Abstract
The manual counting of colonies on agar plates to estimate the number of viable organisms (so‐called colony‐forming units–CFUs) in a defined sample is a commonly used method in microbiological laboratories. The automation of this arduous and time‐consuming process through benchtop devices with integrated image processing capability addresses the need for faster and higher sample throughput and more accuracy. While benchtop colony counter solutions are often bulky and expensive, we investigated a cost‐effective way to automate the colony counting process with smart devices using their inbuilt camera features and a server‐based image processing algorithm. The performance of the developed solution is compared to a commercially available smartphone colony counter app and the manual counts of two scientists trained in biological experiments. The comparisons show a high accuracy of the presented system and demonstrate the potential of smart devices to displace well‐established laboratory equipment.
Keywords: Colony counter, Digital image processing, Laboratory automation, Scientific app, Smart devices
Abbreviations
- CFU
colony‐forming unit
- FTP
file transfer protocol
- IP
internet protocol
- ROI
region of interest
- SSL
secure sockets layer
- TCP
transmission control protocol
1. Introduction
More than 20 years after IBM introduced their personal communicator “Simon”, which is said to be the father of all smartphones, the technology, and software constantly evolved into today's smartphones that do not have a lot in common anymore with its ancient predecessor 1. While “Simon” was already able to receive and send faxes and e‐mails, to browse the internet and even had a touchscreen user interface and basic applications installed, modern smartphones are, in addition to these standard features, full of sensors, and advanced wireless technologies and their processing capability stands up to even most demanding tasks. Due to constant innovations, miniaturized hardware became both powerful and affordable and handheld smart devices are now a substantial part of our life.
The number of smartphone users worldwide currently exceeds more than two billion people 2. In addition to that also wearables like smartwatches, and smart glasses are rising stars that more and more pervade and accompany our everyday life. The open and universal programming interfaces of most smart devices open the door to create not only software applications (so called “apps”) that serve as “daily little helpers,” but also diverse professional apps. The quickly evolving mobile hard‐ and software technologies make the technical concept of “ubiquitous computing” more and more a reality 3.
Within the last years many scientists began to harness the possibilities mobile devices offered to them whether to support them during experiments or to perform whole experiments independent of conventional laboratory devices 4, 5. The era of “scientific apps” began 6.
1.1. Scientific apps
Currently a lot of research effort is put into using smart devices for scientific purposes. Not only the use of tablets as modern lab journals is evaluated to digitally document lab processes and to make old‐fashioned paper notes obsolete, but also the built‐in sensors of smartphones and other wearables are used to record experimental data and physical quantities 7, 8, 9, 10, 11, 12.
Scientific apps in the area of life science research can roughly be divided into two areas. One big area summarizes apps that function as “little helpers” in the lab. They range from apps that help to easily access relevant information, for example restriction sites within a given DNA sequence, to apps that help with buffer design, that align DNA sequences and even apps that allow to control devices within the laboratory 5.
The other area summarizes apps that are able to displace conventional laboratory devices. Some of them just use the unmodified inbuilt sensors, for example the camera for protein‐ and DNA‐gel analysis 13, 14. Others take advantage of additionally installed constructions that expand the capabilities of the inbuilt sensors or add new sensors to the smart device.
Examples for apps that harness modified smartphone hardware are frequently found in the field of diagnostic sensing. Coskun et al. developed a cell phone module and an app that can be used to detect and quantify Albumine in urine samples 15. Other cell phone constructions were used to detect allergens in food, for the quantification of glucose in urine and for pathogen DNA detection 16, 17, 18, 19, 20, 21. Also a lot of effort was put into the establishment of efficient smartphone‐based microscopes by several research groups 22, 23, 24, 25. In addition to smartphones, also emerging technologies like smart glasses gained the interest of the scientific community. So far applications for smart glasses as a reader platform for diagnostic lateral flow devices and for the determination of the chlorophyll content of plant leafs were published 26, 27.
1.2. Applications of colony counting
The counting of colonies on agar plates is a both tedious and error‐prone, but absolutely essential task in laboratories with a focus on microbiological work. The approach is a common technique to estimate the concentration of viable cells within a given liquid sample 28. In order to do this, a dilution series is performed of a given culture suspension, which afterwards is dropped and spread onto agar plates 29. After a specific incubation time under specific conditions, the grown colonies can be counted and used to calculate the concentration of colony‐forming units (CFUs) within the original cultivation liquid.
