Abstract
Rationale: Pneumococcal pneumonia remains a global health problem. Colonization of the nasopharynx with Streptococcus pneumoniae (Spn), although a prerequisite of infection, is the main source of exposure and immunological boosting in children and adults. However, our knowledge of how nasal colonization impacts on the lung cells, especially on the predominant alveolar macrophage (AM) population, is limited.
Objectives: Using a controlled human infection model to achieve nasal colonization with 6B serotype, we investigated the effect of Spn colonization on lung cells.
Methods: We collected BAL from healthy pneumococcal-challenged participants aged 18–49 years. Confocal microscopy and molecular and classical microbiology were used to investigate microaspiration and pneumococcal presence in the lower airways. AM opsonophagocytic capacity was assessed by functional assays in vitro, whereas flow cytometry and transcriptomic analysis were used to assess further changes on the lung cellular populations.
Measurements and Main Results: AMs from Spn-colonized individuals exhibited increased opsonophagocytosis to pneumococcus (11.4% median increase) for approximately 3 months after experimental pneumococcal colonization. AMs also had increased responses against other bacterial pathogens. Pneumococcal DNA detected in the BAL samples of Spn-colonized individuals were positively correlated with nasal pneumococcal density (r = 0.71; P = 0.029). Similarly, AM-heightened opsonophagocytic capacity was correlated with nasopharyngeal pneumococcal density (r = 0.61, P = 0.025).
Conclusions: Our findings demonstrate that nasal colonization with pneumococcus and microaspiration prime AMs, leading to brisker responsiveness to both pneumococcus and unrelated bacterial pathogens. The relative abundance of AMs in the alveolar spaces, alongside their potential for nonspecific protection, render them an attractive target for novel vaccines.
Keywords: Streptococcus pneumoniae nasopharyngeal colonization, microaspiration, alveolar macrophages, interferon-γ, CD4+ T cells
At a Glance Commentary
Scientific Knowledge on the Subject
Pneumococcal pneumonia rates remain high globally, affecting the youngest and the oldest. Colonization of the human nasopharynx with Streptococcus pneumoniae, although a prerequisite of infection, is the predominant source of immunizing exposure and immunological boosting in both children and adults. However, our knowledge of how nasal colonization affects the lung cellular populations, especially the predominant alveolar macrophage population, is limited.
What This Study Adds to the Field
During colonization of the human nasopharynx with pneumococcus, lung cells are exposed to the pathogen through microaspiration. In healthy young adults, this process triggers T-helper cell type 1 responses and associates with alveolar macrophage–increased opsonophagocytic capacity against both pneumococcus and other respiratory pathogens.
Streptococcus pneumoniae (the pneumococcus, Spn) is a leading cause of severe infection, responsible annually for the death of up to a million children worldwide (1). Pneumonia is the most frequent manifestation of pneumococcal disease (2), and, despite current vaccination strategies, the burden of pneumococcal pneumonia remains very high globally (3), affecting disproportionally the very young and very old throughout the world (4).
Despite its pathogenicity, Spn commonly colonizes the human nasopharynx, a state known as pneumococcal colonization or carriage (4). Pneumococcal colonization rates in the absence of disease range from 40% to 95% in infants and approximately 10% to 25% among adults (5, 6). In humans, exposure to pneumococcus through nasopharyngeal colonization is an immunizing event, as it elicits humoral and cellular immune responses, both systemically and in the nasal mucosa (7–9).
However, lung mucosal immune responses to pneumococcus are not well understood in humans. It is believed that protection against development of pneumonia relies on a successful regulation of colonization in the nasopharynx and a brisk alveolar macrophage (AM)-mediated immune response in the lung (10). The AM—an innate-type resident lung cell—is an integral component of lung immunity (11) and the first cell type to combat pneumococci during early infection (12). It also plays a key role in shaping the adaptive immunity through their effects on dendritic cells and T cells (13). In murine models, it has been shown that AMs are mainly self-maintained, although during lung insult or as a result of aging (14), peripheral monocytes contribute to their replenishment (15, 16).
A clear understanding of the mechanisms that underlie brisk but controlled lung immune responses at the early stages of the infection is essential to inform us why high rates of pneumonia persist in the high-risk groups (infants, elderly, and immunocompromised). These high-risk groups are characterized by underdeveloped or defective adaptive immunity. Although current immunization strategies to pneumococcal diseases target B cell–dependent immunity exclusively, the recently described memory properties of innate cells, including natural killer cells and monocytes, indicate that innate immune cells could be considered as a promising alternative or complementary vaccine target (17–19).
In this study, we investigated the effect of antecedent pneumococcal colonization on AM function in healthy adults. We showed for the first time that nasal human colonization in absence of disease leads to pneumococcal aspiration to the lower respiratory tract, which enhanced AM opsonophagocytic capacity against a range of bacterial respiratory pathogens, likely through an IFN-γ–mediated mechanism.
Some of the results of this study have been previously reported in the form of an abstract (20).
Methods
Study Design and BAL Collection
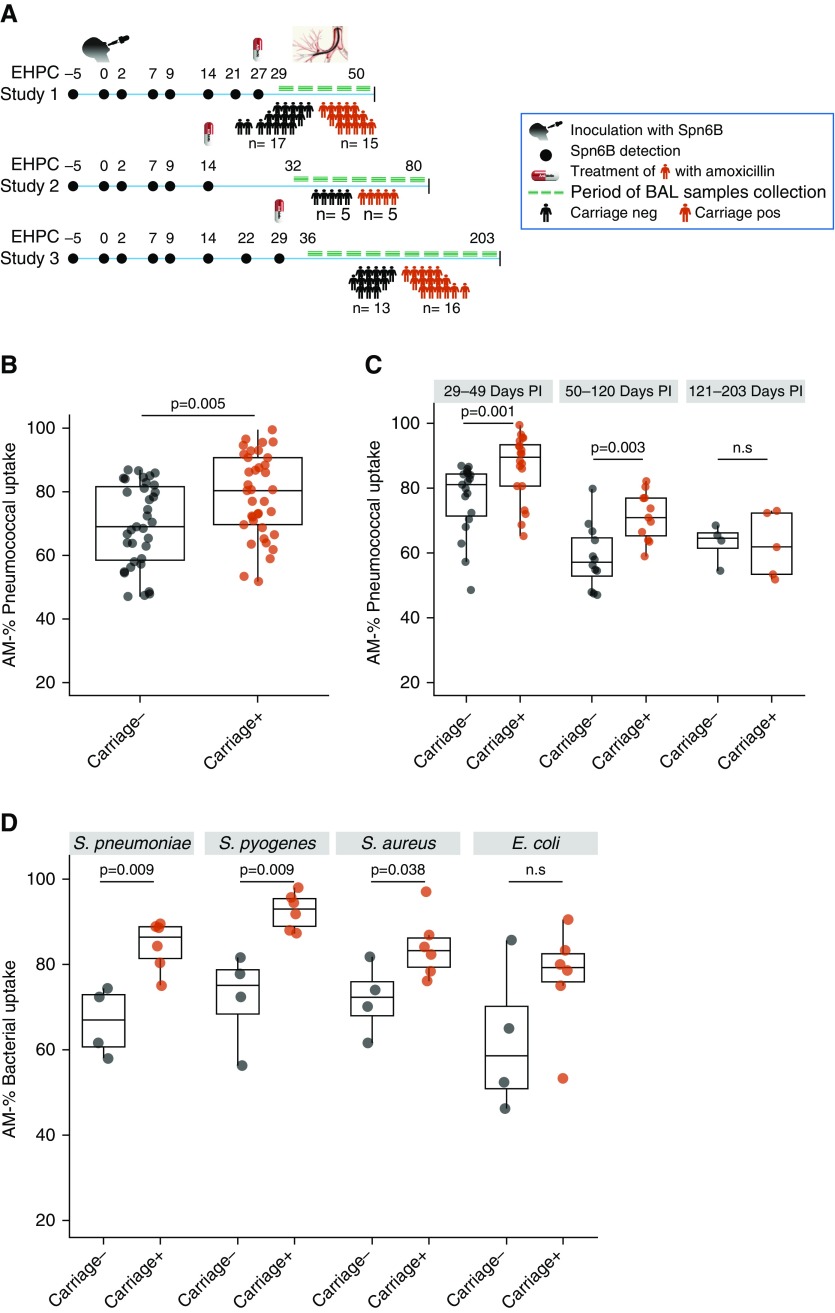
Healthy, nonsmoking, adult volunteers aged from 18 to 49 years, enrolled in one of the Experimental Human Pneumococcal Challenge studies (21) between 2015 and 2018 (22, 23), underwent a one-off research bronchoscopy, as previously described (24, 25). Experimental human pneumococcal challenge was conducted as previously described (21, 26), and 80,000 cfu of serotype 6B (strain BHN418) were instilled into each nostril of participants. Pneumococcal colonization was detected by classical microbiology methods, and individuals were defined as Spn-colonized, if any nasal wash culture following experimental challenge grew Spn serotype 6B (Spn6B). BAL samples were obtained from 29 to 203 days after the intranasal inoculation (Figure 1A). Spn-colonized individuals received three doses of amoxicillin at the end of the clinical trial (at Day 14, 27, or 29) before the bronchoscopy (Figure 1A). In addition, a nasopharyngeal swab (NPS) was collected in skim milk, tryptone, glucose, and glycerine on the day of the bronchoscopy, and clearance of colonization was assessed by both classical microbiology and molecular methods.
