Abstract
The red-crowned crane (Grus japonensis) is an endangered, large-bodied crane native to East Asia. It is a traditional symbol of longevity and its long lifespan has been confirmed both in captivity and in the wild. Lifespan in birds is known to be positively correlated with body size and negatively correlated with metabolic rate, though the genetic mechanisms for the red-crowned crane’s long lifespan have not previously been investigated. Using whole genome sequencing and comparative evolutionary analyses against the grey-crowned crane and other avian genomes, including the long-lived common ostrich, we identified red-crowned crane candidate genes with known associations with longevity. Among these are positively selected genes in metabolism and immunity pathways (NDUFA5, NDUFA8, NUDT12, SOD3, CTH, RPA1, PHAX, HNMT, HS2ST1, PPCDC, PSTK CD8B, GP9, IL-9R, and PTPRC). Our analyses provide genetic evidence for low metabolic rate and longevity, accompanied by possible convergent adaptation signatures among distantly related large and long-lived birds. Finally, we identified low genetic diversity in the red-crowned crane, consistent with its listing as an endangered species, and this genome should provide a useful genetic resource for future conservation studies of this rare and iconic species.
Keywords: genome, longevity, red-crowned crane
INTRODUCTION
The red-crowned crane (Grus japonensis) is one of the rarest cranes in the world and is a symbol of longevity in East Asia. It has a maximum lifespan of 30 years in the wild and 65 years in captivity (Rasmussen and Engstrom, 2004), which is substantially longer than the average lifespan recorded in 144 other avian species (Supplementary Table S1). It is also one of the largest avian species (Chen et al., 2012), measuring on average 150–158 cm long with a 220–250 cm wingspan (del Hoyo et al., 1996), and weighing on average 8.9 kg (John and Dunning, 2008) (ranging from 4.8 to 10.5 kg). Since avian body sizes are known to be positively correlated to lifespan and negatively correlated to metabolic rate (McKechnie and Wolf, 2004; Scholander et al., 1950), it has previously been suggested that the longevity of the red-crowned crane is related to its low metabolic rate (Speakman, 2005).
There are currently two primary red-crowned crane populations, including a non-migratory population in northern Japan and a migratory continental group that ranges across Southeastern Russia, Northeastern China, Eastern Mongolia, and Korea (IUCN, 2017). It relies on wetlands for breeding and the loss and pollution of these habitats has resulted in severe population declines (Wang et al., 2011). It has been classified as threatened since 2000 and is classified as ‘endangered’ on the International Union for Conservation of Nature Red List (Yu et al., 2001). With only estimated 3,000 individuals in 2017, coupled with long generational lengths (12 years), it is one of the most endangered avian species (IUCN, 2017). Conservation genetic studies have primarily focused on microsatellite and mitochondrial markers, and an absence of genomic-level sequence data has precluded more comprehensive demographic modeling.
To infer the population history and evolutionary adaptations of this endangered species, we sequenced the first red-crowned crane whole genome and compared it to 18 other avian species. The Avian Phylogenomics Project (http://avian.genomics.cn/) previously published 48 avian reference genomes (Zhang et al., 2014), including the grey-crowned crane (Balearica regulorum) and the common ostrich (Struthio camelus). The grey-crowned crane belongs to the family of Gruidae together with the red-crowned crane, though the body mass of this crane (weighing up to 4 kg, height up to 1.4 m) (Walkinshaw, 1973) is smaller than that of red-crowned crane and the lifespan, particularly in captivity, is shorter (Wasser and Sherman, 2010). Conversely, the common ostrich is a member of the infraclass Palaeognathae and is deeply divergent from the red-crowned crane (111 million years ago, Mya) (Hedges et al., 2015). It is the largest Aves (up to 156.8 kg, up to 1.7 m in height) with a lifespan closer to the red-crowned crane (maximum longevity of 50 years in captivity) (Wasser and Sherman, 2010). To identify common evolutionary signatures associated with lifespan in birds, we compared our red-crowned crane genome to the genomes of the common ostrich (large/long-lived, but belonging to different family) and grey-crowned crane (belonging to the same family, but small/short-lived).
MATERIALS AND METHODS
Sample preparation, genomic DNA extraction and sequencing
The protocol for blood sample preparation was carried out in accordance with guidelines of Korean Association for Bird Protection under the Cultural Heritage Administration (Korea) permit. All experimental protocols were approved by the Genome Research Foundation. All methods were carried out in accordance with relevant guidelines and regulations. A blood sample was secured from a single female red-crowned crane collected upstream of Gunnam dam, Gyeonggi-do, Republic of Korea (Lat = 38°06′24.8″N and Long = 127°01′17.1″E). Genomic DNA was extracted using a QIAamp DNA Mini Kit (Qiagen, USA) following the manufacturer’s instructions. The DNA concentration was measured using the Qubit dsDNA assay kit (Invitrogen, USA) and an Infinite 200 PRO Nanoquant system (Tecan, Germany). Fragmentation of high-molecular weight genomic DNA was carried out with a Covaris S2 Ultrasonicator (Covaris, USA), generating 500 bp fragments. Whole-genome shotgun libraries were prepared using a TruSeq library sample prep kit (Illumina, USA). Aliquots were analyzed on an Agilent 2100 Bioanalyzer (Agilent Technologies, USA) to determine the library concentration and size. Sequencing was performed on an Illumina HiSeq 2000 sequencer (Illumina), using the TruSeq Paired-End Cluster Kit v3 (Illumina) and the TruSeq SBS HS Kit v3 (Illumina) for 200 cycles.
