Abstract
Physical entrapment of enzymes within a porous matrix is a fast and gentle process to immobilize biocatalysts to enable their recycling and long‐term use. This study introduces the development of a biocompatible 3D‐printing material suitable for enzyme entrapment, while having good rheological and UV‐hardening properties. Three different viscosity‐enhancing additives have been tested in combination with a poly(ethylene glycol) diacrylate‐based hydrogel system. The addition of polyxanthan or hectorite clay particles results in hydrogels that degrade over hours or days, releasing entrapped compounds. In contrast, the addition of nanometer‐sized silicate particles ensures processability while preventing disintegration of the hydrogel. Lattice structures with a total height of 6 mm consisting of 40 layers were 3D‐printed with all materials and characterized by image analysis. Rheological measurements identified a shear stress window of 200 < τ < 500 Pa at shear rates of 25 s−1 and 25°C for well‐defined geometries with an extrusion‐based printhead. Enzymes immobilized in these long‐term stable hydrogel structures retained an effective activity of approximately 10% compared to the free enzyme in solution. It could be shown that the reduction of effective activity is not caused by a significant reduction of the intrinsic enzyme activity but by mass transfer limitations within the printed hydrogel structures.
Keywords: 3D‐bioprinting, Biocatalysis, Biocompatible hydrogel, Mass transfer limitations, Physical entrapment
Abbreviations
- BMA
Bentone MA
- DXG
Deuteron XG
- LRD
Laponite RD
- ONP
ortho‐nitrophenol
- ONPG
O‐nitrophenyl‐β‐d‐galactopyranoside
- PEG‐DA
poly(ethylene glycol) diacrylate
1. Introduction
In recent years 3D‐printing has moved beyond its classical applications in architecture and design to become increasingly popular in the fields of biotechnology and bioengineering. Individualized modular concepts such as printable adapters 1 or fittings 2, 3 have been reported for simplification of daily laboratory work. The individualization of biotechnological products by 3D‐printing has mostly been demonstrated in the field of medicine 4, 5, 6, where metal implants or models for surgical planning are tailor‐made by 3D‐printing according to patient data. Another emerging application of 3D‐printing is tissue engineering. For this, artificial materials are combined with biological components such as cells or growth factors to mimic a living system, with the aim of building up artificial organs in the future. Hydrogels are a special focus of ongoing research. They offer an aqueous environment with restricted movement of biological macromolecules, while small substrates and products diffuse through the hydrogel network 7, 8. These properties can be used to stimulate the 3D growth of cells within the material as well as to continuously release small molecules such as pharmaceuticals 9.
The use of enzymes as catalysts for highly specific reactions is a typical application of biological molecules. If the enzymes are cheap, single use batch processes in solution can be used 10, although these typically must be followed by a separation step in order to remove the enzyme from the product 11. When enzymes are not abundantly available, they are often immobilized on surfaces or in porous support material 12. The process of immobilization increases costs, but the reuse in consecutive process cycles saves money in the long term. Enzymes can be immobilized covalently on surfaces such as membranes or particles, but this requires optimization of the immobilization conditions for any change of the matrix‐enzyme combination. The use of tagged enzymes with specific end‐groups that bind to appropriately activated carriers simplifies the search for suitable coupling conditions but requires genetically modified enzymes and in many cases more demanding surface chemistries. Physical entrapment to immobilize enzymes has the disadvantage of possible leakage 13 if the matrix disintegrates and also brings a diffusion limitation that is avoided in solution 14. Nevertheless, the simplicity and universal applicability of enzyme entrapment into hydrogels is attractive, especially if the enzyme‐loaded hydrogel can be 3D‐printed into shapes optimized for the required application. The 3D‐printing of hydrogels with physically entrapped biological components can make the immobilization process itself highly flexible, automatable, and fast in comparison to covalent immobilization. However, the incorporation of enzymes into 3D‐printed highly porous carriers is challenging: for broad applicability, the hydrogel material must be able to withstand harsh chemicals and elevated temperatures that may occur in later reactions 2. In addition, long‐term mechanical stability, easy printability, and fine structures resulting in high porosities are desirable properties that are hard to combine.
Besides biobased choices like alginate or gelatin hydrogels, synthetic PEG‐based hydrogels appear to meet these requirements best. Its biocompatibility has already been demonstrated in the area of tissue engineering 15, where it was shown that the polymer is inert as well as difficult to cleave once reacted. Last but not least, the materials for synthesis can be purchased at reasonable cost. For 3D‐printing, PEG can be functionalized with acrylic side groups and combined with a water‐soluble photoinitiator. This allows the production of PEG hydrogel structures by several 3D‐printing techniques, including inkjet, stereolithographic, and extrusion‐based methods 16, 17, 18, 19, since the material can be photocured locally and in a layer‐by‐layer fashion with UV light within seconds. The most cost‐effective method makes use of simple extrusion, applying single use cartridges that offer a maximum of freedom for the user. When a change in product such as the entrapped enzyme is necessary, the components that were in contact with the processed material can be quickly disposed. However, in order to process poly(ethylene glycol) diacrylate (PEG‐DA) hydrogels with extrusion‐based 3D‐printers, the viscosity of the printing material, also called the “bioink,” has to be increased to avoid dripping and to achieve consistent hardening properties 8, 20. In this context, we present the screening of PEG‐DA based hydrogels with different viscosity increasing additives. Adjusting the viscosity enables extrusion‐based printing of uniform filaments followed by crosslinking via UV‐light. Within a systematic approach, first, the influences of the hydrogel forming components as well as of the shear during printing onto the intrinsic enzyme activity were determined independently. Afterwards, the effective enzyme activity within printed hydrogels is measured and mass transfer effects are estimated by help of the resulting catalyst efficiencies and Thiele moduli.