Figure 1.
Alveolar macrophages (AMs) display an increased opsonophagocytic activity against bacterial pathogens for a prolonged period after nasal pneumococcal carriage. (A) Defined time period of BAL sample collection from Streptococcus pneumoniae (Spn) colonized (carriage+) and noncolonized (carriage−) individuals within three independent Experimental Human Pneumococcal Challenge studies. Individuals were purposively sampled according to colonization state. After the final nasal wash (Day 27, 14, or 29, based on study), Spn-colonized individuals received a 3-day course of antibiotics. (B) Percentage of pneumococcal uptake by AMs after in vitro infection in carriage− (n = 35) and carriage+ (n = 37) groups. P = 0.005 by Mann-Whitney test. Multiplicity of infection used was 1:100. (C) Chronological representation of all BAL samples (n = 72) collected from 1 to 6 months after intranasal pneumococcal inoculation divided into three consecutive time periods. T1 (29–49 d PI), P = 0.001; T2 (50–120 d PI), P = 0.003; and T3 (121–203 d PI), P = 0.82 by Mann-Whitney test. (D) Percentage of bacterial uptake by AMs after in vitro infection with Spn6B, Staphylococcus aureus, Streptococcus pyogenes, or Escherichia coli. P = 0.009, P = 0.009, P = 0.038, and P = 0.067, respectively, by Mann-Whitney test. Boxplots and individual subjects are depicted with carriage− in black dots and carriage+ in red dots. EHPC = Experimental Human Pneumococcal Challenge; neg = negative; n.s = not significant; PI = postinoculation; pos = positive.
Ethics Statement
All volunteers gave written informed consent, and research was conducted in compliance with all relevant ethical regulations. Ethical approval was given by the North West National Health Service Research Ethics Committee (Ethics Committee reference numbers: 15/NW/0146, 14/NW/1460 and 15/NW/0931, and Human Tissue Authority licensing number: 12548).
BAL Processing and AM Isolation
BAL samples were processed (24, 25), and AMs were routinely separated from other cell populations using adherence step, as previously described (9). In the experiments in which highly pure AM population was requested, AMs were purified from the whole BAL sample through cell sorting (fluorescence-activated cell sorter ARIAIII), following seeding on 96-well plate and overnight incubation at 37°C, 5% CO2 (full details in online supplement).
AM Opsonophagocytic Assay
AM opsonophagocytic capacity was evaluated as previously described, with minor modifications (9, 27, 28). Briefly, live Spn serotype 6B, Streptococcus pyogenes, Staphylococcus aureus, or Escherichia coli were opsonized with human intravenous immunoglobulin (IVIG, Gamunex; Grifols, Inc.) at 37°C for 15 minutes. An opsonized bacterial strain, baby rabbit complement (Mast Group), and isolated AMs were incubated at 37°C for 1 hour. Following incubation, 10 μl of reaction mixture was plated, in triplicate, onto blood agar (Oxoid) or Luria-broth agar plates and incubated at 37°C, 5% CO2 overnight. Colony-forming units from cell supernatants were counted the following day. Multiplicity of infection used was 1:100 for all the gram-positive bacteria, except assays where AMs isolated by cell sorting were used. Opsonophagocytic killing assay for E. coli was modified as described elsewhere (multiplicity of infection = 1:20 for 30 min) (27). Due to the lack of readout of AM lysosome acidification, the term pneumococcal uptake rather than pneumococcal killing was used (full details in online supplement).
Bacterial DNA Extraction and Quantification of Pneumococcal DNA in Nasal and Lung Samples
Extraction of bacterial DNA from NPS and BAL samples was performed as previously described with minor modifications (29). Presence of pneumococcal DNA in NPS and BAL samples was determined using primers and probe specifically designed for 6B serotype, targeting a capsular polysaccharide gene known as wciP, the rhamnosyl transferase gene. The primers and probe sequences were forward primer 5′-GCTAGAGATGGTTCCTTCAGTTGAT-3′; reverse primer 5′-CATACTCTAGTGCAAACTTTGCAAAAT-3′; and probe 5′-[FAM] ACT GTC TCA TGA TAA TT [MGBEQ]-3′, as previously published (30) (full details in online supplement).
Confocal Microscopy
Fresh BAL cells were washed and stained with antihuman CD14 (cluster of differentiation 14) (Texas red) or CD45 (magenta). Cells were permeabilized and incubated with 6B pneumococcal antisera (Statens Serum Institute) for 30 minutes and then with secondary conjugated antibody (anti-rabbit 488) for another 30 minutes. Pneumococcal antisera against serotype 23F (Statens Serum Institute) was used as control antibody. After washing, cells were cytospun onto microscope slides. DAPI solution was applied directly on the spun cells for 5 minutes. After washing, samples were mounted using Aqua PolyMount (VWR International). Images were captured using an inverted TissueFAXS Zeiss Confocal Microscope. Z-stacks were recorded at 1-μm intervals at either 40× oil or 63× oil objectives.
In thawed BAL samples, fixed and permeabilized cells were incubated in blocking solution (phosphate-buffered saline, 5% goat serum), following 1 hour incubation with primary antibodies and 45 minutes with secondary antibody solution, following rinsing and then mounting with DAPI. Antipneumococcal capsule anti-6B serum (Statens Serum Institute) was used to stain bacteria, whereas macrophages were labeled with anti-human CD169 (Thermoscientific). Combinations of Alexafluor-conjugated antibodies (Thermoscientific) were used as secondary antibodies (488 and 568 with different host specificity). Images were acquired in Olympus FluoView 1000 confocal laser- scanning microscope using 40× objectives. For the bacterial localization assays, Alexafluor 633–conjugated wheat germ agglutinin was used before membrane permeabilization. Z-stack was created from microscope images, elaborated using Huygens Essential deconvolution software version 16 (Scientific Volume Imaging) and viewed in Imaris 3D reconstruction software 9.4 (Bitplane).
Flow Cytometry Assays
In each flow cytometry assay, the corresponding cell population was stained with predetermined optimal concentration of fluorochrome-conjugated monoclonal antibodies against human cell surface proteins or intracellular cytokines (full details in online supplement).
Luminex Analysis of BAL Fluid
The acellular BAL fluid was collected after centrifugation of the whole BAL sample (400 g for 10 min at 4°C), divided to 1-ml aliquots and stored at −80°C until analysis. On the day of the analysis, samples were concentrated 10× (1 ml of BAL supernatant concentrated to 100 μl using rotary vacuum concentrator RVC2-18), following acquisition using a 30-plex magnetic Luminex cytokine kit (ThermoFisher) and analyzed on a LX200 with xPonent3.1 software following manufacturer’s instructions. Samples were analyzed in duplicates, and BAL samples with a coefficient of variation > 50% were excluded.
AM Gene Analysis Using Nanostring Platform
Nanostring for AM gene analysis was used as previously described (22) (full details in online supplement).
Quantification and Statistical Analysis
Statistical analyses were performed using GraphPad Prism (Version 6; GraphPad Software) and R software (version 3.5.1), including Bioconductor packages. P values are two-tailed. For parametric group comparisons, t test was used for unpaired and paired groups. For nonparametric group comparisons, a Mann-Whitney or Wilcoxon test was used for unpaired and paired groups, respectively. For gene expression and Luminex analysis, P values were corrected by applying multiple correction testing (Benjamini-Hochberg). To quantify association between groups, Pearson or Spearman correlation test was used for parametric or nonparametric groups, respectively. Differences were considered significant at P ≤ 0.05.
