Abstract
Polysialic acid (polySia), consisting of α‐(2,8)‐linked N‐acetylneuraminic acid monomers plays a crucial role in many biological processes. This study presents a novel process for the production of endogenous polySia using Escherichia coli K1 in a disposable bag reactor with wave‐induced mixing. Disposable bag reactors provide easy and fast production in terms of regulatory requirements as GMP, flexibility, and can easily be adjusted to larger production capacities not only by scale up but also by parallelization. Due to the poor oxygen transfer rate compared to a stirred tank reactor, pure oxygen was added during the cultivation to avoid oxygen limitation. During the exponential growth phase the growth rate was 0.61 h−1. Investigation of stress‐related product release from the cell surface showed no significant differences between the disposable bag reactor with wave‐induced mixing and the stirred tank reactor. After batch cultivation a cell dry weight of 6.8 g L−1 and a polySia concentration of 245 mg L−1 were reached. The total protein concentration in the supernatant was 132 mg L−1. After efficient and time‐saving downstream processing characterization of the final product showed a protein content of below 0.04 mgprotein/gpolySia and a maximal chain length of ∼90 degree of polymerization.
Keywords: Disposable bag reactor, Escherichia coli K1, Oxygen limitation, Polysialic acid, Stirred tank reactor
Abbreviations
- CDW
cell dry weight
- DP
degree of polymerization
- DMB
1,2‐diamino‐4,5‐methylenedioxybenzene
- DO
dissolved oxygen
- MWCO
molecular weight cut‐off
- polySia
polysialic acid
- Rpm
rocks per minute
- STR
stirred tank reactor
1. Introduction
Polysialic acid (polySia) is a polysaccharide present in prokaryotes as α‐(2,8)‐ and/or α‐(2,9)‐ and in eukaryotic cells as α‐(2,8)‐linked N‐acetylneuraminic acid monomers 1, 2. Some bacteria (e.g., Escherichia coli K1 or Neisseria meningitidis B 3) use a glycocalyx of polySia, linked by a lipid anchor to the cell membrane 4, to mimic the surface of vertebrate cells 5. The polySia capsule protects the bacteria to a certain degree from antibodies, macrophages, and the alternative complement pathway 6, 7.
In mammalians, polySia has different functions. The polymer is involved in the development of the nervous system 8. It also accumulates on the surface of cancer cells 9. PolySia acts as an anti‐adhesive and is involved in the interaction between cells as well as in cell‐matrix and molecular interactions at cell surfaces 1, 8, 10.
The various biological functions make polySia an interesting molecule for different applications. For instance, low molecular weight polySia interacts with SIGLEC‐11, a sialic acid binding immunoglobulin‐like lectin receptor that suppresses the immune defense through recognition of host cells sialic acid decoration 11. It also has an anti‐inflammatory effect on human THP1 macrophages 11. That makes polySia a potential molecule for the treatment of age‐related neurodegenerative diseases (e.g., Alzheimer's disease or age‐related macular degeneration) 11, 12, 13, 14.
The good biocompatibility as well as the targeted degradation by specific enzymes make polySia furthermore interesting for tissue engineering to support the growth of specific human nervous cells 15, 16. PolySia‐based hydrogels are potential scaffold materials 16 and polySia can also be used for coating of scaffold materials 17, 18. PolySia can be also used in vaccine development and cancer immunotherapy 1, 10, 19, 20, 21. Furthermore, stability and the half‐life in the human organism of certain drugs (e.g., insulin or asparaginase) can be improved by polysialylation, as this potentially improves the efficiency of those drugs 1, 22, 23, 24, 25, 26.
The production of polySia is commonly realized by using bacteria (e.g., E. coli K1 or N. meningitidis B) 1, 27, 28, 29. The use of bacteria for the biotechnological polySia production has the advantages of being virus free and a fast cell growth with high productivities on cheap media compared to the production with mammalian cell culture.
The cultivation of microorganisms in single‐use bioreactor systems has several advantages, especially if the products are used for medical application. The systems are generally provided certified and pre‐sterilized by the manufacturer. Thus, time‐consuming implementation of, for example, cleaning steps and the documentation of these steps is omitted, which eases processing according to GMP regulations 30. Those systems also provide flexibility. Different production processes can be run in one single‐use reactor, each using the same equipment 30. Additionally, the acquisition cost of a single‐use bioreactor system is usually lower than of bioreactor systems based on stainless steel with the same volume and the capacity of the disposable reactor systems can easily be adjusted to new conditions and larger scales.
Nevertheless, single‐use systems have disadvantages compared to the established reusable bioreactor systems, such as the recurring consumable costs, because the reactor containers (bags) need to be exchanged after each cultivation and the volume of such disposable bag reactors is limited to a volume range of 2000–3000 L 31. Single‐use systems are mostly suitable for high‐value products, which are only produced in small amounts with varying capacities.
Disposable bag reactors with wave‐induced mixing (maximal working volume below 500 L 31) have a special concept of mixing and aerating the liquid phase and are often used for inoculum production. In a stirred tank reactor (STR) an impeller is used for mixing. STRs are also available as single‐use systems and have become widely accepted for product expression 32. Aeration is usually realized by a ringsparger close to the bottom of the reactor. In a disposable bag reactor aeration is only realized through the head space and the mixing is wave‐induced by rocking motion. Due to the wave‐induced mixing shear stress is minimized and homogenous mixing is possible 33. However, the oxygen transfer in a disposable bag reactor with wave‐induced mixing, compared to an STR, is lower. The volumetric oxygen transfer coefficient value in such reactors is in the range of 47 h−1 compared to 392 h−1 in an STR 34. Disposable bag reactors are commonly used for shear stress sensitive microorganisms with a comparatively low growth rate (e.g., mammalian cells or shear stress sensitive basidiomycetes Flammulina velutipes and Pleurotus sapidus) and not for aerobic bacteria with high oxygen demand and growth rates 33.