Red-crowned crane genome assembly by reference mapping
Low quality DNA reads with phred quality scores < 20 and/or ambiguous nucleotides ‘N’ ratios > 10% were filtered out. Clean reads were aligned to the grey-crowned crane genome sequence using BWA-aln 0.6.2 (Li and Durbin, 2009) at default settings. Polymerase chain reaction duplicates from the reads were removed using ‘rmdup’ command of SAMtools 0.1.18 (Li et al., 2009) at default settings. Complete consensus sequences were determined by SAMtools mpileup, Bcftools view, and SAMtools vcfutils.pl vcf2fq pipelines from the SAMtools 0.1.19 suite (Li et al., 2009). The consensus sequence included all alignment depths. Single nucleotide variant (SNV) calling was conducted based on the consensus genome sequences (McKenna et al. 2010; Van der Auwera et al., 2013). Indels were called using the GATK 3.3 UnifiedGenotyper with the -dcov 1000 option. The indels were marked by VariantFiltration in GATK 3.3 with the following criteria: (1) hard to validate, MQ0 ≥ 4 && ((MQ0 / (1.0 × DP)) > 0.1); (2) quality filter, QUAL < 10.0; (3) depth filter, DP < 5. The SnpEff 3.3 software was used to predict the effects of the indels. Heterozygous SNV rate was used as the nucleotide diversity value. The average avian nucleotide diversity was calculated by averaging the values of the American flamingo (0.00372), Anna’s hummingbird (0.00288), common ostrich (0.00176), downy woodpecker (0.00455), grey-crowned crane (0.00202), peregrine falcon (0.00112), white-throated tinamou (0.00560), and white-tailed tropicbird (0.00162). Coding sequence (CDS) were selected by filtering out the following conditions: (1) there is an ambiguous nucleotide ‘N’ in a CDS; (2) there is a premature stop codon in a CDS. Among the 14,173 CDSs, 13,407 CDSs were selected and used in subsequent analyses. K-mers (K = 17) were obtained using the JELLYFISH 2.2.4 (Marcais and Kingsford, 2011) program with the command “jellyfish count -m 17 -o output -c 32 -s 8589934592 -t 64”. We calculated the genome size from the total K-mer frequencies.
Orthologous gene clustering
The protein sequences of the red-crowned crane were predicted using the gene model of the grey-crowned crane. We downloaded 18 high quality avian reference genomes from the GigaDB dataset (Zhang et al., 2014) (http://gigadb.org/dataset/101000). Orthologous gene clusters were constructed using OrthoMCL 2.0.9 (Li et al., 2003). Self-to-self BLASTP searches were conducted for all the protein sequences of the red-crowned crane and reference genomes with an E-value of 1E-5. The Markov clustering algorithm was used to define the cluster structure between the proteins with inflation values (-I) of 1.5. Lastly, the OrthoMCL program was used to determine orthologous gene clusters with an e-value exponent cutoff value of 1.0 × e−5 and ‘percentMatchCutoff’ value of 50%.
Evolutionary analyses
We applied the tree topology and divergence times of the 18 avian reference genomes represented from Jarvis et al. (2015) to the positively selected gene (PSG) analysis. The divergence time of the red-crowned crane and grey-crowned crane was taken from TimeTree database (Hedges et al., 2015). The white-throated tinamou’s divergence time was also taken from TimeTree (Hedges et al., 2015). The multiple sequence alignment of orthologous genes was constructed using PRANK alignment program package (Loytynoja and Goldman, 2010), and the rates of synonymous (dS) and non-synonymous substitutions (dN) and the dN/dS ratio (nonsynonymous substitutions per nonsynonymous site to synonymous substitutions per synonymous site, ω) were estimated using the CODEML program in PAML 4.5 (Yang, 1997). The branch-site test of positive selection was conducted with a 10% false discovery rate criterion. The red-crowned and common ostrich were used as the foreground branch for each analysis accordingly, respectively, and the other species were used as background branches. Functional effects of the species-specific amino acid (SSAA) changes were predicted using PolyPhen-2 (Adzhubei et al., 2010) and PROVEAN (Choi et al., 2012) programs using the default cutoff values. Gene enrichment analysis was conducted with the database for annotation, visualization, and integrated discovery (DAVID) database (Huang et al., 2008) and network drawn by Cytoscape (ver. 3.6.1) (Shannon et al., 2003).
Demographic history
Whole genome sequencing data can be used to accurately estimate the population history (Li and Durbin, 2011; Osada, 2014). We inferred the demographic history of the red-crowned crane using a pairwise sequentially Markovian coalescent (PSMC) analysis with scaffolds of length ≥ 50 kb. For grey-crowned crane, we downloaded illumina short reads from National Center for Biotechnology Information (Acc. PRJNA212879) and used in the PSMC analysis. The PSMC analysis was performed with 100 bootstrapping rounds, with 9.59×10−9 substitutions per site per generation and generation time of 12.3 years, as previously reported (IUCN, 2017).
RESULTS AND DISCUSSION
The red-crowned crane genome
Genomic DNA from a red-crowned crane individual was sequenced using the Illumina HiSeq2000 platform. We produced a total of 989 million paired-end reads with a 100 bp read length and an insert size of 216 bp (Supplementary Table S2, Supplementary Fig. S1). After trimming the low-quality reads, we obtained 750 M (75.86%) reads, yielding 75 Gb and a depth of coverage of 66x. A K-mer analysis with K = 17–21 was performed and the red-crowned crane’s genome size was estimated to be 1.146 Gb (Supplementary Table S3, Supplementary Fig. S2), which is comparable to other avian species (0.96–2.2 Gb) (Zhang et al., 2014). Its genome was assembled by mapping the short reads to the grey-crowned crane (Balearica regulorum) reference (Zhang et al., 2014) (Supplementary Table S4), which has a 20 Mya divergence time (Hedges et al., 2015). A total of 610 M red-crowned crane reads (81.08%) were mapped to the 1.13 Gb grey-crowned crane reference (Supplementary Table S4). We obtained 94.1% genomic regions with more than 10× sequencing depth (Supplementary Fig. S3). The reads covered 99.77% of CDS region with a mapping depth of at least 1×, and 99.02% of the CDS regions had higher than a 5× sequencing depth (Supplementary Table S4). The total GC content was 41.75% (Supplementary Table S4). Using the grey-crowned crane reference genome, approximately 36 million SNVs and 3.6 million small insertions/deletions (indels) were identified (Supplementary Table S5). These SNVs were comprised of 34,733,024 (96%) homozygous and 1,433,466 (4%) heterozygous SNVs, yielding a nucleotide diversity of 1.29 × 10−3 nucleotide substitutions per site; which is lower than nucleotide diversity of the grey-crowned crane (1.88 × 10−3). Compared with a median nucleotide diversity of 2.49 × 10−3 in least concern avian species (Zhang et al., 2014), the red-crowned crane possesses a low level of nucleotide diversity and is genetically consistent with an endangered species. The red-crowned crane consensus sequences, covering a total of 14,173 genes (Supplementary Table S4), were created by substituting the red-crowned crane SNVs into the grey-crowned crane reference genome (see Materials and Methods section).
