ABSTRACT
Shortening of mRNA poly(A) tails (deadenylation) to trigger their decay is mediated mainly by the CCR4-NOT deadenylase complex. While four catalytic subunits (CNOT6, 6L 7, and 8) have been identified in the mammalian CCR4-NOT complex, their individual biological roles are not fully understood. In this study, we addressed the contribution of CNOT7/8 to viability of primary mouse embryonic fibroblasts (MEFs). We found that MEFs lacking CNOT7/8 expression [Cnot7/8-double knockout (dKO) MEFs] undergo cell death, whereas MEFs lacking CNOT6/6L expression (Cnot6/6l-dKO MEFs) remain viable. Co-immunoprecipitation analyses showed that CNOT6/6L are also absent from the CCR4-NOT complex in Cnot7/8-dKO MEFs. In contrast, either CNOT7 or CNOT8 still interacts with other subunits in the CCR4-NOT complex in Cnot6/6l-dKO MEFs. Exogenous expression of a CNOT7 mutant lacking catalytic activity in Cnot7/8-dKO MEFs cannot recover cell viability, even though CNOT6/6L exists to some extent in the CCR4-NOT complex, confirming that CNOT7/8 is essential for viability. Bulk poly(A) tail analysis revealed that mRNAs with longer poly(A) tails are more numerous in Cnot7/8-dKO MEFs than in Cnot6/6l-dKO MEFs. Consistent with elongated poly(A) tails, more mRNAs are upregulated and stabilized in Cnot7/8-dKO MEFs than in Cnot6/6l-dKO MEFs. Importantly, Cnot6/6l-dKO mice are viable and grow normally to adulthood. Taken together, the CNOT7/8 catalytic subunits are essential for deadenylation, which is necessary to maintain cell viability, whereas CNOT6/6L are not.
KEYWORDS: The CCR4-NOT complex, CNOT6/6L, CNOT7/8, deadenylation, cell viability
Introduction
Regulation of mRNA decay in the cytoplasm is important for proper gene expression, and its dysregulation causes various disorders. Shortening of mRNA poly(A) tails by deadenylation is the initial, rate-limiting step in the exonucleolytic mRNA decay pathway, which is also relevant to translational suppression [1,2]. mRNA deadenylation occurs via two distinct steps. First, PAN2-PAN3 trims long poly(A) tails to ~110 nucleotides (nt) and subsequently, the CCR4-NOT complex completes deadenylation [3,4].
Deadenylation activity of the CCR4-NOT complex was first identified in yeast [5]. CCR4-NOT is a multi-protein complex, consisting of at least eight core subunits (Ccr4p, Caf1p, Caf40p, Caf130p, Not1p, Not2p, either Not3p or Not5p, and Not4p) in yeast, and eight subunits (CNOT1, CNOT2, CNOT3, either CNOT7 or CNOT8, either CNOT6 or CNOT6L, CNOT9, CNOT10 and CNOT11) in mammals [6–8]. Consequently, in mammals, there are four possible catalytic and six non-catalytic subunits. The catalytic subunits include DEDD (Asp-Glu-Asp-Asp) family proteins (CNOT7 and CNOT8) and exonuclease-endonuclease-phosphatase (EEP) family proteins (CNOT6 and CNOT6L). CNOT7/8 and CNOT6/6L are orthologs of yeast Caf1p and Ccr4p, respectively, [6–9]. CNOT1, CNOT2, CNOT3, CNOT9, CNOT10, and CNOT11 are non-catalytic subunits that are involved in complex formation, regulation of catalytic activity, and recruitment of the CCR4–NOT complex to target mRNAs [10–17]. Structural analyses have revealed that CNOT1 is a scaffold subunit responsible for assembly of the whole complex [11,12,18]. When CNOT1 is suppressed in HeLa cells, expression of most other subunits also decreases, resulting in severe reduction of deadenylase activity [10]. Therefore, CNOT1 is critical not only for assembly but also for maintenance of the CCR4–NOT complex. A given CCR4-NOT complex includes either CNOT6 or CNOT6L and either CNOT7 or CNOT8 [9,19]. CNOT1 binds directly to CNOT7/8 through its MIF4G domain [11,18]. CNOT7/8 are required for incorporation of CNOT6/6L into the complex via protein–protein interaction between CNOT7/8 and the leucine-rich repeat (LRR) domain of CNOT6/6L [20].
The CCR4-NOT complex is critical to a variety of physiological functions in mammals, including energy metabolism, B-cell development, heart function, and liver maturation, via mRNA deadenylation and subsequent degradation [14,21–23]. Mouse embryonic development also requires CCR4-NOT complex activity, as mice lacking the Cnot1 or Cnot3 genes (Cnot1-KO or Cnot3-KO mice, respectively) show embryonic lethality [14,22,24]. Furthermore, suppression of the complex leads to various forms of cell death, such as apoptosis and necroptosis, indicating the importance of mRNA decay in cell viability [10,21–23,25,26].
Several studies have described different roles of the catalytic subunits in deadenylase activity and biological processes. While an EEP family member, Ccr4p, is largely associated with deadenylase activity in yeast [5], DEDD family proteins, Ccf1 and CNOT7/8, are critical in C. elegans and mammals, respectively, [27,28]. Suppression of Cnot6l or both Cnot6/6l in human cancer cell lines leads to proliferation arrest and cell death in a deadenylase activity-dependent manner, indicating the importance of CNOT6/6L in mammals [26,29]. In the same study, it was shown that CNOT6/6L and CNOT7/8 regulate distinct groups of genes [26]. Therefore, CNOT6/6L and CNOT7/8 serve different functions, depending on cell types, biological processes, and target genes. Importantly, recent studies show that CNOT6/6L and CNOT7/8 display distinct biochemical activities in deadenylation. These differences explain why two different types of ribonuclease occur in a single complex [30,31]. The two paralogs, CNOT6/6L and CNOT7/8, in mammals have been considered basically redundant. Cnot6l-KO or Cnot7-KO mice are viable and normally grow to adulthood, supporting their functional redundancy, at least in mouse embryonic development and in embryo-to-adult viability [32–34]. On the other hand, while CNOT7 and CNOT8 have overlapping roles in some cases, as in growth of human breast cancer (MCF7) cells [35], they also have unique roles in other contexts. For instance, Cnot7-KO mice manifest defects in spermatogenesis that cannot be compensated by CNOT8 [32,33]. To better understand the different biological roles of the four catalytic subunits, further analyses are necessary to clarify their distinctive functions in various biological contexts.
In this study, we used primary MEFs prepared from mice lacking CCR4-NOT complex subunits. Since Cnot1-KO, Cnot3-KO, and Cnot8-KO mice are embryonically lethal (14, 22, 24 and this study), we prepared MEFs from mice possessing conditional alleles, in which loxP sequences were inserted, and we deleted the corresponding targets using Cre-mediated somatic recombination. Given that primary MEFs lacking CCR4-NOT complex-dependent deadenylation undergo cell death [25], roles of the catalytic subunits in cell viability were investigated. We found that CNOT7/8 is essential for viability of MEFs, whereas CNOT6/6L are not. Gene expression analysis using RNA-sequencing (RNA-seq) showed that upregulation and stabilization of mRNAs were more pronounced in Cnot7/8-dKO MEFs than in Cnot6/6l-dKO MEFs. Thus, we propose that CNOT7/8-, but not CNOT6/6L-mediated mRNA decay regulates MEF viability and that CNOT8 is required in mouse embryonic development.
Results
Cnot8-KO mice die in embryo, whereas Cnot6-KO mice display no obvious abnormality
We generated Cnot6-KO and Cnot8-KO mice to examine physiological roles of these subunits in mouse embryonic development and to prepare primary MEFs. We inserted loxP sequences and the neomycin-resistance gene cassette between frt sequences into Cnot6 or Cnot8 gene loci in order to generate either null or conditional alleles (Supplementary Fig. 1A). Conditional alleles could be used to examine tissue-specific roles in future studies. In KO alleles, exons 8 or 3 are removed in the Cnot6 or Cnot8 genes, respectively. We found that Cnot6-KO mice were born live at the predicted mendelian frequency (Supplementary Fig. 1B). They grew to adulthood and were fertile, at least under standard breeding conditions. By crossing Cnot6-KO (generated in this study) and Cnot6l-KO mice [34], Cnot6-KO/Cnot6l-heterozygous (Het) mice were generated. Cnot6-KO/Cnot6l-Het mice were also normal and fertile, although litter sizes in Cnot6-KO/Cnot6l-Het pairs were slightly smaller than those of controls (Supplementary Fig. 1B). Furthermore, Cnot6/6l-dKO mice were born at the predicted mendelian frequencies and grew to adulthood. These data indicate that CNOT6/6L are dispensable for mouse embryonic development. Unexpectedly, Cnot8-KO mice died before embryonic day 10.5 (Supplementary Fig. 1B), and CNOT7 could not compensate for the lack of CNOT8 during mouse embryonic development.
