Abstract
Cancer is a worldwide increasing burden and its therapy is often challenging and causes severe side effects in healthy tissue. If drugs are loaded into nanoparticles, side effects can be reduced, and efficiency can be increased via the enhanced permeability and retention effect. This effect is based on the fact that nanoparticles with sizes from 10 to 200 nm can accumulate in tumor tissue due to their leaky vasculature. In this work, we produced polycaprolactone (PCL) in the sizes 1.8, 5.4, and 13.6 kDa and were able to produce spherical shaped nanoparticles with mean diameters of 64 ± 19 nm out of the PCL5.4 and 45 ± 8 nm out of the PCL13.6 reproducibly. By encapsulation of paclitaxel the diameter of that nanoparticles did not increase, and we were able to encapsulate 73 ± 7 fmol paclitaxel per 1000 particles in the PCL5.4‐nanoparticles and 35 ± 8 fmol PTX per 1000 PCL13.6‐nanoparticles. Furthermore, we coupled the aptamer S15 to preformed PCL5.4‐nanoparticles resulting in particles with a hydrodynamic diameter of 153 nm. This offers the opportunity to use these nanoparticles for targeted drug delivery.
Keywords: aptamer, nanoparticle, paclitaxel, polycaprolactone
Abbreviations
- DCC
dicyclohexylcarbodiimide
- DLS
dynamic light scattering
- EDC
1‐ethyl‐3‐(3‐dimethylaminopropyl)carbodiimide
- FDA
US Food and Drug Administration
- NHS
N‐hydroxysuccinimide
- NP
nanoparticle
- PCL
polycaprolactone
- PTX
paclitaxel
- TEM
transmission electron microscopy
1. INTRODUCTION
In 2018, there were estimated 18.1 million new cancer cases and 9.6 million cancer deaths worldwide and this number is still growing 4. This shows the importance to optimize chemotherapy to prevent unspecific side effects of chemotherapeutics as well as to improve biopharmaceutical properties. One approach is the utilization of drug delivery systems, where drugs are combined with carriers like micelles, liposomes, or nanoparticles to transport the drug to the target tissue 2. These carrier systems usually have sizes between 10 and 200 nm 5. The determined size limit of particles for internalization into cells via receptor‐mediated endocytosis was 200 nm, consequently the size of carriers should not exceed this size if particles are to be internalized by this uptake route 6. Furthermore, the size between 10 and 200 nm limits the penetration through the endothelial cells in healthy vasculature, but the leaky vasculature in solid tumors enables enrichment of particles in the tumor tissue 5. This effect is called enhanced permeability and retention effect 2. For drug delivery systems polymer nanoparticles are widely used 7. For example, polycaprolactone (PCL) has been used in combination with other polymers for many drug delivery systems 7, 8, 9. PCL is usually produced by ring‐opening polymerization of ε‐caprolactone 10. It is a FDA (US Food and Drug Administration) approved polymer that is biodegradable and non‐toxic, and therefore suitable for clinical applications 10. However, the preparation of nanoparticles out of pure PCL has only been performed in a few studies and resulting nanoparticles had diameters from 190 to 350 nm, although different preparation methods have been used 10, 11, 12, 13. These sizes are not suitable for the use in drug delivery systems especially if targeting ligands should be added that can further increase nanoparticle size 14. By introducing a targeting ligand to a drug delivery system, nanocarriers can specifically bind to target cells, which consequently internalize the particles and therefore allow a drug release inside the cells 5. Aptamers are one group of affinity ligands, which can be used for such targeting purpose 5. Aptamers are short single stranded DNA or RNA oligonucleotides that are selected against their targets in a process called systematic evolution of ligands by exponential enrichment 15. They fold in secondary and tertiary structures, and based on the resulting 3D structure they are able to bind their targets with high affinity and specificity 15.
The aim of this work is to develop a suitable platform to encapsulate hydrophobic drugs like paclitaxel (PTX). PTX (dissolved in polyethoxylated castor oil and ethanol) is FDA approved for treatment of solid tumors, but the formulation causes severe side effects 9. We chose polycaprolactone as a biocompatible and FDA approved polymer for the generation of nanoparticles (NP) with a size below 100 nm. PCL is hydrophobic and therefore can transport hydrophobic chemotherapeutics, e.g. PTX, and a reduction of side effects compared to the standard formulation could be expected. The aptamer S15 which specifically binds to non‐small cell lung cancer cells will be coupled to the nanoparticles to show that adding a targeting moiety is possible for the developed system 16.