Results
AMs Exhibit Augmented Responsiveness to Bacteria for 3 Months after Experimental Pneumococcal Colonization
We coupled the experimental human pneumococcal challenge model with research bronchoscopy to investigate whether and how nasopharyngeal pneumococcal colonization affects AM function in healthy adults. BAL sample was collected from both pneumococcal (Spn) colonized and noncolonized healthy adults (aged from 18 to 49 yr) between 1 and 7 months (29–203 d) after bacterial challenge (Figure 1A). Antecedent pneumococcal colonization was associated with 11.4% increase in AM capacity to take up pneumococci in vitro, ranging in noncolonized (carriage−) from 58.0% to 82.1% (median = 69.0%) and in the Spn-colonized (carriage+) group from 69.1% to 90.7% (median = 80.4%) (P = 0.005; Figure 1B). The observed differential AM opsonophagocytic activity (OPA) was reproducible between studies and persisted for approximately 2–3 months (median = 76 d) following the intranasal pneumococcal inoculation (Figure 1C). Later after challenge (120–203 d), uptake was similar between the two groups, although we were insufficiently powered to detect any statistical differences (n = 4 per group).
We also sought to examine whether this enhanced activity was specific to pneumococcus or whether AM responses to other pathogens were similarly increased. AMs from Spn-colonized individuals had greater capacity to take up the respiratory pathogens S. pyogenes and S. aureus (increased by 18% and 11%, respectively), when compared with AMs isolated from noncolonized individuals (P = 0.009 and P = 0.038, respectively, Figure 1D). For the gram-negative bacterium E. coli there was a nonsignificant increase in AM OPA in the Spn-colonized group (median, 20.6% increase; P = 0.067; Figure 1D).
Spn Can Be Detected in the Lung after Clearance of Nasal Colonization
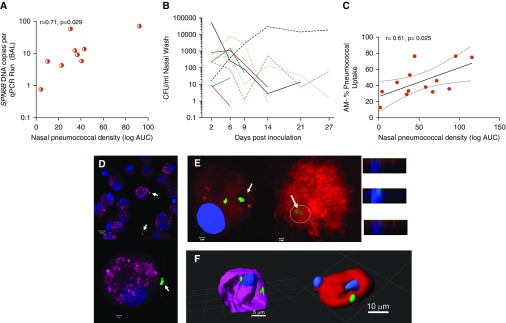
To investigate whether pneumococcus is the stimulus of the enhanced AM responses in the pulmonary mucosa after nasal colonization, we sought to find evidence of presence of the pneumococcal challenge strain (Spn6B) in the alveolar spaces. For the detection of pneumococcus in the BAL samples, we utilized classical microbiology and/or molecular methods targeting a capsular polysaccharide gene specific to Spn6B (wciP). In volunteers who underwent bronchoscopy between 29 and 49 days after the challenge, Spn6B DNA was detected in the BAL of 41% (9/22) of Spn-colonized individuals by quantitative PCR (Table E1 in online supplement), 1–3 weeks following the clearance of nasal colonization. None of the noncolonized individuals had detectable Spn6B DNA in their BAL sample. Additionally, BAL samples collected from volunteers between 50 and 120 days after the challenge were negative for presence of Spn6B DNA, irrespective of colonization status. Nasal pneumococcal density positively correlated with the copies of pneumococcal DNA detected in BAL samples (Figure 2A). Spn-colonized individuals differed in both density and duration of the colonization episode (Figure 2B). In addition, AM capacity to take up pneumococci correlated positively with nasal pneumococcal density (Figure 2C). Additionally, confocal microscopy was used to qualitatively visualize the location of pneumococci in the AMs. Pneumococci (Spn6B) were found associated with the surface of AMs or internalized by them, a phenomenon only observed in the Spn-colonized group (Figures 2D–2F and Video E1). These data suggest that during asymptomatic pneumococcal colonization of the nasopharynx, aspiration of pneumococci can occur, modulating the pulmonary immunological responses.
Figure 2.
Evidence of pneumococcal presence in the lung of nasopharyngeal Streptococcus pneumoniae (Spn)-colonized individuals. (A) Positive correlation between the nasal pneumococcal density, expressed as the logarithm of the area under the curve (log AUC), and the copies of pneumococcal DNA (Spn6B) detected in the BAL fluid of carriage+ individuals. r = 0.71 and P = 0.029 by Pearson correlation test. (B) Duration and density of nasal colonization per individual with detected Spn6B DNA in the BAL fluid (9 in 22 Spn-colonized). The end of each colored line indicates the time point that the individual cleared colonization, assessed by classical microbiology. (C) Positive correlation between the nasal pneumococcal density (log AUC) and sorted alveolar macrophage (AM) opsonophagocytic activity (n = 13). Pearson correlation test results and linear regression line with 95% confidence interval are shown. (D and E) Representative images taken by confocal microscope showing pneumococci (arrows) around AMs (D) and internalized pneumococci (arrows) by AMs derived from Spn-colonized individuals (E). CD45 (cluster of differentiation 45) is represented by magenta, CD14 by red, nucleus by blue (DAPI), and Spn6 capsule by green. Scale bars: D, top panel, 10 μm; bottom panel, 2 μm; E, left and right panels, 2 μm. Inset panels represent Z-stack images of the cell with internalized pneumococcus (circle). (F) Three-dimensional reconstruction of deconvolved Z-stack confocal images of AMs. The samples were stained with either wheat germ agglutinin (on the left; magenta) or anti-CD169 monoclonal antibody (on the right; red). Spn6 in green and nuclei in blue (DAPI). Video images of the three-dimensional reconstructions are available as an online supplement file. A scale bar is shown on the images. CFU = colony-forming units; qPCR = quantitative PCR; Rxn = reaction.
CD4+ T-Helper Cell Type 1 Skewed Responses Rapidly Prime AMs
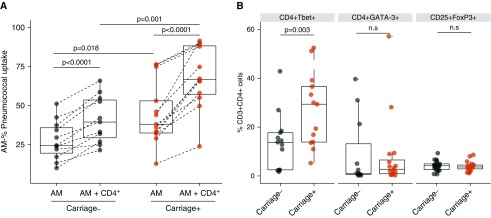
To investigate whether the observed augmented AM capacity to take up pneumococci in vitro was dependent on lung lymphocytes, we coincubated AMs with autologous CD3+CD4+ T cells during in vitro infection with Spn6B. The presence of CD3+CD4+ T cells enhanced the basal AM opsonophagocytic capacity in both noncolonized (1.6-fold; P < 0.0001) and Spn-colonized individuals (1.8-fold; P < 0.0001) (Figure 3A). AM opsonophagocytic capacity differed between the two groups at baseline (before lung-derived autologous CD3+CD4+ T-cell addition) and remained higher in Spn-colonized individuals when autologous CD4+ T cells were present (Figure 3A).
Figure 3.
Alveolar macrophage (AM) cross-talk and priming by autologous CD4+ (cluster of differentiation 4–positive) T subsets. (A) Comparison of phagocytic activity between sorted AMs and sorted AMs plus autologous BAL isolated CD4+ T cells from both carriage− (n = 11) and carriage+ (n = 13). Multiplicity of infection = 1:20. AM and CD4+ T cells were used in a 10:1 ratio. P < 0.0001 in both groups by paired t test. Comparison of AM basal opsonophagocytic activity between the two groups. P = 0.018 by unpaired t test with Welch’s correction. Comparison of AM opsonophagocytic activity in the presence of lung CD4+ T cells between carriage− and carriage+ group. P = 0.001 by unpaired t test with Welch’s corrections. Boxplots and individual subjects are depicted with carriage− in black and carriage+ in red, with paired samples connected by a dashed line. (B) Intracellular staining of CD4+ T cells for T-bet, GATA-3, and FoxP3 transcription factors, expressed as percentage of CD3+ CD4+ BAL lymphocytes. P = 0.003, P = 0.85, and P = 0.33, per CD4+ T-cell subset, respectively, by unpaired t test with Welch’s correction. Boxplots and individual subjects are depicted with carriage− in black dots and carriage+ in red dots. n.s = not significant.
To elucidate the mechanism underlying this increased boosting of AM function by CD4+ T cells from Spn-colonized individuals, we stained lung lymphocytes intracellularly for T-bet (T-box transcription factor expressed in T cells), GATA-3 (GATA-binding protein-3), and FoxP3 (Forkhead box P3) transcription factors (Figure E1). In the Spn-colonized group the levels of CD4+ T-bet–expressing cells were twice as high than in the noncolonized group (P = 0.003), indicating T-helper cell type 1 (Th1) polarization (Figure 3B). There were no significant differences in the levels of either CD4+ GATA-3–expressing or CD4+ FoxP3–expressing T cells between the two groups (Figure 3B).