Simultaneous suppression of Cnot7 and Cnot8 causes marked cell death in MEFs
Based on embryonic phenotypes of mice lacking each catalytic subunit in the CCR4-NOT complex, we prepared MEFs from mice with Cnot8-flox alleles (Cnot8-flox MEFs) and subsequently deleted the Cnot8 gene with Cre-mediated recombination to generate Cnot8-KO MEFs. We employed recombinant retrovirus to introduce Cre in MEFs. Cnot6-KO, Cnot6l-KO, Cnot7-KO [32], and Cnot6/6l-dKO MEFs were prepared from the corresponding KO mouse embryos. We also prepared Cnot7-KO/Cnot8-flox MEFs by crossing Cnot7-Het/Cnot8-flox pairs. Using Cre-mediated recombination, we generated Cnot7/8-dKO MEFs. For controls, mock-infected MEFs were used. MEFs were also prepared from mice with Cnot1-flox alleles followed by Cre-mediated deletion of the Cnot1 gene (Cnot1-KO MEFs). Immunoblot analysis confirmed that expression of CNOT1 protein was substantially decreased (Fig. 1A, B). CNOT1 protein was still detected after Cnot1 gene deletion (Fig. 1A). We previously observed a similar result in Cnot3-KO MEFs [25]. We speculate that cells which had comparably high levels of CNOT1 protein before Cre-mediated gene deletion survived until the time of our analysis, while cells with reduced CNOT1 levels started to die. It is also possible that some cells escaped gene deletion due to insufficient Cre expression, because random integration of the Cre gene into the host genome leads to heterogeneous cell populations that vary in Cre expression. Amounts of other subunits in the CCR4-NOT complex also decreased in Cnot1-KO MEFs, resulting in decreased expression of the whole complex, as shown by immunoprecipitation using anti-CNOT3 antibody (Fig. 1A, B). Furthermore, the appearance of floating cells and a significant increase in the number of propidium iodide (PI)-incorporated cells indicated that Cnot1-KO MEFs underwent cell death (Fig. 1C, D). Thereafter, we used Cnot1-KO MEFs as positive controls, because they largely lack the CCR4-NOT complex.
Figure 1.
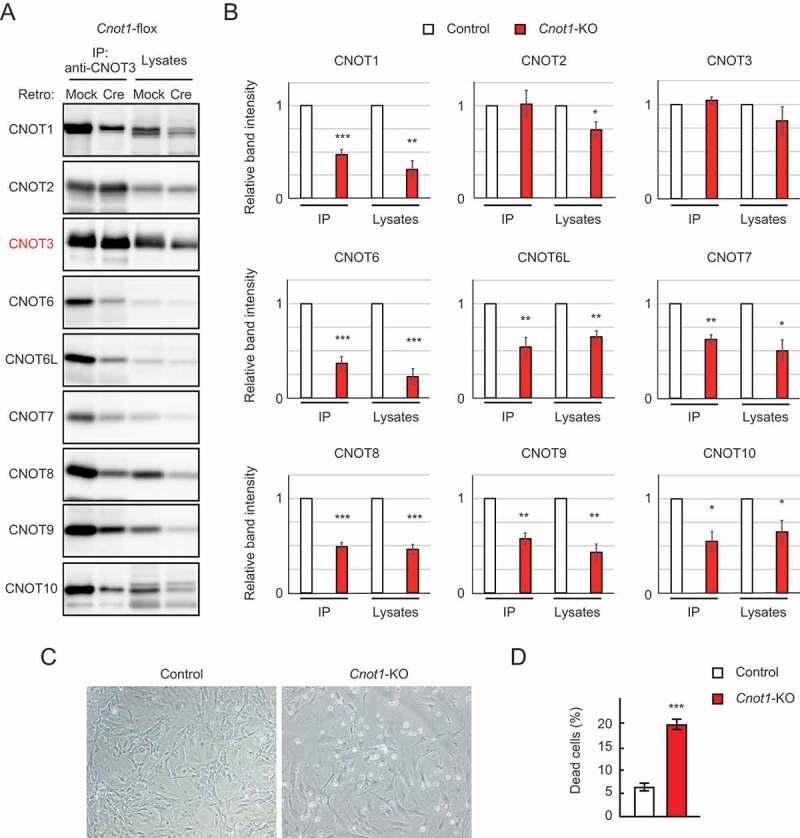
Cnot1-KO MEFs undergo cell death. (A) Lysates were prepared from Cnot1-flox MEFs that were infected with mock or Cre-expressing retrovirus and subjected to immunoprecipitation with anti-CNOT3 antibody. CNOT3 are shown in red to indicate a precipitated molecule. Lysates and immunoprecipitates (IP) were analysed by immunoblot with the indicated antibodies. (B) Quantification of the immunoblot data in Fig. 1A. Relative band intensities normalized to those of IP or lysates in control MEFs are shown (n = 3). Values represent means ± standard error of the mean (S.E.M.). (C) Morphology of Cnot1-flox MEFs infected with mock (Control) or Cre-expressing retrovirus (Cnot1-KO). Photographs are at the same magnification and represent one of the three independent experiments. Dead cells that were about to lose adhesion were observed in Cnot1-KO MEFs. (D) Cell death was assessed by propidium iodide uptake using flow cytometry (n = 3). Values in the graphs represent mean ± S.E.M. *P< 0.05, **P< 0.01, ***P< 0.001.
We then examined viability of Cnot7/8-dKO and Cnot6/6l-dKO MEFs. Cnot7/8-dKO MEFs underwent cell death, while such outcomes were not observed in Cnot7-KO, Cnot8-KO, and mock-infected MEFs (Fig. 2A, B and Supplementary Fig. 2). There was no significant difference in PI-incorporated cells among Cnot6-KO, Cnot6l-KO, Cnot6/6l-dKO, and wild-type (WT) MEFs (Fig. 2A, B and Supplementary Fig. 2). Therefore, as in the case of embryonic development, CNOT6/6L are dispensable for cell viability. In contrast, CNOT7 and CNOT8 have redundant functions in maintaining MEF viability.
Figure 2.
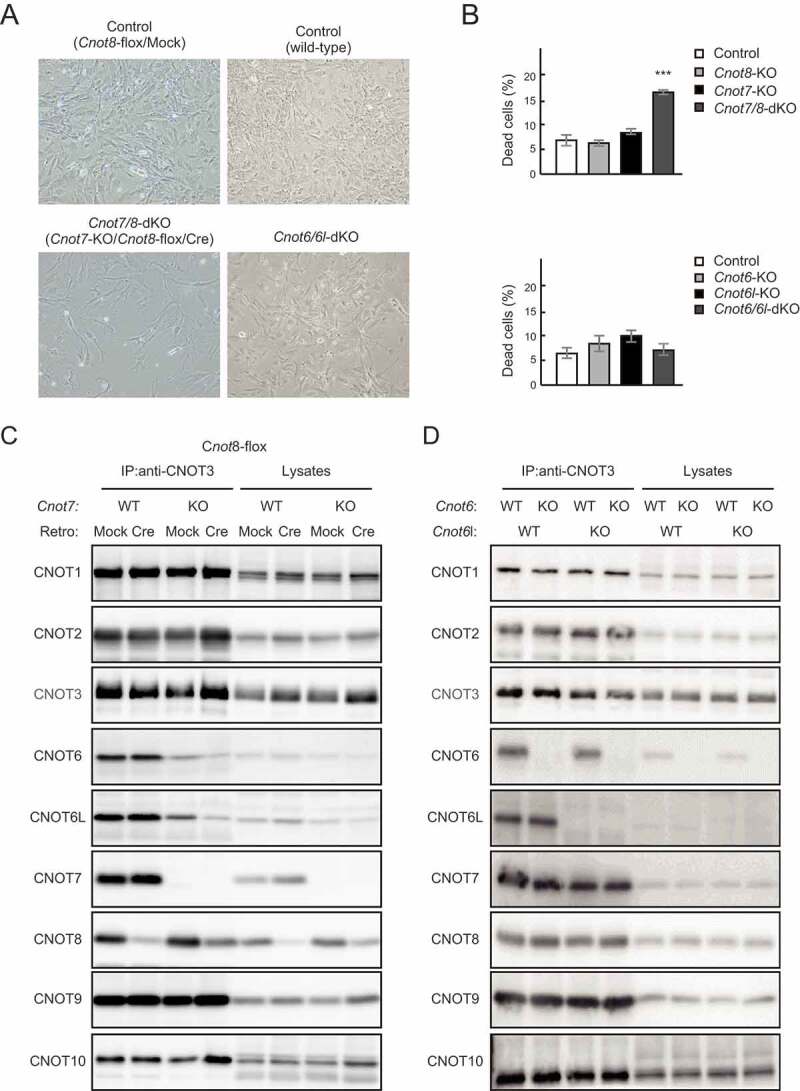
Suppression of CNOT7/8, but not CNOT6/6L, affects viability of MEFs. (A) Morphology of MEFs with the indicated genotypes shown as in Fig. 1B. Mock and Cre represent cells infected with retrovirus. (B) Cell death was assessed as in Fig. 1D (n = 3). Values represent means ± S.E.M. ***P< 0.001. (C) Lysates were prepared from Cnot8-flox MEFs, which were infected with mock or Cre-expressing retrovirus and subjected to immunoprecipitation with anti-CNOT3 antibody. CNOT3 is shown in red to indicate a precipitated molecule. Lysates and IP were analysed by immunoblot with the indicated antibodies. WT: wild-type, KO: knockout. (D) Lysates were prepared from MEFs with the indicated genotypes and subjected to immunoprecipitation with anti-CNOT3 antibody. Lysates and IP were analysed by immunoblot.
