Abstract
Acute myocardial infarction (AMI) is one of the leading causes of death throughout the world. Usual methods for detecting AMI are expensive, time‐consuming and using blood samples as biological samples. Therefore, creating an ultra‐fast, sensitive and non‐invasive diagnostic test is necessary. Herein, a novel ultra‐sensitive, fluorescent, plasmon‐exciton coupling hybrid of a gold nanoparticle‐quantum dot (PQ)‐based aptamer nanobiosensor is presented for the detection of human cardiac troponin I (cTnI), the golden biomarker of AMI, and a preclinical test is performed within saliva. The binding of the cTnI protein to aptamer leads to a fluorescence enhancement of the plexcitonic hybrid system. The response range of this nanobiosensor is 0.4–2500 fM and the limit of detection is 0.3 fM. It seems that this novel design of nanobiosensor in the form of the PQ plexcitonic hybrid system can presents new opportunities for nanobiosensor progress.
Keywords: Aptamer, Gold nanoparticle, Human cardiac troponin I, Nanobiosensor, Quantum dot
Abbreviations
- GNP
gold nanoparticle
- hcTnI
human cardiac troponin I
- PQ
gold nanoparticle‐quantum dot
- QD
quantum dot
1. Introduction
Acute myocardial infarction (AMI) is one of the leading causes of death and a major health problem that affects human health in many countries (WHO). In ischemic heart diseases, blood vessels that supply nutrients and oxygen to the heart muscles (coronary arteries) narrow gradually with atherosclerosis plaques. If the plaque is cracked or ruptured, a blood clot can form on the surface of the plaque, and this clot can completely block the blood flow. Once the flow in the artery is obstacled, AMI will occur. As a result of insufficient blood supply to cardiomyocytes, necrosis of heart muscle cells occurs. In myocardial infarction, the heart pumping function may fail or abnormal heart rhythm may occur. Both of these sequelae can be physiologically devastating and ending in sudden death or major disability of the patients if not diagnosed or treated timely. The most beneficial therapeutic results are seen in the patients who undergone revascularization therapies in the golden period after AMI occurrence. Therefore, early diagnosis and performing early revascularization therapies is an important goal and result in significantly better prognosis and recovery of patients who suffer AMI 1, 2. As a result of necrosis of cardiomyocytes, intracellular macromolecules leak out through damaged cell membrane, these molecules include myoglobin, cardiac troponin T & I, creatine kinase, lactate dehydrogenase and many others 3, 4.
Several biomarkers have been used for early diagnose of AMI 5. Human cardiac troponin I (hcTnI) is recognized as the marker of choice and the gold standard for the diagnosis of AMI 6. As it is solely expressed in cardiomyocytes compared with cardiac troponin T (cTnT) and troponin C (TnC), which are also expressed in skeletal and cardiac muscles 7, 8, and other cardiac biomarkers, such as creatine kinase‐muscle b (CK‐MB), which is also present at low level in skeletal muscles cells 9. Common laboratory tests in emergency departments are measuring CK‐MB and cTnT as an AMI biomarker 10, 11, 12, and patient's blood is used as a biological sample 13; furthermore, common methods are expensive and time‐consuming, like electrochemiluminescence and ELISA 14. Available diagnostic methods are equivocal in about 50% of cases of AMI on arrival to the emergency department, resulted in postponing effective revascularization therapies 15, thus reaching a rapid, non‐invasive, inexpensive and accurate diagnostic assay for the detection of cTnI is an important goal.
Saliva is an exciting, appropriate choice among biological samples for detecting biomarkers, as almost always, sampling is convenient and fast 16, 17. Besides, 99.5% of saliva is water and the other 0.5% consists of enzymes, mucus, glycoprotein, etc. 18, 19. Therefore, saliva has a simpler environment in compare to blood samples due to lower protein diversity, thus it is a more suitable option for biosensing. Two studies have shown that in patients with AMI, cTnI is increased significantly in saliva 20, 21.
In the design of nanobiosensors, recognition elements are essential continent, as these elements bind to the target and lead to recognition 22. Aptamers are short, single‐strand DNA or RNA that bind to the targets with high affinity and specificity 23. Aptamers have many advantages over antibodies, as these molecules are smaller, easily producible via the systematic evolution of ligands by exponential enrichment (SELEX) 24 and can be applied for a wider range of the targets, even nonimmunogenic substances. In addition, they are inexpensive and easily re‐natured after denaturation without the loss of function, while the re‐naturation of antibodies is difficult due to the protein nature 25. As a result, in this study, we use cTnI specific aptamer as a recognition element of the nanobiosensor.
Recently, researchers have interested in applications of plexcitonic hybrid systems in nanobiosensors 26, 27, 28, 29, 30, 31, 32, nanoantennas 33, 34, 35, light emitting diodes (LEDs) 36, 37, 38 and solar cells 39, 40. A plexcitonic hybrid system is formed via the coupling of the localized surface plasmon resonance (LSPR) of metallic nanoparticles with the exciton of a semiconductor nanocrystal. These hybrid systems have many diverse and complex interactions 41, such as metal‐enhanced fluorescence (MEF) 42, 43, 44, 45, to quench the fluorescence of a semiconductor nanocrystal 46, 47, 48, 49. The energy transfer in these systems depends on the type, size 50, 51 and concentration of nanoparticles 51, as well as the dielectric constant of the surrounding environment and distance between them 51, 52.
Herein, we detected cTnI via the novel design of a plexcitonic hybrid system using gold nanoparticles (GNPs) as plasmonic nanoparticles and quantum dots (QDs) as an excitonic semiconductor nanocrystal. GNPs and QDs are bioconjugated with linker DNA, then self‐assembled via DNA hybridization with complementary linkers added to cTnI aptamer.
2. Materials and methods
2.1. Materials
HAuCl4, cetyltrimethylammonium bromide (CTAB), sodium citrate, dithiothreitol (DTT) and CdSeS/ZnS alloyed quantum dots COOH functionalized were purchased from Sigma‐Aldrich. In addition, 1‐ethyl‐3‐(3‐dimethylaminopropyl) carbodiimide (EDC), hydroxy‐2, 3‐dixopyrrolidine‐3‐sulfonic acid sodium salt (Sulfo‐NHS) was acquired from Santa Cruz Biotechnology. Specific single‐strand DNAs were ordered from FAZA Pajooh (FAZA Biotech) with high‐performance liquid chromatography purification, while cTnI was acquired from Life Diagnostics.