2. Materials and methods
2.1. Materials
Poly(ethylene glycol) diacrylate (average Mn 700) (PEG‐DA), 2‐hydroxy‐4'‐(2‐hydroxyethoxy)‐2‐methylpropiophenone, O‐nitrophenyl‐β‐d‐galactopyranoside (ONPG), and ortho‐nitrophenol (ONP) were obtained from Sigma‐Aldrich (Darmstadt, Germany). Ethanol, sodium citrate, NaOH, Na2CO3 (all analytical grade), and nitric acid (suprapur grade) were purchased from VWR (Radnor, Pennsylvania, USA). The viscosity‐enhancing materials Laponite RD (BYK‐Chemie GmbH, Wesel, Germany), Deuteron XG (Deuteron GmbH, Achim, Germany), Bentone MA (Elementis UK Ltd. c/o Elementis GmH, Cologne, Germany) were free samples. Deionized water was prepared with a Milli‐Q Ultrapure system from Merck Millipore (Billerica, USA). All chemicals were used as received.
For demonstration of the suitability of the developed 3D‐printable hydrogels to entrap enzymes β‐galactosidase (EC 3.2.1.23) from Aspergillus oryzae was chosen as a model. β‐Galactosidase plays an important role in carbohydrate metabolism, which primarily cleaves lactose into galactose and glucose 7. Industrially, this reaction is used to produce lactose‐free dairy products 21. The enzyme has a broad substrate spectrum and reaction kinetics can conveniently be determined on the artificial substrate ONPG, by cleaving it into galactose and ONP, which is quantifiable by UV‐vis absorption 22.
2.2. Hydrogel preparation
The hydrogel mixture was prepared as follows: deionized water was mixed with viscosity‐enhancing materials and stored for 16 h for equilibration. To this premixture, liquid PEG‐DA, stock solution of the initiator (100 g/L 2‐hydroxy‐4'‐(2‐hydroxyethoxy)‐2‐methylpropiophenone in 70% w/v ethanol in water), and enzyme solution were added. The components were thoroughly mixed by stirring manually with a spatula followed by vortexing. Final concentrations were 73% w/w water, 22.3% w/w PEG‐DA, 2.2% w/w initiator solution and 2.5% w/w enzyme solution (c = 10 g/L). The amount of viscosity‐enhancing particles w/v in the mixture was varied over a range from 0.5 to 7% of the total amount of liquid. Three different particulate additives were used. Bentone MA (BMA) is a hectorite clay of natural origin formed by plate‐like particles with dimensions of 50 nm × 250 nm × 1 nm. Its chemical composition can be approximated as Na0.74 [Mg5.33 Li0.60] (Si7.98 Al0.02) O20F2.69(OH)1.31 according to the manufacturer 23. Laponite RD (LRD) is a silicate that can be used as a synthetic alternative to natural clay, with a formula of (Na+ 0.7 [(Si8 Mg5.5 Li0.3) O20(OH)4]−0.7) and particle dimensions of 25 nm × 25 nm × 0.92 nm 24, meaning that the main dimension of LRD is about one order of magnitude smaller than that of BMA. Both additives have a crystalline structure that can be described by a flat slab with different ionic charges at the sides and the edges, leading to an ordered 3D structure between several particles. Deuteron XG (DXG), in contrast, is a polyxanthan with defined molecular weight of 2 × 106 g/mol 25. Its branched and highly hydrated side chains lead to increased viscosity of the mixture.
2.3. Rheological measurements
Flow curves of the hydrogel mixtures were obtained at 25°C with a Physica MCR 301 plate rheometer with one grooved plate PP25/Q1 D = 25 mm (Anton Paar GmbH, Graz, Austria). The material was initially sheared at a constant shear rate of 1 s−1 for 40 s, after which a logarithmic shear rate gradient between 0.01 and 300 s−1 was applied over a duration of 160 s while measuring the resulting shear stress. The viscous behavior of the hydrogel mixture is time‐dependent because of the additives 26 and the slow radical polymerization of PEG‐DA that happens even in the absence of UV light. Accordingly, representative samples for the residence time in the storage tank of the 3D‐printer were chosen ranging between 20 and 150 min. Rheological data were monitored at relatively low shear rates to model the behavior when the extrusion process is started and the material crosses the threshold to flowing. Flowing at high shear rates, as applied in Blaeser et al. 27, can be approximated, but is not relevant for 3D‐printing, during which defined strands of material are extruded.
2.4. 3D‐printing
For the production of defined hydrogel structures, the desired lattice structures were designed with the software BioCAD (regenHU, Villaz‐St‐Pierre, Switzerland) and printed with a pneumatic extrusion‐based 3D‐printer (3D Discovery, regenHU, Villaz‐St‐Pierre, Switzerland). Briefly, the printing pathway is designed as user‐determined coordinates and the parameters for the printheads are set within the software. The allocated information is converted by the software in the G‐code needed to control the printer, including commands to open and close valves or combine different printheads. Alternatively, the desired geometry can be designed in any custom CAD software and converted into G‐code that is readable by the 3D‐printer.
Extrusion was done through cannulas with an inner diameter of 200 or 250 μm and with a xy‐speed of 35 mm/s across the printing platform. The layer height was adapted to the extruded amount of material between 120 and 150 μm to get defined lattice structures with strand thickness of 700–900 μm, grid width of 1500 μm and varying total heights of the structure. The resulting extrusion pressure depended on the material but was about 0.120 MPa. Each layer was prehardened with the built‐in UV light for 10 s, followed by a complete hardening of 10 min after completing the print. The built‐in UV light has a spot size of 12 mm in diameter, greater geometries were exposed to UV‐light by moving the lamp to several positions followed by 10 min exposition with a VL‐8.L lamp (Vilber Lourmat, Marne‐la‐Vallée, France) to be able to start the next 3D‐print in parallel. A lattice with dimensions of 12 mm × 13.5 mm and a height of 6 mm can be printed within 20 min and hardened completely within 10 extra minutes. The lattice structures were stored in deionized water at 5°C for 18–24 h prior to use. Hereby, the hydrogels could equilibrate in solution to have comparable starting conditions for the kinetic measurements of the enzyme.
