Abstract
The application of PAT for in‐line monitoring of biopharmaceutical manufacturing operations has a central role in developing more robust and consistent processes. Various spectroscopic techniques have been applied for collecting real‐time data from cell culture processes. Among these, Raman spectroscopy has been shown to have advantages over other spectroscopic techniques, especially in aqueous culture solutions. Measurements of several process parameters such as glucose, lactate, glutamine, glutamate, ammonium, osmolality and VCD using Raman‐based chemometrics models have been reported in literature. The application of Raman spectroscopy, coupled with calibration models for amino acid measurement in cell cultures, has been assessed. The developed models cover four amino acids important for cell growth and production: tyrosine, tryptophan, phenylalanine and methionine. The chemometrics models based on Raman spectroscopy data demonstrate the significant potential for the quantification of tyrosine, tryptophan and phenylalanine. The model for methionine would have to be further refined to improve quantification.
Keywords: Amino acids, Cell culture, In‐line monitoring, PAT, Raman spectroscopy
Abbreviations
- QbD
Quality by Design
- VCD
viable cell density
1. Introduction
The QbD (Quality by Design) paradigm is an innovative framework established by the United States Federal Drug Administration (US FDA) to increase efficiency in pharmaceutical development, manufacturing and quality assurance. The ultimate goal in applying QbD tools is to render pharmaceutical manufacturing processes more consistent, robust and flexible by designing and developing the process such that predefined quality requirements are ensured throughout the process 1. One key component in QbD paradigm is the application of in‐line process monitoring and control of processes to avoid the risks and delays associated with offline methods. Furthermore, real‐time monitoring provides better process understanding and can lead to designing more robust processes. A real‐time sensing technology should be able to determine the concentration of a particular metabolite without interference from other metabolites. Many different types of sensors have been used for the control of processes 2, 3. Optical sensors such as Fourier Transform Infra‐red (FTIR), Near Infra‐red(NIR), Mid Infra‐red (MIR), UV‐visible spectroscopy and Raman spectroscopy all have the potential for effective in‐line monitoring 4, 5, 6, 7, 8, 9, 10. However, Raman spectroscopy provides advantages over other spectroscopic methods as it has no interference from water, plastic and glass 11, 12, and is sensitive to fundamental vibrations in any spectral region from infrared to ultra‐violet.
Raman spectroscopy has been previously used to measure important cell culture process parameters such as glucose, lactate, glutamine, and ammonium concentrations, osmolality, viable cell density (VCD), and protein aggregation 13, 14, 15, 16, 17. First described in 1928 by Indian physicist CV Raman, Raman spectroscopy is a fundamental vibrational spectroscopy technique that provides information about molecular vibrations and relies on the inelastic scattering of monochromatic light by vibrations in molecules 18. When the sample is irradiated with a monochromatic source of light, most of the radiation scatters off the sample at the same wavelength as the input light, known as Rayleigh scattering. However, a small portion of the scattered light shifts its energy from the original laser wavelength. Plotting the intensity of the inelastic scattered light against the frequency gives the Raman spectra. Due to the tremendous amount of spectral data generated by Raman spectroscopy, chemometrics methods of data analysis are ideal, as they can be applied to extract useful information out of vast amounts of data 6, 19, 20.
Amino acids are major components in cell culture media as they play a key role in antibody production and glycosylation 21. Additionally, amino acids are building blocks of the proteins and serve as intermediates in many metabolic pathways. Amino acids are provided in the media to fulfill the nutritional requirements of the cells. Amino acid concentrations are continuously changing in the media during the cell culture process as some amino acids are being consumed and others are being released by the cells, whereas the ideal conditions for the cells require a well‐controlled balance of amino acids concentration profile. Hence, monitoring the continuous change in amino acid concentrations can help to more accurately control their concentration, which in turn will ensure improved productivity and more consistent product quality.
Currently, amino acid analysis for bioreactor samples is performed using HPLC methods at the conclusion of a batch. There are several techniques available for amino acid analyses ‐including liquid chromatography, HPLC, TLC, GC‐MS spectrometry, NMR and capillary electrophoresis 22, 23, 24, 25, 26, 27, 28. Chromatography‐based techniques are the most accurate amino acid analysis methods, but are time consuming and require comprehensive sample processing 29.
