Abstract
Imaging of biofilms on opaque surfaces is a challenge presented to researchers especially considering pathogenic bacteria, as those typically grow on living tissue, such as mucosa and bone. However, they can also grow on surfaces used in industrial applications such as food production, acting as a hindrance to the process. Thus, it is important to understand bacteria better in the environment they actually have relevance in. Stainless steel and titanium substrata were line structured and dotted surface topographies for titanium substrata were prepared to analyze their effects on biofilm formation of a constitutively green fluorescent protein (GFP)‐expressing Escherichia coli strain. The strain was batch cultivated in a custom built flow cell initially for 18 h, followed by continuous cultivation for 6 h. Confocal laser scanning microscopy (CLSM) was used to determine the biofilm topography. Biofilm growth of E. coli GFPmut2 was not affected by the type of metal substrate used; rather, attachment and growth were influenced by variable shapes of the microstructured titanium surfaces. In this work, biofilm cultivation in flow cells was coupled with the most widely used biofilm analytical technique (CLSM) to study the time course of growth of a GFP‐expressing biofilm on metallic surfaces without intermittent sampling or disturbing the natural development of the biofilm.
Keywords: biofilm, flow cell, microstructure, stainless steel, titanium
Abbreviations
- 3D
three‐dimensional
- CLSM
confocal laser scanning microscopy
- GFP
green fluorescent protein
- PHLIP
phobia laser scanning microscopy imaging processor
1. INTRODUCTION
Microorganisms present in natural ecosystems usually do not exist as single planktonic cells; they prefer to live in communities attached to surfaces or to each other. This community is commonly referred to as biofilm 1. In general, solid–liquid and air–liquid interfaces have been found to be optimal locations for microbial attachment and biofilm formation. Biofilms have many advantages such as protection of the cells from the environment, nutrient availability, and metabolic cooperativity as well as acquisition of new genetic traits (horizontal gene transfer) 2. Cells within the biofilm can develop resistance to antimicrobial substances, heavy metals and other xenobiotics. Mitra et al. 3 reviewed the ecological roles and biotechnological applications of intertidal microbial biofilms like production of antimicrobial or antifouling compounds, exopolysaccharides, pigments, vitamins, and biotechnologically important enzymes. Immobilization of cells in the form of biofilms can be beneficial for various bioprocesses such as wastewater treatment 4, environmental bioremediation 5 or microbial fuel cells 6. It has been discovered that the surface topography of the substratum also plays an important role in microbial cell attachment to the surface 7, 8, 9. Aluminum, copper, and stainless steel were reported to be favorable for microbial cell attachment and biofilm formation for certain species 10. Another example is titanium, which exhibits several advantages such as corrosion resistance, low specific weight, and low toxicity. Moreover, the formation of an oxide film (TiO2) over the surface makes titanium highly biocompatible 11. Additionally, substrata have been examined for several surface microstructures and modifications such as changes of surface roughness and wettability 12, 13, 14.
Confocal laser scanning microscopy (CLSM) is a powerful and the most widely applied tool for non‐invasive three‐dimensional (3D) sectioned imaging of microbial biofilms 15. It can also be used in combination with other techniques such as catalyzed reporter deposition fluorescence in situ hybridization (CARD‐FISH) to monitor bacterial communities 16. In contrast to classical methods, which relied on offline sample processing 17, a continuous approach provides an opportunity to study the development of microbial biofilms in an undisturbed state. Previous works of Crusz et al. 18 and Tolker‐Nielsen and Sternberg 19, 20 described flow cell biofilm cultivation in combination with CLSM analysis for transparent substrata. However, techniques for continuous monitoring of biofilm growth on structured non‐transparent metallic substrata embedded in flow cells have not yet been described. Such a technique would be very useful in light of the observed effects of different metallic surfaces on diverse biofilm growth patterns. The effects of equally structured stainless steel and titanium substrata as well as differentially structured titanium substrata on biofilm growth were examined in the flow cell system. Here we present the coupling of CLSM, the most widely applied biofilm analytical technique to date, with flow cell cultivation for continuous monitoring of biofilm growth of the fluorescent GFPmut2 E. coli strain on opaque surfaces.
PRACTICAL APPLICATION
In this study a novel way of assessing biofilm growth patterns on opaque metallic surfaces is reported. This method provides a valuable tool for monitoring biofilm growth on industrially and medically used metallic materials such as V4A stainless steel and cp‐titanium. With further refinement of the method, it allows for precise online monitoring of biofilm growth on these surfaces. Additionally, it provides insight into how biofilms behave on differentially structured surfaces, thus allowing to modify surfaces in a way to either increase biofilm productivity and/or thickness or inhibit biofilm growth for medical applications.