1.3. Automated colony counting systems
Due to the time‐consuming and monotonous nature of manual colony counting, automated approaches to facilitate this job were developed. These solutions process the images taken from colony agar plates and can be subdivided into fully automated systems and semi‐automated systems. The difference between the two approaches is that semi‐automated systems need to be set up by the user in terms of specific parameters like plate dimension, the maximal colony diameter or contrast, whereas fully automated systems do not need user input and have built‐in algorithm steps to determine those characteristics 30, 31. Many automated colony counting systems include a suitable underground illuminated setup for image acquisition to ensure a reproducible and uniform Petri dish illumination and image quality. An example for a fully automated colony counting system is the PetriJet platform, which is an automated Petri dish handling and content analyzing device that is able to successively analyze plates within stacks of up to 20 Petri dishes. Another fully automated solution is the apparatus presented by Chiang et al. that is based on a illumination chamber concept to block external light sources 30, 32.
1.4. Colony counting apps
Currently at least five different Apps are listed in Google's Android Play Store, which enable the use of smart devices as colony counters (search word: colony counter). The “Promega Colony Counter” is a fully automated colony counter whose detection algorithms are based on Open Computer Vision (OpenCV) library programming functions 33. Other Apps are “Colony Count BETA,” “Colony Counter,” “Colony Counter (automated),” and “APD Colony Counter App PRO.” Of the mentioned apps only the “APD Colony Counter App PRO” offers a functioning feature for the import and analysis of previously taken agar plate images or images acquired by other devices. The App was developed by Wong et al. in 2016 and is based on a watershed and threshold algorithm provided by the OpenCV library 34.
This study aims at the development of a fully automated colony counting app for the processing of images of E. coli LB agar plates acquired at nonstandardized conditions. The application should be developed for the use with smartphones, tablets as well as smart glasses. The developed “TCI colony counter” Android application is compared to a commercially available colony counter app and quantitative and qualitative differences are shown.
2. Materials and methods
2.1. Chemicals
All used bulk chemicals were purchased from Sigma–Aldrich Chemie GmbH (Taufkirchen, Germany). Deionized water was prepared with a Sartorius Arium® device (Sartorius Stedim Biotech, Göttingen, Germany).
2.2. Flask and plate cultivations
The flask precultivations for the CFU determinations were performed in 100 mL baffled shake flasks containing 10 mL of lysogenic broth (LB) kanamycine medium consisting of yeast extract (10 L−1), tryptone (10 g/L), NaCl (5 g/L), and kanamycine (25 mg/L) set to a pH of 7.5. For the inoculation of the medium either 1 mL of Escherichia coli BL21 (DE3) (Novagen, Germany) cryo culture or a single colony of Escherichia coli BL21 (DE3) picked from a previously prepared agar plate was used. Flask cultivations were done with a rotary shaker set to 250 min−1 at 37°C in darkness. The preculture was cultivated until an OD600 of 0.04 was reached. Afterwards dilutions of the preculture were prepared (Dilutions: 1/100, 1/1000, 1/2000, 1/4000, 1/8000, and 1/16000) and 50 μL of the dilutions were dropped onto Petri dishes, containing LB kanamycine agar medium consisting of yeast extract (10 L−1), tryptone (10 g/L), NaCl (5 g/L), Agar (15 g/L), and kanamycine (25 mg/L) set to a pH of 7.5. Thereafter the cell suspension was spread homogeneously across the agar medium using a Drigalski spatula. A total of nine biological replicates of the precultivations and the following dilutions were done, whereas four resulting bacterial plates were discarded due to no colony growth (probably due to too high dilutions and inconsistent bacterial growth). After a short drying period the plates were incubated at 37°C for 24 h. Subsequently images were taken by several smart devices using the developed colony counter application.
2.3. Image acquisition
The images of the E. coli colony plates, which were processed within this study, were acquired using a Samsung Galaxy S4 mini (Samsung Electronics Co., Ltd., South Korea) using the developed colony counter application. The colony plates were placed onto a black polyoxymethylene plate to increase the contrast. The images were acquired under standard laboratory light conditions. It was tried to minimize reflections due to a nonstandardized light environment. The taken images had a resolution of 2,048 × 1,152 pixels and an average file size of 456 ± 28.5 kilobyte.
2.4. Method comparison
The comparison of the automated app counting results was done to the gold standard of manual counts. The acquired high resolution pictures were analyzed independently by two scientists. Individual colonies were manually marked within a picture processing program (GIMP 2.9.4, http://www.gimp.org) and afterwards manually counted. The means of the specific manual counts were used as a comparison with app achieved counting results. The “APD Colony Counter App PRO” was chosen as comparison and as a benchmark for the developed app, since its “Lite” version was one of the most downloaded colony counters within Google's Play Store and it received the highest ratings of all colony counting apps. It also has the feature implemented to process previously taken pictures of colony agar plates, which enabled the processing of the same image data with both apps. To determine the accuracy of the colony detection of the developed app and the benchmark app the following equation was used:
| (1) |
= Mean of manually counted colonies on a plate; = App counted colonies on a plate
A linear regression graph of the respective counting results was plotted using OriginPro 2017G (OriginLab Corporation, USA) (see Fig. 3). A Bland‐Altman diagram was plotted to depict differences of the respective apps in terms of counting deviation (see Fig. 4) 35.