2. MATERIALS AND METHODS
2.1. Materials
ε‐Caprolactone, methanesulfonic acid, dicyclohexylcarbodiimide (DCC), and 1‐ethyl‐3‐(3‐dimethylaminopropyl)carbodiimid (EDC) were purchased from Sigma Aldrich GmbH, Germany. Triethylamine, ethanol, agarose, and Roti®Gelstain, as well as sodiumcarbonate and sodiumhydrogencarbonate that were used to prepare the carbonate buffer, were purchased from Carl Roth GmbH, Karlsruhe, Germany. Sodium borate buffer was produced with boric acid (Sigma Aldrich GmbH, Germany) and sodium hydroxide (AppliChem GmbH, Darmstadt, Germany). Trizma®‐base purchased from Sigma Aldrich GmbH, Germany, glacial acetic acid purchased from AppliChem GmbH, Darmstadt, Germany, and EDTA purchased from Fluka Chemie AG (Swiss) were used for TAE buffer for agarose gel electrophoresis. 5x Green GoTaq® reaction buffer was acquired from Promega GmbH, Mannheim, Germany. Acetonitrile was purchased from VWR international, Darmstadt, Germany. N‐Hydroxysuccinimide (NHS) was purchased from Fluka Chemie AG (Swiss). Paclitaxel (PTX) was purchased from Cayman Chemical Company, USA. The S15‐aptamer with a 5′ terminal amino‐C6‐linker was purchased from BioSpring Gesellschaft für Biotechnologie mbH, Frankfurt/Main, Germany, and it has the following sequence: ACGCTCGGATGCCACTACAGGCTATCTTATGGAAATTTCGTGTAGGGTTTGGTGTGGCGGGGCTACTCATGGACGTGCTGGTGAC
PRACTICAL APPLICATION
In developed countries cancer is the leading cause of death 1. Cancer treatment often causes severe side effects usually due to the unspecific distribution of administered drug in the patient's body 2. Furthermore, some drugs have unfavorable properties, e.g. poor water‐solubility, and application is challenging 3. Drug delivery systems (DDS) can mitigate these disadvantages by encapsulating the drugs in carrier particles which deliver drugs to target tissue 3. In this work we demonstrate the production of small polycaprolactone nanoparticles with a narrow size distribution and a mean diameter of 35–65 nm. In addition, it was shown that the drug paclitaxel can be encapsulated inside these nanoparticles. Furthermore, we evaluate different strategies to functionalize the nanoparticles with a targeting ligand such as an aptamer. Due to the small size and the addition of a targeting ligand we made advances towards polycaprolactone‐based targeted drug delivery systems based on pure polycaprolactone.
2.2. Preparation and characterization of polymers
PCL with a terminal carboxylic group with different sizes was produced by ring‐opening reaction of ε‐caprolactone as reported in Talom et al. 17. Briefly, 45 mmol ε‐caprolactone was dissolved in a toluene–water mixture with methanesulfonic acid. The reactions were performed at 50°C and were quenched by addition of triethylamine, and then the solvents were removed by rotary evaporation. By changing the reaction conditions, polymers with different sizes were produced: a size of 1775 Da was obtained by mixing 45 mmol ε‐caprolactone with 10 mL toluene, 9 µL water, and 0.59 mmol methanesulfonic acid for 1.5 h and quenching with 9 µL triethylamine. For a polymer size of 5536 Da, 45 mmol ε‐caprolactone was incubated with 20 mL toluene, 18 µL water, and 1.15 mmol methanesulfonic acid for 1 h; for quenching, 50 µL triethylamine was used. The size of 13598 Da was obtained by incubation of 45 mmol ε‐caprolactone with 20 mL toluene, 18 µL water, and 1.15 mmol methanesulfonic acid for 1.5 h; for quenching, 50 µL triethylamine was used. Purification was performed by dissolving the solid in 30 mL dichloromethane and subsequently precipitating the polymer in 100 mL cold methanol. After centrifugation (7000 × g, 10 min) the supernatant was removed and the remaining solid was washed with 100 mL cold methanol. The polymers were dried and analyzed by 1H‐NMR (BRUKER‐Advance‐400 at 400 mHz, CDCl3). According to Claesson et al., the degree of polymerization of the PCL was calculated by comparing the integrals of the protons on the methylene group next to the hydroxyl group with the protons of the methylene group next to the carbonyl carbon 18. The resulting polymers had 13.4, 36, and 117 degrees of polymerization, which correspond to molecular weights of 1775, 5436, and 13598 Da, respectively.