2. MATERIALS AND METHODS
2.1. Reactor construction
A custom‐designed flow cell previously described in Schlegel et al. 21 was used for cultivation (Figure 1). The flow cell consisted of a stainless steel frame (1.4571, EN X6CrNiMoTi17‐12‐2) with polyether ether ketone embedded stainless steel nozzles (d = 1.5 mm; 1.4571, EN X6CrNiMoTi17‐12‐2), the substratum of choice (available surface for growth: 200 mm2) and a glued‐on cover slip (Hecht‐Assistent 20 × 60 mm controlled thickness 0.17 mm ± 0.01 mm, Glaswarenfabrik Karl Hecht GmbH & Co KG, Sondheim/Rhoen, Germany). Twinsil addition‐curing duplicating silicone (Picodent, Wipperfürth, Germany) was used as biocompatible, gas‐tight, and autoclavable adhesive. The flow channel had a volume of 1.76 mL (16 × 2 × 55 mm, W × H × L) and provided a homogeneous flow of up to 5 mL/min, equaling a flow velocity of 8 × 10‐2 m/s 14. This was shown via computational fluid dynamics simulations.
Figure 1.

Custom‐made stainless steel flow‐cell with cover slip used in the present study. The substratum sample is placed in the well in the middle of the chamber. Dimensions of the flow channel are 16 × 2 × 55 mm, W x H x L
2.2. Micro‐structuring of surfaces
Substrates of titanium (commercially pure, Grade 2, type 3.7035/34, ISO 5832‐2) and stainless steel (1.4571, EN X6CrNiMoTi17‐12‐2) were microstructured by micro milling and different shapes and sizes of surface modifications were produced. Micro milling was carried out in a two‐step process with a high‐precision three‐axis computer numerical control milling machine developed at the Institute for Manufacturing Technology and Production System at the TU Kaiserslautern, Germany 8 22. The manufacturing process was described in detail by Fingerle et al. 23.
2.3. Determination of surface properties of the substrata
Tactile determination of surface roughness (R a) was carried out with the roughness probe TKU 300, HommelEtamic T 8000 (Jenoptik, Jena, Germany). The determination of the contact angle (Θ) for evaluation of hydrophobicity was determined by the sessile drop method at the Department of Physics and Research Center OPTIMAS, Kaiserslautern, Germany. The contact angle measuring system G2 (Krüss, Hamburg, Germany) was used to place a 3 μL drop of deionized water on the samples. An image of the drop was taken, whereupon a circle detection program (Drop Shape Analyzer, Krüss, Hamburg, Germany) analyzed the droplet shape and measured the angle between the surface and the droplet.
2.4. Microorganism
In all experiments, an E. coli strain constitutively expressing GFPmut2 was used for biofilm cultivation in flow cells. Electrocompetent E. coli XL‐1 Blue (recA1 endA1 gyrA96 thi‐1 hsdR17 supE44 relA1 lac[F´ proAB lacIqZ∆M15 Tn10 (Tetr)]) obtained from Promega, Mannheim, Germany, was used for transformation of the recombinant plasmid pBAD33 (ATCC/LGC Standards, Wesel, Germany) which encodes the gene for GFPmut2 24. Addition of l‐arabinose induces transcription of PBAD 25 thus expressing GFPmut2, which can be excited at 488 nm. Emission can be measured at 500–530 nm. After transformation and selection, cryostocks were prepared, flash‐frozen in liquid nitrogen and stored at −80°C until use.
2.5. Cultivation medium and culture conditions
All chemicals were reagent grade and purchased from Carl Roth GmbH + Co. KG (Karlsruhe, Germany) or Sigma‐Aldrich (St. Louis, USA) unless stated otherwise. For growth and cultivation, TB medium (pH 7.4) was used, containing: tryptone 12 g/L, yeast extract 24 g/L and glycerol 4 mL/L. After autoclaving and cooling, sterile phosphate buffer (0.17 M KH2PO4 and 0.72 M K2HPO4) was added to an end concentration of 100 mM. Precultures were prepared by growing the cryo‐preserved cells in 30 mL TB medium containing 30 μg/mL chloramphenicol at 37°C and 160 rpm in an orbital shaker for 7 h. The connection was made by autoclavable silicone tubing (d = 1.6 mm, Idex Health and Science, Lake Forest, USA). For biofilm cultivation, the flow cells were first run as a batch culture in a loop (see Figure 2, dotted line). The corresponding P & ID is shown in Figure 2. Handling of all components and media as well as inoculation and cultivation was carried out under strict sterile or monoseptic conditions, respectively. Batch processes were carried out in 50 mL TB medium containing 30 μg/mL chloramphenicol in 250 mL conical flasks (flask B). Cultivation was started by inoculation with mid‐exponential cells to an OD600 of 0.01. A downstream integrated rotating displacement pump (IPC‐N 8 peristaltic pump; Idex Health and Science) was used and the flow rate was maintained at 5 mL/min with an intermittent operation (15 min pumping, 60 min break) for cell attachment and biofilm formation 21, 26. Flask B was incubated at 37°C and 120 rpm in a shaking incubator for approximately 3 h until the OD600 reached 0.8. Then, expression of GFPmut2 was induced by sterile addition of l‐arabinose to a final concentration of 0.2 mg/mL