Figure 3.
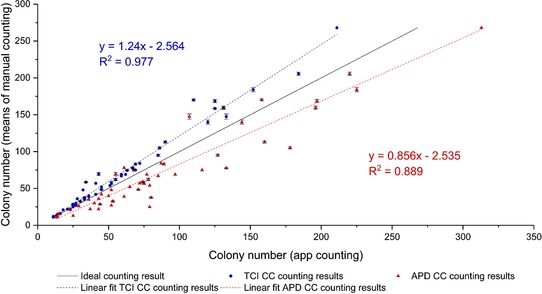
Correlation between manual counted colonies (means of two datasets) and colonies counted by the developed TCI colony counter (TCI CC – blue circles) and the commercially available “APD Colony Counter App PRO” (APD CC – red triangles). The black line represents an ideal line (means of manual counting on both x‐ and y‐axis). The dashed lines in the respective color show the linear fits of the data. Whiskers represent the respective standard deviations of the manual counts.
Figure 4.
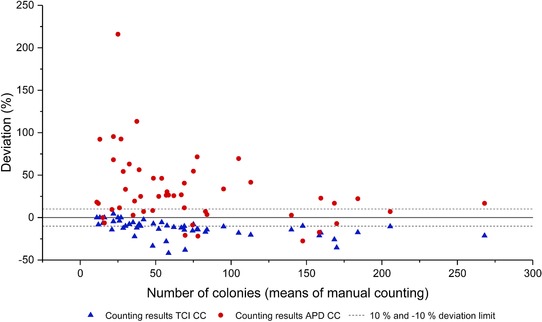
Deviation between the means of the manual counting and the results of the counting performed by the developed TCI colony counter (TCI CC—blue) and the commercially available “APD Colony Counter App PRO” (APD CC—red). The dashed lines represent the deviation of the app‐counted colonies from manual counted colonies within a 10% and – 10% limit.
2.5. Smart devices
The created application was installed and tested on different smartphones including LG G3 (LG Corporation, South Korea), Huawei Ascend Mate 7 (Huawei Technologies Co. Ltd, China) Samsung Galaxy S4 mini (Samsung Electronics Co., Ltd., South Korea) and smart glasses Vuzix M100 (Vuzix Corporation, USA). The data shown in this study were acquired by using the Samsung Galaxy S4 mini.
2.6. Application development
The application was programmed using the MIT App Inventor 2 development environment on a Windows 7 Professional 64‐Bit desktop PC. A modified version of the Java‐based application with a trimmed and voice triggered user interface was programmed to fit the needs of Vuzix M100 smart glasses (Vuzix Corporation, USA). Data transport between the app and the image processing server was realized using the file transfer protocol (FTP).
The CFUs/mL concentration is calculated within the app by using the user‐entered parameters (dilution factor and the volume dropped onto the agar plate) and the counting data, which is provided by the counting server. The following equation was used:
| (2) |
To enhance the usability of the smart glasses based client app, only the CFUs of the current plate are shown, which does not require the previous input of experimental parameters.
2.7. Server‐based image processing algorithm
The image processing algorithm was programmed in C# using the Microsoft .NET Framework 4.6 (Microsoft Corporation, USA). For basic algorithm routines (e.g. edge detection and thresholding) the AForge.Imaging .NET library was used, whereas more complex algorithm routines were self‐developed. The data exchange between client mobile devices and the image processing server was established using the FTP. FTP access for external clients was realized by opening and forwarding custom FTP ports (default FTP network TCP ports are 20 and 21) and the use of a Dynamic Domain Name Service (DynDNS) service. The use of specific name filters within the server software (FileZilla Server 0.9.60.2, filezilla‐project.org) ensures that only specific files can be up‐ and downloaded. The described setup runs 24/7 on a designated desktop computer (Intel i5 6500T CPU, 8 GB of RAM, 256 GB Samsung SSD) running with Windows 7 Professional 64‐Bit OS (Microsoft Corporation, USA).