This study aims at the development of a novel process using a disposable bag reactor system with wave‐induced mixing for the production of α‐(2,8)‐linked polySia with E. coli K1. The developed process is compared to the state‐of‐the‐art processing in conventional stainless steel reactor setups 28, 29. The produced polySia was investigated regarding product yield, quality, and chain length.
2. Materials and methods
2.1. Bacterial strains and stock cultures
Escherichia coli B2032/82 serotype K1 was used for the experiments. This wild‐type strain is an original clinical isolate 28. The stock cultures were stored in 50% v/v glycerol at −80°C.
2.2. Chemicals and growth media
All bulk chemicals were purchased from Sigma–Aldrich (Taufkirchen, Germany). Oxygen was purchased from Linde (Pullach, Germany). Deionized water was prepared with Arium® (Sartorius Stedim Biotech, Göttingen, Germany). For the bioreactor cultivation, a basic defined medium was used as reported previously for a batch cultivation process 29.
2.3. Cultivation of E. coli K1
2.3.1. Shake flask cultivation and preculture
Precultures were performed in 500 mL baffled shake flasks with 100 mL complex medium consisting of yeast extract (10 g L−1), tryptone (10 g L−1), and NaCl (5 g L−1) at pH 7.3. The E. coli K1 cultures were incubated on a rotary shaker at 37°C, 130 min−1 for 10–12 h. This cell suspension was used as inoculum for the bioreactor cultivations.
2.3.2. Disposable bag reactor
The bioreactor cultivation was carried out on a rocking platform (BIOSTAT® CultiBag RM, Sartorius Stedim Biotech) in a 20 L disposable bag reactor (BIOSTAT® CultiBag RM 20 optical, Sartorius Stedim Biotech). Ten liters basic defined medium was filled in the reactor via a sterile filter (Sartoban® P 300, Sartorius Stedim Biotech). The remaining 10 L reactor volume was used as head space. The basic parameters for the cultivation were as follows: platform angle 10°, pH 7.5, temperature 37°C, and gas supply 1 L min−1 (0.1 vvm, volume per volume per minute). The pH was controlled automatically with 1 M HCl and 25% NH4OH v/v. The set point for dissolved oxygen (DO) was 50%. Variable parameters such as rocking motion, oxygen supply, and internal pressure of the reactor system were investigated and are shown in Section 3. Inoculation volume was 3% v/v of total cultivation volume.
2.3.3. Offline sampling
For the offline analysis described below 10 mL sample was taken out through the sample port of the bioreactor every 2 h.
2.3.3.1. Optical density
The optical density was measured at 600 nm with a photometer (Libra S80, Biochrom, Cambridge, UK).
2.3.3.2. Cell dry weight
The cell dry weight (CDW) was measured by weight difference after centrifugation (13 000 rpm for 5 min), removal of the supernatant, and drying. Drying was performed for 48 h at 80°C.
2.3.3.3. Glucose concentration
The glucose concentration was determined by YSI 2300 STAT Glucose & Lactate Analyzer (Yellow Springs Institutes, OH, USA).
2.3.3.4. Cell disruption
An ultrasonic homogenizer was used for cell disruption (LABSONIC® M, Sartorius Stedim Biotech). The cell disruption was performed in three 30 s intervals, followed each by a 30 s cooling step. The amplitude was set to 100% and the active period was set to 0.6 s−1.
2.4. Downstream process
2.4.1. Cell separation
Directly after harvesting, the cells were separated by continuous centrifugation (Heraeus Contifuge Stratos, Thermo Scientific, Waltham, MA, USA). The cell separation was performed at 17 000 rpm (25 000 g), 4°C, and a flow rate of 100 mL min−1.
2.4.2. Product concentration via cross‐flow ultrafiltration
The supernatant of the continuous centrifugation was approximately 20‐fold concentrated by cross‐flow ultrafiltration. A filter cassette with a molecular weight cut‐off (MWCO) of 10 kDa was used for the ultrafiltration (Hydrosart®, Sartorius Stedim Biotech). The inlet pressure toward the filter cassette was regulated to maximal 4 bar by the flow rate of the pump (SartoJet, Sartorius Stedim Biotech).
2.4.3. Precipitation with ethanol
The retentate of the cross‐flow ultrafiltration was used for precipitation with ethanol. The precipitation was performed at room temperature in three steps. In the first step, the retentate was precipitated by 80% v/v ethanol, in the second step by 85% v/v ethanol, and in the final step by 90% v/v ethanol. After each step, the precipitates were spun down at 4500 rpm and 4°C for 15 min (Multifuge X3 FR, Thermo Scientific). The pellet was dissolved in deionized water after each step.
2.4.4. Purification with clay minerals
The clay mineral EX M 1753 (experimental product name 1753, Clariant, Moosburg, Germany) was used for protein adsorption. EX M 1753 is a saponite with a cation exchange capacity of 20–30 meq 100 g−1, a BET surface of 124 m2 g−1, a cumulative pore volume of 0.12 cm3 g−1, and an average pore diameter of 6.06 nm 35. EX M 1753 was equilibrated in buffer (100 mM trisodium phosphate, pH 7) for 1 h at 130 rpm on a shaker. After the equilibration, the product solution was added and shaken for 3 h under the same conditions. Afterwards EX M 1753 was removed by centrifugation (4500 rpm, 4°C for 15 min).