2.3. Functionalization of polymers
Coupling of polymers with the amino‐aptamer was performed with DCC and NHS as coupling reagents. An amount of 2.5 nmol S15‐aptamer was mixed with 34.4 µL 70 mM carbonate buffer (70 mM sodium carbonate and sodium hydrogen carbonate, pH 10). Then 250 nmol PCL5.4, 12.5 µmol DCC, and 12.5 µmol NHS were dissolved in 100 µL DMSO at 70°C and the solution was added to the aptamer. After 2 min of ultrasonication, the reaction volume was incubated at 20°C, 500 rpm for 6 h. Every 60 min fresh DCC and NHS (each 12.5 µmol in 40 µL DMSO) were added followed by ultrasonication for 2 min. Then the solvents were removed under reduced pressure and the solid was resuspended in water. The aptamer‐PCL5.4 conjugate was purified with a RP‐HPLC (Hitachi Chromaser, VWR; C‐18 Kinetex 2,6 u, 100 × 4,6 nm column, Phenomenex; gradient elution with triethylammoniumacetate solution (0.1 M triethylamine, 0.1 M glacial acetic acid) from 5 to 50% ACN over 25 min; 1 mL fractions were collected).
2.4. Preparation and characterization of nanoparticles
Nonfunctional NPs were prepared by a solvent evaporation method. The polymer was dissolved in ACN to get a 70 µM polymer solution; if PTX was encapsulated PTX was added to the ACN to get a 140 µM PTX solution. To ensure that the polymer is completely dissolved, the solution was heated at 50°C. An amount of 400 µL of this solution was added to 800 µL 100% ethanol and mixed at 1000 rpm for 30 min, then the solution was diluted with 4.8 mL deionized water resulting in a final PCL concentration of 5.6 µM. Later, the solvents were removed by rotary evaporation and the resulting NP solution was characterized by dynamic light scattering (DLS) (Litesizer™ 500, Anton Paar), nanoparticle tracking analysis (Nanoparticle Analysis System, Firefly, NanoSight), and transmission electron microscopy (TEM). TEM samples were prepared on a carbon coated mesh copper grid. Therefore, a droplet of nanoparticles in water was dropped on the copper grid. After drying, the measurements were done (FEI Tecnai G2 F20 TMP‐TEM with a 120 kV FEG in bright field mode).
2.5. Functionalization of nanoparticles
Coupling of aptamers to preformed nanoparticles was done with EDC as coupling reagent. Nanoparticles were concentrated under reduced pressure (rotary evaporator) and subsequently mixed with S15‐aptamer in a molar ratio of 1:10000 (NP to S15) in 700 µL 10 mM sodium borate buffer (10 mM boric acid, pH adjusted with sodium hydroxide to 7.4) resulting in concentrations of 352 nM S15 and 35.2 pM particles. After 5 min incubation 189 µL of a 10 mg/mL EDC solution in 10 mM sodium borate buffer was added and mixed for 24 h at room temperature. Then the reaction was terminated by adding 50 mM sodium borate buffer (50 mM boric acid, pH adjusted with sodium hydroxide to 8.3). Here a volume of sodium borate buffer was used that is equal to the volume of the reaction mixture and incubation was performed for 1 h. To determine the amount of uncoupled aptamer, a sample of the reaction mixture was filtrated with a 100 kDa cut‐off membrane and washed with 200 µL sodium borate buffer (50 mM, pH 8.3) eight times. The flow through was dried under reduced pressure. After solving in 10 µL water, the amount of uncoupled aptamer was determined densitometrically by agarose gel electrophoresis (1% agarose gel with Roti®Gelstain in TAE buffer [40 mM Trizma®‐Base, 20 mM glacial acetic acid, 1 mM EDTA, pH 8], 100 V, 30 min, different concentrations of the S15‐aptamer as calibration, analysis with ImageJ). Purification of the reaction mixture was performed by dialysis against deionized water with a 100 kDa cut‐off dialysis tube at 4°C to remove unbound aptamers, EDC, and buffer components from the PCL5.4‐S15 nanoparticles.