3. Results and Discussion
3.1. Android application for colony counting
Once the developed “TCI colony counter” application is started, the user is asked to either take a picture of an agar plate or to pick an image of an already taken image stored on the smart device. After clicking the designated button, the image is either taken or selected. Afterwards the image is shown in a preview and the dilution factor and the used volume for the plate inoculation can be entered. These values are necessary to calculate the CFUs/mL within the original sample. After entering those values and after clicking the “calculate CFUs/mL” button, the image gets uploaded to a remote server via the inbuilt WiFi function or a mobile internet connection and is directly processed. The user subsequently receives information about the counted colonies that includes the number of single colonies and colonies in clusters, as well as the calculation time and the CFUs/mL within the original sample.
A cropped image that shows the region of interest (ROI) with colorized counted colonies is displayed as well. The whole workflow is visualized in Fig. 1.
Figure 1.
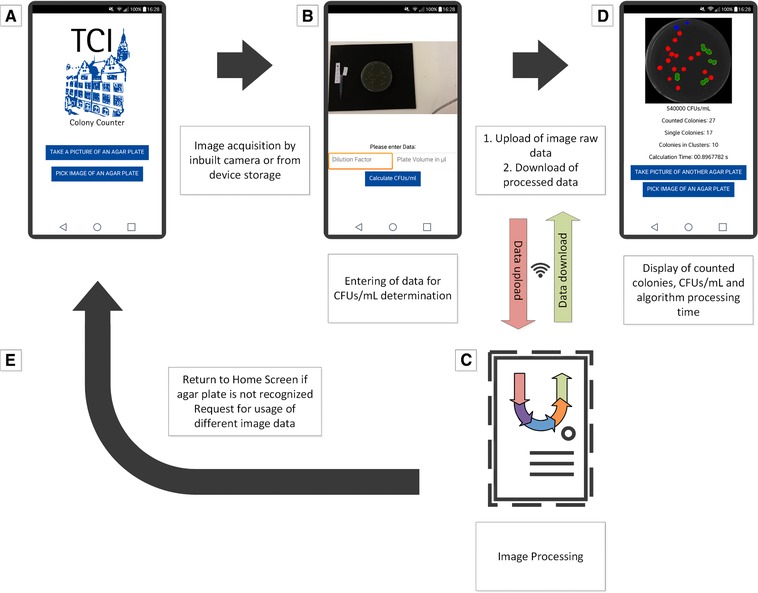
Screen flow chart of the developed Android application. (A) Welcome screen and buttons for image acquisition from storage or inbuilt camera. (B) Preview of taken or selected picture, input of dilution factor and used volume for plate inoculation. (C) Data upload and image processing by server‐based algorithm and download of results. (D) Display of counted colonies and processed image, calculation of CFUs/mL. (E) Return to home screen if image data is not sufficient. Screenshots were taken from actual smartphone screens and used for the visualization with Microsoft Visio Professional 2013 (Microsoft Corporation, USA).
3.2. Server‐based image processing algorithm
Although the processing power of some of today's tablets and smartphones is sufficient for complex image processing, the computational power of smart glasses (like Google Glass or the Vuzix M100) or older devices still often reaches its limits even while running comparatively low demanding tasks.
To ensure a rapid image processing, the corresponding algorithm was outsourced to a remote server. This enables the fast processing of image data independent from the hardware specifications of the used mobile device. Prerequisites that need to be met are either a WiFi internet connection or a mobile internet connection to ensure the upload and download of image data.
The image processing algorithm is basically divided into five consecutive steps (see Fig. 2). As soon as an image is uploaded to the server, the algorithm software handles the image data and determines the plate dimensions by using edge detection algorithms. The area of the image occupied by the detected plate is defined as ROI. If this crucial step is unsuccessful and no plate can be detected the algorithm is programmed to abort. A protocol file is then saved and the app asks the user to take another image under different light conditions or from another angle. In case of a successful recognition of the agar plate, the image gets cropped within in the border of the ROI. Only the ROI is used for further analyses. Within step 2 of the algorithm the optimal binarization threshold for colony detection is defined based on the roundness of the detected objects by using an iterative thresholding approach. Afterwards in step 3, based on the previously defined threshold, the ROI is examined for objects which than will be divided into single colonies and colony clusters by using a classification algorithm. Step 4 includes the segregation of colony clusters into single colonies by making use of a Hough circle transformation 36, 37. The last part of the algorithm is dedicated to the finding of colonies that could not be detected within the previous steps. For this the sizes of previously found colonies are compared with other objects that could be found on the plate. If these objects fit into the size range of previously found colonies and have as suitable roundness, they are recognized as a colony. The sum of all found single colonies and colony clusters as well as the calculation time and a result image of the analyzed plate is then saved and afterwards downloaded by the colony counting app. Within the result image single colonies are colored red, colony clusters are colored green and colonies found in the last step of the algorithm are colored blue.