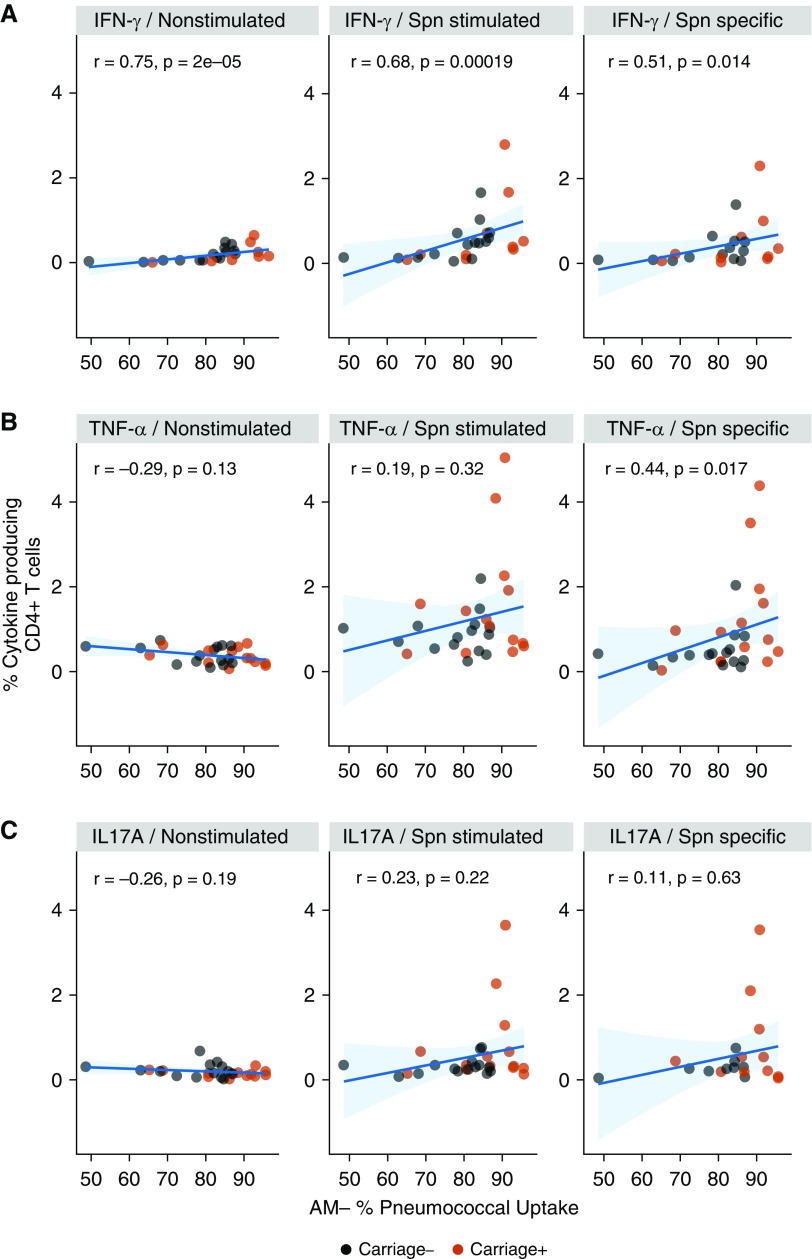
In parallel, lymphocytes from both Spn-colonized and noncolonized volunteers were stimulated with pneumococcal antigen (heat-inactivated-Spn6B). Cytokine (IFN-γ, TNF-α [tumor necrosis factor α], or IL-17A)- producing CD4+ T cells were subsequently detected by flow cytometry (Figure E2). AM OPA correlated with cytokine-producing CD4+ T cells, classified as spontaneous (unstimulated) or pneumococcal-responding cells (Figure 4). Increased levels of IFN-γ–producing CD4+ T cells, both pneumococcal-specific and spontaneous responding, positively correlated with AM ability to take up live pneumococci in vitro (Figure 4A). On the other hand, AM OPA correlated positively with only the pneumococcal-specific TNF-α–producing CD4+ T cells (Figure 4B), whereas IL-17A–producing CD4+ T cells did not correlate with AM OPA in any condition (Figure 4C).
Figure 4.
Correlations of alveolar macrophage (AM) opsonophagocytic activity with CD4+ (cluster of differentiation 4–positive) T-helper cell type 1 (Th1) and Th17 responses. (A) From left to right are illustrated significant correlations of the levels of IFN-γ–expressing CD4+ T cells at baseline (nonstimulated), total IFN-γ–expressing CD4+ T cells after stimulation with heat-inactivated Streptococcus pneumoniae (Spn) 6B, and the Spn-specific responding CD4+ T cells (nonstimulated condition subtracted from Spn-stimulated condition) with AM capacity to take up pneumococci. Spearman rho and P values are shown. (B) Significant correlation of Spn-specific, TNF-α (tumor necrosis factor α)-expressing CD4+ T cells with AM opsonophagocytic activity. Spearman rho and P value are shown. (C) From left to right are illustrated the levels of IL-17A–expressing CD4+ T cells at baseline and the levels of total and Spn-specific, IL-17A–expressing CD4+ T cells in association with AM opsonophagocytic activity. There are no significant correlations shown. Spearman correlation test results and linear regression line with 95% confidence interval (light blue shading) are shown.
Together, these results indicate that skewing of CD4+ T cells and production of IFN-γ are associated with the increased capacity of AMs to take up bacteria.
Alterations of Lung Cytokine Milieu after Nasal Colonization and the Effect of IFN-γ on AM Opsonophagocytic Capacity
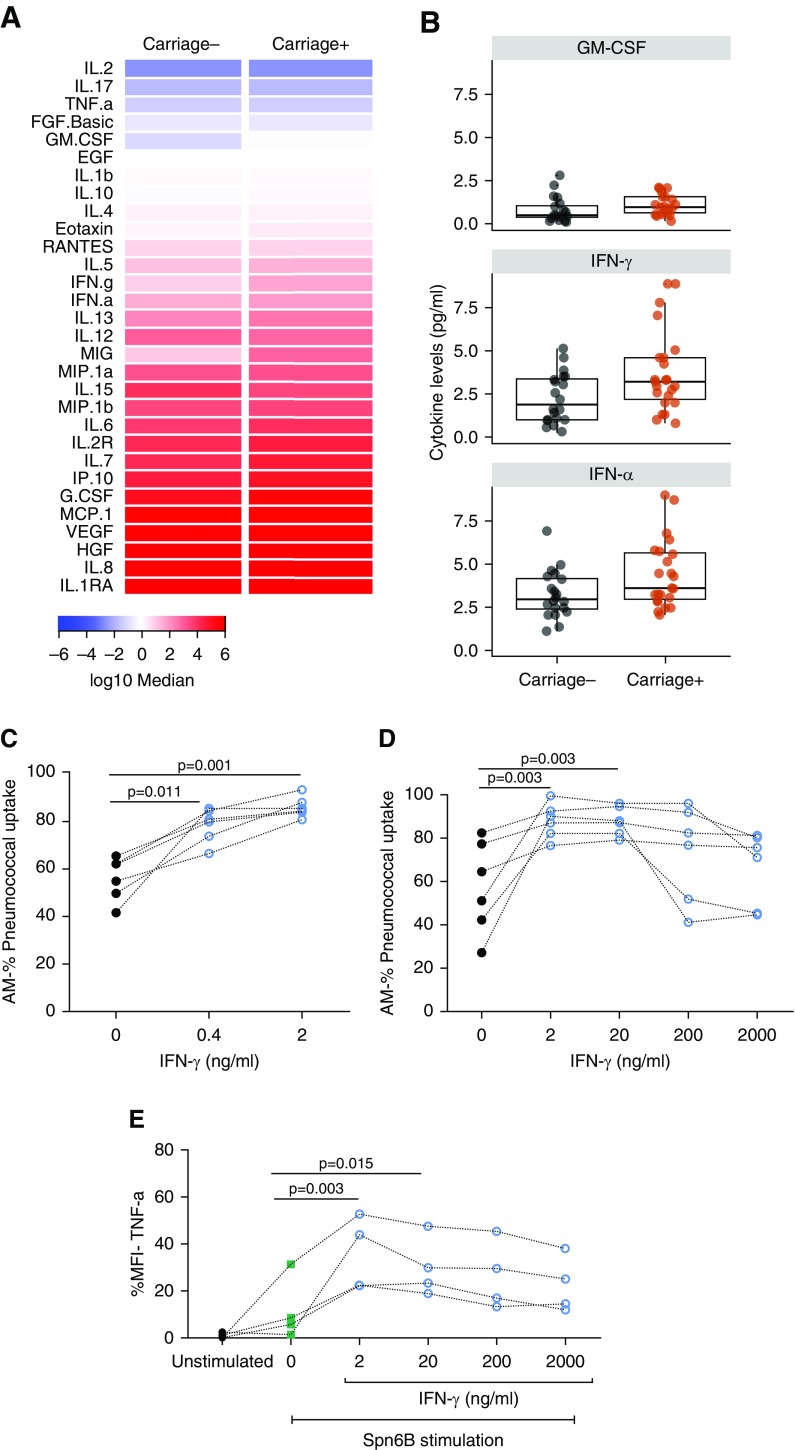
The alveolar microenvironment is crucial for cell signaling, shaping how local cells respond to different stimuli (31). To assess alterations of the alveolar cytokine milieu induced by nasal pneumococcal colonization, we measured levels of 30 cytokines in the BAL fluid retrieved from both Spn-colonized and noncolonized individuals (Figure 5A and Table E2). Levels of granulocyte–macrophage colony–stimulating factor (median 0.96 pg/ml vs. 0.49 pg/ml), IFN-γ (median 3.92 pg/ml vs. 1.88 pg/ml), and IFN-α (median 2.96 pg/ml vs. 3.61 ng/ml) were higher in the BAL fluid of Spn-colonized individuals than the noncolonized group, although statistical significance was lost upon correcting for multiple testing (Figure 5B and Table E2). As our results on lung-derived CD4+ T-cell polarization profile in Spn-colonized individuals, together with the strong correlation between AM opsonophagocytic capacity and levels of IFN-γ–producing CD4+ T cells, pointed toward IFN-γ, we sought to address its effect on AM function. Directly before the OPA, AMs were stimulated with exogenous IFN-γ, either with a concentration that would better reflect the average in vivo levels of IFN-γ in the lung-lining fluid of the Spn-colonized group (0.4 ng/ml) or with 2 ng/ml. Sole addition of IFN-γ enhanced significantly the AM capacity to take up pneumococci in both concentrations (median: 1.37-fold change using 0.4 ng/ml and 1.45-fold change using 2 ng/ml of recombinant IFN-γ) (Figure 5C). We also investigated further the role of IFN-γ upon AM activation by stimulating them with 10-fold increasing concentrations of exogenous IFN-γ. The lowest tested titers of IFN-γ (2 and 20 ng/ml) augmented AM OPA, resulting both in 1.5-fold increase (1.5× median; interquartile range,1.2×–2.1×) in AM pneumococcal uptake, whereas no significant increase was seen with the highest used concentrations (200 and 2,000 ng/ml) (Figure 5D). These results were verified when AM response was assessed using a flow cytometric cytokine production assay (Figure E3). AMs produced increased levels of TNF-α in response to stimulation with heat-inactivated Spn6B only at the lower prestimulation doses of IFN-γ (Figure 5E). The mechanism seems to have a threshold, as demonstrated by the data, with IFN-γ signaling being beneficial for AM function at lower doses in vitro, but not at higher concentration.