2.4.5. Product concentration via ultracentrifugation (MWCO 10 kDa)
Centrifuge tubes with an MWCO of 10 kDa and a volume of 20 mL (Vivaspin 20, Sartorius Stedim Biotech) were used for the product concentration before the dialysis. The supernatant after treatment with EX M 1753 was centrifuged at 4500 rpm and 4°C until a fifth of the starting volume was left. With an MWCO of 10 kDa polySia with an approximate degree of polymerization (DP) over 35 units remained in the concentrate.
2.4.6. Dialysis
The product concentrate (50 mL) was dialysed against 5 L deionized water with 5.5 g NaCl for 24 h and afterwards twice against 5 L deionized water 24 h for each (every time pH 9 set with NaOH) using a Visking dialysis membrane (Carl Roth GmbH & Co. KG, Karlsruhe, Germany) with a typical MWCO of 14 kDa.
2.4.7. Lyophilization
Freeze drying of the solution after the dialysis was performed at 0.04 mbar until all of the liquid was removed (Alpha 1–4 LSC, Martin Christ Gefriertrocknungsanlagen GmbH, Osterode, Germany).
2.5. Analysis
2.5.1. Analysis of the polySia concentration
The concentration of polySia was measured colorimetrically using a modified thiobarbituric acid assay as reported in previous studies 28, 29. The calibration curve ranged from 0 to 0.75 g L−1 polySia. The sample volume was 50 μL. First polySia was hydrolyzed with 200 μL 50 mM H3PO4 at 70°C for 18 h. The reaction was stopped with 100 μL 0.1 M NaOH. Afterwards 100 μL oxidation solution (0.2 M periodic acid in 0.5% H3PO4 v/v) was added and the mixture was incubated at 37°C for 30 min. After the incubation 500 μL reduction solution (0.38 M NaAsO2 and 0.5 M Na2SO4 in 2% H2SO4 v/v) was added followed by the addition of 500 μL thiobarbituric acid solution (0.2 M thiobarbituric acid in 1.2% NaOH v/v). The absorption of the emerging pink dye was measured at 549 nm (Multiskan Spectro, Thermo Scientific) after 13 min of incubation at 95°C.
2.5.2. Analysis of the protein concentration
The protein concentration was determined by the Bradford method 36. Twenty microliters sample was mixed with 300 μL Bradford reagent [0.1 g L−1 Coomassie brilliant blue, 50 mL L−1 ethanol (96 % v/v), and 100 mL L−1 phosphoric acid (85% v/v)]. The samples were incubated for 10 min at room temperature and the protein concentration was measured afterwards at 595 nm (Multiskan Spectro, Thermo Scientific).
2.5.3. Analysis of the DNA concentration
The DNA concentration was determined by a UV‐Vis Spectrometer at 260 nm (NanoDrop 2000, Thermo Scientific).
2.5.4. Analysis of the endotoxin concentration
The endotoxin concentration was determined using the Endosafe®‐PTS™ system (Endosafe®‐PTS™, Charles River Laboratories, Boston, MA, USA).
2.5.5. Chain length characterization of polySia by DMB‐HPLC analysis
As reported in previous studies, the DP of the produced polySia was analyzed by 1,2‐diamino‐4,5‐methylenedioxybenzene (DMB) HPLC analysis 29. Therefore, 50 μL sample with a polySia concentration of approximately 1 g L−1 was incubated with 20 μL deionized water, 80 μL TFA (20 mM) and 80 μL DMB solution (7 mM DMB, 12.5 mM Na2SO3, and 0.8 M β‐mercaptoethanol) for 24 h at 10°C. The reaction was stopped by the addition of 20 μL 1 M NaOH. The DMB‐labeled polySia samples were separated by a DNAPac PA‐100 column (Dionex, Idstein, Germany). The injection volume was 25 μL. For the detection, a fluorescence detector (Jasco FP‐1520, Gross‐Umstadt, Germany) was used at 372 nm for extinction and 456 nm for emission. The eluents were deionized water (E1) and 4 M ammonium acetate (E2) at a flow rate of 1 mL min−1. The elution gradient was used as previously reported 29.
3. Results and discussion
The production process is shown in Fig. 1. In the following, first the cultivation is presented followed by the downstream processing and analysis of the final product.
Figure 1.
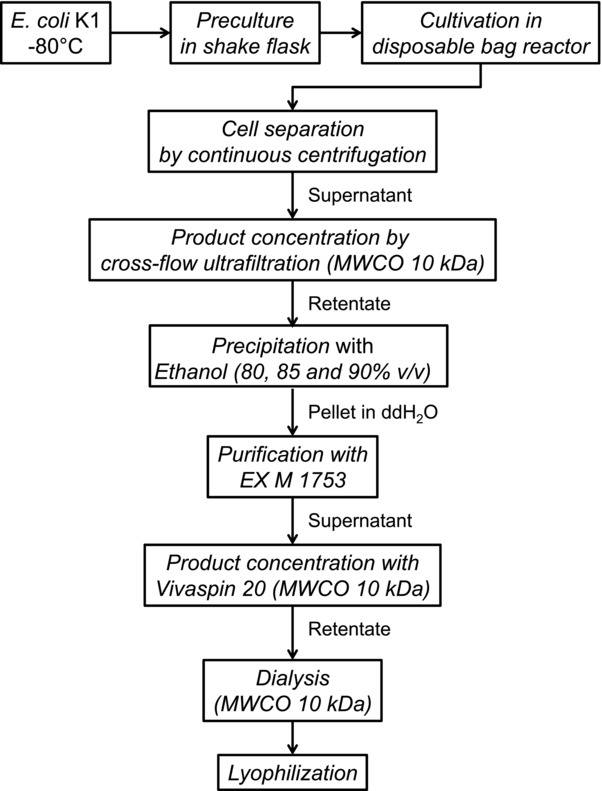
Production process. Each step of the production process is shown. Alongside the arrows the starting material for following step is shown.