2.2. Design of single‐strand DNA sequences
The aptamer sequence against cTnI was selected from the Changill Ban 2012 US patent 53 and analyzed with the Mfold web server 54. Aptamer was synthesized with two 9 nt oligonucleotide sequences positioned at 5´and 3´terminus of the aptamer, and these terminus oligonucleotide are not complements to each other or to the aptamer. In addition, two 9‐oligonuelotide (L1, L2) complementary linkers with specific functionalized ends were designed. The sequences are shown in Table 1.
Table 1.
Sequence of aptamer and oligonucleotide linkers L1 and L2
| Name | Sequences (5′ → 3′) |
|---|---|
| Apt | ACCACCACCCGCATGCCAAACGTTGCCTCATAGTTCCCTCCCCGTGTCCTTCTTCTTC |
| L1 | GGTGGTGGT‐ C6 Amine |
| L2 | Thiol‐GAAGAAGAA |
2.3. Synthesis of 15 nm GNPs
GNP synthesis was performed via the reduction of HAuCl4 by sodium citrate according to modified Mirkin et al.’s method 55 by Azizi et al. 56. In the first step, three solutions were prepared; including: 250 μL of HAuCl4 (0.01 M); 1.5 mg of sodium citrate in 500 μL of deionized distilled water (DDW) and 3 mg of NaBH4 in 2 mL of DDW. Then after, the first solution was added to 9750 μL of DDW and then 250 μL of second solution and 75 μL of third solution was subjoined, respectively. The solution was stirred for 10 min. In the second step, 250 μL of HAuCl4 (0.01 M) was added in 9750 μL of DDW and 0.3 g of CTAB was dissolved. Then, 500 μL of prepared vitamin C solution (35.2 mg of Vitamin C in 2 mL of DDW) was added, ultimately 1 mL of the final solution of the first step was added and the ending solution was stirred for 10 min.
2.4. Preparation and purification of oligo‐GNP bioconjugate
According to Azizi et al.’s method 56, GNPs were bioconjugated with a 5′‐thiol terminated oligonucleotide linker 2 (L2) (Fig. 1A). In the first phase, 10 μL of L2 oligonucleotide (5 μM) added to 40 μL solution of HEPES (4‐(2‐Hydroxyethyl) piperazine‐1‐ethanesulfonic acid, N‐(2‐Hydroxyethyl) piperazine‐N‐(2‐ethanesulfonic acid) 50 mM, and 10 μL solution of DTT (1‐mg DTT in 20‐μL sodium acetate 0.01 M pH = 5.2) was subjoined and then, after 1 h, DTT solution was removed by ethyl acetate 50 μL. In this study, a reduction of disulfide bond is tested by DTNB (5,5′‐dithiobis‐(2‐nitrobenzoic acid) solution (0.004g DTNB /1‐mL PBS buffer). 10 μL of L2 solution was added to 40 μL of DTNB solution and as the color became dark yellow the test reduction of disulfide bond had occurred (Supporting Information Fig. S1).
Figure 1.
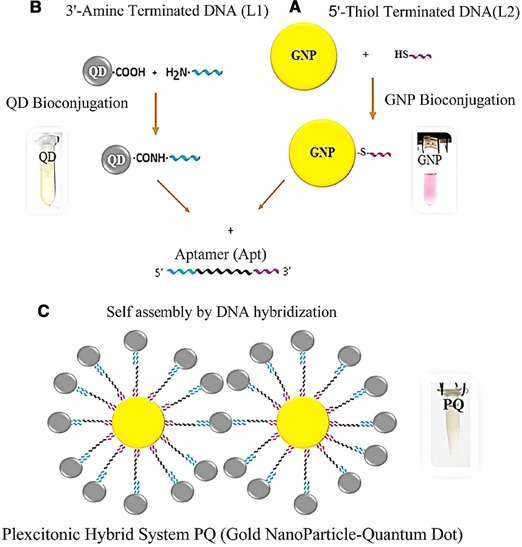
Schematic representation of the formation of the plexcitonic hybrid system. (A) Bioconjugation of GNPs. (B) Bioconjugation of QDs. (C) Self‐assembly of the plexcitonic hybrid system (GNP‐QD).
In the second phase, 15 μL of L2 oligonucleotide (1 μM) was added to 485 μL synthesized GNP solution with OD = 1 (0.158 mM) then 5‐μL of SDS 0.1% was adjoined and the solution was sonicated for 40 s. After 1 h, 12.5 μL of NaCl solution (1 M) was added and again sonication was done for 20 s. The process of adding NaCl was repeated two other times. At the end, 5 μL of SDS 0.1% was added. After 1 h, bioconjugated GNPs were purified from free‐thiolated oligonucleotide and centrifuged at 14 000 rpm for 10 min; then, the supernatant was discarded and the precipitate was dissolved in the 250 μL of PBS (1 M, pH = 8).
2.5. Preparation and purification of oligo‐QD bioconjugate
Quantum dots were bioconjugated with a 3'‐amine terminated oligonucleotide linker 1 (L1) via an EDC, Sulfo‐NHS crosslinking reaction through modified Wu et al. method 57 (Fig. 1B). For oligonucleotide‐QD bioconjugation, three solutions were prepared, including: 400 μL of QDs (5 mg/mL) in 1100 μL of DDW; 60‐mg EDC in 200 μL of PBS buffer (25 mM, pH = 6.5) and 30 mg of Sulfo‐NHS in 200 μL of PBS buffer, then, respectively, 500 μL of QDs solution was added to prepared EDC and Sulfo‐NHS solutions and it was sonicated for 20 s. After 1 h, 65 μL of L2 oligonucleotide (10 μM) was adjoined and under gone 2‐min sonication. Then, bioconjugated QDs were purified by centrifuge at 14 000 rpm for 10 min. The supernatant was discarded and the yellowish pellet was suspended in the 250‐μL PBS buffer (1 M, pH = 8).