Figure 5.
Lung cytokine milieu alterations after nasal colonization and the effect of IFN-γ on alveolar macrophage (AM) opsonophagocytic function. (A) Heatmap of the 30 cytokine levels, expressed as log10 median (pg/ml), measured in the BAL fluid (carriage−; n = 20 and carriage+; n = 22). (B) Levels of different cytokines between the two groups, expressed as pg/ml: granulocyte–macrophage colony–stimulating factor, IFN-γ, and IFN-α with P = 0.032, P = 0.047, and P = 0.043, respectively, analyzed by Mann-Whitney test. Statistical significance was lost upon correction for multiple testing. Boxplots and individual subjects are depicted with carriage− in black dots and carriage+ in red dots. (C) Treatment of AMs with 0.4 ng/ml (average IFN-γ concentration measured in the BAL fluid of carriage+) and 2 ng/ml of exogenous IFN-γ. (D) The effect of 10-fold increasing doses of exogenous IFN-γ (2–2,000 ng/ml) on the capacity of AMs to take up pneumococcus (live Streptococcus pneumoniae 6B used, multiplicity of infection = 1:100). AMs were isolated from six nonchallenged subjects. Individual samples are depicted and connected by dashed lines. P values by Friedman test followed by Dunn’s multiple comparison are shown. (E) TNF-α (tumor necrosis factor α) production from AMs, pretreated or not with exogenous IFN-γ (2–2,000 ng/ml) after stimulation with heat-inactivated Streptococcus pneumoniae serotype 6B. AMs were isolated from four nonchallenged subjects. Individual samples are depicted and connected by dashed lines. P values by Friedman test followed by Dunn’s multiple comparison are shown. GM-CSF = granulocyte–macrophage colony–stimulating factor; MFI = mean fluorescence intensity.
Pneumococcal Colonization May Promote Monocyte-to-Macrophage Differentiation in the Alveolar Spaces
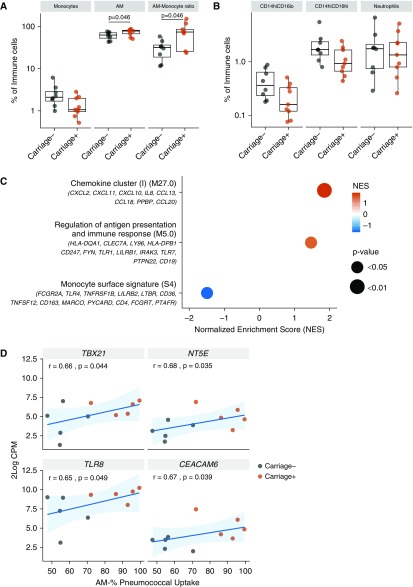
Previously, we have demonstrated that AM phenotype is not altered by nasopharyngeal pneumococcal colonization, as defined by classical monocyte polarization surface markers (24). However, given the increased capacity of AMs to take up pneumococci, we extended our assessment to other lung myeloid cell populations and neutrophils to determine whether recent pneumococcal carriage alters the distribution of these cells in the airway (Figure E4). Spn-colonized individuals displayed significantly greater AM levels (1.2-fold increase; P = 0.04) and higher AM/monocyte ratio (2.3-fold increase; P = 0.04) in the lung compared with noncolonized individuals (Figure 6A). On the other hand, monocyte levels, both total and CD14hiCD16lo and CD14hiCD16hi subsets, had no significant difference between the two groups, despite their trend for increased presence in the noncolonized group (Figures 6A and 6B). Similarly, no difference in neutrophil levels was observed between the two groups (Figure 6B), indicating that nasal carriage in absence of disease does not lead to neutrophil recruitment to the lung.
Figure 6.
Pneumococcal colonization may promote monocyte-to-macrophage differentiation. (A) Levels of monocytes and alveolar macrophages (AMs) in the BAL of carriage− (n = 8) and carriage+ (n = 9), expressed as percentage of CD45+ (cluster of differentiation 45–positive) cells. Significant comparison of AM levels and AM:monocytes ratio between the two study groups, P = 0.046 by Mann-Whitney test. Boxplots and individual subjects are depicted with carriage− in black and carriage+ in red. (B) Monocytes and neutrophils analyzed based on their CD14 and CD16 expression. In monocytes, CD16 expression levels divided them into two subsets, CD14hiCD16lo and CD14hiCD16hi. Boxplots and individual subjects are depicted with carriage− in black and carriage+ in red. (C) Top pathways after gene set enrichment analysis for pathways of differentiation and function applied on 2logFC (n = 5 subjects per group). Leading edge of genes per pathway is shown. Normalized Enrichment Score (NES) is presented in gradient color. Red shades indicate pathways overpresented, whereas blue depicts the pathway underpresented in the carriage-positive group. P values as shown on the graph. (D) Correlations between AM opsonophagocytic activity and 2log counts per million (CPM) of TBX21, NT5E, TLR8, and CEACAM6. Spearman correlation test results and linear regression line with 95% confidence interval (light blue shading) are shown.
