Abstract
Saccharomyces cerevisiae is a popular expression system for recombinant proteins. In most cases, production processes are performed as carbon‐limited fed‐batch cultures to avoid aerobic ethanol formation. Especially for constitutive expression systems, the specific product formation rate depends on the specific growth rate. The development of optimal feeding strategies strongly depends on laboratory‐scale cultivations, which are time and resource consuming, especially when continuous experiments are carried out. It is therefore beneficial for accelerated process development to look at alternatives. In this study, S. cerevisiae AH22 secreting a heterologous endo‐polygalacturonase (EPG) was characterized in microwell plates with an enzyme‐based fed‐batch medium. Through variation of the glucose release rate, different growth profiles were established and the impact on EPG secretion was analyzed. Product formation rates of 200–400 U (gx h)−1 were determined. As a reference, bioreactor experiments using the change‐stat cultivation technique were performed. The growth‐dependent product formation was analyzed over dilution rates of D = 0.01–0.35 with smooth change of D at a rate of 0.003 h−2. EPG production was found to be comparable with a qp of 400 U (gx h)−1 at D = 0.27 h−1. The presented results indicate that parallel miniaturized fed‐batch cultures can be applied to determine product formation profiles of putative production strains. With further automation and parallelization of the concept, strain characterization can be performed in shorter time.
Keywords: Change‐stat, Fed‐batch, High‐throughput, Polygalacturonase, Recombinant protein
Abbreviations
- EPG
endo‐polygalacturonase
- OD600
optical density at 600 nm
1. Introduction
Yeasts are utilized for large‐scale production of proteins and small molecules. One popular host organism is Saccharomyces cerevisiae due to its well‐characterized genome, robust growth characteristics and the GRAS (generally regarded as safe) status. Since it is a Crabtree‐positive yeast, industrial production processes exclusively apply the fed‐batch technique 1. Selection and evaluation of production strains, however, is mostly done in batch cultures. This may cause problems during industrialization of production processes 2. Ideally, the cultivation conditions should remain as similar as possible during scale‐up 3.
Strain variants obtained from mutant libraries or strain collections are commonly screened in parallel batch cultures and evaluated from end‐point measured data. Under these conditions, oxygen limitation, aerobic ethanol formation and medium pH instability often occur, which can severely affect the outcome of strain screening experiments 4.
In the last decade, a number of key enabling technologies for consistent bioprocess development have been commercialized, such as microwell plates equipped with sensors 5, 6 and lids for improved aeration 7, minibioreactor systems 8 and fed‐batch media for small‐scale cultures 9, 10. These technologies are expected to significantly improve bioprocess development and scale‐up 11.
One major factor for yeast fed‐batch development is the determination of feeding profiles, which avoid excessive ethanol formation, while maintaining high product formation rates within the mass‐transfer boundaries of the bioreactor. The classical approach for investigations of the productivity at different dilution rates is to perform a series of chemostat experiments, which is very tedious and has been recently scaled down to mini‐bioreactors 12. Another option for accelerated data collection from continuous cultures is the change‐stat technique. With a smooth change of the dilution rate the culture is kept in a quasi‐steady‐state and a wide range of growth rates can be investigated. The technique has been successfully applied to bacteria 13, 14 and yeasts 15, 16, but so far there has been no attempt to investigate recombinant protein production in yeast 17. The change‐stat technique has been successfully applied for the investigation of protein secretion at different growth rates. The technology could be combined with miniaturized continuous cultivation systems 12 in order to characterize production strains, e.g. for improved productivity with processes at higher growth rates 18.
The aim of this study was to determine the specific product formation rate (qP) in relation to the corresponding specific growth rate (μ) of a putative production strain significantly faster than using chemostat cultures. Microwell plate cultures with parallel enzyme‐based fed‐batch experiments were performed at different glucose release rates to characterize the production strain. The results were compared to change‐stat continuous cultivations.
2. Materials and methods
2.1. Yeast strain and culture storage
Cultivations were carried out using Saccharomyces cerevisiae AH22 (leu2‐1, leu2‐112, his4‐519, can1, cir+, mating type a) harboring the plasmid pPG6, which was constructed for heterologous expression of polygalacturonase from Aspergillus niger 19, 20.