To achieve faster response times without compromising accuracy, Raman techniques are employed to develop chemometrics models for the quantitation of four amino acids, namely tyrosine, tryptophan, phenylalanine, and methionine. These amino acids are found to be important in cell culture media and controlling their concentrations below set limits are shown to improve cell growth and productivity 30. Three of these amino acids (tryptophan, phenylalanine and methionine) are categorized as essential amino acids.
Partial Least Squares (PLS) models for these four amino acids are calibrated using bench and pilot scale data. Previous studies have demonstrated the feasibility of monitoring cell culture metabolites such as glucose, lactate, glutamine, glutamate, osmolality and VCD using Raman spectroscopy. Moreover, publications from other groups have reported Raman spectra of various amino acids 31, 32 as part of the characterization of amino acids peaks. However to the best of our knowledge there are no publications on quantitation of amino acids in mammalian cell culture processes using a combination of Raman spectroscopy with chemometrics methods with the purpose of monitoring cell culture bioreactor runs. These models can later be utilized for real‐time monitoring and control of amino acids in cell culture processes, which, in turn, can help in process optimization and product quality control.
2. Materials and methods
2.1. Materials
All the chemicals used were of analytical grade (Sigma‐Aldrich, St. Louis, MO). Kaiser RXN2 Raman system (Kaiser Optics, MI) was used to collect the spectra from the samples. Data were processed to build PLS models using SIMCA 13.0.3 software (MKS, San Jose, CA). All the stock solutions for amino acids (250 mM) were prepared in deionized water. To solubilize the amino acids (tryptophan, phenylalanine and methionine), pH of the stock solutions was adjusted by 10N H2SO4. Tyrosine was dissolved in deionized water directly. All other samples were prepared from the above mentioned stock solution. All the samples were stored at 2–8°C.
2.2. Cell culture
The cell culture samples for the amino acid data collection was collected by using four different CHO monoclonal antibody (mAb) producing cell lines derived from the same host. All the cell lines were transfected with proprietary DNA. All the proprietary media used for the culture were developed by Pfizer. The fed batch cell culture experiments were performed at laboratory and pilot scale. The VCD range for the batch runs used in this study has been 25–280 E5 cells/mL, while glucose and lactate concentrations has been in the range of 0.5–6 and 0–4.5 g/L, respectively. The amino acid data collected from the 57 samples from the above‐mentioned batches was used for model calibration. Independent data set from the lab scale and pilot scale batch was used for model validation. The Cells were cultured by using 3L working volume Applikon bioreactors (Applikon, Inc., Schiedam, Netherlands) operating with BioNet modular controllers (Broadley‐James Corp., Irvine, CA, USA) that included pump and gas mass flow controller modules. A combination of sodium bicarbonate and carbon‐di‐oxide was used to control the pH at 7. Micro‐bubble sparging of pure oxygen and overlay of 7% carbon dioxide /air was used to maintain dissolved oxygen in the range of 25–50%. The temperature was controlled between 31 and 37°C with electric heating blankets. Calculated power per unit volume of approximately 50W/m3 was supplied to agitate the culture.
2.3. Analytical methods
For in‐line Raman data collection from cell culture, an immersion probe coupled with fiber optics was integrated with the bioreactors and iCRaman 4.1 software (Kaiser Optical Systems Inc., Ann Arbor, MI) was used to collect the data. To minimize the fluorescence, a laser excitation wavelength of 785 nm with 200 mW power at the probe tip was used to generate Raman spectra from the cell culture. Cosmic ray and dark spectrum were removed to acquire Raman spectra with 75 scans and an exposure of 10 seconds per scan. The spectral data (.spc files) used for model calibration and validation were collected from eight different batches performed at both pilot and bench scales. The data was divided in two sets. One set including seven batches (57 data points) was used for model calibration and the other set including one batch (ten data points) was utilized to perform model validation. Spectra were collected from 100 to 3400 cm−1. For peak identification, concentrated samples (20 mM and 100 mM) of the four amino acids were used. Amino acid solutions were prepared in deionized water using the 250 mM stock solution. To eliminate the stray light, the samples were analyzed within a dark chamber. 20 mL of each sample were used to collect spectra. All the samples were run in duplicates. The spectra were pre‐processed (second derivative and SNV) and analyzed for different peaks using SIMCA software.