2.6. Characterization of PTX encapsulation
To determine the amount of encapsulated PTX ultrafiltration was used to separate the solution containing residual PTX from the PTX‐containing NPs. Therefore, 2 mL of the prepared NP solution was centrifuged on a 100 kDa membrane at 5000 × g for overall 8 min. Then possibly remaining free PTX was washed from the membrane with 500 µL water. The flow through was dried under reduced pressure and then resuspended in 50 µL ACN. Possibly remaining free polymers were also able to permeate the membrane. To make sure that they are not contained in the ACN‐PTX solution, a centrifugation at 4°C, 5000 × g was performed for 10 min, two times. Preliminary studies showed that at this temperature PCL precipitates and therefore, can be removed from the PTX containing supernatant which enables an interference free measurement of PTX concentration. The absorption of the supernatant at 227 nm was measured by spectral photometry in triplicate. The encapsulated amount of PTX was calculated using the following equation:
| (1) |
3. RESULTS AND DISCUSSION
Aim of the work was the preparation of a nanocarrier system to encapsulate the hydrophobic drug paclitaxel. As carrier material we used the polymer PCL with different sizes. Furthermore, the nanocarrier was modified with a targeting moiety. For this purpose we utilized the aptamer S15.
3.1. Polymer preparation
Polymers were prepared by ring‐opening reaction of ε‐caprolactone. A terminal carboxylic group was introduced by methanesulfonic acid to enable later functionalization of the polymers. Successful polymer preparation was confirmed by 1H‐NMR, as shown in Figure 1 (for the spectra of the other polymers see Supporting Information S1). The degree of polymerization was determined by comparing the integrals of the protons on the methylene group next to the hydroxyl group with the protons of the methylene group next to the carbonyl carbon. Calculated molecular weights are shown in Table 1, as well as the nomenclature of the used polymers and the resulting nanoparticle diameters.
Figure 1.
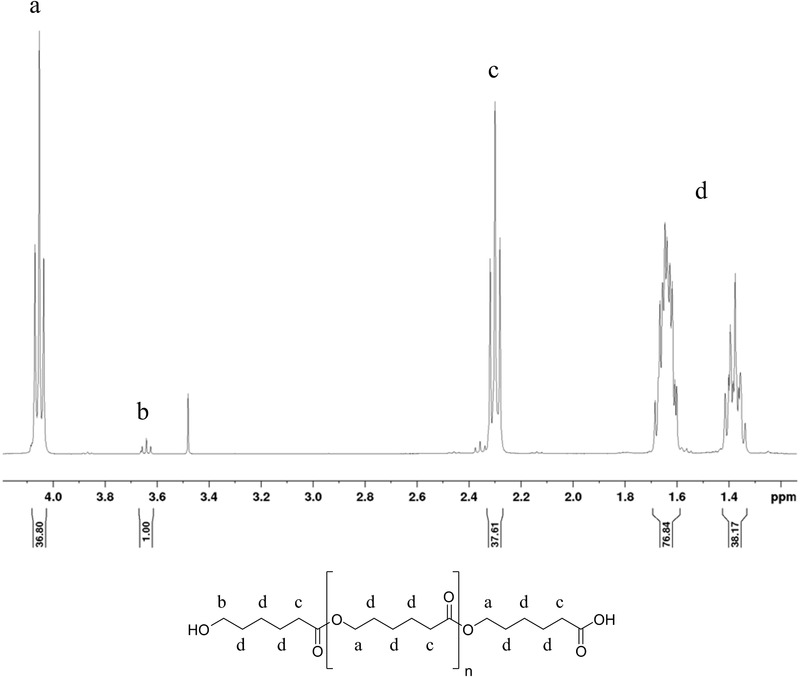
1H‐NMR of PCL; structure and corresponding peaks in the NMR diagram
Table 1.
Denominations of synthesized polymers, their molecular weights, and the resulting nanoparticle sizes
| Denomination of polymers* | Molecular weight [Da] | Mean hydrodynamic diameter of NP ± SD [nm] | PCL‐NP concentration ± SD [particles/mL] | Mean hydrodynamic diameter of PTX‐PCL‐NP ± SD [nm] |
|---|---|---|---|---|
| PCL1.8 | 1775 | 78 ± 112 | 1.94 × 108 ± 0.7 × 108 | 287 ± 202 |
| PCL5.4 | 5436 | 64 ± 19 | 0.89 × 108 ± 0.05 × 108 | 37 ± 3 |
| PCL13.6 | 13598 | 45 ± 8 | 1.72 × 108 ± 0.5 × 108 | 55 ± 8 |
*
Subscript indicates the size of the used polymer (molecular weight in kDa).