2.6. Formation and purification of plexcitonic hybrid system
250 μL of prepared bioconjugated GNPs and 250 μL of bioconjugated QDs solutions were mixed; and subsequently, 25 μL of cTnI specific aptamer (apt) (5 μM) was added. The solution was placed in the thermoblock in 95°C for 10 min. Then, cooling was done to 25°C and the self‐assembly of the plexcitonic hybrid system (PQ) was achieved (Fig. 1C). For purification, the plexcitonic hybrid system was centrifuged at 14 000 rpm for 10 min and the supernatant is discarded. The brownish plate was dissolved in the 500 μL of PBS (1 M, pH = 8). Then after, it was sonicated for 2 min and kept at 4°C until use. For fluorescence spectroscopy measurements, the plexcitonic hybrid system was diluted (100 μL of PQ hybrid system solution in 200 μL of PBS buffer, repeated for three times).
2.7. Collection of saliva and sample and preparation
Saliva was collected with the method previously described by Mirzaii‐Dizgah 20 from a healthy donor. First, the mouth was rinsed with water, then after 5 min, without moving the tongue, or speaking, saliva was collected in 2‐mL micro‐tubes and kept in –80°C for 20 min. Subsequently, the saliva sample had undergone thawing for cleavage of mucoproteins. The prepared sample was centrifuged at 10 000 rcf for 20 min and ultimately 1 mL of supernatant was collected in micro tubes and kept in 4°C till use.
2.8. Preparation of proteins
Human cTnI has a molecular weight of 24 kDa. The Stock is 1.55 mg/mL solution (64.58 uM). The primary solution was diluted to get a concentration of 100 nM (1.548 μL in 998.452 μL of buffer [50 mM Tris‐HCL, 10 mM EDTA, pH = 8]). Then, subsequent dilutions can prepare from 1 nM stock (10 μL of 100 nM solution in 990 μL of buffer) and 1 pM stock (1 μL of 1 nM solution in 999 μL of buffer).
Lysozyme has a molecular weight of 14.3 kDa. The stock is 5 mg/mL (0.3495 mM). To prepare 100 nM solution, the stock was diluted (0.286 μL in 999.713 μL of PBS buffer) and other dilutions were prepared from 1 nM stock (10 μL of 100 nM solution in 990 μL of buffer) and 1 pM stock (1 μL of 1 nM solution in 999 μL of buffer).
2.9. Binding studies
Fluorescence and UV‐Vis spectroscopy studies of the plexcitonic hybrid system were performed in the saliva. Human cardiac troponin I was spiked to the saliva samples in determined concentrations. All sample volumes were 250 and 1 μL saliva was included in each sample, for example, in order to prepare of 0.4 fM cTnI solution, 0.1‐μL cTnI from 1000 fM solution, 1.9‐μL buffer and 1‐μL saliva was added to 247 μL of final prepared PQ hybrid solution and for preparation of 400 fM cTnI solution, 0.1‐μL cTnI from 1 nM solution, 1.9‐μL buffer and 1‐μL saliva was added to 247‐μL of final prepared PQ hybrid solution and other cTnI concentrations were prepared in this fashion. Studies were done in PBS buffer for evaluation of nanobiosensor photo stability response. Also, non‐specific binding of nanobiosensor is evaluated by adding determined concentration of lysozyme.
2.10. Transmission electron microscopy measurement
GNPs were sonicated for 15 min with a probe sonicator and the size of GNPs was measured by transmission electron microscopy (TEM) (TE 2000 Zeiss TEM). Characterization of the plexcitonic hybrid system also was performed by TEM imaging.
2.11. Dynamic light scattering measurement
GNPs were filtered with a 0.2‐μm filter. Determination of the dispersity of GNPs and estimation of the size of GNPs and bioconjugated GNPs was done by dynamic light scattering (DLS) analysis technique (Malvern, USA).
2.12. UV‐Vis spectroscopy measurement
GNPs and QDs before and after bioconjugation were characterized with UV‐Vis spectrophotometer (Agilent Technologies Cary spectrophotometer, USA) in UV‐Vis range.
2.13. Fluorescence spectroscopy measurement
Fluorescence spectroscopy was carried out on the single‐beam fluorimeter (Perkin Elmer, USA). Fluorescence study of the PQ plexcitonic hybrid system in comparison with bioconjugated QDs was done at λ 200–600 scan range, and the photostability behavior of 250 μL of the final prepared PQ plexcitonic hybrid system was assessed. In the next step, the fluorescence response of the plexcitonic hybrid system, in the presence of 1 μL of saliva in determined concentrations of cTnI (0.4–2500 fM) in λ ex220–λ em350 was evaluated. Then, the non‐specific binding of the sensor was investigated in the presence of 2500 fM cTnI and 2500 fM lysozyme (as the control) in the presence of saliva.
3. Results and discussion
3.1. Characterization analysis
The result of UV‐Vis spectroscopy indicates that the maxima of localized surface plasmon resonance absorption intensity of GNPs are near 525 nm (Fig. 2A). TEM imaging (Fig. 2B) represents that the GNP size seems to be near 15 nm. On the other hand, GNPs hydrodynamic size by DLS analysis is illustrated to be 42 nm. As, the DLS analysis measure hydrodynamic size of particles (particle core size along with any coating material and the solvent layer attached to the particle), so DLS hydrodynamic size measuring will be larger than core particle size estimation by TEM imaging 58. DLS study also exhibit that the polydispersity index of GNPs before and after bioconjugation are, respectively, 0.21 and 0.22, representing, though the particles are not monodisperse but, particle size dispersity are in the acceptable range (below 0.7) according to Malvern DLS user manual (Fig. 3A). DLS study also demonstrates that the hydrodynamic size of GNPs is incriminated after bioconjugation from 44.2 to 44.8 nm (Fig. 3B). The maxima of LSPR absorption intensity, decreased after bioconjugation of GNPs with 5′‐thiol‐terminated DNA (Fig. 3C). The reduction of GNPs LSPR, following bioconjugation is due to the sensitivity of LSPR of GNPs to trace changes in the refractive index of surrounding environment due to the bioconjugation 56, 59. In addition, in this study the LSPR of GNPs in presence of variable concentrations of linker DNA was assessed. The results demonstrate that following bioconjugation, the LSPR is decreased (Supporting Information Fig. S2).
Figure 2.