Ethyl methanesulfonate mutagenesis was applied with subsequent mutant selection on pectin agar plates. One colony, which showed improved pectin hydrolysis was isolated from the mutagenesis experiment (strain pPG6 M27). The cell bank was stored at –80°C in minimal medium containing 20% glycerol.
2.2. Media
The cultivations were performed in a minimal medium based on WMVIII 19 containing glucose or 20 g/L EnPump glucose polymer (BioSilta Ltd., Cambridge, UK). Glucose is released from the EnPump polymer, when Reagent A is added. The composition of the modified WMVIII (mWM8) was as follows: NH4H2PO4 0.25 g/L, NH4Cl 5.48 g/L, MgCl2·6 H2O 0.25 g/L, CaCl2·2 H2O 0.1 g/L, KH2PO4 2.0 g/L, MgSO4·7 H2O 0.55 g/L, myo‐inositol 75 mg/L, sodium glutamate 1.5 g/L ZnSO4·7H2O 1.75 mg/L, FeSO4·7H2O 0.5 mg/L, CuSO4· 5 H2O 0.1 mg/L, MnCl2·4 H2O 0.1 mg/L, Na2MoO4·2 H2O 0.1 mg/L, nicotinic acid 10 mg/L, pyridoxin‐HCl 25 mg/L, thiamine HCl 10 mg/L, biotin 2.5 mg/L, calcium panthotenate 50 mg/L, histidine 100 mg/L.
2.3. A‐stat cultivation
The preculture was inoculated from a cryo vial into 100 mL mWM8 containing 50 g/L glucose in a 500 mL UltraYield flask™ covered with AirOtop Enhance Seal™ (Thompson Instrument Company, Oceanside, USA), which was incubated at 30°C and 180 rpm overnight in a shaken incubator (Kühner LT‐X, Kühner AG, Basel, Switzerland). The continuous culture was carried out in a 3.7 L bioreactor (KLF 2000, Bioengineering AG, Wald, Switzerland) with a working volume of 1.5 L at 30°C. The preculture was transferred into the bioreactor containing 1.5 L mWM8 with 15 g/L of glucose. The pH was controlled at 6.0 by addition of 10% H3PO4 and 25% ammonia. After an initial batch phase, the continuous culture was initialized by feeding medium into the reactor via a voltage‐controlled pump and harvesting culture broth through an overflow. The culture was stabilized at a dilution rate of 0.07 h−1 for at least five retention times to ensure steady‐state conditions. Samples were taken for the analysis of optical density (OD600), biomass, medium composition and EPG activity. The dilution rate was then increased, and in a subsequent experiment decreased, by an acceleration factor of 0.003 h−2 according to the following formula:
| (1) |
This acceleration factor has been previously used in a study by Adamberg et al. 13, which served as the main reference for this study.
2.4. Strain characterization in 24 well plates
Precultures were grown in 125 mL UltraYield flasks with 20 mL of mWM8 containing 40 g/L glucose. Cultures were incubated for 48 h at 30°C and 250 rpm (25 mm amplitude) on an orbital shaker. For the main culture, mWM8 with 20 g/L of EnPump polysaccharide (BioSilta Ltd.) was inoculated with the volume of the preculture corresponding to an initial OD600 of 0.1 and glucose release was initiated by the addition of 1 U/L of reagent A (BioSilta Ltd.). The main cultivation was performed in 24 well sensor plates (OxoDish, HydroDish, PreSens) with a filing volume of 1.1 mL covered with ‘System Duetz’ lids (Enzyscreen B.V., Heemstede, The Netherlands) and shaken at 300 rpm (50 mm amplitude). After an initial overnight phase, variations of the glucose‐release were introduced by supplementing duplicate wells in each sensor plate with 1–30 U/L of reagent A. After an adaptation phase of 3 h, samples for OD600 and EPG measurement were taken using a liquid handling robot (Hamilton Microlab Star, Hamilton Bonaduz AG, Bonaduz, Switzerland). The OD600 values were converted to biomass with a predetermined factor of 0.38 and the growth rate and product formation rates were calculated using (2) and (3):
| (2) |
| (3) |
X = Biomass, EA = volumetric enzyme activity, t = sampling time.