Cnot7/8 suppression results in loss of catalytic subunits from the CCR4-NOT complex
Decreased levels of intact CCR4-NOT complex result in decreased MEF viability (25, and Fig. 1). We performed co-immunoprecipitation experiments to examine the effects of CNOT6/6L or CNOT7/8 suppression on complex formation. CNOT8 protein was still detected after Cnot8 gene deletion (Fig. 2C), probably for the same reasons described in the case of Cnot1-KO MEFs (Fig. 1A, see above). Importantly, immunoprecipitation of lysates from Cnot7/8-dKO MEFs with anti-CNOT3 antibody showed that levels of co-immunoprecipitated CNOT6 and CNOT6L decreased substantially compared to control MEFs (Fig. 2C and Supplementary Fig. 3A), indicating that few catalytic subunits were associated with core subunits (CNOT1-3) of the CCR4-NOT complex in Cnot7/8-dKO MEFs. This is consistent with a study demonstrating CNOT7/8 bridge interaction between CNOT6/6L and CNOT1 [20]. We found that CNOT1, CNOT2, CNOT3, CNOT7, CNOT8, CNOT9, and CNOT10 existed at similar levels in anti-CNOT3 immunoprecipitates among WT, Cnot6-KO, Cnot6l-KO and Cnot6/6l-dKO MEFs (Fig. 2D and Supplementary Fig. 3B). These data suggest that loss of CNOT6/6L does not affect association of other CCR4-NOT complex subunits.
CNOT7 catalytic activity is essential to maintain MEF viability
MEFs without CNOT6/6L in the CCR4-NOT complex rarely underwent cell death, indicating that CNOT7/8 are sufficient for MEF viability (Fig. 2A, B). To examine whether CNOT7/8 catalytic activity is critically involved in MEF viability, we introduced either FLAG epitope-tagged CNOT7-WT or CNOT7 mutants into Cnot7/8-dKO MEFs, using a recombinant retrovirus. CNOT7 mutants used included a catalytically negative mutant (CN) in which Asp40 and Glu42 were replaced with Ala, and a dominant negative mutant (DN), which lacked both catalytic activity and the capacity to bind CNOT6/6L (Cys67 and Leu71 were replaced with Glu, in addition to the catalytically negative mutation) [36]. We first characterized CNOT7 mutants by overexpressing them in WT-MEFs. Results of immunoprecipitation with anti-FLAG antibody confirmed that CNOT7-WT and CNOT7-CN, but not CNOT7-DN, interacted with CNOT6/6L, whereas CNOT1, CNOT3 and CNOT9 were co-precipitated with all CNOT7 constructs (Supplementary Fig. 4A). We found that both CNOT7-CN and CNOT7-DN induced death in WT-MEFs (Supplementary Fig. 4B), suggesting that these mutants have a dominant negative effect on MEF viability. In the series of complementation experiments, we used a recombinant adenovirus expressing Cre to delete the Cnot8 gene. When CNOT7-WT was reintroduced into Cnot7/8-dKO MEFs, cell viability was comparable to that of control virus-infected MEFs (Fig. 3A, B). Expression of either CNOT7-CN or CNOT7-DN failed to recover viability, but further induced cell death (Fig. 3A, B). This effect was likely caused by a dominant negative effect of CNOT7-CN and CNOT7-DN as both mutants prevented endogenous CNOT8, residually expressed after Cre-mediated recombination, from binding to CNOT1, leading to formation of a non-functional complex. Immunoblot analyses revealed the successful expression of FLAG-CNOT7-WT and mutants in Cnot7/8-dKO MEFs (Fig. 3C, D). We found that the amount of CNOT6/6L in anti-CNOT3 immunoprecipitates from Cnot7/8-dKO MEFs expressing CNOT7-WT or CNOT7-CN increased compared to that in Cnot7/8-dKO MEFs and those expressing CNOT7-DN (Fig. 3C, D). These data suggest that the catalytic activity of DEDD family proteins (CNOT7/8) in the CCR4-NOT complex is essential for cell viability.
Figure 3.
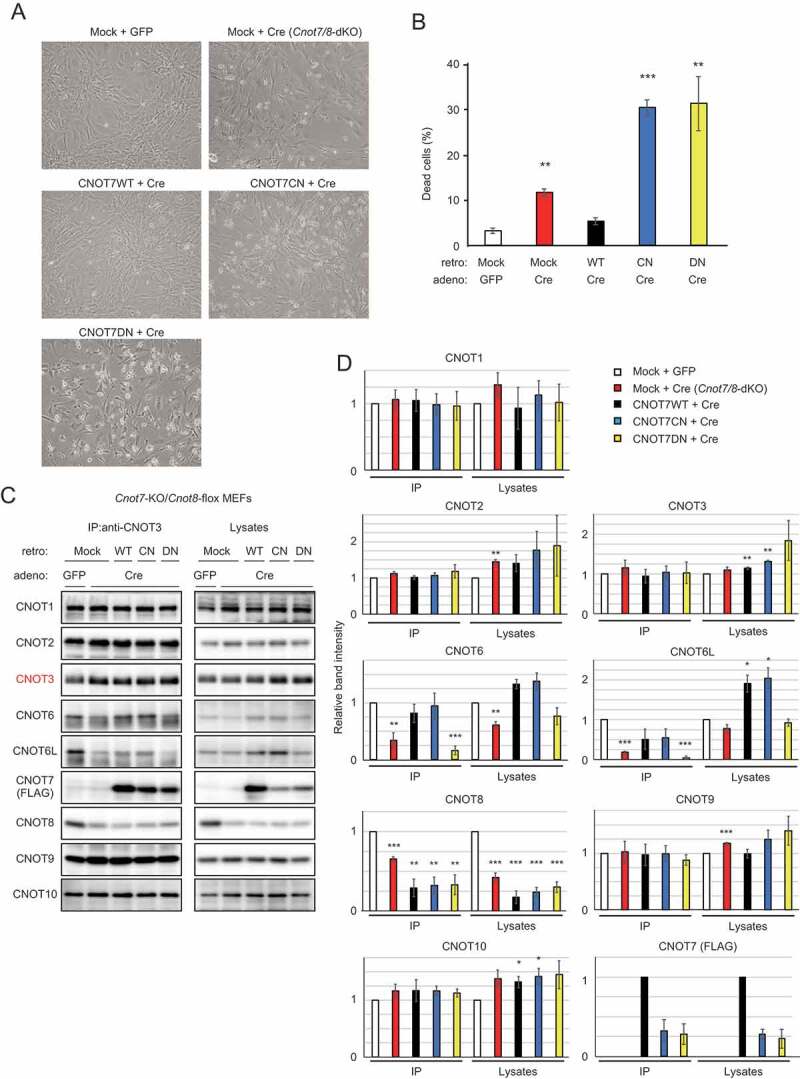
CNOT7 catalytic activity is sufficient to maintain MEF viability. (A) Morphology of Cnot7-KO/Cnot8-flox MEFs infected with the indicated (retro + adeno) viruses. (B) Cell death was assessed as in Fig. 1D (n = 3). (C) Lysates were prepared from Cnot7-KO/Cnot8-flox MEFs infected with the indicated viruses and subjected to immunoprecipitation with anti-CNOT3 antibody. CNOT3 is shown in red to indicate a precipitated molecule. Lysates and IP were analysed by immunoblot. WT: CNOT7 wild-type, CN: CNOT7 lacking catalytic activity, DN: CNOT7 dominant negative mutant which lacks catalytic activity and an ability to bind to CNOT6/6L. (D) Quantification of the immunoblot data in Fig. 3C. Relative band intensities normalized to those of IP or lysates in control MEFs are shown (n = 3). Values represent means ± S.E.M. *P< 0.05, **P< 0.01, ***P< 0.001.
In addition, we examined whether exogenous overexpression of CNOT6L can restore viability of Cnot7/8-dKO MEFs. The results of cell death analysis showed that exogenous overexpression of CNOT6L partly, but significantly, restored viability of Cnot7/8-dKO MEFs (Supplementary Fig. 5A, B). When we performed immunoprecipitation experiments, exogenously overexpressed CNOT6L existed in anti-CNOT3 immunoprecipitates (Supplementary Fig. 5C, D). This could explain the recovery of viability. We speculate that residual expression of CNOT8 enabled exogenously expressed CNOT6L to exist in the CCR4-NOT complex.
Poly(A) tail length of bulk RNAs is elongated in Cnot1-KO and Cnot7/8-dKO MEFs
We next compared deadenylase activity in Cnot1-KO, Cnot7/8-dKO, and Cnot6/6l-dKO MEFs with their respective controls (see Fig. 4 legend), and then examined poly(A) tail lengths of bulk RNAs. In Cnot1-KO and Cnot7/8-dKO MEFs, the population of mRNAs with poly(A) tail lengths longer than 50 nt increased, while the population of mRNAs with poly(A) tail lengths shorter than 50 nt decreased (Fig. 4A, B). On the other hand, the distribution of poly(A) tail lengths was similar in control and Cnot6/6l-dKO MEFs (Fig. 4C). We observed a slight increase in the population of mRNAs with poly(A) tail lengths between 50 and 100 nt (Fig. 4C), which is reminiscent of what occurs in poly (A)-binding protein (PABP)-mediated prevention of deadenylation, though the effect was smaller than in human cancer cell lines [30]. These data suggest that deadenylase activity is largely impaired in Cnot1-KO and Cnot7/8-dKO, but not in Cnot6/6l-dKO MEFs.
