ABSTRACT
Mitophagy, which is a conserved cellular process for selectively removing damaged or unwanted mitochondria, is critical for mitochondrial quality control and the maintenance of normal cellular physiology. However, the precise mechanisms underlying mitophagy remain largely unknown. Prior studies on mitophagy focused on the events in the mitochondrial outer membrane. PHB2 (prohibitin 2), which is a highly conserved membrane scaffold protein, was recently identified as a novel inner membrane mitophagy receptor that mediates mitophagy. Here, we report a new signaling pathway for PHB2-mediated mitophagy. Upon mitochondrial membrane depolarization or misfolded protein aggregation, PHB2 depletion destabilizes PINK1 in the mitochondria, which blocks the mitochondrial recruitment of PRKN/Parkin, ubiquitin and OPTN (optineurin), leading to an inhibition of mitophagy. In addition, PHB2 overexpression directly induces PRKN recruitment to the mitochondria. Moreover, PHB2-mediated mitophagy is dependent on the mitochondrial inner membrane protease PARL, which interacts with PHB2 and is activated upon PHB2 depletion. Furthermore, PGAM5, which is processed by PARL, participates in PHB2-mediated PINK1 stabilization. Finally, a ligand of PHB proteins that we synthesized, called FL3, was found to strongly inhibit PHB2-mediated mitophagy and to effectively block cancer cell growth and energy production at nanomolar concentrations. Thus, our findings reveal that the PHB2-PARL-PGAM5-PINK1 axis is a novel pathway of PHB2-mediated mitophagy and that targeting PHB2 with the chemical compound FL3 is a promising strategy for cancer therapy.
Abbreviations: AIFM1: apoptosis inducing factor mitochondria associated 1; ATP5F1A/ATP5A1: ATP synthase F1 subunit alpha; BAF: bafilomycin A1; CALCOCO2/NDP52: calcium binding and coiled-coil domain 2; CCCP: chemical reagent carbonyl cyanide m-chlorophenyl hydrazine; FL3: flavaglines compound 3; HSPD1/HSP60: heat shock protein family D (Hsp60) member 1; LC3B/MAP1LC3B: microtubule associated protein 1 light chain 3 beta; MEF: mouse embryo fibroblasts; MPP: mitochondrial-processing peptidase; MT-CO2/COX2: mitochondrially encoded cytochrome c oxidase II; MTS: mitochondrial targeting sequence; OA: oligomycin and antimycin A; OPTN: optineurin; OTC: ornithine carbamoyltransferase; PARL: presenilin associated rhomboid like; PBS: phosphate-buffered saline; PGAM5: PGAM family member 5, mitochondrial serine/threonine protein phosphatase; PHB: prohibitin; PHB2: prohibitin 2; PINK1: PTEN induced kinase 1; PRKN/Parkin: parkin RBR E3 ubiquitin protein ligase; Roc-A: rocaglamide A; TOMM20: translocase of outer mitochondrial membrane 20; TUBB: tubulin beta class I.
KEYWORDS: Mitophagy, PARL, PGAM5, PHB2, PINK1-PRKN
Introduction
Macroautophagy/autophagy is a dynamic cell survival mechanism, that allows cells to survive under certain stressful conditions through the lysosomal degradation pathway [1]. Mitophagy, which is a selective form of autophagy, is the major pathway for the degradation of dysfunctional or superfluous mitochondria in eukaryotic cells [2]; therefore, mitophagy plays a crucial role in maintaining cellular homeostasis, including matching metabolic demand, orchestrating mitochondrial quality control, and protecting cells against the deleterious effects of damaged mitochondria [3,4]. Defects in mitophagy may contribute to a variety of human disorders such as cancer, neurodegenerative, cardiovascular and liver diseases [5].
Parkinson disease (PD) is a common neurodegenerative disorder that is characterized by the selective loss of dopaminergic neurons [6]. Oxidative stress originating from mitochondria and impaired mitophagy may contribute to the pathogenesis of PD [7]. PINK1 (PTEN induced kinase 1) and PRKN/Parkin (parkin RBR E3 ubiquitin protein ligase), which are two genes associated with autosomal recessive PD, were linked to mitochondrial quality control [8]. PINK1 is a serine/threonine kinase localized at mitochondria [9], while PRKN is an E3 ubiquitin ligase that is localized in the cytosol under normal condition [10]. In healthy cells, PINK1 is continuously processed and degraded by mitochondrial proteases, including mitochondrial inner protease PARL (presenilin associated rhomboid like), or cooperatively with m-AAA, i-AAA [11–14]. Upon mitochondrial damaged or uncoupling, however, PINK1 proteolysis is inhibited, resulting in the accumulation of PINK1 in the mitochondrial outer membrane, where PINK1 recruits the cytosolic E3 ubiquitin protein ligase PRKN to the mitochondrial outer membrane to carry out the ubiquitination of several mitochondrial outer membrane proteins, thereby mediating the autophagic elimination of damaged mitochondria [15–17]. It has been reported that certain mitochondrial proteins, including TOMM7 and PGAM5, can retain and stabilize PINK1 in the mitochondrial outer membrane [17,18]. TOMM7, which is a component of the protein translocase of outer mitochondrial membrane (TOMM) complex, stabilizes PINK1 on the outer membrane of damaged mitochondria in a manner that is unrelated to the efficiency of mitochondrial protein import [17]. PGAM5 is a serine/threonine protein phosphatase that is located to the mitochondria through its N-terminal TM domain [19]. PGAM5 stabilizes PINK1 and regulates PINK1-PRKN-mediated mitophagy. In addition, the genetic deficiency in PGAM5 in mice causes a PD-like phenotype [18]. SAMM50, which is a key component of the SAM complex, is also associated with PINK1 import and processing [20]. However, the detailed mechanisms of PINK1 degradation and stabilization remain unclear.
During mitophagy, certain autophagy receptors bind certain ubiquitinated mitochondrial outer membrane proteins, such as TOMM20; then, MAP1LC3B/LC3B-coated phagophores surround the damaged mitochondria and deliver it to the lysosome for degradation [21]. SQSTM1/p62 (sequestosome 1), NBR1, CALCOCO2/NDP52 (calcium binding and coiled-coil domain 2), TAX1BP1 (Tax1 binding protein 1), and OPTN (optineurin) serve as mitochondrial outer membrane receptors, which bind to MAP1LC3B to mediate mitophagy [22]. Additionally, cardiolipin, which is an inner mitochondrial membrane phospholipid, can also relocate to the mitochondrial outer membrane where it serves as a receptor for mitophagy in neuronal cells [23]. Notably, the mitochondrial outer membrane protein FUNDC1 (FUN14 domain containing 1) was identified as a specific receptor of mitophagy under hypoxia [4,24]. In addition, recently, PHB2 (prohibitin 2), which is a conserved mitochondrial inner membrane scaffold protein, was identified as a novel inner mitochondrial membrane mitophagy receptor that plays a critical role in PINK1-PRKN-mediated mitophagy [25]. Moreover, the proteasome-dependent mitochondrial outer membrane rupture is required for the PHB2-MAP1LC3B interaction during mitophagy [25]. However, whether and how PHB2 contacts and cooperates with the PINK1-PRKN-induced rupture of the mitochondrial outer membrane is still unknown and warrants further exploration.
PHB2 and PHB (prohibitin) assemble into a ring-like macromolecular structure that is known as the prohibitin complex at the mitochondrial inner membrane [26]. Prohibitins can regulate mitochondrial membrane protein processing and degradation by modulating mitochondrial proteases including the m-AAA protease [27]. Mitochondrial proteases play a key role in mitochondrial protein homeostasis. PARL regulates the processing of PINK1 and PGAM5, and OMA1 and YME1L (YME1 like 1 ATPase) cooperate to control OPA1 processing and degradation [12,18,28]. However, the relationship between the mitochondrial internal protein quality control (protein homeostasis regulated by mitochondrial proteases) and the overall mitochondrial quality control (mitophagy) remains poorly understood.
Cancer cells predominantly utilize glycolysis but not oxidative phosphorylation for energy production, even in the presence of oxygen, which is known as the ‘warburg effect’ [29]. However, certain mitochondrial functions are still essential for cancer proliferation and survival [30]. Mitophagy may serve to eliminate dysfunctional mitochondria to alleviate oxidative stress and prevent tumorigenesis. Conversely, mitophagy can be activated by oncogenic signaling pathways to protect cancer cells from cell death and promote tumor cell survival under certain adverse conditions [31]. Therefore, targeting mitophagy has been considered as a promising therapeutic strategy for treating advanced tumors, which have already switched to glycolysis metabolism but still require functional mitochondria for other metabolic functions [32].
Here, we demonstrate that, in addition to acting as a mitophagy receptor, PHB2 can promote PINK1-PRKN-mediated mitophagy by stabilizing PINK1 and increasing the mitochondrial recruitment of PRKN. Importantly, we show that PHB2 regulates PINK1 processing by regulating the activity of the mitochondrial protease PARL. Moreover, PHB2 stabilizes PINK1 through the PARL-PGAM5 axis when mitochondria are depolarized. In addition, we show that a known ligand of prohibitins, namely, FL3, inhibits PHB2-mediated mitophagy and blocks cancer cell growth at nanomolar concentrations. Therefore, these data suggest that FL3 is a promising candidate for cancer therapy due to its inhibitory effect on mitophagy.
