Abstract
We present a novel protocol that uses single‐cell force spectroscopy to characterize the bacteria‐to‐surface interactions involved in early steps of biofilm formation. Bacteria are immobilized as a monolayer by electrostatic interactions on a polyethylenimine‐coated silica bead, and the Escherichia coli‐bead complex is then glued on a tipless cantilever. We validated our new protocol by comparing to earlier published methods using single bacteria, but in contrast to these, which carry out bacterial attachment to the bead after fixation to the cantilever, our protocol results in more reliable production of usable cell probes. Measurements of interactions of E. coli with bio‐inspired surfaces by single‐cell force spectroscopy yielded comparable detachment forces to those found with the previous methods.
Keywords: Anti‐adhesive surfaces, Bacterial adhesion force quantification, Biofilm, Bioinspired structural materials, Single‐cell force spectroscopy
Abbreviations
- AFM
atomic force microscopy
- CLSM
confocal laser scanning microscopy
- GFP
green fluorescence protein
- PBS
phosphate buffered saline
- PEI
polyethylenimin
1. Introduction
Nearly all surfaces both natural and synthetic, where interfaces exist between fluid phases, gas and fluid, or liquid and solid phases, provide fertile ground for the growth of biofilms. Biofilms consist of a cellular assemblage, mostly comprising bacteria irreversibly attached to a substrate and enclosed in a matrix of extracellular polymeric substances 1. Examples of their current beneficial uses in biotechnology include the production of chemicals 2 or extracellular enzymes 3, 4. In contrast, however, biofilms are generally considered deleterious in medicine 1, 5, 6 and in industrial technology 7, 8, 9. Therefore, efforts are underway to develop anti‐adhesive surfaces and coatings for machinery that is susceptible to biofilm growth 10, 11, 12.
We focus on the discovery and development of surfaces that act as biofilm preventives, using biomimetic and bio‐inspired approaches. Some natural materials that have been investigated for this purpose include leaf surfaces 13, 14, shark skin 15 and the cuticles of springtails 16, 17. The topography of insect wings 18, 19 has been explored as a potential inspiration for the generation of synthetic anti‐fouling surfaces. We recently investigated nanostructured surfaces with profiles mimicking the natural cicada wing, with the aim of ultimately applying it in industrial processing to reduce biofouling 20.
Because the irreversible attachment of microbes to a surface constitutes the critical first step in biofilm formation, our studies focus on elucidating the mechanisms of these early bacterium‐to‐surface interactions. To do this, we developed a modular microfluidic flow cell system in which bacterial adhesion and initial biofilm formation can be observed under continuous liquid flow conditions by optical microscopy 20, 21, 22.
In addition to these microscopic observations, information on forces operating at the nanoscale can be derived from single‐cell force spectroscopy (SCFS). Using optical microscopy combined with atomic force microscopy (AFM), by which the deflection of the cantilever is an indicator of interaction forces 23, bacteria immobilized on the cantilever are brought into close apposition with a surface to assess particular interactions 24.
Generally, the cantilever is immersed in a drop of a concentrated bacterial cell suspension prior to force measurements 25, 26, 27. In an alternative approach, albeit requiring precise handling by a well‐trained operator, a single living bacterium is taken up from a diluted sample and immobilized on a modified cantilever 28. A disadvantage of all of these methods is that they yield large numbers of unusable probes, and the bacterial viability and distribution on the cantilever can be determined only after assembly of the entire probe 27, 29.
In this work, we present a new method that addresses the aforementioned difficulties in probe production. Here, the pre‐complexed E. coli loaded PEI‐bead is glued to the cantilever (hereafter referred to as “cell probe”). Because they are loaded onto the bead before attachment to the cantilever, the viability and distribution of the bacteria can be easily assessed by fluorescence microscopy, and only optimally complexed beads chosen for attachment to the cantilever and subsequent measurement of force‐distance curves.
2. Materials and methods
2.1. Strains and culture conditions
GFP‐tagged E. coli SM2029 strain was kindly provided by Søren Molin (Technical University of Denmark) 30.