Figure 4.
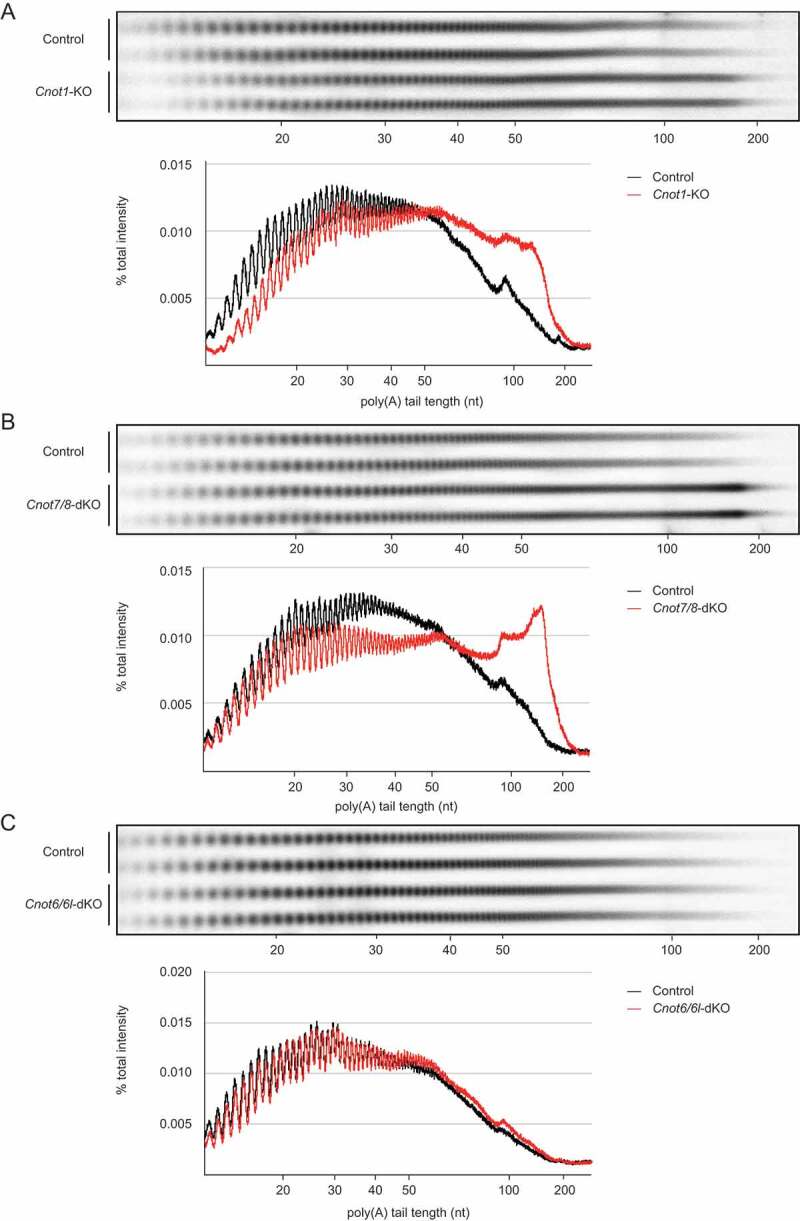
RNAs in Cnot1-KO and Cnot7/8-dKO MEFs have longer polyA tails compared to those in Cnot6/6l-dKO MEFs. (A-C) Poly(A) tail lengths of bulk RNA in Cnot1-KO (A), Cnot7/8-dKO (B) and Cnot6/6l-dKO MEFs (C) (n = 2 for each genotype). Cnot1-flox MEFs infected with mock retrovirus, Cnot7+/+; Cnot8-flox MEFs infected with mock retrovirus, and WT MEFs were used as controls, respectively. Densitograms of poly(A) tail lengths are shown below each image. The signal intensity normalized to total intensity (%) was calculated. The mean of two independent experiments was used.
Upregulation and stabilization of mRNAs in Cnot1-KO and Cnot7/8-dKO MEFs compared to Cnot6/6l-dKO MEFs
To examine the effects of impaired deadenylation on global gene expression, we performed total RNA-seq on Cnot1-KO, Cnot7/8-dKO, Cnot6/6l-dKO MEFs and their respective controls (see Methods). We found that 8257, 1344, and 710 mRNAs were upregulated more than twofold in Cnot1-KO, Cnot7/8-dKO, and Cnot6/6l-dKO MEFs, respectively, compared with controls (Fig. 5A). The number of upregulated mRNAs in Cnot7/8-dKO MEFs was much smaller than that in Cnot1-KO MEFs. This can be partly explained by residual expression of CNOT8 in Cnot7/8-dKO MEFs (Fig. 2C). In addition, only a few mRNAs decreased in Cnot1-KO MEFs, whereas 662 mRNAs decreased in Cnot7/8-dKO MEFs when compared with their respective controls (Fig. 5A). Similar results were observed in MCF7 [35]. These data suggest that CNOT7/8 have deadenylation-independent, CNOT1-independent roles. The difference in gene expression between control and Cnot6/6l-dKO MEFs was less prominent (Fig. 5A).
Figure 5.
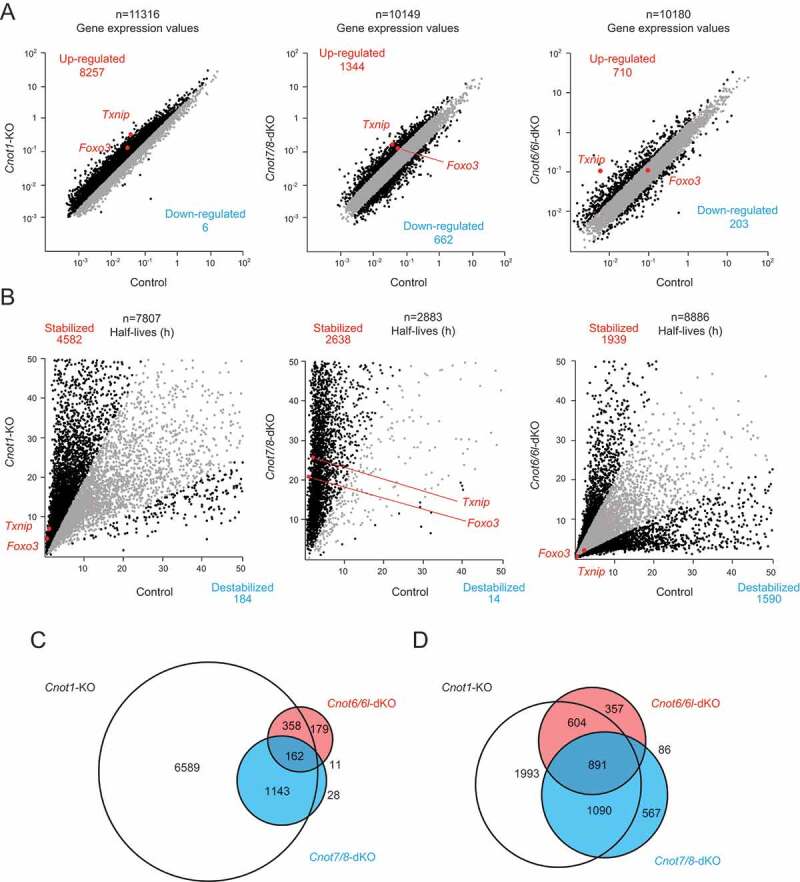
Gene expression and mRNA stability differ among Cnot1-KO, Cnot7/8-dKO and Cnot6/6l-dKO MEFs. RNA-seq analysis of Cnot1-KO, Cnot7/8-dKO and Cnot6/6l-dKO MEFs compared to controls: Cnot1-flox MEFs infected with mock retrovirus, Cnot8-flox MEFs infected with mock retrovirus and WT MEFs, respectively (n = 2). (A, B) Scatter plots comparing gene expression values (A) or mRNA half-lives (B). mRNAs showing expression (A) or half-lives (B) that differed more than twofold are displayed in black. All values represent the means of two independent experiments. (C, D) Venn diagrams showing overlap of upregulated (C) or stabilized genes (D) (more than twofold) in Cnot1-KO, Cnot7/8-dKO and Cnot6/6l-dKO MEFs compared to controls.
We next assessed the stability of mRNAs based on the calculation of mRNA half-lives following actinomycin D (Act. D) chase. The number of stabilized mRNAs was much greater than that of destabilized mRNAs in both Cnot1-KO and Cnot7/8-dKO MEFs (Fig. 5B). While around 2000 mRNAs were stabilized in Cnot6/6l-dKO MEFs, a similar number of mRNAs was destabilized (Fig. 5B). More than 70% of upregulated mRNAs in Cnot7/8-dKO or Cnot6/6l-dKO MEFs overlapped with those in Cnot1-KO MEFs (Fig. 5C). Similar results were observed in stabilized mRNAs (Fig. 5D). In contrast, less than 50% of upregulated or stabilized mRNAs in Cnot7/8-dKO were observed in Cnot6/6l-dKO MEFs (Fig. 5C, D). These data, together with MEF viability data, suggest that the upregulated and stabilized mRNAs common to both Cnot1-KO and Cnot7/8-dKO, but not Cnot6/6l-dKO MEFs, are relevant to death of MEFs. Gene ontology (GO) analysis indicated that stabilized mRNAs overlapping between Cnot1-KO and Cnot7/8-dKO, but not Cnot6/6l-dKO MEFs showed significant enrichment for molecules pertaining to ‘response to DNA damage stimulus’ and ‘DNA damage repair’ (Supplementary Fig. 6 and Supplementary Table 1).