to the culture. Afterwards, the batch cultivation was operated at 30°C for 15 h to allow for O2 dependent maturation of the produced GFP. Hereafter, the setup was switched to the continuous process through a sterile mounted single‐use three‐way valve (Discofix, B. Braun Melsungen AG, Melsungen, Germany). Fresh medium was supplied at a constant flow rate of 1 mL/min from a separate feed vessel (flask C) (see Figure 2, solid line). Flask C was connected with a main feed bottle F via silicone tubing, to maintain an uninterrupted flow of the fresh medium. The entire setup of flask B and C was placed in a shaking incubator at 37°C and 120 rpm. The flow cell (Figure 2, FC) was kept in an incubation chamber at 37°C equipped with the confocal laser scanning microscope. The outlet of the flow cell was switched to a waste bottle (Figure 2, W) through another sterile three‐way valve for the continuous process. Two sets of experiments were performed to study the comparative analysis of biofilm growth between the micro‐milled line structured stainless steel (SS_line) and line structured titanium substrata (T_line1) as well as between line structured titanium (T_line2) and dot structured titanium substrata (T_dot) both with variable interspace (Table 1). All the substrata were successively cleaned with acetone, isopropanol, and ultrapure water for 10 min in an ultra‐sonic bath at maximum energy input. The titanium samples were treated with oxygen plasma to remove residual organic components according to 23. Each set of experiments was performed as biological triplicate.
Figure 2.
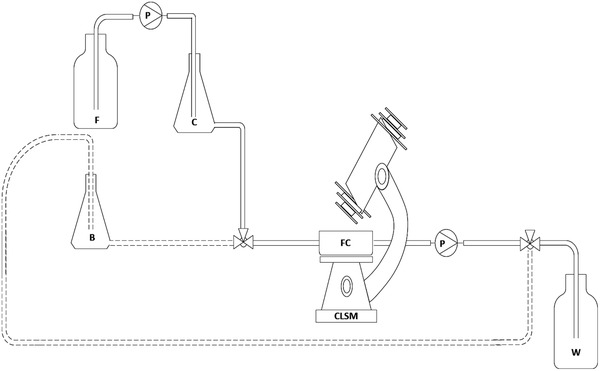
P&I diagram for analysis of biofilm growth in the flow cell cultivated in batch (dotted line) and continuous process (solid line); B: batch flask; C: continuous flask; F: feed bottle; FC: flow cell; P: pump; W: waste; CLSM: confocal laser scanning microscope
Table 1.
Structural details and surface properties of micro‐milled stainless steel and titanium substrata used in this study
| Metallic substrate | Structural details | Mean roughness R a (μm) with error | Contact angle Θ (°) with error |
|---|---|---|---|
| SS_line | Longitudinally parallel, 56 × 5 μm (W × D), 250 μm interspace | 49.1 ± 1.9 | 86.7 ± 4.1 |
| T_line1 | Longitudinally parallel, 50 × 5 μm (W × D), 250 μm interspace | 33.5 ± 4.4 | 56.0 ± 5.8 |
| T_line2 | Longitudinally parallel, 50 × 5 μm (W × D), 50/100/250/500 μm interspace (4‐array) | 35.6 ± 3.3 | 46.1 ± 14.3 |
| T_dot | Pockets, 5 μm deep, 50/100/250/500 μm interspace (4‐array) | 334.1 ± 15.4 | 45.2 ± 14.8 |
2.6. Analysis of biofilm growth
Biofilm growth was monitored with a Leica SP5 II confocal laser scanning microscope equipped with HCX PL Fluotar 10x/0.30 NA dry objective and a XYZ electronic translator platform to ensure reproducible measurements. The used magnification provides a wide field of view, thus allowing to scan a larger area of the sample at once to determine the biofilm coverage area. Image stacks were taken from the substratum surface up to a suitable height in order to record the complete z‐dimension of each biofilm. LAS AF software was used to export the data into individual image files (JPEG/TIFF). During the continuous monitoring, the flow cell was kept on the stage of the microscope. Biofilm growth on each substratum was monitored every hour for 6 h of continuous cultivation. Reflection mode at 488 nm was used to determine the surface position. Biofilm cells were studied by detecting the GFPmut2 expression with excitation at 488 nm and emission at 500–530 nm. All image stacks were taken under identical conditions at a system optimized auto generated z‐interval of 2.39 μm. After the CLSM measurement, the image stacks were post processed with the ImageJ 1.51 (US National Institutes of Health, Bethesda, MD, USA, http://imagej.nih.gov/ij/) software to obtain a complete depiction of biofilm development on different surfaces. The 3D biofilm architectural parameters such as mean thickness, substratum coverage, and roughness coefficient were determined by Phobia Laser Scanning Microscopy Imaging Processor (PHLIP) software (http://phlip.sourceforge.net) 27. The image stacks were processed using Auto‐PHLIP‐ML (http://sourceforge.net/projects/auto-phlip-ml). Biofilm mean thickness, representing the spatial size and upper extent of the biofilm, was calculated as the average distribution of maximum pixel height in z‐direction of the image stack 27, 28. Substratum coverage represents the area occupied by biomass in the first image stack and denotes how efficiently bacteria colonize the substratum 29. The roughness coefficient provides a measure of variations in biofilm thickness and is an indicator of biofilm heterogeneity 30. Data were exported in HTML format and statistically analyzed using Origin 2016G. ImageJ with Volume Viewer 1.31 plugin (https://imagej.nih.gov/ij/plugins/volume-viewer.html) was used for processing of generated image stacks 5, 31.