To test whether antecedent pneumococcal colonization led to monocyte differentiation and AM activation, we sought to identify the differential gene signatures of Spn-colonized and noncolonized volunteers. We isolated AMs by cell sorting from a subset of BAL samples and performed NanoString expression analysis of 594 immunological genes. Gene set enrichment analysis was performed on all genes, ranked from high to low expressed in the Spn-colonized individuals compared with the noncolonized group, using published blood transcriptional modules (Table E3) (32). Purified AMs from Spn-colonized individuals showed an enrichment in pathways of cell differentiation and function, revealing underpresentation of monocyte surface markers and overpresentation of antigen-presentation markers in the Spn-colonized group (Figure 6C). Some of the genes in these sets that were most increased or decreased in Spn-colonized individuals included HLA-DQA1 (major histocompatibility complex, class II, DQ α1), CLEC7A (C-type lectin domain family 7 member A or Dectin-1), LY96 (lymphocyte antigen 96), and HLA-DPB1 (major histocompatibility complex, class II, DP α1) (antigen presentation module M5.0) and PTAFR (platelet-activating factor receptor), FCGRT (Fc fragment of IgG receptor and transporter), and CD4 (monocyte surface signature module S4). This finding complements the previous observation that nasal Spn colonization may lead to monocyte–AM differentiation. We then looked at whether individual AM gene expression was associated with colonization and/or function. Between the two groups, 34 genes were altered, although none remained significant after correction for multiple testing (potentially due to the small sample size of five per group) (Table E4). Moreover, when the AM OPA per individual was compared with gene expression (log counts per million) measured for each of the 594 genes, 34 genes were positively correlated with AM function to take up the bacteria (Table E5). Four genes were both positively correlated with AM OPA and increased in Spn-colonized individuals: TBX21 (T-box 21), NT5E (ecto-5′-nucleotidase), CEACAM6 (Carcinoembryonic antigen-related cell adhesion molecule 6), and TLR8 (Toll-like receptor 8) (Figure 6D).
Discussion
This study provides insights into the immune responses elicited at the human pulmonary mucosa after a pneumococcal carriage episode. Using our experimental human pneumococcal challenge model, we demonstrated that nasopharyngeal pneumococcal colonization results in bacterial aspiration to the lower airspaces, leading to a brisker AM opsonophagocytic capacity against both pneumococcus and other bacterial pathogens. Aspirated pneumococci most likely act as the stimulus that leads to enhanced AM responsiveness mediated by AM–CD4+ T-cell cross-talk and Th1 cytokine secretion. Systemic effects of colonization may have also contributed to the heightened lung immune responses. However, the systemic profile of immune cells usually does not reflect the mucosal sites, due to compartmentalization (33). Also, in the past we described that the effect of pneumococcal colonization on cognate T cells is more pronounced in the lung than in blood (9). Despite temporary influences from blood, the human lung exhibits a unique microenvironment that adapts to environmental challenges, and so do AMs adapt to accommodate the ever-changing needs of the tissue (31).
The lung mucosa is not the sterile environment previously thought (34, 35). By employing classical microbiology, molecular, and visualization methods, we demonstrated that pneumococcal aspiration occurs during nasal pneumococcal colonization, a phenomenon that was previously observed only in pneumonia cases (36, 37). The positive correlation between AM OPA and nasal pneumococcal density suggested pneumococcal cell trafficking from the nasopharynx to the lung airways. Spn-colonized volunteers received amoxicillin treatment before the bronchoscopy to clear pneumococcal colonization. This was a prerequisite in sampling the human lung without artificially introducing pneumococci; however, it is possible that bacterial fragments generated during the antibiotic treatment might trigger host responses both systemically and at the respiratory mucosal sites. Although it is difficult to assess the direct effect of this intervention on the AM responsiveness, our findings indicate that AM function is equally increased in those Spn-colonized volunteers who had naturally cleared colonization 1–2 weeks before the antibiotic treatment.
Interestingly, pneumococci were detected inside AMs or isolated alive from the BAL sample of Spn-colonized individuals when colonization had cleared a few days before the bronchoscopy. This observation merits deeper investigation into the aspects of pneumococcal survival inside AMs, analytical characterization of the AM subsets in the healthy human lung, and molecular signatures that drive differential AM function. Linked to the recent finding that pneumococcus has the ability to survive and replicate within murine splenic macrophages (38), there is an urgent need to analyze human AMs to single cell level. AM subpopulations that retain pneumococci or internalize them utilizing receptors that could lead to insufficient bacterial lysis could stand as a potential pneumococcal reservoir.
In addition, the increased opsonophagocytic capacity displayed by AMs after colonization was a nonspecific response to pneumococcal stimulus. AMs responded with equal efficacy to both Spn and other gram-positive respiratory pathogens in vitro. By contrast, we did not see significant enhancement of AM OPA against E. coli, although the small sample size used might have limited the detection of a less pronounced difference between the two experimental groups.
Our observation shares some similarities with the findings of emerging studies on “trained immunity” (or innate immune memory) (19, 39), which reported increased responsiveness of innate immune cells to microbial stimuli, caused by epigenetic changes, after their activation by varying stimuli (e.g., bacillus Calmette-Guérin or measles vaccination). Similarly to our observation, this augmented functional state persisted for weeks to months and additionally conferred; resistance to reinfection or heterologous infection (17, 18, 40, 41). Further controlled human infection studies, including pre– and post–pneumococcal challenge BAL sampling and studies focusing on AM epigenetic and metabolic changes, will be able to address whether human AMs acquire a “trained immunity” phenotype as a response to pneumococcal exposure. It will also enable comparisons of immune responses before and after colonization on an individual level.
Our findings on CD4+ Th1 skewed responses, and exogenous IFN-γ effect on AM antimicrobial function supported the idea that Th1-type responses and IFN are crucial in controlling bacteria at the early stages of infection. Increased rates of pneumococcal colonization in children and clinical cases of pneumonia in adults have been associated with a reduction in systemic circulating Th-1 (IFN-γ secreting) CD4+ T cells (42, 43). Polymorphisms in the adaptor Mal, which regulates IFN-γ signaling (44), have been associated with altered susceptibility to a number of infectious diseases including severe pneumococcal disease (45). Moreover, we observed a rapid priming of AMs when cocultured with autologous lung-derived CD4+ T cells in vitro. A very recent study in mice described a similar mechanistic link between adaptive and innate immune system, suggesting that effector CD8+ T cells, in the context of respiratory adenoviral infection, are able to prime AMs and render innate memory via IFN-γ (46).
Our study highlighted that IFN-γ has a dose-dependent effect on human AM function, which offers an explanation to the contradictory reports around this topic. For instance, in murine models, high production of IFN-γ during influenza infection impaired phagocytosis and killing of Spn by AMs (47, 48). In contrast, much other evidence suggests that induction of IFN-γ secretion related to nonacute viral infection is beneficial for innate immune cells, promoting a range of antimicrobial functions, plus macrophage polarization and activation (46, 49, 50). The dose-dependent effect of IFN-γ on AM OPA could also explain why HIV-infected adults are still at increased risk of developing pneumococcal pneumonia, despite the preserved Th1 responses against Spn (51).
By assessing AM gene expression levels, we found that AM population derived from Spn-colonized individuals was characterized by increased antigen presentation and decreased monocyte surface marker signature. Our flow-based data corroborated this result by showing greater AM levels and increased AM-to-monocyte ratio in the Spn-colonized individuals. The positive correlation of AM OPA with genes such as NT5E (or CD73) (52) and TBX21 (a master regulator of Th1 responses) indicates at some degree that monocyte-to-macrophage differentiation and AM polarization to a more active functional state occur in the human lung after interacting with the pneumococcus. Studies on human monocytes/macrophages have reported detectable expression of CD73 in only M(LPS-TNF) polarized cells and increase of T-bet mRNA displayed by M1-polarized macrophages (53, 54). Also, our finding on increased expression of TLR8 in the Spn-colonized group might be the readout of IFN-γ signaling, as TLR8 is responsive to IFNs. Increase in TLR8 levels might lead to enhanced viral sensing and thus have a beneficial effect upon viral infection, such as influenza.
In conclusion, this study emphasizes the effect that nasopharyngeal pneumococcal colonization has upon the pulmonary innate immune system, describing the potential pathways for the development of a robust immune response. It is well accepted that colonization is a prerequisite for disease. However, disease is more likely caused immediately after acquisition of colonization (55, 56). During longer colonization episodes, AMs display the increased functionality we described, which in turn could lead to reduced risk of bacterial respiratory infections. It is also important to note that our findings derive from young healthy adults, which are not at high risk of pneumococcal disease. The elderly are at an increased risk of infection, and these are colonized less frequently. The lack of repeated colonization and associated AM (and other immunological) boosting could contribute to their increased susceptibility and explain the paradox of low colonization and high disease in the elderly.
The seeding of human lung with activated AMs that exert prolonged and enhanced opsonophagocytic properties has potential implications for vaccine development. Pneumococcal vaccines that focus solely on inducing a robust Th17 response may not be the best strategy for vaccine-targeting serotype-independent protection against pneumonia. On the other hand, such a nonspecific boosting of innate lung immunity may be an alternative strategy to successful pneumonia prevention, especially for newborns, whose immune system is still developing, or for the elderly, whose acquired immunity is beginning to wear off. In particular, the elderly, who have been described as an age group with high incidence of community-acquired pneumonia cases, would benefit from the boosting effect that mucosal stimulation with whole-cell pneumococcus confers to the lung immune cells. These results, in combination with our previous finding on increased frequency of pneumococcal-specific CD4+ Th17 cells in human lung after nasal colonization (9), suggest that a nasally administered live-attenuated pneumococcal vaccine could augment the pulmonary immune responses and confer serotype-independent protection against development of pneumococcal pneumonia.