2.5. Analysis methods
2.5.1. Biomass determination
Samples taken from microwell plate cultures were measured in 0.9% NaCl or EPG assay buffer using flat‐bottom 96‐well plates (Greiner Bio‐One, Frickenhausen, Germany) for OD600 determination in the microplate reader. One OD unit of the plate reader corresponds to a cell dry weight of 380 mg/L. Shake flask and bioreactor samples were diluted in 0.9% NaCl solution and measured in a cuvette spectrophotometer (Ultrospec 2100 pro, Amersham Biosciences, Glattburg, Switzerland). Dry biomass was determined from bioreactor samples as follows: 2 mL of culture broth were centrifuged in pre‐dried Eppendorf tubes at 21 500 g, the supernatant was discarded and the cell pellet was dried at 75°C until a constant weight was recorded.
2.5.2. Analysis of medium composition
The culture supernatant was analyzed for glucose, ethanol, ammonia, and glutamate levels with enzymatic test kits. Calibration curves for the respective concentration ranges were prepared for all assays.
Glucose was determined using the Hexokinase FS test (DiaSys, Holzheim, Germany) in 96‐well plates or cuvettes. In microwell plates, 10 μL of sample were added to 190 μL of test solution and incubated for 15 min until read‐out at 340 nm. In cuvettes, 1 mL of test solution was applied to 10 μL of sample. Ethanol, ammonia and glutamate were measured using test kits for 1 mL cuvettes according to the manufacturer's instructions (R‐Biopharm, Darmstadt, Germany).
2.5.3. EPG activity assay
The determination of EPG activity was performed in 96‐well plate format using the liquid handling robot. A colorimetric assay using 3‐methyl‐2benzothiazolinonehydrazone (MBTH), which was developed for test tubes 21, 22 was adapted to the 96‐well format. Polygalacturonic acid (20 g/L) in 100 mM sodium acetate buffer (pH 4.5) served as a substrate. After an incubation time of 15 min, the reducing ends of the released galacturonic acid were quantified using 7 mM MBTH in a two‐step reaction. The first step was carried out at 65°C for 15 min in a thermal cycler, while the second step required the addition of acidic Fe3+ solution (10 mM NH4Fe(SO4)2·12 H2O, 51.5 mM sulfamic acid in 250 mM HCl) and took place for 15 min at room temperature. The color complex was detected at 620 nm and galacturonic acid served as a standard. One unit of enzyme releases 1 μmole of reducing sugar from the substrate per minute.
3. Results
State‐of‐the‐art characterization of a production strain with regards to its product formation profile is typically carried out in continuous fermentation systems (Fig. 1), which results in the acquisition of data from a limited number of steady states. A significant reduction of experimental efforts can be achieved using the change‐stat method. Here, the whole growth space can be analyzed, which leads to a more detailed description of the strain in shorter time. Finally, individual fed‐batch experiments can be carried out at different feed‐rates. The most time‐efficient option is to run the cultures in parallel and sample during the transition phase of exponential to glucose‐limited growth.
Figure 1.
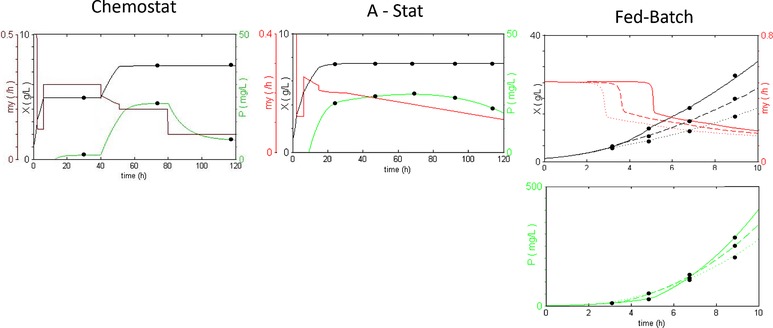
Schematic overview of methods to determine growth‐rate dependent product formation. In a chemostat experiment, product formation can be measured at a limited number of steady. The adapta‐stat is a chemostat with a smooth change of the dilution rate, which allows measurements in a quasi‐steady‐state. In fed‐batch cultures with constant feeding, the growth rate decreases abruptly once substrate limitation is reached. When monitoring cell density and product accumulation over time, a relation between μ and qP can be drawn.