Results
PHB2 regulates PRKN-mediated mitophagy in a manner that is dependent or independent of binding to MAP1LC3B
Prohibitins serve as scaffold proteins and are localized to the mitochondrial inner membrane [33]. PHB and PHB2 assemble and form a prohibitin complex, and deficiencies in either PHB or PHB2 result in the loss of both prohibitin proteins and the disruption of the prohibitin complex [34]. To identify the role of prohibitins in PINK1-PRKN-mediated mitophagy, we performed PHB2 knockdown using two unbiased short hairpin RNA mediated RNA interference targeting PHB2 (shPHB2-1 and shPHB2-2) in GFP-PRKN stably expressed HeLa cells. To induce PINK1-PRKN-mediated mitophagy, we treated HeLa cells expressing GFP-PRKN with the chemical reagent carbonyl cyanide m-chlorophenyl hydrazone (CCCP) or a mixture of oligomycin and antimycin A (OA), which exacerbate PINK1-dependent mitophagy by uncoupling the mitochondrial proton gradient or inhibiting the mitochondrial electron transport chain complex [15]. Immunostaining analysis revealed that shPHB2 remarkably inhibited CCCP or OA-induced the reduction of mitochondrial proteins including the outer membrane protein TOMM20 and the matrix protein HSPD1/HSP60 in GFP-PRKN-expressed cells (Figures S1a–e). In addition, western blotting data showed that, upon OA treatment for 16 h or 24 h, the levels of TOMM20, HSPD1, and MT-CO2/COX2 (mitochondrial inner membrane protein) were significantly decreased in the control cells; however, the reduction of mitochondrial proteins was blocked in PHB2 knockdown cells (Figures S1F–1I). These results indicated that PHB2 knockdown may inhibit mitophagy, consisting with the recent report that PHB2 promotes mitophagy [25]. To further confirm the role of PHB2 in mitophagy, we used mito-Keima, a useful tool in the assessment of single mitophagic events to evaluate mitophagy [35]. Upon treatment with DMSO, limited red fluorescence was detected in the mito-Keima and FLAG-PRKN expressed control or shPHB2 (using shPHB2-1) HeLa cells (Figure 1a,b). However, in response to OA, a remarkable increase in red fluorescence was detected in the control cells; in contrast, the intensity of the red fluorescence was clearly reduced in shPHB2 HeLa cells (Figure 1a,b), suggesting that OA-induced mitophagy is inhibited in the PHB2 deficient cells.
Figure 1.

PHB2 controls PINK1-PRKN-mediated mitophagy. (a) HeLa cells stably co-expressing FLAG-PRKN and mito-Keima were infected with control (scrambled shRNA) or shPHB2 lentiviral particles. Four days later, cells were treated with OA (10 mM oligomycin plus 4 mM antimycin-A) or DMSO for 16 h. Intracellular mito-Keima excited at 448 nm (measuring mitochondria with a neutral pH) was shown in green color, while red color indicated the mito-Keima fluorescence excited at 552 nm (measuring mitochondria with an acidic pH) in the same cell. (b) Quantification of the relative ratio of fluorescence intensity (552 nm:448 nm) of the cells described in (a). Student’s t-test was used to determine p-values, ***p < 0.001. (c) HeLa cells with or without GFP-PRKN expression were transfected with control, PHB2-FLAG or PHB2[mLIR]-FLAG for 72 h, then immunostained with anti-HSPD1, anti-FLAG antibodies, and visualized by confocal microscopy. (d) Quantification of the percentage of decreased HSPD1 cells as described in (c). All data represent the means ± SD of 3 independent experiments (100 cells per independent experiment). Statistical significance was assessed by student’s t-test, *p < 0.05, **p < 0.01 versus control. (e) HeLa cells stably expressing GFP-PRKN were co-transfected with mitochondrial-targeted Red (mito-Red) and control, PHB2-FLAG or PHB2[mLIR]-FLAG. 24 h later, cells were incubated with bafilomycin A1 (BAF, 1 μM) for 6 h, and then immunostained with anti-MAP1LC3B, anti-FLAG antibodies, and visualized by confocal microscopy (right panels). Left panels showed the pixel intensity of mito-Red and MAP1LC3B from a line.
Next, to explore whether exogenous PHB2 could promote mitophagy, we overexpressed PHB2 in HeLa cells with or without the expression of GFP-PRKN (Figure S2a). The overexpression of PHB2 led to a decrease in the mitochondrial protein HSPD1 in GFP-PRKN-expressed but not PRKN-absent cells (Figure 1c,d). Furthermore, PHB2 overexpression directly induced the relocation of LC3B/MAP1LC3B to the mitochondria in GFP-PRKN-expressed HeLa cells (Figure 1e). These data suggested that the overexpression of PHB2 promoted PRKN-dependent mitophagy. We then assessed whether the overexpression of a PHB2 LIR (LC3-interacting region) mutant (mLIR, PHB2Y121A,L124A), which lacks the ability to bind to MAP1LC3B (Figure S2b and S2c), could also promote mitophagy. Interestingly, the overexpression of the PHB2 LIR mutant (PHB2[mLIR]), which does not bind to MAP1LC3B, also induced mitophagy (Figure 1c,D). Additionally, as with PHB2 wild-type, PHB2[mLIR] also promoted the localization of MAP1LC3B to the mitochondria (Figure 1e). These data suggest that, in addition to acting as an inner membrane mitophagy receptor, PHB2 can mediate mitophagy through another pathway that is independent of binding to MAP1LC3B.
PHB2 promotes the mitochondrial recruitment of PRKN
Because overexpressed PHB2 and PHB2[mLIR] were shown to promote mitophagy (Figure 1c,d), we investigated how PHB2[mLIR], which lacks the ability to interact with MAP1LC3B, can still induce mitophagy. We then assessed whether PHB2 regulates the PINK1-PRKN pathway. The mitochondrial recruitment of PRKN was analyzed in HeLa cells that stably express GFP-PRKN. In response to CCCP or OA treatment for 1 h, most PRKN was recruited to the mitochondria in the control cells but not in the shPHB2 cells (Figure 2a,b), suggesting that PHB2 deficiency inhibits the mitochondrial recruitment of PRKN. Mitochondrial PRKN may mediate the ubiquitination of certain mitochondrial outer membrane proteins, such as TOMM20, leading to the subsequent proteasomal degradation of these mitochondrial proteins [15–17]. We found that compared with the control, both PHB2 and PHB2[mLIR] overexpression resulted in decreased levels of TOMM20 (Figure S3a). However, treatment with MG132, a potent proteasome inhibitor, but not with bafilomycin A1 (BAF, a potent inhibitor of autophagy), blocked the PHB2 or PHB2[mLIR]-induced reduction of TOMM20 (Figure S3a), further suggesting that PHB2 regulates the mitochondrial recruitment of PRKN and subsequent degradation of some mitochondrial outer membrane proteins. The accumulation of unfolded proteins in the mitochondrial matrix may induce the mitochondrial recruitment of PRKN independent of mitochondrial membrane potential [36]. Thus, we further investigated the role of PHB2 in the mitochondrial unfolded protein accumulation-induced mitochondrial recruitment of PRKN. We overexpressed the mitochondrial-localized mutant OTC (ornithine carbamoyltransferase; ΔOTC) in HeLa cells stably expressing GFP-PRKN to induce the mitochondrial unfolded protein response. Immunofluorescence analysis revealed that the transient expression of ΔOTC greatly increased the mitochondrial recruitment of PRKN (Figure 2c,d). However, PHB2 knockdown markedly inhibited the ΔOTC-induced mitochondrial recruitment of PRKN (Figure 2c,d). In addition, we treated HeLa cells expressing GFP-PRKN with actinonin, which stalls mitochondrial translation and induces mitochondrial protein misfolding [37]. Actinonin treatment resulted in the accumulation of GFP-PRKN in focal spots on mitochondrial subdomains in the control cells (Figures S3b,3c), while actinonin treatment failed to induce the recruitment of GFP-PRKN to the mitochondria in shPHB2 cells (Figures S3b,3c), further confirming that PHB2 depletion inhibited the mitochondrial unfolded protein accumulation-induced mitochondrial recruitment of PRKN.
Figure 2.

PHB2 regulates the mitochondrial recruitment of PRKN. (a) HeLa cells stably expressing GFP-PRKN were infected with control or shPHB2, treated with DMSO, CCCP (10 μM) or OA for 1 h, and then immunostained with an anti-TOMM20 antibody and DAPI, and analyzed by confocal microscopy. (b) Quantification of colocalization between PRKN and mitochondria in cells described in (a). (c-d) Control or shPHB2 HeLa cells stably expressing GFP-PRKN were transiently transfected with WT-OTC or △OTC. Cells were immunostained 48 h later with anti-OTC and anti-HSPD1 antibodies, and then imaged by confocal microscopy (c). Quantification of the mitochondrial PRKN-positive cells is shown in (d). (e) HeLa cells expressing GFP-PRKN were infected with control or shPHB2 lentiviral particles. Five days later, cells were treated with OA for 2 h, and then immunostained with anti-TOMM20 and anti-Ubiquitin antibodies. Cells were then visualized by confocal microscopy. (f) Quantification of the mitochondrial ubiquitin-positive cells described in (e). (g-h) HeLa cells expressing GFP-PRKN were treated with OA for 2 h, and immunostained with anti-OPTN and anti-TOMM20 antibodies. Cells were then assessed by confocal microscopy. Quantification of the mitochondrial OPTN-positive cells is shown in (h). Results shown were representative of at least 3 independent experiments. Error bars represent the mean ±SD (n = 3, 100 cells per independent experiment), Statistical significance was assessed by student’s t-test, *p < 0.05, **p < 0.01, ***p < 0.001.
PRKN, which is an E3 ubiquitin ligase, is activated by PINK1 and then ubiquitinates certain mitochondrial outer membrane proteins to form polyubiquitin chains. Mitochondrial outer membrane mitophagy receptors bind to these polyubiquitin chains and induce mitophagy [22,38]. OPTN and NDP52 are the primary receptors for PINK1 and PRKN-mediated mitophagy [22]. We next analyzed the ubiquitination of mitochondrial outer membrane proteins in the shPHB2 HeLa cells. We used an anti-ubiquitin antibody to detect the localization of endogenous ubiquitin in cells with or without OA treatment. Under normal conditions, ubiquitin was localized ubiquitously throughout cells (Figure 2e,f). However, most ubiquitin was recruited to the mitochondria following OA treatment; in contrast, this mitochondrial recruitment of ubiquitin was dramatically decreased in the shPHB2 cells (Figure 2e,f). In addition, we examined the mitochondrial localization of the mitophagy receptor OPTN; and showed that, upon OA or OA plus MG132 treatment, mitochondrial OPTN was significantly decreased in shPHB2 mitochondria compared with the control mitochondria (Figure 2g-h, and S3D). Taken together, these results demonstrate that PHB2 promotes mitophagy by increasing the mitochondrial recruitment of PRKN and the mitochondrial outer membrane mitophagy receptors.
It has been reported that PRKN mediates proteasome-dependent degradation of the mitochondrial outer membrane proteins and induces rupture of the mitochondrial outer membrane upon membrane depolarization [39], and the rupture of mitochondrial outer membrane is required for PHB2 binding MAP1LC3B during mitophagy [25]. Therefore, we investigated whether the mitochondrial membrane is ruptured upon membrane depolarization. Transmission electron microscopy (TEM) analysis revealed that, in the control HeLa cells expressing GFP-PRKN, the mitochondrial double membranes were intact, but upon OA treatment, some part of the mitochondrial outer membrane was ruptured (Figure S3e); importantly, the OA-induced mitochondrial outer membrane rupture was remarkably inhibited in shPHB2 cells (Figure S3e). These data suggest that PHB2 knockdown inhibits mitochondrial outer membrane rupture upon membrane depolarization.