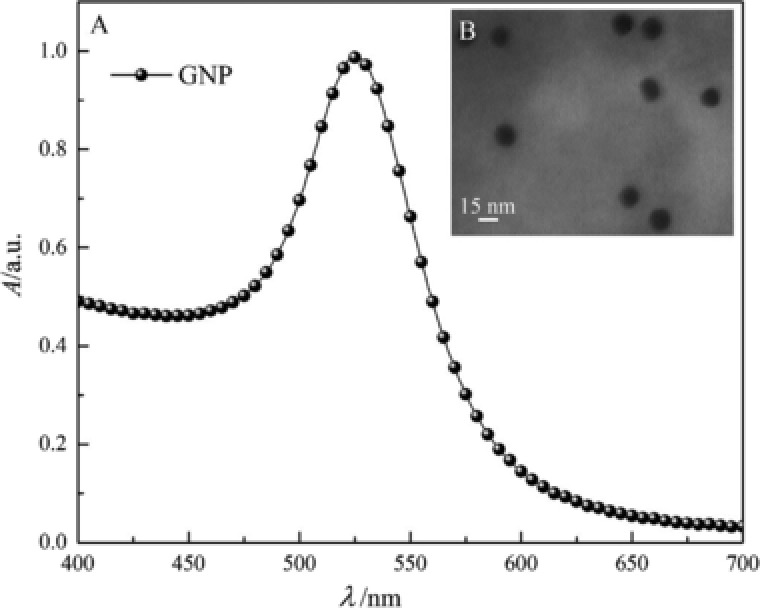
(A) UV‐Vis spectroscopy of synthesized GNPs. (B) TEM image of GNPs.
Figure 3.
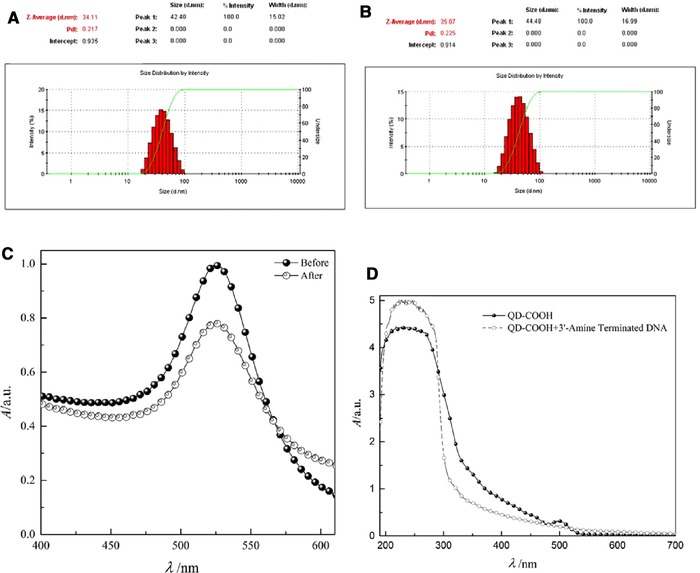
DLS analysis of GNPs. (A) Before and (B) after bioconjugation. (C) UV‐Vis spectroscopy of GNPs before and after bioconjugation with 5′‐thiol‐terminated DNA. (D) UV‐Vis spectroscopy of QDs before and after bioconjugation with 3′‐amine‐terminated DNA.
The UV‐Vis spectra of bioconjugated QDs in compare to nonconjugated particle, increment at λ 190–300 and decline at λ 300–500 (Fig. 3D). Following QDs bioconjugation with 3′‐amine‐terminated DNA, different HOMO‐LUMO levels of DNA nucleobases are coupled with excitonic states of QDs, cause an energy‐charge transfer that yield to alteration of the absorption spectrum 60.
The TEM image depicts the construction of the PQ plexcitonic hybrid system is formed. The image also displays that the QDs are seen as a hypodense thin shell surrounding GNPs (Fig. 4A).
Figure 4.
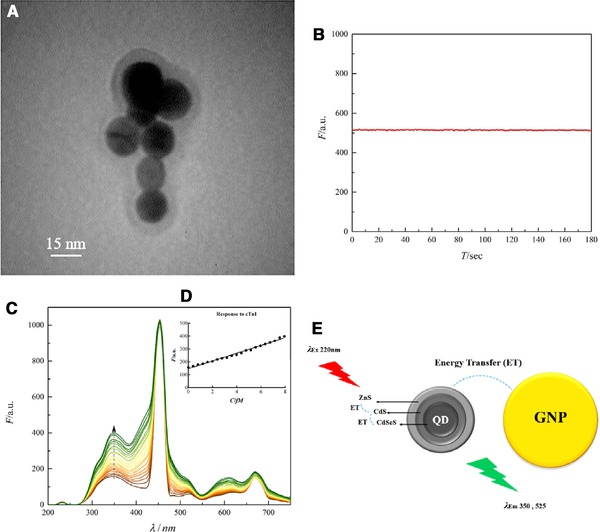
(A) TEM of plexcitonic hybrid system made by GNPs and quantum dots QDs. (B) Photo stability of plexcitonic hybrid system made by PQ. (C) Fluorescence study of plexcitonic hybrid system made by PQ in the different cTnI concentrations in saliva. (D) The response of the PQ sensor at 350 nm emission wavelength. (E) Schematic representation of energy transfer between different parts of the PQ plexcitonic hybrid system.
3.2. PQ hybrid system spectroscopy
Fluorescence scans of bioconjugated QD at λ 220–600 indicated that QDs are entirely tunable and could be excited at different excitation wavelengths (see the Supporting Information Figs. S3–S5). Fluorescence scans of the PQ hybrid system at λ 220–600 are also illustrated in the Supporting Information Figs. S6–S8. Fluorescence emission of the PQ hybrid system in comparison with bioconjugated QD showed the behavior of the system changed; these alterations in the form of the spectra are related to the formation of a QD‐GNP hybrid system, which is the LSPR of the GNP couples with the exciton of the QD, and the optics of QDs are significantly changed 26, 31. In addition, it leads the PQ hybrid system becomes strongly sensitive to small changes in the dielectric constant of the surrounding environment 26, making the PQ hybrid system a proper choice for biosensing purposes 41. As the PQ hybrid system exhibit higher fluorescence intensity at λ 220 (see the Supporting Information Fig. S6) excitation at λ 220 is selected for the fluorescence study.
3.3. Nanobiosensor photo stability
The PQ hybrid system response to cTnI is assessed through time. As the diagram represents (Fig. 4B), the sensor has a stable signal (no bleaching), after adding the cTnI and the intensity of fluorescence response remains in a steady state (plateau). Therefore, for the other measurements, the response is studied at 30 s after adding the biomarker.