3. RESULTS
3.1. Surface properties of substrata
Deterministically patterned surfaces of titanium, as well as stainless steel with defined microstructures, were fabricated through micro milling, as well as polishing to different grades of roughness. The values of mean roughness and contact angle for all substrata are given in Table 1. Compared to the microstructured titanium substrata, the stainless steel surface was found to be significantly more hydrophobic.
3.2. Biofilm formation and determination of biofilm architectural parameters
In this study, CLSM‐based continuous monitoring of biofilm growth on non‐transparent metallic surfaces is reported for the first time. To enable proper penetration of the laser beam necessary for CLSM studies, the batch process was shifted to a continuous process in which fresh medium was supplied at a flow rate of 1 mL/min in order to minimize artifacts and disturbances such as increased turbidity due to planktonic cells. Biofilm growth underwent 6 h of continuous cultivation. No significant differences in the development of the biofilm and its 3D architectural parameters were found between the line structured stainless steel (SS_line) and titanium surfaces (T_line1). In the beginning of the continuous phase, biofilm thickness was 135 ± 43 μm for SS_line and 132 ± 73 μm for T_line1 substrata. For titanium, this did not increase significantly with time, while for stainless steel no significant changes in biofilm thickness were observed during the first 5 h of cultivation. Final values for biofilm thickness were 246 ± 28 μm and 260 ± 61 μm, respectively, after 6 h of continuous cultivation (Figure 3). Substratum coverages on SS_line and T_line1 surfaces were 34 ± 23% and 46 ± 31% at 0 h, respectively. This remained comparable over 6 h of cultivation (60 ± 10% and 62 ± 15%, respectively) (Figure 3). Roughness coefficients for the biofilms grown on stainless steel and titanium surfaces showed a decrease over time, indicating a more homogeneous biofilm was progressively developing. The values were 0.48 ± 0.05 μm and 0.40 ± 0.12 μm initially at 0 hours and 0.14 ± 0.06 μm and 0.17 ± 0.04 μm at 6 h for SS_line and T_line1 surface respectively (Figure 3).
Figure 3.
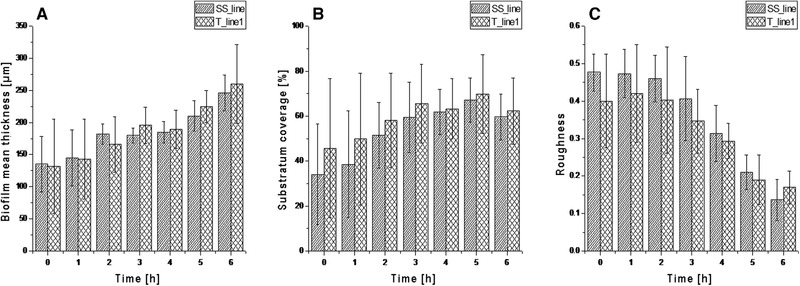
3D biofilm architectural parameters with respective standard deviations (n = 3) for the biofilms grown on stainless steel (SS_line) and titanium (T_line1) substrata. A being the biofilm mean thickness, B the surface coverage and C the biofilm roughness. Data were extracted from CLSM images taken with Leica SP5 II, HCX PL Fluotar 10x/0.3 NA dry objective
It was observed that E. coli_GFPmut2 attaches and develops robust biofilms on both stainless steel and titanium substrata. Additionally, prominent irregular voids, which are believed to be water or nutrient channels 32 were observed on both surfaces (Figures 4 and 6); therefore, sufficient oxygen supply was assumed 33. Expression of GFPmut2 was shown for both surfaces.
Figure 4.