For amino acid reference data used to generate the models, daily offline samples were collected from the bioreactor and were centrifuged at 1000 rpm for 10 min and the supernatant was stored at ‐80°C. The reference data points for amino acid concentration were generated by using Acquity UPLC‐H class bio‐system. Amino acid data was analyzed by using Empower 2 (Waters, Milford MA).
2.4. Calibration models
Data for chemometrics models were collected from seven different batches (at bench and pilot scale). Spectral data for 57 different data points from these seven batches was generated. Combinations of different pre‐processing methods were assessed to evaluate the best possible combination, leading to maximum signal‐to‐noise ratio. The pre‐processing combination, which includes mean centering, first derivative, standard normal variate (SNV) and Savitzkz‐Golay (SG) filter, was chosen for model calibration. The first derivative was used to eliminate the baseline drift between the samples. Next, SNV was used to correct the differences caused by path length changes. Finally, SG was used as a smoothing step. The spectral regions used for individual models were determined based on the unique peaks for particular amino acids as mentioned in Table 1. The performances of each model were assessed using correlation coefficient (R2) and root mean square error of evaluation (RMSEE).
Table 1.
Regions of peaks for the four amino acids
| A.A. | Raman shift |
|---|---|
| Tyrosine | 643 / 831 / 852 / 1174 / 1223 / 1263 / 1332 / 1603 / 3051 |
| Tryptophan | 577 / 747 / 759 / 772 / 879 / 1013 / 1261 / 1319 / 1344 / 1363 / 1374 / 1430 / 1449 / 1463 / 1473 / 1554 / 1566 |
| Phenylalanine | 485 / 618 / 623 / 750 / 766 / 821 / 857 / 993 / 1005 / 1032 / 1185 / 1210 / 1586 / 1589 / 1607 |
| Methionine | 653 / 700 / 723 / 1428 |
R2 represents the variability captured by the model and RMSEE and is generally calculated by the formula,
Where yi is the observed value, Y calculated is the value calculated by the models and N indicates the number of samples used for model building.
2.5. Model validation
An independent dataset from a different bench scale batch with 10 data points was used to validate all chemometrics models. The data variability captured by the model can be evaluated by R 2 while the prediction ability of the model was evaluated by the parameter Q2Y, which is fraction of variation of Y variables predicted by the model.
Root mean square error (RMSE) is frequently used to measure the difference between experimental values and model prediction. Root mean square values were calculated by using the following equations. Root mean square error for cross validation (RMSEcv) gives an estimate of how the built model performs for unknown samples.
Where y i is the observed value, Y cvpred is the value predicted by the model by seven‐fold cross validation and N indicates the number of samples used for model building. Root mean square error for prediction (RMSEP) is calculated as
Where y i is the observed value and Y pred is the value predicted by the model for an external data set and N indicates the number of samples used for model building.
3. Results and discussion
The primary goal of the current work was to investigate the feasibility of in‐line amino acid monitoring in cell culture processes using Raman spectroscopy and multivariate calibration models. Four amino acids (tyrosine, tryptophan, phenylalanine and methionine), having a significant impact on the product of interest, were considered for the present study. Initially, the individual peaks for amino acids were analyzed in aqueous solution to minimize any interference from any impurities. The peaks analyzed from the aqueous solution were employed for the amino acid model calibration with the cell culture samples. All four models were validated by using an independent set of 10 data points.