3.1. A‐stat Fermentation
For an efficient quantification of EPG‐secretion at a wide range of dilution rates, the change‐stat technique is known to be very beneficial. We performed cultivations with a working volume of 1.5 L that started as a batch culture with a subsequent chemostat for five residence times. Then, the dilution rate was smoothly increased (accellerostat, A‐stat) or decreased (deceleration‐stat, De‐stat) with a linear profile until D = 0.01 h−1 for De‐stat and wash‐out for A‐stat.
The De‐stat experiment was performed under glucose‐limitation at an initial D of 0.12 h−1, which was below the D, at which aerobic ethanol formation was expected. From the EPG activity data obtained, it could be concluded that from the initial dilution rate until D ≈ 0.07 h−1, the product formation rate remained constant in the interval between a dilution rate D of 0.12 h−1 and 0.07 h−1. A further decrease of D caused a drastic decrease of qP and the experiment was ended at qP < 70 U (gx h)−1. To compare the different cultivation protocols, a second change‐stat experiment was carried out starting from a chemostat at D = 0.07 h−1. The dilution rate was increased with the same acceleration factor. From at‐line enzymatic ethanol and glucose measurements, an onset of ethanol formation at D ≈ 0.2 h−1 was detected, while glucose accumulation was visible at D ≈ 0.28 h−1 (Fig. 2). The biomass concentration remained constant at 6.5 g/L until D = 0.2 h−1 and then gradually decreased until D = 0.35 h−1, where at a biomass concentration of 1.2 g/L the cultivation was stopped. EPG was detected at all dilution rates, while at D< 0.05, productivity decreased significantly. Dilution rates of 0.08–0.11 resulted in an EPG production rate of 250–400 U (gx h)−1 in the De‐stat experiment. In the A‐stat experiment, a maximum product formation rate of 400 U (gx h)−1 at D = 0.27 h−1 was found.
Figure 2.
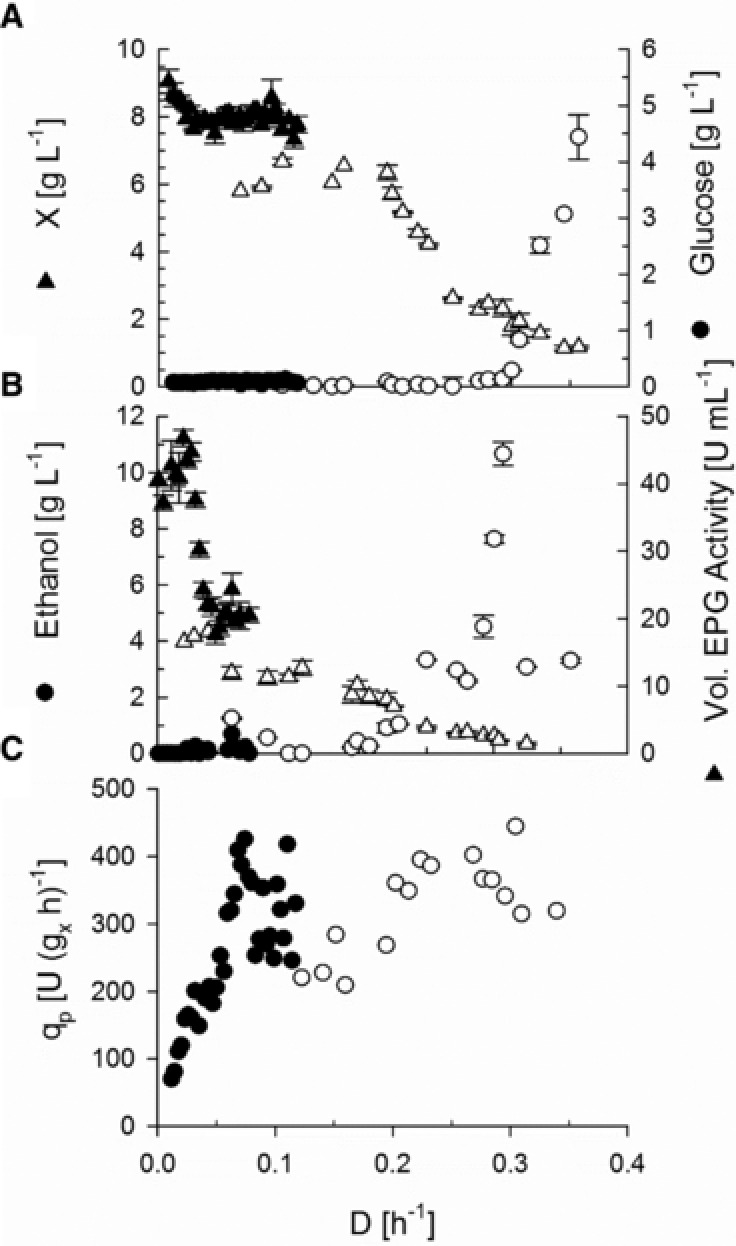
Growth space characterization of S. cerevisiae AH22 pPG6 M27 in change‐stat cultivations. A‐stat (open symbols) and De‐stat (closed symbols) cultivations were performed. (A) Biomass and glucose concentrations. (B) Ethanol concentration and volumetric EPG activity. (C) Specific EPG formation rate (qP).