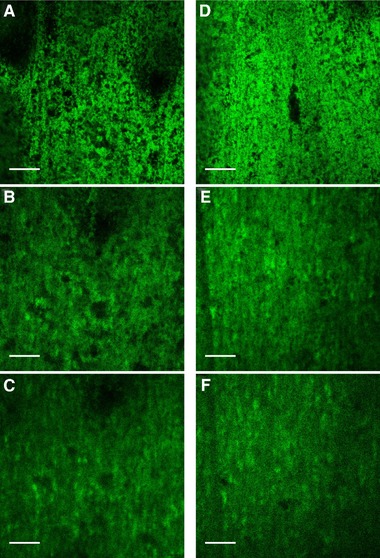
CLSM images of E. coli biofilms observed for SS_line (A‐C) and T_line1 (D‐F) after 0 h, 4 h and 6 h of continuous cultivation in flow cell, respectively. Images were taken of one distinct spot using a Leica SP5 II, HCX PL Fluotar 10x/0.3 NA dry objective. Scale bars indicate 250 μm
Figure 6.
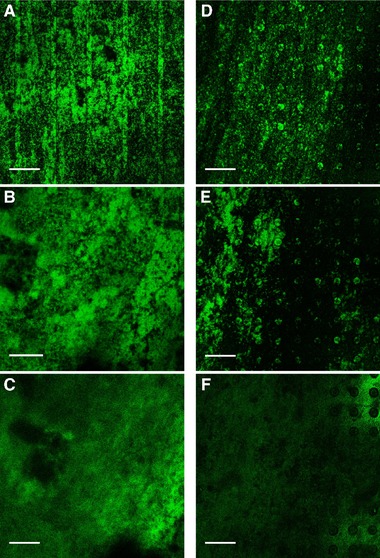
CLSM images of E. coli biofilms observed for T_line2 (A‐C) and T_dot (D‐F; 100 μm interspace) after 0 h, 2 h and 6 h of continuous cultivation in flow cell, respectively. Images were taken of one distinct spot using a Leica SP5 II, HCX PL Fluotar 10x/0.3 NA dry objective. Scale bars indicate 250 μm
Differentially microstructured titanium substrata were also analyzed for biofilm formation by E. coli_GFPmut2. From the PHLIP analysis, it was inferred that the mean thickness and substratum coverage of the biofilms were significantly higher on the line structured surface (T_line2) than the dot structured surface (T_dot). Initially at 0 h, biofilm mean thickness was 164 ± 28 μm and 86 ± 44 μm on T_line2 and T_dot surfaces, respectively. The biofilms became thicker with time on the T_line2 with a final value of 302 ± 25 μm at 6 h, but the thickness did not increase significantly with time on the T_dot surface with a final value of 85 ± 22 μm at 6 h (Figure 5). From the start of the continuous cultivation up to 1 h, substratum coverage was similar for the lined and dotted substrata with initial values of 39 ± 18% and 26 ± 1%. Over time, the value of the said parameter increased significantly for the T_line2 with a final value of 65 ± 7% after 6 h. No significant increase was observed for the T_dot substratum, with a final value of 29 ± 12% (Figure 5). The roughness coefficient for the biofilms grown on the line structured substratum was initially 0.47 ± 0.03 and showed a significant decrease with time (0.18 ± 0.09 after 6 h), signifying the development of a stable and homogenous biofilm. In contrast, biofilm formation was more heterogeneous on the dotted substratum, as shown by the respective roughness value of 0.54 ± 0.06 at 0 h and 0.45 ± 0.17 at 6 hours (Figure 5). It is evident from the PHLIP as well as ImageJ analyses (Figure 6) that biofilm formation was favored on the T_line2 and not on the T_dot substratum. The biofilms were evenly spread and growth occurred consistently with time on the T_line2. On the contrary, the latter surface was not conducive to biofilm formation as evidenced by significantly less biofilm accumulation (Figure 6).
Figure 5.
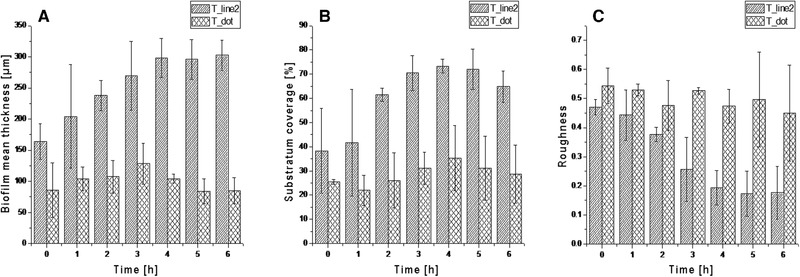
3D biofilm architectural parameters with respective standard deviations (n = 3) for the biofilms grown on lined (T_line2) and dotted (T_dot) titanium substrata. A being the biofilm mean thickness, B the surface coverage and C the biofilm roughness. Data were extracted from CLSM images taken with Leica SP5 II, HCX PL Fluotar 10x/0.3 NA dry objective
4. DISCUSSION
This study presents an approach for continuous monitoring of fluorescent E. coli biofilms during cultivation on various metallic substrata, as well as the assessment of their structure from CLSM data. While each of the techniques used in this study has been independently described before 20, 34, this is the first time that they have been combined to study biofilm growth on metallic substrata continuously during cultivation. Analyzing growth on metallic surfaces is a challenge by itself due to the highly reflective surface and the fact that the substrata are usually opaque, only allowing them to be observed from above. This is further complicated by the aim of monitoring living biofilms, which in this study were grown in custom‐built flow cells and thus posed other challenges in developing the experimental setup with several limitations (such as microscopy with lower magnification due to flow chamber height). In spite of these challenges, we successfully demonstrated the applicability of the system by continuously monitoring the growth of GFP‐expressing E. coli biofilms in flow cells on metallic substrata of different type and structure.