3.2. Nanoparticle preparation
Nanoparticles were prepared by solvent evaporation method in triplicate using ACN as a solvent. Polymer solutions were mixed with ethanol for 30 min and then diluted with water (Figure 2F). Solvents were evaporated and the resulting nanoparticles were analyzed with DLS. Using the PCL1.8 leads to very high variation in size distributions and none of them is in the desired size range between 30 and 100 nm (Figure 2A). Obviously, the chosen method is not suitable to prepare NPs out of the PCL1.8 in a reproducible manner, therefore the SD of the hydrodynamic diameter is very large (Figure 2D). A reason for this could be the different CMCs for polymers with different sizes. Letchford et al. showed that for mPEG‐PCL block copolymers with increasing PCL size, the CMC decreases 19. This means that for smaller polymers the CMC is higher than for larger polymers. Arguably, the concentration of 5.6 µM PCL1.8 is below the CMC and therefore the particle formation cannot occur properly. The PCL5.4 forms nanoparticles with a mean diameter of 64 nm, while the mean diameter of the PCL13.6‐NP is 45 nm with an additional population of approximately 2500 nm in one measurement, both size distributions look very similar for the triplicate (Figure 2B and C). Furthermore, TEM analysis of PCL5.4‐NP shows that the nanoparticles have a spherical shape and no aggregates occur. These analyses show that PCL5.4 and PCL13.6 can be used to reproducibly produce spherical sized NPs within the desired size range.
Figure 2.
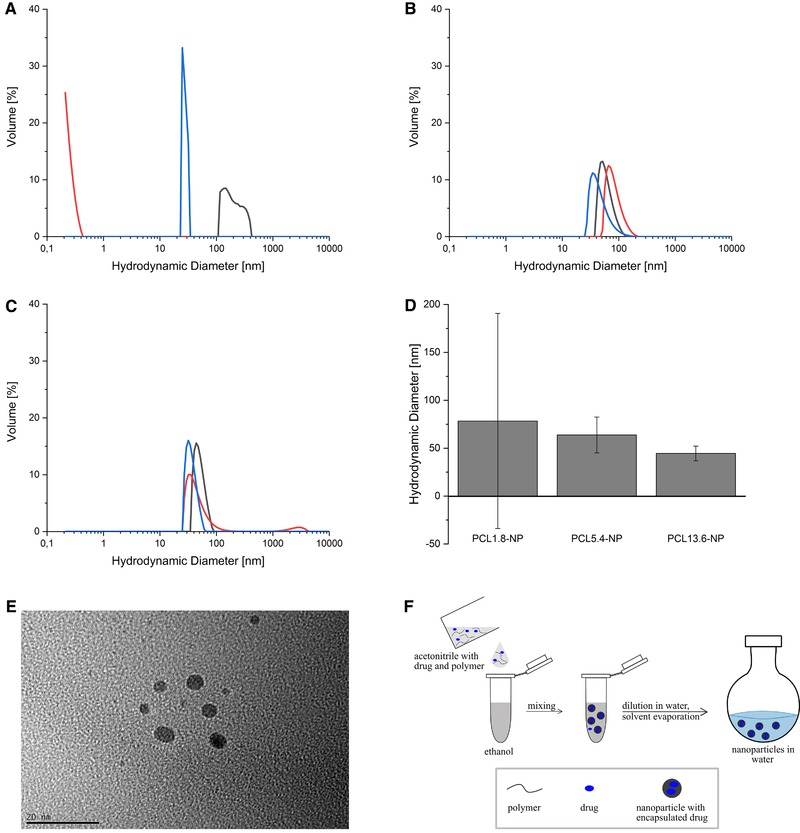
Size distribution of nanoparticles generated from different PCL‐length polymers. (A) PCL1.8‐NP, (B) PCL5.4‐NP, (C) PCL13.6‐NP, (D) mean diameters with SD, (E) TEM image of PCL5.4‐NP, (F) scheme of NP preparation
3.3. Encapsulation of PTX
It could be demonstrated that the chosen method is suitable to produce nanoparticles from PCL in a size range from 45 to 80 nm. To show that the nanoparticles are suitable for drug encapsulation, PTX was encapsulated as a model drug during the nanoparticle preparation. Here, we investigate the effect of the polymer size on the amount of encapsulated PTX. The PTX was encapsulated into the nanoparticles by adding the drug into the acetonitrile polymer solution during the nanoparticle preparation. Again, nanoparticles generated from the PCL1.8 with PTX show high variations in size distribution; therefore, the used method is proven not to yield in nanoparticles with a reproducible size with this polymer (Figure 3A). However, the PCL5.4‐PTX‐NP shows homogenous size distribution with a mean diameter of 37 ± 3 nm (size distributions for all polymers see Supporting Information S2). The PCL13.6‐PTX‐NP with a mean diameter of 55 ± 8 nm also shows homogeneous size distribution. For all polymers, no effect of the PTX encapsulation on the final size of nanoparticles was observed. For the PCL5.4‐PTX‐NP and the PCL13.6‐PTX‐NP the mean diameter is reproducible between 30 and 60 nm.