Various peaks for the four amino acids were identified by analyzing high concentration samples of each individual amino acid in water. Two spectra from each concentration 20 and 100 mM were collected and analyzed using SIMCA multivariate data analysis tool. The spectra collected in the 100–3400 cm−1 region were trimmed (500–2000 cm‐1) to exclude noise and were pre‐processed using second derivative and SNV to maximize a signal to noise ratio prior to analysis. Table 1 summarizes all the observable peaks for the four amino acids in the aqueous solution.
The peaks observed for all four amino acids are in confirmation with the work reported in the literature 32. The ranges considered for chemometric models were based on the peak regions observed from the individual amino acid solutions. It has been observed that tyrosine, tryptophan and phenylalanine have more distinguished peaks as compared to methionine that has several weak or medium intensity peaks signals, with no strong peaks observed (as shown in Fig. 1).
Figure 1.
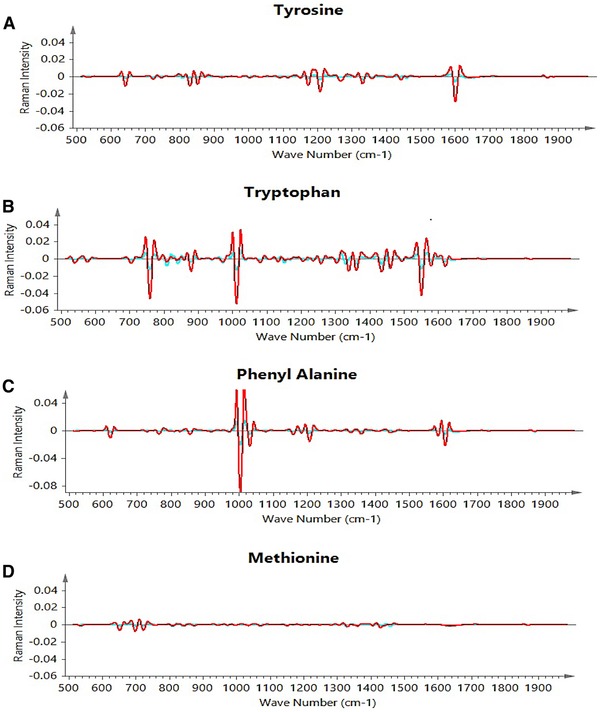
The individual amino acid spectral peaks (A) tyrosine, (B) tryptophan, (C) phenylalanine, (D) methionine.
The training set used a total of 57 samples from seven different batches at bench and pilot scales, and the validation set contained 10 samples from an independent batch at bench scale. The data dispersion was random for calibration and validation sets (as shown in Table 2).
Table 2.
Data ranges for calibration and validation sets mM
| Data set/AA | Tyrosine | Tryptophan | Phenylalanine | Methionine |
|---|---|---|---|---|
| Calibration | 0.19–4.97 | 0.19–2.20 | 1.20–4.35 | 0.59–3.80 |
| Validation | 0.28–4.05 | 0.29–1.81 | 1.23–3.05 | 1.70–2.50 |
All the amino acid concentrations are in micromolar.
PLS models were calibrated individually for each amino acid. Various combinations of pre‐processing treatments and spectral ranges were investigated to optimize the developed models. In the final best performing models, the combination of mean centering, first derivative, SNV and SG was used to calibrate the models. Different spectral regions for model calibration were decided based on the unique peak regions for individual amino acids (Table 1).
Model calibration and validation parameters are summarized in Table 3. The parameters reported for the developed models are the number of samples and principal components, correlation coefficient (R2Y) and standard errors of calibration and prediction. Relative performances of the models were assessed by correlation coefficient (R2), root mean square error of evaluation (RMSEE) and root mean square error of cross validation (RMSEcv). Since extrapolation in regression models can provide biased results, it has been ensured to include the operating concentration of the amino acid concentrations samples in calibration sets.
Table 3.
