Abstract
Background
Hepatocellular carcinoma (HCC) is the third major cause of cancer-related death. Mounting evidence shows that microRNAs play critical roles in the initiation and progression of HCC and may potentially serve as diagnostic markers for HCC.
Methods and Results
In the present study, we explored the biological effects of miR-885-5p on HCC progression. We performed flow cytometry analyses of miR-885-5p in HCC cell lines and identified miR-885-5p as a recurrence-related microRNA. Overexpression of miR-885-5p significantly inhibited cell migration, invasion, proliferation, angiogenesis and EMT. Then, the correlation of miR-885-5p and AEG1 were confirmed by using luciferase assays, quantitative real-time PCR analysis and Western blotting. It was subsequently confirmed that Astrocyte Elevated Gene1 (AEG1) was a direct target gene of miR-885-5p.
Conclusion
miR-885-5p likely acts as a tumor suppressor by regulating AEG1, suggesting that miR-885-5p may be a potential biomarker and can be targeted in therapeutic strategies against HCC in the future.
Keywords: miR-885-5p, AEG1, biomarker, EMT, hepatocellular carcinoma
Introduction
Hepatocellular carcinoma (HCC) is one of the most common malignancies in the world, and a highly invasive tumor with frequent distant metastasis in China.1,2 The underlying pathogenesis of HCC may be very complicated, though most HCC cases may be caused by hepatitis B or hepatitis C virus infection and cirrhosis.3,4 Unfortunately, despite recent advances in HCC early diagnosis and developed treatment approaches, HCC patients still suffer from poor prognosis and high recurrence rate.5 Consequently, it is of crucial importance to identify molecular pathways regulating HCC metastasis in order to develop novel treatment strategies.
MicroRNAs, small non-coding endogenous RNAs, act as post-transcriptional regulators by binding to 3′-untranslated region (3′-UTR) of the target mRNAs and have been demonstrated to be associated with a wide range of pathological processes, including cell differentiation, proliferation, apoptosis, invasion and migration.6,7 In the past few years, there has been growing evidence focused on the function of miR-885-5p in tumorigenesis. MiR-885-5p has been identified as a tumor suppressor in glioblastomas, neuroblastomas and pancreatic cancer.8–10 Although Zhang et al reported that miR-885-5p contributes to suppression of HCC metastasis,11 whether miR-885-5p may also be dysregulated and function as miR-885-5p-sponging epigenetic regulators in HCC is not clear.
In this study, aberrant down regulation of miR-885-5p expression was detected in HCC tissues. We provide the first evidence that miR-885-5p can promote apoptosis and reverse EMT in HCC via inhibiting AEG1. Our results demonstrate that miR-885-5p may potentially act as a novel biomarker and potentially effective therapeutic target for HCC.
Methods
Cell Culture and Transfection
Human cancer cell lines HepG2 and Hela cells purchased from Type Culture Collection of Chinese Academy of Sciences (Shanghai, China) were maintained in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (GIBCO, Carlsbad, CA, USA), 100 U/mL penicillin G and 100 μg/mL streptomycin at 37 °C in a humidified incubator containing 5% CO2. HepG2 cells were transfected with either miR-885-5p mimics (5ʹ-UCC AUU ACA CUA CCC UGC CUC U-3ʹ) or negative control (5ʹ-UUC UCC GAA CGU GUC ACG UTT-3ʹ) (RiboBio, Guangzhou, China) using LipofectamineTM RNA iMax transfection reagent according to the manufacturer’s protocols.
Cell Viability and Flow Cytometry
The cell viability of HepG2 was assessed at 72 hrs after transfection with either miR-885-5p mimics or negative control, using a Cell Counting Kit-8 (Dojino, Japan), following the manufacturer’s instructions. Absorbance was then measured at 450 nm using Elx800 Reader (Bio-Tek, USA).