Cells from a preculture grown at 30°C in a shaker flask containing 20 g/L LB‐medium with 50 mg/L kanamycin (Lennox, Carl Roth GmbH) for 12–14 h at 50 rpm in a Compact Shaker KS 15 A (Edmund Bühler GmbH, Germany) were harvested in the exponential growth phase, washed three times in pH 7.4 PBS (9 g/L NaCl, 0.98 g/L Na2HPO4·2H2O, 0.16 g/L KH2PO4), centrifuged at 5000 x g and resuspended by vortexing. Finally the cell concentration was adjusted to OD600 = 5.
2.2. Substrate surfaces
The microstructured surface, hereafter referred to as “structured” was a porous anodized aluminum oxide membrane with well‐defined surface nanopatterns having pore diameters of 350 ± 20 nm and a pitch of 360 nm fabricated by SmartMembranes GmbH using electrochemical precision etching 20. Planar aluminum oxide was used as a reference surface (referred to as the “planar” surface).
2.3. SCFS measurements with PEI‐beads carrying multiple E. coli
Sedimented aminated silica beads (PSI‐20.0NH2) were from Kisker Biotech GmbH, Germany.
To prepare the coating solution, 50 μL of polyethyleneimine (PEI) (analytical standard; 50% (w/v) in H2O; Sigma‐Aldrich Chemie GmbH, Germany) were added to 5 mL PBS in a 12 mL conical centrifuge tube (VWR International GmbH, Germany).
Coating of the beads was performed every day just prior to their use in experiments. For coating, 10 μL of beads were transferred into coating solution and shaken manually and subsequently horizontally at 150 rpm for 50 min on a Compact Shaker KS 15 A (Edmund Bühler GmbH, Germany). Next, PEI‐beads were left standing for 5 min to sediment and washed three times with PBS and gently manually mixed to avoid agglomeration.
Approximately 10 μL of the PEI‐beads were transferred to a 3.5 mL Polystyrene sample tube (Sarstedt AG & Co., Germany) filled with 1 mL of E. coli suspension (OD600 = 5). The suspension was mixed by inverting the tube a few times. The beads were then left to sediment for 3 min, the supernatant was replaced with 2.7 mL PBS and tube was slowly inverted by hand for 5 min to avoid detachment of cells. The washing procedure was repeated three times.
PEI‐beads loaded with E. coli (hereafter referred to as “E. coli‐PEI‐beads”) were used within 3 h for AFM analysis.
The distribution of GFP‐tagged bacteria on the E. coli‐PEI‐beads was visualized using Axioplan 2 Imaging (Carl Zeiss Microscopy GmbH, Germany, Zeiss set 38). To investigate viability of bacteria in addition samples were stained with propidium iodide (2 mg/L) and examined with a LSM 780/FLIM (Carl Zeiss Microscopy GmbH, Germany, Zeiss sets 38 and 43). Z‐stack images of each of five to seven E. coli‐PEI‐beads were taken four times independently and 3D images were generated using the image processing software arivis Vision4D v. 2.12.3 (Arivis AG, Germany). The average projection area of a single bacterium was determined and the ratio of living to dead cells was counted with Fiji (ImageJ). The coverage of E. coli on PEI‐beads [%] was calculated from the ratio of area occupied by attached bacteria and bead surface.
E. coli on the PEI‐bead and positioning of the cantilever was observed with a NanoWizard® II AFM (JKP Instruments) including an Axio Observer D.1 fluorescence microscope (Carl Zeiss Microscopy GmbH, Germany). A V‐shaped tipless cantilever (PNP‐TR‐TL‐Au (200 μm), NanoWorld AG, Switzerland) was assembled in the AFM. The cantilever´s spring constant and the resonant frequency were calibrated by the thermal noise method according to the user`s manual 31.
To glue an E. coli‐PEI‐bead on the cantilever, UHU Plus Schnellfest (UHU GmbH & Co. KG; Germany) with a curing time of 5 min was applied.
A thin layer of glue was spread on one third of a coverslip (24 mm x 24 mm) in a petri dish (diameter 40 mm; VWR International GmbH, Germany) and positioned under the AFM head. The cantilever apex was dipped for approx. 5 s in the glue and pulled sideways towards the clean area of the coverslip to remove surplus glue.