PHB2 stabilizes PINK1 in the mitochondria
The mitochondrial recruitment of PRKN was impaired in shPHB2 cells (Figure 2a–d); therefore, we examined the stability of PINK1 in PHB2 deficient cells. Upon CCCP or OA treatment, endogenous PINK1 or exogenous PINK1-GFP accumulated in the control HeLa cells. In contrast, the accumulation of PINK1 or PINK1-GFP was markedly reduced in shPHB2 cells (Figure 3a and S4A-B). We also examined the effect of shPhb2 and shPhb on the stabilization of PINK1 in MEFs. shPhb2 and shPhb, which disrupt the prohibitin complex, markedly inhibited the accumulation of PINK1-GFP in MEFs induced by CCCP or OA treatment (Figure 3b,C). In addition, shPHB2 resulted in a significant decrease in the mitochondrial membrane potential (Figures S4c-S4e), which is thought to promote PINK1 accumulation, surprisingly, shPHB2 blocked the accumulation of PINK1 (Figure 3a–c), suggesting that the role of shPHB2 in PINK1 stabilization is independent of mitochondrial membrane potential. Furthermore, ΔOTC overexpression, which is independent of mitochondrial membrane potential [36], markedly induced the accumulation of PINK1 (Figure 3d); however, the △OTC overexpression-induced accumulation of PINK1 was also inhibited in shPHB2 cells (Figure 3d). These data demonstrate that PHB2 stabilizes PINK1 in a manner that is independent of mitochondrial membrane depolarization. To rule out the possibility of off-target gene silencing, we recovered PHB2-FLAG in shPHB2 HeLa cells. PHB2 depletion dramatically inhibited the OA-induced accumulation of PINK1 (Figure 3e); in contrast, PINK1 accumulation was restored in the presence of exogenous PHB2-FLAG (Figure 3e). Moreover, the overexpression of PHB2-FLAG spontaneously induced the accumulation of PINK1 and promoted OA-induced PINK1 accumulation in HeLa cells (Figure 3f). Furthermore, the mitochondrial recruitment of PRKN, which is dependent upon PINK1 accumulation, was markedly increased in the PHB2-FLAG-overexpressed cells (Figure S5a,5b). Overall, our data demonstrate that PHB2 stabilizes PINK1 in the mitochondria.
Figure 3.
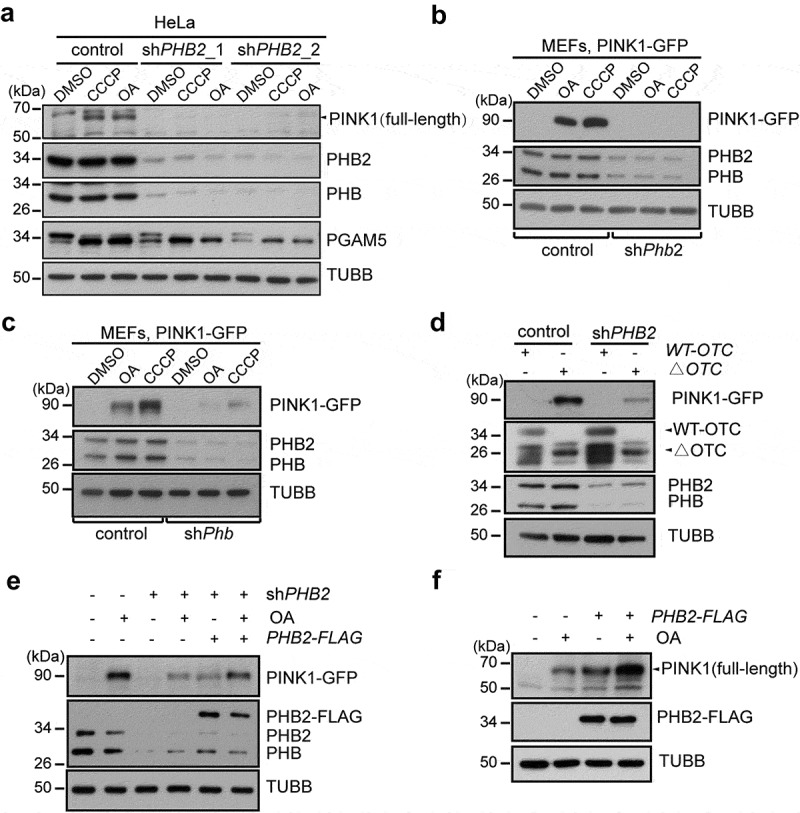
PHB2 stabilizes PINK1 in mitochondria. (a) HeLa cells were infected with control, shPHB2_1 or shPHB2_2 lentiviral particles, and cultured for 5 days. Cells were then incubated with OA or CCCP for 2 h. Cell lysates were analyzed by western blotting with antibodies against PINK1, PGAM5, PHB2, PHB, or TUBB/β-Tubulin. The black arrowhead indicates the full-length PINK1. (b-c) Control, shPhb2 (b), or shPhb (c) MEFs stably expressing PINK1-GFP were treated with CCCP (10 μM) or OA for 3 h. Cell lysates were subjected to western blotting using the indicated antibodies. (d) HeLa cells expressing PINK1-GFP were infected with control or shPHB2 lentiviral particles, 4 days later, cells were then transiently transfected with WT-OCT or ΔOTC for additional 3 days. Cells lysates were analyzed by western blotting with the indicated antibodies. The black arrowheads pointed to the indicated protein. (e) HeLa cells expressing PINK1-GFP were infected with control or shPHB2 lentiviral particles, 4 days later, cells were transiently transfected with empty vector (control) or PHB2-FLAG (2 nucleotides of PHB2 cDNA in shPHB2 target sequence were mutated, but PHB2 amino acids remain unchanged) for an additional 2 days, and then the cells were treated with OA for 2 h. Cells lysates were analyzed by western blotting with the indicated antibodies. (f) HeLa cells were transiently transfected with control (empty vector) or PHB2-FLAG. Cells were treated 48 h later with or without OA for 3 h, and then cell lysates were analyzed by western blotting using the indicated antibodies.
PHB2 regulates the PINK1-PRKN pathway independent of interacting with MAP1LC3B
PHB2 can bind to MAP1LC3B and act as a mitochondrial inner membrane mitophagy receptor [25]. To investigate whether the PHB2-regulated PINK1-PRKN activity is dependent upon interacting with MAP1LC3B, we assessed the mitochondrial recruitment of PRKN in HeLa cells expressing PHB2[mLIR], which lacks the ability to bind to MAP1LC3B (Figure S2b). The overexpression of PHB2[mLIR] significantly promoted the recruitment of PRKN to the mitochondria (Figure 4a,b). In addition, the expression of PHB2[mLIR] in shPHB2 cells also restored the PINK1 accumulation induced by OA treatment (Figure 4c). Moreover, PHB2[mLIR] also spontaneously promoted the accumulation of PINK1 (Figure 4d). Furthermore, PHB2[mLIR] restored the mitochondrial PRKN induced by OA treatment in shPhb2 MEFs cells (Figure 4e). These data suggest that PHB2 regulates the PINK1-PRKN pathway independent of interacting with MAP1LC3B.
Figure 4.

PHB2[mLIR] mutant also induce mitophagy. (a) HeLa cells expressing GFP-PRKN were transiently transfected with control (empty vector) or PHB2[mLIR]-FLAG. After 24 h transfection, cells were immunostained with anti-TOMM20 and anti-FLAG antibodies, and then analyzed and imaged by confocal microscopy. (b) Quantification of the mitochondrial PRKN-positive cells described in (a). Error bars represent the mean ±SD (n = 3, 100 cells per independent experiment), statistical significance was assessed by student’s t-test, ***p < 0.001. (c) Control or shPHB2 HeLa cells stably expressing PINK1-GFP were transiently transfected with control (empty vector) or PHB2[mLIR]-FLAG. After 48 h transfection, cells were treated with or without OA for 2 h, cell lysates were then analyzed by western blotting using antibodies against GFP, PHB, PHB2, or TUBB/β-Tubulin. (d) HeLa cells were transiently transfected with control (empty vector) or PHB2[mLIR]-FLAG. 48 h later, cells were treated with or without OA for 3 h, cells lysates were analyzed by western blotting with the indicated antibodies. (e) MEFs expressing GFP-PRKN cells were infected with lentiviral particles containing control and shPhb2. Four days later, cells were transiently transfected control (empty vector), PHB2-FLAG, or PHB2[mLIR]-FLAG for an additional 2 days, then treated with or without OA for 2 h, and immunostained with the indicated antibodies. Cells were visualized and imaged by confocal microscopy.