3.4. Nano biosensor response in the presence of target protein
The fluorescence response of the PQ hybrid system in the presence of determined increasing concentration of cTnI in the saliva illustrates an increment of fluorescence intensity, along with increasing cTnI concentration that which obeyed a linear relation (Fig. 4C and D). This increment may be related to metal‐enhanced fluorescence (MEF) via the fluorescence energy transfer (FRET) 44, 45 mechanism (Fig. 4E). In this study, the QD consisted of three layers: the core is CdSeS, inner shell is CdS and the outer shell is ZnS and ZnCdS, where λ em350 and λ em525 are near the band gap energy of ZnS (5.8×10–19 J) and CdS (3.9×10–19 J), respectively (Table 2). In this novel design, an interesting phenomenon has happened, as the PQ hybrid system, can be excited at λ 220 and λ 340. Excitation at λ 220 leads to emissions at both λ 350 and λ 525, which overlaps with GNPs λ ex at λ 525. As a result of the overlaps between excitation and emission wavelengths of QDs and GNPs, the energy transfer between different layers of QDs and GNPs has occurred, which may cause MEF and the high sensitivity of the PQ hybrid system as cTnI binds to the target aptamer (Fig. 5A and B). According to FRET mechanism, energy transfer is proportional to distance (1/d6) thus it seems that following binding of the target protein to aptamer, probably a conformational change in the linker DNA, not the aptamer occurs (Supporting Information Fig. S9) and yields to reduction of the distance between GNPs and QDs which intensifies energy transfer and enhancement of fluorescence emission of the system. Also, it should be mentioned that circular dichroism (CD) study of the aptamer in the presence of different concentrations of cTnI illustrates no conformational change in the aptamer (Supporting Information Fig. S9). It seems that in regard to cTnI structure, two α‐helix act as a clamp which leads to shorten the distance between two linker oligonucleotides, thus yields to reduction of the distance between GNPs and QDs. The PQ hybrid system exhibits increment of fluorescence response in 0.4–2500 fM of cTnI concentration (Fig. 5C). In addition, the limit of detection is calculated to be 0.3 fM (Supporting Information calculation) and considering limit of detection of current cTnI biosensors, which sense in the fM range, this new biosensor possess the lowest limit of detection (Table 3, adapted from Fathil et al.’s review) 61.
Table 2.
Summary of the properties of elements of the PQ plexcitonic hybrid system
| QDs | PQ plexcitonic hybrid system | GNP | |||
|---|---|---|---|---|---|
| Band Gap Energy[J] | λ Band Gap[nm] | λem[nm] | λex[nm] | λ max ex[nm] | |
| ZnS | ∼5.8×10−19 | ∼344.400 | 320PBS,350Saliva | 220 | |
| CdS | ∼3.9×10−19 | ∼512.333 | 520PBS,525Saliva | 340 | 525 |
| CdSe | ∼2.7×10−19 | ∼716.670 | |||
Figure 5.
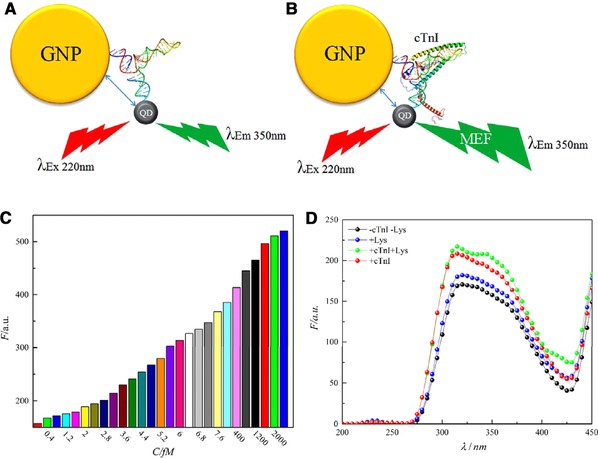
Schematic representation of the probable mechanism of enhancement of the PQ sensor (A) in the absence and (B) in the presence of cTnI. (C) The response range of the sensor. (D) The specificity of the sensor.
Table 3.
Comparison between cTnI biosensors (adapted from Fathil et al.'s review) 61
| Type of biosensor | Recognition element | Detection limit [fM] |
|---|---|---|
| Functionalized SnO2 nanobelt FETs | Ab | 83 330 |
| ELISA | Ab | 79 170 |
| Fiber‐optic‐based SPR sensor | Ab | 58 330 |
| Aptamer‐based biosensor | Aptamer | 49 740 |
| Optical‐based‐SPR | Ab | 10 420 |
| Fluoro‐microbead guiding chip (FMGC) | Ab | 4170 |
| SiNW FETs CMOS | Ab | 3830 |
| Chemiluminometric EOC | Ab | 1130 |
| PDMS‐GNP composite film | Ab | 416.67 |
| Mitsubishi PATHFAST® | Ab | 125 |
| Single PANI nanowire | Ab | 10.42 |
| Monolithic graphene sheet | Ab | ∼4.17 |
| PQ plexcitonic | Aptamer | 0.3 (this work) |
3.5. Nanobiosensor selectivity
The non‐specific binding of the sensor is also investigated in the presence of the cTnI biomarker and lysozyme as a control protein (Fig. 5D). Results indicated the sensor hasn't displayed non‐specific binding. As, the aptamer is highly specific for cTnI with interaction similar to antibody–antigen binding, the sensor demonstrates high affinity and selectivity for the target protein.
4. Concluding remarks
The study of Mirzaii‐Dizgah and Riahi illustrates that salivary troponin I is an indicator of myocardial infarction and the salivary cTnI concentration is correlated to blood concentration. This study also remarks that salivary concentration of cTnI is significantly higher in patients with AMI at both 12 and 24 h [16.67 (7.08–25.83) fM and 29.58 (21.67–44.58) fM, respectively] following the onset of myocardial infarction 20; thus, designed nanobiosensor in our study has an impactful limit of detection (0.3 fM) and range of response (0.4 to 2500 fM).
For further application of the sensor in clinical settings, more preclinical‐ and clinical‐based studies should be carried out, especially in determining cut off point of cTnI in the saliva upon severity of coronary artery disease proved by the current diagnostic method including: infarction (ST elevation MI and non ST elevation MI) vs. ischemia vs. transient coronary artery spasm; in different age groups, gender etc. As few studies have examined salivary cTnI in patients who suffer AMI, new studies also should be done on pattern of blood and salivary concentration of cTnI post AMI occurrence.