The change‐stat experiments confirmed that growth‐rates below the onset of the Crabtree effect are necessary for the efficient production of EPG, as there is no considerable benefit from cultivations at growth rates above the threshold level of ethanol production. Depending on the selected cultivation mode, the product formation profile differs, which indicates an influence of the cell's history on the obtained results. In‐depth analysis of the cell's metabolic state would be necessary to investigate this matter more closely.
3.2. Strain characterization in parallel fed‐batch cultures at the mL scale
We propose to determine the specific product formation rate (qP) at a wide range of specific growth rates (μ) with parallel miniaturized fed‐batch cultures to obtain the same results in shorter time. This is now possible due to the advances in automation and miniaturization. A 24‐well‐plate system with online DO and pH measurement and improved aeration was applied. Glucose feeding was performed with the enzyme‐based glucose delivery system (EnBase) combined with the mWM8 medium. After an overnight phase with 1 U/L of reagent A for constant glucose release, the cultures were supplemented with 1–30 U/L to introduce variations in the growth pattern (Fig. 3). Samples were analyzed for biomass and EPG activity at‐line, i.e. during the experiment, with the robotic platform.
Figure 3.
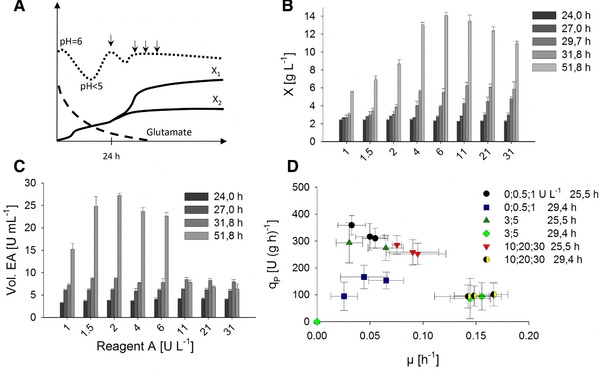
Strain characterization in parallel fed‐batch cultures. (A) Schematic overview of the experimental set‐up: After an initial batch phase, indicated by a pH drop (dotted line), glutamate consumption stabilized pH (dashed line). Additional reagent A supplementation (arrow) induces a short batch phase, after which samples are taken for OD600 and EPG measurements (dashed arrows). (B) Biomass data from cultures grown with 1–31 U/L reagent A. (C) Volumetric EPG activity of the culture supernatant. (D) Correlation of product formation rate (q P) and specific growth rate.