PHB2 negatively modulates PARL activity
In healthy mitochondria, PINK1 is firstly recognized and cleaved by MPP (mitochondrial-processing peptidase) to remove mitochondrial targeting sequence (MTS), then undergoes cleavage by the mitochondrial protease PARL in the inner membrane and is subsequently degraded [12]. To investigate how PHB2 regulates PINK1 stability, we firstly tested whether the instability of PINK1 in shPHB2 cells is related to PARL. We used CRISPR/Cas9 technology to construct a parl knockout (KO) HCT116 cell line, which harbors a 13-bp deletion in the coding region of the PARL gene, resulting in a frameshift mutation (Figure S6a). This truncated form of PARL contains only 28 amino acids (while WT PARL contains 379 amino acids). In healthy cells, little PINK1 protein is detected because it is continuously subjected to proteolysis and proteasomal degradation [12,40]. However, compared to HCT116 wild-type (WT) cells, increased endogenous processed PINK1 protein, which is the production of PINK1 cleavage by MPP (indicated by the red arrowhead), was detected by western blotting in the parl KO cells in the absence of OA, although the level of PINK1 was lower compared with cells treated with OA (Figure 5a and S6b), consisting with the previous report [12]. Following the treatment with CCCP or OA, the level of full-length PINK1 (indicated by the green arrowhead) was elevated and exhibited a similar level in both HCT116 WT and parl KO cells (Figure 5a and S6b). Furthermore, endogenous or exogenous PINK1 protein accumulated following treatment with OA or CCCP in HCT116 WT cells; however, PINK1 accumulation was inhibited in response to shPHB2 (Figure 5a,b and S6C). In contrast, shPHB2 failed to inhibit PINK1 accumulation induced by CCCP or OA treatment in parl KO cells (Figure 5a,b), suggesting that the PHB2-mediated regulation of PINK1 stability is dependent on PARL. Therefore, we then investigated the relationship between PHB2 and PARL. PGAM5, which is a substrate of PARL, can be cleaved by PARL, leading to a short form of PGAM5 (S-PGAM5) [41]. We found that the cleavage of PGAM5 was dramatically promoted in shPHB2 HeLa and HCT116 cells (Figures 3a and 5a), suggesting that the activity of the mitochondrial inner membrane protease PARL is promoted in response to shPHB2. Moreover, PINK1 processing by PARL was also promoted in shPHB2 cells, indicating that the shPHB2-promoted PARL activity is independent of shPHB2-induced mitochondrial membrane depolarization, as mitochondrial membrane depolarization inhibits PINK1 import and processing by PARL [12]. To clarify why PHB2 depletion activates PARL, we detected the protein level of PARL in the control and shPHB2 cells. Both endogenous and exogenous PARL protein were markedly increased in shPHB2 cells (Figure 5c-f), indicating that the knockdown of PHB2 stabilizes the PARL protein. Furthermore, upon the treatment of cycloheximide (CHX, a protein synthesis inhibitor), the PARL protein was more stable in shPHB2 cells than that in control cells (Figure 5g), further suggesting that the PARL protein is stabilized upon PHB2 knockdown. In addition, compared with control, shPHB2 had no effect on the mRNA level of PARL (Figure S6d). PHB and PHB2 are mitochondrial inner membrane scaffold proteins that interact with the mitochondrial m-AAA protease and regulate its activity [27]. We next used a co-immunoprecipitation (co-IP) assay to investigate whether PHB2 or PHB binds to PARL. PARL-FLAG successfully precipitated endogenous PHB and PHB2 (Figure 5h), indicating that PARL interacts with these prohibitins. Interestingly, the interaction between PARL and the prohibitins was markedly increased in CCCP-treated cells (Figure 5h), suggesting that mitochondrial membrane depolarization promotes the interaction of PARL with the prohibitins. In addition, STOML2/SLP2, which is another mitochondrial inner membrane scaffold protein that regulates PARL activity [42], remained unchanged in response to PHB2 depletion (Figure S6e), indicating that PHB2 regulates PARL activity in a manner that is independent of STOML2. Taken together, these results indicate PHB2 regulates the stability of PINK1 by destabilizing PARL and negatively regulating the activity of PARL.
Figure 5.

PHB2 regulates PARL activity. (a-b) HCT116 Wild type (WT) or parl KO cells were infected by lentiviral particles containing control or shPHB2, and further cultured for 5 days. Cells were then treated with DMSO, CCCP or OA for 3 h. Cell lysates were examined by western blotting using the indicated antibodies (a). The black arrowhead indicated the full-length PINK1 and red arrowhead point to the band of PINK1 with MTS deleted after MPP cleaved. The asterisk indicated a nonspecific band. The relative protein levels were evaluated by densitometry analysis using ImageJ software and were quantified for the ratio of PINK1:TUBB/β-Tubulin (b). Error bars indicate the mean ±SD of 3 independent experiments, ***p < 0.001, n.s indicates none significance. (c) HCT116 WT and parl KO cells were infected with control and shPHB2 contained lentiviral particles. Five days after infection, cells were treated with or without CCCP for 4 h, and then harvested for the isolation of mitochondria. Mitochondria were lysed and analyzed by western blotting with the indicated antibodies. The asterisk indicates a nonspecific band. HSPD1 served as mitochondrial loading control. (d) Densitometric quantification of PARL:HSPD1 in (c). Error bars indicate the mean ±SD of 3 independent experiments, **p < 0.01. (E) HCT116 parl KO cells stably expressing PARL-FLAG were infected with control and shPHB2 contained lentiviral particles. Five days after infection, cells were treated with or without CCCP (10 μM) for 2 h. Cell lysates were analyzed by western blotting with anti-FLAG, anti-PHB, anti-PHB2, anti-PGAM5 or anti-TUBB antibodies. (f) Densitometric quantification of PARL-FLAG:TUBB in (e). Error bars indicate the mean ±SD of three independent experiments, **p < 0.01. (g) HCT116 parl KO cells complemented with PARL-FLAG were infected with control or shPHB2 contained lentiviral particles. 5 days after infection, cells were treated with fresh Cycloheximide (CHX, 50 μg/ml) for the indicated time. Cell lysates were analyzed by western blotting with anti-FLAG, anti-PHB, anti-PHB2, or anti-TUBB antibodies. (h) HCT116 parl KO cells or parl KO cells stably expressing PARL-FLAG were treated with or without CCCP for 2 h; cell lysates were then immunoprecipitated with anti-FLAG M2 affinity gel, followed by western blotting using anti-FLAG, anti-PHB, or anti-PHB2 antibodies.
PARL-PGAM5 axis is required for the PHB2-mediated PINK1 stabilization in depolarized mitochondria
In depolarized mitochondria, PINK1 cannot be integrated into the inner membrane and cleaved by PARL [12]. We determined that shPHB2 inhibited the accumulation of PINK1 in depolarized mitochondria, and this inhibition was blocked in parl KO cells (Figure 5a, B, and S6C), indicating that, in addition to regulating the cleavage of PINK1, PARL also regulates PHB2-mediated PINK1 stabilization in depolarized mitochondria though an unknown mechanism, which we next investigated. PGAM5, which is a mitochondrial serine/threonine protein phosphatase that is cleaved by PARL, can stabilize PINK1 by interacting with PINK1 on damaged mitochondria [18]. Consistently, we found that PINK1 failed to accumulate on mitochondria in the PGAM5 knockdown cells (Figure 6a). Moreover, the overexpression of PGAM5 stabilized PINK1-GFP (Figure 6b). Additionally, full-length PINK1-GFP was sensitive to protease K, whereas PGAM5 was protected (Figure 6c), indicating that PGAM5 is located at the mitochondrial inner membrane. Furthermore, the co-IP assay demonstrated that PGAM5 interacted with PINK1 in the depolarized mitochondria; this interaction was decreased upon CCCP treatment, likely due to the increased processing of PGAM5 (Figure 6d). In addition, PGAM5 interacted with prohibitins (PHB and PHB2) in healthy mitochondria, but such interactions were weak in depolarized mitochondria where the short form of PGAM5 (S-PGAM5) predominated (Figure 6e,F). Moreover, PHB2 bound to and dissociated from PGAM5 in a time-dependent manner in response to CCCP, whereas the PHB2-PARL interaction was increased in a manner inversely correlated with the PHB2-PGAM5 interaction (Figure 6g), suggesting that the degree to which PHB2 interacted with either PGAM5 or PARL was affected by the mitochondrial membrane depolarization. In addition, we proposed that the ability of S-PGAM5 stabilizing PINK1 might be lower than that of full-length PGAM5 (L-PGAM5). To test this hypothesis, we constructed S-PGAM5 fused with an N-terminal mitochondrial targeting sequence (MTS) of AIFM1 (apoptosis inducing factor mitochondria associated 1, a mitochondrial inner membrane protein), which lacks PARL cleavage site and could directly produce S-PGAM5 after MPP cleavage. As shown in Figure 6(h,i), L-PGAM5 markedly increased the ratio of full-length to cleaved PINK1-GFP; whereas, S-PGAM5 resulted in a significantly decreased ratio. Our results indicate that compared with L-PGAM5, S-PGAM5 has a decreased ability to stabilize full-length PINK1. Thus, PHB2 may bind to PGAM5 to protect it from processing by PARL. In contrast, upon mitochondria depolarization, PHB2 tends to bind to PARL instead of interacting with PGAM5, and the released PGAM5 retains PINK1 at the mitochondrial outer membrane and subsequently initiates mitophagy. However, upon PHB2 depletion, PARL is activated to cleave PGAM5. In the absence of full-length PGAM5, PINK1 fails to be retained at the mitochondrial outer membrane; rather, PINK1 is imported into the mitochondrial inner membrane for processing by PARL and the other proteases and is finally degraded. These data suggest that the PARL-PGAM5 axis is required for PHB2-mediated PINK1 stabilization.
Figure 6.

The PARL-PGAM5 axis is involved in PHB2-mediated PINK1 stabilization. (a) HeLa cells were transfected with negative control (NC) or siRNA against PGAM5 for 72 h, and then treated with OA for 2 h. Cell lysates were analyzed by western blotting with the antibodies against PINK1, PGAM5, or TUBB. (b) HeLa cells stably expressing PINK1-GFP were transiently transfected with control (empty vector) or PGAM5-MYC. Cells were treated 48 h later with or without OA for 2 h. Then cell lysates were analyzed by western blotting with anti-GFP, anti-MYC, or anti-TUBB antibodies. (c) 293T cells were transiently co-transfected with PGAM5-MYC and PINK1-GFP. Cells were then harvested 48 h later for the isolation of mitochondria. Mitochondria were treated with the indicated doses of proteinase K for 30 min on the ice and then were analyzed by western blotting with anti-GFP, anti-MYC, anti-TOMM20, or anti-TIMM23 antibodies. (d) 293T cells were transiently co-transfected with control (empty vector) and PINK1-GFP, or PGAM5-MYC and PINK1-GFP. Cells were treated 48 h later with CCCP (10 μM) for 4 h. Cell lysates were used for immunoprecipitation with Dynabeads Protein G pre-coupled with anti-MYC antibody at 4°C overnight, followed by western blotting with anti-GFP or anti-MYC antibodies. (e) 293T cells were transiently transfected with control (empty vector) or PGAM5-MYC for 48 h, and then treated with CCCP for 4 h. Cells lysates were used for immunoprecipitation with anti-MYC antibody coupled Dynabeads Protein G at 4°C overnight, followed by western blotting using anti-PHB, anti-PHB2, or anti-MYC antibodies. (f) 293 cells were treated with or without CCCP for 4 h. Cells lysates were used for immunoprecipitation with Dynabeads Protein G coupled with anti-PHB2 antibody at 4°C overnight, followed by western blotting with the indicated antibodies. (g) 293T cells were transiently transfected with empty vector or plasmids coding for PHB2-FLAG and PGAM5-MYC for 48 h, and then treated with CCCP (10 μM) for the indicated time. Cells lysates were used for immunoprecipitation with anti-FLAG M2 affinity gel at 4°C overnight. Immunoprecipitates and cells lysates were analyzed by western blotting with anti-FLAG, anti-MYC, or anti-PARL antibodies. (h-i) HeLa cells expressing PINK1-GFP were transfected with control (empty vector), PGAM5-MYC, or MTS (AIFM1)-S-PGAM5-MYC for 48 h, and then treated with OA for 2 h. Cell lysates were analyzed by western blotting with anti-GFP, anti-MYC, or anti-TUBB antibodies. (i) Quantification of the ratio between full-length PINK1-GFP and cleaved-PINK1-GFP. Error bars indicate the mean ±SD of 3 independent experiments, **p < 0.01.