2.5. Printing fidelity analysis via morphological characterization
To check the fidelity between the theoretical CAD model and the 3D‐printed lattice, pictures of the structures were taken just after printing. A similar study was done by Kang et al. who tested the settings of the 3D‐printer 17 in terms of extrusion of one strand. The authors in this study concentrated on deviations between design and experiment that were associated with delayed extrusion, e.g. because of pressure increase within the extruder, affecting the line width of extruded strands. In contrast, we put an emphasis on the behavior of the bioink when printed in structures of at least 10 layers on top of each other. The top view of the printed lines is especially relevant, as the spreading of the material and the consistency of the lines can be evaluated. For this, photos of lattice structures with 2.4 mm grid width were converted into 8‐bit graphics and thresholded into black‐and‐white pictures with ImageJ (open‐source software, available on https://imagej.net). The function “plot profile” was applied that approximates an integral of the black pixels of the pore. The slope of the approximated integral is compared to the ideal structure designed on the computer that represents a rectangular function. The resulting relative shape fidelity was highlighted with a color‐coded plot. Grid squares filled with hydrogel were color‐coded with red, as they inhibit the mass transfer by enlarged diffusion distance and irregular flow pattern. Areas with strong printing errors were excluded from the analysis and sorted into an extra category. To analyze the microstructure and surface morphology, hydrogel structures were dried and analyzed with an environmental scanning electron microscope (ESEM; XL30 ESEM‐FEG, Philips, Netherlands). The 3D‐printed hydrogel samples consisting of only a few printed layers were predried at ambient room conditions (22°C, humidity 40%) before being freeze‐dried (Alpha 1–4 LDplus, Martin Christ, Osterode am Harz, Germany) at 5 × 10−3 mbar and gas ballast level 1 for 24 h. Freeze‐dried structures were coated with a silver‐platinum alloy before ESEM measurements.
2.6. Bioreaction kinetics of free and immobilized enzymes
For determination of the batch kinetics, plain enzyme solutions, solutions containing mixtures of free enzyme and individual components of the hydrogel, as well as printed hydrogel structures (weight about 400 mg) with entrapped enzyme were tested regarding their enzymatic activity. The tests were done in 6 mL of citrate buffer (333 mM sodium citrate titrated with NaOH to pH 4.6) containing the substrate ONPG at a concentration of 2.2 mmol/L. Through the catalytic activity of the enzyme, ONPG is hydrolyzed into ONP and galactose. The experiments were done at 37°C in an Eppendorf thermoshaker (Eppendorf, Hamburg, Germany) at 750 rpm. Samples of 25 or 50 μL were taken over a period of one hour. For absorption measurement of ONP at 420 nm in microtiter plates, these aliquots were added to 100 μL of 1 M sodium carbonate solution and citrate buffer was added to a final volume of 200 μL 28. As the effective extinction coefficient may change in presence of solutes and dispersed particles, individual calibration curves for each solvent composition were done in triplicate. To compare the activities, data were normalized with the amount of enzyme used.
2.7. Leaching tests using ICP‐OES analysis
Inductively coupled plasma optical emission spectrometry (ICP‐OES) (Perkin Elmer Optima 8300 DV ICP‐OES, Shelton, CT, USA) was used to detect dissolved metal ions that were released from the hydrogel structure over a period of seven days. For this, printed structures with LRD that had long‐term stability were immersed into 6 mL of citrate buffer (333 mM sodium citrate titrated with NaOH to pH 4.6). Analyses were carried out on the supernatant after removal of the hydrogel structures and appropriate acidification with nitric acid.
Lithium was chosen as a tracer for leaching of additives because it only occurs in the particles of the additive LRD. The buffer system was analyzed as a control group. Further control solutions included the raw materials PEG‐DA, the initiator solution, and pure LRD. For all solutions, a volume similar to that present in the hydrogel structures was used. The concentration of lithium in the samples was determined at three different wavelengths, 267.6, 242.8, and 208.2 nm with a plasma flow of 12 L/min.
3. Results and discussion
The aim of this study was to develop an enzyme friendly PEG‐DA based hydrogel for extrusion‐based bioprinting. The material should be able to permanently entrap enzymes and to be extrusion printed into defined structures, which last long enough to be hardened by UV light in their original form. Within this respect, different additives with defined particle size or molecular weight were tested. The additives were selected based on their properties but also with consideration to their availability in bulk quantities. Three different types of possible additives were used, with two examples of clays and one polysaccharide.
3.1. Rheology
Rheological measurements were done to analyze the shear forces that occur during 3D‐printing through an extrusion‐based printing nozzle. For an optimal extrusion process in a 3D‐printer, shear‐thinning bioinks are used. One factor that is especially important for the extrusion process is the behavior at low shear rates, which determines the flow of the hydrogel mixture in the printhead at the start of the printing process. In this initial phase, it is desirable that high shear forces are required to move the fluid, as this avoids dripping when the printhead is not used. In contrast, at higher shear rates facilitated flow for a continuous extrusion of material is favorable to prevent mechanical damage to the biological components and to enable uniform deposition of the material. Figure 1 shows rheological data of hydrogels with three different concentrations for each additive material. The identified suitable concentrations represent the range of the lower viscosity limit for printing, the range of good printing fidelity and the limit at which the high viscosity of the bioink tends to clog the printing nozzle.
Figure 1.
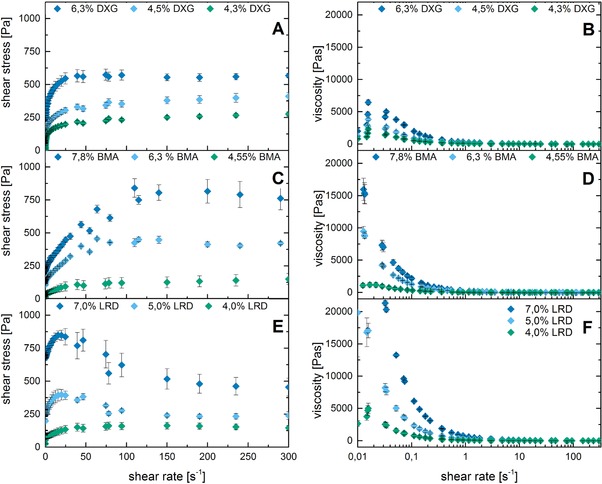
Flow curves of PEG‐DA hydrogel mixtures with different additives to modify the rheological behavior (DXG (A, B), BMA (C, D), LRD (E, F)). All material compositions are shown with three different percentages of additives: the lower value is a composition that is 3D‐printable, but that tends to drip during the printing process, the middle is for optimal printing results, and the high value results in a highly viscous material, which tends to clog the printhead.