Summary of model calibration and validation
| A.A. | N | # Factors | R2Y | Q2 | RMSEE | RMSEcv | RMSEP |
|---|---|---|---|---|---|---|---|
| Tyrosine | 56 | 2 | 0.90 | 0.86 | 0.41 | 0.53 | 0.35 |
| Tryptophan | 55 | 2 | 0.82 | 0.78 | 0.24 | 0.27 | 0.07 |
| Phenylalanine | 56 | 2 | 0.79 | 0.64 | 0.35 | 0.47 | 0.32 |
| Methionine | 57 | 2 | 0.21 | 0.01 | 0.27 | 0.29 | 0.68 |
Figure 2 shows model calibration results by comparing the experimental values from the calibration dataset with the values predicted by the developed models. As shown in Table 3, the RMSEE values for developed models are 0.41, 0.24, 0.35, and 0.27 for tyrosine, tryptophan, phenylalanine and methionine, respectively. RMSEcv values for the same models are 0.53, 0.27, 0.47, and 0.29, respectively. The proximity of RMSEE and RMSEcv values is a positive sign indicating model robustness. The models were validated using a validation dataset including data from a separate batch at bench scale. Figure 3 shows the model validation results by comparing the experimental values with the values from model predictions. It is observed that all the developed models, except the methionine model, predict well. R 2 values for tyrosine, tryptophan and phenylalanine models are 0.90, 0.82, and 0.79, respectively, compared to 0.21 for the methionine model, indicating that a significant amount of variability in data was captured by the first three models. The predictive abilities of the models were evaluated from prediction parameter, Q2, and RMSEP. For the developed models, Q2 is 0.86, 0.78, 0.64 and 0.01 respectively and RMSEP for the same models is 0.35, 0.07, 0.32 and 0.68 respectively. The RMSEE, RMSEcv and RMSEP values for all the models are comparable and in the same range, indicating the stability of the developed models. These results show that except for methionine, the predictive power of the developed models is significant.
Figure 2.
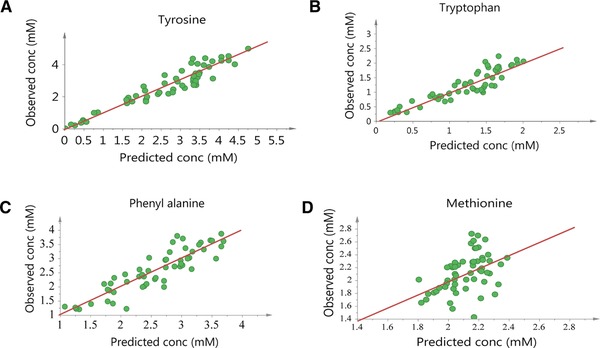
Calibration results showing actual versus predicted concentrations of (A) tyrosine (B) tryptophan (C) phenylalanine and (D) methionine.
Figure 3.
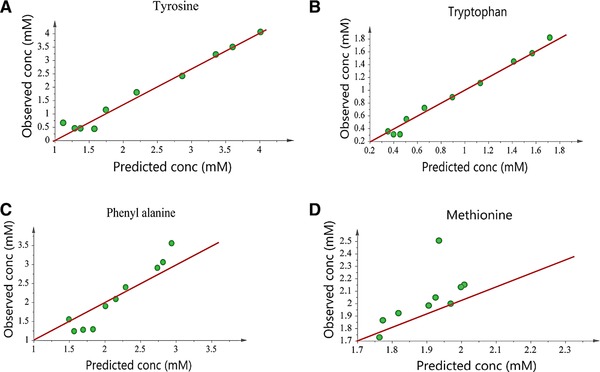
Validation results for Raman models for (A) tyrosine (B) tryptophan (C) phenylalanine and (D) methionine.
The methionine model has an RMSEP value of 0.68 which is larger compared to other models and shows difficulty in quantitation of methionine using Raman‐based chemometric models. This result is also confirmed by low R 2 and Q 2 values (R 2 = 0.21 and Q 2 = 0.01). Our analysis shows that the main reason for this is because methionine does not have any strong Raman peaks and the identified Raman peaks are weak and insignificant as shown in Fig. 1. In addition, the 653, 699 and 723 cm−1 peaks of methionine are very close to cysteine peaks, as both are sulfur‐containing amino acids 33. Moreover, the bands generated by di‐sulphide and C‐S bonds in methionine (655 cm−1 and 724 cm−1), which are supposed to be significant as they are medium signal peaks, are masked by the liberational broad band of water 34. All these factors in combination leads to lower accuracy for methionine models in comparison to the other investigated amino acids. We continue working on improving model predictions for methionine by considering a better combination of preprocessing methods and improving the spectral regions used for model fitting.