Figure 2.
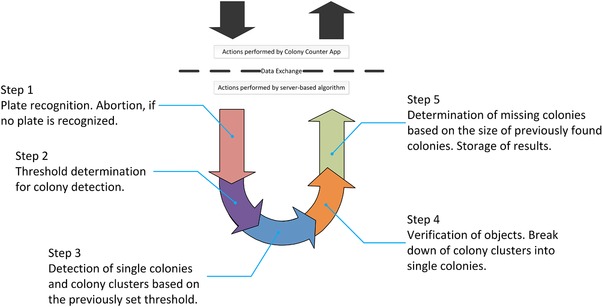
Proceeding of the 5‐step server‐based image processing algorithm after image takeover. Visualization was done using Microsoft Visio Professional 2013 (Microsoft Corporation, USA).
Many static colony counting solutions utilize a similar sequence of image preprocessing and colony detecting steps (e.g. binarization, circle detection, and colony segmentation) and require the manual selection of the Petri dish dimensions or only process an ROI that is defined by the hardware conditions of the experimental setup 38, 39. The developed solution makes use of a preceding step that involves the automatic detection of the dish for further processing. This step especially focuses on the handling of diverse variations within the input images and is crucial to offer a user‐friendly and fully automatic colony counting solution under dynamic image acquisition conditions.
3.3. App performance and accuracy
To determine the performance and the accuracy of the developed application 50 agar plates incubated with E. coli colonies were prepared and analyzed by the developed “TCI colony counter” app. Subsequently the images of the same plates were analyzed by two scientists trained in biological experiments and present colonies were counted manually. To see how the developed app benchmarks against other available counting solutions, the taken images were analyzed by a commercially available app for smart devices, the “APD Colony Counter App PRO.” Different from the application that is presented in this study the “APD Colony Counter PRO” is following a semi‐automatic approach. Besides the manual cropping of the ROI, also the threshold for colony detection as well as brightness parameters need to be set manually 34.
The mean number of colonies as determined by manual counting of scientist 1 resulted in a mean of 72.76 colonies and a median of 57.5 colonies. The manual counting of scientist 2 resulted in an overall colony mean of 72.4 and a median of 56.5 colonies. The number of colonies on the 50 plates was ranging from 11 to 268 as determined by the manual countings.
Figure 3 shows the comparisons between the results of the manual counting method and the outcomes of the analyses of the developed app and the commercially available “APD Colony Counter App PRO” app. Here, the developed “TCI colony counter” conducted a correlation coefficient of R 2 = 0.977 and a regression slope of 1.24, whereas the APD app resulted in a correlation coefficient of R 2 = 0.889 and a regression slope of 0.8561.
Considering this, it can be said that the developed Android app tends to detect not all present colonies, especially on agar plates with high colony densities. This is probably due to higher colony densities in the plate rim area of agar plates with high colony numbers and the fact that the plate rim area often evokes image obstructions due to light reflections and the uneven geometry of the Petri dish edge. However, the APD app seems to recognize noncolony objects as colonies, which results in a higher counting result than the manual countings.
Figure 4 shows a Bland‐Altman plot that displays the deviations between the means of the manual counting results and the counting results acquired by the respective apps 35. The deviation data suggests that the developed colony counter app is missing colonies, whereas the APD app is detecting colonies where there are none. This confirms the previously made statement, that the developed app is slightly undercounting and the benchmark app tends to overcount. Over all the mean value of the absolute deviation between the counting results of the developed app and the means of the manual countings was 13.24 ± 9.67%, while 42% of the automatic counting results had a an absolute deviation of ≤ 10% (see Fig. 4). The absolute deviation mean between the means of the manual countings and the APD Colony Counter App PRO was 35.8 ± 37.8%, while only 24% of the automated counting results had an absolute deviation of ≤ 10%.
The trend of the deviation shows that the developed app has a higher sensitivity in detecting lower amounts of colonies with a decreasing sensitivity for the detection of the correct colony numbers on agar plates with high amounts of colonies. The benchmark APD app seems to have a lower sensitivity than the developed app when it comes to the detection of lower amounts of colonies. Yet it seems to gain more sensitivity with an increased density of colonies on the agar plate. Since dense plating of microorganisms is usually avoided with the intention to prevent nutrient and space competition between neighboring colonies, the TCI colony counter seems to be a more reliable choice 28.
The average accuracy of the developed application lies at 86.76 ± 9.76%, whereas the average accuracy of the commercially available app for this experimental setup lies at 66.79 ± 28.2%. This shows a clear tendency toward a higher accuracy of the developed app.
