Abstract
The oleaginous yeast Rhodosporidium toruloides AS 2.1389 is viewed as desirable industrial microorganisms that can accumulate a high content of lipids for biodiesel production. In this study, we attempted to improve lipid accumulation in the yeast Rhodosporidium toruloides by UV irradiation mutagenesis and selection based on lithium chloride tolerance or ethanol–H2O2 tolerance. The biomass concentration, lipid yield and glucose consumption of mutant R. toruloides were determined. The transcription levels of lipid accumulation‐related genes in the wild‐type and mutant strains were also determined. The lithium chloride‐tolerant strain R‐ZL2 and the ethanol–H2O2‐resistant strain R‐ZY13 were generated by UV mutagenesis. The two mutant strains showed greater lipid productivity and lipid yield compared to the wild type. Transcriptional analysis revealed that IDP1, GPD1 and GND were expressed at significantly higher levels in the two high‐lipid‐producing mutants. In conclusion, lipid productivity and lipid yield in R. toruloides were successfully improved via UV mutagenesis and selection. We also identified some lipid accumulation‐related genes for improving lipid productivity through genetic engineering.
Keywords: microbial lipid, oleaginous yeast, Rhodosporidium toruloides, UV mutagenesis
1. INTRODUCTION
Taking advantage of renewable resources for energy production has become a hot topic worldwide given the decreasing fossil fuel supply and growing environmental problems 1, 2. Biodiesel fuel as one of the most important renewable and clean energy alternatives to fossil fuels, has received wide attention 3. The first generation biodiesel fuel mainly produced from plant oils (e.g., rapeseed, sunflower, soybean, coconut, peanut, jatropha sp., and jojoba oil) 4, 5. However, the prices of plant oils are rapidly increasing as the biodiesel consumption increases worldwide 6. Moreover, the competition between the cultivation of oil crops and food crops for nutrients, water resources and arable land is also indeed worrisome 7. Therefore, alternative resources for biodiesel production, which are economically competitive, renewable, and do not compete with food crops, needs to be developed.
Utilization of microbial oils as alternative resources for biodiesel industries has received considerable attention as its chemical component is similar to plant oils used as feedstock for biodiesel 8. Some microorganisms (e.g., yeast, bacteria, and algae) can synthesize and accumulate intracellularly oils more than 20% of their biomass. These microbes are called oleaginous microorganisms, and the oils they produce are termed as microbial oils 9. Microbial oils for biodiesel production are advantageous over traditional plant oils that include non‐polluting, short life‐cycles, not occupy large amounts of arable land, and unaffected by environmental factors 8. However, the high cost of biodiesel production restricts its wide commercialization 10. Those costs mainly include expensive fermentation substrates and complex oil extraction processes (cell rupture, extraction, and phase separation) 11. Although the current price of biodiesel from microbial oils is higher than first generation biodiesel, it is expected to decrease as the technology matures 12, 13.
Oleaginous yeast is an important microbial oil producer, it belongs to heterotrophic microorganisms that needs to convert sugar‐based substrate to obtain energy for growth and oil accumulation 14, 15. Although it cannot accumulate oils through photosynthesis, it is feasible to decrease costs by using zero or negative value waste substrate as nutrient source 9, 16. In addition, oleaginous yeasts offer advantages over other oleaginous microorganisms, such as faster growth, higher oil accumulation, shorter cultivation cycle, stronger ability to use more carbon sources, and better ability to adapt environmental changes 9, 17. Therefore, use of oleaginous yeasts to produce more microbial oil is thus very important to promote the development of second generation biodiesel industry 18.
In order to produce microbial oil in a more effective way, much effort has been focused on optimizing the culture conditions of oleaginous yeasts, evaluating the effects of different cultivation modes on oil production by oleaginous yeasts, and investigating the metabolic mechanisms and physiological characteristics of oleaginous yeasts 19, 20, 21. In fact, random mutagenesis in oleaginous yeast by mutagens such as mutagenic agents and UV light may also be a promising strategy for improving lipid productivity. Zhang 22 reported that the yields of β‐galactosidases in Aspergillus oryzae were improved by UV mutagenesis and selection using LiCl. The arachidonic acid in Mortierella alpinahas has also been improved by UV mutagenesis and selection using LiCl 23. Yamada 1 reported that the lipid productivity in Rhodosporidium toruloides NBRC 8766 was improved by UV mutagenesis and selection using ethanol–H2O2. Therefore, random mutagenesis and selection could be a promising strategy for improving lipid production by some oleaginous microorganisms.
The aim of this study was to improve lipid accumulation in the yeast Rhodosporidium toruloides by UV irradiation mutagenesis and selection. R. toruloides was irradiated by UV, and mutants were selected based on tolerance to lithium chloride (LiCl) or ethanol–H2O2. Subsequently, the cell growth, lipid production, and glucose consumption of mutant R. toruloides were determined. Finally, the transcription levels of oil accumulation‐related genes in the wild type and mutant strains were evaluated.