The parallel fed‐batch cultures showed different optima for biomass and EPG production. At glucose release rates from 1.5 to 4 U/L, volumetric yield of active EPG was highest, while peak biomass formation was detected with 6 U/L. From the first three data points, μ and qP were calculated and it was found that the range was comparable to the A‐stat results (Fig. 3D)
As an example, a maximal production rate of 218.8 U (gx h)−1 at a growth rate of 0.095 h−1 was identified for cultures grown with 1.5 U/L. However, due to the rapid decrease of the growth rate at constant glucose release rates, the range of growth rates was narrower than in the continuous cultures. Moreover, the optimum for EPG secretion was found to be at μ = 0.05–0.1, which is considerably lower than in the A‐stat experiments.
4. Discussion
Choosing the right feeding strategy for the best space time yield is key for the development of a biotechnological production process. Traditionally, the determination of the relation of μ and qP is done in chemostat cultures exclusively 23, 24, 25, 26. Due to the long experimental time to reach steady state, even the characterization of a single strain is very labor intensive, which usually rules out the chance to evaluate several putative production strains. In an effort to reduce experimental times, change‐stat methods have been proposed.
In this study, the use of parallel fed‐batch cultures is compared to the change‐stat method to further reduce experimental time and costs. First, the change‐stat technique was applied to characterize the entire growth space. The concept of determining product formation at different growth profiles was then applied to parallel fed‐batch cultures in 24‐well plates with at‐line OD600 and EPG determination allowed an estimation of the μ‐dependent specific EPG‐production rate.
Growth‐dependent product formation was detected with a stable secretion level of 200–400 U (gx h)−1 over a wide range of dilution rates. However, the product formation profiles indicate that the operation mode of the change‐stat culture has an influence on the strain behavior. This observation was confirmed by the subsequent microwell plate experiments, in which the product formation profile indicated a lower optimum than in the continuous cultures.
The fed‐batch approach offers significant experimental time reduction and a simplified experimental set‐up. While it is very demanding to perform chemostat cultures in micro‐ and milliliter reactors, reproducible fed‐batch fermentations are easy to perform and only require minimal expenses for material and chemicals, compared to bioreactor experiments. Moreover, the determination of μ‐dependency of qP in fed‐batch cultures provides good information about the dynamic changes, which are not obtained from chemostat experiments. Further development of this approach through a combination with mechanistic models will provide a real breakthrough for process development 27.
For future studies, automation can be further advanced using on‐line biomass sensing or automated cell separation, as they are used by others 28, 29. The μ‐dependent protein secretion could be performed in pH‐controlled minibioreactor systems, which could improve the predictive power of the obtained results 30. In conclusion, fed‐batch process development can be accelerated with small‐scale fed‐batch cultures which may replace the need for chemostat and change‐stat experiments.
Practical application
When designing fed‐batch processes, the optimal specific growth rate for efficient production is a key target parameter, which needs to be experimentally determined. Instead of the traditional method, using serial chemostat experiments, we have applied change‐stat cultures to determine the optimal growth rate for efficient product secretion of a S. cerevisiae AH22 strain expressing fungal polygalacturonase. For even faster evaluation of the strain, parallel fed‐batch cultivations with enzyme‐based glucose delivery were performed. From these small‐scale experiments, we could investigate influence of the specific growth rate (μ) on the specific production rate (qP), which leads to results comparable to the bioreactor scale obtained in two weeks instead of several months.
The authors have declared no conflict of interest.
Acknowledgments
The authors would like to thank BioSilta Ltd. and PreSens GmbH for technical support and Rick Nolte, Franziska Jehle and Michael Heiser for carrying out parts of the experimental work. The financial support by the German Federal Ministry of Education and Research (BMBF) within the Framework Concept “Research for Tomorrow's Production” (project no. 02PJ1150, AUTOBIO project) was greatly acknowledged.