Prohibitin ligand FL3 inhibits PINK1-PRKN-mediated mitophagy
To further explore the role of prohibitins in mitophagy, we examined whether a known PHBs ligand, FL3 [43,44], affects PINK1-PRKN-mediated mitophagy. We treated HeLa cells expressing GFP-PRKN with FL3 for 24 h and then incubated cells with CCCP or OA for an additional 24 h or 16 h to induce mitophagy. As illustrated in Figure 7a-d, FL3 treatment potently inhibited the elimination of mitochondria, as assessed by quantitative image analysis of the mitochondrial proteins TOMM20 and HSPD1, or by western blotting analysis of the degradation of TOMM20, MT-CO2/COX2, ATP5F1A/ATP5A1 (mitochondrial matrix protein), and HSPD1. Furthermore, FL3 treatment inhibited the mitochondrial recruitment of PRKN and PINK1 accumulation in depolarized mitochondria (Figure 7e-G). These results demonstrate that FL3 strongly blocks PINK1-PRKN-mediated mitophagy.
Figure 7.

FL3 inhibits PINK1-PRKN mediated mitophagy by targeting PHBs. (a) HeLa cells expressing GFP-PRKN were treated with DMSO, CCCP (10 μM), OA for the indicated time periods, or treated with FL3 (50 nM) prior 24 h and then incubated with DMSO, CCCP, or OA for the indicated time periods. Cells were then immunostained with anti-TOMM20 and anti-HSPD1 antibodies, and analyzed by confocal microscopy. (b-c) Quantification of TOMM20 (b) or HSPD1 (c) puncta in cells described in (A). Error bars represent the mean ±SD (n = 3, 100 cells per independent experiment), *p < 0.05, **p < 0.01, ***p < 0.001. (D) HeLa cells expressing GFP-PRKN were pre-treated without or with FL3 (50 nM) for 24 h, and then were incubated with CCCP (10 μM) or OA for the indicated time periods. Cell lysates were subjected to western blotting using the indicated antibodies. (TOMM20, mitochondrial out membrane; MT-CO2, mitochondrial inner membrane; ATP5F1A and HSPD1, mitochondrial matrix). (e) HeLa cells expressing GFP-PRKN were pre-treated with FL3 (50 nM) for 24 h, and then were incubated with CCCP or OA for 1 h. Cells were stained with DAPI and immunostained with anti-TOMM20 antibody, and then visualized and imaged by confocal microscopy. (f) Quantification of mitochondrial PRKN-positive cells described in (e). Error bars represent the mean ±SD (n = 3, 100 cells per independent experiment), statistical significance was assessed by student’s t-test, ***p < 0.001. (g) HeLa cells stably expressing PINK1-GFP were pre-treated with or without FL3 (50 nM) for 24 h, then treated with CCCP for the indicated time periods. Cell lysates were analyzed by western blotting using antibodies against GFP or anti-TUBB. (h) parl KO HCT116 cells stably expressing PARL-FLAG were treated with DMSO, FL3 (50 nM) or Roc-A (50 nM) for 24 h. Cell lysates were then analyzed by western blotting with anti-FLAG, anti-PHB or anti-PHB2 antibodies. (i) parl KO HCT116 cells expressing PARL-FLAG were treated with DMSO, FL3 (50 nM) or RocA (50 nM) for 24 h, and then incubated with or without CCCP for 4 h. Cell lysates were immunoprecipitated with IgG (control) or anti-FLAG M2 affinity gel, the input and IP protein samples were analyzed by western blotting with anti-FLAG, anti-PHB or anti-PHB2 antibodies.
We next investigated how FL3 regulates PINK1-PRKN-mediated mitophagy. PHB2 knockdown resulted in the upregulation of mitochondrial protease PARL (Figure 5c-F). Therefore, we used western blotting to analyze the level of PARL in cells treated with FL3 or rocaglamide (Roc-A, another prohibitin ligand). FL3 or Roc-A treatment markedly increased the protein level of PARL but had no effect on the protein levels of PHB and PHB2 (Figure 7h). Moreover, the interactions between PARL and PHBs were highly impaired upon FL3 or Roc-A treatment (Figure 7i). Therefore, FL3 affects PINK1-PRKN-mediated mitophagy by selectively targeting prohibitins.
Mitophagy has been reported to be linked to tumorigenesis in cancer therapy [32,45,46]. We therefore examined the cytotoxicity of FL3 in human cancer cells. We showed that a low dose of FL3 (50 nM) significantly inhibited the proliferation of cancer cell lines, including HeLa (a cervical cancer cell line), H1299 (a non-small cell lung cancer cell line, p53 null) and HCT116 (a colorectal cancer cell line, p53 wild-type) (Figures S7a-S7c). Our data are consistent with previous reports that FL3 displays potent in vivo anticancer effects without causing any major adverse effect [47–49]. Taken together, our results reveal that targeting PHB2-mediated mitophagy using the PHB2 ligand FL3 is a potential and promising strategy for cancer therapy.
Discussion
Mitophagy eliminates damaged mitochondria by delivering them to the lysosome for degradation and this process is essential for mitochondrial quality control [4]. In this study, we demonstrated that PHB2 regulates PINK1-PRKN-mediated mitophagy through a novel pathway that is independent of binding to MAP1LC3B. PHB2 promotes PINK1 stabilization through the PARL-PGAM5-PINK1 axis.
Beth Levine and collaborators recently reported that PHB2 binds to MAP1LC3B and serves as a mitochondrial inner membrane mitophagy receptor to mediate PINK1-PRKN-dependent mitophagy, but this event requires the rupture of the mitochondrial outer membrane [25]. It is still unknown how the inner and outer membrane mitophagy receptors simultaneously and synergistically promote mitophagy. In the present study, our results reveal a novel pathway for PHB2-mediated mitophagy. We demonstrate that PHB2 modulates the mitochondrial inner membrane protease PARL to stabilize PINK1 in damaged mitochondria and, subsequently, PINK1 recruits PRKN to the mitochondria then carry out the ubiquitination and degradation of certain mitochondrial outer membrane proteins (Figure 2–3), leading to the initiation of mitophagy (Figure 1). Our findings indicate that the sites of rupture of the mitochondrial outer membrane are likely spatially coordinated with PHB2 localization in the inner membrane, which facilitates the binding of PHB2 to MAP1LC3B in spatial to initiate mitophagy. Furthermore, a PHB ligand, FL3, effectively inhibited PHB2-mediated mitophagy and blocked cancer cell growth (Figure S7). These data confirm that targeting PHB2 is a promising therapeutic target for treating mitochondrial diseases, including cancer.
The ubiquitin-proteasome system is critical for PRKN-dependent mitophagy [50]. Proteasome-dependent mitochondrial membrane rupture is necessary for PHB2-mediated mitophagy, as PHB2 needs to binds to MAP1LC3B, which is localized in the cytosol [25]. It has been reported that PRKN mediates proteasome-dependent protein degradation and the rupture of the outer mitochondrial membrane [39]. Therefore, we explored the role of PHB2 in PRKN activity during mitophagy. Our data demonstrate that, in damaged mitochondria, PHB2 stabilizes PINK1 through the PARL-PGAM5 axis. Consequently, the stabilized PINK1 phosphorylates and activates PRKN, which promotes the ubiquitination of mitochondrial proteins, recruits outer membrane mitophagy receptors, and then mediates the proteasome-dependent degradation of mitochondrial outer membrane proteins and the rupture of the outer membrane. Subsequently, MAP1LC3B gets across the outer membrane to interact with PHB2 to initiate mitophagy (Figure 8a). In contrast, depletion of PHB2 promotes the activation of the mitochondrial proteases PARL, leading to the cleavage of PGAM5. Full-length PGAM5 stabilizes PINK1 (Figure 6a,b), thus, the PHB2 depletion-mediated processing of PGAM5 by PARL inhibits PINK1-PRKN-dependent mitophagy (Figure 8b). Our findings reveal that PHB2 may precisely determine the site of the mitochondrial outer membrane rupture, which facilitates the binding of PHB2 to the autophagosomal protein MAP1LC3B to initiate mitophagy.
Figure 8.

Model of a novel signal pathway for PHB2-mediated mitophagy. (a) PHBs interacts with PARL and regulates its protease proteolytic activity. Upon mitochondria depolarization or damage, PHBs binds to PARL to protect PGAM5 from cleavage. Full-length PINK1 protected by PGAM5 moves to OMM and then recruits PRKN to mitochondria. Mitochondrial outer membrane proteins are then ubiquitinated and degraded by the proteasome, resulting in mitochondrial OMM rupture. Then, exposed PHB2 is thus detected by MAP1LC3B in phagophores to initiate mitophagy. OMM, outer mitochondrial membrane; IMS, inner membrane space; IMM, inner mitochondrial membrane; ΔOTC, the mitochondrial-localized mutant ornithine carbamoyltransferase. (b) The depletion of PHBs promotes PARL proteolytic activity and the cleavage of its substrate, PGAM5. Without the help of full-length PGAM5 in the insertion of PINK1 to OMM, PINK1 imports into IMM and gets cleaved and degraded. Therefore, PINK1-PRKN-mediated mitophagy is inhibited.