Supplementary Material
Acknowledgments
Acknowledgment
The authors would like to thank all volunteers for participating in this study, as well as all staff of the Clinical Research Unit at the Royal Liverpool Hospital and the clinical staff of the Respiratory Infection Group at the Liverpool School of Tropical Medicine. The authors also thank Dr. Caroline Weight for operating the confocal microscope. The Escherichia coli strain (NCTC86) was a kind gift from Dr. Adam Roberts.
Footnotes
Supported by the Bill and Melinda Gates Foundation (OPP1117728) (awarded to D.M.F.) and Medical Research Council Grants MR/M011569/1 and MR/M003078/1 (awarded to S.B.G. and M.R.O., respectively). Flow cytometric acquisition was performed on a BD LSRII and cell sorting was performed on a BD fluorescence-activated cell sorter ARIAIII funded by WelIcome Trust Multi-User Equipment Grant 104936/Z/14/Z.
Author Contributions: E.M.: conception and design, data analysis and interpretation of experiments, and manuscript writing and drafting. B.C., J. Reiné, E.N., C.S., S.P., and M.D.S.C.: manuscript writing and drafting, sample processing, and data analysis. J. Rylance, S.Z., V.C., A.M.C., H.H., and A.D.H.-W.: manuscript writing and drafting, assisting in procedures, and recruitment. A.S.-S., A.S., K.C.J., M.R.O., and S.B.G.: manuscript writing and drafting. S.P.J. and D.M.F.: conception and design, data analysis and interpretation experiments, and manuscript writing and drafting.
This article has an online supplement, which is accessible from this issue’s table of contents at www.atsjournals.org.
Originally Published in Press as DOI: 10.1164/rccm.201903-0607OC on October 18, 2019
Author disclosures are available with the text of this article at www.atsjournals.org.
References
- 1.Liu L, Oza S, Hogan D, Chu Y, Perin J, Zhu J, et al. Global, regional, and national causes of under-5 mortality in 2000-15: an updated systematic analysis with implications for the sustainable development goals. Lancet. 2016;388:3027–3035. doi: 10.1016/S0140-6736(16)31593-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Brien KL, Wolfson LJ, Watt JP, Henkle E, Deloria-Knoll M, McCall N, et al. Hib and Pneumococcal Global Burden of Disease Study Team. Burden of disease caused by Streptococcus pneumoniae in children younger than 5 years: global estimates. Lancet. 2009;374:893–902. doi: 10.1016/S0140-6736(09)61204-6. [DOI] [PubMed] [Google Scholar]
- 3.Wilson R, Cohen JM, Reglinski M, Jose RJ, Chan WY, Marshall H, et al. Naturally acquired human immunity to pneumococcus is dependent on antibody to protein antigens. PLoS Pathog. 2017;13:e1006137. doi: 10.1371/journal.ppat.1006137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bogaert D, De Groot R, Hermans PW. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004;4:144–154. doi: 10.1016/S1473-3099(04)00938-7. [DOI] [PubMed] [Google Scholar]
- 5.Goldblatt D, Hussain M, Andrews N, Ashton L, Virta C, Melegaro A, et al. Antibody responses to nasopharyngeal carriage of Streptococcus pneumoniae in adults: a longitudinal household study. J Infect Dis. 2005;192:387–393. doi: 10.1086/431524. [DOI] [PubMed] [Google Scholar]
- 6.Hussain M, Melegaro A, Pebody RG, George R, Edmunds WJ, Talukdar R, et al. A longitudinal household study of Streptococcus pneumoniae nasopharyngeal carriage in a UK setting. Epidemiol Infect. 2005;133:891–898. doi: 10.1017/S0950268805004012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferreira DM, Neill DR, Bangert M, Gritzfeld JF, Green N, Wright AK, et al. Controlled human infection and rechallenge with Streptococcus pneumoniae reveals the protective efficacy of carriage in healthy adults. Am J Respir Crit Care Med. 2013;187:855–864. doi: 10.1164/rccm.201212-2277OC. [Published erratum appears in Am J Respir Crit Care Med 187:1153.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McCool TL, Cate TR, Moy G, Weiser JN. The immune response to pneumococcal proteins during experimental human carriage. J Exp Med. 2002;195:359–365. doi: 10.1084/jem.20011576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wright AK, Bangert M, Gritzfeld JF, Ferreira DM, Jambo KC, Wright AD, et al. Experimental human pneumococcal carriage augments IL-17A-dependent T-cell defence of the lung. PLoS Pathog. 2013;9:e1003274. doi: 10.1371/journal.ppat.1003274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jambo KC, Sepako E, Heyderman RS, Gordon SB. Potential role for mucosally active vaccines against pneumococcal pneumonia. Trends Microbiol. 2010;18:81–89. doi: 10.1016/j.tim.2009.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marriott HM, Dockrell DH. The role of the macrophage in lung disease mediated by bacteria. Exp Lung Res. 2007;33:493–505. doi: 10.1080/01902140701756562. [DOI] [PubMed] [Google Scholar]
- 12.Gordon SB, Read RC. Macrophage defences against respiratory tract infections. Br Med Bull. 2002;61:45–61. doi: 10.1093/bmb/61.1.45. [DOI] [PubMed] [Google Scholar]
- 13.Lambrecht BN. Alveolar macrophage in the driver’s seat. Immunity. 2006;24:366–368. doi: 10.1016/j.immuni.2006.03.008. [DOI] [PubMed] [Google Scholar]
- 14.Morales-Nebreda L, Misharin AV, Perlman H, Budinger GR. The heterogeneity of lung macrophages in the susceptibility to disease. Eur Respir Rev. 2015;24:505–509. doi: 10.1183/16000617.0031-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hashimoto D, Chow A, Noizat C, Teo P, Beasley MB, Leboeuf M, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38:792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guilliams M, De Kleer I, Henri S, Post S, Vanhoutte L, De Prijck S, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. J Exp Med. 2013;210:1977–1992. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Netea MG, Joosten LA, Latz E, Mills KH, Natoli G, Stunnenberg HG, et al. Trained immunity: a program of innate immune memory in health and disease. Science. 2016;352:aaf1098. doi: 10.1126/science.aaf1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quintin J, Cheng SC, van der Meer JW, Netea MG. Innate immune memory: towards a better understanding of host defense mechanisms. Curr Opin Immunol. 2014;29:1–7. doi: 10.1016/j.coi.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 19.Cheng SC, Quintin J, Cramer RA, Shepardson KM, Saeed S, Kumar V, et al. mTOR- and HIF-1α-mediated aerobic glycolysis as metabolic basis for trained immunity. Science. 2014;345:1250684. doi: 10.1126/science.1250684. [Published erratum appears in Science 346:aaa1503.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitsi E, Carniel B, Jochems SP, Rylance J, Reine J, Schanoski AS, et al. Cross-talk of alveolar macrophages and T-cells boosts the lung immunity post nasopharyngeal pneumococcal colonisation [abstract]. Presented at the 11th International Symposium on Pneumococci and Pneumococcal Diseases. April 15–19, 2018, Melbourne, Australia. [Google Scholar]
- 21.Gritzfeld JF, Wright AD, Collins AM, Pennington SH, Wright AK, Kadioglu A, et al. Experimental human pneumococcal carriage. J Vis Exp. 2013;(72):50115. doi: 10.3791/50115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jochems SP, Marcon F, Carniel BF, Holloway M, Mitsi E, Smith E, et al. Inflammation induced by influenza virus impairs human innate immune control of pneumococcus. Nat Immunol. 2018;19:1299–1308. doi: 10.1038/s41590-018-0231-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jochems SP, Piddock K, Rylance J, Adler H, Carniel BF, Collins A, et al. Novel analysis of immune cells from nasal microbiopsy demonstrates reliable, reproducible data for immune populations, and superior cytokine detection compared to nasal wash. PLoS One. 2017;12:e0169805. doi: 10.1371/journal.pone.0169805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mitsi E, Kamng’ona R, Rylance J, Solórzano C, Jesus Reiné J, Mwandumba HC, et al. Human alveolar macrophages predominately express combined classical M1 and M2 surface markers in steady state. Respir Res. 2018;19:66. doi: 10.1186/s12931-018-0777-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zaidi SR, Collins AM, Mitsi E, Reiné J, Davies K, Wright AD, et al. Single use and conventional bronchoscopes for Broncho alveolar lavage (BAL) in research: a comparative study ( NCT 02515591) BMC Pulm Med. 2017;17:83. doi: 10.1186/s12890-017-0421-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Collins AM, Wright AD, Mitsi E, Gritzfeld JF, Hancock CA, Pennington SH, et al. First human challenge testing of a pneumococcal vaccine: double-blind randomized controlled trial. Am J Respir Crit Care Med. 2015;192:853–858. doi: 10.1164/rccm.201503-0542OC. [DOI] [PubMed] [Google Scholar]