Another important detail that needs to be mentioned is the time that needs to be spent for the use of the apps to get the counting results. While the average manual counting time was over 2 min, the automated processing took maximum 2 s. Taking the picking of a picture from the devices memory or the image acquisition via built‐in camera into account, the whole process from image acquisition to the counting results usually took under 10 s, including the data up‐ and download. Due to the semi‐automated approach of the APD Colony Counter App PRO, the result acquisition took significantly longer. The manual image cropping, the setting of an adequate brightness and the finding of a suitable threshold take a lot of time especially for inexperienced users and were in the range of 60–80 s due to the tedious cropping procedure. In some cases the benchmarking app had problems during the image processing, causing the app to crash on some of the tested smartphones. Another problem was the detection of colony artifacts within the discarded image area after the cropping process. The developed app did not have any stability problems, most likely due to the outsourcing of the demanding image processing procedure to a remote server, which saved processing power and main memory.
4. Concluding remarks
This study shows the applicability of a smart device‐based app for the automated estimation of E. coli colonies on LB agar Petri dishes at nonstandardized image acquisition conditions. It could be shown that the developed application provides more reliable counting results than a similar application within a shorter time. Although the automated app analysis of the colony plates is significantly faster than the manual counting method, its accuracy yet has to be improved. A possible way might be the introduction of an additional algorithm step for the removal of image obstructions caused by light reflections 40. This would enable the detection of colonies covered by those, but requires the recording of multiple images of the plate from different angles. Also the removal of image destructions is a hardware demanding task and even the processing of image sequences with a resolution of 1152 × 648 with up‐to‐date hardware can take up to 20 min.
It must also be said that a potential security issue arises by using FTP for file transport within the internet, which is generally regarded as unsafe for the use in nonprivate networks due to its lack of encryption. FTP over Secure Sockets Layer (SSL) or another secured file transport protocol should be introduced for the communication out of the private network. Apart from that also a virtual private network tunnel could be used to assure a secured server connection, which is a tool often used within professional environments 41. Since open transmission control protocol (TCP) ports are always a risk in terms of security, a way to prevent unwanted hacker attacks would be the opening of those to designated Internet Protocol (IP) addresses only.
Since the recent version of the client app accesses a server, which is also used for other tasks within the laboratory (e.g. data processing, device handling, and programming), and which is not dimensioned to handle an enormous number of requests from external clients, it is not published on an app store so far. This might be done at a later stage after an appropriate infrastructure is provided. Another option would be the individual set up of a server within a research facility that only handles the requests of a specific number of known internal clients.
The outsourcing of the image processing procedure to a well‐equipped remote server enabled a faster image analysis than possible by the internal processing capabilities of modern smart devices and ensures the use of the app even on older, low performing smart devices. The automated fashion of the underlying processing algorithm does not require the setup of parameters by the user and allows the use of the app on devices with limited user interaction possibilities like today's smart glasses. A trimmed and voice command triggered version of the developed app was adjusted to fit the needs of smart glasses and was successfully tested.
In further developments the app could be adapted to other organisms and a manual colony marking function could be implemented to include missed colonies to the counting result. It can be concluded that the shown application shows a high accuracy, although it can probably not compete with designated benchtop devices so far due to the optimized image acquiring solutions they deliver. Current developments in digital image processing as well as hardware innovations could compensate this in the near future, though 40, 42.
To conclude, it can be said that the rise of scientific apps and smart devices within the laboratory environment promises to give researchers independence from traditional laboratory devices and paves the way for inexpensive science experiments for everyone 5.
Practical application
We described a convenient and efficient way for automated colony counting with mobile devices, which offers the potential to replace bulky and costly benchtop colony counters. An Android application was created that uses the internal camera of smartphones and smart glasses to collect data for image processing. A bidirectional wireless communication between a server‐based algorithm and the mobile device ensures the upload and processing of image data and the download of counting results to the mobile front end. Taken images of E. coli agar plates were analyzed in less than 10 s and the counting accuracy was superior to commercial smart device‐based colony counting solutions. The application would be useful in all areas of microbiological work and could be of special interest for laboratories in the field of food safety and infection microbiology.
The authors have declared no conflict of interest.
Acknowledgments
This work was financially supported by the Bavarian Ministry of Economic Affairs and Media, Energy and Technology within the Information and Communications Technology program (Grant number: IUK‐1504‐0006//IUK470/001).
5 References
- 1. Bayus, B. L. , Jain, S. , Rao, A. G. , Too little, too early: introduction timing and product performance in personal digital assistant industry. J. Mark. Res. 1997, 34, 50–63. [Google Scholar]
- 2. Statista, Number of smartphone users worldwide 2014–2020. 2014.