The mitochondrial inner membrane protease PARL cleaves PINK1 and negatively regulates PINK1-PRKN mediated mitophagy [11]. In this study, we demonstrated that PHB2 binds to and destabilizes PARL (Figure 5c-H); moreover, the activity of PARL is promoted upon PHB2 deficiency (Figure 5c-F). Similar to the PHBs, another mitochondrial inner membrane scaffold protein, STOML2, which also belongs to the SPFH (stomatin, prohibitin, flotillin, HflC/K) superfamily, regulates the activity of PARL [42]; however, STOML2 is not involved in the PHB2-mediated regulation of PINK1 (Figure S6e). In damaged mitochondria, PINK1 accumulates at the outer membrane where it cannot be cleaved by PARL. However, PINK1 stabilization is also impaired in response to PHB2 deficiency (Figure 3a-D), suggesting that other mitochondrial inner membrane proteins related to PHB2 are also likely involved in the accumulation of PINK1. Indeed, PGAM5, a mitochondrial inner membrane protein that is processed by PARL, positively regulates the anchoring of PINK1 in the mitochondrial outer membrane (Figures 5 and 6). Lu et al reported that PGAM5 transiently associates with full-length PINK1, which enables the transfer of PINK1 to the mitochondrial outer membrane and the initiation of mitophagy [18]. Upon PHB2 deletion, PGAM5 was cleaved by PARL, generating the short form of PGAM5 (Figure 3a); thus, the PGAM5-mediated insertion of PINK1 into the mitochondrial outer membrane was impaired, leading to the subsequent processing and degradation of PINK1 even under mitochondrial membrane depolarization or misfolded protein aggregation (Figure 3a-D). Thus, the PHB2-PARL-PGAM5-PINK1 axis plays an important role in PHB2-mediated PINK1 stabilization. Therefore, we propose two PHB2-dependent events that stabilize PINK1. One is through binding to PARL to prevent it from directly processing PINK1 in the mitochondrial inner membrane, and the other is through binding to and maintaining PGAM5 in the long form to retain PINK1 in the mitochondrial outer membrane. We think two PHB2-dependent events are interconnected. PHB2 knockdown results in PARL activation and PGAM5 cleavage, and these two events cooperate to inhibit PINK1 accumulation in the mitochondrial outer membrane. PHB2 binds to both PARL and PGAM5 (Figure 5h, 6E and 6F), moreover, PGAM5 cleavage depends on PARL activation (Figure 5a). Without PARL, L-PGAM5 is still exist even in response to shPHB2, thus, shPHB2 could not inhibit PINK1 accumulation (Figure 5a,C). In addition, certain other proteins may also be involved in PHB2-mediated PINK1 stabilization, and we will focus on identifying these proteins in our future work. Taken together, our results reveal an important role for PHB2 in regulating mitochondrial quality control by modulating the mitochondrial inner membrane protease PARL.
Mitophagy maintains normal cellular physiology by removing damaged mitochondria, but this process has been considered a double-edged sword for tumors. Specific defects in mitophagy have been linked to tumorigenesis through the inactivation of mitophagy regulators such as PRKN and BNIP3 [32,51]. However, mitophagy facilitates cancer cell survival under conditions of energetic stress through eliminating ROS and maintaining valuable nutrients (such as oxygen) [45,46]. In addition, mitophagy has been associated with drug resistance, and the inhibition of autophagy has been reported to sensitize cancer cells to the cytotoxicity of anticancer drugs [52]. Therefore, targeting mitophagy may be an effective strategy to inhibit tumor progression to malignancy. PHB2 is an inner membrane mitophagy receptor [25], that promotes PINK1-PRKN-mediated mitophagy by modulating the mitochondrial protease PARL (Figure 5a-F). Additionally, PHB2 is positively correlated with the malignant progression of certain cancers, such as prostate cancer [53]. Thus, targeting PHB2 may offer opportunities to selectively inhibit tumor proliferation and progression. Indeed, FL3, a synthetic PHB ligand, strongly inhibits PHB2-mediated mitophagy (Figure 7) and, importantly, blocks cancer cell proliferation (Figure S7). Together, our findings reveal that FL3, which targets PHB2 to inhibit mitophagy, is a potent and promising anticancer agent.
Materials and methods
Cell culture and reagents
HeLa cells, HCT116 cells, 293T cells, and MEFs were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Thermo Fisher, 12,800–017) supplemented with 10% (v:v) fetal bovine serum (PAN, P30-3302), 100 U/mL penicillin (Gibco, 15,140–122) and 100 mg/mL streptomycin (Gibco, 15,140–122) at 37°C with 5% (v:v) CO2. Tetramethylrhodamine methyl ester (TMRM) was obtained from Molecular Probes (Thermo Fisher, T668). FL3 was synthesized by our group, as previously described [44]. Rocaglamide (HY-19,356) and z-VAD-FMK (HY-16,658) were purchased from MedChemExpress. Carbonyl cyanide 3-chlorophenylhydrazone (CCCP; C2759), MG132 (474,787), bafilomycin A1 (88,899–55-2) and antimycin A (A8674) were purchased from Sigma-Aldrich. Oligomycin was obtained from Calbiochem (1404–19-9). FL3 (50 nM), rocaglamide (Roc-A, 50 nM), CCCP (10 μM), oligomycin (10 μM) together with antimycin A (4 μM) (OA), and MG132 (30 μM) were used for all experiments. Long treatment durations of both oligomycin-antimycin A and CCCP were also supplemented with the apoptosis inhibitor 20 mM z-VAD-FMK to prevent cell death.
Plasmids and RNA interference
Human PINK1 and PRKN cDNA were gifts from Dr. Quan Chen (Chinese Academy of Sciences). The inserts were amplified by PCR and cloned into a lentiviral vector containing a C-terminal GFP and N-terminal GFP tag, respectively. The cDNAs of Human PARL, PHB2, and PHB2 LIR domain mutation (PHB2Y121A,L124A) were amplified by PCR and subcloned into the lentiviral vector pHAGE-puro (Addgene, 118,692, deposited by Christopher Vakoc) containing a C-terminal 3× FLAG tag. PGAM5-MYC was a gift from Quan Chen and was subcloned into pMSCV-puro constructs (Addgene, K1062-1, deposited by clontech). Wild-type OTC and its deletion mutant (Δ30-114; named ΔOTC) were a gift from Bin Lu (Wenzhou Medical University).
In brief, the shRNA target sequences against human PHB2 (shPHB2-1: 5ʹ-CCAGAATATCTCCAAGACGAT-3ʹ; and shPHB2-2: 5ʹ- AAGAACCCTGGCTACATCAAA-3) were subcloned into the lentiviral vector pLKO.1 (Addgene, 8453, deposited by Bob Weinberg) to drive the expression of shRNAs. Cells infected with lentivirus were selected with 1 µg/mL puromycin (Sigma, P8833) for 2 days in 6-well plates. shRNA-mediated silencing of Phb and Phb2 in MEFs was performed as previously described [54]. The scrambled shRNA against luciferase (target sequence: 5´-AACGCTGCTTCTTCTTATTTA-3´) were used as control. siRNA-mediated knockdown of PGAM5 was performed as previously described [55], and the siRNA target sequences against human PGAM5 were: siPGAM5-1, 5′-CCATAGAGACCACCGATAT-3′; siPGAM5-2: 5′-AACCACTGTCTCTGATCAA −3′.
Generation of knockout cells
HCT116 cells lacking PARL were generated using CRISPR/Cas9 gene editing. Briefly, for gene targeting DNA fragments specific for human PARL DNA (5´-CAGGCGTGGGGTGCGTCGGT-3´), was synthesized, and subcloned into LentiCRISPR plasmid (Addgene, 49,535, deposited by Feng Zhang). To produce lentivirus, recombinant LentiCRISPR were transfected in 293T cells together with plasmids psPAX2 and VSV-G using 40-kDa linear polyethylenimine (Polysciences, 24,765). The medium containing lentiviral particles was collected and was used for infecting HCT116 cells in the presence of polybrene (5 μg/mL; Sigma-Aldrich, H9268). The infected cells were selected with puromycin (2 μg/mL), and the single cells were sorted into 96-well dishes for the screen of PARL knockout lines. The surviving clones were picked, expanded and selected on the basis of PARL expression by western blotting. As a secondary screen of some knockout cell lines, genomic DNA was isolated from the cells and the genomic regions of interest were amplified by PCR (Primer: Forward, 5´-AAGTAGTTGCGCAGCTGGGG-3´, Reverse, 5´-GAAGGGGCAGGTAAGGTGAA-3´) to confirm the presence of frameshifting in the gene of PARL.
Mito-Keima mitophagy assay
Mito-Keima mitophagy assay was performed as previously described [20]. Briefly, HeLa cells stably expressing FLAG-PRKN were infected with a lentivirus harboring the mito-Keima vector (a gift from Michael Lenard). Then, HeLa cells were infected with a lentivirus containing shPHB2 and grown for several days. Next, cells were treated with or without OA in fresh growth medium for 16 h, and subsequently analyzed by confocal microscopy. Live cells were cultured in glass-bottom dishes. After treatment with OA, the cells were scanned and images were collected using a Leica SP8 confocal microscopy (× 63 oil objective NA 1.35) using an argon laser (448 nm, mito-Keima at neutral pH) and (552 nm, mito-Keima at acidic pH). Ratiometric (552 nm:448 nm) analysis was performed using ImageJ software.
Western blotting analysis and co-immunoprecipitations
Western blotting and co-immunoprecipitation (co-IP) analyses were performed as previously described [56]. Briefly, cells were harvested with RIPA buffer (50 mM Tris-HCl, 150 mM NaCl, 1% Nonidet P-40 [Fluka, 74,385], 0.5% sodium deoxycholate [Sigma-Aldrich, D6750], 1 mM EDTA [Sigma-Aldrich, 03609], 0.1% SDS [Sigma-Aldrich, L5750] and complete protease inhibitor [Roche, 04693132001]). Equal amounts of proteins were loaded onto an SDS-polyacrylamide gel, separated by electrophoresis and blotted onto a PVDF membrane (Merck Millipore, IPVH00010). The following antibodies were used: anti-FLAG (F1804), anti-MAP1LC3B (L7543) were obtained from Sigma-Aldrich; anti-PINK1 (Novus Biologicals, BC100-494); anti-GFP (SC-9996), anti-MYC (SC-40) and anti-HSPD1 (SC-13,115) were obtained from Santa Cruz Biotechnology; anti-TOMM20 (11,802–1-AP), anti-PHB (10,787–1-AP), anti-PHB2 (12,295–1-AP), anti-ATP5F1A (14,676–1-AP), and anti-OTC (26,470–1-AP) were obtained from Proteintech; anti-TUBB/β-tubulin (GNI4110-BT), and anti-ACTB (GNI4110-BA) were obtained from GNI. The antibody against PGAM5 was a gift from Quan Chen. For the co-IP, all steps were performed at 4°C. The cells were solubilized with IP buffer (150 mM NaCl [Vetec, 793,566], 10% glycerol [Sinopharm chemical reagent company, 10,010,618], 20 mM Tris-HCl pH 7.4 [Biofroxx, 1115GR500; Sinopharm chemical reagent company, 10,011,018], 2 mM EDTA, 0.5% Nonidet P-40, 0.5% Triton X-100 [Vetec, V900502] and complete protease inhibitor) for 1 h. Detergent-solubilized membrane proteins were cleared by centrifugation at 12,000 × g for 15 min and then subsequently incubated with anti-FLAG M2 affinity gel (Sigma-Aldrich, A2220) or Dynabeads Protein G (Thermo Fisher, 10004D) with antibody at 4℃ overnight. The resin was washed 5 times with lysis buffer, and then, the proteins were recovered by boiling the beads in SDS sample buffer and analyzed on SDS-PAGE followed by western blotting analysis.