Highlights
-
‐
R. toruloides mutants were selected based on LiCl and ethanol–H2O2 tolerance
-
‐
UV & LiCl tolerance increased lipid production by 43.6% over the wild type
-
‐
UV & LiCl tolerance increased lipid productivity by 43.2% over the wild type
-
‐
UV & ethanol–H2O2 tolerance increased lipid production by 37.8% over the wild type
-
‐
UV & ethanol–H2O2 tolerance increased lipid productivity by 37.5% over the wild type
2. MATERIALS AND METHODS
2.1. Strains, media, and cultivation
The yeast strain R. toruloides 2.1389 was purchased from China General Microbiological Culture Collection Center and used for oil production and mutagenesis. R. toruloides cells were cultivated in yeast extract peptone dextrose medium (20 g/L peptone, 20 g/L dextrose and 10 g/L yeast extract) or in fermentation (FM) medium (2 g/L (NH4)2SO4, 1 g/L KH2PO4, 1 g/L MgSO4·7H2O, 1 g/L CaCl2·2H2O, 1 g/L NaCl, 0.5 g/L yeast extract, 30 g/L dextrose). R. toruloides strains were grown at 28°C in an air shaker at 150 rpm. The monoclonal cell strain was inoculated from agar medium and grown overnight in 5 mL medium in φ16 × 125‐mm2 test tubes. The overnight cultures were inoculated in 100 mL of medium using a 500‐mL flask and grown for 96 h. Cell growth was started at an optical density (OD 600 nm) of 0.1–0.2 and continued for 24, 48, 72, 96, 120, and 144 h. We used a correlation factor to convert OD to dry cell weight (DCW) per liter.
2.2. Mutagenesis and selection
The R. toruloides cell strains were inoculated into 50 mL of medium using a 250‐mL flask and cultured for 24 h. The yeast suspensions were then irradiated by UV light at 15 W/m2 for 8 min, which corresponded to a fatality rate of approximately 95%. Next, 1 × 106 cell strains were inoculated into fresh yeast extract peptone dextrose agar medium containing a certain concentration of ethanol and H2O2 or LiCl. After five cycles of compound mutagenesis and cultivation, the dominant strains (those that showed a high oil production rate and produced a large amount of oil) were cultivated on FM medium. The concentration ratio of ethanol %v/v to H2O2 (5 mmol/L) was 1:1. The concentration of LiCl was 5 mol/L. The lipid concentration was measured using the sulfo‐phospho‐vanillin reaction method.
2.3. Real‐time polymerase chain reaction analysis
Total RNA was extracted from the yeast cells using a Yeast RNA Kit R6870 (OMEGA) following the manufacturer's instructions. First‐strand cDNA was synthesized using a PrimeScript™ II First‐strand cDNA Synthesis Kit (TaKaRa) and adopted as a template for reverse transcription PCR amplification. The transcription levels of lipid production‐related genes 24 shown in Figure 1 were determined using real‐time quantitative PCR (RT‐qPCR) with specific primers. The primers for RT‐qPCR were designed using Primer Premier 5.0 software and are listed in Supporting Information Table 1. RT‐qPCR analysis of genes was performed using a LightCycler® 480 Real‐Time PCR System with SYBR Green Real‐Time PCR Master Mix (Toyobo). The β‐actin gene was used as an internal reference gene for all experiments 25. The expression levels of the genes were calculated using the 2−ΔΔCt method. Data are presented as the average of three independent experiments. SPSS statistics software was used to analyze the statistical significance; p < 0.05 was considered to indicate statistical significance.
Figure 1.
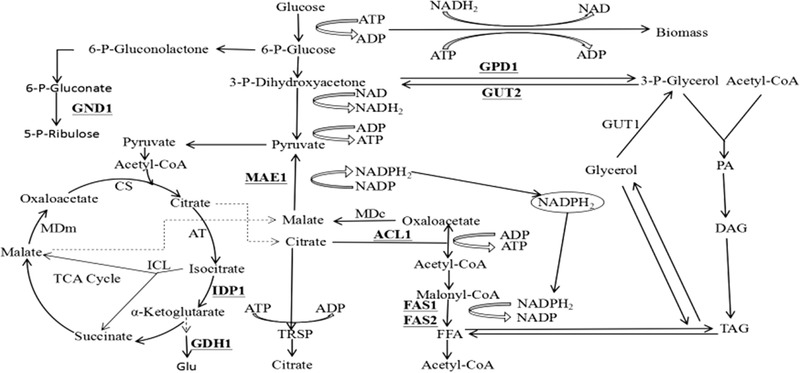
Lipid synthesis related pathway in R. toruloides. Abbreviations for genes are as follows: ACL1 ATP citrate lyase, FAS1 and FAS2 fatty acid synthase 1/2, GDH1 glutamate dehydrogenase, GND1 6‐phosphogluconate dehydrogenase, GPD1 glycerol‐3‐phosphate dehydrogenase 1, GUT2 glycerol‐3‐phosphate dehydrogenase, IDP1 isocitrate dehydrogenase, MAE1 mitochondrial malic enzyme
2.4. Analytical methods
DCW was determined using gravimetric analysis. Each sample of culture broth (10 mL) was harvested by centrifugation at 8000 g/min for 5 min and then washed three times with distilled water. The total DCW was determined after drying at 85°C for 24 h.