At the bottom of another petri dish a coverslip (24 mm x 24 mm) was fixed with UHU Plus Schnellfest and the microstructured and reference surfaces > 5 mm2 were glued on top, 3 mL PBS were added, and the assembly was placed under the AFM head. 10 μL of E. coli‐PEI‐beads were added at fixed distances away from the surface. Under the Axio Observer D.1 microscope one of the sedimented beads with a suitable monolayer of bacteria was chosen and the cantilever apex was moved towards it and contact was applied with a force of 10 nN for 30 s. All following steps were performed without removing the cantilever from the liquid.
The cantilever with the attached E. coli‐PEI‐bead was transferred to the substrate with an approach and retraction speed of 10 μm/s. Force‐distance curves were recorded using a loading force () of 10 nN and a contact time () of 10 s, on at least 3 randomly chosen areas, each with 3 × 3 measurement points 33.3 μm apart. After every batch of measurements, the status of attached cells was checked assessing GFP‐fluorescence.
2.4. SCFS measurements with PEI‐beads carrying a single E. coli
A published protocol for cell probe preparation 28 was altered as follows: 5 μm silica beads PSi‐5.0NH2 (Kisker Biotech GmbH, Germany) were fixed on the cantilever using a UV‐curable glue (Dymax Light Weld® 429; Dymax Europe GmbH, Germany), and the attached beads were coated in a drop of PEI‐ solution, rinsed in PBS, and stored under dry conditions. E. coli were used instead of Lactobacillus plantarum, and the measurements were done at 10 μm/s approach and retraction speed, with = 2 nN applied and = 2 s.
3. Results and discussion
In the following, we present measurements aimed at showing proof of principle for the cell probe preparation protocol described here, by comparing detachment forces required to detach the bacteria(um) presented by either probe type from a given surface, either structured or planar, as described below. For both types of preparation, these detachment forces were measured using SCFS. In SCFS, a laser beam reflected from the upper side of the cantilever facilitates the transduction of mechanical cantilever bending into an electrical signal. Deflection of the cantilever occurs due to attractive or repulsive forces between the surface and a cell and is a measure for forces in the nano‐Newton range.
3.1. Optimization of cell probe preparation
In Table1, we present a comparison of the advantages and disadvantages between different methods that have been described in the literature for measuring adhesion, primarily of bacteria, by SCFS using cantilevers of various configurations.
Table 1.
Cantilever types used in SCFS studies of bacterial adhesion
| Cantilever | Advantages | Disadvantages | References |
|---|---|---|---|
| Multiple bacteria | |||
| Tipless |
|
|
35 |
| Plateau tip |
|
|
25, 36 |
| Single bacteria | |||
| Tipless |
|
|
36 |
| Plateau tip |
|
|
37 |
| Pyramidal tip (spherical or sharp) |
|
|
[26, 27, 28, 38] |
| tipless, with attached single bead which is then loaded with one cell |
|
For this study, we chose to work with a setup where the bead is attached to the tipless cantilever, in part because this allowed us to turn to established protocols for guidance. In our experiment, however, in contrast to the existing protocols, we used beads loaded with multiple bacteria instead of single bacteria (for details see below).
Different approaches for the immobilization of cells on a cantilever have been described 25, 26, 27, 28. In contrast to receptor‐ligand interactions 23, cells can be immobilized via a wide range of nonspecific adhesion mechanisms. Cationic adhesives constituting amine groups such as polylysine 27, 29, 32 or imines in PEI 25, 26 mediate electrostatic interactions with negatively charged cell membranes.
Prefabricated amino‐functionalized silica beads were found to produce insufficient binding efficiency to obtain a monolayer of bacteria (data not shown). To overcome this, beads were coated with polymeric cationic PEI (Fig. 1).
Figure 1.
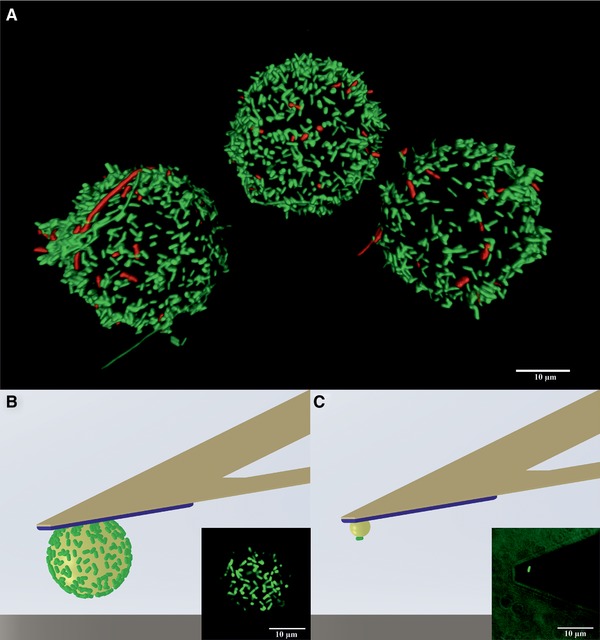
(A) 3D fluorescence image of E. coli‐PEI‐beads (Ø 20 μm) stored for 6 h in PBS. Living cells appear green, dead cells appear red.