- 27.Abbanat D, Davies TA, Amsler K, He W, Fae K, Janssen S, et al. Development and qualification of an opsonophagocytic killing assay to assess immunogenicity of a bioconjugated Escherichia coli vaccine. Clin Vaccine Immunol. 2017;24:e00123–17. doi: 10.1128/CVI.00123-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pennington SH, Pojar S, Mitsi E, Gritzfeld JF, Nikolaou E, Solórzano C, et al. Polysaccharide-specific memory B cells predict protection against experimental human pneumococcal carriage. Am J Respir Crit Care Med. 2016;194:1523–1531. doi: 10.1164/rccm.201512-2467OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Connor V, German E, Pojar S, Mitsi E, Hales C, Nikolaou E, et al. Hands are vehicles for transmission of Streptococcus pneumoniae in novel controlled human infection study. Eur Respir J. 2018;52:1800599. doi: 10.1183/13993003.00599-2018. [DOI] [PubMed] [Google Scholar]
- 30.Tarragó D, Fenoll A, Sánchez-Tatay D, Arroyo LA, Muñoz-Almagro C, Esteva C, et al. Identification of pneumococcal serotypes from culture-negative clinical specimens by novel real-time PCR. Clin Microbiol Infect. 2008;14:828–834. doi: 10.1111/j.1469-0691.2008.02028.x. [DOI] [PubMed] [Google Scholar]
- 31.Hussell T, Bell TJ. Alveolar macrophages: plasticity in a tissue-specific context. Nat Rev Immunol. 2014;14:81–93. doi: 10.1038/nri3600. [DOI] [PubMed] [Google Scholar]
- 32.Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sathaliyawala T, Kubota M, Yudanin N, Turner D, Camp P, Thome JJ, et al. Distribution and compartmentalization of human circulating and tissue-resident memory T cell subsets. Immunity. 2013;38:187–197. doi: 10.1016/j.immuni.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011;184:957–963. doi: 10.1164/rccm.201104-0655OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017;15:259–270. doi: 10.1038/nrmicro.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Albrich WC, Madhi SA, Adrian PV, van Niekerk N, Mareletsi T, Cutland C, et al. Use of a rapid test of pneumococcal colonization density to diagnose pneumococcal pneumonia. Clin Infect Dis. 2012;54:601–609. doi: 10.1093/cid/cir859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenberg D, Givon-Lavi N, Newman N, Bar-Ziv J, Dagan R. Nasopharyngeal carriage of individual Streptococcus pneumoniae serotypes during pediatric pneumonia as a means to estimate serotype disease potential. Pediatr Infect Dis J. 2011;30:227–233. doi: 10.1097/INF.0b013e3181f87802. [DOI] [PubMed] [Google Scholar]
- 38.Ercoli G, Fernandes VE, Chung WY, Wanford JJ, Thomson S, Bayliss CD, et al. Intracellular replication of Streptococcus pneumoniae inside splenic macrophages serves as a reservoir for septicaemia. Nat Microbiol. 2018;3:600–610. doi: 10.1038/s41564-018-0147-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saeed S, Quintin J, Kerstens HH, Rao NA, Aghajanirefah A, Matarese F, et al. Epigenetic programming of monocyte-to-macrophage differentiation and trained innate immunity. Science. 2014;345:1251086. doi: 10.1126/science.1251086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kleinnijenhuis J, Quintin J, Preijers F, Joosten LA, Ifrim DC, Saeed S, et al. Bacille Calmette-Guerin induces NOD2-dependent nonspecific protection from reinfection via epigenetic reprogramming of monocytes. Proc Natl Acad Sci USA. 2012;109:17537–17542. doi: 10.1073/pnas.1202870109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Netea MG, Quintin J, van der Meer JW. Trained immunity: a memory for innate host defense. Cell Host Microbe. 2011;9:355–361. doi: 10.1016/j.chom.2011.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Q, Bagrade L, Bernatoniene J, Clarke E, Paton JC, Mitchell TJ, et al. Low CD4 T cell immunity to pneumolysin is associated with nasopharyngeal carriage of pneumococci in children. J Infect Dis. 2007;195:1194–1202. doi: 10.1086/512617. [DOI] [PubMed] [Google Scholar]
- 43.Kemp K, Bruunsgaard H, Skinhøj P, Klarlund Pedersen B. Pneumococcal infections in humans are associated with increased apoptosis and trafficking of type 1 cytokine-producing T cells. Infect Immun. 2002;70:5019–5025. doi: 10.1128/IAI.70.9.5019-5025.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ní Cheallaigh C, Sheedy FJ, Harris J, Muñoz-Wolf N, Lee J, West K, et al. A common variant in the adaptor Mal regulates interferon gamma signaling. Immunity. 2016;44:368–379. doi: 10.1016/j.immuni.2016.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khor CC, Chapman SJ, Vannberg FO, Dunne A, Murphy C, Ling EY, et al. A Mal functional variant is associated with protection against invasive pneumococcal disease, bacteremia, malaria and tuberculosis. Nat Genet. 2007;39:523–528. doi: 10.1038/ng1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yao Y, Jeyanathan M, Haddadi S, Barra NG, Vaseghi-Shanjani M, Damjanovic D, et al. Induction of autonomous memory alveolar macrophages requires T cell help and is critical to trained immunity. Cell. 2018;175:1634–1650, e17. doi: 10.1016/j.cell.2018.09.042. [DOI] [PubMed] [Google Scholar]
- 47.Mina MJ, Brown LA, Klugman KP. Dynamics of increasing IFN-γ exposure on murine MH-S cell-line alveolar macrophage phagocytosis of Streptococcus pneumoniae. J Interferon Cytokine Res. 2015;35:474–479. doi: 10.1089/jir.2014.0087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun K, Metzger DW. Inhibition of pulmonary antibacterial defense by interferon-gamma during recovery from influenza infection. Nat Med. 2008;14:558–564. doi: 10.1038/nm1765. [DOI] [PubMed] [Google Scholar]
- 49.Matsuzawa T, Fujiwara E, Washi Y. Autophagy activation by interferon-γ via the p38 mitogen-activated protein kinase signalling pathway is involved in macrophage bactericidal activity. Immunology. 2014;141:61–69. doi: 10.1111/imm.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.MacMicking JD. Interferon-inducible effector mechanisms in cell-autonomous immunity. Nat Rev Immunol. 2012;12:367–382. doi: 10.1038/nri3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Peno C, Banda DH, Jambo N, Kankwatira AM, Malamba RD, Allain TJ, et al. Alveolar T-helper 17 responses to Streptococcus pneumoniae are preserved in ART-untreated and treated HIV-infected Malawian adults. J Infect. 2018;76:168–176. doi: 10.1016/j.jinf.2017.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Eichin D, Laurila JP, Jalkanen S, Salmi M. CD73 activity is dispensable for the polarization of M2 macrophages. PLoS One. 2015;10:e0134721. doi: 10.1371/journal.pone.0134721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez FO, Gordon S, Locati M, Mantovani A. Transcriptional profiling of the human monocyte-to-macrophage differentiation and polarization: new molecules and patterns of gene expression. J Immunol. 2006;177:7303–7311. doi: 10.4049/jimmunol.177.10.7303. [DOI] [PubMed] [Google Scholar]
- 54.Bachmann M, Scheiermann P, Härdle L, Pfeilschifter J, Mühl H. IL-36γ/IL-1F9, an innate T-bet target in myeloid cells. J Biol Chem. 2012;287:41684–41696. doi: 10.1074/jbc.M112.385443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ghaffar F, Friedland IR, McCracken GH., Jr Dynamics of nasopharyngeal colonization by Streptococcus pneumoniae. Pediatr Infect Dis J. 1999;18:638–646. doi: 10.1097/00006454-199907000-00016. [DOI] [PubMed] [Google Scholar]
- 56.Robinson J. Colonization and infection of the respiratory tract: what do we know? Paediatr Child Health. 2004;9:21–24. doi: 10.1093/pch/9.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.