4. Concluding remarks
This research presents a platform for a potential application in achieving PAT goals, which are enhancing the understanding, monitoring, and control of manufacturing process by use of real‐time monitoring of amino acids in cell culture media. Over the last few years, Raman spectroscopy has been successfully employed in the biopharmaceutical industry for in‐line monitoring and control of cell culture processes. In‐line monitoring of amino acid concentrations can potentially be used to control the nutrient feed rate and thereby improve process productivity and control of product quality attributes. Accurate Raman‐based calibration models eliminate the need for offline sampling and manual control in manufacturing processes and render more robust and automated processes. Models for measurement of glucose, lactate, glutamine, glutamate, ammonium, osmolality and VCD are previously developed, demonstrating the potential of Raman spectroscopy for real‐time in‐line measurement of these cell culture process parameters. Here, we present a proof of concept for application of the Raman‐based calibration models for real‐time monitoring of amino acids concentration in culture media. The developed models can predict the concentration of three amino acids, namely tyrosine, tryptophan and phenylalanine, with acceptable accuracy, while the developed model for measurement of methionine has lower performance than the other three models. Our analysis shows this is due to the fact that most Raman peaks for methionine are weak peaks, while stronger peaks coincidence with cysteine peaks and di‐sulphide bond peaks are masked by the liberational broad band of water 29, 30.
In summary the results of this study show that the combination of Raman spectroscopy with chemometrics models is a suitable method for real‐time measurement of amino acids in cell culture processes. The presented PAT platform can be seen as a step towards the real‐time monitoring and control of product quality attributes by regulating the amino acid feed in cell culture media 35, 36. The developed models can be further optimized to reduce the errors. Future research shall include the modeling of the rest of the amino acids and the inline control of amino acids in cell cultures.
Practical application
Process analytical technology (PAT) is a paradigm introduced by FDA to ensure the robust and consistent processes in biopharmaceutical industry. On‐line or at –line control method is one key factor to achieve the PAT goals. At line measurement in cell culture processes is gaining importance and has been applied for many cell culture components. This paper presents the feasibility of Raman spectroscopy to monitor the amino acid during the process. The amino acid measurement in cell culture media during the process can potentially reduce the need for off line sampling.
Acknowledgments
The authors have declared no conflict of interest.
5 References
- 1. US‐FDA , Guidance for Industry PAT – A Framework for Innovative Pharmaceutical Development, Manufacturing, and Quality Assurance U.S.D.o.H.a.H. Services, Editor. 2004, Pharmaceutical CGMPs., FDA, Editor. 2004.
- 2. Becker, T. , Hitzmann, B. , Muffler, K. , Pörtner, R. et al., Future aspects of bioprocess monitoring, in white biotechnology, in: Ulber R. and Sell D., (Ed.), Springer, Berlin Heidelberg: 2007, pp. 249–293. [DOI] [PubMed] [Google Scholar]
- 3. Ulber, R. F. J. , Beutel, S. , Optical sensor systems for bioprocess monitoring. Anal. Bioanal. Chem. 2003, 376, 342–348. [DOI] [PubMed] [Google Scholar]
- 4. Card, C. H. B. , Smith, T. , Hirsch, J. , Near Infrared spectroscopy for rapid, simultaneous monitoring of multiple components in mammalian cell culture. BioProcess. Int. 2008, 6, 58–67. [Google Scholar]
- 5. Rogge, P. , Laukel, M. , and Dudziak, G. , Disposable Downstream Processing for Clinical Manufacturing, in Bioprocess International. 2011.