(B and C) Schematic illustrations and corresponding fluorescence images of cell probes: (B) with a monolayer of E. coli, bead diameter 20 μm; (C) with a single E. coli, bead diameter 5 μm (according to 28).
The 3D image shown in Fig. 1A shows a typical distribution of E. coli obtained on PEI‐beads. On average, 26 ± 8% of the surface was covered by well‐separated bacteria, enabling SCFS‐measurements. Propidium iodide staining showed that after five to 6 h of storage, 93 ± 3% of the bacteria on the PEI‐beads were alive, while this had decreased to 75 ± 11% 2 h later (data not shown).
In accordance with Ong et al. 25, as well as similar data on Lactobacillus lactis 26, we conclude that PEI provides an effective bead coating, in that it supports bacterial adherence as well as survival.
To fix an E. coli‐PEI‐bead on the cantilever (Fig. 1B) a 2‐component epoxy glue was preferred 27. We chose UHU Plus Schnellfest because it cures consistently, can be applied in liquid, leaching of solvents is negligible, and it has no intrinsic fluorescence that would interfere with the viability assay. However, due to its working time of 5 min, a well‐structured experimental plan is required.
In comparison to an earlier protocol 28 (Table 1 and Fig. 1C), our approach presents multiple advantages. Among these are that we coat the PEI bead with multiple bacteria before attachment to the cantilever. This avoids the necessity of picking cells from a glass surface in liquid, which is made difficult by E. coli´s moti‐lity and lack of sedimentation. Additionally, the movement of the cantilever towards the bacteria can cause an unintended flow of the liquid phase and displace cells from the glass surface 28. Finally, due to the small size of bacteria, an AFM operator requires extensive training to be able to pick up a single bacterium, and if the bacterium is not fixed in the right position the cell probe has to be rejected 28.
3.2. Principle of SCFS measurements
Typically loading forces () applied in experiments with living cells are in a range of 0.1 to 30 nN depending on whether scanning or contact AFM mode is used, as well as on the cell type 31. We found that = 10 nN and = 10 s were sufficient to establish contact to the surface without damaging the E. coli.
In the force‐distance curves (Fig. 2) the blue line indicates the approach of the cell probe toward the surface. Where it comes into close proximity with the surface, a sudden increase in measured force is seen. After reaches a maximum of 10 nN (not shown on the curve) and has passed, retraction starts, during which time the detachment force () is recorded (red line). The difference between the minimum and the base line corresponds to the force required to separate the cell probe from the surface. In SCFS it is assumed that this force is equivalent to the adhesion force () 28, 29, 32, 33.
Figure 2.
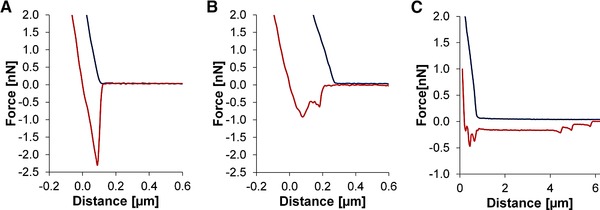
Force‐distance curves of (blue) and (red). (A) Sudden rupture from a structured surface. (B) Stepwise detachment from an unstructured surface. (C) Stepwise detachment with a rupture length of several micrometers indicating a measurement failure.
Several types of interactions were observed from the force‐distance curves: (i) sudden (Fig. 2A) or (ii) stepwise (Fig. 2B) bond ruptures. We speculate that the latter could be caused by membrane proteins on the bacterial surface, which can vary in elasticity and therefore influence the detachment behavior 31, 34. Rupture length is another parameter used to describe detachment behavior, where bond ruptures with multiple steps generally exhibit longer rupture lengths compared to sudden bond ruptures.