Comparative evolutionary analyses of the red-crowned crane
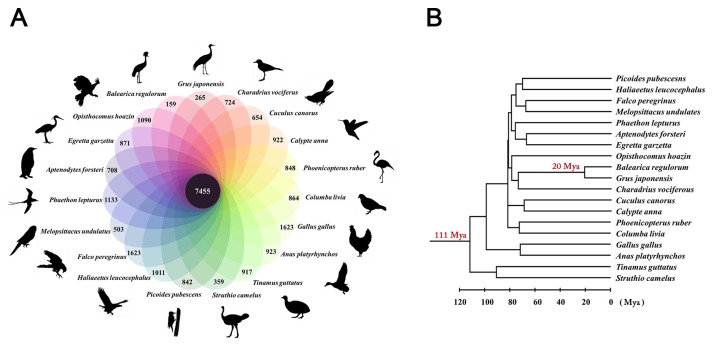
To investigate the environmental adaptations of the red-crowned crane and the genetic basis of its longevity (Table 1) (Ji and DeWoody, 2017), we analyzed the red-crowned crane consensus sequence compared to 18 other avian genomes (Supplementary Table S6). Among these genomes, we identified a total of 31,795 orthologous genes, 7,455 of which were conserved across all 18 of the avian genomes (Fig. 1A). Of these, 5,878 orthologous families had one-to-one relationships among the 19 avian genomes. We used a phylogenetic tree topology which was constructed by Bayesian phylogenomic analysis (Zhang et al., 2014) (Fig. 1B) and estimated the divergence time with one-to-one orthologous genes. It estimated the divergence time between the red-crowned and grey-crowned cranes to be 20 Mya. The reported divergence time between the red-crowned crane and the common ostrich (used as an outgroup in this study) is roughly 111 Mya (Hedges et al., 2015).
Table 1.
Reference genomes and biological traits
| Latin name | Common name | Abbreviation | Height (cm) | Body weight (kg) | Metabolic rate (W/kg) | Lifespan (y) | ||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| Wild | Captivity | |||||||
| 1 | Phoenicopterus ruber ruber | American flamingoc | PHORU | 120–145 | 2.8 | 15.254d | - | - |
| 2 | Calypte anna | Anna’s hummingbirdb | CALAN | 10–11 | 0.004 | - | 8.5 | - |
| 3 | Haliaeetus leucocephalus | Bald eagleb | HALEU | 70–102 | 5.6 (female) 4.1 (male) |
- | - | - |
| 4 | Melopsittacus undulatus | Budgerigar (parakeet)b | MELUN | 18 | 0.03–0.04 | 9.8e | - | 21.0 |
| 5 | Cuculus canorus | Common cuckoob | CUCCA | 32–34 | 0.11–0.13 | 0.838d | 12.9 | - |
| 6 | Struthio camelus australis | Common ostrichb | STRCA | 170–280 | 63–145 | 6.305d | - | 70.0 |
| 7 | Picoides pubescens | Downy woodpeckerb | PICPU | 14–17 | 0.02–0.03 | 0.383d | 11.9 | - |
| 8 | Aptenodytes forsteri | Emperor penguinb | LEPDI | 110–130 | 23 | 42.871d | - | 23.4 |
| 9 | Balearica regulorum gibbericeps | Grey-crowned cranec | BALRE | 100 | 3.5 | - | - | 27.2 |
| 10 | Ophisthocomus hoazin | Hoatzinb | OPHHO | 65 | 0.8 | - | - | - |
| 11 | Charadrius vociferus | Killdeerb | CHAVO | 23–27 | 0.09 | 0.416d | 10.9 | - |
| 12 | Egretta garzetta | Little egretb | EGRGA | 55–65 | 0.35–0.55 | - | 22.34 | - |
| 13 | Anas platyrhynchos domestica | Peking duckb | ANAPL | 50–76 | 1.6–2.3 | 4.068d | 23.4 | - |
| 14 | Falco peregrinus | Peregrine falconb | FALPE | 34–58 | 0.33–1.5 | - | - | - |
| 15 | Gallus gallus | Chickena | GALGA | 30–45 | 1–3 | 6.005d | - | 30.0 |
| 16 | Columba livia | Rock pigeonb | COLLI | 32–37 | 0.36 | 1.714d | - | - |
| 17 | Phaethon lepturus | White-tailed tropicbirdc | PHALE | 71–80 | 0.33 | - | - | - |
| 18 | Tinamus guttatus | White-throated tinamoub | THGUT | 32–36 | - | - | - | - |
| 19 | Grus japonensis | Red-crowned crane | GRJAP | 150–158 | 8.9 | - | 30.0 | 65.0 |
-, data not available.
Sanger,
high-coverage genomes,
low-coverage genomes.
Basal metabolic rate values were obtained from the literature
Fig. 1. Orthologous gene clusters of the red-crowned crane compared to the other avian species.