Altogether, this study introduces a novel design of the plexcitonic hybrid system based upon GNP‐QD and a highly specific aptamer for cTnI that is able to detect cTnI rapidly and in low concentration in saliva. This novel platform not only will open up new opportunities for the development of nanobiosensors with favorable characteristics such as low limit of detection, high accuracy and rapid responding; but also the clinical application of rapid, highly specific cTnI sensors will improve patient therapeutic management (application of revascularization therapies) and affect post AMI prognosis.
Practical application
Human cardiac troponin I is the gold standard biomarker for the detection of AMI. In this study, we designed a novel fluorescence, plasmon‐exciton coupling nanobiosensor aptamer‐based for ultrasensitive and fast detection of cTnI. This nanobiosensor showed selectivity toward cTnI with a limit of detection 0.3 fM for cTnI. The designed nanobiosensor was successfully used to detect cTnI in the saliva.
Nomenclature
| A | [a.u.] | Absorption intensity |
| C | [fM] | Concentration |
| F | [a.u.] | Fluorescence intensity |
| Τ | [Sec] | Time |
Greek symbols
| λ | [nm] | Wavelength |
4.1.
The authors would like to thank the Research Council of Tarbiat Modares University for financial support.
The authors have declared no conflicts of interest.
Supporting information
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Figure S1. DTNB Test
Figure S2. GNPs LSPR in the presence of increasing concentrations of linker DNA (0–80 nM). The results demonstrate that following bioconjugation, the LSPR is decreased.
Figure S3. Fluorescence study of the bioconjugated quantum dots at λexc220–250.
Figure S4. Fluorescence study of the bioconjugated quantum dots at λexc300–460.
Figure S5. Fluorescence study of the bioconjugated quantum dots at λexc480–600.
Figure S6. Fluorescence study of the PQ plexcitonic hybrid system at λexc220–250.
Figure S7. Fluorescence study of the PQ plexcitonic hybrid system at λexc300–460.
Figure S8. Fluorescence study of the PQ plexcitonic hybrid system at λexc480–600.
Figure S9. Circular Dichroism study of aptamer (5 μM) in the presence of different concentrations cTnI (fM)
Figure S10. TEM images of pure QDs
5 References
- 1. Ambrose, J. A. , Singh, M. , Pathophysiology of coronary artery disease leading to acute coronary syndromes. F1000Prime. Rep. 2015, 7, 8–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kumar, V. , Abbas, A. K. , Fausto, N. , Mitchell, R. , Robbins Basic Pathology, Elsevier Health Sciences, Philadelphia: 2007. [Google Scholar]
- 3. Singh, V. , Martinezclark, P. , Pascual, M. , Shaw, E. S. et al., Cardiac biomarkers ‐ the old and the new: A review. Coron. Artery Dis. 2010, 21, 244–256. [DOI] [PubMed] [Google Scholar]
- 4. Daubert, M. A. , Jeremias, A. , The utility of troponin measurement to detect myocardial infarction: Review of the current findings. Vasc. Health Risk Manage. 2010, 6, 691–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chan, D. , Ng, L. L. , Biomarkers in acute myocardial infarction. BMC Med. 2010, 8, 34–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hasić, S. , Kiseljaković, E. , Jadrić, R. , Radovanović, J. et al., Cardiac troponin I: The gold standard in acute myocardial infarction diagnosis. Bosnian. J. Basic Med. Sci. 2003, 3, 41–44. [DOI] [PubMed] [Google Scholar]
- 7. Sharma, S. , Jackson, P. G. , Makan, J. , Cardiac troponins. J. Clin. Pathol. 2004, 57, 1025–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Schreier, T. , Kedes, L. , Gahlmann, R. , Cloning, structural analysis, and expression of the human slow twitch skeletal muscle/cardiac troponin C gene. J. Biol. Chem. 1990, 265, 21247–21253. [PubMed] [Google Scholar]
- 9. Schwartz, J. G. , Prihoda, T. J. , Stuckey, J. H. , Gage, C. L. et al., Creatine kinase MB in cases of skeletal muscle trauma. Clin. Chem. 1988, 34, 898–901. [PubMed] [Google Scholar]
- 10. Kim, Y. , Kim, H. , Kim, S. Y. , Lee, H. K. et al., Automated heart‐type fatty acid‐binding protein assay for the early diagnosis of acute myocardial infarction. Am. J. Clin. Pathol. 2010, 134, 157–162. [DOI] [PubMed] [Google Scholar]
- 11. Budak, Y. U. , Huysal, K. , Bulut, M. , Polat, M. , Evaluation in an emergency department of rapid separator tubes containing thrombin for serum preparation prior to hs‐cTnT and CK‐MB analyses. BMC Clin. Pathol. 2013, 13, 20–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Collinson, P. O. , Gaze, D. C. , Morris, F. , Morris, B. et al., Comparison of biomarker strategies for rapid rule out of myocardial infarction in the emergency department using ACC/ESC diagnostic criteria. Ann. Clin. Biochem. 2006, 43, 273–280. [DOI] [PubMed] [Google Scholar]
- 13. Jaffe, A. S. , Babuin, L. , Apple, F. S. , Biomarkers in acute cardiac disease: The present and the future. J. Am. Coll. Cardiol. 2006, 48, 1–11. [DOI] [PubMed] [Google Scholar]
- 14. Qureshi, A. , Gurbuz, Y. , Niazi, J. H. , Biosensors for cardiac biomarkers detection: A review. Sensors Actuators B Chem. 2012, 171–172, 62–76. [Google Scholar]