For flow cytometry studies, HepG2 cells were transfected with either miR-885-5p mimics or negative control for 48 h, respectively. Then, the cells were double-stained with Annexin V or propidium iodide, according to the manufacturer’s instructions. Cells were analyzed using FACS Calibur flow cytometer (BD Biosciences, USA), followed by flowjo7.6 software analysis. Experiments were conducted using three independent biological samples.
Transwell Migration and Tube Formation Assay
Transwells with 8-μm pore size filters covered with (BD Biosciences, USA) were inserted into 24-well plates for invasion or migration assays, respectively. Medium (600 μL) containing 10% FBS was added to the lower chamber, and 100 μL of a serum free cell suspension (1 × 105 cells) was placed onto the upper chamber. The cells that did not migrate or invade after 24 h of incubation were removed from the upper face of the filters by scrubbing with a cotton swab. Membranes were then fixed with 4% formaldehyde for 30 min at room temperature and stained with DAPI. In each sample, the number of migrating or invading cells was counted at 20× magnification from five different fields for each filter and analyzed to determine statistically significance. For invasion assays, cells were seeded onto a matrigel-coated chamber and were incubated at 37 °C.
HUVEC purchased from Type Culture Collection of Chinese Academy of Sciences (Shanghai, China) was seeded onto a 96-well plate coated with Matrigel (BD Biosciences, USA) followed by transfection with miR-885-5p. Peak tube formation was observed at 8 h post-treatment. The number of tubes for each treatment was quantified.
Immunofluorescence
Cells seeded on glass coverslips in 6-well plates were fixed in 4% of formaldehyde solution and permeabilized with 1% of Triton/PBS. Cells were blocked with 1% of BSA/PBS for 1 h at room temperature and incubated with E-cadherin or Vimentin antibodies (Proteintech, USA) for 2 h at room temperature, followed by the incubation with fluorescent-dye conjugated secondary antibody (Cell Signaling Technology, USA) for 1 h. Samples were stained with DAPI and images were taken under an inverted fluorescence microscope.
Western Blotting and Real-Time PCR
The expression of cellular proteins was analyzed by Western blotting. The following antibodies were used in our study: AEG1, PD-L1 and EGFR (Abacm, USA), β-actin (Proteintech, USA) and the secondary antibodies (Cell Signaling Technology, USA).
Total RNA was extracted using RNA Trizol (Invitrogen, USA) according to the manufacturer’s instructions. PrimeScript RT reagent kit (Takara, Japan) was used to synthesize cDNA according to the manufacturer’s instructions. Real-time PCR (qRT-PCR) and Stem-loop qRT-PCR were performed using SYBR Mixture (Vazyme Biotech, China) in 96 well optical plates at 95 °C for 5 mins, followed by 40 cycles of 95 °C for 10 seconds and 60 °C for 30 min. After the reactions, the cycle threshold (Ct) data were determined using the default threshold settings. The relative gene expression levels were normalized to GAPDH. The primers for PCR were listed (5ʹ-3ʹ): AEG1 forward: CCT GGC CTT GCT GAA GAA TC; AEG1 reverse: GGC TGC TTT GCT GTT ACA CT; PD-L1 forward: GGC ATT TGC TGA ACG CAT; PD-L1 reverse: CAA TTA GTG CAG CCA GGT; EGFR forward: TCT ACA ACC CCA CCA CGT AC; EGFR reverse: TTC CGT TAC ACA CTT TGC GG; GAPDH forward: CAT GAG AAG TAT GAC AAC AGC CT; GAPDH reverse: AGT CCT TCC ACG ATA CCA AAG T. The relative miR-885-5p expression was normalized to U6 snRNA. The primers for PCR were listed (5ʹ-3ʹ): miR-885-5p forward: GTC CAT TAC ACT ACC CTG CCT C; miR-885-5p reverse: CGC GAG CAC AGA ATT AAT ACG; U6 snRNA forward: CTC GCT TCG GCA GCA CA; U6 snRNA reverse: AAC GCT TCA CGA ATT TGC GT.