5 References
- 1. Porro, D. , Sauer, M. , Branduardi, P. , Mattanovich, D. , Recombinant protein production in yeasts. Mol. Biotechnol. 2005, 31, 245–259. [DOI] [PubMed] [Google Scholar]
- 2. Neubauer, P. , Cruz, N. , Glauche, F. , Junne, S. et al., Consistent development of bioprocesses from microliter cultures to the industrial scale. Eng. Life Sci. 2013, 13, 224–238. [Google Scholar]
- 3. Siurkus, J. , Panula‐Perälä, J. , Horn, U. , Kraft, M. et al., Novel approach of high cell density recombinant bioprocess development: Optimisation and scale‐up from microliter to pilot scales while maintaining the fed‐batch cultivation mode of E. coli cultures. Microb. Cell Fact. 2010, 9, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Scheidle, M. , Jeude, M. , Dittrich, B. , Denter, S. et al., High‐throughput screening of Hansenula polymorpha clones in the batch compared with the controlled‐release fed‐batch mode on a small scale. FEMS Yeast Res. 2010, 10, 83–92. [DOI] [PubMed] [Google Scholar]
- 5. Arain, S. , John, G. T. , Krause, C. , Gerlach, J. et al., Characterization of microtiterplates with integrated optical sensors for oxygen and pH, and their applications to enzyme activity screening, respirometry, and toxicological assays. Sensors Actuators, B Chem. 2006, 113, 639–648. [Google Scholar]
- 6. Glauche, F. , John, G.T. , Arain, S. , Knepper, a. et al., Toward microbioreactor arrays: A slow‐responding oxygen sensor for monitoring of microbial cultures in standard 96‐well plates. J. Lab. Autom. 2015, 20, 438–446. [DOI] [PubMed] [Google Scholar]
- 7. Duetz, W. a. , Rüedi, L. , Hermann, R. , O'Connor, K. et al., Methods for intense aeration, growth, storage, and replication of bacterial strains in microtiter plates. Appl. Environ. Microbiol. 2000, 66, 2641–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Puskeiler, R. , Kusterer, A. , John, G. T. , Weuster‐Botz, D. , Miniature bioreactors for automated high‐throughput bioprocess design (HTBD): Reproducibility of parallel fed‐batch cultivations with Escherichia coli . Biotechnol. Appl. Biochem. 2005, 42, 227–235. [DOI] [PubMed] [Google Scholar]
- 9. Panula‐Perälä, J. , Siurkus, J. , Vasala, A. , Wilmanowski, R. et al., Enzyme controlled glucose auto‐delivery for high cell density cultivations in microplates and shake flasks. Microb. Cell Fact. 2008, 7, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Jeude, M. , Dittrich, B. , Niederschulte, H. , Anderlei, T. et al., Fed‐batch mode in shake flasks by slow‐release technique. Biotechnol. Bioeng. 2006, 95, 433–445. [DOI] [PubMed] [Google Scholar]
- 11. Long, Q. , Liu, X. , Yang, Y. , Li, L. et al., The development and application of high throughput cultivation technology in bioprocess development. J. Biotechnol. 2014, 192, 323–338. [DOI] [PubMed] [Google Scholar]
- 12. Schmideder, A. , Severin, T. S. , Cremer, J. H. , Weuster‐Botz, D. , A novel milliliter‐scale chemostat system for parallel cultivation of microorganisms in stirred‐tank bioreactors. J. Biotechnol. 2015, 210, 19–24. [DOI] [PubMed] [Google Scholar]
- 13. Adamberg, K. , Lahtvee, P.‐J. , Valgepea, K. , Abner, K. et al., Quasi steady state growth of Lactococcus lactis in glucose‐limited acceleration stat (A‐stat) cultures. Antonie Van Leeuwenhoek 2009, 95, 219–226. [DOI] [PubMed] [Google Scholar]
- 14. Nahku, R. , Valgepea, K. , Lahtvee, P.‐J. , Erm, S. et al., Specific growth rate dependent transcriptome profiling of Escherichia coli K12 MG1655 in accelerostat cultures. J. Biotechnol. 2010, 145, 60–65. [DOI] [PubMed] [Google Scholar]