Finally, we validated the results of RNA-seq by performing quantitative PCR (qPCR) analysis. We first examined levels of Foxo3 and Txnip mRNAs, which are stabilized in both Cnot1-KO and Cnot7/8-dKO, but not Cnot6/6l-dKO MEFs in our RNA-seq results (Fig. 5B). Foxo3 and Txnip mRNAs were significantly upregulated in Cnot1-KO and Cnot7/8-dKO MEFs, but not in Cnot6/6l-dKO MEFs, compared to controls (Fig. 6A). We next assessed their stability using total RNAs from Act. D-treated cells. Consistent with the results of RNA-seq, both Foxo3 and Txnip mRNAs had significantly elongated half-lives in Cnot1-KO and Cnot7/8-dKO MEFs, compared to controls (Fig. 6B). Elongation of their half-lives was less obvious in Cnot6/6l-dKO MEFs (Fig. 6B).
Figure 6.
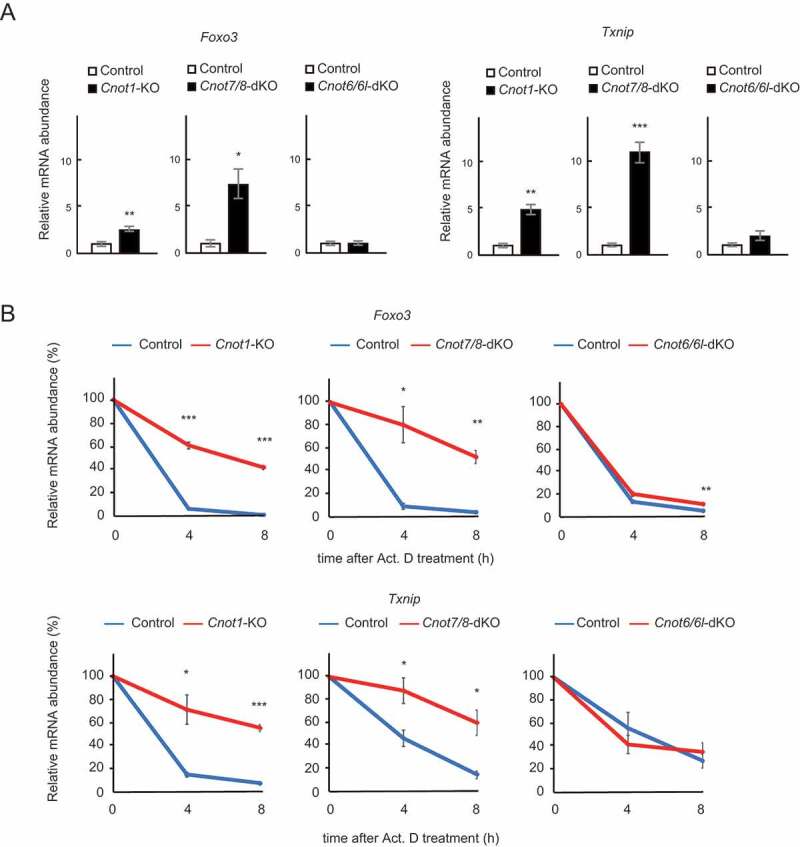
Foxo3 and Txnip mRNAs are upregulated and stabilized in Cnot1-KO and Cnot7/8-dKO MEFs. (A) qPCR analysis of the indicated mRNAs in Cnot1-KO, Cnot7/8-dKO and Cnot6/6l-dKO MEFs together with their controls, as in Fig. 5. Relative mRNA levels were determined by qPCR and normalized to the Gapdh mRNA level (n = 3). (B) Decay curves of mRNAs. Total RNAs were prepared from the indicated MEFs treated with Act. D (0, 4 or 8 h). Relative mRNA levels were determined as in (A). mRNA levels without Act. D treatment (0 h) were set to 100% (n = 3). All values represent means ± S.E.M. *P< 0.05, **P< 0.01, ***P< 0.001.
Discussion
Suppression of the CCR4-NOT complex causes severe defects in cytoplasmic deadenylation and subsequent degradation of mRNA [5–7]. Although many studies have addressed its function in various organisms, it was not clear whether its different catalytic subunits serve distinct biological functions. In this study, we showed that CNOT7/8, but not CNOT6/6L, are essential for MEF viability. It is important to emphasize that we evaluated the roles of the CCR4-NOT complex in normal cells, by using primary fibroblasts with a low risk of mutation, in contrast to cancer cell lines. The latter usually contain genetic mutations that promote tumorigenesis, with the consequence that malignant cells are governed by different molecular mechanisms and signalling pathways than normal cells.
The decreased viability of Cnot7/8-dKO MEFs was concomitant with a lack of all the catalytic subunits in the CCR4-NOT complex (Fig. 2). While reintroduction of CNOT7-WT in Cnot7/8-dKO MEFs recovered cell viability, expression of CNOT7-CN and CNOT-DN mutants did not (Fig. 3), suggesting that CNOT7 catalytic activity is critical in maintenance of viability. Thus, a complex composed of CNOT6/6L, catalytically inactive CNOT7, CNOT1 and other subunits fail to function as an intact deadenylation complex for maintenance of cell viability. We also found that overexpression of CNOT6L in Cnot7/8-dKO MEFs rescued the cell death phenotype (Supplementary Fig. 5). However, in our Cnot7/8-dKO MEFs, incomplete deletion of CNOT8 may have recruited exogenously expressed CNOT6L to the complex, resulting in enough deadenylase activity to maintain cell viability. Whether CNOT6L overexpression can overcome the absence of CNOT7/8 for cell viability remains unaddressed and will require a different experimental system.
Gene expression profiling using RNA-seq revealed a larger contribution of CNOT7/8 than CNOT6/6L in regulation of global gene expression in MEFs. Similar results have been observed in human cell lines, trypanosomes and Drosophila [35,37,38]. Remarkably, the catalytic effect of CNOT7/8 was more prominent than that of CNOT6/6L, as is evident from the results of bulk poly(A) tail analysis, which are clearly reflected in mRNA expression (26, 37 and this study). Absence of all known catalytic subunits could explain, at least in part, the greater differences in gene expression and mRNA stability in Cnot7/8-dKO MEFs than in Cnot6/6l-dKO MEFs, as in cell viability. Although CNOT6/6L are not part of the CCR4-NOT complex in Cnot7/8-dKO MEFs, genes affected by Cnot6/6l-dKO were little represented in the set of mRNAs affected by Cnot7/8-dKO. A similar observation was reported in a previous study [26]. It is possible that CNOT6/6L might be able to function independently of the complex to deadenylate mRNAs, because purified CNOT6/6L and CNOT7/8 exhibit deadenylase activity in vitro [17,39,40]. It would be intriguing to examine whether there are any biological contexts where CNOT6/6L function independently in vivo.
While target mRNAs and biological processes vary by species (human or mouse) or cellular status (transformed or non-transformed), there seems to be a common mechanism by which CNOT6/6L and CNOT7/8 regulate distinct groups of mRNAs and biological processes. In MCF7, CNOT7/8 is required for cell proliferation, whereas CNOT6/6L are required for cell viability [26]. Furthermore, upon CNOT7/8 suppression, upregulated or stabilized mRNAs showed limited overlap with those upregulated or stabilized by CNOT6/6L suppression. A recently proposed model of deadenylation considers the critical involvement of PABP on mRNAs, suggesting distinct roles of CNOT6/6L and CNOT7/8. Specifically, CNOT7/8 deadenylate PABP-free poly(A) tails until they encounter PABP, and then CNOT6/6L take over, removing PABP and subsequently deadenylating mRNAs [30,31]. Importantly, PABP local availability varies according to cell type and cell culture conditions [30]. This may explain, at least in part, why CNOT6/6L and CNOT7/8 execute context-dependent deadenylation. In S. cerevisiae, mRNAs have higher Pab1 (PABP in yeast) occupancy and consequently, Ccr4p, rather than Caf1p, contributes significantly to deadenylation [31]. Similarly, mRNAs are saturated with PABP in HeLa cells and are deadenylated mainly by CNOT6/6L [30]. Active translation state is relevant to the abundance of PABP on mRNA. Binding of PABP to mRNA is important for translation initiation and elongation, because the translation initiation factor complex and PABP interact on mRNAs to form closed-loop messenger ribonucleoprotein complexes [41]. On the other hand, microRNA-mediated translational repression causes dissociation of PABP from mRNA, leading to mRNA degradation [42]. It is possible that CNOT6/6L or CNOT7/8 preferentially target mRNAs which are translationally active (abundant PABP) or silent (less PABP), respectively, resulting in different biological roles. Global analysis of translation status in MEFs or mouse embryos using ribosome profiling should be useful to determine whether CNOT6/6L and CNOT7/8 activities are dependent on the mRNA translation status.