Dual Luciferase Reporter Assay
The 3′-UTR region of AEG1 with the potential binding sites for intact or mutant sites of miR-885-5p were generated, followed by the fusion into pGL3 promoter vector (Promega, USA) named as AEG-1 3ʹ-UTR (AEG1-WT) or a mutated 3ʹ-UTR of AEG-1 (AEG1-MU). Sequences were amplified according to the following primers: AEG1-WT forward: 5′- CTA GTC TAG AAT TAC CTC CCC CAC CCC CAA GAA CAC TT −3′, and reverse: 5′- CCG GAA TTC GGA ACA TTA TTA GCT GCT ACA CTA AGC −3ʹ; AEG1-MU forward: 5′- TCT TCT CAA ATC GAA TTA CCT CAC CTC TAA CAG TGT CTG TC −3′, and reverse: 5′- AGG TAA TTC GAT TTG AGA AGA TCC TAA CAG AAT CTC AAA AT-3ʹ. Luciferase reporter experiments were conducted in 96 well plates by using Dual Luciferase Reporter Assay kit (Promega, USA), and SpectraMax M5 instrument software (Molecular Devices, USA) was used to analyze the results. AEG1-WT or AEG1-MU was co-transfected with either miR-885-5p mimics or the negative control (miR-NC) using 50 nM Lipofectamine 2000 (Life Technologies, USA). After 48 h, cells were analyzed using a Dual-Luciferase Reporter Assay kit in accordance with the manufacturer’s guidance. Data were normalized using the relative luciferase activity was normalized to Renilla luciferase activity.
Statistical Analysis
All experiments were performed in triplicate. All the data are expressed as the mean ± SEM and were analyzed using GraphPad Prism 6 software (GraphPad Software, Inc, USA). Differences between individual groups were analyzed by Student’s t-tests. p < 0.05 was considered statistically significant.
Results
Overexpression miR-885-5p Inhibits Proliferation as Well as Cell Cycle Progression and Promotes Cell Apoptosis in HCC Cells
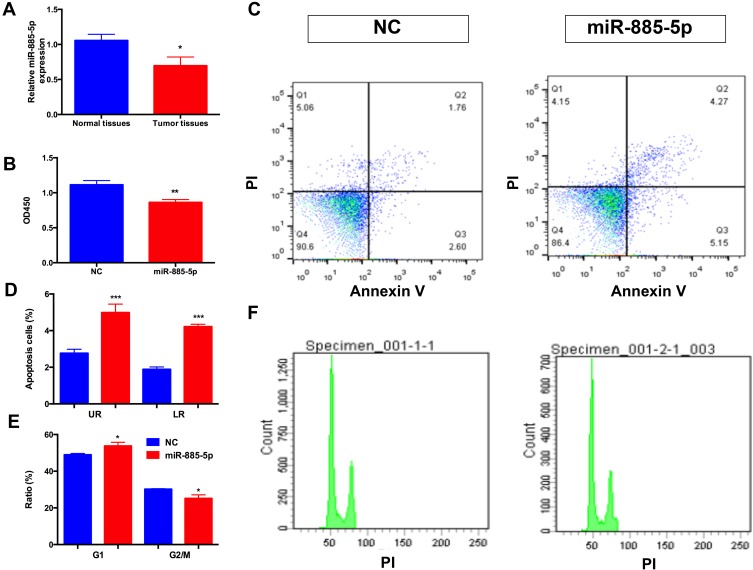
The relative expression levels of miR-885-5p in HCC samples and adjacent normal tissue samples were determined by qRT-PCR. As presented in Figure 1A, the expression of miR-885-5p in tumor tissue samples were reduced compared with adjacent normal tissues, respectively. Next, to explore the function of miR-885-5p on metastasis of HCC cells, we used HepG2 cells were transfected with miR-885-5p mimics, respectively by using miR-NC as a negative control (NC). We performed CCK-8 assay to determine the effect of miR-885-5p on the proliferative capacity of HepG2 cells. As demonstrated in Figure 1B, miR-885-5p significantly decreased the proliferation of HepG2 cells at 3 days post-transfection. Then we evaluated the effect of miR-885-5p on cell apoptosis and cell cycle progression, as shown in Figure 1C–F, flow cytometry analysis indicated that miR-885-5p significantly promoted apoptosis and caused cell cycle arrest at G1 phase in HepG2 cells.