- 15. Tataris, K. , Kivlehan, S. , Govindarajan, P. , National trends in the utilization of emergency medical services for acute myocardial infarction and stroke. West J. Emerg. Med. 2014, 15, 744–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Malon, R. S. P. , Sadir, S. , Balakrishnan, M. , Córcoles, E. P. , Saliva‐based biosensors: Noninvasive monitoring tool for clinical diagnostics. BioMed. Res. Int. 2014, 2014, 962903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wei, F. , Wong, D. T. W. , Point‐of‐care platforms for salivary diagnostics. Chin. J. Dent. Res. 2012, 15, 7–15. [PubMed] [Google Scholar]
- 18. Mirzaii‐Dizgah, I. , Riahi, E. , Salivary troponin I as an indicator of myocardial infarction. Indian J. Med. Res. 2013, 138, 861–865. [PMC free article] [PubMed] [Google Scholar]
- 19. Rahim, M. A. A. , Rahim, Z. H. A. , Ahmad, W. A. W. , Hashim, O. H. , Can saliva proteins be used to predict the onset of acute myocardial infarction among high‐risk patients? Int. J. Med. Sci. 2015, 12, 329–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Carpenter, G. H. , The secretion, components, and properties of saliva. Annu. Rev. Food Sci. Technol. 2013, 4, 267–276. [DOI] [PubMed] [Google Scholar]
- 21. de Almeida, P. D. V. , Grégio, A. M. T. , Machado, M. A. N. , de Lima, A. A. S. et al., Saliva composition and functions: A comprehensive review. J. Contemp. Dent. Pract. 2008, 9, 72–80. [PubMed] [Google Scholar]
- 22. Chambers, J. P. , Arulanandam, B. P. , Matta, L. L. , Weis, A. et al., Biosensor recognition elements. Curr. Issues Mol. Biol. 2008, 10, 1–12. [PubMed] [Google Scholar]
- 23. Mayer, G. , The chemical biology of aptamers. Angew. Chem. Int. Ed. 2009, 48, 2672–2689. [DOI] [PubMed] [Google Scholar]
- 24. Stoltenburg, R. , Reinemann, C. , Strehlitz, B. , SELEX–a (r)evolutionary method to generate high‐affinity nucleic acid ligands. Biomol. Eng. 2007, 24, 381–403. [DOI] [PubMed] [Google Scholar]
- 25. Ozer, A. , Pagano, J. M. , Lis, J. T. , New technologies provide quantum changes in the scale, speed, and success of SELEX methods and aptamer characterization. Mol. Ther. Nucleic Acids 2014, 3, e183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hatef, A. , Sadeghi, S. M. , Boulais, É. , Meunier, M. , Quantum dot‐metallic nanorod sensors via exciton‐plasmon interaction. Nanotechnology 2013, 24, 015502. [DOI] [PubMed] [Google Scholar]
- 27. Huang, D. , Niu, C. , Ruan, M. , Wang, X. et al., Highly sensitive strategy for Hg2+ detection in environmental water samples using long lifetime fluorescence quantum dots and gold nanoparticles. Environ. Sci. Technol. 2013, 47, 4392–4398. [DOI] [PubMed] [Google Scholar]
- 28. Choi, Y. , Cho, Y. , Kim, M. , Grailhe, R. et al., Fluorogenic quantum dot‐gold nanoparticle assembly for beta secretase inhibitor screening in live cell. Anal. Chem. 2012, 84, 8595–8601. [DOI] [PubMed] [Google Scholar]
- 29. Li, M. , Wang, Q. , Shi, X. , Hornak, L. A. et al., Detection of mercury(II) by quantum dot/DNA/gold nanoparticle ensemble based nanosensor via nanometal surface energy transfer. Anal. Chem. 2011, 83, 7061–7065. [DOI] [PubMed] [Google Scholar]
- 30. Liu, J. , Lee, J. H. , Lu, Y. , Quantum dot encoding of aptamer‐linked nanostructures for one‐pot simultaneous detection of multiple analytes. Anal. Chem. 2007, 79, 4120–4125. [DOI] [PubMed] [Google Scholar]
- 31. Lee, J. , Hernandez, P. , Lee, J. , Govorov, A. O. et al., Exciton‐plasmon interactions in molecular spring assemblies of nanowires and wavelength‐based protein detection. Nat. Mater. 2007, 6, 291–295. [DOI] [PubMed] [Google Scholar]
- 32. Oh, E. , Hong, M. Y. , Lee, D. , Nam, S. H. et al., Inhibition assay of biomolecules based on fluorescence resonance energy transfer (FRET) between quantum dots and gold nanoparticles. J. Am. Chem. Soc. 2005, 127, 3270–3271. [DOI] [PubMed] [Google Scholar]
- 33. Sadeghi, S. M. , Hatef, A. , Meunier, M. , Quantum detection and ranging using exciton‐plasmon coupling in coherent nanoantennas. Appl. Phys. Lett. 2013, 102, 203113. [Google Scholar]
- 34. Kinkhabwala, A. , Yu, Z. , Fan, S. , Avlasevich, Y. et al., Large single‐molecule fluorescence enhancements produced by a bowtie nanoantenna. Nat. Photonics 2009, 3, 654–657. [Google Scholar]
- 35. Gong, H. M. , Zhou, L. , Su, X. R. , Xiao, S. et al., Illuminating dark plasmons of silver nanoantenna rings to enhance exciton‐plasmon interactions. Adv. Funct. Mater. 2009, 19, 298–303. [Google Scholar]
- 36. Chen, J. , Huang, Q. , Du, Q. G. , Zhao, D. et al., Localized surface plasmon resonance enhanced quantum dot light‐emitting diodes via quantum dot‐capped gold nanoparticles. RSC Adv. 2014, 4, 57574–57579. [Google Scholar]
- 37. Gu, X. , Qiu, T. , Zhang, W. , Chu, P. K. , Light‐emitting diodes enhanced by localized surface plasmon resonance. Nanoscale Res. Lett. 2011, 6, 199–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Cho, C. Y. , Lee, S. J. , Song, J. H. , Hong, S. H. et al., Enhanced optical output power of green light‐emitting diodes by surface plasmon of gold nanoparticles. Appl. Phys. Lett. 2011, 98, 051106. [Google Scholar]
- 39. Pathak, N. K. , Ji, A. , Sharma, R. P. , Study of efficiency enhancement in layered geometry of excitonic‐plasmonic solar cell. Appl. Phys. A 2013, 115, 1445–1450. [Google Scholar]
- 40. Paz‐Soldan, D. , Lee, A. , Thon, S. M. , Adachi, M. M. et al., Jointly tuned plasmonic‐excitonic photovoltaics using nanoshells. Nano Lett. 2013, 13, 1502–1508. [DOI] [PubMed] [Google Scholar]
- 41. Hatef, A. , Sadeghi, S. M. , Fortin‐Deschênes, S. , Boulais, E. et al., Coherently‐enabled environmental control of optics and energy transfer pathways of hybrid quantum dot‐metallic nanoparticle systems. Opt. Express 2013, 21, 5643–5653. [DOI] [PubMed] [Google Scholar]