Several lines of evidence show that in addition to its function as a deadenylase, the CCR4-NOT complex has nuclear roles in transcription and mRNA export [8]. In S. cerevisiae, the CCR4-NOT complex interacts with TATA-binding protein (TBP) and TBP-associated factor (TAF) to suppress transcription initiation [43,44]. On the other hand, it stimulates transcription elongation through its association with RNA polymerase II complexes [45]. Moreover, the CCR4-NOT complex is involved in transcriptional regulation in mammals [24,32,46–49]. We previously showed that dysregulation of transcriptional mechanisms occurs in liver development and adipocyte homoeostasis upon suppression of CNOT3 or CNOT1 [23,50]. In this study, not all upregulated mRNAs in Cnot1-KO MEFs can be explained on the basis of mRNA stabilization (Fig. 5). These data suggest that suppression of transcription and other nuclear roles of the CCR4-NOT complex could be responsible for upregulation of a subset of genes. We hypothesize that the decrease of a significant number of mRNA species in Cnot7/8-dKO is possibly due to loss of the catalytic activity-independent function of CNOT7/8, which positively affects transcription. Indeed, CNOT7/8 upregulate transcription mediated by the nuclear receptors, ERα and RARβ [32,51,52].
We did not observe any obvious abnormalities in mouse embryonic development, growth to adulthood, or MEF viability in the absence of CNOT6/6L, suggesting that CNOT6/6L do not target mRNAs encoding molecules involved in those processes. It is also possible that CNOT7/8-mediated deadenylation is sufficient to regulate mRNAs in those processes, because CNOT7/8 could deadenylate PABP-bound mRNAs, albeit with low efficiency [30]. Yet, CNOT6/6L-mediated mRNA decay is still central to certain biological events. For example, CNOT6/6L are critical to survival of MCF7 [26]. In NIH3T3 mouse fibroblasts, knockdown of CNOT6L causes a severe reduction in proliferation, concomitant with significant stabilization of p27kip1 mRNA [29]. We found that mRNAs encoding molecules involved in cell proliferation were enriched among stabilized mRNAs in Cnot6/6l-dKO MEFs. Therefore, CNOT6/6L are likely to regulate proliferation of fibroblasts without affecting viability. Furthermore, Cnot6l-KO mice exhibit resistance to diet-induced obesity, enhanced energy expenditure, and improved insulin sensitivity [34].
While both Cnot7-KO and Cnot8-KO MEFs are viable, Cnot7/8-dKO MEFs undergo death. This is consistent with functional redundancy of CNOT7 and CNOT8, owing to their high amino acid sequence similarity [35]. In this study, we found that Cnot8-KO mice die in embryo. A specific developmental role of CNOT8 is also observed in zebrafish [53]. Attenuation of Cnot8 in zebrafish caused upregulation of developmental control genes and early lethality [53]. Reciprocally, Cnot7-KO male mice have defects in spermatogenesis, even in the presence of CNOT8 [32,33]. Together, despite the functional redundancy of CNOT7 and CNOT8 in some contexts, as in MEFs (in this study) and MCF7 cells [35], they play distinct roles [32,33,53]. Differential expression of CNOT7 and CNOT8 could be relevant to their specific roles, as previously reported in mouse neural tissues [54]. Furthermore, SMG5/7 proteins, which function in nonsense-mediated mRNA decay (NMD), preferentially interact with CNOT8 rather than CNOT7 [55], suggesting different biological roles. Importantly, NMD is essential for embryonic development and viability [56]. On the other hand, we cannot exclude the possibility that CNOT8 functions in zebrafish or mouse embryonic development independently of its catalytic activity in the CCR4-NOT complex. Further analyses are necessary to address the molecular basis for the distinctive functions of CNOT7 and CNOT8.
Our data suggest that maintenance of cell viability is one of the fundamental roles of the CCR4-NOT complex, which is mediated mainly by the catalytic activity of the CNOT7/8. Their vital importance in regulating global mRNA expression is clearly reflected in our RNA-seq results. On the other hand, CNOT6/6L are dispensable in embryonic development and for MEF viability. However, relatively few biological processes have been examined to date. Mouse strains generated with null or conditional alleles for catalytic subunits of the CCR4-NOT complex are valuable tools to analyse functions of each catalytic subunit in various tissues and biological contexts.
Methods
Mice
Cnot6 and Cnot8 conditional mice (Accession No. CDB0794K and CDB0584K: http://www2.clst.riken.jp/arg/mutant%20mice%20list.html) were generated with TT2 ES cell lines as described previously (http://www2.clst.riken.jp/arg/Methods.html). To generate conditional alleles (floxed alleles) or KO alleles (null alleles) for targeted alleles, mice with targeted alleles were crossed with mice expressing FLP (Jackson #009086) or those expressing Cre under control of the CAG promoter (CAG-Cre mice) [57], respectively. Primers for genotyping PCR are listed in Supplementary Table 2. The absence of FLP knock-in alleles in mice with floxed alleles was also confirmed by PCR. Cnot1-flox and Cnot8-flox represent Cnot1loxP/loxP and Cnot8loxP/loxP, respectively. While Cnot1-KO and Cnot8-KO mice represent null alleles, Cnot1-KO and Cnot8-KO MEFs represent Cnot1-flox and Cnot8-flox MEFs, together with Cre expression. Cnot6-KO, Cnot6l-KO, and Cnot7-KO represent null alleles (Cnot6−/-, Cnot6l−/-, and Cnot7−/-). Cnot6l-KO and Cnot7-KO mice have been described previously [32,34]. Note that we removed the β-galactosidase-neomycin cassette in the Cnot6l-KO allele [32] by crossing with CAG-Cre mice. We backcrossed these mouse strains with C57BL/6J mice for at least eight generations. Mice were maintained on a 12 h light/dark cycle in a temperature-controlled (22°C) barrier facility with free access to water and a normal diet (NCD, CA-1, CLEA Japan, Inc.). Mouse experiments were approved by the Animal Care and Use Committee in Okinawa Institute of Science and Technology Graduate University, and by Institutional Animal Care and Use Committee in RIKEN Kobe Branch.
Cell culture
MEFs were prepared as described previously [25] and were cultured at 37°C in Dulbecco’s modified Eagle’s medium supplemented with 10% foetal bovine serum. We used Cnot1-flox MEFs infected with mock retrovirus and Cnot7+/+; Cnot8-flox MEFs (littermates of Cnot7-KO; Cnot8-flox MEFs) infected with mock retrovirus, as controls for Cnot1-KO and Cnot7/8-dKO MEFs, respectively. We used WT MEFs with the same passage number as controls for Cnot6/6l-dKO MEFs.
Antibodies
Rabbit polyclonal antibodies against CNOT1 (14,276-1-AP) and CNOT2 (34,214) were purchased from Proteintech and Cell Signalling Technology, respectively. Mouse monoclonal antibodies against CNOT3, CNOT6L, and CNOT8 were generated by immunizing mice in cooperation with Bio Matrix Research Incorporation [25]. A mouse monoclonal antibody against CNOT6 was raised against a recombinant Autographa californica multiple nuclear polyhedrosis virus displaying a fusion protein containing amino acids 141–190 of human CNOT6, as previously described [58]. Mouse polyclonal antibody against CNOT7 (H00029883-M01) was obtained from Abnova. Antibody against CNOT10 (A304-899A) was from Bethyl Laboratories.
Virus infection
Infection of MEFs with retrovirus (mock or Cre) was performed as described previously [59]. Retroviruses (mock and Cre) were produced by transfecting Plat-E packaging cells with 2 μg empty pMX-puro plasmid or 2 μg pMX-puro-Cre plasmid [25] using TransIT-LT1 transfection reagent (Takara). Two days after transfection, cell culture supernatants containing the retroviruses were filtered (MILLEX GV 0.22 μm, Millipore) and polybrene (0.5 μg/mL, Sigma) was added. The resultant viral solutions were used for infection of MEFs that were seeded at 8.5 × 105 cells per 10 cm dish the day before infection. Two days after retroviral infection, cells were diluted following trypsinization and cultured in the presence of puromycin (1 µg/mL) for additional 2 days to select infected cell populations and subsequently used for analyses. In the series of rescue experiments: CNOT7-WT, CNOT7-mutants (from T. Fujiwara) and CNOT6L [29] cDNA fragments were inserted into pMX-vectors and used for retrovirus production and subsequent infection to MEFs. Adenovirus infection [control (GFP) and Cre] was performed 2 days after retrovirus infection at MOI 7.5. Two days later, adenovirus-infected MEFs were used for immunoprecipitation. For cell death analysis, MEFs were cultured in the presence of puromycin (1 µg/mL) for additional 2 days.
Immunoprecipitation and immunoblotting
MEFs were lysed with TNE lysis buffer (1% NP-40, 50 mM Tris–HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonylfluoride, and 10 mM NaF). We used 5% of each lysate for an expression check (lysate lanes). The remainder of each lysate was incubated with anti-CNOT3 antibody for 1 h at 4°C with rotation, followed by incubation with Protein G Sepharose (GE Healthcare) for 2 h at 4°C with rotation. Half (one-fifth only in Supplementary Fig. 5) of each immunoprecipitated product was loaded per lane and analysed by immunoblot, as described previously [25].
Measurement of cell survival rate
Cell viability was measured by the ability of cells to exclude propidium iodide (PI). Cells were treated with 0.1% trypsin, collected by centrifugation, washed once with phosphate-buffered saline (PBS), and resuspended in PBS containing 5 μg/mL of PI (Sigma). Levels of PI incorporation were quantified using a FACS Calibur (Becton Dickinson).