Figure 1.
Overexpression miR-885-5p inhibits the metastasis of HCC cells.
Notes: (A) Relative miR-885-5p expressions measured by qRT-PCR in liver tumor tissues and paired normal tissues (*p < 0.05). (B) CCK8 assays were applied to determine the effect of miR-885-5p on HCC cells proliferation ability (**p < 0.01). (C) (D) Flow cytometry assays were employed to exam the function of miR-885-5p on the apoptosis capacity of HCC cells (***p < 0.001). (E, F) Flow cytometry assays were employed to exam the function of miR-885-5p on the cell cycle capacity of HCC cells (*p < 0.05).Abbreviations: CCK-8, cell counting kit-8; HCC, hepatocellular carcinoma; qRT-PCR, quantitative real-time PCR; NC, negative control.
Overexpression of miR-885-5p Inhibits Tumor Angiogenesis and EMT
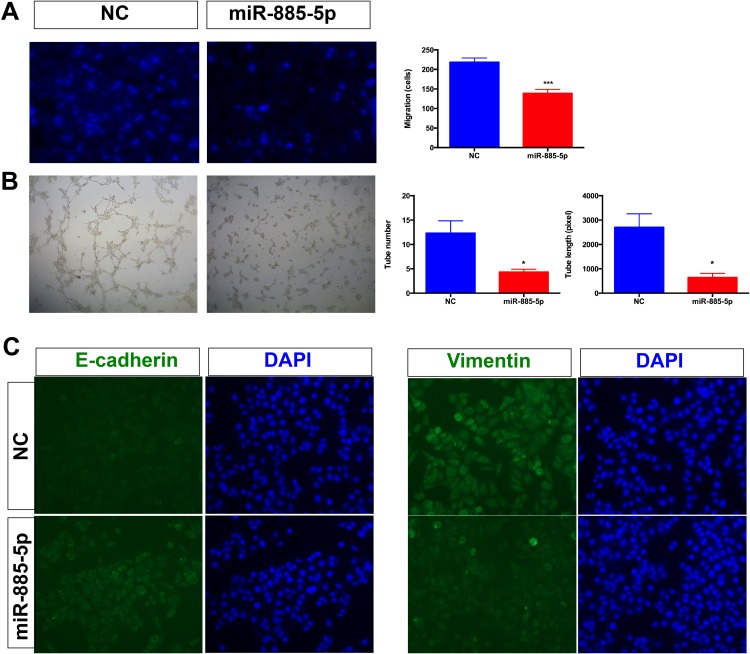
To examine whether miR-885-5p functionally contributed to migratory capacities of HCC cells, transwell assay was performed in HCC cells after transfection with miR-885-5p mimics or miR-NC. The results demonstrated that overexpression of miR-885-5p potentially suppresses the migratory ability of HCC cells (Figure 2A). To evaluate the effects of miR-885-5p on angiogenesis, we conducted angiogenesis assay by assessing the tubes formation of HUVEC cells in vitro for angiogenesis assays. Our results demonstrated that miR-885-5p reduces the angiogenic tubes formation of HUVEC cells. Furthermore, miR-885-5p also inhibited the formation of new blood vessels in the matrix gel (Figure 2B). It is established that EMT process can promote the migratory capacities of cancer cells. Next, we examined the EMT markers in HCC cells. Immunofluorescence analysis was employed to examine the levels of epithelial marker (E-cadherin) and mesenchymal marker (Vimentin) in cells transfected with miR-885-5p mimics or miR-NC. As shown in Figure 2C, the result of immunofluorescence hinted that the overexpression of miR-885-5p in HepG2 cells increased level of epithelial marker (E-cadherin) and decreased level of mesenchymal (Vimentin) marker. These data indicated that miR-885-5p reduces migratory ability of HCC cells through inhibiting the EMT pathway.
