren, Douglas Stinson, Berend C. Stoel, Juerg Tschirren, Edwin Van Beek, Bram van Ginneken, Eva van Rikxoort, George Washko, Carla G. Wilson, Robert Jensen, Douglas Everett, Jim Crooks, Camille Moore, Matt Strand, John Hughes, Gregory Kinney, Katherine Pratte, Kendra A. Young, Surya Bhatt, Jessica Bon, Carlos Martinez, Susan Murray, Xavier Soler, Russell P. Bowler, Katerina Kechris, Farnoush Banaei-Kashani, Jeffrey L. Curtis, Carlos H. Martinez, Perry G. Pernicano, Nicola Hanania, Philip Alapat, Mustafa Atik, Venkata Bandi, Aladin Boriek, Kalpatha Guntupalli, Elizabeth Guy, Arun Nachiappan, Amit Parulekar, R. Graham Barr, John Austin, Belinda D’Souza, Gregory D. N. Pearson, Anna Rozenshtein, Byron Thomashow, Neil MacIntyre, Jr., H. Page McAdams, Lacey Washington, Charlene McEvoy, Joseph Tashjian, Robert Wise, Robert Brown, Nadia N. Hansel, Karen Horton, Allison Lambert, Nirupama Putcha, Richard Casaburi, Alessandra Adami, Matthew Budoff, Hans Fischer, Janos Porszasz, Harry Rossiter, William Stringer, Amir Sharafkhaneh, Charlie Lan, Christine Wendt, Brian Bell, Eugene Berkowitz, Gloria Westney, Richard Rosiello, David Pace, Gerard Criner, David Ciccolella, Francis Cordova, Chandra Dass, Gilbert D’Alonzo, Parag Desai, Michael Jacobs, Steven Kelsen, Victor Kim, A. James Mamary, Nathaniel Marchetti, Aditi Satti, Kartik Shenoy, Robert M. Steiner, Alex Swift, Irene Swift, Maria Elena Vega-Sanchez, Mark Dransfield, William Bailey, Anand Iyer, Hrudaya Nath, J. Michael Wells, Joe Ramsdell, Paul Friedman, Andrew Yen, Alejandro P. Comellas, Karin F. Hoth, Brad Thompson, Joanne Billings, Abbie Begnaud, Tadashi Allen, Frank Sciurba, Divay Chandra, Carl Fuhrman, Joel Weissfeld, Antonio Anzueto, Sandra Adams, Diego Maselli-Caceres, and Mario E. Ruiz
References
- 1.Mannino DM, Buist AS. Global burden of COPD: risk factors, prevalence, and future trends. Lancet. 2007;370:765–773. doi: 10.1016/S0140-6736(07)61380-4. [DOI] [PubMed] [Google Scholar]
- 2.Vestbo J, Edwards LD, Scanlon PD, Yates JC, Agusti A, Bakke P, et al. ECLIPSE Investigators. Changes in forced expiratory volume in 1 second over time in COPD. N Engl J Med. 2011;365:1184–1192. doi: 10.1056/NEJMoa1105482. [DOI] [PubMed] [Google Scholar]
- 3.Bhatt SP, Soler X, Wang X, Murray S, Anzueto AR, Beaty TH, et al. Association between functional small airway disease and FEV1 decline in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2016;194:178–184. doi: 10.1164/rccm.201511-2219OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Castaldi PJ, Dy J, Ross J, Chang Y, Washko GR, Curran-Everett D, et al. Cluster analysis in the COPDGene study identifies subtypes of smokers with distinct patterns of airway disease and emphysema. Thorax. 2014;69:415–422. doi: 10.1136/thoraxjnl-2013-203601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burgel P-R, Paillasseur J-L, Caillaud D, Tillie-Leblond I, Chanez P, Escamilla R, et al. Initiatives BPCO Scientific Committee. Clinical COPD phenotypes: a novel approach using principal component and cluster analyses. Eur Respir J. 2010;36:531–539. doi: 10.1183/09031936.00175109. [DOI] [PubMed] [Google Scholar]
- 6.Ross JC, Castaldi PJ, Cho MH, Chen J, Chang Y, Dy JG, et al. A Bayesian nonparametric model for disease subtyping: application to emphysema phenotypes. IEEE Trans Med Imaging. 2017;36:343–354. doi: 10.1109/TMI.2016.2608782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross JC, Castaldi PJ, Cho MH, Hersh CP, Rahaghi FN, Sánchez-Ferrero GV, et al. Longitudinal modeling of lung function trajectories in smokers with and without chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2018;198:1033–1042. doi: 10.1164/rccm.201707-1405OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bragman FJS, McClelland JR, Jacob J, Hurst JR, Hawkes DJ. Manifold learning of COPD. Med Image Comput Comput Assist Interv. 2017;10435:46–54. doi: 10.1007/978-3-319-66179-7_6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harmouche R, Ross JC, Diaz AA, Washko GR, Estepar RSJ. A robust emphysema severity measure based on disease subtypes. Acad Radiol. 2016;23:421–428. doi: 10.1016/j.acra.2015.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Young AL, Oxtoby NP, Daga P, Cash DM, Fox NC, Ourselin S, et al. Alzheimer’s Disease Neuroimaging Initiative. A data-driven model of biomarker changes in sporadic Alzheimer’s disease. Brain. 2014;137:2564–2577. doi: 10.1093/brain/awu176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fonteijn HM, Modat M, Clarkson MJ, Barnes J, Lehmann M, Hobbs NZ, et al. An event-based model for disease progression and its application in familial Alzheimer’s disease and Huntington’s disease. Neuroimage. 2012;60:1880–1889. doi: 10.1016/j.neuroimage.2012.01.062. [DOI] [PubMed] [Google Scholar]