Previous studies on other microorganisms have proven an influence of the surface topography and/or the material used on attachment and growth, as well as production of compounds by biofilms 21, 35, 36. Furthermore, Singh et al. 37 suggested a relationship between surface roughness and bacterial attachment in that they are inversely related beyond a specific threshold. The dotted titanium surface in our study had the highest mean roughness of the substrata used and the lowest biofilm growth with approximately half the values for thickness and surface coverage of the biofilms grown on the line structured titanium surface 2 and higher biofilm roughness, indicating stunted growth on that surface. This could be due to the irregularity of the surface, generating small turbulences that make it harder for the bacteria to attach. The hydrophobicity of the surfaces seems to have no influence in this case, as the contact angles were almost identical for the line structured titanium surface 2 and the dotted titanium surface. Considering the properties of stainless steel and cp‐titanium, this was expected. When comparing biofilm growth on line structured surfaces of stainless steel and titanium (SS_line and T_line1), no significant differences in either biofilm thickness, surface coverage, or biofilm roughness were detected, even though the materials differ in surface roughness and hydrophobicity, further supporting this assumption. A similar observation has been made by Koseki et al. 38 for S. epidermidis biofilms, where the biofilm coverage rates were almost identical on both stainless steel and titanium after 6 h cultivation time.
As indicated, the general biofilm structure regarding channeling, voids and porosity justified the assumption that oxygen as well as nutrients were not limited, as shown by, e.g., Lewandowski and de Beer et al. 32, 33.
The apparent influence of surface topography on cell attachment and biofilm growth of various bacteria, including E. coli, has been shown for other microtopographies and surface modifications 39, 40, 41, corroborating the present findings that biofilm growth on identically structured stainless steel and titanium do not differ. The present results indicate that the topography is the main factor in biofilm growth and development. For example, cells attach preferably to grooves and crevices 42. In accordance to the findings of Mitik‐Dineva et al. 42, this was also shown for the dotted surface T_dot, but subsequent biofilm development was inhibited compared to the line structured titanium. A possible explanation may be the occurrence of microturbulences at those pits. All line structured substrata are oriented in the direction of media flow, which ensures a steady, laminar flow all throughout the flow cell (flow cell CFD analysis, e.g., in 14). Future studies should include substrata with perpendicular grooves to investigate the influence of surface structuring on micro fluid mechanics and thus biofilm development.
Overall, this approach enables researchers from basic to medical research and industrial applications to further their understanding of biofilm development on opaque surfaces.
CONFLICT OF INTEREST
The authors have declared no conflicts of interest.
ACKNOWLEDGMENTS
This work was supported by the German Research Foundation (DFG, Grant UL 170/14‐1) and within the Collaborative Research Center (SFB) 926 on “microscale morphology of component surfaces” (MICOS).
Kleine D, Chodorski J, Mitra S, et al. Monitoring of biofilms grown on differentially structured metallic surfaces using confocal laser scanning microscopy. Eng Life Sci. 2019;19:513–521. 10.1002/elsc.201800176
REFERENCES
- 1. Costerton, J. W. , Lewandowski, Z. , de Beer, D. , Caldwell, D. , Korber, D. , James, G. , Biofilms, the customized microniche. J. Bacteriol. 1994, 176, 2137–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davey, M. E. , O'toole, G. A. , Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 2000, 64, 847–867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Mitra, S. , Sana, B. , Mukherjee, J. , Ecological roles and biotechnological applications of marine and intertidal microbial biofilms. Adv. Biochem. Eng. Biotechnol. 2014, 146, 163–205. [DOI] [PubMed] [Google Scholar]
- 4. Jensen, H. , Biggs, C. A. , Karunakaran, E. , The importance of sewer biofilms. WIREs Water. 2016, 3, 487–494. [Google Scholar]