Lipid concentration was determined using the sulfo‐phospho‐vanillin reaction method 1. First, 1 mL of culture broth was centrifuged (8000 g/min for 5 min). The wet yeast cells were washed three times with sterile water and resuspended in 0.5 mL of sterile water. Subsequently, 100 µL of cell suspension was mixed with 2 mL of 98% sulfuric acid and incubated in a boiling water bath for 10 min. After cooling to room temperature, 5 mL of phospho‐vanillin regent (6 g/L vanillin, 10% ethanol, and 68% phosphoric acid) was added, and the reaction mixture was incubated at 37°C for 15 min. After cooling to room temperature, the reaction mixture was centrifuged at 6000 g/min for 5 min, and the absorbance of the supernatant was measured at 530 nm. The lipid concentration was determined from a calibration curve constructed using the conventional gravimetric method.
Lipid composition was analyzed by GC–MS using an Agilent 5975 series MSD and Agilent 7890A instrument equipped with an HP‐5 column (30 m × 0.25 mm, film thickness of 0.25 m; HP). Mealy yeast (15 mg) was used to extract fatty acids, and 10 µL of nonadecanoic acid was added as an internal standard (Sigma–Aldrich). The mealy yeast was added with 2 mL of 1 N HCl in methanol, vortexed for 50 min with glass beads, placed in a water bath at 80°C for 60 min for methylation, and then cooled to room temperature. Next, 2 mL of 0.9% NaCl was added to the solution. After vortexing for 30 s, 1.5 mL of hexane was added followed by vortexing for 5 min. The solution was then centrifuged at 6000 r/min for 10 min. The upper hexane layer was transferred to a clean GC vial. The program used during GC/MS analysis was as follows: 140°C for 2 min; heat to 180°C at a rate of 5°C/min; hold at 180°C for 5 min; heat to 230°C at 5°C/min; hold at 230°C for 6 min with electron ionization of 70 eV 26. The differences in the fatty acid contents of wild‐type R. toruloides and its mutants were statistically analyzed by t test with p < 0.05 considered to indicate statistical significance.
The physical and chemical properties of biodiesel fuel were roughly evaluated by lipid composition according to the method of Tanimura and Hoekman 27, 28.
3. RESULTS AND DISCUSSION
3.1. Selection of high‐lipid‐producing mutant yeasts based on LiCl and ethanol–H2O2 tolerance
High‐lipid‐producing mutants were selected from LiCl and ethanol–H2O2 stress‐tolerant mutants based on the relative lipid productions of approximately 100 mutants compared to the wild type. And the LiCl and ethanol–H2O2 stress tolerant mutants, those exhibiting the highest lipid production were selected and named R‐ZL2 and R‐ZY13, respectively.
As shown in Figure 2A, approximately 80% of LiCl‐tolerant mutants produced larger amounts of lipid than the wild type. By using LiCl as a selection reagent, mutants with higher lipid productivity compared to the original strain were obtained. The combination of UV mutagenesis and LiCl‐based selection has previously been successfully applied for mutant breeding. The yields of β‐galactosidases in Aspergillus oryzae 22 and arachidonic acid in Mortierella alpinahas 23 were improved by UV mutagenesis and selection using LiCl. Therefore, LiCl can be a promising selection regent for generating high lipid producing mutants from oleaginous yeasts.
Figure 2.
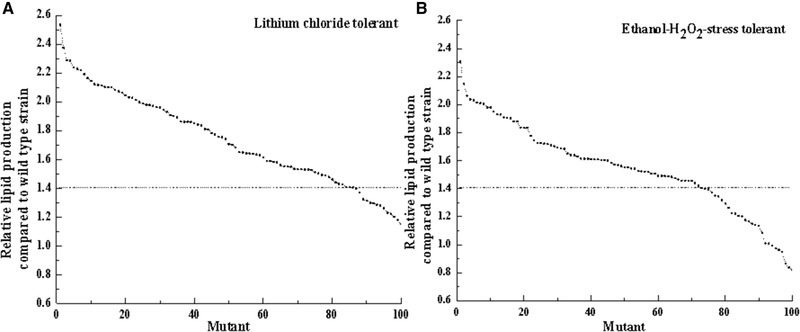
Relative lipid production in mutants compared to the wild type strain R. toruloides 2.1389. (A) Lithium chloride tolerant mutants and (B) ethanol‐H2O2 stress tolerant mutants are shown
As shown in Figure 2B, approximately 72% of ethanol–H2O2 stress‐tolerant mutants produced larger amounts of lipid than the wild type. In this study, the use of ethanol–H2O2 for selection also led to the successful generation of mutants with higher lipid productivity than the original strain. Some studies have shown that stress tolerance can influence the lipid productivity of yeast 29. The clear correlation between lipid productivity and stress tolerance in yeast has not been established, but the multiple stress tolerances are more conducive to improving lipid productivity than a single stress tolerance 1. Therefore, high‐lipid‐producing yeast strains can be successfully created through the use of ethanol–H2O2 combined with an efficient mutagenesis procedure.
The stability of mutants is very important in industrial practice, and the results of lipid production of the mutant R‐ZL2 and R‐ZY13 for eight generations are presented in Table 1. As shown in Table 1, the lipid production of the mutant R‐ZL2 for eight generations ranged from 2.24–2.30 g/L and the lipid production of the mutant R‐ZY13 for eight generations ranged from 2.12–2.18 g/L, indicating a weak influence of passage times on lipid production of the two mutants. The results of the hereditary stability test of mutant strains showed that mutant R‐ZL2 and R‐ZY13 were stable isolates.
Table 1.