The rheological differences between the three materials are noticeable. Although BMA and LRD are both clay particles, their behavior differed significantly. Hydrogels containing LRD show a lower viscosity (1.2 Pa s) at high shear rates than hydrogels containing the other tested additives, resulting in easily flowing material. As a consequence, the printed structures do not retain their shape exactly, especially at 90° corners and edges. However, the low viscosity is an advantage in terms of fast production, as continuous lines can be produced at maximum printing speed of the printhead. LRD particles seem to induce stronger shear thinning behavior and decrease the tendency for clogging of the nozzle even at high particle content compared to the BMA clay particles. This might be explained by the smaller size of LRD particles. The third tested additive, the polyxanthan DXG, is a branched polysaccharide that induces a 3D network stabilized by hydrogen bonds. Hydrogel mixtures with varying additive content of DXG and BMA showed similar shape of their shear stress curve despite the chemical differences. Initial increase in the shear rate produced a steep ascent of the shear stress, after which the curve flattened to a plateau.
The average dynamic viscosity for shear rates from 150 s−1 to 300 s−1 is about 1.7 Pa s for BMA and 1.9 Pa s for DXG at the material composition that was optimal for extrusion‐based 3D‐printing. The printing process results in structures with small strand thickness, defined edges, and highly stable shapes of aspect ratios greater than five, as flowing of the material is minimized compared to LRD with average viscosity of 1.2 Pa s. Comparing the rheological measurements, it can be stated that the hydrogel properties can be altered dramatically by changing the proportion of rheology‐modifying materials, as some of them do not increase viscosity linearly.
3.2. Optical analysis for shape fidelity
For a more detailed view of the hydrogel surface, ESEM measurements were done with samples dried according to Section 2.5. The conventional method of preparing wet samples for ESEM measurements, namely to dehydrate the hydrogels by storing them in solutions with increasing ethanol content up to 100% ethanol as a predrying step was also tested, but was found to be unsuitable: The solvent evaporated almost instantaneously during freeze‐drying and the structures collapsed. A look at the junctions of the printed strands (Fig. 2) shows the degree of confluence between printed strands. While BMA and DXG hydrogels have defined structures with strands that seem to stretch between the linking points, the flow behavior of LRD hydrogels results in aligned junctions where single strands are no longer visible. The results match the rheological data. The photos of the surfaces at higher resolution reveal completely different surface morphologies, depending on the type of viscosity‐enhancing additive used. BMA hydrogels have a large specific surface area, while LRD has serpentine bulges. DXG has a very smooth surface compared to the other materials. It has to be noted that some of the surface appearance is a result of the drying technique, such as the sheets that peel off the LRD structure.
Figure 2.
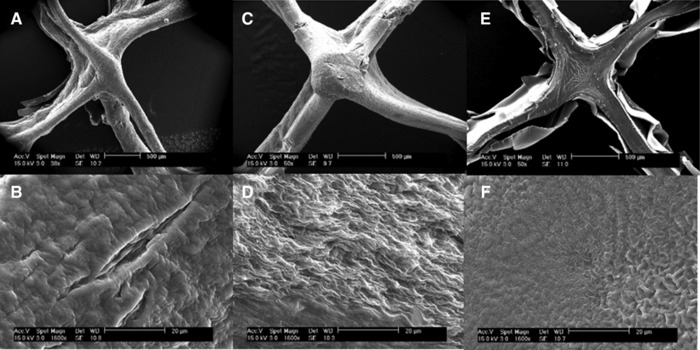
ESEM pictures of printed hydrogel structures with different additives for viscosity enhancement: DXG (A, B); BMA (C, D); LRD (E, F). The upper row shows one junction of a stack of strands printed in an alternating horizontal and vertical mode (scale bar: 500 μm). The lower row shows the magnification of the dried surface (scale bar: 20 μm).
To check the printing quality of the printed structures, top view pictures were compared with the original design (see Fig. 3). In general, the hydrogel structure matches the desired structure better in the center and printing errors occur more often on the edges. This can be partly explained by the deceleration of the printheads at curves of the printing path, which results in an increased material deposition at edges of the structure. All materials are extruded to a lesser extent at the beginning of each layer (upper left corner), as the shear stress in the printhead cannula has to surpass the yield point to start the flow. This may be corrected by pausing the cannula at the starting point for a few milliseconds to preextrude material.
Figure 3.
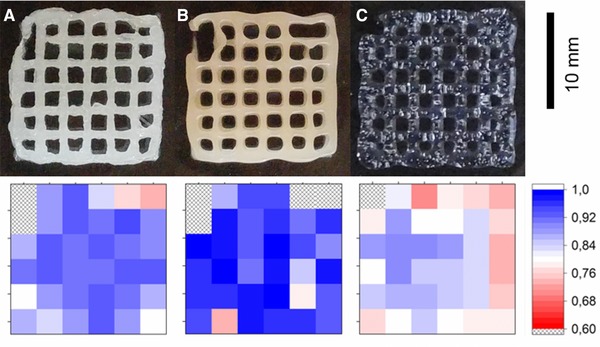
Quality control of the printed lattice structures. The upper row shows top view images of the printed hydrogels using the additives DXG (A), BMA (B), and LRD (C). The bottom row shows the shape fidelity of the comparison with the desired structure, graded with a color code. Blue shades depict a high accuracy of printing, red a low conformity. Areas with printing errors such as hydrogel‐filled pores and broken connections are marked with an “x.”
3.3. Enzyme activity of printed hydrogels with embedded β‐galactosidase
The biocompatibility of the hydrogels and their suitability as enzyme carrier were demonstrated with enzyme kinetic studies using β‐galactosidase and its synthetic substrate ONPG. First, the kinetics was tested for dissolved enzyme in solutions containing the hydrogel precursors. These results were compared to kinetics of hydrogel mixtures before and after UV‐curing. This allowed us to identify the critical process steps for enzyme inactivation as well as the degree of mass transfer limitations.