- 12.Oxtoby NP, Alexander DC EuroPOND consortium. Imaging plus X: multimodal models of neurodegenerative disease. Curr Opin Neurol. 2017;30:371–379. doi: 10.1097/WCO.0000000000000460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donohue MC, Jacqmin-Gadda H, Le Goff M, Thomas RG, Raman R, Gamst AC, et al. Alzheimer’s Disease Neuroimaging Initiative. Estimating long-term multivariate progression from short-term data. Alzheimers Dement. 2014;10(Suppl):S400–S410. doi: 10.1016/j.jalz.2013.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bilgel M, Prince JL, Wong DF, Resnick SM, Jedynak BM. A multivariate nonlinear mixed effects model for longitudinal image analysis: application to amyloid imaging. Neuroimage. 2016;134:658–670. doi: 10.1016/j.neuroimage.2016.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang X, Sontag D, Wang F. Unsupervised learning of disease progression models; Presented at the 20th ACM SIGKDD International Conference on Knowledge Disovery and Data Mining. 2014. p. 85–94. [Google Scholar]
- 16.Young AL, Marinescu RV, Oxtoby NP, Bocchetta M, Yong K, Firth NC, et al. Genetic FTD Initiative (GENFI); Alzheimer’s Disease Neuroimaging Initiative (ADNI) Uncovering the heterogeneity and temporal complexity of neurodegenerative diseases with Subtype and Stage Inference. Nat Commun. 2018;9:4273. doi: 10.1038/s41467-018-05892-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bragman FJS, Young AL, Hawkes DJ, Alexander DC, Hurst JR. Disease progression patterns in COPD. Eur Respir J. 2018;52:OA2139. [Google Scholar]
- 18.Regan EA, Hokanson JE, Murphy JR, Make B, Lynch DA, Beaty TH, et al. Genetic epidemiology of COPD (COPDGene) study design. COPD. 2010;7:32–43. doi: 10.3109/15412550903499522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vestbo J, Anderson W, Coxson HO, Crim C, Dawber F, Edwards L, et al. ECLIPSE investigators. Evaluation of COPD longitudinally to identify predictive surrogate end-points (ECLIPSE) Eur Respir J. 2008;31:869–873. doi: 10.1183/09031936.00111707. [DOI] [PubMed] [Google Scholar]
- 20.Galbán CJ, Han MK, Boes JL, Chughtai KA, Meyer CR, Johnson TD, et al. Computed tomography-based biomarker provides unique signature for diagnosis of COPD phenotypes and disease progression. Nat Med. 2012;18:1711–1715. doi: 10.1038/nm.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nakano Y, Wong JC, de Jong PA, Buzatu L, Nagao T, Coxson HO, et al. The prediction of small airway dimensions using computed tomography. Am J Respir Crit Care Med. 2005;171:142–146. doi: 10.1164/rccm.200407-874OC. [DOI] [PubMed] [Google Scholar]
- 22.Coxson HO, Dirksen A, Edwards LD, Yates JC, Agusti A, Bakke P, et al. Evaluation of COPD Longitudinally to Identify Predictive Surrogate Endpoints (ECLIPSE) Investigators. The presence and progression of emphysema in COPD as determined by CT scanning and biomarker expression: a prospective analysis from the ECLIPSE study. Lancet Respir Med. 2013;1:129–136. doi: 10.1016/S2213-2600(13)70006-7. [DOI] [PubMed] [Google Scholar]
- 23.Filley GF, Beckwitt HJ, Reeves JT, Mitchell RS. Chronic obstructive bronchopulmonary disease: II. Oxygen transport in two clinical types. Am J Med. 1968;44:26–38. doi: 10.1016/0002-9343(68)90234-9. [DOI] [PubMed] [Google Scholar]
- 24.Hogg JC, Macklem PT, Thurlbeck WM. Site and nature of airway obstruction in chronic obstructive lung disease. N Engl J Med. 1968;278:1355–1360. doi: 10.1056/NEJM196806202782501. [DOI] [PubMed] [Google Scholar]
- 25.Barnes PJ. Inflammatory mechanisms in patients with chronic obstructive pulmonary disease. J Allergy Clin Immunol. 2016;138:16–27. doi: 10.1016/j.jaci.2016.05.011. [DOI] [PubMed] [Google Scholar]
- 26.Fletcher C, Peto R. The natural history of chronic airflow obstruction. BMJ. 1977;1:1645–1648. doi: 10.1136/bmj.1.6077.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tantucci C, Modina D. Lung function decline in COPD. Int J Chron Obstruct Pulmon Dis. 2012;7:95–99. doi: 10.2147/COPD.S27480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rennard SI, Drummond MB. Early chronic obstructive pulmonary disease: definition, assessment, and prevention. Lancet. 2015;385:1778–1788. doi: 10.1016/S0140-6736(15)60647-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sashidhar K, Gulati M, Gupta D, Monga S, Suri S. Emphysema in heavy smokers with normal chest radiography: detection and quantification by HCRT. Acta Radiol. 2002;43:60–65. doi: 10.1080/028418502127347457. [DOI] [PubMed] [Google Scholar]
- 30.Srinakarin J, Thammaroj J, Boonsawat W. Comparison of high-resolution computed tomography with pulmonary function testing in symptomatic smokers. J Med Assoc Thai. 2003;86:522–528. [PubMed] [Google Scholar]
- 31.Mets OM, de Jong PA, van Ginneken B, Gietema HA, Lammers JWJ. Quantitative computed tomography in COPD: possibilities and limitations. Hai. 2012;190:133–145. doi: 10.1007/s00408-011-9353-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.