Figure 2.
Overexpression of miR-885-5p inhibits tumor angiogenesis and EMT.
Notes: (A) Transwell assays were employed to exam the function of miR-885-5p on the migration capacity of HCC cells (***p < 0.001). (B) Tube number and tube length in HUVEC cells were performed to investigate the function of miR-885-5p on angiogenesis (*p < 0.05). (C) Immunofluorescence was performed to investigate the function of miR-885-5p on EMT formation of HCC cells.
Abbreviations: HCC, hepatocellular carcinoma; EMT, Epithelial-Mesenchymal Transition; NC, negative control.
AEG1 Is Identified as a Target of miR-885-5p
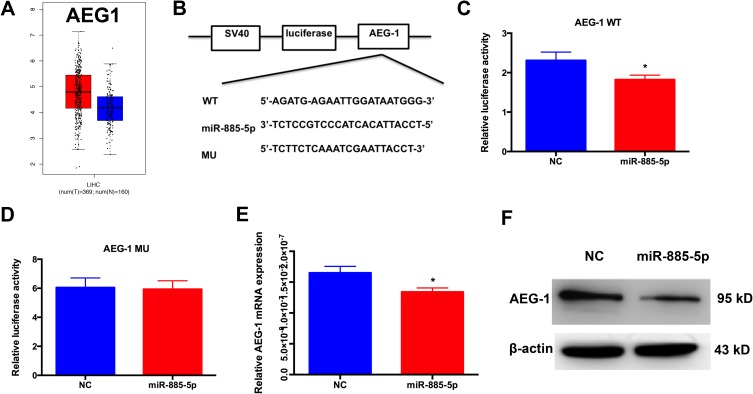
Overexpression of AEG1 has been documented to be involved in many tumors’ biological processes, including tumor proliferation, metastasis, EMT.12–15 To evaluate the possibility that AEG1 is important for HCC, we examined AEG1 expression in corresponding noncancerous tissues and HCC using Gene Expression Profiling Interactive Analysis (GEPIA) website based on TCGA database.16 Notably, AEG1 expression in HCC samples was significantly higher compared to normal samples (Figure 3A). To confirm whether AEG1 mediated the function of miR-885-5p, we used two bioinformatic analysis tools (TargetScan and RNA22), and along with luciferase reporter assays were utilized to confirm the binding sites and regulation relationship between miR-885-5p and AEG1. As presented in Figure 3B–D, miR-885-5p could bind to the 3′-UTR of AEG1, and miR-885-5p mimics reduced the luciferase activity of wild-type (WT) AEG1 reporter vector but not that of mutant reporter vector (MU). What’s more, we found that miR-885-5p overexpression decreased the AEG1 mRNA and protein levels decrease at 72h-post transfection (Figure 3E and F). Our findings indicated that miR-885-5p directly targets the AEG1 3′UTR, thus reducing AEG1 expression in HCC cells.
Figure 3.
AEG1 is identified as a target of miR-885-5p.
Notes: (A) AEG1 messenger RNA expression was higher in HCC tissues than in normal tissues. (B–D) the binding sites between miR-885-5p and AEG1; miR-885-5p mimics decreased the luciferase reporter analysis of AEG1 (*p < 0.05). (E) Real-time PCR results for endogenous AEG1 infected with miR-885-5p or NC (*p < 0.05). (F) Western blotting for endogenous AEG1 infected with miR-885-5p or NC.