3.3.1. Biocompatibility of the components of the hydrogel
The individual components of the hydrogels (PEG‐DA, the 70% ethanol used to dissolve the initiator, initiator in 70% ethanol, viscosity‐enhancing additives) were tested to determine their impact on enzyme activity in solution (Fig. 4). For the individual components (PEG‐DA, ethanol, initiator), the same concentrations than the ones in printed hydrogel structures were used (Fig. 4A); for the viscosity‐enhancing additives (Fig. 4B), a low concentration of 0.5% was used to retain a sufficiently low viscosity for mixing and activity tests. The data show a slight reduction of activity when PEG‐DA‐oligomer is added to the solution. The presence of additives normally decreases the effective activity in comparison to the control solution because of decreased mass transfer as noted for the solutions containing BMA or LRD. However, presence of the polyxanthan DXG enhanced ONPG cleavage in comparison to the solution without additives. A possible explanation may be that the active state of the enzyme is stabilized by the surrounding anionic DXG polymer chains, as recently proposed by Zhao et al. 29, where negatively charged DNA‐nanocages resulted in a strongly bound hydration layer that enhanced enzyme activity.
Figure 4.
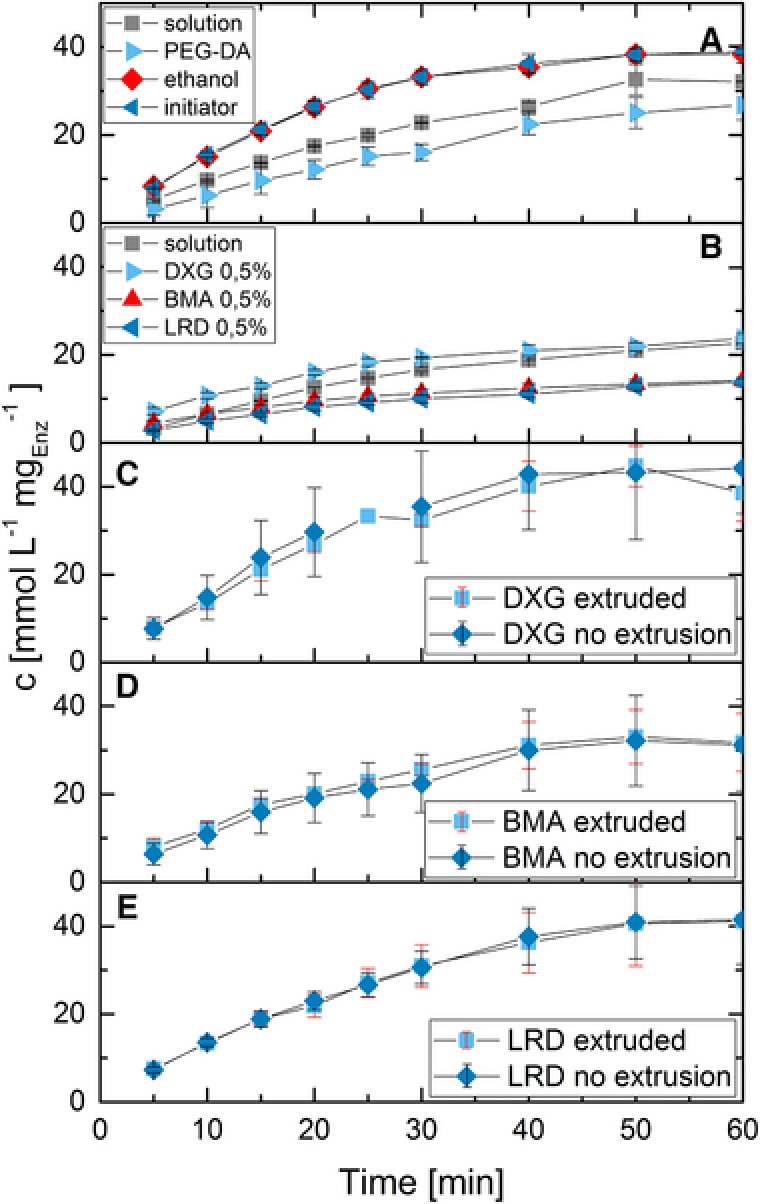
Comparison of enzyme kinetics in hydrogel precursor solutions and hydrogels passing through the extrusion process. (A) Kinetics in the presence of individual components of PEG‐DA hydrogels; (B) Kinetics with the addition of viscosity‐enhancing particles into suspension; (C–E) Activity of enzymes in a ready to use hydrogel mixture before and after extrusion through the printhead of the 3D‐printer. UV‐hardening was omitted and the hydrogel mixture was redissolved in buffer. Resulting concentration of the individual compounds in the runs are as follows (enzyme concentration 10 mg/L in all runs): (A) PEG‐DA 22.3% v/v, ethanol 2.2% v/v, initiator 2.2% v/v; (B) DXG, BMA, and LRD 0.5% w/v. The hydrogel mixtures used in C–E contained 22.3% PEG‐DA, 2.2% initiator solution and 2.5% enzyme stock solution as well as 4.7% DXG (C) respectively 4.7% BMA (D) or 5.0% LRD (E). About 400 mg hydrogel were dissolved per run.
The extrusion of the hydrogel mixture in the printhead of the 3D‐printer exerts shear forces that could potentially denature the enzyme. Figure 4C–E show kinetic data of extruded hydrogel mixtures and control samples that were stored in a closed vial for the time of the extrusion process. Amounts of about 400 mg of those mixtures were resuspended in buffer in order to allow activity tests. No significant differences were detected between the extruded hydrogel and the control. This result is in accordance with previously published results of Blaeser et al., where extrusion of hydrogels was not detrimental for entrapped cells 27. This indicates that the PEG‐DA hydrogel system can be used for biotechnological applications without substantial activity loss due to material properties or the printing procedure.