(A) The Venn diagram shows the number of unique (number in the ovals) and shared (the number in the center black circle) gene families analyzed by OrthoMCL (Li et al., 2003). It was drawn manually using the online photo editor (https://pixlr.com/editor/). (B) Divergence time of the red-crowned-crane and grey-crowned crane (Hedges et al., 2015).
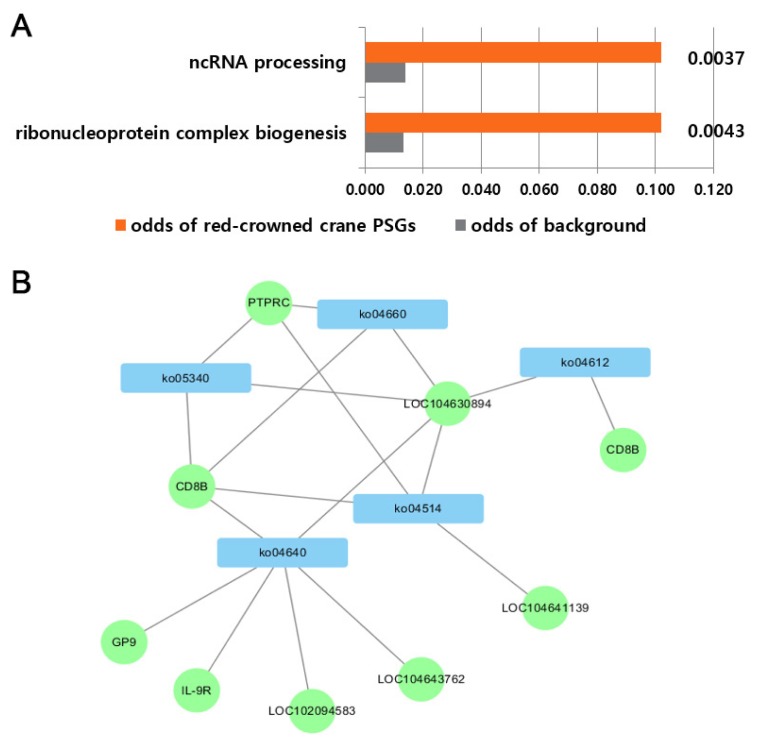
Avian body sizes are known to be positively correlated with lifespan (Supplementary Table S1) and negatively correlated with metabolic rate (Zhang et al., 2014). In order to investigate the contribution of genes in these pathways to the longevity of the red-crowned crane, PSGs were identified among the 5,878 one-to-one orthologues shared by all 19 species. A total of three and 162 genes were identified as putative PSGs in the red-crowned crane by applying the branch-site model and branch model, respectively (Supplementary Tables S7 and S8). With the PSG set, a functional enrichment analysis of the red-crowned crane PSGs was conducted using the DAVID functional annotation tool (Huang et al., 2008) for Gene Ontology (GO) categories (Fig. 2A, Supplementary Table S9) and the Fisher’s exact tests for the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways (Fig. 2B, Supplementary Table S10). Two statistically enriched GO terms included “ribonucleoprotein complex biogenesis” (GO:0022613; EBNA1BP2, PHAX, PDCD11, LSM6, and FTSJ2, P = 3.7 × e−3) and “ncRNA processing” (GO:0034470; PDCD11, LSM6, RG9MTD1, FTSJ2, and RPP14, P = 4.3 × e−3) (Fig. 2A).
Fig. 2. Functional analysis of the red-crowned crane PSGs.
(A) Red-crowned crane PSGs enriched in GO categories. The values to the right of the bar represent the p-values from the enrichment test analyzed on the DAVID website (Huang et al., 2008). (B) Network diagram of the red-crowned crane PSGs associated with KEGG pathways. The blue rectangles represent pathway IDs from the KEGG database and the green circles represent the red-crowned crane PSGs. Each edge represents the association between the pathways and the genes.
Telomere length has been proposed to be a ‘molecular clock’ that underlies organismal aging (Collins, 2008; Collins and Mitchell, 2002). Interestingly, the PHAX (phosphorylated adaptor for RNA export) gene plays an essential role in the human telomerase RNA (hTR) export to Cajal body (Boulon et al., 2004) enabling hTR maturation and telomerase localization (Egan and Collins, 2012; Schmidt and Cech, 2015). In addition, numerous reports indicate that immune functions are associated with aging (Accardi and Caruso, 2018) and we identified eight red-crowned crane PSGs (CD8B, GP9, IL-9R, PTPRC, LOC102094583, LOC104630894, LOC104643762, and LOC104641139) which were significantly enriched in immunity associated pathways (P value < 0.01); including hematopoietic cell lineage (KO04640), cell adhesion molecules (KO04514), primary immunodeficiency (KO05340), antigen processing and presentation (KO04612), and T cell receptor signaling pathway (KO04660) (Fig. 2B).
We further investigated red-crowned crane PSGs involved in metabolic pathways, which are associated with longevity (Speakman, 2005), by searching against the KEGG database (Kanehisa and Goto, 2000). We could identify seven red-crowned crane PSGs (HNMT, HS2ST1, NDUFA5, NDUFA8, NUDT12, PPCDC, and PSTK) that were predicted by the branch-site model (Table 2). Of these, two genes were associated with energy metabolism, including NDUFA5 (ω average: 0.19652, ω red-crowned crane: 2.2221) and NDUFA8 (ω average: 0.13284, ω red-crowned crane: 1.1281). NDUFA5 and NDUFA8 encode the Q-module and ND1 module of NADH-ubiquinone oxidoreductase (Complex 1; EC 1.6.5.3), respectively (Supplementary Fig. S4). Complex 1 functions in the transfer of electrons derived by NADH to the respiratory chain, and is a major source of damaging reactive oxygen species (ROS) that are known to be involved in aging and energy metabolism processes (Stroud et al., 2016). Previous studies of NDUFA5 mutants in Caenorhabditis elegans have shown this gene to be involved in lifespan extension in the nematode and the regulation of lifespan in yeast (Chen et al., 2012; Valentine and Valentine, 2014). Furthermore, it was reported that expression levels of both NDUFA5 and NDUFA8 are decreased in the mitochondria of older humans (Ghosh et al., 2011).