3.1. Cultivation of E. coli K1 in a disposable bag reactor
Escherichia coli K1 was cultivated in a 20 L disposable bag reactor with wave‐induced mixing. In total four cultivations were performed. While in an STR a sparger system is used to supply the air directly into the liquid phase, the air in the disposable bag reactor is supplied solely through the head space. In the disposable bag reactor a relatively low gas flow of 1 L min−1 (0.1 vvm) was used (STR 10 L min−1, 1 vvm). A higher gas flow would cause an internal pressure excess in the bag system (limited to 50 mbar). Due to the low gas flow the internal pressure could be kept under 15 mbar throughout the cultivation. A comparison of all relevant cultivation process parameters is listed in Table 1.
Table 1.
Comparison of the cultivation conditions in the reactor systems
The disposable bag reactor was inoculated with 300 mL preculture (3% v/v of the bioreactor working volume). An oxygen limitation was observed already after 5.3 (±1) h at low bacteria concentrations (CDW approximately 1 g L−1) when using air (gas flow: 0.1 vvm) for aeration and controlling the DO concentration (setpoint 50%) by increasing the rocking motion (maximum: 42 rocks per minute [Rpm]). Thus, the aeration flow was supplied with pure oxygen to establish a DO value of 50%. The increase of pure oxygen supply in the aeration flow is shown as solid line in Fig. 2. At the end of the cultivation the gas mixture consisted of almost pure oxygen to achieve a DO value of 50%, allowing the E. coli K1 to grow up to maximal 7.15 g L−1 without oxygen limitation in the disposable bag reactor. All online data of a typical cultivation are shown in Fig. 2.
Figure 2.
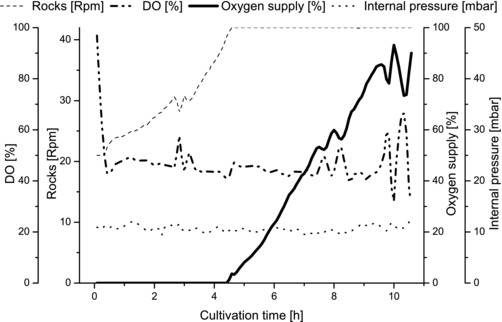
Online data of a typical cultivation in a disposable bag reactor. The dissolved oxygen level (line: dash dot dot) was controlled by the rocking motion (line: dash). After 4.5 h, when the rocking capacity of the system reached its maximum (42 Rpm), oxygen supply (line: solid) started. The oxygen supply increased during the cultivation, because of the cell activity. During the whole process no oxygen limitation was observed and the internal pressure (line: dot) was below the maximal pressure of 50 mbar.
The cultivation was stopped after 11.3 (±0.7) h when all glucose (carbon source) was consumed. At the end of the cultivation a CDW of 6.8 (±0.4) g L−1 was reached, which is in the range of previous batch cultivation results in a 10 L STR (6.5–8.4 g L−1 28, 29). Using the disposable bag reactor a biomass yield (cCDW/cglucose) of 0.34 (±0.02) gCDW/gglucose was reached. The polySia concentration in the supernatant was at 245 (±56) mg L−1, which is also in the range of previous studies (200–370 mg L−1 28, 29). Higher polySia concentrations might be obtained due to fed‐batch process and further cultivation after glucose consumption 27, 28. In the process shown in this work, the cultivation was immediately stopped after glucose consumption to avoid cell lysis and host cell protein release into the supernatant.
To compare the polySia concentration in the supernatant with the total polySia concentration the cells were disrupted. Analysis of the total polySia concentration after cell disruption showed a polySia content of 456 (±113) mg L−1. Previous studies showed that around 60% of the capsule polysaccharides are partially released from the cell surface into the medium most probably either due to friction and shear stress or self‐cleavage in a conventional STR 28, 37. Due to the gentle wave‐induced mixing in the disposable bag reactor less shear stress‐related product release from the cell surface was expected. Nevertheless 54 (±8)% of the polySia could be found in the supernatant at the end of the batch cultivation. Thus, the gentle wave‐induced mixing had no significant effect on the product release from the cell surface compared to the rough stirred mixing in a conventional stainless steel reactor. In general 20–50% of the polySia are covalently bound to the cell wall while the rest is bound due to ionic interactions 29. The product yield of the polySia in the supernatant (cpolySia/cglucose) was 0.012 (±0.003) gpolySia/gglucose. The protein concentration in the supernatant was 132 (±30) mg L−1 at the end of the batch cultivation, which is slightly lower than the 200 mg L−1 observed in previous studies in an STR 29. The gentle wave‐induced mixing could have an effect on the host cell protein release related to cell lysis due to shear stress.
Protein content could be further decreased by using a defined preculture medium instead of medium with large amounts of complex components. All relevant cultivation results in comparison to the STR are summarized in Table 2. The maximal growth rate (μmax) was at 0.61 (±0.08) h−1 during the exponential growth phase. The offline data of a typical cultivation are shown in Fig. 3.
Table 2.