A high number of small rupture events in combination with an extended rupture length of several micrometers are an indicator of measurement failure 28, in this case (Fig. 2C) due to detachment of the surface or a defective cell probe.
3.3. Bacterial adhesion in micrometer and nanometer scale
In previous experiments, which were also carried out on cicada wing‐mimicking aluminum oxide surfaces under continuous liquid flow conditions, we saw a reduction in the surface area covered by E. coli on the wing‐like surface of up to 87% compared to a control reference surface 20. Our current experiments reveal an explanation for this drastic reduction in bacterial coverage, in that the structured surface affords a much smaller area for potential attachment, totaling only 28% of that present in the non‐structured, planar surface (compare Fig. 3B with 3A). The difference in available surface area is in accordance with the reduction in bacterial attachment observed earlier.
Figure 3.
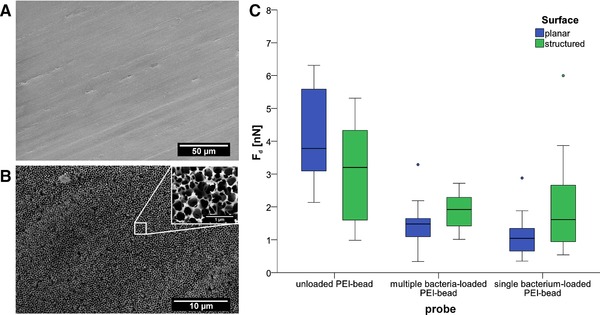
Scanning electron microscopy images of (A) planar and (B) structured aluminum oxide surfaces. (C) Box–whisker plots of on the two surfaces. Left: unloaded cell probe (n = 7); middle: cell probe with a multiple E. coli on a bead (diameter 20 μm) (n = 25); right: cell probe with a single E. coli on a bead (diameter 5 μm) (according to 28) (n = 18).
In order to measure the attachment force of bacteria to the structured and unstructured surfaces, SCFS data were collected on bacterial detachment trials using either unloaded PEI‐beads as a control (blank), or PEI‐beads prepared in the traditional way such that a single bacterium is attached, or PEI beads prepared with our new method where the bead carries multiple E. coli. These preparations are compared in Fig. 3C, showing box–whisker plots of their respective ' values.
The unloaded PEI‐bead control showed the highest median value (Fig. 3C, left), indicating a strong interaction of the PEI with the surface, compared to beads that were loaded with bacteria using either the previous or the current method. Moreover, the unloaded PEI‐bead displayed a stronger on the planar surface than on the structured surface, which has a much smaller potential contact area than the planar surface (3.8 nN on the planar versus 3.2 nN on the structured surface).
We went on to compare the binding strengths of the different cell probes to both surfaces, using either the established method with single bacterium, or the multiple bacteria‐loaded beads described here.
Surprisingly, although the area covered by bacteria is less in total on the structured surface (as found previously), we found that the adhesion of these bacteria to the surface using either kind of cell probe, reflected by the , is stronger than that on the smooth surface (a multiple bacteria‐loaded bead resulted in 1.8 nN for the structured surface versus 1.5 nN for the planar surface (Fig. 3C, middle)). The current PEI bead‐attachment protocol yielding single bacteria gave values of 1.6 nN compared to 1.0 nN (Fig. 3C, right).
We can exclude the possibility that the measurements might have derived from adhesion between naked areas of PEI and the surface, because of the lower found between both cell probe types and the surfaces, compared to the naked PEI bead control reference. If naked PEI had had an influence on the adhesion in the cell loaded cases, the measurements would have been expected to be higher.
The relatively large errors in the measurements indicate that more data are needed (> 50 measurements per surface point) in order to be able to draw conclusions about the influence of surface structure on and therefore on , with confidence.
Additionally, it should be noted that each measurement comparing the differently prepared cell probes on the structured surface was carried out by necessity on a different area of a relatively heterogeneous surface. This heterogeneity, which can be clearly seen in Fig. 3B on both the micrometer and the nanometer length scale, comes about during the etching process, in which pores are widened, yielding a somewhat irregular distribution of peaks and holes. This irregularity could lead to a comparatively large variability in individual measurements, compared to measurements done on the planar surface (blue bars vs. green bars in Fig. 3) giving a larger spread in the data.