Table 2.
PSGs involved in pathways related to metabolism in the red-crowned crane
| KEGG pathways | Branch model PSGs | dN/dS of all avian means | dN/dS of the red-crowned crane |
|---|---|---|---|
| Histidine metabolism | HNMT | 0.3162 | 1.5981 |
| Oxidative phosphorylation | NDUFA8 | 0.1328 | 1.1281 |
| NDUFA5 | 0.1965 | 2.2221 | |
| Glycosaminoglycan biosynthesis - heparan sulfate / heparin | HS2ST1 | 0.0437 | 1.1207 |
| Metabolic pathways | NUDT12 | 0.0304 | 1.0047 |
| NDUFA8 | 0.1328 | 1.1281 | |
| NDUFA5 | 0.1965 | 2.2221 | |
| PPCDC | 0.1664 | 1.7130 | |
| Nicotinate and nicotinamide metabolism | NUDT12 | 0.0304 | 1.0047 |
| Pantothenate and CoA biosynthesis | PPCDC | 0.1664 | 1.7130 |
| Selenocompound metabolism | PSTK | 0.3001 | 1.1039 |
Comparative genetic analyses of metabolic genes among long-lived Aves
The common ostrich has a lifespan comparable to the red-crowned crane (Wasser and Sherman, 2010), and it is possible that the longevity of these species could be due in part to convergent polymorphisms affecting the rate of energy metabolism. To identify any shared adaptations for longevity-related physiological characteristics, we compared the PSGs of the red-crowned crane with that of the common ostrich. We identified 36 and 76 ostrich PSGs using branch-site and branch model, respectively (Supplementary Tables S11 and S12). Six of the ostrich PSGs (KIAA1009, NOVA1, SOD3, SOX14, TMEM80, and UBXN11) were also positively selected in the red-crowned crane (Table 3). SOD3 is associated with the superoxide metabolic process (GO:0006801) GO molecular function (Ashburner et al., 2000). The free-radical theory of aging is one of the major hypotheses of biological aging, and suggests that aging is a result of accumulated damage from ROS (Speakman, 2005). Organisms have a variety of protection mechanisms to mitigate the deleterious effects of the ROS (Held, 2012), and SOD3 is an antioxidant enzyme that plays a critical role in converting the superoxide anion radical into hydrogen peroxide (Supplementary Fig. S5A) (Gentschew et al., 2013; Held, 2012). In addition, the overexpression of SOD3 results in increased cell growth and proliferation, and the inhibition of apoptosis (Mendez et al., 2005). We speculate that the SOD3 gene product may help protect the tissues and organs from oxidative stress, consequently increasing longevity.
Table 3.
PSGs shared in both red-crowned crane and common ostrich
| PSGs | Statistics of PSGs prediction of red-crowned crane | Statistics of PSGs prediction of common ostrich | ||||||
|---|---|---|---|---|---|---|---|---|
|
|
|
|||||||
| Branch models | Branch-site models | Branch models | Branch-site models | |||||
|
|
|
|
|
|||||
| M0:one-ratioa | M1:free-ratiob | 2ΔLc | P value | M0:one-ratioa | M1:free-ratiob | 2ΔLc | P value | |
| KIAA1009 | 0.4014 | 1.0295 | - | - | - | - | 10.8273 | 0.0155 |
| NOVA1 | 0.1016 | 1.5792 | - | - | 0.1016 | 1.8746 | 7.9132 | 0.0489 |
| SOD3 | 0.1454 | 1.2380 | - | - | 0.1454 | 1.4134 | - | - |
| SOX14 | 0.0186 | 1.4472 | - | - | 0.0186 | 1.6574 | - | - |
| TMEM80 | 0.2138 | 2.1448 | - | - | 0.2138 | 2.6535 | - | - |
| UBXN11 | 0.4847 | 2.1552 | - | - | 0.4847 | 1.3068 | - | - |
-, no statistical significance.
M0 denotes dN/dS for all the species in the study.
M1 denotes dN/dS for the lineage leading to the target species.
2ΔL: Likelihood ratio tests (LRT) were used to detect positive selection.
The SSAAs with possible function-change can provide evolutionary information to given species. We hypothesized that the common genes with function altering SSAAs in both red-crowned crane and common ostrich are probably associated with longevity by comparing common ostrich SSAAs to the other 18 avian species with an average lifespan. We could identify 10,126 SSAAs for the common ostrich from 3,551 genes (Supplementary Table S13). Among them, 2,001 genes contained at least one function altering SSAA (in a total of 3,698 ostrich SSAAs). Interestingly, 435 genes with ostrich function-altering SSAAs were shared with the red-crowned crane, suggesting the possibility of some level of convergent evolution (Supplementary Table S14). The permutation test of these genes identified an enrichement of “cilium morphogenesis” (P value of 1.2 × e−4) and “centriole” (P value 1.2 × e−2). Notably, cilium assembly and dis-assembly are unique mechanisms that regulate cell physiology during proliferation and morphogenesis (Eggenschwiler and Anderson, 2007; Loncarek and Bettencourt-Dias, 2017; Stroud et al., 2016), and are associated with replicative aging such as the aging of skin and the hemopoietic system (Tkemaladze and Chichinadze, 2005). To further examine the role of these genes in aging, we queried the red-crowned crane and common ostrich predicted function-altering PSGs in the GeneAge database (Tacutu et al., 2013), and identified a total of eleven genes (BUB1B, MDM2, ERCC8, RPA1, NCOR2, CTH, MTBP, SOD3, NCOR2, CIT, and NCOR1) for the red-crowned crane and seven genes in the common ostrich (ALG6, TKT, CTH, MDH2, GCLC, SDHB, and COQ3). Interestingly, the only common gene, CTH (Cystathionine gamma lyase), encodes a cytoplasmic enzyme in the trans-sulfuration pathway that converts cystathione derived from methionine into cysteine (Supplementary Fig. S5B). Glutathione is a well-known anti-oxidant known to contribute to longevity, and glutathione synthesis in the liver is dependent upon the availability of cysteine. Higher levels cystathionine are associated with aging and produce an augmented oxidant effect in elderly humans (Hernanz et al., 2000). In addition, it was reported that RNAi-mediated knockdown of CTH-1 and CTH-2 in C. elegans caused median lifespan reductions (up to ~15%) (Hine et al., 2015). These biological data observed in other models raise the intriguing possibility that similar mechanisms may increase longevity in the red-crowned crane and common ostrich.