3.3.2. Mass transfer effects on effective enzyme activity
When operating a biocatalytic process with entrapped enzyme, the productivity is dependent on the activity and accessibility of the enzyme as well as the diffusion coefficient of the substrate and of the product within the hydrogel. To get a first idea of mass transfer limitations, enzyme kinetics was characterized with polymerized hydrogel structures immersed in substrate solution. Comparing the initial slopes of product formation per mg of enzyme in Figs. 4 and 5 it can be seen that the effective activity of ß‐galactosidase entrapped in our 3D‐printed hydrogel structure is roughly an order of magnitude lower than that of the freely dissolved enzyme. The reduction of effective enzyme activity therefore seems to be mainly caused by mass transport limitations after gel formation. In order to verify this hypothesis, 3D‐printed hydrogel lattices were cut with a scalpel into small pieces of about 1 mm3, resulting in an increase of the surface of roughly a factor of two. The amounts of product produced by the respective DXG, BMA, and LRD hydrogel fragments is about 1.5–2.5 times the amount produced with the original lattice samples, which is in line with the increase in surface area of the fragments. In order to get a quantitative approximation of the degree of mass transfer limitation within the hydrogel, the kinetic data of the enzymatic reaction within the hydrogel fragments (Fig. 5) is compared to the one in free solution. The resulting effectiveness factor (normalized catalytic efficiency) is found to be 16, 13, and 13% for DXG, BMA, and LRD (details on the calculation of the effectiveness factor and following characteristic numbers can be found in the Supporting Information). The low effectiveness factor also expresses itself in high numbers of around 20 of the Thiele modulus, describing the ratio between the reaction rate and mass transport due to diffusion in the enzyme carrier. The strong mass transfer limitation is caused by a comparably high enzyme concentration in combination with a reduced substrate diffusivity within the hydrogel. Given the Thiele modulus and first‐order reactions kinetics of substrate limited enzymatic reactions, the effective diffusion coefficient within the hydrogel can be estimated to be in a range of 3–5·10−12 m²/s, which is in the range reported for other hydrogels 30, 31.
Figure 5.
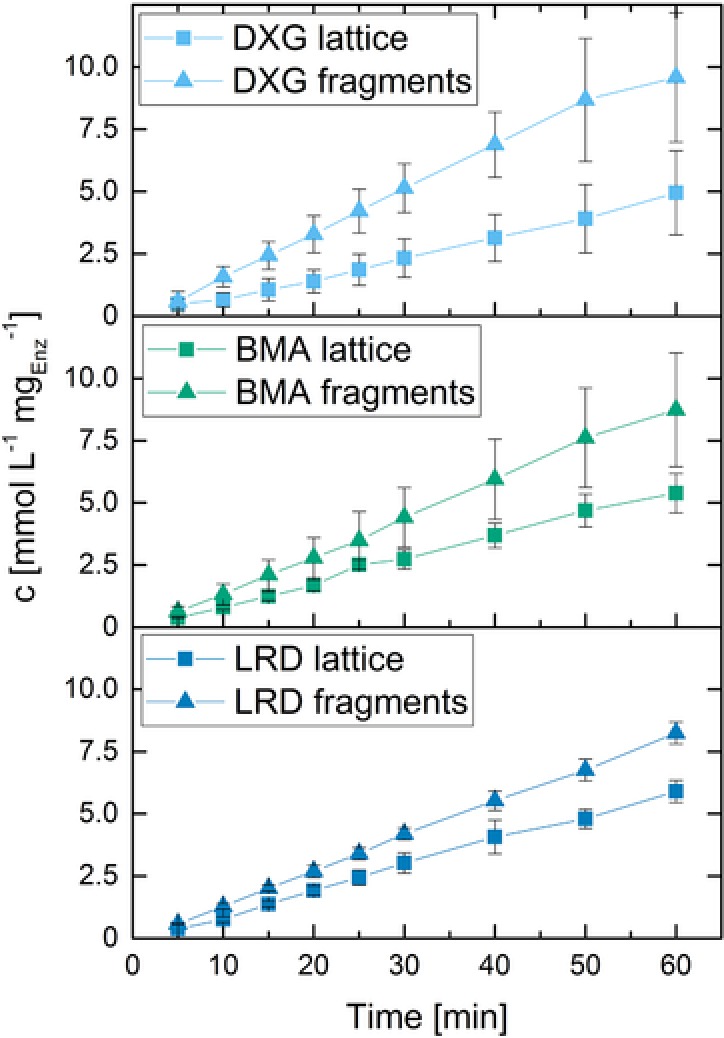
ONP concentration in the supernatant for enzymes entrapped in polymerized hydrogel structures and chopped hydrogel structures. All hydrogels contained 22.3% PEG‐DA, 2.2% initiator solution and 2.5% enzyme stock solution. DXG content was 4.7%, BMA 4.7%, and LRD 5.0%. BMA content is at the lower limit of 3D‐printability to produce stable hydrogel structures.
As written in Section 2.4 the lattice structures were stored in deionized water at 5°C for 18–24 h prior to use. During this time, it can be expected that a fraction of the enzymes located in the region of the hydrogel surface leaches into the storage solution. In order to quantify this fraction, the enzyme concentration in the storage solution is measured using the activity test described in Section 2.6 (see Supporting Information Fig. S3 for the resulting activity plots showing the measured ONP concentration versus time). Product formation caused by released enzymes does not exceed 18.9, 13.4, and 7.7% of the product formation observed for free enzyme in case of hydrogels using BMA, DXG, or LRD as additive. This shows on the one hand, that enzyme leaching is more pronounced for less stable hydrogels, and on the other hand that LRD hydrogels loose less than 8% of the originally entrapped enzyme during storage. Before applying the hydrogel structures in the tests regarding mass transfer limitations discussed above, the storage solution is discarded in order to remove the leached enzyme. Nevertheless, a small quantity of enzyme may leach again influencing the activity tests with hydrogels. However, a worst case estimation shows that this influence is less than 3% and therefore negligible (see Supporting Information).
3.4. Structure stability and additive leaching
Hydrogels with LRD as a rheology modifier result in lattice structures that have long‐term stability in water over a period of weeks. In contrast, structures containing BMA or DXG degraded over a few days. Hydrogels with high content of BMA additive showed especially rapid degradation within hours, making them an interesting candidate for dissolvable support. However, due to our focus on 3D‐printed enzyme immobilisates, our main interest was in long‐term stable structures. We therefore developed a very sensitive way to test for the stability of LRD hydrogels against erosion and dissolution in aqueous environments by monitoring the release of dissolved lithium atoms in storage solution over 7 days (Fig. 6). While the freshly released amount of lithium decreases for longer storage, the total amount of the released lithium after 7 days is about 90% of the material used in the printing. Nevertheless, the lattice structures are mechanically stable and no breakage of the hydrogel strands was observed. This can be explained by the polymerization of PEG‐DA, which forms a nondegradable network that does not break or disintegrate when the LRD particles are released. The high structure fidelity is an advantage for the application of the hydrogels in technical applications, as the mechanically stable system is able to retain embedded enzymes and the blocking of subsequent filters or enzyme carriers by eroded hydrogel fragments can be avoided.