In summary, we have devised an improved method of probe preparation for the measurement of bacterial adhesion to surfaces by SCFS, especially of interest for biofilm‐resistance applications. We find that, although biofilm coverage is hindered by the wing‐like structured surface, the actual binding force of bacteria, measured either with the traditional PEI‐bacteria coating method, or our new multiple bacteria‐loaded PEI probes, was preliminarily found to be stronger on the wing‐like surface.
Nevertheless adhesion force of bacteria is evidently not the only important factor, in determining the biofilm preventing qualities of a surface material. In designing such materials, it will be helpful to obtain more detailed information on the mechanisms of adhesion involved, and in this regard the SCFS measurements with the cell probes described here will be the subject of further study.
4. Concluding remarks
Understanding the influence of surface structures on bacterial adhesion is becoming increasingly important, both to acquire fundamental data concerning industrial biofilm formation and to facilitate the development of surfaces with anti‐adhesive, easy‐to‐clean properties. In this regard, SCFS is an effective tool to study cell‐to‐surface interactions in the nanoscale range. Here, we demonstrated how a PEI‐bead can be first loaded with multiple E. coli and attached to a cantilever, using an easy and reproducible method, and we have optimized the protocol for its application showing proof of principle. Importantly, the adhesion forces measured with this new method are comparable to those obtained with the previous existing protocol. It should be noted, however, that adhesion forces here are derived from detachment forces measured after pressing bacteria to the surface and lifting them off, rather than from attachment forces that must be playing a role in determining initial bacterial interaction with a given surface. Therefore, in future it will be important to additionally focus on these attachment forces active during the approach phase of bacterial adhesion. The results moreover reveal interesting aspects of bacterial attachment to the structured surface.
Practical application
Despite their beneficial uses in biotechnology, biofilms are generally considered deleterious in various branches of industrial technology, such as pharmaceutics and food processing. To combat this, major efforts are underway to develop anti‐adhesive, easy‐to‐clean surfaces and coatings for industrial components that are susceptible to biofilm growth.
Examples from nature are now serving as an emerging source of inspiration for solutions to the challenge of preventing biofilm initiation and thus their inhibition. In addition to macroscopic and microscopic features of biofilms, an important aspect of our understanding of their formation must come from nanoscale observations of microbial behavior in the context of surface structure. Single‐cell force spectroscopy is an effective tool for the elucidation of such questions.
The authors have declared no conflict of interest.
Nomenclature
|
|
[nN] | detachment force | |
|
|
[nN] | applied loading force | |
|
|
[nN] | adhesion force | |
|
|
[s] | contact time |
Acknowledgments
We acknowledge the valuable help in CLSM provided by Dr. Ruth Hans and Dr. Hella Hartmann of the Light Microscopy Facility at the Biotechnology Center of the TU Dresden. This work was financially supported by the Central Innovation Program for SMEs (ZIM) in the context of the government‐funded project KF2049818.