Abbreviations: HCC, hepatocellular carcinoma; MU, mutant; WT, wild-type; NC, negative control.
AEG1 Silencing Inhibits the PD-L1 and EGFR Expression in HCC
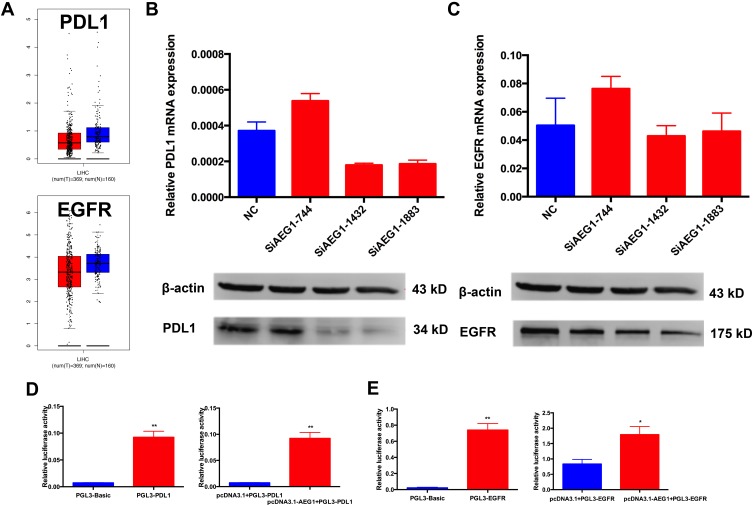
To investigate the functions of AEG1 in HCC metastasis, we transfected HepG2 cells with SiAEG1 into HepG2 cells and measured the PD-L1 and EGFR mRNA and protein levels using qRT-PCR and Western blotting. We found that when AEG-1 was down-regulated, the levels of both PD-L1 and EGFR mRNA and protein both decreased (Figure 4B and C). Next, we cloned the PD-L1 and EGFR gene promoter regions into a luciferase reporter construct (pGL3-basic). The luciferase activity of PD-L1 and EGFR increased when AEG1 was overexpressed compared with controls (Figure 4D and E). These findings indicated that AEG1 transcriptionally activates PD-L1 and EGFR.
Figure 4.
AEG1 silencing inhibits the PD-L1 and EGFR expression in HCC.
Notes: (A) There is no difference of PD-L1 and EGFR messenger RNA expression between HCC tissues and normal tissues. (B) qRT-PCR assay and Western blot assay to measure PD-L1 mRNA and protein levels when AEG1 was down-regulated, and that the siRNA efficiency of AEG1 was most marked at the 744 and 1883 sites in HCC cells. (C) qRT-PCR assay and Western blot assay to measure EGFR mRNA and protein levels when AEG1 was down-regulated, and that the siRNA efficiency of AEG‐1 was most marked at the 744 and 1883 sites in HCC cells. (D) Luciferase assay showing activity for co-transfection of AEG1 and PDL1 promoter vector in HCC cells (**p < 0.01). (E) Luciferase assay showing activity for co-transfection of AEG1 and EGFR promoter vector in HCC cells (**p < 0.01; *p < 0.05).
Abbreviations: HCC, hepatocellular carcinoma; MU, mutant; WT, wild-type; qRT-PCR, quantitative real-time PCR; NC, negative control.