- 15. Paalme, T. , Elken, R. , Vilu, R. , Korhola, M. , Growth efficiency of Saccharomyces cerevisiae on glucose/ethanol media with a smooth change in the dilution rate (A‐stat). Enzyme Microb. Technol. 1997, 20, 174–181. [Google Scholar]
- 16. Van Sluis, C. Der , Westerink, B. H. , Dijkstal, M. M. , Castelein, S. J. et al., Estimation of steady‐state culture characteristics during acceleration‐stats with yeasts. Biotechnol. Bioeng. 2001, 75, 267–275. [DOI] [PubMed] [Google Scholar]
- 17. Valgepea, K. , Vilu, R. , Adamberg, K. , Advanced continuous cultivation methods for systems microbiology. Microbiology 2015, 161, 1707–1719. [DOI] [PubMed] [Google Scholar]
- 18. Klein, C. J. L. , Rasmussen, J. J. , Rønnow, B. , Olsson, L. et al., Investigation of the impact of MIG1 and MIG2 on the physiology of Saccharomyces cerevisiae. J. Biotechnol. 1999, 68, 197–212. [DOI] [PubMed] [Google Scholar]
- 19. Lang, C. , Looman, a. C. , Efficient expression and secretion of Aspergillus niger RH5344 polygalacturonase in Saccharomyces cerevisiae. Appl. Microbiol. Biotechnol. 1995, 44, 147–156. [DOI] [PubMed] [Google Scholar]
- 20. Lang, C. , Göllnitz, C. , Popovic, M. , Stahl, U. , Optimization of fungal polygalacturonase synthesis by Saccharomyces cerevisiae in fed‐batch culture. Chem. Eng. J. 1997, 65, 219–226. [Google Scholar]
- 21. Honda, S. , Nishimura, Y., Chiba, H., Kakehi, K., Determination of carbohydrates by condensation with 3‐methyl‐2‐benzothiazolinonehydrazone. Anal. Chim. Acta. 1981, 131, 293–296. [Google Scholar]
- 22. Anthon, G. E. , Barrett, D. M. , Determination of reducing sugars with 3‐methyl‐2‐benzothiazolinonehydrazone. Anal. Biochem. 2002, 305, 287–289. [DOI] [PubMed] [Google Scholar]
- 23. Kocharin, K. , Nielsen, J. , Specific growth rate and substrate dependent polyhydroxybutyrate production in Saccharomyces cerevisiae . AMB Express 2013, 3, 18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Vos, T. , de la Torre Cortés, P. , van Gulik, W. M. , Pronk, J. T. et al., Growth‐rate dependency of de novo resveratrol production in chemostat cultures of an engineered Saccharomyces cerevisiae strain. Microb. Cell Fact. 2015, 14, 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu, Z. , Hou, J. , Martínez, J. L. , Petranovic, D. et al., Correlation of cell growth and heterologous protein production by Saccharomyces cerevisiae . Appl. Microbiol. Biotechnol. 2013, 97, 8955–8962. [DOI] [PubMed] [Google Scholar]
- 26. Rebnegger, C. , Graf, A. B. , Valli, M. , Steiger, M. G. et al., In Pichia pastoris, growth rate regulates protein synthesis and secretion, mating and stress response. Biotechnol. J. 2014, 9, 511–525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cruz Bournazou, M. N. , Barz, T. , Nickel, D. , Lopez Cárdenas, D. et al., Online optimal experimental re‐design in robotic parallel fed‐batch cultivation facilities for validation of macro‐kinetic growth models using E. coli as an example. Biotechnol. Bioeng. 2016, 114, 1–29. [DOI] [PubMed] [Google Scholar]
- 28. Unthan, S. , Radek, A. , Wiechert, W. , Oldiges, M. et al., Bioprocess automation on a mini pilot plant enables fast quantitative microbial phenotyping. Microb. Cell Fact. 2015, 14, 216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Rohe, P. , Venkanna, D. , Kleine, B. , Freudl, R. et al., An automated workflow for enhancing microbial bioprocess optimization on a novel microbioreactor platform. Microb. Cell Fact. 2012, 11, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hortsch, R. , Weuster‐Botz, D. , Milliliter‐Scale Stirred Tank Reactors For The Cultivation Of Microorganisms, 1st ed., Elsevier, Amsterdam, 2010. [DOI] [PubMed] [Google Scholar]