Figure 6.
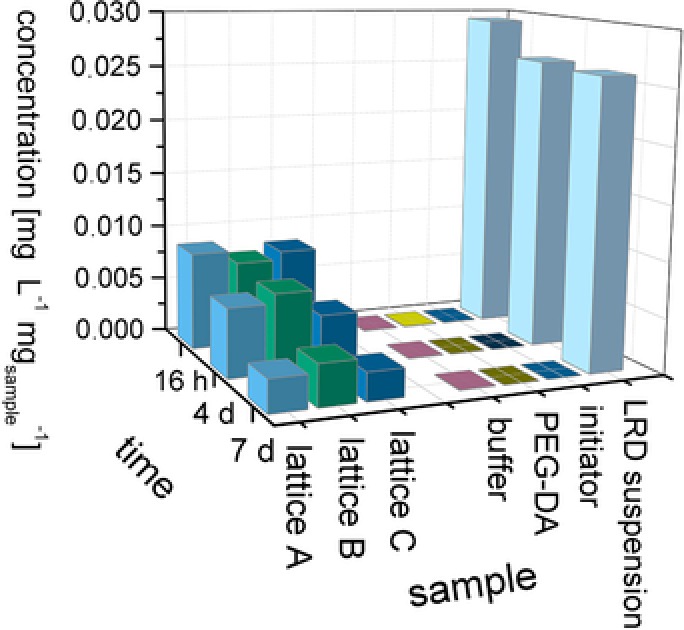
Lithium concentration in the supernatant of different samples at three timepoints. Hydrogel structures A, B, and C (triplicates) immersed in buffer are compared to the respective amount of raw material in solution.
4. Concluding remarks
For an efficient application of 3D‐printed components in bioprocess development, biocompatible materials with defined and stable printing properties are essential. Using a PEG‐DA hydrogel, which can be rapidly hardened by UV‐light as basis, several 3D printing materials were developed and characterized by the use of different viscosity enhancing additives. Varying this component changes the properties of the hydrogel from stable structures to hydrogels that degrade in water within hours. Printing tests in combination with rheological measurements led to the definition of a printhead‐specific operation window in terms of viscous behavior, spanning a shear stress range of 200–500 Pa at an angular frequency of 25 s−1 for extrusion‐based printers with dispensing tips having a 200 μm aperture. Enzyme entrapment into the different hydrogel systems has been tested for the example of the enzyme β‐galactosidase. The hydrogel components themselves had only a minor influence onto the intrinsic enzyme activity, however, effective activity of the enzyme entrapped in the hydrogel structures reached only about 7–10% of the activity of the free enzyme. The reason for this reduction has been identified as mass transfer limitation, resulting in a Thiele modulus of more than 20 and an effective diffusivity within the hydrogel of about 3·10−12 m²/s. In order to improve the effective activity of the entrapped enzymes, either hydrogel structures would have to be decreased to strands having a diameter of around 200 μm, or enzyme concentration within the hydrogel must be strongly reduced. Nevertheless, no disintegration of the LRD hydrogels was observed, making this hydrogel variant widely deployable for enzyme immobilization and long‐term activity tests. Applications could be batch screenings of immobilized enzymes as well as long‐term tests of immobilized enzymes in small flow‐through bioreactors.
Practical application
The use of tailored 3D‐printed components offers a new way to apply flow‐through equipment for small‐scale processes, such as biocatalytic reactions with immobilized enzymes. Prerequisite for the 3D‐printing of enzymatically active parts is a set of new, biocompatible, and user friendly materials. We present the screening and application of such a bioink, which on the one hand does not affect the intrinsic enzyme activity, and on the other hand offers rheological and UV‐hardening properties, which allow reliable printing using commercial extrusion printers. Based on this material 3D‐printed enzymatically active parts can be quickly generated and tested, e.g. for process development of enzymatic cascades. To characterize the bioink, the manuscript delivers rheological data as well as information about the degree of mass transfer limitation of embedded enzymes and how this limitation depends on the intrinsic enzyme activity, the structural dimensions of the printed part and the applied enzyme concentration.
The authors have declared no conflict of interest.
Supporting information
Supporting information.
Acknowledgments
The authors would like to acknowledge the Project “Molecular Interaction Engineering: From Nature's Toolbox to Hybrid Technical Systems,” which is funded by the German Federal Ministry of Education and Research (BMBF), funding code 031A095, for their financial support.