Immunostaining
HeLa cells stably expressing GFP-PRKN were infected with a lentivirus against PHB2 and then treated with other reagents. Next, the cells were fixed with 4% formaldehyde [Sigma-Aldrich, P6148] in PBS (1 mM KH2PO4 [Vetec, V900041], 155 mM NaCl, 2.97 mM Na2HPO4-7H2O [Sigma-Aldrich, 431,478]) for 15 min at 37°C. After being washed with PBS three times, the cells were incubated with PBS plus 0.1% Triton X-100 buffer for 10 min. Next, the cells were blocked with PBS plus 10% FBS at room temperature (RT) for 1 h. Then, cells were incubated with the primary antibody for 2 h at RT and subsequently washed with PBS buffer three times. Then, the cells were incubated with PBS buffer containing the secondary antibody (Jackson immunoresearch, Cy3 affinipure goat anti-rabbit IgG (H + L) [111–165-003], alexa fluor 647 affinipure donkey anti-mouse IgG (H + L) [715–605-150]) for 1 h in the dark at RT. Finally, the cells were washed three times with PBS, and the slides were analyzed using a Leica confocal microscopy (Leica microsystem, Germany).
Mitochondria isolation and proteinase K treatment
Mitochondria were isolated from cells as previously described [57]. Briefly, cells were harvested and washed with PBS, and then cell pellets were homogenized with buffer A (83 mM sucrose [Vetec, V900116], 10 mM MOPS [Sangon biotech, mb0360], pH 7.2). After adding an equal volume of buffer B (250 mM sucrose, 30 mM MOPS [pH 7.2]), the nuclei and unbroken cells were removed by centrifugation at 1000 × g for 5 min. Mitochondria were collected from the supernatant by centrifugation at 12,000 × g for 5 min and washed once under the same conditions with buffer C (320 mM sucrose, 1 mM EDTA, 10 mM Tris-HCl, pH 7.4).
For the proteinase K (PK; Sigma-Aldrich, P4032) treatment, mitochondria were treated with various concentrations of PK for 30 min on ice. The PK digestion of mitochondria was terminated with 1 mM PMSF (Roche, 10,837,091,001). Samples were analyzed by SDS-PAGE and immunoblotting.
Evaluation of mitochondrial membrane potential (δψm)
Cells were cultured in glass-bottom dishes, 24 h later, cells were stained with 200 nM Tetramethylrhodamine methyl ester (TMRM) with complete medium at 37°C for 15 min. Cells were washed three times with DMEM and analyzed by confocal microscopy with a 552 nm argon laser.
Cryofixation and electron microscopy
HeLa cells were digested with trypsin (Gibco, 25,200–072) and the cell pellets were suspended using 20% BSA (Sigma-Aldrich, A1933). Then, cells were fixed by using Leica EM ICE according the manufacturer’s instructions. Next, cells were transferred in liquid nitrogen to the freeze‐substitution apparatus (Leica EM AFS) where eppendorf tubes filled with medium (the mixture of 1% osmium tetroxide [Ted Pella, Inc, 18,451] and 0.1% acetone [Sinopharm chemical reagent company, 10,000,418]) are precooled, followed by the programmed dehydration. 1 h later, the specimens are washed three times in anhydrous acetone in 0°C. The samples were then infiltrated sequentially in 1:1 (vol:vol) propylene oxide (Sinopharm chemical reagent company, 80,059,118):epoxy resin (Spi supplies, 90,529–77-4) (4 h), 1:2 propylene oxide:epoxy resin (overnight), 100% epoxy resin (4 h) and finally 100% epoxy resin (48 h) at 60°C for polymerization. The sections were supported on copper grids. The 80-nm sections were post stained in Sato lead (Sinopharm chemical reagent company; Lead nitrate [80,073,616], Sodium citrate [XW00680421]) for 1 min, and the stained sections were imaged onto negatives using a JEM-1400 plus electron microscope operated at 100 kV (Joel Ltd, Tokyo, Japan).
Quantitative real-time PCR analysis
RNA from the control and shPHB2 cells was extracted using Trizol (Life Technologies, 15,596–026) followed by DNase (Promega, M610A) treatment, and cDNA was synthesized using the RevertAid Synthesis Kit (Thermo Scientific, K1622) according to the manufacturer’s instructions. The cDNA samples were used as templates for quantitative real-time quantitative PCR (Q-RT-PCR) analysis using SYBR Green Supermix (Roche, 06924204001) and the iCycler real-time PCR Detection System (Bio-Rad). The fold change of target mRNA expression was calculated using the 2-ΔΔCT method. The primers used in this study were as followed:
PARL: forward, 5ʹ-TGTCAGTTACGTGGGTAAAGTTG-3ʹ, and reverse, 5ʹ-TCTGGGATCTTAGTGCAGACAG-3ʹ;
Tubulin: forward, 5ʹ-GACCTGACTGACTACCTCATGAAGAT-3ʹ, and reverse, 5ʹ- GTCACACTTCATGATGGAGTTGAAGG-3ʹ.
Statistical analysis
Densitometry was performed using ImageJ software for the quantitative analysis of the bands on the western blots. Data were presented as the mean ± S.D. Student’s t-test was used to calculate P-values. Statistical significance is displayed as *p < 0.05, **p < 0.01, ***p < 0.001.
Funding Statement
This work was supported by the National Natural Science Foundation of China [31671393, 91854107, 31471264, and 81673296], and the Fundamental Research Funds for the Central Universities (2042017kf0197 and 2042017fk0242).
Acknowledgments
We gratefully thank Dr. Quan Chen for the GFP-PRKN/parkin plasmid. This work is supported by National Natural Science Foundation of China (31671393, 91854107, 31471264, and 81673296), and the Fundamental Research Funds for the Central Universities (2042017kf0197 and 2042017kf0242).
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary materials
The supplemenmtary material for this article can be accessed here.
References
- [1].Singh R, Cuervo AM.. Autophagy in the cellular energetic balance. Cell Metab. 2011. May 4;13(5):495–504. PubMed PMID: 21531332; PubMed Central PMCID: PMC3099265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Georgakopoulos ND, Wells G, Campanella M.. The pharmacological regulation of cellular mitophagy. Nat Chem Biol. 2017. January 19;13(2):136–146. PubMed PMID: 28103219. [DOI] [PubMed] [Google Scholar]
- [3].Tatsuta T, Langer T. Quality control of mitochondria: protection against neurodegeneration and ageing. Embo J. 2008. January 23;27(2):306–314. PubMed PMID: 18216873; PubMed Central PMCID: PMCPmc2234350. eng. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011. January;12(1):9–14. PubMed PMID: 21179058; PubMed Central PMCID: PMCPMC4780047. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Redmann M, Dodson M, Boyer-Guittaut M, et al. Mitophagy mechanisms and role in human diseases. Int J Biochem Cell Biol. 2014. August;53:127–133. PubMed PMID: 24842106; PubMed Central PMCID: PMC4111979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Damier P, Hirsch EC, Agid Y, et al. The substantia nigra of the human brain. II. Patterns of loss of dopamine-containing neurons in Parkinson’s disease. Brain. 1999. August;122(Pt 8):1437–1448. PubMed PMID: 10430830; eng. [DOI] [PubMed] [Google Scholar]
- [7].Lin MT, Beal MF. Mitochondrial dysfunction and oxidative stress in neurodegenerative diseases. Nature. 2006. October 19;443(7113):787–795. PubMed PMID: 17051205. . [DOI] [PubMed] [Google Scholar]
- [8].Pickrell Alicia M, Youle Richard J. The roles of PINK1, Parkin, and mitochondrial fidelity in parkinson’s disease. Neuron. 2015;85(2):257–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Zhou C, Huang Y, Shao Y, et al. The kinase domain of mitochondrial PINK1 faces the cytoplasm. Proc Natl Acad Sci U S A. 2008. August 19;105(33):12022–12027. PubMed PMID: 18687899; PubMed Central PMCID: PMC2575334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Matsuda N, Sato S, Shiba K, et al. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J Cell Biol. 2010;189(2):211–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Meissner C, Lorenz H, Hehn B, et al. Intramembrane protease PARL defines a negative regulator of PINK1- and PARK2/Parkin-dependent mitophagy. Autophagy. 2015;11(9):1484–1498. PubMed PMID: 26101826; PubMed Central PMCID: PMC4590680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Jin SM, Lazarou M, Wang C, et al. Mitochondrial membrane potential regulates PINK1 import and proteolytic destabilization by PARL. J Cell Biol. 2010. November 29;191(5):933–942. PubMed PMID: 21115803; PubMed Central PMCID: PMC2995166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Greene AW, Grenier K, Aguileta MA, et al. Mitochondrial processing peptidase regulates PINK1 processing, import and Parkin recruitment. EMBO Rep. 2012. April;13(4):378–385. PubMed PMID: 22354088; PubMed Central PMCID: PMC3321149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Sekine S, Youle RJ. PINK1 import regulation; a fine system to convey mitochondrial stress to the cytosol. BMC Biol. 2018. January 10;16(1):2. PubMed PMID: 29325568; PubMed Central PMCID: PMC5795276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Vives-Bauza C, Zhou C, Huang Y, et al. PINK1-dependent recruitment of Parkin to mitochondria in mitophagy. Proc Natl Acad Sci U S A. 2010. January 5;107(1):378–383. PubMed PMID: 19966284; PubMed Central PMCID: PMCPMC2806779. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Narendra D, Tanaka A, Suen DF, et al. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J Cell Biol. 2008. December 1;183(5):795–803. PubMed PMID: 19029340; PubMed Central PMCID: PMCPMC2592826. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Hasson SA, Kane LA, Yamano K, et al. High-content genome-wide RNAi screens identify regulators of parkin upstream of mitophagy. Nature. 2013. December 12;504(7479):291–295. PubMed PMID: 24270810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Lu W, Karuppagounder SS, Springer DA, et al. Genetic deficiency of the mitochondrial protein PGAM5 causes a Parkinson’s-like movement disorder. Nat Commun. 2014. September 15;5:4930. PubMed PMID: 25222142; PubMed Central PMCID: PMC4457367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Lo SC, Hannink M. PGAM5 tethers a ternary complex containing Keap1 and Nrf2 to mitochondria. Exp Cell Res. 2008. May 1;314(8):1789–1803. PubMed PMID: 18387606; PubMed Central PMCID: PMCPmc2409987. eng. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jian F, Chen D, Chen L, et al. Sam50 Regulates PINK1-Parkin-mediated mitophagy by controlling PINK1 stability and mitochondrial morphology. Cell Rep. 2018;23(10):2989–3005. [DOI] [PubMed] [Google Scholar]