RNA analysis
Total RNA was extracted with Isogen II, according to the manufacturer’s protocol (Nippon Gene). To measure mRNA stability, we treated cells with actinomycin D (2.5 μg/mL) 4 days after viral infection, including the 2 days of puromycin selection (see virus infection above). Total RNA was extracted at 0, 4 and 8 h after addition of actinomycin D. RNA purity and concentration were evaluated by spectrophotometry using a NanoDrop ND-2000 (ThermoFisher). Total RNA (1 μg) was used for reverse transcription with oligo(dT)12-18primer (Invitrogen) using the SuperScript III First-Strand Synthesis System (Invitrogen). qPCR reactions were carried out using TB Green Premix Ex Taq (Takara) and the ViiA 7 Real-Time PCR System (Applied Biosystems). Gapdh mRNA levels were used for normalization. Relative mRNA expression was determined by the ∆∆CT method. Primers for qPCR reactions are listed in Supplementary Table 2.
RNA sequencing
The quality of RNA was assessed using the Agilent 2100 Bioanalyzer microfluidics-based platform (Agilent Technologies, Inc.). The RNA Integrity Number (RIN) was over 9.0 in all samples. RNA-seq was performed by the DNA Sequencing Section at Okinawa Institute of Science and Technology Graduate University for two biological replicates per condition. One hundred nano grams of total RNA were used for RNA-seq library preparation with a TruSeq Stranded mRNA Library Prep Kit for NeoPrep (NP-202-1001, Illumina), which allows polyA-oligo(dT)-based purification of mRNA, according to the manufacturer’s protocol, with minor modifications and optimization as follows. Custom dual index adaptors were ligated at the 5ʹ and 3ʹ-ends of the library, and PCR was performed for 11 cycles. 150-base-pair pair-end read RNA-seq was performed using a Hiseq 3000/4000 PE Cluster Kit (PE-410-1001; Illumina) and a Hiseq 3000/4000 SBS Kit (300 Cycles) (FC-410-1003; Illumina) on a Hiseq4000 (Illumina), according to the manufacturer’s protocol. Sequence data are available through ArrayExpress under the accession number [E-MTAB-8287].
RNA-sequencing data analysis
Paired-end RNA-seq data were mapped to the Mus musculus reference strain mm10 UCSC using Strand NGS software (Strand Genomics). Transcript abundance for each replicate was measured in Fragments Per Kilobase Mapped (FPKM). Genes with FPKM < 0.1 were removed, and only ‘protein-coding genes’ were considered. For comparison, we used gene expression values defined as Gene FPKMs normalized with Gapdh FPKM. Fold changes in gene expression values (KO against control MEFs) were determined. Half-lives were calculated based on the decay rate of each transcript after suppression of transcription. Total RNA was prepared at 0, 4, and 8 h after Act. D treatment, and subjected to RNA-seq. To calculate the mRNA degradation rate constant (), gene expression values were plotted on a semilogarithmic scale as a function of time (t). The line that best fit the data was identified using linear regression, then was obtained from the slope. The t1/2 was calculated by substitution in the equation:
mRNAs having half-lives less than 0 h or more than 50 h were excluded as unreliable. Cnot1-flox or Cnot7+/+/Cnot8-flox MEFs infected with mock virus were used as controls for Cnot1-KO and Cnot7/8-dKO MEFs, respectively. WT MEFs were used as controls for Cnot6/6l-dKO MEFs. mRNAs having more than twofold longer half-lives compared to controls were considered stabilized genes. GO enrichment analysis was performed with DAVID Bioinformatics Resources 6.8 (https://david.ncifcrf.gov).
Bulk poly(A) tail assay
We confirmed that RINs were over 9.0 in all samples using the Agilent 2100 Bioanalyzer microfluidics-based platform. Total RNA (10 µg) was labelled with [5′-32P] pCp (cytidine 3′,5′-bis[phosphate]) (0.11 pmol/μL in a total reaction volume of 30 μL) (PerkinElmer; NEG019A) using T4 RNA ligase 1 (NEB, M0204S) at 16°C overnight. Labelled RNAs were incubated at 85°C for 5 min and placed on ice. Then, labelled RNAs were digested with Ribonuclease A (50 ng/μL, Sigma) and Ribonuclease T1 (1.25 U/μl, ThermoFisher Scientific) at 37°C for 2 h in digestion buffer (100 mM Tris-HCl [pH7.5], 3M NaCl, 0.5 μg/mL yeast tRNA). Reactions were stopped by adding 5x stop solution (10 mg/mL Proteinase K, 0.125 M EDTA, 2.5% SDS) and subsequently incubating at 37°C for 30 min. After adding 400 μL of RNA precipitation buffer (0.5 M NH4OAc, 10 mM EDTA), digested RNA samples were purified by phenol-chloroform extraction and isopropanol precipitation. Final products (10 μL) were mixed with RNA Gel loading Dye (NEB, R0641) and incubated at 95°C for 2 min. Then, samples were fractionated on 8 M urea-10% polyacrylamide denaturing gels (0.8 mm thick). Markers (Prestained Markers for small RNA Plus, BioDynamics Laboratory DM253) were also loaded. The gel was analysed using a Typhoon FLA 9500 Fluorescence Imager (GE Healthcare). Band intensity was quantified using Image J.
Funding Statement
This work was supported by the Japan Society for the Promotion of Science [21229006]; Japan Society for the Promotion of Science [18K06975]; Japan Society for the Promotion of Science [17K07292]; Japan Society for the Promotion of Science [18K07079].
Acknowledgments
We thank J Miyazaki for the CAG-Cre mice and T Fujiwara for providing the CNOT7 mutant cDNAs. We also thank the DNA Sequencing Section in Okinawa Institute of Science and Technology Graduate University (OIST) for preparing sequence libraries and for performing Hiseq sequencing.
Author contributions
DM, AY, and TS performed the experiments. DM, AT, and TS analyzed the RNA-seq results. AT and TK maintained and crossed the mouse strains for preparing MEFs. T. Yamaguchi and KK helped preparation of MEFs. YK prepared recombinant adenoviruses. TA and YF generated mouse strains. DM, T. Yamamoto, and TS designed the study. DM, T. Yamamoto, and TS wrote the manuscript.
Disclosure statement
No potential conflict of interest was reported by the authors.
Statistical analysis
Differences between groups were examined for statistical significance using Student’s t-test (two-tailed distribution with two-sample equal variance). We considered a p-value of <0.05 statistically significant.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Garneau NL, Wilusz J, Wilusz CJ.. The highways and byways of mRNA decay. Nat Rev Mol Cell Biol. 2007;8:113–126. [DOI] [PubMed] [Google Scholar]
- [2].Parker R. RNA Degradation in Saccharomyces cerevisae. Genetics. 2012;191:671–702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Yamashita A, Chang T-C, Yamashita Y, et al. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nat. Struc. Mol. Biol. 2005;12:1054. [DOI] [PubMed] [Google Scholar]
- [4].Wahle E, Winkler GS. RNA decay machines: deadenylation by the Ccr4–not and Pan2–pan3 complexes. Biochim Biophys Acta. 2013;1829:561–570. [DOI] [PubMed] [Google Scholar]
- [5].Tucker M, Valencia-Sanchez MA, Staples RR, et al. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 2001;104:377–386. [DOI] [PubMed] [Google Scholar]
- [6].Collart MA, Panasenko OO. The Ccr4-Not complex. Gene. 2012;492:42–53. [DOI] [PubMed] [Google Scholar]
- [7].Bartlam M, Yamamoto T. The structural basis for deadenylation by the CCR4-NOT complex. Protein Cell. 2010;1:443–452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Collart MA. The Ccr4-Not complex is a key regulator of eukaryotic gene expression. Wiley Interdiscip Rev RNA. 2016;7:438–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Lau N-C, Kolkman A, van Schaik Frederik MA, et al. Human Ccr4–not complexes contain variable deadenylase subunits. Biochem J. 2009;422:443–453. [DOI] [PubMed] [Google Scholar]
- [10].Ito K, Takahashi A, Morita M, et al. The role of the CNOT1 subunit of the CCR4-NOT complex in mRNA deadenylation and cell viability. Protein Cell. 2011;2:755–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Petit AP, Wohlbold L, Bawankar P, et al. The structural basis for the interaction between the CAF1 nuclease and the NOT1 scaffold of the human CCR4-NOT deadenylase complex. Nucleic Acids Res. 2012;40:11058–11072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Boland A, Chen Y, Raisch T, et al. Structure and assembly of the NOT module of the human CCR4-NOT complex. Nat Struct Mol Biol. 2013;20:1289–1297. [DOI] [PubMed] [Google Scholar]
- [13].Ito K, Inoue T, Yokoyama K, et al. CNOT2 depletion disrupts and inhibits the CCR4-NOT deadenylase complex and induces apoptotic cell death. Genes Cells. 2011;16:368–379. [DOI] [PubMed] [Google Scholar]
- [14].Morita M, Oike Y, Nagashima T, et al. Obesity resistance and increased hepatic expression of catabolism-related mRNAs in Cnot3± mice. Embo J. 2011;30:4678–4691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Chen Y, Boland A, Kuzuoğlu-Öztürk D, et al. A DDX6-CNOT1 complex and W-binding pockets in CNOT9 reveal direct links between miRNA target recognition and silencing. Mol Cell. 2014;54:737–750. [DOI] [PubMed] [Google Scholar]