5 References
- 1. Donlan, R. M. , Biofilms: microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Halan, B. , Buehler, K. , Schmid, A. , Biofilms as living catalysts in continuous chemical syntheses. Trends Biotechnol. 2012, 30, 453–465. [DOI] [PubMed] [Google Scholar]
- 3. Govender, S. , Pillay, V. L. , Odhav, B. , Nutrient manipulation as a basis for enzyme production in a gradostat bioreactor. Enzyme Microb. Technol. 2010, 46, 603–609. [Google Scholar]
- 4. Gross, R. , Schmid, A. , Buehler, K. , Catalytic biofilms: a powerful concept for future bioprocesses, in: Lear G., Lewis G. (Eds.), Microbial Biofilm: Current Research and Applications, Caister Academic Press, Norfolk, UK: 2012, pp. 193–222. [Google Scholar]
- 5. James, G. A. , Swogger, E. , Wolcott, R. , Pulcini, E. d. et al., Biofilms in chronic wounds. Wound Repair Regen. 2008, 16, 37–44. [DOI] [PubMed] [Google Scholar]
- 6. Beech, I. B. , Sunner, J. A. , Arciola, C. R. , Cristiani, P. , Microbially‐influenced corrosion: damage to prostheses, delight for bacteria. Int. J. Artif. Organs 2006, 29, 443–452. [DOI] [PubMed] [Google Scholar]
- 7. Van Houdt, R. , Michiels, C. W. , Role of bacterial cell surface structures in Escherichia coli biofilm formation. Res. Microbiol. 2005, 156, 626–633. [DOI] [PubMed] [Google Scholar]
- 8. Srey, S. , Jahid, I. K. , Ha, S.‐D. , Biofilm formation in food industries: a food safety concern. Food Control 2013, 31, 572–585. [Google Scholar]
- 9. Chamberland, J. , Lessard, M. H. , Doyen, A. , Labrie, S. et al., Biofouling of ultrafiltration membrane by dairy fluids: characterization of pioneer colonizer bacteria using a DNA metabarcoding approach. J. Dairy Sci. 2017, 100, 981–990. [DOI] [PubMed] [Google Scholar]
- 10. Truong, V. K. , Lapovok, R. , Estrin, Y. S. , Rundell, S. et al., The influence of nano‐scale surface roughness on bacterial adhesion to ultrafine‐grained titanium. Biomaterials 2010, 31, 3674–3683. [DOI] [PubMed] [Google Scholar]
- 11. Priha, O. , Raulio, M. , Cooke, K. , Fisher, L. et al., Microbial populations on brewery filling hall surfaces ‐ Progress towards functional coatings. Food Control 2015, 55, 1–11. [Google Scholar]
- 12. Banerjee, I. , Pangule, R. C. , Kane, R. S. , Antifouling coatings: recent developments in the design of surfaces that prevent fouling by proteins, bacteria, and marine organisms. Adv. Mater. 2011, 23, 690–718. [DOI] [PubMed] [Google Scholar]
- 13. Bhushan, B. , Jung, Y. C. , Niemietz, A. , Koch, K. , Lotus‐like biomimetic hierarchical structures developed by the self‐assembly of tubular plant waxes. Langmuir 2009, 25, 1659–1666. [DOI] [PubMed] [Google Scholar]
- 14. Bhushan, B. , Jung, Y. C. , Natural and biomimetic artificial surfaces for superhydrophobicity, self‐cleaning, low adhesion, and drag reduction. Prog. Mater. Sci. 2011, 56, 1–108. [Google Scholar]
- 15. Su, Y. W. , Ji, B. H. , Huang, Y. , Hwang, K. C. , Nature's design of hierarchical superhydrophobic surfaces of a water strider for low adhesion and low‐energy dissipation. Langmuir 2010, 26, 18926–18937. [DOI] [PubMed] [Google Scholar]
- 16. Helbig, R. , Nickerl, J. , Neinhuis, C. , Werner, C. , Smart skin patterns protect springtails. PLoS One 2011, 6, e25105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nickerl, J. , Helbig, R. , Schulz, H. J. , Werner, C. et al., Diversity and potential correlations to the function of Collembola cuticle structures. Zoomorphology 2013, 132, 183–195. [Google Scholar]
- 18. Fang, Y. , Sun, G. , Wang, T. Q. , Cong, Q. et al., Hydrophobicity mechanism of non‐smooth pattern on surface of butterfly wing. Chin. Sci. Bull. 2007, 52, 711–716. [Google Scholar]
- 19. Pogodin, S. , Hasan, J. , Baulin, V. , Webb, H. et al., Biophysical model of bacterial cell interactions with nanopatterned cicada wing surfaces. Biophys. J. 2013, 104, 835–840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Mulansky, S. , Goering, P. , Ruhnow, M. , Lenk, F. et al., A modular flow cell system for studying biomimetic and bioinspired anti‐adhesive and antimicrobial surfaces. Heat Transfer Eng. 2017, 38, 805–817. [Google Scholar]