Figure 3.
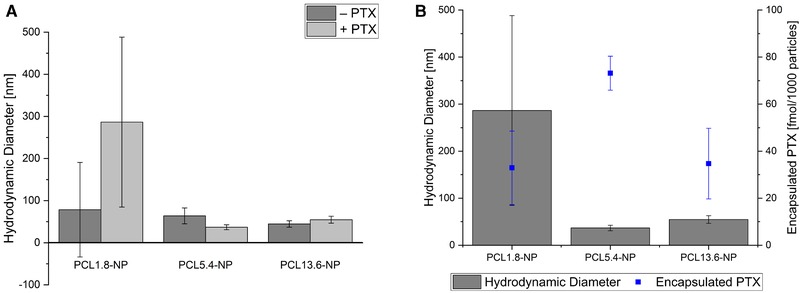
Comparison of nanoparticle diameter. Nanoparticles were prepared from polymers with different molecular weights with and without encapsulated PTX (A); encapsulated amount of PTX is shown in (B)
Despite no apparent effect of the PTX encapsulation on the diameter of the NPs was observed, we investigated the effect of polymer size on the amount of encapsulated PTX. For this, the free drug was removed from the prepared nanoparticles by filtration with a 100 kDa membrane. This 100 kDa cutoff corresponds to a pore size of about 8 nm and therefore, NPs cannot penetrate the membrane while the PTX (0.8 kDa) can pass through it 20. Subsequently, the flow through was dried by vacuum concentration and the contained PTX was resolved in ACN. Remaining free polymer was removed by centrifugation. The concentration of PTX in the supernatant was determined by absorption measurement at 227 nm with a spectral photometer. Particle concentration was determined with nanoparticle tracking analysis and the amount of encapsulated PTX per nanoparticle was calculated (Figure 3B).
Successful encapsulation of PTX was shown for all prepared polymer sizes. However, the PCL5.4‐NPs contain the highest amount of PTX (73.2 ± 7.2 fmol per 1000 particles). Therefore, PCL5.4 was chosen for the following functionalization with the aptamer S15.
3.4. Functionalization of PCL5.4‐NPs with S15‐aptamer
By adding the aptamer S15 as a targeting moiety to a drug delivery system, cellular uptake can be promoted 21. One way to produce aptamer‐modified NPs is to prepare NPs first and subsequently couple the aptamers to the preformed nanoparticles. Coupling was performed with EDC as coupling reagent. After coupling unreacted EDC, buffer compounds and free aptamers were removed from the reaction mixture by dialysis with a 100 kDa membrane. Subsequent PCL5.4‐S15‐NPs were analyzed by DLS, resulting in a hydrodynamic diameter of 153 nm (Figure 4). To determine the amount of unbound aptamer, a sample of the reaction mixture was taken before purification and filtrated with a 100 kDa membrane. The size of the S15‐aptamer is 14.2 kDa, so it can pass through the membrane easily. Flow through fractions were subsequently dried by vacuum concentration and the containing S15‐aptamer was redissolved in water for quantification. By comparison with a calibration of S15 with different concentrations, the amount of unbound S15 was calculated. We were able to couple about 9330 aptamers per nanoparticle.
Figure 4.