- [21].Stolz A, Ernst A, Dikic I. Cargo recognition and trafficking in selective autophagy. Nat Cell Biol. 2014. June;16(6):495–501. PubMed PMID: 24875736. . [DOI] [PubMed] [Google Scholar]
- [22].Lazarou M, Sliter DA, Kane LA, et al. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 2015. August 20;524(7565):309–314. PubMed PMID: 26266977; PubMed Central PMCID: PMCPMC5018156. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Chu CT, Ji J, Dagda RK, et al. Cardiolipin externalization to the outer mitochondrial membrane acts as an elimination signal for mitophagy in neuronal cells. Nat Cell Biol. 2013. October;15(10):1197–1205. PubMed PMID: 24036476; PubMed Central PMCID: PMC3806088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Liu L, Feng D, Chen G, et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nat Cell Biol. 2012. January 22;14(2):177–185. PubMed PMID: 22267086. [DOI] [PubMed] [Google Scholar]
- [25].Wei Y, Chiang WC, Sumpter R Jr. et al. Prohibitin 2 is an inner mitochondrial membrane mitophagy receptor. Cell. 2017. January 12;168(1–2):224–238 e10. PubMed PMID: 28017329; PubMed Central PMCID: PMC5235968. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Artal-Sanz M, Tavernarakis N. Prohibitin and mitochondrial biology. Trends Endocrinol Metab. 2009. October;20(8):394–401. PubMed PMID: 19733482. . [DOI] [PubMed] [Google Scholar]
- [27].Steglich G, Neupert W, Langer T. Prohibitins regulate membrane protein degradation by the m-AAA protease in mitochondria. Mol Cell Biol. 1999. May;19(5):3435–3442. PubMed PMID: 10207067; PubMed Central PMCID: PMC84136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].MacVicar T, Langer T. OPA1 processing in cell death and disease - the long and short of it. J Cell Sci. 2016. June 15;129(12):2297–2306. PubMed PMID: 27189080; eng. . [DOI] [PubMed] [Google Scholar]
- [29].Warburg O. On the origin of cancer cells. Science. 1956. February 24;123(3191):309–314. PubMed PMID: 13298683; eng. [DOI] [PubMed] [Google Scholar]
- [30].Wallace DC. Mitochondria and cancer. Nat Rev Cancer. 2012. October;12(10):685–698. PubMed PMID: 23001348; PubMed Central PMCID: PMC4371788. . [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Lu H, Li G, Liu L, et al. Regulation and function of mitophagy in development and cancer. Autophagy. 2013. November 01;9(11):1720–1736. PubMed PMID: 24091872. [DOI] [PubMed] [Google Scholar]
- [32].Chourasia AH, Boland ML, Macleod KF. Mitophagy and cancer. Cancer Metab. 2015;3:4. PubMed PMID: 25810907; PubMed Central PMCID: PMC4373087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Tatsuta T, Langer T. Prohibitins. Curr Biol. 2017. July 10;27(13):R629–R631. PubMed PMID: 28697355. . [DOI] [PubMed] [Google Scholar]
- [34].Kasashima K, Ohta E, Kagawa Y, et al. Mitochondrial functions and estrogen receptor-dependent nuclear translocation of pleiotropic human prohibitin 2. J Biol Chem. 2006. November 24;281(47):36401–36410. PubMed PMID: 17008324; eng. [DOI] [PubMed] [Google Scholar]
- [35].Sun N, Malide D, Liu J, et al. A fluorescence-based imaging method to measure in vitro and in vivo mitophagy using mt-Keima. Nat Protoc. 2017. August;12(8):1576–1587. PubMed PMID: 28703790. [DOI] [PubMed] [Google Scholar]
- [36].Jin SM, Youle RJ. The accumulation of misfolded proteins in the mitochondrial matrix is sensed by PINK1 to induce PARK2/Parkin-mediated mitophagy of polarized mitochondria. Autophagy. 2013. November 1;9(11):1750–1757. PubMed PMID: 24149988; PubMed Central PMCID: PMC4028334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Richter U, Lahtinen T, Marttinen P, et al. A mitochondrial ribosomal and RNA decay pathway blocks cell proliferation. Curr Biol. 2013. March 18;23(6):535–541. PubMed PMID: 23453957; eng. [DOI] [PubMed] [Google Scholar]
- [38].Jin SM, Youle RJ. PINK1- and Parkin-mediated mitophagy at a glance. J Cell Sci. 2012. February 15;125(Pt 4):795–799. PubMed PMID: 22448035; PubMed Central PMCID: PMC3656616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Yoshii SR, Kishi C, Ishihara N, et al. Parkin mediates proteasome-dependent protein degradation and rupture of the outer mitochondrial membrane. J Biol Chem. 2011. June 03;286(22):19630–19640. PubMed PMID: 21454557; PubMed Central PMCID: PMC3103342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Yamano K, Youle RJ. PINK1 is degraded through the N-end rule pathway. Autophagy. 2013. November 01;9(11):1758–1769. PubMed PMID: 24121706; PubMed Central PMCID: PMC4028335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Sekine S, Kanamaru Y, Koike M, et al. Rhomboid protease PARL mediates the mitochondrial membrane potential loss-induced cleavage of PGAM5. J Biol Chem. 2012. October 05;287(41):34635–34645. PubMed PMID: 22915595; PubMed Central PMCID: PMC3464569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Wai T, Saita S, Nolte H, et al. The membrane scaffold SLP2 anchors a proteolytic hub in mitochondria containing PARL and the i-AAA protease YME1L. EMBO Rep. 2016. December;17(12):1844–1856. PubMed PMID: 27737933; PubMed Central PMCID: PMC5283581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Polier G, Neumann J, Thuaud F, et al. The natural anticancer compounds rocaglamides inhibit the Raf-MEK-ERK pathway by targeting prohibitin 1 and 2. Chem Biol. 2012. September 21;19(9):1093–1104. PubMed PMID: 22999878. [DOI] [PubMed] [Google Scholar]
- [44].Thuaud F, Bernard Y, Turkeri G, et al. Synthetic analogue of rocaglaol displays a potent and selective cytotoxicity in cancer cells: involvement of apoptosis inducing factor and caspase-12. J Med Chem. 2009. August 27;52(16):5176–5187. PubMed PMID: 19655762. [DOI] [PubMed] [Google Scholar]
- [45].Kulikov AV, Luchkina EA, Gogvadze V, et al. Mitophagy: link to cancer development and therapy. Biochem Biophys Res Commun. 2017. January 15;482(3):432–439. PubMed PMID: WOS:000393270000008; English. [DOI] [PubMed] [Google Scholar]
- [46].Bernardini JP, Lazarou M, Dewson G. Parkin and mitophagy in cancer. Oncogene. 2017. March;36(10):1315–1327. PubMed PMID: 27593930; eng. [DOI] [PubMed] [Google Scholar]
- [47].Ribeiro N, Thuaud F, Bernard Y, et al. Flavaglines as potent anticancer and cytoprotective agents. J Med Chem. 2012. November 26;55(22):10064–10073. PubMed PMID: 23072299. [DOI] [PubMed] [Google Scholar]
- [48].Yurugi H, Marini F, Weber C, et al. Targeting prohibitins with chemical ligands inhibits KRAS-mediated lung tumours. Oncogene. 2017. October 19;36(42):5914. PubMed PMID: 28846116. [DOI] [PubMed] [Google Scholar]
- [49].Li-Weber M. Molecular mechanisms and anti-cancer aspects of the medicinal phytochemicals rocaglamides (=flavaglines). Int J Cancer. 2015. October 15;137(8):1791–1799. PubMed PMID: 24895251. [DOI] [PubMed] [Google Scholar]
- [50].Chan NC, Salazar AM, Pham AH, et al. Broad activation of the ubiquitin-proteasome system by Parkin is critical for mitophagy. Hum Mol Genet. 2011. May 1;20(9):1726–1737. PubMed PMID: 21296869; PubMed Central PMCID: PMCPmc3071670. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Matsuda S, Nakanishi A, Minami A, et al. Functions and characteristics of PINK1 and Parkin in cancer. Front Biosci (Landmark Ed). 2015. January 1;20:491–501. PubMed PMID: 25553463; eng. [DOI] [PubMed] [Google Scholar]
- [52].Hu YL, Jahangiri A, Delay M, et al. Tumor cell autophagy as an adaptive response mediating resistance to treatments such as antiangiogenic therapy. Cancer Res. 2012. September 1;72(17):4294–4299. PubMed PMID: 22915758; PubMed Central PMCID: PMCPmc3432684. eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shen Y, Gao Y, Yuan H, et al. Prohibitin-2 negatively regulates AKT2 expression to promote prostate cancer cell migration. Int J Mol Med. 2018. February;41(2):1147–1155. PubMed PMID: 29207197; eng. [DOI] [PubMed] [Google Scholar]
- [54].Zhang K, Li H, Song Z. Membrane depolarization activates the mitochondrial protease OMA1 by stimulating self-cleavage. EMBO Rep. 2014;15(5):576–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Wu H, Xue D, Chen G, et al. The BCL2L1 and PGAM5 axis defines hypoxia-induced receptor-mediated mitophagy. Autophagy. 2014. October 01;10(10):1712–1725. PubMed PMID: 25126723; PubMed Central PMCID: PMC4198357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Li H, Ruan Y, Zhang K, et al. Mic60/Mitofilin determines MICOS assembly essential for mitochondrial dynamics and mtDNA nucleoid organization. Cell Death Differ. 2016. March;23(3):380–392. PubMed PMID: 26250910; eng. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Acin-Perez R, Fernandez-Silva P, Peleato ML, et al. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008. November 21;32(4):529–539. PubMed PMID: 19026783. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