Lipid production of Mutant Strains for eight Generations
| Passage timeslipid production (g/L) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Strain | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| R‐ZL2 | 2.24 ± 0.10 | 2.30 ± 0.15 | 2.28 ± 0.12 | 2.25 ± 0.10 | 2.26 ± 0.08 | 2.25 ± 0.05 | 2.28 ± 0.06 | 2.24 ± 0.04 |
| R‐ZY13 | 2.15 ± 0.05 | 2.12 ± 0.06 | 2.18 ± 0.02 | 2.15 ± 0.03 | 2.12 ± 0.08 | 2.12 ± 0.05 | 2.16 ± 0.02 | 2.15 ± 0.04 |
3.2. Cultivation of high‐lipid‐producing mutant yeasts
To evaluate lipid production over time, the wild type RC (2.1389) and its mutants R‐ZL2 and R‐ZY13 were cultivated in FM medium in a flask. All strains showed similar glucose consumption rates (Figure 3A) and DCWs (Figure 3B). The maximum amount of lipid produced by the wild type RC was 1.56 g/L after 96 h of cultivation (Figure 3C; Table 2). The corresponding values for mutants R‐ZL2 and R‐ZY13 were 2.24 and 2.15 g/L after 96 h of cultivation, respectively. Therefore, the R‐ZL2 and R‐ZY13 mutants exhibited higher rates of lipid production (23.323 ± 1.8 and 22.396 ± 2.5 mg/L/h, respectively) than the wild type RC (16.291 ± 1.6 mg/L/h). Continue to prolong the time of culturing, the sugar depletion resulted in cellular lipid degradation, which eventually led to the decline of lipid production at the end of fermentation 18. Meanwhile, the biomass of yeasts also declined in the later stage of fermentation. Similar results were reported in Yamada's study 1.The results showed that the biomass of the mutant strain was similar to that of the original strain in the same incubation time. The ultimate cause of increase of the lipid production of the mutant strain might be the increase of the lipid content.
Figure 3.
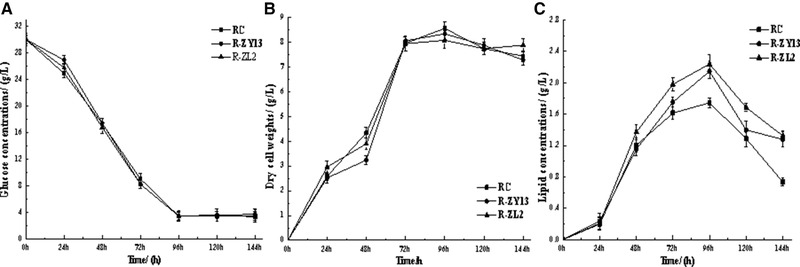
Time course analysis of glucose and lipid concentration and dry cell weight in wild type mutant strains. (A) Glucose concentrations, (B) dry cell weights (DCWs), and (C) lipid concentrations are shown. Data are presented as the average of three independent experiments. Error bars represent means ± standard deviation
Table 2.
Summary of lipid production in the wild‐type R. toruloides 2.1389 strain and the derived mutants
| Strain | Maximum lipid production (g/L) | Maximum lipid production time (h) | Maximum lipid production rate (mg/L/h) |
|---|---|---|---|
| RC (2.1389) | 1.56 ± 0.1 | 96 | 16.291 ± 1.6 |
| R‐ZL2 | 2.24 ± 0.2** | 96 | 23.323 ± 1.8 ** |
| R‐ZY13 | 2.15 ± 0.08** | 96 | 22.396 ± 2.5 ** |
Data shown are averages of three independent experiments. Significant differences were determined by t test (**p < 0.01, *p < 0.05).
3.3. Relative transcription levels of lipid production‐related genes in mutants
To evaluate the relative transcription levels of lipid production‐related genes in the mutants, the wild‐type strain RC and its mutants R‐ZL2 and R‐ZY13 were cultivated in FM medium for 96 h, and real‐time PCR analysis was performed. In the R‐ZL2 mutant, the transcription levels of IDP1, GPD1, GND1, and ACL1 were significantly higher than in the wild type (Figure 4A). In the R‐ZY13 mutant, the transcription levels of IDP1, GPD1, GND1, and FAS1 were significantly higher than in the wild type (4B).
Figure 4.