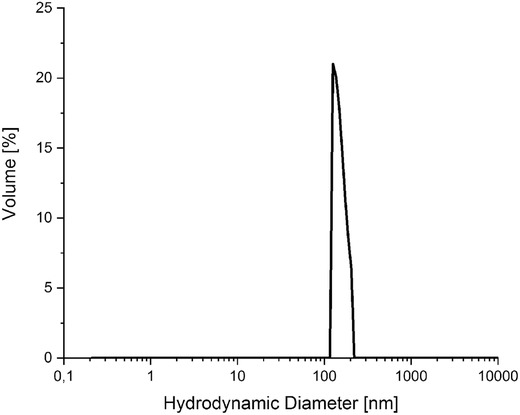
Size distribution of PCL5.4‐S15‐NP; nanoparticles were prepared prior to coupling with the aptamer S15
3.5. Functionalization of PCL5.4 with S15 aptamer prior to NP formation
An alternative route to formation of aptamer‐modified PCL nanoparticles is the functionalization of the PCL polymer prior to NP formulation. This way is less commonly used when compared to functionalization of preformed NPs. The reason is that PCL‐NPs are stable in water, and the aptamer can be dissolved in water. In contrast, the aptamer and the PCL5.4 are not soluble in the same solvent; therefore, this procedure entails some challenges during the production of PCL5.4 aptamer conjugates. In this work, we were able to adapt a method from Talom et al. to couple the aptamer with PCL5.4 utilizing NHS and DCC as coupling reagents for the amino modified aptamer to the carboxylic group of PCL5.4 in a one‐step coupling 17. PCL5.4, as well as the DCC and NHS, was dissolved in DMSO whereas the aptamer was dissolved in carbonate buffer. By ultrasonication, a large surface area was introduced into the two‐phase system to enhance coupling efficiency. Subsequent purification of aptamer PCL5.4 conjugate (S15‐PCL5.4) was performed via preparative RP‐HPLC (Chromatogram in Supporting Information S3). The coupling efficiency of the aptamer to the polymer was 81%. By mixing the conjugate with unconjugated PCL5.4 it is easy to control the amount of aptamer presented on the nanoparticle surface. Another advantage of this method is that the aptamer is hydrophilic and therefore, increases the ambition of the polymer to form micelles with a hydrophobic core, as already been shown for other oligonucleotide‐polymer conjugates 22.
4. CONCLUDING REMARKS
The aim of this work was the development of a platform to encapsulate PTX in polycaprolactone nanoparticles in a size range that is suitable for drug delivery. Nanoparticles were successfully produced with and without encapsulated PTX with sizes between 40 and 70 nm for the PCL5.4‐NP and PCL13.6‐NP, whereas the encapsulation of PTX seems to have negligible effect on the final particle size. In addition the aptamer S15 was added to the system by coupling the aptamer S15 to preformed nanoparticles, resulting in hydrodynamic diameters of 153 nm. As an alternative route for aptamer introduction, a method for coupling of the hydrophilic aptamer with the hydrophobic PCL was developed successfully.
CONFLICT OF INTEREST
The authors have declared no conflict of interest. The manuscript does not contain experiments using animals or human studies.
Supporting information
Supporting Information
ACKNOWLEDGMENTS
We would like to thank Dr. Gerrit Jürjens (Helmholtz Zentrum München) for 1H‐NMR measurements and the Institute for Physical Chemistry and Electrochemistry (Leibniz University Hannover) for access to the Nanoparticle Tracking Analysis (NTA). Furthermore, the authors thank Bilal Temel for TEM measurement and the Laboratory of Nano and Quantum Engineering (LNQE) of the Leibniz University of Hannover for the TEM instrument. This work was funded by the Lower Saxony Ministry for Science and Culture (MWK).
Witt S, Scheper T, Walter J‐G. Production of polycaprolactone nanoparticles with hydrodynamic diameters below 100 nm. Eng Life Sci. 2019;19:658–665. 10.1002/elsc.201800214
REFERENCES
- 1. Jemal, A. , Bray, F. , Center, M. M. , Ferlay, J. et al., Global cancer statistics. CA Cancer J. Clin. 2011, 61, 69–90. [DOI] [PubMed] [Google Scholar]
- 2. Torchilin, V. P. , Passive and active drug targeting: drug delivery to tumors as an example, in: Schäfer‐Korting M. (Ed.), Handbook of Experimental Pharmacology, Springer‐Verlag, Berlin Heidelberg, 2010. [DOI] [PubMed] [Google Scholar]
- 3. Farokhzad, O. C. , Langer, R. , Impact of nanotechnology on drug delivery. ACS Nano 2009, 3, 16–20. [DOI] [PubMed] [Google Scholar]
- 4. Bray, F. , Ferlay, J. , Soerjomataram, I. , Siegel, R. L. et al., Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [DOI] [PubMed] [Google Scholar]
- 5. Zhang, H. , Zhou, L. , Zhu, Z. , Yang, C. , Recent progress in aptamer‐based functional probes for bioanalysis and biomedicine. Chemistry 2016, 22, 9886–9900. [DOI] [PubMed] [Google Scholar]