- 5. Mitra, S. , Pramanik, A. , Banerjee, S. , Haldar, S. et al., Enhanced biotransformation of fluoranthene by intertidally derived Cunninghamella elegans under biofilm‐based and niche‐mimicking conditions. Appl. Environ. Microbiol. 2013, 79, 7922–7930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Read, S. T. , Dutta, P. , Bond, P. L. , Keller, J. , Rabaey, K. , Initial development and structure of biofilms on microbial fuel cell anodes. BMC Microbiol. 2010, 10, 98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Edwards, K. J. , Rutenberg, A. D. , Microbial response to surface microtopography: the role of metabolism in localised mineral dissolution. Chem. Geol. 2001, 180, 19–32. [Google Scholar]
- 8. Whitehead, K. A. , Colligon, J. , Verran, J. , Retention of microbial cells in substratum surface features of micrometer and sub‐micrometer dimensions. Coll. Surf. B. 2005, 41, 129–138. [DOI] [PubMed] [Google Scholar]
- 9. Whitehead, K. A. , Rogers, D. , Colligon, J. , Wright, C. , Verran, J. , Use of the atomic force microscope to determine the effect of substratum surface topography on the ease of bacterial removal. Coll. Surf. B. 2006, 51, 44–53. [DOI] [PubMed] [Google Scholar]
- 10. AlAbbas, F. M. , Kakpovbia, A. , Olson, D. L. , Mishra, B. , Spear, J. R. , The role of bacterial attachment to metal substrate and its effects on microbiologically influenced corrosion (MIC) in transporting hydrocarbon pipelines, in: Narayan R., Bose S., Bandyopadhyay A. (Eds.), Biomaterials Science: Processing, Properties and Applications II: Ceramic Transactions, John Wiley & Sons, Hoboken, NJ, 2010. [Google Scholar]
- 11. Oldani, C. , Dominguez, A. , Titanium as a biomaterial for implants, in: Fokter S. (Ed.), Recent Advances in Arthroplasty, InTech; 2010. [Google Scholar]
- 12. Perera‐Costa, D. , Bruque, J. M. , González‐Martín, M. L. , Gómez‐García, A. C. , Vadillo‐Rodríguez, V. , Studying the influence of surface topography on bacterial adhesion using spatially organized microtopographic surface patterns. Langmuir. 2016, 16, 4633–4641. [DOI] [PubMed] [Google Scholar]
- 13. Kim, L. , Pagaling, E. , Zuo, Y. Y. , Yan, T. , Impact of substratum surface on microbial community structure and treatment performance in biological aerated filters. J. Appl. Environ. Microbiol. 2014, 80, 177–183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Schlegel, C. , 2016. Productive Biofilms on Structured Metal Surfaces. Cuvillier‐Verlag, Göttingen, Germany, 2016 [Google Scholar]
- 15. Neu, T. R. , Lawrence, J. R. , Investigation of microbial biofilm structure by laser scanning microscopy. Adv. Biochem. Eng. Biotechnol. 2014, 146, 1–51. [DOI] [PubMed] [Google Scholar]
- 16. Lupini, G. , Proia, L. , Di Maio, M. , Amalfitano, S. , Fazi, S. , CARD‐FISH and confocal laser scanner microscopy to assess successional changes of the bacterial community in freshwater biofilms. J. Microbiol. Methods. 2011, 86, 248–51. [DOI] [PubMed] [Google Scholar]
- 17. Nivens, D. E. , Chambers, J. Q. , Anderson, T. R. , White, D. C. , Longterm, online monitoring of microbial biofilms using a quartz crystal microbalance. Anal. Chem. 1993, 65, 65–69. [Google Scholar]
- 18. Crusz, S. A. , Popat, R. , Rybtke, M. T. , Cámara, M. et al., Bursting the bubble on bacterial biofilms: a flow cell methodology. Biofouling. 2012, 28, 835–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sternberg, C. , Tolker‐Nielsen, T. , Growing and analyzing biofilms in flow cells. Curr. Protoc. Microbiol. 2012, 1, Unit 1B.2. [DOI] [PubMed] [Google Scholar]
- 20. Tolker‐Nielsen, T. , Sternberg, C. , Methods for studying biofilm formation: flow cells and confocal laser scanning microscopy. Methods Mol. Biol. 2014, 1149, 615–29. [DOI] [PubMed] [Google Scholar]
- 21. Schlegel, C. , Chodorski, J. , Huster, M. , Davoudi, N. et al., Analyzing the influence of microstructured surfaces on the lactic acid production of Lactobacillus delbrueckii lactis in a flow‐through cell system. Eng. Life Sci. 2017, 17, 865–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Reichenbach, I. G. , Bohley, M. , Micromachining of CP‐titanium on a desktop machine‐study on bottom surface quality in micro end milling. Adv. Mater. Res. 2014, 769, 53–60. [Google Scholar]
- 23. Fingerle, M. , Köhler, O. , Rösch, C. , Kratz, F. et al., Cleaning of titanium substrates after application in a bioreactor. Biointerphases. 2015, 10, 019007. [DOI] [PubMed] [Google Scholar]
- 24. Cormack, B. , Valdivia, R. , Falkow, S. , FACS‐optimized mutants of the green fluorescent protein (GFP). Gene 1996, 173, 33–38 [DOI] [PubMed] [Google Scholar]