Comparison of the cultivation results in the reactor systems
| Result | Disposable bag reactor | STR 28, 29 |
|---|---|---|
| CDW | 6.8 (±0.4) g L−1 | 6.5–8.4 g L−1 |
| Chain length | ∼90 DP | 110 DP |
| PolySia | 245 (±56) mg L−1 | 200–370 mg L−1 |
| Protein | 132 (±30) mg L−1 | 200 mg L−1 |
| YP/S | 0.012 (±0.003) gpolySia/gglucose | 0.011–0.019 gpolySia/gglucose |
| YX/S | 0.34 (±0.02) gCDW/gglucose | 0.35–0.42 gCDW/gglucose |
YP/S, product yield; YX/S, biomass yield.
Data are presented as mean ± SD.
Figure 3.
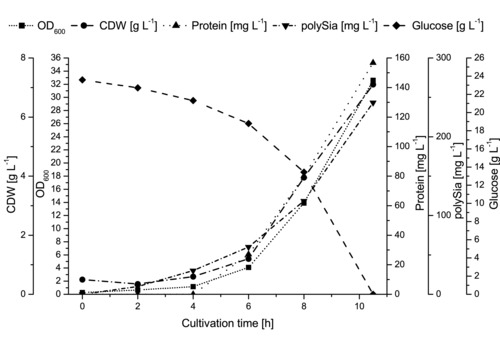
Offline data of a typical cultivation in a disposable bag reactor. After approximately 4 h of lag phase, the bacteria entered the exponential growth phase, noticeable by the OD600 (symbol: square, line: short dot) and the CDW (symbol: circle, line: dash dot). The exponential growth phase took place between 4 and 8 h of cultivation time. Connected to the bacteria concentration, the protein concentration (symbol: triangle, line: dot) and the polySia concentration (symbol: triangle, line: short dash dot) increased. The glucose (symbol: square, line: dash) was consumed after 10.5 h. At the end of the cultivation CDW reached 7.1 g L−1, the protein concentration 160 mg L−1 and the polySia concentration 243 mg L−1.
3.2. Purification of the produced polySia
At the end of the cultivation the cells were immediately separated by continuous centrifugation. The supernatant had a composition of approximately 0.5 gprotein/gpolySia. In the next step, the supernatant was concentrated by cross‐flow ultrafiltration with an MWCO of 10 kDa, to reduce the working volume and ease the handling of the subsequent purification steps. Peptides and small proteins with a molecular weight under 10 kDa were partially removed with the permeate stream in this step, while a large amount of the polySia remained in the retentate. PolySia with an approximate DP above 35 units cannot pass the membrane (MWCO of about 10 kDa) 29. Thus, polySia with chain lengths under 35 DP was lost in this step. The ratio after cross‐flow ultrafiltration was approximately 0.4 gprotein/gpolySia.
In the next downstream step, the retentate of the cross‐flow filtration was precipitated stepwise with ethanol; the ethanol concentration was subsequently altered from 80% up to 90% v/v. Due to this procedure, the polySia loss could be minimized. Nevertheless, 22% of the polySia content was lost in the step, but also the protein concentration was reduced efficiently (ratio approximately 0.4 gprotein/gpolySia).
After the precipitation step an adsorption step was implemented. Therefore, EX M 1753 was used, a clay mineral consisting of magnesium‐aluminium silicate with a particle size of ≥45 μm and a surface structure and size which is suitable to bind unspecifically high amounts of protein 29, 38. After this step only negligible amounts of the initial protein concentration were left and nearly no loss of polySia was observed (ratio below 0.04 gprotein/gpolySia).
After the treatment with EX M 1753, the solution was concentrated in the fifth downstream step in centrifuge tubes (Vivaspin 20 with a MWCO of 10 kDa), to reduce the sample volume. Afterwards, only low residual amounts of the initial protein were detectable and another 6% of the remaining polySia was lost (ratio below 0.04 gprotein/gpolySia).
To remove the remaining salts, dialysis was performed followed by lyophilization of the product. The final product had a protein content below 40 mgprotein/gpolySia compared to 500 mgprotein/gpolySia before downstream processing. This downstream processing method shown in the production process scheme (Fig. 1) focused mainly on the efficient removal of protein. The developed method is time saving and the handling is easy. As shown in Fig. 4, the protein content was effectively depleted during the purification process. The overall recovery yield of this purification process was 26%. Around 40% of the polySia have been lost during the cross‐flow ultrafiltration and Vivaspin 20 concentrating steps. This result was due to binding of the polySia to the membranes and the loss of polySia with short chain length (under ∼35 DP) in the permeate stream. Furthermore, 20% have been lost during the precipitation with ethanol, since not all of the polySia get precipitated. Reported purification yields range from 20 to 56% for precipitation‐based downstream processing methods 28, 39.
Figure 4.
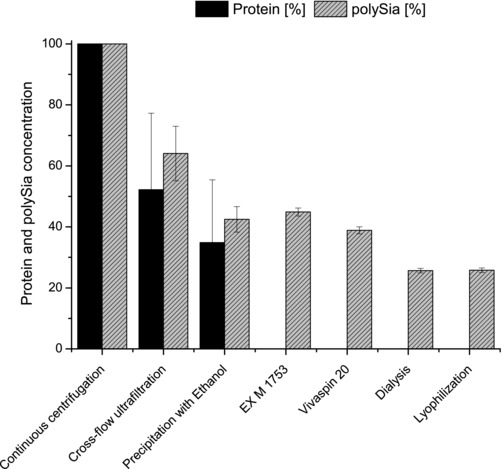
Protein and polySia concentration during the purification process. During the purification process the amount of protein (black bars) was efficiently reduced. After the final step of lyophilization, a recovery rate of 26% was observed for polySia (gray bars). The protein concentration was below 0.04 gprotein/gpolySia after the purification process. Data are presented as mean ± SD.