- 42. Bian, J. C. , Yang, F. , Li, Z. , Zeng, J. L. et al., Mechanisms in photoluminescence enhancement of ZnO nanorod arrays by the localized surface plasmons of Ag nanoparticles. Appl. Surf. Sci. 2012, 258, 8548–8551. [Google Scholar]
- 43. Jin, Y. , Gao, X. , Plasmonic fluorescent quantum dots. Nat. Nanotechnol. 2009, 4, 571–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Pompa, P. P. , Martiradonna, L. , Torre, A. D. , Sala, F. D. et al., Metal‐enhanced fluorescence of colloidal nanocrystals with nanoscale control. Nat. Nanotechnol. 2006, 1, 126–130. [DOI] [PubMed] [Google Scholar]
- 45. Song, J. H. , Atay, T. , Shi, S. , Urabe, H. et al., Large enhancement of fluorescence efficiency from CdSe/ZnS quantum dots induced by resonant coupling to spatially controlled surface plasmons. Nano Lett. 2005, 5, 1557–1561. [DOI] [PubMed] [Google Scholar]
- 46. Pons, T. , Medintz, I. L. , Sapsford, K. E. , Higashiya, S. et al., On the quenching of semiconductor quantum dot photoluminescence by proximal gold nanoparticles. Nano Lett. 2007, 7, 3157–3164. [DOI] [PubMed] [Google Scholar]
- 47. Nikoobakht, B. , Burda, C. , Braun, M. , Hun, M. et al., The quenching of CdSe quantum dots photoluminescence by gold nanoparticles in solution. Photochem. Photobiol. 2007, 75, 591–597. [DOI] [PubMed] [Google Scholar]
- 48. Dyadyusha, L. , Yin, H. , Jaiswal, S. , Brown, T. et al., Quenching of CdSe quantum dot emission, a new approach for biosensing. Chem. Commun. 2005, 3201–3203. [DOI] [PubMed] [Google Scholar]
- 49. Gueroui, Z. , Libchaber, A. , Single‐molecule measurements of gold‐quenched quantum dots. Phys. Rev. Lett. 2004, 93, 166108. [DOI] [PubMed] [Google Scholar]
- 50. Viste, P. , Plain, J. , Jaffiol, R. , Vial, A. et al., Enhancement and quenching regimes in metal‐semiconductor hybrid optical nanosources. ACS Nano 2010, 4, 759–764. [DOI] [PubMed] [Google Scholar]
- 51. Wang, Q. , Wang, H. , Lin, C. , Sharma, J. et al., Photonic interaction between quantum dots and gold nanoparticles in discrete nanostructures through DNA directed self‐assembly. Chem. Commun. 2010, 46, 240–242. [DOI] [PubMed] [Google Scholar]
- 52. Li, Y. Q. , Guan, L. Y. , Zhang, H. L. , Chen, J. et al., Distance‐dependent metal‐enhanced quantum dots fluorescence analysis in solution by capillary electrophoresis and its application to DNA detection. Anal. Chem. 2011, 83, 4103–4109. [DOI] [PubMed] [Google Scholar]
- 53. Ban, C. , Song, K. M. , Jeong, W. , DNA aptamer specifically binding to human cardiac troponin I. US Patent 8404448, 2012.
- 54. Zuker, M. , Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003, 31, 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Mirkin, C. A. , Letsinger, R. L. , Mucic, R. C. , Storhoff, J. J. , A DNA‐based method for rationally assembling nanoparticles into macroscopic materials. Nature 1996, 382, 607–609. [DOI] [PubMed] [Google Scholar]
- 56. Azizi, A. , Ranjbar, B. , Moghadam, T. T. , Bagheri, Z. , Plasmonic circular dichroism study of DNA–gold nanoparticles bioconjugates. Plasmonics 2013, 9, 273–281. [Google Scholar]
- 57. Wu, C. S. , Cupps, J. M. , Fan, X. , Compact quantum dot probes for rapid and sensitive DNA detection using highly efficient fluorescence resonant energy transfer. Nanotechnology 2009, 20, 305502. [DOI] [PubMed] [Google Scholar]
- 58. Lim, J. , Yeap, S. P. , Che, H. X. , Low, S. C. , Characterization of magnetic nanoparticle by dynamic light scattering. Nanoscale Res. Lett. 2013, 8, 381–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Anker, J. N. , Hall, W. P. , Lyandres, O. , Shah, N. C. et al., Biosensing with plasmonic nanosensors. Nat. Mater. 2008, 7, 442–453. [DOI] [PubMed] [Google Scholar]
- 60. Goodman, S. M. , Singh, V. , Ribot, J. C. , Chatterjee, A. et al., Multiple energy exciton shelves in quantum‐dot–DNA nanobioelectronics. J. Phys. Chem. Lett. 2014, 5, 3909–3913. [DOI] [PubMed] [Google Scholar]
- 61. Fathil, M. F. M. , Md Arshad, M. K. , Gopinath, S. C. B. , Hashim, U. et al., Diagnostics on acute myocardial infarction: Cardiac troponin biomarkers. Biosens. Bioelectron. 2015, 70, 209–220. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
As a service to our authors and readers, this journal provides supporting information supplied by the authors. Such materials are peer reviewed and may be re‐organized for online delivery, but are not copy‐edited or typeset. Technical support issues arising from supporting information (other than missing files) should be addressed to the authors.
Figure S1. DTNB Test
Figure S2. GNPs LSPR in the presence of increasing concentrations of linker DNA (0–80 nM). The results demonstrate that following bioconjugation, the LSPR is decreased.
Figure S3. Fluorescence study of the bioconjugated quantum dots at λexc220–250.
Figure S4. Fluorescence study of the bioconjugated quantum dots at λexc300–460.
Figure S5. Fluorescence study of the bioconjugated quantum dots at λexc480–600.
Figure S6. Fluorescence study of the PQ plexcitonic hybrid system at λexc220–250.
Figure S7. Fluorescence study of the PQ plexcitonic hybrid system at λexc300–460.
Figure S8. Fluorescence study of the PQ plexcitonic hybrid system at λexc480–600.
Figure S9. Circular Dichroism study of aptamer (5 μM) in the presence of different concentrations cTnI (fM)
Figure S10. TEM images of pure QDs