- 25. Lee, N. , Francklyn, C. , Hamilton, E. P. Arabinose‐induced binding of AraC protein to araI2 activates the araBAD operon promoter. Proc. Natl. Acad. Sci. USA. 1987, 84, 8814–8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lüdecke, C. , Jandt, K. D. , Siegismund, D. , Kujau, M. J. et al., Reproducible biofilm cultivation of chemostat‐grown escherichia coli and investigation of bacterial adhesion on biomaterials using a non‐constant‐depth film fermenter. PLOS One. 2014, 9, e84837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mueller, L. N. , De Brouwer, J. F. C. , Almeida, J. S. , Stal, L. J. , Xavier, J. B. , Analysis of a marine phototrophic biofilm by confocal laser scanning microscopy using the new image quantification software PHLIP. BMC Ecol. 2006, 6, 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Da Silva, W. J. , Seneviratne, J. , Samaranayake, L. P. , Del Bel Cury, A. A. , Bioactivity and architecture of Candida albicans biofilms developed on poly(methyl methacrylate) resin surface. J. Biomed. Mater. Res. B Appl Biomater. 2010, 94, 149–156. [DOI] [PubMed] [Google Scholar]
- 29. Heydorn, A. , Nielsen, A. T. , Hentzer, M. , Sternberg, C. et al., Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000, 146, 2395–2407. [DOI] [PubMed] [Google Scholar]
- 30. Bridier, A. , Dubois‐Brissonnet, F. , Boubetra, A. , Thomas, V. , Briandet, R. , The biofilm architecture of sixty opportunistic pathogens deciphered using a high throughput CLSM method. J. Microbiol. Methods. 2010, 82, 64–70. [DOI] [PubMed] [Google Scholar]
- 31. Mitra, S. , Gachhui, R. , Mukherjee, J. , Enhanced biofilm formation and melanin synthesis by the oyster settlement‐promoting Shewanella colwelliana is related to hydrophobic surface and simulated intertidal environment. Biofouling. 2015, 31, 283–296. [DOI] [PubMed] [Google Scholar]
- 32. Lewandowski, Z. , Structure and function of biofilms. In: Evans L. V. (Ed.), Biofilms: Recent Advances in Their Study and Control, Harwood Academic Publishers, Amsterdam, The Netherland, 2000. [Google Scholar]
- 33. de Beer, D. , Stoodley, P. , Roe, F. , Lewandowski, Z. , Effects of biofilm structures on oxygen distribution and mass transport. Biotechnol. Bioeng. 1994, 43, 1131–1138. [DOI] [PubMed] [Google Scholar]
- 34. Ross, S. S. , Tu, M. H. , Falsetta, M. L. , Ketterer, M. R. et al., Quantification of confocal images of biofilms grown on irregular surfaces. J. Microbiol. Meth. 2014, 100, 111–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Huster, M. , Müller‐Renno, C. , Ziegler, C. , Schlegel, C. et al., Chloroperoxidase production by Caldariomyces fumago biofilms. Eng. Life. Sci. 2016, 16, 88–92. [Google Scholar]
- 36. Truong, V. K. , Lapovok, R. , Estrin, Y. S. , Rundell, S. et al., The influence of nano‐scale surface roughness on bacterial adhesion to ultrafine‐grained titanium. Biomaterials. 2010, 31, 3674–3683. [DOI] [PubMed] [Google Scholar]
- 37. Singh, A. V. , Vyas, V. , Patil, R. , Sharma, V. et al., Quantitative characterization of the influence of the nanoscale morphology of nanostructured surfaces on bacterial adhesion and biofilm formation. PLOS One. 2011, 6, e25029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Koseki, H. , Yonekura, A. , Shida, T. , Yoda, I. et al., Early Staphylococcal biofilm formation on solid orthopaedic implant materials: In vitro study. PLOS One. 2014, 9, e107588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Almaguer‐Flores, A. , Olivares‐Navarrete, R. , Wieland, M. , Ximénez‐Fyvie, L. A. et al., Influence of topography and hydrophilicity on initial oral biofilm formation on microstructured titanium surfaces in vitro. Clin. Oral. Implants Res. 2012, 23, 301–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Hsu, L. C. , Fang, J. , Borca‐Tasciuc, D. A. , Worobo, R. W. , Moraru, C. I. , Effect of micro‐and nanoscale topography on the adhesion of bacterial cells to solid surfaces. Appl. Environ. Microbiol. 2013, 79, 2703–2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lorenzetti, M. , Dogša, I. , Stošicki, T. , Stopar, D. et al., The influence of surface modification on bacterial adhesion to titanium‐based substrates. ACS Appl. Mater. Interfaces. 2015, 7, 1644–1651. [DOI] [PubMed] [Google Scholar]
- 42. Mitik‐Dineva, N. , Wang, J. , Stoddart, P. R. , Crawford, R. J. , Ivanova, E. P. , Nano‐structured surfaces control bacterial attachment. Int Conf Nanosci Nanotechnol (ICONN), Melbourne 2008, 113–116.