- 3. Campbell, A. , Choudhury, T. , From smart to cognitive phones. IEEE Pervasive Comput. 2012, 11, 7–11. [Google Scholar]
- 4. Libman, D. , Huang, L. , Chemistry on the Go: review of chemistry apps on smartphones. J. Chem. Educ. 2013, 90, 320–325. [Google Scholar]
- 5. Gan, S. K.‐E. , Poon, J.‐K. , The world of biomedical apps: their uses, limitations, and potential. Sci. Phone Apps Mob. Devices 2016, 2, 6. [Google Scholar]
- 6. Young, H. A. , Scientific Apps are here (and more will be coming). Cytokine 2012, 59, 1–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Perkel, J. M. , The internet of things comes to the lab. Nature 2017, 542, 125–126. [DOI] [PubMed] [Google Scholar]
- 8. Guerrero, S. , Dujardin, G. , Cabrera‐Andrade, A. , Paz‐y‐Miño, C. et al., Analysis and implementation of an electronic laboratory notebook in a biomedical research institute. PLoS One 2016, 11 10.1371/journal.pone.0160428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Roda, A. , Michelini, E. , Cevenini, L. , Calabria, D. et al., Integrating biochemiluminescence detection on smartphones: mobile chemistry platform for point‐of‐need analysis. Anal. Chem. 2014, 86, 7299–7304. [DOI] [PubMed] [Google Scholar]
- 10. Yu, H. , Tan, Y. , Cunningham, B. T. , Smartphone fluorescence spectroscopy. Anal. Chem. 2014, 86, 8805–8813. [DOI] [PubMed] [Google Scholar]
- 11. Barbosa, A. I. , Gehlot, P. , Sidapra, K. , Edwards, A. D. et al., Portable smartphone quantitation of prostate specific antigen (PSA) in a fluoropolymer microfluidic device. Biosens. Bioelectron. 2015, 70, 5–14. [DOI] [PubMed] [Google Scholar]
- 12. Lopez‐Ruiz, N. , Curto, V. F. , Erenas, M. M. , Benito‐Lopez, F. et al., Smartphone‐based simultaneous pH and nitrite colorimetric determination for paper microfluidic devices. Anal. Chem. 2014, 86, 9554–9562. [DOI] [PubMed] [Google Scholar]
- 13. Sim, J.‐Z. , Nguyen, P.‐V. , Lee, H.‐K. , Gan, S. K. , GelApp: mobile gel electrophoresis analyser. Nat. Methods Appl. Notes 2015, 1–2. 10.1038/an964 [DOI] [Google Scholar]
- 14. Nguyen, P.‐V. , Verma, C. S. , Gan, S. K.‐E. , DNAApp: a mobile application for sequencing data analysis. Bioinformatics 2014, 30, 3270–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Coskun, A. F. , Nagi, R. , Sadeghi, K. , Phillips, S. et al., Albumin testing in urine using a smart‐phone. Lab Chip 2013, 13, 4231–4238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Coskun, A. F. , Wong, J. , Khodadadi, D. , Nagi, R. et al., A personalized food allergen testing platform on a cellphone. Lab Chip 2013, 13, 636–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ludwig, S. K. J. , Zhu, H. , Phillips, S. , Shiledar, A. et al., Cellphone‐based detection platform for rbST biomarker analysis in milk extracts using a microsphere fluorescence immunoassay. Anal. Bioanal. Chem. 2014, 406, 6857–6866. [DOI] [PubMed] [Google Scholar]
- 18. Ludwig, S. K. J. , Tokarski, C. , Lang, S. N. , Van Ginkel, L. A. et al., Calling biomarkers in milk using a protein microarray on your smartphone. PLoS One 2015, 10, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jia, M. Y. , Wu, Q. S. , Li, H. , Zhang, Y. et al., The calibration of cellphone camera‐based colorimetric sensor array and its application in the determination of glucose in urine. Biosens. Bioelectron. 2015, 74, 1029–1037. [DOI] [PubMed] [Google Scholar]
- 20. Bates, M. , Zumla, A. , Rapid infectious diseases diagnostics using Smartphones. Ann. Transl. Med. 2015, 3, 215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Walker, F. M. , Ahmad, K. M., Eisenstein, M. , Soh, H. T. , Transformation of personal computers and mobile phones into genetic diagnostic systems. Anal. Chem. 2014, 86, 9236–9241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Contreras‐Naranjo, J. C. , Wei, Q. , Ozcan, A. , Mobile phone‐based microscopy, sensing, and diagnostics. IEEE J. Sel. Top. Quantum Electron. 2016, 22, 1–14 [Google Scholar]