- [16].Bawankar P, Loh B, Wohlbold L, et al. NOT10 and C2orf29/NOT11 form a conserved module of the CCR4-NOT complex that docks onto the NOT1 N-terminal domain. RNA Biol. 2013;10:228–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Raisch T, Chang C-T, Levdansky Y, et al. Reconstitution of recombinant human CCR4-NOT reveals molecular insights into regulated deadenylation. Nat Commun. 2019;10:3173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Basquin J, Roudko VV, Rode M, et al. Architecture of the nuclease module of the yeast Ccr4-not complex: the Not1-Caf1-Ccr4 interaction. Mol Cell. 2012;48:207–218. [DOI] [PubMed] [Google Scholar]
- [19].Goldstrohm AC, Wickens M. Multifunctional deadenylase complexes diversify mRNA control. Nat Rev Mol Cell Biol. 2008;9:337–344. [DOI] [PubMed] [Google Scholar]
- [20].Clark LB, Viswanathan P, Quigley G, et al. Systematic mutagenesis of the leucine-rich repeat (LRR) domain of CCR4 reveals specific sites for binding to CAF1 and a separate critical role for the LRR in CCR4 deadenylase activity. J Biol Chem. 2004;279:13616–13623. [DOI] [PubMed] [Google Scholar]
- [21].Inoue T, Morita M, Hijikata A, et al. CNOT3 contributes to early B cell development by controlling Igh rearrangement and p53 mRNA stability. J Exp Med. 2015;212:1465–1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Yamaguchi T, Suzuki T, Sato T, et al. The CCR4-NOT deadenylase complex controls Atg7-dependent cell death and heart function. Sci Signal. 2018;11:eaan3638. [DOI] [PubMed] [Google Scholar]
- [23].Suzuki T, Kikuguchi C, Nishijima S, et al. Postnatal liver functional maturation requires Cnot complex-mediated decay of mRNAs encoding cell cycle and immature liver genes. Development. 2019;146. dev.168146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Neely GG, Kuba K, Cammarato A, et al. A global in vivo Drosophila RNAi screen identifies NOT3 as a conserved regulator of heart function. Cell. 2010;141:142–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Suzuki T, Kikuguchi C, Sharma S, et al. CNOT3 suppression promotes necroptosis by stabilizing mRNAs for cell death-inducing proteins. Sci Rep. 2015;5. srep14779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mittal S, Aslam A, Doidge R, et al. The Ccr4a (CNOT6) and Ccr4b (CNOT6L) deadenylase subunits of the human Ccr4–not complex contribute to the prevention of cell death and senescence. Mol Biol Cell. 2011;22:748–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Piao X, Zhang X, Wu L, et al. CCR4-NOT deadenylates mRNA associated with RNA-induced silencing complexes in human cells. Mol Cell Biol. 2010;30:1486–1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Nousch M, Techritz N, Hampel D, et al. The Ccr4-Not deadenylase complex constitutes the main poly(A) removal activity in C. elegans. J Cell Sci. 2013;126:4274–4285. [DOI] [PubMed] [Google Scholar]
- [29].Morita M, Suzuki T, Nakamura T, et al. Depletion of mammalian CCR4b deadenylase triggers elevation of the p27Kip1 mRNA level and impairs cell growth. Mol Cell Biol. 2007;27:4980–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Yi H, Park J, Ha M, et al. PABP cooperates with the CCR4-NOT complex to promote mRNA deadenylation and block precocious decay. Mol Cell. 2018;70:1081–1088. [DOI] [PubMed] [Google Scholar]
- [31].Webster MW, Chen Y-H, Stowell JAW, et al. mRNA deadenylation is coupled to translation rates by the differential activities of Ccr4-Not nucleases. Mol Cell. 2018;70:1089–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Nakamura T, Yao R, Ogawa T, et al. Oligo-asthenoteratozoospermia in mice lacking Cnot7, a regulator of retinoid X receptor beta. Nat Genet. 2004;36:528–533. [DOI] [PubMed] [Google Scholar]
- [33].Berthet C, Morera AM, Asensio MJ, et al. CCR4-associated factor CAF1 is an essential factor for spermatogenesis. Mol Cell Biol. 2004;24:5808–5820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Morita M, Siddiqui N, Katsumura S, et al. Hepatic post-transcriptional network comprising of CCR4-NOT deadenylase and FGF21 maintains systemic metabolic homeostasis. Proc Natl Acad Sci USA. 2019;116:7973–7981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Aslam A, Mittal S, Koch F, et al. The Ccr4–not deadenylase subunits CNOT7 and CNOT8 have overlapping roles and modulate cell proliferation. Mol Biol Cell. 2009;20:3840–3850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Mishima Y, Tomari Y. Codon usage and 3ʹUTR length determine maternal mRNA stability in zebrafish. Mol Cell. 2016;61:874–885. [DOI] [PubMed] [Google Scholar]
- [37].Temme C, Zaessinger S, Meyer S, et al. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. Embo J. 2004;23:2862–2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Schwede A, Ellis L, Luther J, et al. A role for Caf1 in mRNA deadenylation and decay in trypanosomes and human cells. Nucl Acids Res. 2008;6:3374–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Horiuchi M, Takeuchi K, Noda N, et al. Structural basis for the antiproliferative activity of the Tob-hCaf1 complex. J Biol Chem. 2009;284:13244–13255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Wang H, Morita M, Yang X, et al. Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity. Embo J. 2010;29:2566–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Wells SE, Hillner PE, Vale RD, et al. Circularization of mRNA by eukaryotic translation initiation factors. Mol Cell. 1998;2:135–140. [DOI] [PubMed] [Google Scholar]
- [42].Zekri L, Kuzuoğlu-Öztürk D, Izaurralde E. GW182 proteins cause PABP dissociation from silenced miRNA targets in the absence of deadenylation. Embo J. 2013;32:1052–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Collart MA, Struhl K. NOT1(CDC39), NOT2(CDC36), NOT3, and NOT4 encode a global-negative regulator of transcription that differentially affects TATA-element utilization. Genes Dev. 1994;8:525–537. [DOI] [PubMed] [Google Scholar]
- [44].Badarinarayana V, Chiang YC, Denis CL. Functional interaction of CCR4-NOT proteins with TATA-binding protein (TBP) and its associated factors in yeast. Genetics. 2000;155:1045–1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Kruk JA, Dutta A, Fu J, et al. The multifunctional Ccr4-not complex directly promotes transcription elongation. Genes Dev. 2011;25:581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Winkler GS, Mulder KW, Bardwell VJ, et al. Human Ccr4-not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. Embo J. 2006;25:3089–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Garapaty S, Mahajan MA, Samuels HH. Components of the CCR4-NOT complex function as nuclear hormone receptor coactivators via association with the NRC-interacting factor NIF-1. J Biol Chem. 2008;283:6806–6816. [DOI] [PubMed] [Google Scholar]
- [48].Hu G, Kim J, Xu Q, et al. A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 2009;23:837–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Cejas P, Cavazza A, Yandava CN, et al. Transcriptional regulator CNOT3 defines an aggressive colorectal cancer subtype. Cancer Res. 2017;77:766–779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Takahashi A, Takaoka S, Kobori S, et al. The CCR4–NOT deadenylase complex maintains adipocyte identity. Int J Mol Sci. 2019;20:5274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Morel AP, Sentis S, Bianchin C, et al. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. J Cell Sci. 2003;116:2929–2936. [DOI] [PubMed] [Google Scholar]
- [52].Prevot D, Morel AP, Voeltzel T, et al. Relationships of the antiproliferative proteins BTG1 and BTG2 with CAF1, the human homolog of a component of the yeast CCR4 transcriptional complex: involvement in estrogen receptor alpha signaling pathway. J Biol Chem. 2001;276:9640–9648. [DOI] [PubMed] [Google Scholar]
- [53].Koch P, Löhr HB, Driever W. A mutation in cnot8, component of the Ccr4-not complex regulating transcript stability, affects expression levels of developmental regulators and reveals a role of Fgf3 in development of caudal hypothalamic dopaminergic neurons. PloS One. 2014;9:e113829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Chen C, Ito K, Takahashi A, et al. Distinct expression patterns of the subunits of the CCR4-NOT deadenylase complex during neural development. Biochem Biophys Res Commun. 2011;411:360–364. [DOI] [PubMed] [Google Scholar]
- [55].Loh B, Jonas S, Izaurralde E. The SMG5–SMG7 heterodimer directly recruits the CCR4–NOT deadenylase complex to mRNAs containing nonsense codons via interaction with POP2. Genes Dev. 2013;27:2125–2138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Hwang J, Maquat LE. Nonsense-mediated mRNA decay (NMD) in animal embryogenesis: to die or not to die, that is the question. Curr Opin Genet Dev. 2011;21:422–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Sakai K, Miyazaki J. A transgenic mouse line that retains Cre recombinase activity in mature oocytes irrespective of the cre transgene transmission. Biochem Biophys Res Commun. 1997;237:318–324. [DOI] [PubMed] [Google Scholar]
- [58].Tanaka T, Takeno T, Watanabe Y, et al. The generation of monoclonal antibodies against human peroxisome proliferator-activated receptors (PPARs). J Atheroscler Thromb. 2002;9:233–242. [DOI] [PubMed] [Google Scholar]
- [59].Suzuki T, K-Tsuzuku J, Ajima R, et al. Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell proliferation and transformation. Genes Dev. 2002;16:1356–1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