The DNA content of the product was 5.5 (±0.5) mgDNA/gpolySia and the endotoxin content was above 1000 EU mg−1. DNA and endotoxin content could be further reduced by using anion membrane adsorbers to gain a product that fulfills the requirements in terms of low DNA and endotoxin concentration for medical application as shown previously 29. However, the obtained product quality was suitable for further product characterization.
3.3. Product characterization
As for certain applications high‐chain lengths are required, an important characteristic of the produced polySia is the chain length. The chain length distribution was measured by DMB‐HPLC with a detection threshold of 1.4 fmol 40.
The chain length distribution of the produced polySia in a disposable bag reactor, purified with the downstream processing above is shown in Fig. 5. The obtained maximal chain length with this method was ∼90 DP. This fits well into the previously observed chain lengths under batch conditions in a 10 L STR (maximum 110 DP 29) and is higher than the chain length of commercially available polySia (Sigma‐Aldrich), which was determined as ∼70 DP. However, no estimation of the quantitative chain length distribution was possible with this method. Larger chain lengths might be achieved by decreased growth rates (e.g., by lower temperature or carbon source limitation in a fed‐batch process) 29, 41.
Figure 5.
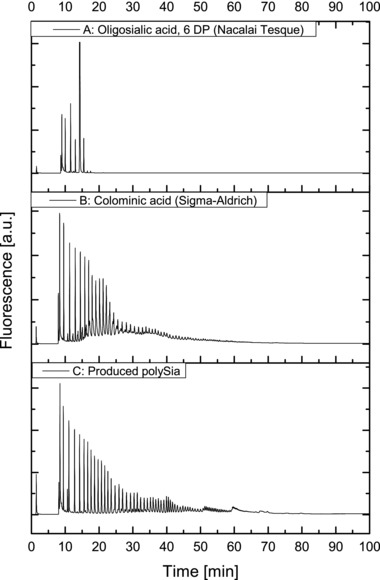
Chain length characterization of the produced polySia. The maximal chain length of the produced polySia was determined ∼90 DP with DMB‐HPLC. The chain length of commercially available polySia (Sigma‐Aldrich), was determined ∼70 DP. Oligosialic acid with a polymerization of 6 DP (Nacalai Tesque, Kyoto, Japan) was used as a control. Oligosialic acid and polySia was partially hydrolyzed during the derivatization with DMB. Therefore a quantitative distribution of the chain length was not possible.
A list of all relevant data comparing the polySia production in a disposable bag reactor with wave‐induced mixing to the process in an STR is shown in Tables 1 and 2.
4. Concluding remarks
This study showed that aerobic batch cultivation of E. coli K1 for the production of polySia is possible in a disposable bag reactor with wave‐induced mixing. Comparable results in terms of cultivation time, CDW, and polySia concentration to the established process in an STR were obtained. It was shown that the gentle wave‐induced mixing in the disposable bag reactor had no significant effect on the polySia release from the cell surface compared to the stirrer mixing in a conventional stainless steel reactor. Due to the low volumetric oxygen transfer coefficient value of the single‐use reactor system a subsequent increase of the rocking motion of the reactor system up to its maximum and an increase of the oxygen content in the gas supply was applied to establish a DO value of 50%. After the cultivation, an efficient downstream processing for the purification of the produced polySia was established. With this downstream processing method the polySia was purified in a short time and the product was suitable for further analysis. The observed DP was also comparable to previous results in an STR. For medical application, the DNA and endotoxin content of the product has to be decreased (e.g., by using membrane adsorbers 29).
The disposable bag reactors provided easy handling and easy and fast production in terms of regulatory requirements as GMP, due to limited process steps. Further benefits are the flexibility and an easy scale up to larger production capacities. Especially, the production of potential biological drugs such as polySia needs to be in accordance to common regulatory requirements. Therefore, the disposable bag reactor with wave‐induced mixing is a suitable bioreactor system, although the low oxygen transfer rate could be limitation, when higher concentrations of bacteria are required.
Practical application
This study presents a novel approach for the production of endogenous long‐chain polysialic acid by E. coli K1 cultivation in disposable reactor systems. The disposable reactor system facilitates the compliance of regulatory requirements as GMP and eases processing by ready to use presterilized reactor containments, so‐called bags. Escherichia coli K1 was cultivated in a disposable bag reactor with wave‐induced mixing to evaluate the performance of the disposable reactor in comparison to conventional stirred tank reactors. Pure oxygen was supplied to avoid oxygen limitation. No significant effect on the product release from the cell surface due the gentle wave‐induced mixing was observed, as 54% of the polySia was released into the medium. With a gas flow of 0.1 vvm a CDW of 6.8 g L−1 and a polySia concentration of 245 mg L−1 with a chain length of ∼90 DP was achieved, which is comparable to the established process in an STR.
Acknowledgments
This work was financially supported by the German Research Foundation (DFG, Funding numbers: DFG‐NE507/14‐1 and DFG‐SCHE279/35‐1) and the German Federal Ministry of Education and Research (BMBF, Funding numbers: BMBF‐03VP00271 and BMBF‐03VP00273). We acknowledge all associates for helpful discussions and technical support.
Drs. Jens Kopatz and Harald Neumann are named inventors on patent applications related to the use of polysialic acid for neurodegenerative diseases filed by the Universities of Bonn and Cologne.