Discussion
Recently, more and more evidences have showed that microRNAs play crucial roles in tumorigenesis, Searching a microRNA signature can potentially be effective clinical tools for diagnosis, prognosis, and therapy in HCC.17 MiR-885-5p was studied in the context of cancer for several years; however, its effect on HCC still needs to be investigated. Afanasyeva et al reported that miR-885-5p was down-regulated upon deletion of 3p25.3 region in neuroblastoma. They showed that miR-885-5p has a tumor-suppressive role in neuroblastoma via interfering with cell cycle progression and cell survival;8 Yan et al demonstrated that miR-885-5p reduces the levels of MMP-9 expression and inhibits cellular invasion in glioma cells;9 Cybula et al illustrated that miR-885-5p may act as a tumor suppressor genes, and it is an indicator for the classification of laryngeal cancer tissue and normal mucosa.18 Such contradictory effects of miR-885-5p prompted us to investigate its role in HCC. Currently, the clinical significance of miR-885-5p was disclosed in HCC. We found that miR-885-5p inhibited proliferation, migration and reduced EMT in HCC cells. Therefore, miR-885-5p potentially serves as a promising biomarker for the prognosis of patients with HCC.
AEG1 has a crucial role in tumor progression, including transformation, evasion of apoptosis, invasion, metastasis and chemoresistance.19 Notably, the present study also identified that AEG1 silencing inhibits the expression of cyclin D1, c‐Myc and MMP9. Previous studies suggested that cyclin D1 and c‐Myc are associated with cell proliferation, and are involved in tumorigenesis and deterioration.20,21 In addition, AEG1 activates various signaling factors involved in mediating EMT, including sonic hedgehog protein, transforming growth factor-β and protein Wnt signaling.22–24 To explore whether AEG1 was involved in the miR-885-5p-mediated regulation in HCC cells, we first utilized the bioinformatics and dual-luciferase report assays to confirm the potential binding sites in miR-885-5p and AEG1 in HCC cells. Next, we measured the expression level of AEG1 in HCC cells. Furthermore, qRT-PCR and Western blotting were employed to examine the function of miR-885-5p on the expression levels of AEG1. We found that up-regulated miR-885-5p could remarkably suppress the level of AEG1 in both mRNA and protein levels. Furthermore, luciferase assays were performed to confirm the regulatory interaction between miR-885-5p and AEG1.
In summary, this study revealed that overexpression of miR-885-5p significantly inhibits the metastatic ability and reverses the EMT to MET in HCC cells. Our data suggested that the function of miR-885-5p in HCC cells is partially exerted through targeting AEG1, and the signal pathway including miR-885-5p/AEG1 might be a potential target for treating HCC.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Finn RS. Current and future treatment strategies for patients with advanced hepatocellular carcinoma: role of mTOR inhibition. Liver Cancer. 2012;1(3–4):247–256. doi: 10.1159/000343839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262 [DOI] [PubMed] [Google Scholar]
- 3.de Martel C, Ferlay J, Franceschi S, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13(6):607–615. doi: 10.1016/S1470-2045(12)70137-7 [DOI] [PubMed] [Google Scholar]
- 4.Perz JF, Armstrong GL, Farrington LA, Hutin YJ, Bell BP. The contributions of hepatitis B virus and hepatitis C virus infections to cirrhosis and primary liver cancer worldwide. J Hepatol. 2006;45(4):529–538. doi: 10.1016/j.jhep.2006.05.013 [DOI] [PubMed] [Google Scholar]
- 5.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604. doi: 10.1038/s41575-019-0186-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004;5(7):522–531. doi: 10.1038/nrg1379 [DOI] [PubMed] [Google Scholar]
- 7.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5 [DOI] [PubMed] [Google Scholar]