Genetic diversity and population history
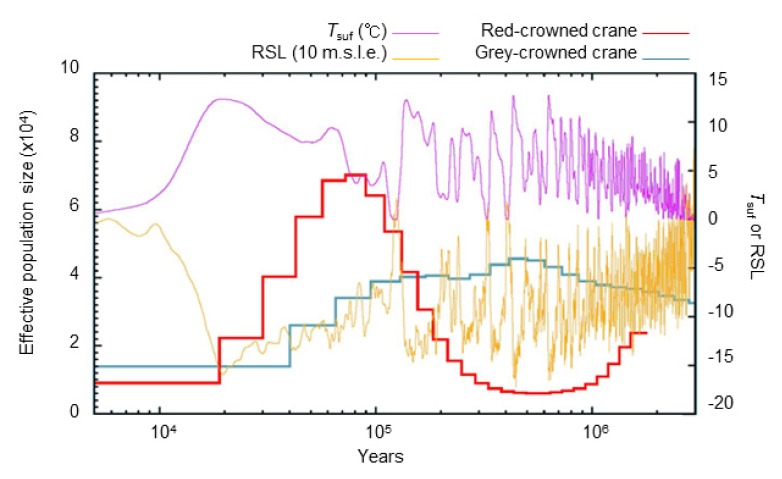
A PSMC analysis was used to model the historical population size fluctuations of the red-crowned crane and the closely related grey-crowned crane (Fig. 3) (Li and Durbin, 2011). We hypothesized that historical population size of these two cranes helps understand crane’s adaptation associated with environmental condition. Based on the genomic data of the grey-crowned crane (Balericinae) and red-crowned crane (Gruinae), we estimated effective population size (Ne) (Fig. 3). We discovered a striking difference in the effective population size between the two species during last glacial age. While red-crowned crane’s effective population size appears to have peaked about 80,000 years ago in the last glacial age, the grey-crowned crane effective population size was at its maximum roughly 500,000 years ago. The Ne of the grey-crowned crane increased from 1 Mya, and peaked at 0.4 Mya, with an estimated Ne ~4.4 × 104. Conversley, we observed a Ne decrease in the red-crowned crane from the late Pliocene to the early Pleistocene (~0.4 Mya). This is consistent with the fossil records, which is biased towards the Gruinae subfamily (Krajewski et al., 2010). Thus, we can infer that the environment was less favorable for the Gruinae at this time. The Ne of the red-crowned crane was increased at late Plestocene (0.7–0.1 Mya), at the end of last ice age. The population size of the red-crowned crane is predicted to be Ne ~7.0 × 104, and was the highest roughly 80,000 years ago (Kya). While it is possible that the red-crowned crane’s northern habitat naturally suppresses their effective population size, it is also known that habitat loss and pollution have contributed to severe population declines.
Fig. 3. Demographic history of the red-crowned crane and grey-crowned crane.
PSMC demographic modeling of the red-crowned crane, using a generation time (g = 12.3 years) and mutation rate per generation time (μ = 9.66 × 10−9), and the grey-crowned crane, with a generation time (g = 15.1) and mutation rate (μ = 1.18 × 10−8). Tsuf, atmospheric surface air temperature; RSL, relative sea level; 10 m.s.l.e., 10 m sea level equivalent.
In conclusion, we generated and analyzed the first whole genome sequence data for the red-crowned crane. Evolutionary genomic analyses of 19 avian species, including the long-lived red-crowned crane and common ostrich, yielded PSGs and function-altering SSAA candidate genes associated with longevity. Functional annotations and enrichment analyses were conducted using GO, KEGG pathways, and GeneAge databases. Both PSGs and function-altering SSAA analyses showed candidate genes responsible for longevity in red-crowed crane by using bioinformatic analysis and discussed longevity-related biological and physiological features previously identified in both human and animal models. Demographic modeling of the red-crowned crane revealed low genetic diversity and a trend of population decline. Taken together, these findings further highlight the importance of continued monitoring and management of this endangered species. Despite the SNV based genetic differences predicted by PSGs and SSAAs which provided us with some speculative interpretations on how such variations affected certain longevity related genes in the red-crowned crane species, it should be noted that the distinct phenotypic differences between the grey-crowned crane reference and whole genome sequences of red-crowned crane that are mapped to the reference may not be fully explained by SNPs only. SNPs are usually functionally relevant in expressed genes and longevity as a biological phenomenon is a whole species evolution and regulation problem that involves numerous regulatory mechanisms of transcriptional, translational, and even epigenetic factors which are often associated with the genome structure itself. Therefore, it will be absolutely necessary to produce a high quality red-crowned crane de novo assembly in the future to compare the structural and copy number variations between the two species’ references with long DNA read generation methods such as PacBio SMRT and Oxford Nanopore sequencing. Nevertheless, we hope the consensus genome presented here will be a valuable resource for aging research and future conservation genetic studies of this iconic species.