5 References
- 1. Lin, B.‐X. , Qiao, Y. , Shi, B. , Tao, Y. , Polysialic acid biosynthesis and production in Escherichia coli: Current state and perspectives. Appl. Microbiol. Biotechnol. 2016, 100, 1–8. [DOI] [PubMed] [Google Scholar]
- 2. Angata, T. , Varki, A. , Chemical diversity in the sialic acids and related alpha‐keto acids: An evolutionary´ perspective. Chem. Rev. 2002, 439–469. [DOI] [PubMed] [Google Scholar]
- 3. Baumann, H. , Brisson, J. R. , Michon, F. , Pon, R. et al., Comparison of the conformation of the epitope of alpha(2‐8) polysialic acid with its reduced and N‐acyl derivatives. Biochemistry 1993, 32, 4007–4013. [DOI] [PubMed] [Google Scholar]
- 4. Dumitriu, S. , Polysaccharides: Structural Diversity and Functional Versatility (2nd ed.), Marcel Dekker, New York: 2005. [Google Scholar]
- 5. Mühlenhoff, M. , Eckhardt, M. , Gerardy‐Schahn, R. , Polysialic acid: Three‐dimensional structure, biosynthesis and function. Curr. Opin. Struct. Biol. 1998, 8, 558–564. [DOI] [PubMed] [Google Scholar]
- 6. Gristina, A. , Costerton, J. , Bacterial adherence and the glycocalyx and their role in musculoskeletal infection. OTD 2011, 9, 23–25. [PubMed] [Google Scholar]
- 7. Devine, D. A. , Roberts, A. P. , K1, K5 and O antigens of Escherichia coli in relation to serum killing via the classical and alternative complement pathways. J. Med. Microbiol. 1994, 41, 139–144. [DOI] [PubMed] [Google Scholar]
- 8. Schnaar, R. L. , Gerardy‐Schahn, R. , Hildebrandt, H. , Sialic acids in the brain: Gangliosides and polysialic acid in nervous system development, stability, disease, and regeneration. Physiol. Rev. 2014, 94, 461–518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Wang, X. , Li, X. , Zeng, Y.‐N. , He, F. et al., Enhanced expression of polysialic acid correlates with malignant phenotype in breast cancer cell lines and clinical tissue samples. Int. J. Mol. Med. 2016, 37, 197–206. [DOI] [PubMed] [Google Scholar]
- 10. Colley, K. J. , Kitajima, K. , Sato, C. , Polysialic acid: Biosynthesis, novel functions and applications. Crit. Rev. Biochem. Mol. Biol. 2014, 49, 498–532. [DOI] [PubMed] [Google Scholar]
- 11. Shahraz, A. , Kopatz, J. , Mathy, R. , Kappler, J. et al., Anti‐inflammatory activity of low molecular weight polysialic acid on human macrophages. Sci. Rep. 2015, 5, 16800. doi: 10.1038/srep16800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Spence, S. , Greene, M. K. , Fay, F. , Hams, E. et al., Targeting Siglecs with a sialic acid‐decorated nanoparticle abrogates inflammation. Sci. Transl. Med. 2015, 7, 303ra140. [DOI] [PubMed] [Google Scholar]
- 13. Claude, J. , Linnartz‐Gerlach, B. , Kudin, A. P. , Kunz, W. S. et al., Microglial CD33‐related Siglec‐E inhibits neurotoxicity by preventing the phagocytosis‐associated oxidative burst. J. Neurosci. 2013, 33, 18270–18276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Linnartz, B. , Neumann, H. , Microglial activatory (immunoreceptor tyrosine‐based activation motif)‐ and inhibitory (immunoreceptor tyrosine‐based inhibition motif)‐signaling receptors for recognition of the neuronal glycocalyx. Glia 2013, 61, 37–46. [DOI] [PubMed] [Google Scholar]
- 15. Haastert‐Talini, K. , Schaper‐Rinkel, J. , Schmitte, R. , Bastian, R. et al., In vivo evaluation of polysialic acid as part of tissue‐engineered nerve transplants. Tissue Eng. Part A 2010, 16, 3085–3098. [DOI] [PubMed] [Google Scholar]
- 16. Berski, S. , van Bergeijk, J. , Schwarzer, D. , Stark, Y. et al., Synthesis and biological evaluation of a polysialic acid‐based hydrogel as enzymatically degradable scaffold material for tissue engineering. Biomacromolecules 2008, 9, 2353–2359. [DOI] [PubMed] [Google Scholar]
- 17. Haile, Y. , Haastert, K. , Cesnulevicius, K. , Stummeyer, K. et al., Culturing of glial and neuronal cells on polysialic acid. Biomaterials 2007, 28, 1163–1173. [DOI] [PubMed] [Google Scholar]
- 18. Steinhaus, S. , Stark, Y. , Bruns, S. , Haile, Y. et al., Polysialic acid immobilized on silanized glass surfaces: A test case for its use as a biomaterial for nerve regeneration. J. Mater. Sci. Mater. Med. 2010, 21, 1371–1378. [DOI] [PubMed] [Google Scholar]
- 19. Tan, L. K. , Carlone, G. M. , Borrow, R. , Advances in the development of vaccines against Neisseria meningitidis . N. Engl. J. Med. 2010, 362, 1511–1520. [DOI] [PubMed] [Google Scholar]