- 6. McGovern, A. C. , Broadhurst, D. , Taylor, J. , Kaderbhai, N. et al., Monitoring of complex industrial bioprocesses for metabolite concentrations using modern spectroscopies and machine learning: application to gibberellic acid production. Biotechnol. Bioeng. 2002, 78, 527–538. [DOI] [PubMed] [Google Scholar]
- 7. Sivakesava, S. I. J. , Ali, D. , Simultaneous determination of multiple components in lactic acid fermentation using FT‐MIR, NIR, and FT‐Raman spectroscopic techniques. Process Biochem. 2001, 37, 371–378. [Google Scholar]
- 8. Sivakesava, S. I. J. , Demirci, A. , Monitoring a bioprocess for ethanol production using FT‐MIR and FT‐Raman spectroscopy. J. Ind. Microbiol. Biotechnol. 2001, 26, 185–190. [DOI] [PubMed] [Google Scholar]
- 9. Cannizzaro, C. , Rhiel, M. , Marison, I. , von Stockar, U. , On‐line monitoring of Phaffia rhodozyma fed‐batch process with in situ dispersive Raman spectroscopy. Biotechnol. Bioeng. 2003, 83, 668–680. [DOI] [PubMed] [Google Scholar]
- 10. Kara, S. , Mueller, J. J. , Liese, A. , Online analysis methods for monitoring of bioprocesses. Chimica Oggi‐chemistry today. 2011, 29, 38–41.
- 11. Adar, F. G. R. , Noonana, J. , Raman spectroscopy for process/ quality control. Appl. Spectrosc. Rev. 1997, 31, 45–101. [Google Scholar]
- 12. Li, B. , Ryan, P. W. , Ray, B. H. , Leister, K. J. et al., Rapid characterization and quality control of complex cell culture media solutions using raman spectroscopy and chemometrics. Biotechnol. Bioeng. 2010, 107, 290–301. [DOI] [PubMed] [Google Scholar]
- 13. Abu‐Absi, N. R. et al., Real time monitoring of multiple parameters in mammalian cell culture bioreactors using an in‐line Raman spectroscopy probe. Biotechnol Bioeng 2011, 108, 1215–1221. [DOI] [PubMed] [Google Scholar]
- 14. Moretto, J. S. J. , Cuellar, M. , Doane, A. , Ryll, T. et al., Process Raman spectroscopy for in‐line CHO cell culture monitoring. Am. Pharmaceut. Rev. 2011. http://www.americanpharmaceuticalreview.com/Featured-Articles/37040-Process-Raman-Spectroscopy-for-In-Line-CHO-Cell-Culture-Monitoring/. [Google Scholar]
- 15. Mungikar, A. K. M. , Use of IN‐line Raman spectroscopy as a non‐destructive and rapid analytical technique to monitor aggregation of a therapeutic protein. Am. Pharmaceut. Rev. 2010. [Google Scholar]
- 16. Xu, Y. , Ford, J. F. , Mann, C. K. , Vickers, T. J. et al., Raman measurement of glucose in bioreactor materials in: Biomedical Sensing, Imaging and Tracking Technologies II. San Jose, CA, 1997. [Google Scholar]
- 17. Mehdizadeh, H. , Lauri, D. , Karry, K. M. , Moshgbar, M. et al., Generic Raman‐based calibration models enabling real‐time monitoring of cell culture bioreactors. Biotechnol. Progress 2015, 31, 1004–1013. [DOI] [PubMed] [Google Scholar]
- 18. Raman, C. K. K. , A new type of secondary radiation. Nature 1928, 121, 501–502. [Google Scholar]
- 19. El‐Gindy, A. and Hadad, G. M. , Chemometrics in pharmaceutical analysis: an introduction, review, and future perspectives. (1060‐3271 (Print)). J AOAC Int. 2012, 95, 609–623. [DOI] [PubMed] [Google Scholar]
- 20. Xia, J. , Psychogios, N. , Young, N. , Wishart, D. S. , MetaboAnalyst: a web server for metabolomic data analysis and interpretation. Nucleic Acids Research 2009, 37, W652–W660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Fan, Y. , Jimenez Del Val, I. , Müller, C. , Wagtberg Sen, J. et al., Amino acid and glucose metabolism in fed‐batch CHO cell culture affects antibody production and glycosylation. Biotechnol. Bioeng. 2015, 112, 521–535. [DOI] [PubMed] [Google Scholar]