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by PGI of Genome Research Foundation and Clinomics Ltd. internal research funds and the No.10075262 of National Center for standard Reference Data. This research was also supported by Ulsan National Institute of Science & Technology (UNIST) internal research funds. And the National Research Foundation of Korea (2013M3A9A5047052 and 2017M3A9A5048999) research funds.
The authors thank many people not listed as authors who provided analyses, data, feedback, samples, and encouragement. Especially, thanks to Taehyung Kim and Byungchul Kim.
Footnotes
Note: Supplementary information is available on the Molecules and Cells website (www.molcells.org).
Disclosure
O.C., Y.S.C., and J.J. are employees and J.B. is one of the founders of Clinomics Ltd. All other authors have no potential conflicts of interest to disclose.
AVAILABILITY OF DATA AND MATERIALS
Whole-genome sequence data were deposited in the SRA database at NCBI with BioSample accession number SAMN07580860. The data can be accessed via reference number SRX3148473 or through BioProject accession number PRJNA400839. Currently, another genome project of the red-crowned crane was reported (PRJNA400839).
REFERENCES
- Accardi G., Caruso C. Immune-inflammatory responses in the elderly: an update. Immun Ageing. 2018;15:11. doi: 10.1186/s12979-018-0117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adzhubei I.A., Schmidt S., Peshkin L., Ramensky V.E., Gerasimova A., Bork P., Kondrashov A.S., Sunyaev S.R. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M., Ball C.A., Blake J.A., Botstein D., Butler H., Cherry J.M., Davis A.P., Dolinski K., Dwight S.S., Eppig J.T., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S., Verheggen C., Jady B.E., Girard C., Pescia C., Paul C., Ospina J.K., Kiss T., Matera A.G., Bordonne R., et al. PHAX and CRM1 are required sequentially to transport U3 snoRNA to nucleoli. Mol Cell. 2004;16:777–787. doi: 10.1016/j.molcel.2004.11.013. [DOI] [PubMed] [Google Scholar]
- Chen W., Ma J., Zhang H., Li D., Zhang X. Behavioural alterations in domestication process: comparative studies between wild, captive and inbred red-crowned cranes (Grus japonensis) J Anim Vet Adv. 2012;11:2711–2715. doi: 10.3923/javaa.2012.2711.2715. [DOI] [Google Scholar]
- Choi Y., Sims G.E., Murphy S., Miller J.R., Chan A.P. Predicting the functional effect of amino acid substitutions and indels. PLoS One. 2012;7:e46688. doi: 10.1371/journal.pone.0046688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K. Physiological assembly and activity of human telomerase complexes. Mech Ageing Dev. 2008;129:91–98. doi: 10.1016/j.mad.2007.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K., Mitchell J.R. Telomerase in the human organism. Oncogene. 2002;21:564–579. doi: 10.1038/sj.onc.1205083. [DOI] [PubMed] [Google Scholar]
- del Hoyo J., Elliott A., Sargatal J. Handbook of the Birds of the World. Barcelona: Lynx Edicions; 1996. [Google Scholar]
- Egan E.D., Collins K. Biogenesis of telomerase ribonucleoproteins. RNA. 2012;18:1747–1759. doi: 10.1261/rna.034629.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler J.T., Anderson K.V. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gentschew L., Flachsbart F., Kleindorp R., Badarinarayan N., Schreiber S., Nebel A. Polymorphisms in the superoxidase dismutase genes reveal no association with human longevity in Germans: a case–control association study. Biogerontology. 2013;14:719–727. doi: 10.1007/s10522-013-9470-3. [DOI] [PubMed] [Google Scholar]
- Ghosh S., Lertwattanarak R., Lefort N., Molina-Carrion M., Joya-Galeana J., Bowen B.P., Garduno-Garcia Jde J., Abdul-Ghani M., Richardson A., DeFronzo R.A., et al. Reduction in reactive oxygen species production by mitochondria from elderly subjects with normal and impaired glucose tolerance. Diabetes. 2011;60:2051–2060. doi: 10.2337/db11-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedges S.B., Marin J., Suleski M., Paymer M., Kumar S. Tree of life reveals clock-like speciation and diversification. Mol Biol Evol. 2015;32:835–845. doi: 10.1093/molbev/msv037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held P. Application Guide. Winooski: BioTek Instruments; 2012. An Introduction to Reactive Oxygen Species: Measurement of ROS in Cells. [Google Scholar]
- Hernanz A., Fernandez-Vivancos E., Montiel C., Vazquez J.J., Arnalich F. Changes in the intracellular homocysteine and glutathione content associated with aging. Life Sci. 2000;67:1317–1324. doi: 10.1016/S0024-3205(00)00722-0. [DOI] [PubMed] [Google Scholar]
- Hine C., Harputlugil E., Zhang Y., Ruckenstuhl C., Lee B.C., Brace L., Longchamp A., Trevino-Villarreal J.H., Mejia P., Ozaki C.K., et al. Endogenous hydrogen sulfide production is essential for dietary restriction benefits. Cell. 2015;160:132–144. doi: 10.1016/j.cell.2014.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang D.W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2008;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- IUCN (International Union for Conservation of Nature) The IUCN Red List of Threatened Species. IUCN Data. 2017 https://www.iucnredlist.org/species/22692167/93339099.