- 6. Rejman, J. , Oberle, V. , Zuhorn, I.S. , Hoekstra, D. , Size‐dependent internalization of particles via the pathways of clathrinand caveolae‐mediated endocytosis. Biochem. J. 2004, 377, 159–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kumari, A. , Yadav, S.K. , Yadav, S.C. , Biodegradable polymeric nanoparticles based drug delivery systems. Colloids Surf. B Biointerfaces 2010, 75, 1–18. [DOI] [PubMed] [Google Scholar]
- 8. Modrejewski, J. , Walter, J.‐G. , Kretschmer, I. , Kemal, E. et al., Aptamer‐modified polymer nanoparticles for targeted drug delivery. BioNanoMaterials 2016, 17, 1–2. [Google Scholar]
- 9. Kim, S. Y. , Lee, Y. M. , Taxol‐loaded block copolymer nanospheres composed of methoxy poly(ethylene glycol) and poly(ε‐caprolactone) as novel anticancer drug carriers. Biomaterials 2001, 22, 1697–1704. [DOI] [PubMed] [Google Scholar]
- 10. Mei, L. , Zhang, Y. , Zheng, Y. , Tian, G. et al., A novel docetaxel‐loaded poly ( ε‐caprolactone)/pluronic F68 nanoparticle overcoming multidrug resistance for breast cancer treatment. Nanoscale Res. Lett. 2009, 4, 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ahmad, A. , Fauzia, E. , Kumar, M. , Mishra, R. K. et al., Gelatin‐coated polycaprolactone nanoparticle‐mediated naringenin delivery rescue human mesenchymal stem cells from oxygen glucose deprivation‐induced inflammatory stress. ACS Biomater. Sci. Eng. 2019, 5, 683–695. [DOI] [PubMed] [Google Scholar]
- 12. Ajiboye, A. L. , Trivedi, V. , Mitchell, J. C. , Preparation of polycaprolactone nanoparticles via supercritical carbon dioxide extraction of emulsions. Drug Deliv. Transl. Res. 2018, 8, 1790–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Badri, W. , Miladi, K. , Robin, S. , Viennet, C. et al., Polycaprolactone based nanoparticles loaded with indomethacin for anti‐inflammatory therapy: from preparation to ex vivo study. Pharm. Res. 2017, 34, 1773–1783. [DOI] [PubMed] [Google Scholar]
- 14. Askarian, S. , Abnous, K. , Taghavi, S. , Oskuee, R.K. et al., Cellular delivery of shRNA using aptamer‐conjugated PLL‐alkyl‐PEI nanoparticles. Colloids Surf. B Biointerfaces 2015, 136, 355–364. [DOI] [PubMed] [Google Scholar]
- 15. Tuerk, C. , Gold, L. , Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 1990, 249, 505–510. [DOI] [PubMed] [Google Scholar]
- 16. Zhang, Y. , Chen, Y. , Han, D. , Ocsoy, I. et al., Aptamers selected by cell‐SELEX for application in cancer studies. Bioanalysis 2010, 2, 907–918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Talom, R.M. , Fuks, G. , Mingotaud, C. , Gineste, S. et al., Investigation of the reversibility of the unimer‐to‐aggregate transition in block copolymers by surface tension‐measurements. J. Colloid Interface Sci. 2012, 387, 180–186. [DOI] [PubMed] [Google Scholar]
- 18. Cleasson, H. , Malmström, E. , Johansson, M. , Hult, A. , Synthesis and characterization of star branched polyesters with dendritic cores and the effect of structural variations on zero shear rate viscosity. Polymer 2002, 43, 3511–3518. [Google Scholar]
- 19. Letchford, K. , Liggins, R. , Burt, H. , Solubilization of hydrophobic drugs by methoxy poly(ethylene glycol)‐block‐polycaprolactone diblock copolymer micelles: theoretical and experimental data and correlations. J. Pharm. Sci. 2008, 97, 1179–90. [DOI] [PubMed] [Google Scholar]
- 20. Guo, L. , Santschi, P. , Ultrafiltration and its applications to sampling and characterisation of aquatic colloids, in: Wilkinson K. J., Lead J. R. (Eds.), Environmental Colloids and Particles, IUPAC, Zurich, Switzerland: 2007, pp. 159–221. [Google Scholar]
- 21. Engelberg, S. , Modrejewski, J. , Walter, J.‐G. , Livney, Y. et al., Cancer cell‐selective, clathrin‐mediated endocytosis of aptamer decorated nanoparticles. Oncotarget 2018, 9, 20993–21006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Jeong, J. H. , Park, G. T. , Novel polymer‐DNA hybrid polymeric micelles composed of hydrophobic poly(d,l‐lactic‐co‐glycolic acid) and hydrophilic oligonucleotides. Bioconjug. Chem. 2001, 12, 917–923. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