- 8.Afanasyeva EA, Mestdagh P, Kumps C, et al. MicroRNA miR-885-5p targets CDK2 and MCM5, activates p53 and inhibits proliferation and survival. Cell Death Differ. 2011;18(6):974–984. doi: 10.1038/cdd.2010.164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yan W, Zhang W, Sun L, et al. Identification of MMP-9 specific microRNA expression profile as potential targets of anti-invasion therapy in glioblastoma multiforme. Brain Res. 2011;1411:108–115. doi: 10.1016/j.brainres.2011.07.002 [DOI] [PubMed] [Google Scholar]
- 10.Schultz NA, Dehlendorff C, Jensen BV, et al. MicroRNA biomarkers in whole blood for detection of pancreatic cancer. JAMA. 2014;311(4):392–404. doi: 10.1001/jama.2013.284664 [DOI] [PubMed] [Google Scholar]
- 11.Zhang Z, Yin J, Yang J, et al. miR-885-5p suppresses hepatocellular carcinoma metastasis and inhibits Wnt/beta-catenin signaling pathway. Oncotarget. 2016;7(46):75038–75051. doi: 10.18632/oncotarget.12602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Feng S, Yao J, Zhang Z, et al. miR96 inhibits EMT by targeting AEG1 in glioblastoma cancer cells. Mol Med Rep. 2018;17(2):2964–2972. doi: 10.3892/mmr.2017.8227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qureshi R, Arora H, Rizvi MA. EMT in cervical cancer: its role in tumour progression and response to therapy. Cancer Lett. 2015;356(2 Pt B):321–331. doi: 10.1016/j.canlet.2014.09.021 [DOI] [PubMed] [Google Scholar]
- 14.Emdad L, Sarkar D, Su ZZ, et al. Activation of the nuclear factor kappaB pathway by astrocyte elevated gene-1: implications for tumor progression and metastasis. Cancer Res. 2006;66(3):1509–1516. doi: 10.1158/0008-5472.CAN-05-3029 [DOI] [PubMed] [Google Scholar]
- 15.Li M, Dai Y, Wang L, Li L. Astrocyte elevated gene-1 promotes the proliferation and invasion of breast cancer cells by activating the Wnt/beta-catenin signaling pathway. Oncol Lett. 2017;13(4):2385–2390. doi: 10.3892/ol.2017.5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi: 10.1093/nar/gkx247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anwar SL, Lehmann U. MicroRNAs: emerging novel clinical biomarkers for hepatocellular carcinomas. J Clin Med. 2015;4(8):1631–1650. doi: 10.3390/jcm4081631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cybula M, Wieteska Ƚ, Jozefowicz-Korczynska M, Karbownik MS, Grzelczyk WL, Szemraj J. New miRNA expression abnormalities in laryngeal squamous cell carcinoma. Cancer Biomark. 2016;16(4):559–568. doi: 10.3233/CBM-160598 [DOI] [PubMed] [Google Scholar]
- 19.Hu G, Wei Y, Kang Y. The multifaceted role of MTDH/AEG-1 in cancer progression. Clin Cancer Res. 2009;15(18):5615–5620. doi: 10.1158/1078-0432.CCR-09-0049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Casimiro MC, Di Sante G, Crosariol M, et al. Kinase-independent role of cyclin D1 in chromosomal instability and mammary tumorigenesis. Oncotarget. 2015;6(11):8525–8538. doi: 10.18632/oncotarget.v6i11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richart L, Carrillo-de Santa Pau E, Rio-Machin A, et al. BPTF is required for c-MYC transcriptional activity and in vivo tumorigenesis. Nat Commun. 2016;7:10153. doi: 10.1038/ncomms10153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Valcourt U, Kowanetz M, Niimi H, Heldin CH, Moustakas A. TGF-beta and the smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol Biol Cell. 2005;16(4):1987–2002. doi: 10.1091/mbc.e04-08-0658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin SY, Rath O, Zebisch A, Choo SM, Kolch W, Cho KH. Functional roles of multiple feedback loops in extracellular signal-regulated kinase and Wnt signaling pathways that regulate epithelial-mesenchymal transition. Cancer Res. 2010;70(17):6715–6724. doi: 10.1158/0008-5472.CAN-10-1377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karhadkar SS, Bova GS, Abdallah N, et al. Hedgehog signalling in prostate regeneration, neoplasia and metastasis. Nature. 2004;431(7009):707–712. doi: 10.1038/nature02962 [DOI] [PubMed] [Google Scholar]