- Jarvis E.D., Mirarab S., Aberer A.J., Li B., Houde P., Li C., Ho S.Y.W., Faircloth B.C., Nabholz B., Howard J.T., et al. Phylogenomic analyses data of the avian phylogenomics project. GigaScience. 2015;4:4. doi: 10.1186/s13742-014-0038-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji Y., DeWoody J.A. Relationships among powered flight, metabolic rate, body mass, genome size, and the retrotransposon complement of volant birds. Evol Biol. 2017;44:261–272. doi: 10.1007/s11692-016-9405-4. [DOI] [Google Scholar]
- John B., Dunning J. CRC Handbook of Avian Body Masses. Florida: CRC Press; 2008. [Google Scholar]
- Kanehisa M., Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krajewski C., Sipiorski J.T., Anderson F.E. Complete mitochondrial genome sequences and the phylogeny of cranes (gruiformes: gruidae) Auk. 2010;127:440–452. doi: 10.1525/auk.2009.09045. [DOI] [Google Scholar]
- Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. Inference of human population history from individual whole-genome sequences. Nature. 2011;475:493–496. doi: 10.1038/nature10231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. Genome Project Data Processing, S. The sequence alignment/map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L., Stoeckert C.J., Jr, Roos D.S. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loncarek J., Bettencourt-Dias M. Building the right centriole for each cell type. J Cell Biol. 2017;217:823–835. doi: 10.1083/jcb.201704093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loytynoja A., Goldman N. webPRANK: a phylogeny-aware multiple sequence aligner with interactive alignment browser. BMC Bioinformatics. 2010;11:579. doi: 10.1186/1471-2105-11-579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcais G., Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27:764–770. doi: 10.1093/bioinformatics/btr011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKechnie A.E., Wolf B.O. The allometry of avian basal metabolic rate: good predictions need good data. Physiol Biochem Zool. 2004;77:502–521. doi: 10.1086/383511. [DOI] [PubMed] [Google Scholar]
- McKenna A.H., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez J.I., Nicholson W.J., Taylor W.R. SOD isoforms and signaling in blood vessels: evidence for the importance of ROS compartmentalization. Arterioscler Thromb Vasc Biol. 2005;25:887–888. doi: 10.1161/01.ATV.0000164043.24549.50. [DOI] [PubMed] [Google Scholar]
- Osada N. Extracting population genetics information from a diploid genome sequence. Front Ecol Evol. 2014;2:7. doi: 10.3389/fevo.2014.00007. [DOI] [Google Scholar]
- Rasmussen P.C., Engstrom R.T. Threatened birds of Asia: the Birdlife international red data book. Auk. 2004;121:619–622. [Google Scholar]
- Schmidt J.C., Cech T.R. Human telomerase: biogenesis, trafficking, recruitment, and activation. Genes Dev. 2015;29:1095–1105. doi: 10.1101/gad.263863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scholander P.F., Hock R., Walters V., Irving L. Adaptation to cold in arctic and tropical mammals and birds in relation to body temperature, insulation, and basal metabolic rate. Biol Bull. 1950;99:259–271. doi: 10.2307/1538742. [DOI] [PubMed] [Google Scholar]
- Shannon P., Markiel A., Ozier O., Baliga N.S., Wang J.T., Ramage D., Amin N., Schwikowski B., Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman J.R. Body size, energy metabolism and lifespan. J Exp Biol. 2005;208:1717–1730. doi: 10.1242/jeb.01556. [DOI] [PubMed] [Google Scholar]
- Stroud D.A., Surgenor E.E., Formosa L.E., Reljic B., Frazier A.E., Dibley M.G., Osellame L.D., Stait T., Beilharz T.H., Thorburn D.R., et al. Accessory subunits are integral for assembly and function of human mitochondrial complex I. Nature. 2016;538:123. doi: 10.1038/nature19754. [DOI] [PubMed] [Google Scholar]
- Tacutu R., Craig T., Budovsky A., Wuttke D., Lehmann G., Taranukha D., Costa J., Fraifeld V.E., de Magalhaes J.P. Human ageing genomic resources: integrated databases and tools for the biology and genetics of ageing. Nucleic Acids Res. 2013;41:D1027–D1033. doi: 10.1093/nar/gks1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tkemaladze J.V., Chichinadze K.N. Centriolar mechanisms of differentiation and replicative aging of higher animal cells. Biochemistry (Mosc) 2005;70:1288–1303. doi: 10.1007/s10541-005-0261-6. [DOI] [PubMed] [Google Scholar]
- Valentine R.C., Valentine D.L. Human Longevity: Omega-3 Fatty Acids, Bioenergetics, Molecular Biology, and Evolution. Florida: CRC Press; 2014. [DOI] [Google Scholar]
- Van der Auwera G.A., Carneiro M.O., Hartl C., Poplin R., del Angel G., Levy-Moonshine A., Jordan T., Shakir K., Roazen D., Thibault J., et al. From FastQ data to high confidence variant calls: the genome analysis toolkit best practices pipeline. Curr Protoc Bioinformatics. 2013;43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walkinshaw L. Cranes of the World. New York: Winchester Press; 1973. [Google Scholar]
- Wang Z., Li Z., Beauchamp G., Jiang Z. Flock size and human disturbance affect vigilance of endangered red-crowned cranes (Grus japonensis) Biol Conserv. 2011;144:101–105. doi: 10.1016/j.biocon.2010.06.025. [DOI] [Google Scholar]
- Wasser D.E., Sherman P.W. Avian longevities and their interpretation under evolutionary theories of senescence. J Zool. 2010;280:103–155. doi: 10.1111/j.1469-7998.2009.00671.x. [DOI] [Google Scholar]
- Yang Z. PAML: a program package for phylogenetic analysis by maximum likelihood. Comput Appl Biosci. 1997;13:555–556. doi: 10.1093/bioinformatics/13.5.555. [DOI] [PubMed] [Google Scholar]
- Yu J., Liu J., Jin W. Analysis of the environment feature of breeding area and endangered factors of red-crowned crane in China. Chin Geogr Sci. 2001;11:186–191. doi: 10.1007/s11769-001-0041-1. [DOI] [Google Scholar]
- Zhang G., Li C., Li Q., Li B., Larkin D.M., Lee C., Storz J.F., Antunes A., Greenwold M.J., Meredith R.W., et al. Comparative genomics reveals insights into avian genome evolution and adaptation. Science. 2014;346:1311–1320. doi: 10.1126/science.1251385. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.