5 References
- 1. Luecking, T. H. , Sambale, F. , Schnaars, B. , Bulnes‐Abundis, D. et al., 3D‐printed individual labware in biosciences by rapid prototyping: In vitro biocompatibility and applications for eukaryotic cell cultures. Eng. Life Sci. 2015, 15, 57–64. [Google Scholar]
- 2. Dragone, V. , Sans, V. , Rosnes, M. H. , Kitson, P. J. et al., 3D‐printed devices for continuous‐flow organic chemistry. Beilstein J. Org. Chem. 2013, 9, 951–959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amrhein, S. , Schwab, M. L. , Hoffmann, M. , Hubbuch, J. , Characterization of aqueous two phase systems by combining lab‐on‐a‐chip technology with robotic liquid handling stations. J. Chromatogr. A 2014, 1367, 68–77. [DOI] [PubMed] [Google Scholar]
- 4. Bose, S. , Vahabzadeh, S. , Bandyopadhyay, A. , Bone tissue engineering using 3D printing. Mater. Today 2013, 16, 496–504. [Google Scholar]
- 5. Mannoor, M. S. , Jiang, Z. , James, T. , Kong, Y. L. et al., 3D printed bionic ears. Nano Lett. 2013, 13, 2634–2639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rengier, F. , Mehndiratta, A. , von Tengg‐Kobligk, H. , Zechmann, C. M. et al., 3D printing based on imaging data: Review of medical applications. Int. J. Comput. Assist. Radiol. Surg. 2010, 5, 335–341. [DOI] [PubMed] [Google Scholar]
- 7. Zhang, Z. , Zhang, R. , Chen, L. , McClements, D. J. , Encapsulation of lactase (β‐galactosidase) into κ‐carrageenan‐based hydrogel beads: Impact of environmental conditions on enzyme activity. Food Chem. 2016, 200, 69–75. [DOI] [PubMed] [Google Scholar]
- 8. Chia, H. N. , Wu, B. M. , Recent advances in 3D printing of biomaterials. J. Biol. Eng. 2015, 9, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Clark, E. A. , Alexander, M. R. , Irvine, D. J. , Roberts, C. J. et al., 3D printing of tablets using inkjet with UV photoinitiation. Int. J. Pharm. 2017, 529, 523–530. [DOI] [PubMed] [Google Scholar]
- 10. Tamborini, L. , Fernandes, P. , Paradisi, F. , Molinari, F. , Flow bioreactors as complementary tools for biocatalytic process intensification. Trends Biotechnol. 2018, 36, 73–88. [DOI] [PubMed] [Google Scholar]
- 11. Sheldon, R. A. , Enzyme immobilization: The quest for optimum performance. Adv. Synth. Catal. 2007, 349, 1289–1307. [Google Scholar]
- 12. Jesionowski, T. , Zdarta, J. , Krajewska, B. , Enzyme immobilization by adsorption: A review. Adsorpt. Int. Adsorpt. Soc. 2014, 20, 801–821. [Google Scholar]
- 13. Mohamad, N. R. , Marzuki, N. H. C. , Buang, N. A. , Huyop, F. et al., An overview of technologies for immobilization of enzymes and surface analysis techniques for immobilized enzymes. Biotechnol. Biotechnol. Equip. 2015, 29, 205–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brady, D. , Jordaan, J. , Advances in enzyme immobilisation. Biotechnol. Lett. 2009, 31, 1639. [DOI] [PubMed] [Google Scholar]
- 15. Pereira, R. F. , Bártolo, P. J. , 3D bioprinting of photocrosslinkable hydrogel constructs. J. Appl. Polym. Sci. 2015, 132, 42458. [Google Scholar]
- 16. Malda, J. , Visser, J. , Melchels, F. P. , Jüngst, T. et al., 25th anniversary article: Engineering hydrogels for biofabrication. Adv. Mater. 2013, 25, 5011–5028. [DOI] [PubMed] [Google Scholar]
- 17. Chia, H. N. , Wu, B. M. , Improved resolution of 3D printed scaffolds by shrinking. J. Biomed. Mater. Res. Part B Appl. Biomater. 2015, 103, 1415–1423. [DOI] [PubMed] [Google Scholar]
- 18. Urrios, A. , Parra‐Cabrera, C. , Bhattacharjee, N. , Gonzalez‐Suarez, A. M. et al., 3D‐printing of transparent bio‐microfluidic devices in PEG‐DA. Lab Chip 2016, 16, 2287–2294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Radtke, C. P. , Hillebrandt, N. , Hubbuch, J. , The Biomaker: An entry‐level bioprinting device for biotechnological applications. J. Chem. Technol. Biotechnol. 2018, 93, 792–799. [Google Scholar]
- 20. Kang, K. H. , Hockaday, L. A. , Butcher, J. T. , Quantitative optimization of solid freeform deposition of aqueous hydrogels. Biofabrication 2013, 5, 035001. [DOI] [PubMed] [Google Scholar]
- 21. Mlichova, Z. , Rosenberg, M. , Current trends of beta‐galactosidase application in food technology. J. Food Nutr. Res. 2006, 45, 47–54. [Google Scholar]
- 22. Miller, J. H. , Experiments in Molecular Genetics—E. coli, Cold Spring Harbor Laboratory, University of Michigan, NY, 1972. [Google Scholar]
- 23. Elementis UK Ltd . c/o Elementis GmbH, BENTONE® MA Rheological Additive for Water‐Borne Systems General Information. URL: http://www.elementisspecialties.com/esweb/webproducts.nsf/allbydocid/D8B3A518BA5D2701852575F200601264/$FILE/Bentone MA: 2014.pdf. 2017. [Google Scholar]
- 24. BYK‐Gardner GmbH , Laponite Performance Additives. Technical Information B‐RI 21.URL: https://www.byk.com/fileadmin/byk/additives/product_groups/rheology/former_rockwood_additives/technical_brochures/BYK_B-RI21_LAPONITE_EN.pdf. 2017.
- 25. DEUTERON GmbH , Deuteron XG Thickening and thixotropic agent for aqueous systems URL: https://www.deuteron.com/wp-content/uploads/pdf-tech/Deuteron-XG-UK.pdf. 2017.
- 26. Willenbacher, N. , Unusual thixotropic properties of aqueous dispersions of Laponite RD. J. Colloid Interface Sci. 1996, 182, 501–510. [Google Scholar]
- 27. Blaeser, A. , Campos, D. F. D. , Puster, U. , Richtering, W. et al., Controlling shear stress in 3D bioprinting is a key factor to balance printing resolution and stem cell integrity. Adv. Healthcare Mater. 2016, 5, 326–333. [DOI] [PubMed] [Google Scholar]
- 28. Brakowski, R. , Pontius, K. , Franzreb, M. , Investigation of the transglycosylation potential of ß‐galactosidase from Aspergillus oryzae in the presence of the ionic liquid [Bmim][PF6]. J. Mol. Catal. B Enzym. 2016, 130, 48–57. [Google Scholar]
- 29. Zhao, Z. , Fu, J. L. , Dhakal, S. , Johnson‐Buck, A. et al., Nanocaged enzymes with enhanced catalytic activity and increased stability against protease digestion. Nat. Commun. 2016, 7, 10619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Cruise, G. M. , Scharp, D. S. , Hubbell, J. A. , Characterization of permeability and network structure of interfacially photopolymerized poly(ethylene glycol) diacrylate hydrogels. Biomaterials 1998, 19, 1287–1294. [DOI] [PubMed] [Google Scholar]
- 31. Evans, S. M. , Litzenberger, A. L. , Ellenberger, A. E. , Maneval, J. E. et al., A microfluidic method to measure small molecule diffusion in hydrogels. Mater. Sci. Eng. C‐Mater. Biol. Appl. 2014, 35, 322–334. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting information.