- 22. Grant, S. L. , Shulman, Y. , Tibbo, P. , Hampson, D. R. et al., Determination of d‐serine and related neuroactive amino acids in human plasma by high‐performance liquid chromatography with fluorimetric detection. 2006, 844, 278–282. [DOI] [PubMed] [Google Scholar]
- 23. Sentrie, L. M. J. , Boudrant, J. , Germain, P. , Thin‐layer chromatographic method for the simultaneous determination of physiological aromatic amino acids. J. Chromatogr. 1991, 547, 531–537. [Google Scholar]
- 24. Pobozy, E. , Czarkowska, W. , Trojanowicz, M. , Determination of amino acids in saliva using capillary electrophoresis with fluorimetric detection. J. Biochem. Biophys. Methods 2006, 67, 37–47. [DOI] [PubMed] [Google Scholar]
- 25. Blanco, M. C. J. , Iturriaga, H. , Maspoch, S. , Redon, M. , artial least‐squares regression for multicomponent kinetic determinations in linear and non‐linear systems. Anal. Chim. Acta. 1995, 303, 309–320. [Google Scholar]
- 26. Lopez, C. G. R. M. , Ubide, C. , Individual kinetic determinations using partial least squares calibration. Analyst 1997, 122, 519–523. [Google Scholar]
- 27. Read, E. K. , Bradley, S. A. , Smitka, T. A. , Agarabi, C. D. et al., Fermentanomics informed amino acid supplementation of an antibody producing mammalian cell culture. Biotechnol. Prog. 2013, 29, 745–753. [DOI] [PubMed] [Google Scholar]
- 28. Safa, F. , Hadjmohammadi, M. R. , Simultaneous optimization of the resolution and analysis time in micellar liquid chromatography of phenyl thiohydantoin amino acids using Derringer's desirability function. J. Chromatogr. A 2005, 1078, 42–50. [DOI] [PubMed] [Google Scholar]
- 29. Riahi, S. G. M. , Pourbasheer, E. , Divsar, F. , Norouzi, P. et al., Development and validation of rapid chemometrics‐ assisted spectrometry and liquiid chromatography methods for simultaneous determination of phenylalanine, tryptophan and tyrosine in the pharmaceutical products. Curr. Pharm. Anal. 2008, 4, 231–237. [Google Scholar]
- 30. Hiller, G. W. , Mulukutla, B. C. , Method of cell culture. 2017, Google Patents.
- 31. Jenkins, A. L. , Larsen, R. A. , Williams, T. B. , Characterization of amino acids using Raman spectroscopy. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2005, 61, 1585–1594. [DOI] [PubMed] [Google Scholar]
- 32. Zhu, G. , Zhu, X. , Fan, Q. , Wan, X. , Raman spectra of amino acids and their aqueous solutions. Spectrochim. Acta A Mol. Biomol. Spectrosc. 2011, 78, 1187–1195. [DOI] [PubMed] [Google Scholar]
- 33. Lee, H. , Kim, M. S. , Suh, S. W. , Raman spectroscopy of sulphur‐containing amino acids and their derivatives adsorbed on silver. J. Raman Spectrosc. 1991, 22, 91–96. [Google Scholar]
- 34. Eunice Li‐Chan, J. C. , Peter, G. , Applications of Vibrational Spectroscopy in Food Science. Vol. 1 2011: John Wiley & Sons. [Google Scholar]
- 35. Bhatia, H. , Read, E. , Agarabi, C. , Brorson, K. et al., A Design Space Exploration for Control of Critical Quality Attributes of mAb. Int. J. Pharm. 2016, 512. [DOI] [PubMed] [Google Scholar]
- 36. Sha, S. , Agarabi, C. , Brorson, K. , Lee, D. Y. et al., N‐Glycosylation Design and Control of Therapeutic Monoclonal Antibodies. Trends Biotechnol. 2016, 34, 835–846. [DOI] [PubMed] [Google Scholar]