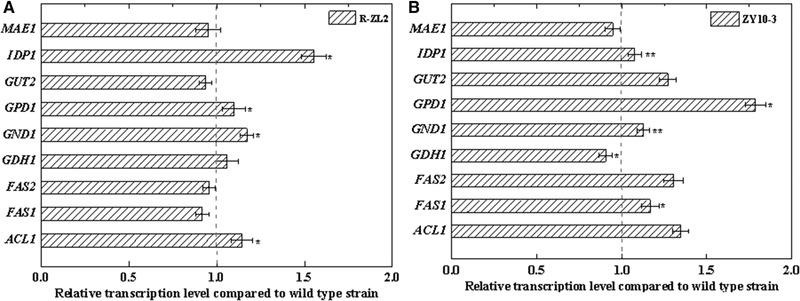
Relative transcription levels of lipid production‐related enzymes in the evaluated mutant strains. (A) The lithium chloride tolerant mutant R‐ZL2 and (B) ethanol and H2O2‐stress tolerant mutant R‐ZY13 are shown. Data are presented as the average of 3 independent experiments. Error bars represent means ± standard deviation. Significant differences were calculated using the t test (**p < 0.01, *p < 0.05)
The transcription levels of three genes, IDP1, GND1, and GPD1, were significantly higher in the two high‐lipid‐producing mutants (Figure 4) generated in this study. IDP1, which encodes isocitrate dehydrogenase, converts isocitrate to α‐ketoglutaric acid to produce nicotinamide adenine dinucleotide phosphate (NADPH) (Figure 1). GND1, which encodes 6‐phosphogluconate dehydrogenase, converts 6‐phos‐phogluconate to ribulose 5‐phosphate, also producing NADPH (Figure 1). Because lipids are highly reduced substances, the production of a reducing agent (i.e., NADPH) is important for lipid production 1, 24. Therefore, the high transcription levels of IDP1 and GND1 might contribute to increasing lipid productivity by providing sufficient reducing power for lipid production in mutant strains. Previous studies have found that the transcription level of IDP1 is increased when the lipid productivity in oleaginous yeasts R. toruloides NP11 24, R. toruloides NBRC8766 1, and Yarrowia lipolytica 30 was increased. Lucero et al. (2017) indicated that the transcription level of GND1 increased under lipid‐productive nitrogen starvation in Ustilago maydis 31. GPD1, which encodes glycerol‐3‐phosphate dehydrogenase, converts 3‐phosphate dihydroxyacetone to 3‐phosphate glycerol to produce acetyl‐CoA, a direct lipid precursor that is important for lipid production. Therefore, the high transcription level of GPD1 might also contribute to enhanced lipid productivity. Wang et al. (2013) reported that the transcription level of GPD1 increased under lipid‐productive nitrogen starvation in oleaginous yeast Y. lipolytica.
In this study, the transcription level of ACL1 was significantly higher than in the wild type only in the R‐ZL2 mutant. ACL1 encodes ATP citrate lyase, which catalyzes the generation of acetyl‐CoA from mitochondrial citrate. Acetyl‐CoA is a vital building block in the endogenous biosynthesis of fatty acids 32, 33. Therefore, ACL1 is important for lipid biosynthesis. Large increases in ACL1 activity have been reported in different studies during lipid accumulation in Trichosporon cutaneum 34, R. toruloides 24, and Y. lipolytica 30. The transcription level of FAS1 was significantly higher than in the wild type only for the R‐ZY13 mutant. FAS1 encodes the β subunit of fatty acid synthase, which catalyzes the NADPH‐dependent condensation of acetyl‐CoA and malonyl‐CoA to produce fatty acid 35. Therefore, the high transcription level of FAS1 in the high‐lipid‐producing mutant might have a positive effect on lipid biosynthesis. Previous studies found that the transcription level of FAS1 is up‐regulated under lipid‐productive culture conditions in the oleaginous yeast R. toruloides 24, 36.
3.4. Lipid composition in wild‐type and mutant yeasts
To evaluate lipid composition, the wild‐type strain RC and its mutants (R‐ZL2 and R‐ZY13) were cultivated in FM medium for 96 h, and the methyl‐esterified lipids were analyzed by GC/MS (each strain was analyzed in triplicate). In the wild type, oleic acid (63.73%) was the major lipid produced followed by palmitic acid (19.2%, Table 3). In the R‐ZL2 and R‐ZY13 mutants, oleic acid (58.26% and 59.89%, respectively) was also the major lipid followed by palmitic acid (23.47% and 23.65%, respectively). It is generally known that the properties of biodiesel fuel (e.g., viscosity, specific gravity, and cetane number) mainly depend on the lipid composition of the feedstocks. Some countries or regions have established relevant standards, such as ASTM D6751 in USA, S50 in China, and EN 14214 in the EU 27, 37. Therefore, lipid composition in oleaginous yeast is important for biodiesel fuel application.
Table 3.
Fatty acid compositions of the wild‐type R. toruloides 2.1389 strain and the derived mutants
| Fatty acid composition (% w/w) | |||||||
|---|---|---|---|---|---|---|---|
| Strain | Myristic (C14:0) | Palmitic (C16:0) | Stearic (C18:0) | Oleic (C18:1) | Linoleic (C18:2) | α‐linolenic (C18:3) | |
| RC (2.1389) | 0.52 ± 0.2 | 19.20 ± 0.4 | 4.8 ± 0.2 | 63.73 ± 2.2 | 7.59 ± 0.3 | 2.95 ± 0.2 | In this study |
| R‐ZL2 | 1.05 ± 0.01 | 23.47 ± 0.01** | 11.79 ± 0.01* | 58.26 ± 0.03 | 4.02 ± 0.01 | 1.23 ± 0.2* | In this study |
| R‐ZY13 | 1.12 ± 0.04 | 23.65 ± 0.02** | 10.07 ± 0.01* | 59.89 ± 0.06 | 4.31 ± 0.03 | 1.14 ± 0.25* | In this study |
| R. toruloides DSM4444 | – | 16.6 ± 1.6 | 10.2 ± 1.0 | 58.5 ± 2.9 | 7.8 ± 1.1 | – | Seraphim 2017 |
| R. toruloides AS 2.1389 | – | 28.5 | 12.9 | 41.3 | 12.8 | – | Xu 2017 |
| Soybean oil | – | 10.05 ± 0.02 | 4.56 ± 0.2 | 26.52 ± 0.03 | 52.38 ± 0.06 | 6.15 ± 0.2 | Sofia 2016 |
Data shown are the average of three independent experiments. C14:0, myristic acid; 16:0, palmitic acid; 18:0, stearic acid; 18:1, oleic acid; 18:2, linoleic acid; 18:3, α‐linolenic acid. Significant differences were determined by t test (**p < 0.01, *p < 0.05).