- 23. Breslauer, D. N. , Maamari, R. N. , Switz, N. A. , Lam, W. A. et al., Mobile phone based clinical microscopy for global health applications. PLoS One 2009, 4 10.1371/journal.pone.0160428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Coulibaly, J. T. , Ouattara, M. , D'Ambrosio, M. V. , Fletcher, D. A. et al., Accuracy of mobile phone and handheld light microscopy for the diagnosis of schistosomiasis and intestinal protozoa infections in Côte d'Ivoire. PLoS Negl. Trop. Dis. 2016, 10, e0004768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gopinath, S. C. B. , Tang, T. H. , Chen, Y. , Citartan, M. et al., Bacterial detection: from microscope to smartphone. Biosens. Bioelectron. 2014, 60, 332–342. [DOI] [PubMed] [Google Scholar]
- 26. Feng, S. , Caire, R. , Cortazar, B. , Turan, M. et al., Immunochromatographic diagnostic test analysis using google glass. ACS Nano 2014, 8, 3069–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Cortazar, B. , Koydemir, H. C. , Tseng, D. , Feng, S. et al., Quantification of plant chlorophyll content using Google Glass. Lab Chip 2015, 15, 1708–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Breed, R.S. , Dotterrer, W. D. , The number of colonies allowable on satisfactory agar plates. J. Bacteriol. 1916, 1, 321–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Barbosa, H. R. , Rodrigues, M. F. A. , Campos, C. C. , Chaves, M. E. et al., Counting of viable cluster‐forming and non cluster‐forming bacteria: a comparison between the drop and the spread methods. J. Microbiol. Methods 1995, 22, 39–50. [Google Scholar]
- 30. Chiang, P.‐J. , Tseng, M.‐J. , He, Z.‐S. , Li, C.‐H. , Automated counting of bacterial colonies by image analysis. J. Microbiol. Methods 2015, 108, 74–82. [DOI] [PubMed] [Google Scholar]
- 31. Niyazi, M. , Niyazi, I. , Belka, C. , Counting colonies of clonogenic assays by using densitometric software. Radiat. Oncol. 2007, 2, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Vogel, M. , Boschke, E. , Bley, T. , Lenk, F. , PetriJet platform technology. J. Lab. Autom. 2015, 20, 447–456. [DOI] [PubMed] [Google Scholar]
- 33. Pulli, K. , Baksheev, A. , Kornyakov, K. , Eruhimov, V. , Real‐time computer vision with OpenCV. Commun. ACM 2012, 55, 61. [Google Scholar]
- 34. Wong, C.‐F. , Yeo, J. Y. , Gan, S. K. , APD colony counter app: using watershed algorithm for improved colony counting. Nat. Methods Appl. Notes 2016, 1–3. 10.1038/an9774 [DOI] [Google Scholar]
- 35. Bland, J. M. , Altman, D. G. , Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 1, 307–310. [PubMed] [Google Scholar]
- 36. Duda, R. O. , Hart, P. E. , Use of the Hough transform to detect lines and cures in pictures. Commun. Assoc. Comput. Mach. 1972, 15, 11–15. [Google Scholar]
- 37. Barber, P. R. , Vojnovic, B. , Kelly, J. , Mayes, C. R., et al., An automated colony counter utilising a compact Hough transform. Proceedings of Medical Image Understanding and Analysis, MIUA, 2000. (ed. by S., Arridge and A., Todd‐Pokropek , 41–44, UCL, London. [Google Scholar]
- 38. Brugger, S. D. , Baumberger, C. , Jost, M. , Jenni, W. et al., Automated counting of bacterial colony forming units on agar plates. PLoS One 2012, 7, e33695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ateş, H. , Gerek, Ö. N. , An image‐processing based automated bacteria colony counter, in: 2009 24th International Symposium on Computer and Information Sciences, ISCIS 2009, 2009, 18–23. 10.1109/ISCIS.2009.5291926 [DOI] [Google Scholar]
- 40. Xue, T. , Rubinstein, M. , Liu, C. , Freeman, W. T. , A computational approach for obstruction‐free photography. ACM Trans. Graph. 2015, 34, 79:1–79:11. [Google Scholar]
- 41. Berger, T. , Analysis of current VPN technologies, in: proceedings—first international conference on availability, reliability and security, ARES 2006, 2006, 108–115. [Google Scholar]
- 42. Debnath, S. , Linke, N. M. , Figgatt, C. , Landsman, K. A. et al., Demonstration of a small programmable quantum computer with atomic qubits. Nature 2016, 536, 63–66 [DOI] [PubMed] [Google Scholar]