- 20. Krug, L. M. , Ragupathi, G. , Hood, C. , George, C. et al., Immunization with N‐propionyl polysialic acid–KLH conjugate in patients with small cell lung cancer is safe and induces IgM antibodies reactive with SCLC cells and bactericidal against group B meningococci. Cancer Immunol. Immunother. 2012, 61, 9–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Heimburg‐Molinaro, J. , Lum, M. , Vijay, G. , Jain, M. et al., Cancer vaccines and carbohydrate epitopes. Vaccine 2011, 29, 8802–8826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lindhout, T. , Iqbal, U. , Willis, L. M. , Reid, A. N. et al., Site‐specific enzymatic polysialylation of therapeutic proteins using bacterial enzymes. Proc. Nat. Acad. Sci. 2011, 108, 7397–7402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fernandes, A. I. , Gregoriadis, G. , The effect of polysialylation on the immunogenicity and antigenicity of asparaginase: Implication in its pharmacokinetics. Int. J. Pharm. 2001, 217, 215–224. [DOI] [PubMed] [Google Scholar]
- 24. Jain, S. , Hreczuk‐Hirst, D. H. , McCormack, B. , Mital, M. et al., Polysialylated insulin: Synthesis, characterization and biological activity in vivo. Biochim. Biophys. Acta 2003, 1622, 42–49. [DOI] [PubMed] [Google Scholar]
- 25. Constantinou, A. , Epenetos, A. A. , Hreczuk‐Hirst, D. , Jain, S. et al., Modulation of antibody pharmacokinetics by chemical polysialylation. Bioconjug. Chem. 2008, 19, 643–650. [DOI] [PubMed] [Google Scholar]
- 26. Constantinou, A. , Epenetos, A. A. , Hreczuk‐Hirst, D. , Jain, S. et al., Site‐specific polysialylation of an antitumor single‐chain Fv fragment. Bioconjug. Chem. 2009, 20, 924–931. [DOI] [PubMed] [Google Scholar]
- 27. Chen, R. , John, J. , Rode, B. , Hitzmann, B. et al., Comparison of polysialic acid production in Escherichia coli K1 during batch cultivation and fed‐batch cultivation applying two different control strategies. J. Biotechnol. 2011, 154, 222–229. [DOI] [PubMed] [Google Scholar]
- 28. Rode, B. , Endres, C. , Ran, C. , Stahl, F. et al., Large‐scale production and homogenous purification of long chain polysialic acids from E. coli K1. J. Biotechnol. 2008, 135, 202–209. [DOI] [PubMed] [Google Scholar]
- 29. Bice, I. , Celik, H. , Wolff, C. , Beutel, S. et al., Downstream processing of high chain length polysialic acid using membrane adsorbers and clay minerals for application in tissue engineering. Eng. Life Sci. 2013, 13, 140–148. [Google Scholar]
- 30. Eibl, R. , Kaiser, S. , Lombriser, R. , Eibl, D. , Disposable bioreactors: The current state‐of‐the‐art and recommended applications in biotechnology. Appl. Microbiol. Biotechnol. 2010, 86, 41–49. [DOI] [PubMed] [Google Scholar]
- 31. Shukla, A. A. , Gottschalk, U. , Single‐use disposable technologies for biopharmaceutical manufacturing. Trends Biotechnol. 2013, 31, 147–154. [DOI] [PubMed] [Google Scholar]
- 32. Jossen, V. , Eibl, R. , Pörtner, R. , Kraume, M. et al., Stirred bioreactors, in: Larroche C., Sanromán M. Á., Du G., Pandey A. (Eds.) Current developments in biotechnology and bioengineering: Bioprocesses, bioreactors and controls, Elsevier, Amsterdam: 2016, pp. 179–215. [Google Scholar]
- 33. Jonczyk, P. , Takenberg, M. , Hartwig, S. , Beutel, S. et al., Cultivation of shear stress sensitive microorganisms in disposable bag reactor systems. J. Biotechnol. 2013, 167, 370–376. [DOI] [PubMed] [Google Scholar]
- 34. Dreher, T. , Husemann, U. , Zahnow, C. , Wilde, D. de. et al., High cell density Escherichia coli cultivation in different single‐use bioreactor systems. Chemie. Ingenieur. Technik 2013, 85, 162–171. [Google Scholar]
- 35. Ralla, K. , Sohling, U. , Suck, K. , Kasper, C. et al., Separation of patatins and protease inhibitors from potato fruit juice with clay minerals as cation exchangers. J. Sep. Sci. 2012, 35, 1596–1602. [DOI] [PubMed] [Google Scholar]
- 36. Bradford, M. M. , A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein‐dye binding. Anal. Biochem. 1976, 72, 248–254. [DOI] [PubMed] [Google Scholar]
- 37. Manzi, A. E. , Higa, H. H. , Diaz, S. , Varki, A. , Intramolecular self‐cleavage of polysialic acid. J. Biol. Chem. 1994, 269, 23617–23624. [PubMed] [Google Scholar]
- 38. Ralla, K. , Anton, F. , Scheper, T. , Kasper, C. , Application of conjoint liquid chromatography with monolithic disks for the simultaneous determination of immunoglobulin G and other proteins present in a cell culture medium. J. Chromatogr. A 2009, 1216, 2671–2675. [DOI] [PubMed] [Google Scholar]
- 39. Liu, J.‐L. , Zhan, X.‐B. , Wu, J.‐R. , Lin, C.‐C. et al., An efficient and large‐scale preparation process for polysialic acid by Escherichia coli CCTCC M208088. Biochem. Eng. J. 2010, 53, 97–103. [Google Scholar]
- 40. Inoue, S. , Lin, S.‐L. , Lee, Y. C. , Inoue, Y. , An ultrasensitive chemical method for polysialic acid analysis. Glycobiology 2001, 11, 759–767. [DOI] [PubMed] [Google Scholar]
- 41. Zheng, Z.‐Y. , Wang, S.‐Z. , Li, G.‐S. , Zhan, X.‐B. et al., A new polysialic acid production process based on dual‐stage pH control and fed‐batch fermentation for higher yield and resulting high molecular weight product. Appl. Microbiol. Biotechnol. 2013, 97, 2405–2412. [DOI] [PubMed] [Google Scholar]