Table 3 shows the fatty acid profiles. Although the lipid composition in mutant yeasts varied from the wild type strain, the major lipids of both the wild type and mutant strains were oleic acid, palmitic acid, linoleic acid, and stearic acid. The dominant fatty acids in yeast lipids are thus similar to those found in R. toruloides DSM4444 18, R. toruloides AS 2.1389 38, and Soybean oil 39, making them an appropriate substitute for biodiesel and other oleochemicals. Furthermore, we roughly evaluated the properties of biodiesel fuel from lipid composition in Table 3 using the method of Hoekman (2012) (summarized in Table 4) 28. And we also compared these properties to those of biodiesel produced from the lipid profiles of oleaginous yeasts R. toruloides NBRC 8766 36 and Lipomyces starkeyi NBRC 10381 27 along with major biodiesel standards S50 (China), ASTM D6751 (USA), and EN 14214 (European Union) 27, 37. The results (Table 4) show that the lipids obtained from R. toruloides and its mutants in this study can serve as alternative oil sources for biodiesel fuel.
Table 4.
Comparison of the properties of biodiesel produced from R. toruloides and its mutants with those of Chinese, U.S. and EU biodiesel standards
| Strain | Viscosity (mm/s2) | Specific gravity | Cloud point (°C) | Cetane number | Iodine number | Higher heating value (MJ/kg) |
|---|---|---|---|---|---|---|
| RC (2.1389) | 4.65 | 0.8774 | 8.27 | 57.02 | 77.98 | 40.05 |
| R‐ZL2 | 4.76 | 0.8764 | 10.65 | 58.21 | 64.76 | 39.77 |
| R‐ZY13 | 4.75 | 0.8765 | 10.39 | 58.08 | 66.21 | 39.80 |
| R. Toruloides NBRC 8766 | 4.64 | 0.8775 | 8.06 | 56.92 | 79.14 | 40.11 |
| L. Starkeyi NBRC 10381 | 4.61 | 0.8778 | 7.38 | 56.58 | 82.93 | 40.20 |
| S50 | 1.9–6.0 | 0.82–0.9 | – | 49 (minimum) | 101 (maximum) | – |
| ASTM D6751 | 1.9–6.0 | – | – | 47 (minimum) | 93 (maximum) | – |
| EN 14214 | 3.5–5.0 | 0.86–0.9 | – | 51 (minimum) | 120 (maximum) | – |
4. CONCLUDING REMARKS
In this study, we successfully demonstrated that the combination of UV mutagenesis and LiCl/ethanol–H2O2 stress were effective to enhance the lipid production of oleaginous yeast R. toruloides. After R. toruloides (2.1389) was mutated by UV and LiCl/ethanol–H2O2 stress, two mutant strains, R‐ZL2 and R‐ZY13, were obtained. The mutant strains R‐ZL2 and R‐ZY13 exhibited lipid yields of 2.24 and 2.15 g/L, respectively, corresponding to 43.6% and 37.8% increases over the wild type. The lipid productivities of mutants R‐ZL2 and R‐ZY13 were 23.323 ± 1.8 and 22.396 ± 2.5 mg/L/h, respectively, corresponding to 43.2% and 37.5% increases over the wild type. Transcription analysis indicated that lipid production might be further improved by regulating some lipid production‐related genes via genetic engineering techniques in future studies. By effectively combining mutagenesis and genetic engineering to improve strain characteristics, yeast strains with low cost and high oil production can be obtained, thus providing cheap raw materials for biodiesel production.
CONFLICT OF INTEREST
The authors have declared no conflict of interest.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
This work was supported by Research project of young and middle‐aged leading scientists, engineers and innovators in Xinjiang production and construction corps (2018CB024); Innovation project for the south‐forward development for Xinjiang production and construction corps (2018DB002).
AUTHOR CONTRIBUTIONS
All authors contributed jointly to all aspects of the work reported in the manuscript. All authors have read and approved the final manuscript.