- 21. Wagner, K. , Friedrich, S. , Stang, C. , Bley, T. et al., Initial phases of microbial biofilm formation on opaque, innovative anti‐adhesive surfaces using a modular microfluidic system. Eng. Life Sci. 2013, 14, 76–84. [Google Scholar]
- 22. Mulansky, S. , Boschke, E. , Bley, T. , Innovative flow chamber for fluorescence microscopic analysis of biofilm formation, in Sensors and Measuring Systems 2014; 17. ITG/GMA Symposium, Nuremberg, Germany: 2014, pp. 1–6. [Google Scholar]
- 23. Helenius, J. , Heisenberg, C. P. , Gaub, H. E. , Muller, D. J. , Single‐cell force spectroscopy. J. Cell Sci. 2008, 121, 1785–1791. [DOI] [PubMed] [Google Scholar]
- 24. Benoit, M. , Gabriel, D. , Gerisch, G. , Gaub, H. E. , Discrete interactions in cell adhesion measured by single‐molecule force spectroscopy. Nat. Cell Biol. 2000, 2, 313–317. [DOI] [PubMed] [Google Scholar]
- 25. Ong, Y. L. , Razatos, A. , Georgiou, G. , Sharma, M. M. , Adhesion forces between E‐coli bacteria and biomaterial surfaces. Langmuir 1999, 15, 2719–2725. [Google Scholar]
- 26. Doan, T. L. L. , Guerardel, Y. , Loubiere, P. , Mercier‐Bonin, M. et al., Measuring kinetic dissociation/association constants between lactococcus lactis bacteria and mucins using living cell probes. Biophys. J. 2011, 101, 2843–2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Pen, Y. , Zhang, Z. J. , Morales‐García, A. L. , Mears, M. et al., Effect of extracellular polymeric substances on the mechanical properties of Rhodococcus. Biochim. Biophys. Acta, Biomembr. 2015, 1848, 518–526. [DOI] [PubMed] [Google Scholar]
- 28. Beaussart, A. , El‐Kirat‐Chatel, S. , Sullan, R. M. A. , Alsteens, D. et al., Quantifying the forces guiding microbial cell adhesion using single‐cell force spectroscopy. Nat. Protoc. 2014, 9, 1049–1055. [DOI] [PubMed] [Google Scholar]
- 29. Chen, Y. , Harapanahalli, A. K. , Busscher, H. J. , Norde, W. et al., Nanoscale cell wall deformation impacts long‐range bacterial adhesion forces on surfaces. Appl. Environ. Microbiol. 2014, 80, 637–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reisner, A. , Haagensen, J. A. J. , Schembri, M. A. , Zechner, E. L. et al., Development and maturation of Escherichia coli K‐12 biofilms. Mol. Microbiol. 2003, 48, 933–946. [DOI] [PubMed] [Google Scholar]
- 31. Nanowizard AFM Handbook version 4.3 ‐ 02/2016, JPK instruments AG 2016.
- 32. Younes, J. A. , van der Mei, H. C. , van den Heuvel, E. , Busscher, H. J. et al., Adhesion forces and coaggregation between vaginal Staphylococci and Lactobacilli . PloS One 2012, 7, 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Friedrichs, J. , Helenius, J. , Muller, D. J. , Quantifying cellular adhesion to extracellular matrix components by single‐cell force spectroscopy. Nat. Protoc. 2010, 5, 1353–1361. [DOI] [PubMed] [Google Scholar]
- 34. Muller, D. J. , Helenius, J. , Alsteens, D. , Dufrene, Y. F. , Force probing surfaces of living cells to molecular resolution. Nat. Chem. Biol. 2009, 5, 383–390. [DOI] [PubMed] [Google Scholar]
- 35. Alsteens, D. , Beaussart, A. , Derclaye, S. , El‐Kirat‐Chatel, S. et al., Single‐cell force spectroscopy of Als‐mediated fungal adhesion. Anal. Methods 2013, 5, 3657–3662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Muller, C. , Ziegler, C. , The scanning force microscope in bacterial cell investigations. Phys. Status Solidi A‐Appl. Mat. 2013, 210, 846–852. [Google Scholar]
- 37. Loskill, P. , "Unraveling the impact of subsurface and surface properties of a material on biological adhesion ‐ a multi‐scale approach," PhD, Naturwissenschaftlich‐Technische Fakultät II ‐ Physik und Mechatronik ‐, Universität des Saarlandes, Saarbrücken, 2012.
- 38. Lower, S. K. , Hochella, M. F. , Beveridge, T. J. , Bacterial recognition of mineral surfaces: nanoscale interactions between Shewanella and α‐FeOOH. Science 2001, 292, 1360–1363. [DOI] [PubMed] [Google Scholar]