Guo M, Cheng S, Chen G, Chen J. Improvement of lipid production in oleaginous yeast Rhodosporidium toruloides by ultraviolet mutagenesis. Eng Life Sci. 2019;19:548–556. 10.1002/elsc.201800203
REFERENCES
- 1. Yamada, R. , Kashihara, T. , Ogino, H. , Improvement of lipid production by the oleaginous yeast Rhodosporidium toruloides through UV mutagenesis. World J. Microbiol. Biotechnol. 2017, 33, 99. [DOI] [PubMed] [Google Scholar]
- 2. González‐García, Y. , Rábago‐Panduro, L. M. , French, T. , Camacho‐Córdova, D. I. , et al., High lipids accumulation in Rhodosporidium toruloides by applying single and multiple nutrients limitation in a simple chemically defined medium. Annals of Microbiol. 2017, 67, 519–527. [Google Scholar]
- 3. Sajid, Z. , Khan, F. , Zhang, Y. , Process simulation and life cycle analysis of biodiesel production. Renewable Energy 2016, 85, 945–952. [Google Scholar]
- 4. Azócar, L. , Ciudad, G. , Heipieper, H. J. , Navia, R. , Biotechnological processes for biodiesel production using alternative oils. Appl. Microbiol. Biotechnol. 2010, 88, 621–636. [DOI] [PubMed] [Google Scholar]
- 5. Wu, H. , Li, Y. , Chen, L. , Zong, M. , Production of microbial oil with high oleic acid content by Trichosporon capitatum . Appl. Energy 2010, 88, 138–142. [Google Scholar]
- 6. Gui, M. M. , Lee, K. T. , Bhatia, S. , Feasibility of edible oil vs. non‐edible oil vs. waste edible oil as biodiesel feedstock. Energy 2008, 33, 1646–1653. [Google Scholar]
- 7. Fakas, S. , Lipid biosynthesis in yeasts: A comparison of the lipid biosynthetic pathway between the model nonoleaginous yeast Saccharomyces cerevisiae and the model oleaginous yeast Yarrowia lipolytica . Eng. Life Sci. 2016, 17, 292–302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Uprety, B. K. , Dalli, S. S. , Rakshit, S. K. , Bioconversion of crude glycerol to microbial lipid using a robust oleaginous yeast Rhodosporidium toruloides ATCC 10788 capable of growing in the presence of impurities. Energy Convers Manag. 2017, 135, 117–128. [Google Scholar]
- 9. Koutinas, A. A. , Chatzifragkou, A. , Kopsahelis, N. , Papanikolaou, S. , Kookos, I. K. , Design and techno‐economic evaluation of microbial oil production as a renewable resource for biodiesel and oleochemical production. Fuel 2014, 116, 566–577. [Google Scholar]
- 10. Huang, C. , Chen, X. F. , Xiong, L. , Chen, X. D. , et al., Single cell oil production from low‐cost substrates: The possibility and potential of its industrialization. Biotechnol. Adv. 2013, 31, 129–139. [DOI] [PubMed] [Google Scholar]
- 11. Martin, G. J. O. , Energy requirements for wet solvent extraction of lipids from microalgal biomass. Bioresour Technol. 2016, 205, 40–47. [DOI] [PubMed] [Google Scholar]
- 12. Jin, M. , Slininger, P. J. , Dien, B. S. , Waghmode, S. et al., Microbial lipid‐based lignocellulosic biorefinery: Feasibility and challenges. Trends Biotechnol. 2015, 33, 43–54. [DOI] [PubMed] [Google Scholar]
- 13. Bonturi, N. , Matsakas, L. , Nilsson, R. , Christakopoulos, P. et al., Single Cell Oil Producing Yeasts Lipomyces starkeyi and Rhodosporidium toruloides: Selection of Extraction Strategies and Biodiesel Property Prediction. Energies 2015, 8, 5040–5052. [Google Scholar]
- 14. Kumar, D. , Singh, B. , Korstad, J. , Utilization of lignocellulosic biomass by oleaginous yeast and bacteria for production of biodiesel and renewable diesel. Renewable Sustainable Energy Rev. 2017, 73, 654–671. [Google Scholar]
- 15. Tchakouteu, S. S. , Kopsahelis, N. , Chatzifragkou, A. , Kalantzi, O. , et al., Rhodosporidium toruloides cultivated in NaCl‐enriched glucose‐based media: Adaptation dynamics and lipid production. Eng. Life Sci. 2017, 17, 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. André, A. , Chatzifragkou, A. , Diamantopoulou, P. , Sarris, D. , et al., Biotechnological conversions of bio‐diesel‐derived crude glycerol by Yarrowia lipolytica strains. Eng. Life Sci. 2009, 9, 468–478. [Google Scholar]
- 17. Lin, H. , Cheng, W. , Ding, H. , Chen, X. , et al., Direct microbial conversion of wheat straw into lipid by a cellulolytic fungus of Aspergillus oryzae A‐4 in solid‐state fermentation. Bioresource Technolog. 2010, 101, 7556–7562. [DOI] [PubMed] [Google Scholar]
- 18. Papanikolaou, S. , Kampisopoulou, E. , Blanchard, F. , Rondags, E. et al., Production of secondary metabolites through glycerol fermentation under carbon‐excess conditions by the yeasts Yarrowia lipolytica and Rhodosporidium toruloides. Eur. J. Lipid Sci. Technol. 2017, 119, 1600507. [Google Scholar]
- 19. Sitepu, I. R. , Garay, L. A. , Sestric, R. , Levin, D. , et al., Oleaginous yeasts for biodiesel: current and future trends in biology and production. Biotechnol. Adv. 2014, 32, 1336–1360. [DOI] [PubMed] [Google Scholar]
- 20. Poli, J. S. , Neres da Silva, M. A. , Siqueira, E. P. , Pasa, V. M. D. , et al., Microbial lipid produced by Yarrowia lipolytica QU21 using industrial waste: a potential feedstock for biodiesel production. Bioresour. Technol. 2014, 161, 320–326. [DOI] [PubMed] [Google Scholar]
- 21. Yen, H. W. , Liu, Y. X. , Application of airlift bioreactor for the cultivation of aerobic oleaginous yeast Rhodotorula glutinis with different aeration rates. Journal of Fermentation and Bioengineering. 2014, 118, 195–198. [DOI] [PubMed] [Google Scholar]
- 22. Zhang, W. , Liu, F. , Yang, M. , Liang, Q. et al., Enhanced β‐galactosidase production of Aspergillus oryzae mutated by UV and LiCl. Preparative Biochemistry and Biotechnology. 2014, 44, 310–320. [DOI] [PubMed] [Google Scholar]
- 23. Cong, L. L. , Ji, X. J. , Nie, Z. K. , Peng, C. , Li, Z. Y. , Breeding of Mortierella alpina for high‐yield arachidonic acid‐rich oil production. Chinese Journal of Bioprocess Engineering. 2012, 10, 34–38. [Google Scholar]
- 24. Zhu, Z. , Zhang, S. , Liu, H. , Shen, H. , et al., A multi‐omic map of the lipid‐producing yeast Rhodosporidium toruloides . Nat. Commun. 2012, 3, 1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wang, Y. , Lin, X. , Zhang, S. , Sun, W. , et al., Cloning and evaluation of different constitutive promoters in the oleaginous yeast Rhodosporidium toruloides. Yeast. 2016, 33, 99–106. [DOI] [PubMed] [Google Scholar]
- 26. Xia, J. , Wang, L. , Zhu, J. B. , Sun, C. J. , et al., Expression of Shewanella frigidimarina fatty acid metabolic genes in E. coli by CRISPR/cas9‐coupled lambda Red recombineering. Biotechnol. Lett. 2016, 38, 117–122. [DOI] [PubMed] [Google Scholar]
- 27. Tanimura, A. , Takashima, M. , Sugita, T. , Endoh, R. , et al., Selection of oleaginous yeasts with high lipid productivity for practical bio‐diesel production. Bioresour Technol. 2014, 153, 230–235. [DOI] [PubMed] [Google Scholar]
- 28. Hoekman, S. K. , Broch, A. , Robbins, C. , Ceniceros, E. , Natarajan, M. , Review of biodiesel composition, properties, and specifications. Renew Sust Energ Rev. 2012, 16, 143–169. [Google Scholar]
- 29. Jamieson, D. J. , Oxidative stress responses of the yeast Saccharomyces cerevisiae . Yeast. 1998, 14, 1511–1527. [DOI] [PubMed] [Google Scholar]
- 30. Wang, Z. P. , Xu, H. M. , Wang, G. Y. , Chi, Z. , Chi, Z. M. , Disruption of the MIG1 gene enhances lipid biosynthesis in the oleaginous yeast Yarrowia lipolytica ACA‐DC 50109. Biochimica et Biophysica Acta‐Molecular and Cell Biology of Lipids. 2013, 1831, 675–682. [DOI] [PubMed] [Google Scholar]
- 31. Romero Aguilar, L. , Pablo Pardo, J. , Montero Lomeli, M. , Luqueno Bocardo, O. I. , et al., Lipid droplets accumulation and other biochemical changes induced in the fungal pathogen Ustilago maydis under nitrogen‐starvation. Arch Microbiol. 2017, 199, 1195–1209. [DOI] [PubMed] [Google Scholar]
- 32. Chypre, M. , Zaidi, N. , Smans, K. , ATP‐citrate lyase: A mini‐review. Biochem. Biophys. Res. Commun. 2012, 422, 1–4. [DOI] [PubMed] [Google Scholar]
- 33. Dulermo, T. , Lazar, Z. , Dulermo, R. , Rakicka, M. , et al., Analysis of ATP‐citrate lyase and malic enzyme mutants of Yarrowia lipolytica points out the importance of mannitol metabolism in fatty acid synthesis. Biochim Biophys Acta. 2015, 1851, 1107–1117. [DOI] [PubMed] [Google Scholar]
- 34. Liu, Z. , Gao, Y. , Chen, J. , Imanaka, T. , et al., Analysis of metabolic fluxes for better understanding of mechanisms related to lipid accumulation in oleaginous yeast Trichosporon cutaneum . Bioresour. Technol. 2013, 130, 144–151. [DOI] [PubMed] [Google Scholar]
- 35. Cui, W. , Liang, Y. , Tian, W. , Ji, M. , Ma, X. , Regulating effect of β‐ketoacyl synthase domain of fatty acid synthase on fatty acyl chain length in de novo fatty acid synthesis. Biochimica et Biophysica Acta‐Molecular and Cell Biology of Lipids. 2016, 1861, 149–155. [DOI] [PubMed] [Google Scholar]
- 36. Yamada, R. , Yamauchi, A. , Kashihara, T. , Ogino, H. , Evaluation of lipid production from xylose and glucose/xylose mixed sugar in various oleaginous yeasts and improvement of lipid production by UV mutagenesis. Biochem. Eng. J. 2017, 128, 76–82. [Google Scholar]
- 37. Knothe, G. , Dependence of biodiesel fuel properties on the structure of fatty acid alkyl esters. Fuel Processing Technol. 2004, 86, 1059–1070. [Google Scholar]
- 38. Xu, J. Y. , Zhao, X. B. , Du, W. , Liu, D. H. , Bioconversion of glycerol into lipids by Rhodosporidium toruloides in a two‐stage process and characterization of lipid properties. Eng. Life Sci. 2017, 17, 303–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maina, S. , Pateraki, C. , Kopsahelis, N. , Paramithiotis, S. , et al., Microbial oil production from various carbon sources by newly isolated oleaginous yeasts. Eng. Life Sci. 2017, 17, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


